Abstract
The crosstalk between NK cells and their surrounding environment is enabled through activating and inhibitory receptors, which tightly control NK cell activity. The co-inhibitory receptor TIGIT decreases NK cell cytotoxicity and is involved in NK cell exhaustion, but has also been associated with liver regeneration, highlighting that the contribution of human intrahepatic CD56bright NK cells in regulating tissue homeostasis remains incompletely understood. A targeted single-cell mRNA analysis revealed distinct transcriptional differences between matched human peripheral blood and intrahepatic CD56bright NK cells. Multiparameter flow cytometry identified a cluster of intrahepatic NK cells with overlapping high expression of CD56, CD69, CXCR6, TIGIT and CD96. Intrahepatic CD56bright NK cells also expressed significantly higher protein surface levels of TIGIT, and significantly lower levels of DNAM-1 compared to matched peripheral blood CD56bright NK cells. TIGIT+ CD56bright NK cells showed diminished degranulation and TNF-α production following stimulation. Co-incubation of peripheral blood CD56bright NK cells with human hepatoma cells or primary human hepatocyte organoids resulted in migration of NK cells into hepatocyte organoids and upregulation of TIGIT and downregulation of DNAM-1 expression, in line with the phenotype of intrahepatic CD56bright NK cells. Intrahepatic CD56bright NK cells represent a transcriptionally, phenotypically, and functionally distinct population of NK cells that expresses higher levels of TIGIT and lower levels of DNAM-1 than matched peripheral blood CD56bright NK cells. Increased expression of inhibitory receptors by NK cells within the liver environment can contribute to tissue homeostasis and reduction of liver inflammation.
Keywords: intrahepatic NK cells, liver organoids, single-cell mRNA analysis, immune tolerance, tissue homeostasis, TIGIT, DNAM-1, PVR/CD155
1. Introduction
Natural Killer (NK) cells are innate lymphocytes that not only play an essential role in the defense against pathogens and tumors but are also involved in modulating immune homeostasis and remodeling of tissues (1–5). Adequate regulation of immune surveillance and function in liver tissues is critical (6), as the liver is exposed to various antigens and metabolites derived from the gastrointestinal tract (7). NK cells represent up to 40% of all intrahepatic lymphocytes, and their role in maintaining immune homeostasis in the liver has received increasing attention (7–9). Intrahepatic NK cells have been associated with a broad spectrum of NK cell functions, ranging from exerting cytotoxicity, producing cytokines to mediating antigen-specific contact hypersensitivity responses (10–13). Based on the expression of the surface markers CD56 and CD16, NK cells can be divided into a CD56bright and a CD56dim subset (14). While CD56dim NK cells predominate in the peripheral blood (pb) (15–17), CD56bright NK cells are the main population of liver-resident (lr) NK cells, as defined by their expression of specific markers regulating liver residency of NK cells in humans, including CXCR6, CD69, CCR5 and CD49a (18–21). CXCR6+ lrNK cells express high levels of the transcription factor Eomes and low levels of Tbet (16, 22), and subsets of lrNK cells also express the transcription factors Hobit (15) and promyelocytic leukemia zinc finger protein (PLZF) (23). These distinct profiles in the expression of transcription factors have been suggested as potential drivers of tissue-specific regulation of NK cell function. Despite differences between CD56bright lrNK cells and in particular CD56dim as well as CD56bright pbNK cells in the expression of transcription factors, the consequences for tissue-specific adaption of NK cell functions are insufficiently understood (18, 20, 23, 24).
NK cell activity is tightly controlled through a plethora of receptors, balancing activating and inhibitory signals. These highly diverse receptors enable the crosstalk between NK cells and their surrounding environment (24, 25). The T cell immunoglobulin and ITIM domain (TIGIT) is a co-inhibitory receptor that was first described to be expressed on T cells and NK cells (26–28). TIGIT is part of the immunoglobulin-superfamily and interacts with ligands of the PVR family, nectin and nectin-like family (26). TIGIT binds CD155 (poliovirus receptor; PVR), CD112 (PVRL2) CD113 (PVRL3), and Nectin4 (PVRL4), although the interaction of TIGIT with PVR showed to have the highest affinity (27, 29). PVR is abundantly expressed on antigen-presenting cells (APC) such as dendritic cells, as well as on T cells and in particular on tumor cells (30–32). Three other receptors, namely CD226 (DNAM-1), CD96 (TACTILE) and KIR2DL5 also interact with PVR, but with lower binding affinity, displaying diverse functions (27, 33–36). Binding of DNAM-1 promotes anti-tumor responses, enhances NK cell cytotoxicity and viral clearance (32, 37–39). CD96 has been shown to be a marker for NK cell exhaustion in patients with hepatocellular carcinoma (HCC) and to limit antitumor functions of NK cells in mice (40, 41). Binding of TIGIT leads to decreased NK cell cytotoxicity and is suggested to ensure self-tolerance, as well as to promote liver regeneration (26, 32, 42). In this study, we performed a detailed characterization of CD56bright NK cells derived from matched peripheral blood and liver tissues on the transcriptional level and by assessing protein expression profiles, and determined their functional activity. The herein described results provide a deeper understanding of the differences between peripheral blood and tissue NK cell subsets, and the impact of the liver environment on CD56bright NK cells.
2. Materials and methods
2.1. Participants
Matched peripheral blood and tumor-free liver samples were obtained from patients at the Asklepios Hospital Barmbek (AKB) undergoing extended liver resection due to liver metastases following colorectal cancer, liver adenoma, cholangiocellular carcinoma or hemangioma (n=17), and at the University Medical Center Hamburg-Eppendorf (UKE) from patients undergoing liver transplantation (n=17) due to end-stage liver disease. Human control peripheral blood samples were obtained from 13 healthy individuals. Furthermore, residual amounts of anonymized peripheral blood samples (buffy coats) from 12 randomly-selected healthy blood donors recruited at the Institute for Transfusion Medicine at the UKE were used. All blood donors gave their general written consent to use their blood samples for scientific studies. The anonymized use of buffy coats complied with a vote by the ethics committee of the German Medical Association. Study protocols (PV4898, PV4081, and PV4780) were approved by the ethics committee of the medical association of Hamburg, and all study participants provided written informed consent. The demographics and clinical characteristics of study participants are summarized in Tables 1 , 2 .
Table 1.
Demographics and clinical characteristics of patients included in this study.
| Figure 1 | Figure 2 | Figure 3 | Figure 4 | Figure 5 | All | |
|---|---|---|---|---|---|---|
| Samples | ||||||
| Liver | 4 | 19 | 8 | – | – | 27* |
| Matched PBMC | 4 | 19 | 0 | – | – | 19* |
| Hepatocyte organoids | – | – | – | 7 | 7** | |
| Demographics | ||||||
| Sex (f/m) | 2/2 | 7/12 | 4/4 | – | 4/3 | 15/19 |
| Median age in years (Range) | 57 (43-68) | 57 (43-68) | 56 (45-65) | – | 57 (50-75) | 57 (43-75) |
| Primary liver disease (LTX) | 17 | |||||
| ALD | 0 | 4 | 0 | – | – | 4 |
| HCV | 0 | 2‡ | 0 | – | – | 2‡ |
| HCC | 0 | 5† | 4† | – | – | 9† |
| PLD | 0 | 0 | 1 | – | – | 1 |
| PSC | 0 | 0 | 1 | – | – | 1 |
| Primary liver disease (AKB) | 17* | |||||
| Colorectal carcinoma | 3 | 7 | 2 | – | 5 | 14* |
| Inflammatory adenoma | 1 | 1 | 0 | – | 0 | 1* |
| Cholangiocellular carcinoma | 0 | 0 | 0 | – | 1 | 1 |
| Hemangioma | 0 | 0 | 0 | - | 1 | 1 |
| CMV serological status | ||||||
| positive/negative/unknown | 4/0/0 | 14/5/0 | 6/1/1 | – | 0/0/7 | 20/6/8 |
*Samples of 4 patients with eiter colorectal carcinoma metastasis or liver adenoma were used in two experiments.
**Hepatocyte organoids from 7 individuals were used for 15 experiments in figure 5.
†HCC was based on: 4x HCV, 1x HBV with HDV, 1x CC, 1x NASH and 2x ALD.
‡One patient suffered from HBV and HCV simultaneously.
AKB, Asklepios Hospital Barmbek liver resection cohort; ALD, alcoholic liver disease; CC, cryptogenic cirrhosis; CMV, cytomegalovirus; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; HDV, hepatitis D virus; LTX liver transplantation cohort; NASH, non-alcoholic steatohepatitis; PBMC, peripheral blood mononuclear cells; PLD, polycystic liver disease; PSC, primary sclerosing cholangitis.
Table 2.
Demographics and characteristics of healthy donors included in this study.
| Healthy donors | Figure 1 | Figure 2 | Figure 3 | Figure 4 | Figure 5 | All |
|---|---|---|---|---|---|---|
| Samples | ||||||
| PBMC | 2 | 5 | – | 8 | – | 13* |
| Demographics | ||||||
| Sex (f/m) | 2/0 | 4/1 | – | 5/3 | – | 9/4 |
| Median age in years (Range) | 27.5 (26-29) | 29 (26-41) | – | 33 (26-43) | – | 33 (26-43) |
| CMV serological status | ||||||
| positive/negative/unknown | 0/2/0 | 0/3/2 | – | 0/3/5 | – | 0/6/7 |
*Samples of 2 healthy donors were used in two experiments.
CMV, cytomegalovirus; PBMC, peripheral blood mononuclear cells.
2.2. Sample processing
Blood sample and liver tissue preparation were performed as previously described (43–45). Peripheral blood mononuclear cells (PBMCs) were obtained using density gradient centrifugation with Ficoll (Biochrom GmbH, Berlin, Germany). Tumor-free liver tissue was dissolved mechanically without enzymatic digestion for the extraction of hepatocytes and intrahepatic NK cells. For liver organoid generation and culture, liver tissue was mechanically and enzymatically digested and cultured as previously described (15, 46). In brief, the liver tissue was cut into small pieces and washed with DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS) and 1% Penicillin/Streptomycin. The tissue pieces were then incubated at 37°C in EBSS (Gibco) with Collagenase D (Sigma-Aldrich) and DNase I (STEMCELL) until single cells were microscopically detectable. The cell solution was then filtered and washed. Single cells were seeded in 50µl droplets of BME2 (Basement Membrane Extract, Type 2, Pathclear) and organoid culture medium (46) and passaged 3 to 4 weeks after the digestion. Further passaging was performed approximately after 7 days of culture. Medium was changed every 2-3 days. PBMCs and liver-derived cells were cryopreserved, stored in liquid nitrogen and thawed for immediate experimental use.
2.3. Antibody staining and flow cytometry
After thawing, liver-derived cells and PBMCs were washed with phosphate buffered saline (PBS) and stained with surface antibodies and the live-dead markers Zombie NIR or Zombie Aqua (BioLegend) for 20 minutes in the dark at room temperature. All antibodies used in this study and corresponding dilutions are listed in Supplementary Table S1 . After washing with PBS, cells were either fixed immediately with 4% paraformaldehyde or fixed and permeabilized using the BD Cytofix/Cytoperm Kit (BD Bioscience), as recommended by the manufacturer, and subsequently stained with intracellular antibodies. The samples were measured using a LSRFortessa™ flow cytometer (BD Bioscience) or a Cytek Aurora (Cytek Biosciences). FlowJo™ version 10.7.1 was used to analyze the data. Gating strategies are shown in the supplementary information ( Figures S1A, B ).
2.4. Single-cell mRNA analysis
Gene expression analyses and data processing were performed as previously described (23, 47, 48). To assess single-cell mRNA expression of selected genes, a Fluidigm platform consisting of C1, Juno and Biomark HD was used. Matched PBMCs and liver-derived cells from four liver resection patients and PBMCs of two healthy donors were sorted for live, CD3−CD14−CD19−CD56brightCD16− NK cells ( Figure S1A ) with a BD FACSAria Fusion (BD Bioscience) into a C1 Single-Cell Preamp Integrated Fluidic Circuit (IFC), 5-10 µm (Fluidigm). Cell lysing, reverse transcription, and preamplification were performed using the C1, according to company protocol (PN 100-4904 K1). 52 primers were used to quantify gene expression and are summarized in the supporting data ( Supplementary Table S2 ). Quantitative PCR was performed using the 96.96 Dynamic Array IFC for Gene Expression in the Biomark HD. All data analysis was performed with the Fluidigm Real-Time PCR Analysis software (Version 4.3.1). The Limit of Detection (LOD) was defined as a Ct value of 24 and subsequently all Ct values above 24 were set to the LOD. Gene expression levels are displayed as 2(LOD-Ct), thus a value of 1 indicates a cell without detectable mRNA expression of the respective gene (49, 50). In the Fluidigm Real-Time PCR Software, all peak detection ranges of the melting curves were defined as the median temperature peak of all non-failed results of the corresponding primer pair +/-1.2°C. Other settings remained unchanged (Peak Sensitivity: 7; Peak Ratio Threshold: 0.8). The Ct value was set to the LOD for all reactions that were marked as failed under these settings.
2.5. Degranulation assay with liver-derived NK cells
Liver-derived cells were thawed in pre-warmed RPMI (Thermo Fisher Scientific) supplemented with 10% FBS (R10) (Biochrom GmbH) and 10 µg/ml DNase. Subsequently, after centrifugation and washing, cells were resuspended at a concentration of 2x106 cells/mL in R10 supplemented with 1 ng/mL IL-15 and rested overnight at 37°C. Cells were washed with PBS and counted before being co-incubated with K562 cells at an effector to target cell (E:T) ratio of 10:1 for 5h at 37°C in the presence of an anti-CD107a-antibody (BV711) and Brefeldin A. Hereafter, cells were stained with surface antibodies and Zombie NIR, as well as stained intracellularly with an anti-TNF-α-antibody (PE-Cy7), as described above.
2.6. Co-culture of peripheral blood NK cells with Huh7 cells
NK cell isolation was performed as recommended by the manufacturer (EasySep Human NK cell Isolation Kit, Stemcell Technologies) with fresh PBMCs from healthy donors. Isolated NK cells were stimulated for 12h with 1 ng/mL IL-15 at 37°C. Afterwards, NK cells were either (co-)incubated with or without Huh7 cells at an E:T ratio of 2:1 in direct contact or in indirect contact using a transwell insert (Sarstedt AG & Co. KG) with pore sizes of 1 µm for 6, 12, 24 and 48h. Finally, cells were stained with antibodies for flow cytometric analysis, as described above. For blocking experiments, Huh7 cells were co-incubated with an anti-PVR blocking antibody (30µg/ml) for 15 min prior to co-culture.
2.7. Co-culture of peripheral blood NK cells and human primary hepatocyte organoids
Hepatocyte organoids were thawed, cultured in 50 µl droplets containing reduced growth factor BME2 and culture medium, as previously described (46). Growing hepatocyte organoids were passaged once before experimental use. Fresh PBMCs from buffy coats were enriched for NK cells using the EasySep™ Human NK Cell Enrichment Kit (Stemcell Technologies) following manufacturers protocols. NK cells were resuspended in RPMI (Thermo Fisher Scientific) supplemented with 10% FBS (Biochrom GmbH) (R10) and 1% Penicillin/Streptomycin and were added to fully-grown hepatocyte organoids. After 24 and 48 hours of co-incubation, cells present in the supernatant or the droplet, respectively, were collected separately, treated with TripLE™ Express Enzyme (Life Technologies GmbH) for organoid dissociation, stained and analyzed by flow cytometry. As controls, NK cells were cultured in R10 supplemented with 1% Penicillin/Streptomycin, but no hepatocyte organoids, and subsequently equally treated with TripLE™ Express Enzyme and stained.
2.8. Multiplex assay
Supernatants from hepatocyte organoids were collected 48h after the last medium change and stored at -80°C until further analysis. The multiplex assay (Bio-Rad) was performed on a Bio-Plex 200 System. Secretion of various chemokines by hepatocyte organoids (CXCL1, CXCL5, CXCL9, CXCL10, CXCL11, CXCL12, CXCL16, CX3CL1, CCL2, CCL5, CCL19, CCL20, and CCL21) was measured.
2.9. Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 and 9 (GraphPad Software, Inc., La Jolla, CA) or R (R 4.0.3 GUI 1.73; R Studio, Version 1.3.1093, lme4, ggplot2, FactoMineR and factoextra packages). Principal component analysis was generated with R using the mRNA expression of 52 genes of 440 single CD56bright ihNK cells or pbNK cells. Wilcoxon matched-pairs signed rank test was used to determine differences between paired samples with assumed non-normal distribution. To assess normal distribution within small sample sizes, the Shapiro-Wilk test was applied and subsequently, the paired t test was used. A mixed-effects model with random intercept, considering intra-sample correlations, was used to compare the single-cell mRNA expression data from different donors (or groups). To limit the detection of false positives, the p-values were adjusted by the Benjamini and Hochberg False Discovery Rate (FDR) method between each comparison groups (cutoff of 0.05). P values <0.05 were considered statistically significant. Statistical analysis for additional data presented in the figures are provided in the respective figure legends.
2.10. Visualization of t-distributed stochastic neighbor embedding (viSNE)
Flow cytometry data were visualized using viSNE (Cytobank, Santa Clara, CA). A tool for visualization of high-dimensional data based on the Barnes-Hut implementation of the t-distributed stochastic neighbor embedding algorithm was used (51). NK cells were equally sampled to randomly select 50 000 cells for analysis. The analysis was performed using the surface expression of CD56, CXCR6, CD69, TIGIT, DNAM-1 and CD96. The following settings were adjusted: 2000 iterations, 20 perplexity, theta 0.5.
3. Results
3.1. Intrahepatic and matched peripheral blood CD56bright NK cells differ in the expression of genes related to liver-residency, migration, and regulatory functions
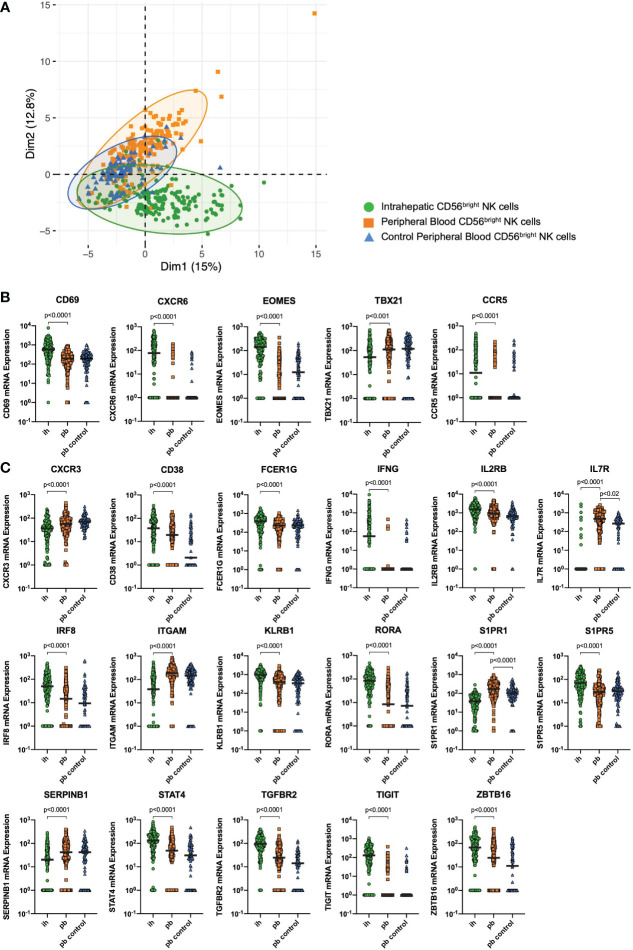
Matched peripheral blood (pb) and intrahepatic (ih) NK cells from patients undergoing liver resection due to either liver metastases (n=3) or liver adenoma (n=1) were used for targeted single-cell mRNA expression analysis in this study. Furthermore, pbNK cells from healthy individuals (n=2) of the Hamburg Healthy Cohort were used as a control. We sorted CD56bright ihNK cells and CD56bright pbNK cells ( Figure S1A ) and investigated the single-cell mRNA expression of 52 representative genes on a total of 440 CD56bright NK cells. All differentially expressed genes can be found in Supplementary Table S3 . Principal component analysis (PCA) revealed divergence of the NK cells depending on their tissue origin. Thus, this unsupervised dimension reduction method showed two partly separating clusters, one for CD56bright ihNK cells and another one for CD56bright pbNK cells, the latter overlapping with the control group of CD56bright pbNK cells ( Figure 1A ). The separation of the PCA-dimension-1 was mainly driven by 18 genes, including genes associated with NK cell activity (e.g. CD38, NCR1, KLRB1) and tissue-residency (e.g. EOMES, TIGIT, CXCR6) ( 16, 22, 52–54). In conclusion, distinct single-cell gene expression discriminates CD56bright pbNK cells and ihNK cells.
Figure 1.
Single-cell mRNA expression. (A) Principal component analysis showing clustering of CD56bright peripheral blood (pb)NK cells from healthy control individuals (blue triangles), CD56bright pbNK cells from liver resection patients (orange squares) and CD56bright intrahepatic (ih)NK cells from liver resection patients (green dots). (B) Quantitative analysis of CD56bright ihNK cells and matched CD56bright pbNK cells from liver resection patients (n=4), and CD56bright pbNK cells from healthy control individuals (n=2) showing single-cell mRNA expression of liver-residency markers CD69, CXCR6, CCR5, Eomes and T-bet (TBX21). (C) Quantitative analysis of CD56bright ihNK cells and matched CD56bright pbNK cells from liver resection patients (n=4), and CD56bright pbNK cells from healthy control individuals (n=2) showing single-cell mRNA expression of selected genes with highly significant differential gene expression comparing ih and pbNK cells. Green dots representing 178 ihNK cells, orange squares representing 173 pbNK cells and blue triangles representing 89 healthy control pbNK cells. A mixed-effects model with random intercept, considering intra-sample correlations, was used to compare the single-cell mRNA expression data from different donors (or groups). Black line indicates median.
We next analyzed whether expression levels of mRNA of the known liver-residency markers CD69, CXCR6, CCR5, Eomes and T-bet differed between CD56bright NK cells derived from livers and matched PBMCs ( Figure 1B ). Compared to CD56bright pbNK cells, CD56bright ihNK cells showed a significantly higher mRNA expression of CD69 (p<0.0001), CXCR6 (p<0.0001), CCR5 (p<0.0001) and the transcription factor EOMES (p<0.0001), while the transcription factor T-bet (TBX21) showed significantly lower (p<0.0001) mRNA expression in CD56bright ihNK cells. This distinct mRNA expression of liver-residency markers in CD56bright ihNK cells is in line with previous results from studies characterizing the phenotype of CD56bright ihNK cells (16, 18–20, 23). In addition to the mentioned differentially expressed known liver-residency markers, we identified 17 genes with a highly significant differential mRNA expression comparing single CD56bright ihNK cells and pbNK cells ( Figure 1C ). CD56bright ihNK cells showed significantly higher mRNA expression of CD38, FCER1G, IFNG, IL2RB, IRF8, KLRB1, RORA, S1PR5, STAT4, TGFBR2, TIGIT and ZBTB16 (all p<0.0001). Most of these genes are primarily associated with regulatory immune functions, anti-inflammatory activity and tissue residency (55–60), while others are involved in pathways of NK cell proliferation (61). In contrast, CD56bright pbNK cells expressed significantly higher levels of CXCR3, IL7R, ITGAM, S1PR1 and SERPINB1 (all p<0.0001), genes that are mainly associated with NK cell migration and maturation (62–67). Interestingly, CD56bright ihNK cells that expressed high mRNA levels of TIGIT also encoded mostly (63%) for CXCR6 and CD69 mRNAs, two markers of liver residency, and also for the chemokine receptor CCR5, associated with tissue homing (see Supplementary Figure S2 ). Taken together, the analysis of gene expression on single NK cells identified significant differences between CD56bright ihNK cells and CD56bright pbNK cells, including the higher expression of genes involved in NK cell proliferation, regulatory functions, and tissue-residency in ihNK cells.
3.2. CD56bright NK cells in liver and peripheral blood differentially express TIGIT, DNAM-1 and CD96
TIGIT is a co-inhibitory receptor that inhibits human NK cell cytotoxicity and has been suggested to be critical in murine liver regeneration (26, 42). The significantly elevated mRNA expression of TIGIT in CD56bright ihNK cells prompted us to seek confirmation for this specific target on protein level by flow cytometry. We included TIGIT’s competing receptors DNAM-1 and CD96 in the analysis. All these receptors bind to the poliovirus receptor (PVR/CD155) and facilitate either immune activation or inhibition (32, 41). We analyzed TIGIT, DNAM-1 and CD96 surface protein expression on CD56bright ihNK cells and matched CD56bright pbNK cells from patients undergoing liver resection or liver transplantation ( Figure 2A ), as well as peripheral blood samples from healthy control individuals ( Figure 2C ). Frequencies of CD56bright and CD56dim NK cell subsets varied between ihNK cells and pbNK cells. CD56bright NK cells were more frequent in ihNK cells compared to pbNK cells or healthy control NK cells (median frequency (%) of CD56bright NK cells: 35%; 3,4%; 6,4%, respectively), while CD56dim NK cells were less frequent in ihNK cells compared to pbNK cells or healthy control NK cells (median frequency (%) of CD56dim NK cells: 40,1%; 88,7%; 82,5%, respectively).
Figure 2.
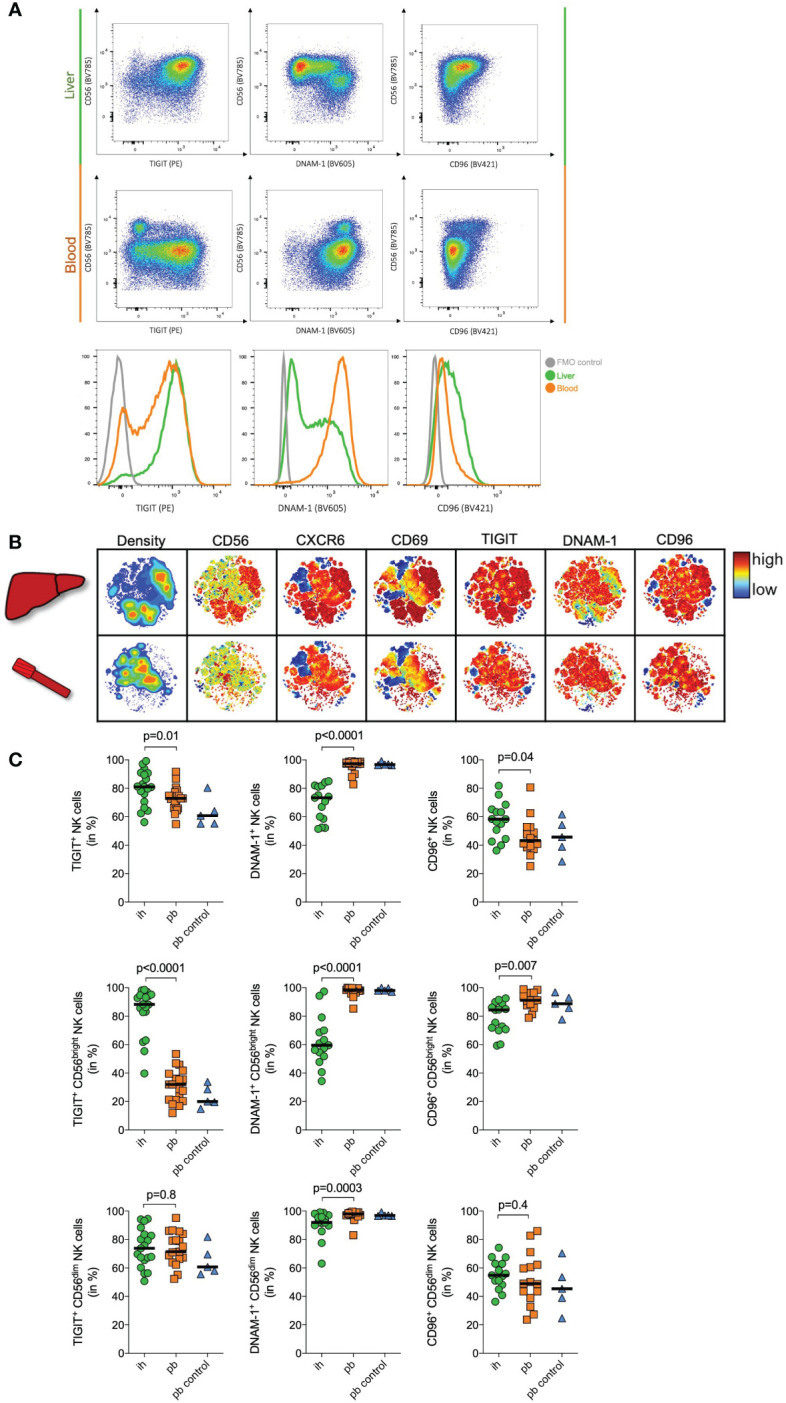
Immunophenotyping of NK cells. (A) Representative plots of flow cytometry data of one donor showing TIGIT, DNAM-1 and CD96 expression on peripheral blood (pb) and intrahepatic (ih) bulk NK cells. Histogram showing TIGIT (left), DNAM-1 (middle) and CD96 (right) expression on bulk NK cells in the liver (green), the blood (orange) and a FMO control (grey). (B) Representative viSNE plots showing density and the expression of CD56, CXCR6, CD69, TIGIT, DNAM-1 and CD96 on ih and pbNK cells from one patient undergoing liver transplantation. Color coding indicates the density and marker expression. viSNE analysis was performed using Cytobank. (C) Quantitative analysis of flow cytometry data of matched ih and pbNK cells from liver resection patients (n=15 for DNAM1- and CD96, n=19 for TIGIT), and control pbNK cells from healthy individuals (n=5) showing TIGIT, DNAM-1 and CD96 expression on bulk, CD56dim and CD56bright NK cells. Green dots representing ihNK cells, orange squares representing pbNK cells and blue triangles representing healthy control pbNK cells. Wilcoxon matched-pairs sign rank test was used to determine statistical differences between ih and pbNKcells in all scatter plots, Mann-Whitney test was used to determine statistical differences between pb and pb control NK cells in all scatter plots. Black line indicates median.
A representative viSNE analysis revealed different clusters when comparing matched CD56bright pbNK cells and ihNK cells from the same individual ( Figure 2B ). We identified clusters with high density and overlapping expression of CD56, liver-residency markers CD69, CXCR6, TIGIT and CD96. However, those clusters showed a distinctly lower expression of DNAM-1. When analyzing both subsets in detail, bulk ihNK cells showed higher TIGIT (p=0.01) and CD96 (p=0.04) expression ( Figure 2C ). In contrast, bulk pbNK cells showed a higher DNAM-1 expression (p<0.0001), which is consistent with a previous study demonstrating significantly higher DNAM-1 expression on pbNK cells (21). Regarding the CD56dim NK cell subset, TIGIT (p=0.8) and CD96 (p=0.4) were equally expressed on pb and ihNK cells ( Figure 2C ). However, CD56dim pbNK cells exhibited a significantly higher expression of DNAM-1 (p=0.0003). Differences in surface protein expression between pb and ihNK cells were most pronounced in the CD56bright NK cell compartment ( Figure 2C ). In line with the corresponding mRNA data, we observed a significantly stronger surface protein expression of the inhibitory receptor TIGIT on CD56bright ihNK cells than in CD56bright pbNK cells (p<0.0001). In contrast, the CD56bright pbNK cells showed significantly higher expression of DNAM-1 (p<0.0001) and CD96 (p=0.007) compared to their intrahepatic counterpart. Taken together, ihNK cells and especially the subset of CD56bright ihNK cells showed higher levels of TIGIT, whereas pbNK cells distinctly exhibited higher expressions of DNAM-1.
3.3. Intrahepatic TIGIT+ CD56bright NK cells exhibit diminished degranulation and TNF-α production
Previous data in mice and humans have suggested that TIGIT+ NK cells are functionally exhausted (68, 69), but the role of TIGIT in human ihNK cells is not well understood. To assess functional differences between TIGIT+ and TIGIT– CD56bright ihNK cells, we next determined tumor necrosis factor alpha (TNF-α) production and degranulation (CD107a expression) after stimulation of intrahepatic cells from patients undergoing liver transplantation (n=6) or liver metastases resection (n=2) with K562 target cells ( Figure 3 ). After stimulation, we observed significantly less degranulation of TIGIT+ CD56bright ihNK cells compared to TIGIT– CD56bright in NK cells (p=0.02). Moreover, the TIGIT+ CD56bright ihNK cells also exhibited a significantly diminished TNF-α response compared to TIGIT– CD56bright ihNK cells (p=0.03). Overall, TIGIT+ CD56bright ihNK cells were significantly less responsive to stimulation than TIGIT– CD56bright ihNK cells, in line with previous findings describing reduced functionality of TIGIT+ NK cells (68, 69).
Figure 3.
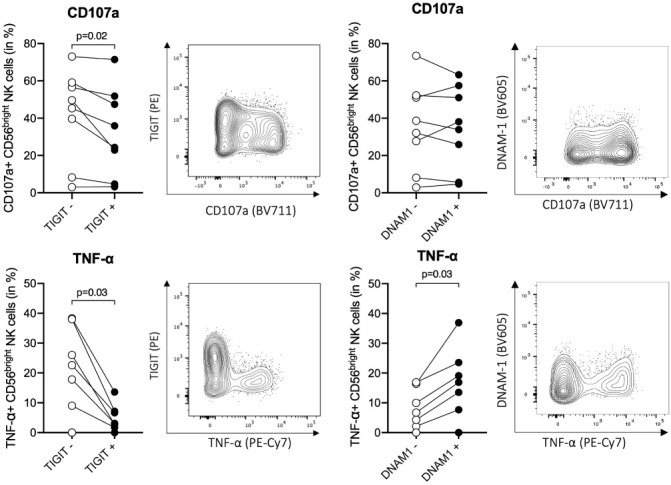
Functional activity of TIGIT+/– and DNAM-1+/– cells. Functional responses of TIGIT+/– and DNAM-1+/– CD56bright ihNK cells, following co-incubation of liver-derived cells with K562 (E:T ratio of 10:1) for 5h at 37°C. Representative flow plots showing distribution of CD107a (top) and TNF-α (bottom) expression on CD56bright ihNK cells depending on their TIGIT or DNAM-1 expression. Graphs on top showing percentage of CD107a+ TIGIT+/– or DNAM-1+/–CD56bright ihNK cells and graphs below showing percentage of TNF-α+ TIGIT+/– or DNAM-1+/–CD56bright ihNK cells. Wilcoxon matched-pairs sign rank test was used to determine statistical differences. Each dot represents one donor (n=8).
3.4. Co-culture of peripheral blood NK cells with human hepatoma cells and hepatocyte organoids induced NK cell migration and an intrahepatic TIGIT+ NK cell phenotype
The above data demonstrate that CD56bright ihNK cells express higher levels of TIGIT on both mRNA and protein levels, and that TIGIT+ NK cells show reduced functionality. We next determined whether the liver environment itself promoted an increased immunotolerance of ihNK cells, as the ligand for TIGIT, PVR/CD155, is expressed by hepatocytes (42). For this purpose, we co-cultured isolated pbNK cells from healthy individuals with the human hepatoma cell line Huh7, which stably expresses PVR ( Figure 4C ). To be able to differentiate between effects of direct receptor binding or soluble stimulation, NK cells were cultured either in direct contact with Huh7 cells or separated via a transwell insert ( Figure 4A ). Subsequently, the expression of TIGIT, DNAM-1 and CD96 was assessed ( Figure 4B ). Regardless of the condition, we did not observe any differences in CD96 expression up to 48 hours of co-culture. DNAM-1 expression in CD56bright NK cells started to decline after 6 hours and continued to decrease up to 48 hours during co-culture, but only when in direct contact with the Huh7 cells (p=0.003). After 48 hours of co-incubation, DNAM-1 expression decreased significantly by almost 40% (p=0.01). However, no effect on the modulation of DNAM-1 expression was observed in the transwell experiments or in the absence of Huh7 cells, suggesting that direct cell-to-cell contact between NK cells and hepatoma cells was required to induce DNAM-1 downregulation. In contrast, TIGIT expression started to increase within 12 hours upon direct contact with Huh7 cells. This increase was even more pronounced after 24 hours of co-culture of CD56bright NK cells in direct contact with Huh7 cells. Again, no effect on the modulation of TIGIT expression was detectable in CD56bright NK cells cultured without Huh7 cells, and only minor changes in TIGIT expression were detected in transwell experiments. To start to elucidate the contribution of different factors to the observed TIGIT upregulation on NK cells, we compared the receptor expression of TIGIT on NK cells after direct co-incubation with Huh7 cells alone or in the presence of an anti-PVR blocking antibody ( Figure 4D ). As observed before, direct co-culture with Huh7 cells resulted in a significant upregulation of TIGIT on NK cells, which was significantly reduced, but not fully suppressed, by addition of the PVR-blocking antibody. Taken together, these data show that CD56bright pbNK cells altered their phenotypes towards CD56bright ihNK cells when cultured together with PVR-expressing human hepatoma cells, by downregulating activating and simultaneously upregulating inhibitory NK cell receptors that bind to PVR. These data suggest a direct impact of the liver environment on the phenotype and functional activity of ihNK cells.
Figure 4.
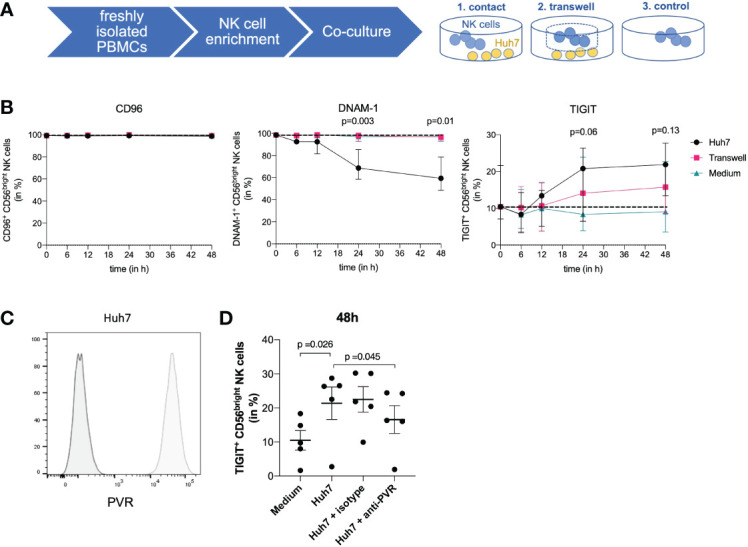
Co-culture of NK cells and Huh7 cells. (A) Co-culture experiments using isolated NK cells from peripheral blood from healthy control individuals and Huh7 cells. NK cells were either cultured in direct contact with Huh7 cells, in a transwell insert with the size of 1 µm to ensure indirect contact with Huh7 cells or with medium only. (B) Graphs are displaying quantitative analysis of CD96, DNAM-1 and TIGIT expression on CD56bright NK cells after 6, 12, 24 and 48 hours. At the timepoints 6, 12, and 24 hours a total of 5 healthy control individuals was used for comparison. At 48 hours, a total of 4 out of the 5 mentioned healthy control individuals were assessed for CD96, DNAM-1 and TIGIT. Black dots representing direct contact, pink squares the transwell insert and blue triangles medium only. Paired t-test was used to determine the displayed statistical differences between direct contact of NK cells and medium control. Each dot represents the median of the individuals; error bars represent the data range. (C) Representative histograms comparing PVR staining (light grey) and unstained control (dark grey) demonstrating PVR expression on Huh7 cells. (D) Graph displays the percentage of TIGIT+ NK cells in different conditions: NK cells only, NK cells in direct contact with Huh7 cells, isotype control and NK cells in direct contact with Huh7 cells including blocking of PVR with an anti-PVR-antibody. Data are shown for 5 paired experiments using NK cells derived from 3 different donors. Statistical analysis for significance was performed using a two-tailed paired T test.
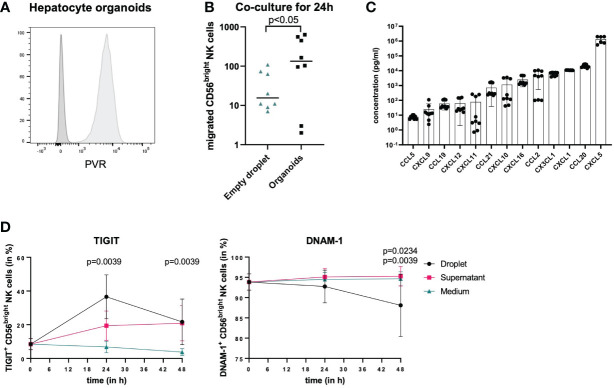
To further determine whether the observed changes in pbNK cells in the presence of hepatoma cells were also observed in response to human primary hepatocytes, we co-cultured hepatocyte organoids, which stably express PVR ( Figure 5A ) and were derived from human liver tissue and were growing within BME2 droplets with pbNK cells. pbNK cells migrate towards hepatocyte organoids, as the number of NK cells entering hepatocyte organoids and the surrounding BME2 droplets was significantly higher in wells with hepatocyte organoids compared to wells only containing empty BME2 droplets after 24 hours ( Figure 5B ). To assess which chemokines attracting NK cells are secreted by hepatocyte organoids, chemokine levels in the cell-culture supernatant from hepatocyte organoids were analyzed ( Figure 5C ). Chemokine levels were detected at different concentrations ranging from mean concentrations below 100 pg/ml (CCL5, CXCL9, CCL19, CXCL12, CXCL11) to concentrations above 700 pg/ml (CCL21, CXCL10, CXCL16, CCL2, CX3CL1, CXCL1, CCL20, CXCL5) ( Figure 5C ). The results from these co-culture studies demonstrate that hepatocyte organoids secrete chemokines that attract NK cells.
Figure 5.
Co-culture of NK cells and hepatocyte organoids. (A) Representative histograms comparing PVR staining (light grey) and unstained control (dark grey) of hepatocyte organoids demonstrating PVR expression on hepatocytes. (B) NK cells isolated from buffy coats were co-cultured with empty BME2 droplets or hepatocyte organoids in BME2 droplets for up to 48h. Graph displaying total cell count of CD56bright NK cells that migrated into BME2 droplets at 24 hours of co-culture with empty droplets (blue triangles) or hepatocyte organoids (black dots). Wilcoxon matched-pairs sign rank test was used for statistical analysis. Black line indicates median. (n=3, triplicates) (C) Chemokine concentrations in hepatocyte organoid culture supernatants are shown in ascending order on a logarithmic scale. Bars represent mean values of three experiments in triplicates for each chemokine and error bars show standard deviations (n=3, triplicates). (D) Migrated CD56bright NK cells (black dots) were phenotypically compared to non-migrated cells (pink squares) and to separately cultured NK cells that did not have any contact to hepatocyte organoids (blue rectangles). Percentage surface expression of TIGIT and DNAM-1 is shown in CD56bright NK cells after 24 hours and 48 hours. For TIGIT, statistical differences represent comparison of migrated NK cells as well as non-migrated NK cells vs. NK cells in medium and for DNAM-1 statistical differences represent migrated NK cells vs. NK cells in medium (top value) or non-migrated cells (lower value). Wilcoxon matched-pairs sign rank test was used for statistical analysis. Each dot represents the median of nine experiments; error bars represent the data range.
To identify phenotypical changes in CD56bright pbNK cells following migration into hepatocyte organoids, we subsequently analyzed TIGIT and DNAM-1 surface expression of CD56bright NK cells upon co-culture with hepatocyte organoids compared to CD56bright NK cells in medium only (controls), and furthermore differentiated those NK cells that entered hepatocyte organoids within BME2 droplets from those NK cells that remained outside of BME2 droplets. Surface expression of TIGIT and DNAM-1 on NK cells was analyzed after 24 and 48 hours of co-culture ( Figure 5D ). Both, CD56bright NK cells that entered BME2 droplets and those that did not showed an increased TIGIT expression after co-culture with hepatocyte organoids compared to control conditions. However, the percentage of TIGIT+ NK cells that migrated into hepatocyte organoids was already significantly higher after 24 hours compared to control conditions without organoids, while the frequency of TIGIT+ NK cells outside of the BME2 droplet (supernatant) increased more slowly ( Figure 5D ). Furthermore, DNAM-1 expression on NK cells continuously decreased only on those NK cells that entered the hepatocyte organoids, reaching significantly lower levels after 48 hours of co-incubation, while no decrease in percentages of DNAM-1 expressing NK cells was observed under the other conditions ( Figure 5D ). These data suggest that TIGIT and DNAM-1 expression on NK cells is preferentially modulated by direct cell-to-cell-contact and that primary human hepatocyte organoids can be used as an in vitro model to study migration of immune cells and their interactions with tissue cells.
4. Discussion
The liver plays a central role in mediating both local and systemic tolerance to self and foreign antigens, and this immune-regulatory capacity has been attributed to specialized liver-resident cells. NK cells make up 40% of total lymphocytes in human livers, and increasing evidence indicates that NK cells play an important role in the regulation of tissue-specific immunity (6). Liver-resident NK cells are mainly CD56bright NK cells that express a distinctive profile of surface receptors and transcription factors (15, 16, 18–20). Unraveling NK cell functions that contribute to tissue homeostasis in the liver is critical to acquire a better understanding of hepatic immune regulation. In this study, we report transcriptional, functional, and phenotypical differences between CD56bright ihNK cells and CD56bright pbNK cells. CD56bright ihNK cells exhibited significant upregulation of genes associated with anti-inflammatory activity and cell proliferation, whereas CD56bright pbNK cells expressed higher levels of genes involved in NK cell migration and development. These data confirm that CD56bright ihNK cells and CD56bright pbNK cells represent two distinct NK cell populations and demonstrate that CD56bright ihNK cells acquire phenotypical and functional characteristics associated with reduced responsiveness, potentially contributing to the tolerogenic environment of the liver.
Data on distinct gene expression profiles of CD56bright ihNK cells presented here are in line with data from gene expression analyses of liver perfusates and peripheral blood (20), as well as with findings of single-cell RNAseq analyses revealing distinct liver NK cell populations (70–73). Our PCA analyses reflected clear divergence of NK cells depending on their tissue origin, while small differences in gene expression of CD56bright pbNK cells between individuals with liver diseases and healthy controls, which might have been impacted by additional variables such as sex, age, or disease status. In ihNK cells, we observed an upregulation of the co-inhibitory receptor TIGIT on CD56bright NK cell populations that also expressed the known liver-residency markers CD69 and CXCR6. The differences in the expression of TIGIT, DNAM-1 and CD96 surface receptors, which all interact with PVR, were most striking when comparing CD56bright ihNK cell to pbNK cell subsets. Consistent with previous findings (40, 68, 74), we observed a functional impairment of TIGIT+ CD56bright ihNK cells after co-incubation with K562 target cells. Importantly, co-culture with PVR-expressing human hepatoma cells or primary hepatocyte organoids enhanced TIGIT expression and reduced DNAM-1 expression on CD56bright pbNK cells, suggesting a potential influence of the liver environment on the phenotype and function of ihNK cells.
Given the significant differences in mRNA expression profiles between CD56bright ihNK cells and CD56bright pbNK cells, our phenotypical and functional studies focused on the co-inhibitory receptor TIGIT and its competing receptors DNAM-1 and CD96. Since the first identification of TIGIT (26–28), multiple studies have shown that TIGIT can promote T cell suppression and that it is also involved in impairing NK cell effector functions (27, 68). A recent study of Doyle and colleagues correlated better liver functions in HCV patients with a greater abundance of CD56bright ihNK cells (75). Additionally, the study demonstrated that portal vein blood contained high plasma levels of IL-10. Interestingly, TIGIT can promote IL-10 production of dendritic cells following engagement of PVR, resulting in a reduction of T cell functions (27, 75). Furthermore, Bi et al. described TIGITs role as a safeguard molecule in liver regeneration by negatively regulating NK-hepatocyte crosstalk in mice (42). In our study, ihNK cells and in particular the subset of CD56bright ihNK cells exhibited higher levels of TIGIT, whereas pbNK cells expressed higher levels of DNAM-1. These differences in receptor profiles resulted in functional differences, since TIGIT+ NK cells were significantly less responsive to stimulation than DNAM-1+ NK cells. While this reduced responsiveness of TIGIT+ CD56bright ihNK cells could contribute to the tolerogenic immune environment of the liver, it might also represent a disadvantage in the context of infections and cancers, resulting in a “sanctuary compartment” within the liver, as observed in the context of chronic infections with hepatotropic viruses. Furthermore, several studies have provided increasing evidence that high TIGIT expression is observed on tumor infiltrating NK cells and is associated with their functional exhaustion (40, 68), and that TIGIT blocking can boost anti-tumor immunity (41, 68).
To better understand the mechanisms underlying TIGIT upregulation and DNAM-1 downregulation on CD56bright ihNK cells, we co-incubated pbNK cells with hepatoma cells or hepatocyte organoids in direct cell-to-cell contact or in a transwell setup, and compared the differential expression levels of TIGIT and DNAM-1 on CD56bright pbNK cells. While we observed a small increase in TIGIT expression on CD56bright pbNK cells co-cultured with hepatoma cells in the transwell condition, CD56bright pbNK cells upregulated TIGIT expression more strongly following direct contact with human hepatoma cells, and this upregulation was significantly reduced, but not fully suppressed, in the presence of an antibody blocking PVR. During co-cultivation with hepatocyte organoids, CD56bright pbNK cells migrated towards the hepatocyte organoids, which released several chemokines that act as chemoattractants to NK cells. The release of CXCL1, CX3CL1, CXCL10 and CCL21 in the cell-culture supernatant of hepatocyte organoids might have contributed to NK cell migration, as these chemokines are established NK cell attractants (76, 77). Particularly, CXCL10 might have attracted CD56bright pbNK cells, as the receptor CXCR3 is involved in NK cell migration and homing and is especially expressed on CD56bright pbNK cells (77). Furthermore, CD56bright NK cells express CCR7 that interacts with CCL21, which therefore might also be involved in the observed migration (76, 77). Additionally, CX3CL1 can contribute to NK cell attraction by serving as a ligand for CX3CR1, which regulates NK cell migration and is expressed, at least at low levels, on CD56bright NK cells (78).
Similar to CD56bright pbNK cells that were co-cultured with hepatoma cells, CD56bright pbNK cells that migrated towards primary hepatocyte organoids preferentially upregulated TIGIT expression. These data indicate that direct cellular interactions with hepatocytes expressing PVR might contribute to the induction of TIGIT expression on NK cells. Furthermore, co-incubation of peripheral blood-derived CD56bright NK cells with Huh7 cells and hepatocyte organoids resulted in a pronounced downregulation of DNAM-1, in line with previous studies showing that physical binding to PVR can downregulate DNAM-1 on NK cells (69, 79). Taken together, our data using primary human NK cells in co-culture with primary hepatocyte organoids show that these 3D organoid systems can be used to study interactions between human immune and tissue cells in vitro, and suggest that TIGIT upregulation and DNAM-1 downregulation are parallel events promoted by direct contact of NK cells with PVR expressed on hepatic cells, representing an adaptation of circulating NK cells that home to the tolerogenic environment of the liver.
Data availability statement
Storage of data is performed by the Leibniz Institute of Virology on an internal server. Raw data will be made available upon request and can be shared after confirming that data will be used within the scope of the originally provided informed consent. Requests to access these datasets should be directed to the corresponding author.
Ethics statement
Study protocols (PV4898, PV4081, and PV4780) were approved by the ethics committee of the medical association of Hamburg. The patients/participants provided their written informed consent to participate in this study.
Author contributions
AZ, LM, PF, MB, SL and MA designed the study. AZ, LM, PF, KH, GM, LH, GR, SML, SB, BP, TP, AN, and SL performed experiments. AZ, LM, PF, and KH analyzed the data. SH gave critical intellectual input. JH and SH helped design and perform mRNA analysis. MR performed statistical analysis of mRNA data and LR gave statistical guidance. AZ wrote the manuscript with the support of PF, LM, MB, SL, and MA. KO, SP, and LF provided patient samples. All authors critically reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
We would like to thank all donors at the University Clinical Center Hamburg-Eppendorf (UKE) and the Asklepios Hospital Barmbek (AKB), as well as all the healthy donors for their participation in our study. Furthermore, we thank all the nurses and doctors at the UKE and the AKB for their involvement. We would also like to thank Arne Düsedau at the LIV Technology Platform Flow Cytometry/FACS.
Funding Statement
Supported by the German Research Foundation (SFB841 to MA, CS, SL; SFB841 graduate school to AZ, LH, SML, and LM; KFO306 to MA, CS, AA, TP, GR) and State Research Funding (LFF FV78 to SB). CS is funded by the Helmut und Hannelore Greve Foundation. The funders had no influence on study design, data collection, and analysis, the decision to publish or contents of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1117320/full#supplementary-material
References
- 1. Schleinitz N, Vély F, Harlé JR, Vivier E. Natural killer cells in human autoimmune diseases. Immunology (2010) 131:451–8. doi: 10.1111/j.1365-2567.2010.03360.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol (2008) 9:503–10. doi: 10.1038/ni1582 [DOI] [PubMed] [Google Scholar]
- 3. Tosello-Trampont A, Surette FA, Ewald SE, Hahn YS. Immunoregulatory role of NK cells in tissue inflammation and regeneration. Front Immunol (2017) 8. doi: 10.3389/fimmu.2017.00301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jost S, Altfeld M. Control of human viral infections by natural killer cells. Annu Rev Immunol (2013) 31:163–94. doi: 10.1146/annurev-immunol-032712-100001 [DOI] [PubMed] [Google Scholar]
- 5. Hanna J, Goldman-Wohl D, Hamani Y, Avraham I, Greenfield C, Natanson-Yaron S, et al. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat Med (2006) 12:1065–74. doi: 10.1038/nm1452 [DOI] [PubMed] [Google Scholar]
- 6. Jenne CN, Kubes P. Immune surveillance by the liver. Nat Immunol (2013) 14:996–1006. doi: 10.1038/ni.2691 [DOI] [PubMed] [Google Scholar]
- 7. Mikulak J, Bruni E, Oriolo F, Di Vito C, Mavilio D. Hepatic natural killer cells: Organ-specific sentinels of liver immune homeostasis and physiopathology. Front Immunol (2019) 10. doi: 10.3389/fimmu.2019.00946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao B, Jeong W-I, Tian Z. Liver: An organ with predominant innate immunity. Hepatology (2008) 47:729–36. doi: 10.1002/hep.22034 [DOI] [PubMed] [Google Scholar]
- 9. Norris S, Collins C, Doherty DG, Smith F, McEntee G, Traynor O, et al. Resident human hepatitis lymphocytes are phenotypically different from circulating lymphocytes. J Hepatol (1998) 28:84–90. doi: 10.1016/S0168-8278(98)80206-7 [DOI] [PubMed] [Google Scholar]
- 10. Highton AJ, Schuster IS, Degli-Esposti MA, Altfeld M. The role of natural killer cells in liver inflammation. Semin Immunopathol (2021) 43:519–33. doi: 10.1007/s00281-021-00877-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peng H, Tian Z. Diversity of tissue-resident NK cells. Semin Immunol (2017) 31:3–10. doi: 10.1016/j.smim.2017.07.006 [DOI] [PubMed] [Google Scholar]
- 12. Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, et al. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J Clin Invest (2013) 123:1444–56. doi: 10.1172/JCI66381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, et al. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol (2010) 11:1127–35. doi: 10.1038/ni.1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poli A, Michel T, Thérésine M, Andrès E, Hentges F, Zimmer J, et al. CD56bright natural killer (NK) cells: An important NK cell subset. Immunology (2009) 126:458–65. doi: 10.1111/j.1365-2567.2008.03027.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lunemann S, Martrus G, Goebels H, Kautz T, Langeneckert A, Salzberger W, et al. Hobit expression by a subset of human liver-resident CD56(bright) natural killer cells. Sci Rep (2017) 7:6676. doi: 10.1038/s41598-017-06011-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harmon C, Robinson MW, Fahey R, Whelan S, Houlihan DD, Geoghegan J, et al. Tissue-resident eomes(hi) T-bet(lo) CD56(bright) NK cells with reduced proinflammatory potential are enriched in the adult human liver. Eur J Immunol (2016) 46:2111–20. doi: 10.1002/eji.201646559 [DOI] [PubMed] [Google Scholar]
- 17. Moroso V, Metselaar HJ, Mancham S, Tilanus HW, Eissens D, van der Meer A, et al. Liver grafts contain a unique subset of natural killer cells that are transferred into the recipient after liver transplantation. Liver Transplant (2010) 16:895–908. doi: 10.1002/lt.22080 [DOI] [PubMed] [Google Scholar]
- 18. Marquardt N, Beziat V, Nystrom S, Hengst J, Ivarsson MA, Kekalainen E, et al. Cutting edge: Identification and characterization of human intrahepatic CD49a+ NK cells. J Immunol (2015) 194:2467–71. doi: 10.4049/jimmunol.1402756 [DOI] [PubMed] [Google Scholar]
- 19.Stegmann KA, Robertson F, Hansi N, Gill U, Pallant C, Christophides T, et al. CXCR6 marks a novel subset of T-bet(lo)Eomes(hi) natural killer cells residing in human liver. Sci Rep (2016) 6:26157. doi: 10.1038/srep26157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hudspeth K, Donadon M, Cimino M, Pontarini E, Tentorio P, Preti M, et al. Human liver-resident CD56(bright)/CD16(neg) NK cells are retained within hepatic sinusoids via the engagement of CCR5 and CXCR6 pathways. J Autoimmun (2016) 66:40–50. doi: 10.1016/j.jaut.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martrus G, Kautz T, Lunemann S, Richert L, Glau L, Salzberger W, et al. Proliferative capacity exhibited by human liver-resident CD49a+CD25+ NK cells. PloS One (2017) 12:e0182532. doi: 10.1371/journal.pone.0182532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cuff AO, Robertson FP, Stegmann KA, Pallett LJ, Maini MK, Davidson BR, et al. Eomeshi NK cells in human liver are long-lived and do not recirculate but can be replenished from the circulation. J Immunol (2016) 197:4283–91. doi: 10.4049/jimmunol.1601424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hess LU, Martrus G, Ziegler AE, Langeneckert AE, Salzberger W, Goebels H, et al. The transcription factor promyelocytic leukemia zinc finger protein is associated with expression of liver-homing receptors on human blood CD56bright natural killer cells. Hepatol Commun (2020) 4:409–24. doi: 10.1002/hep4.1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Björkström NK, Ljunggren HG, Michaëlsson J. Emerging insights into natural killer cells in human peripheral tissues. Nat Rev Immunol (2016) 16:310–20. doi: 10.1038/nri.2016.34 [DOI] [PubMed] [Google Scholar]
- 25. Bryceson YT, Chiang SC, Darmanin S, Fauriat C, Schlums H, Theorell J, et al. Molecular mechanisms of natural killer cell activation. J Innate Immun (2011) 3:216–26. doi: 10.1159/000325265 [DOI] [PubMed] [Google Scholar]
- 26. Stanietsky N, Simic H, Arapovic J, Toporik A, Levy O, Novik A, et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc Natl Acad Sci U.S.A. (2009) 106:17858–63. doi: 10.1073/pnas.0903474106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol (2009) 10:48–57. doi: 10.1038/ni.1674 [DOI] [PubMed] [Google Scholar]
- 28. Boles KS, Vermi W, Facchetti F, Fuchs A, Wilson TJ, Diacovo TG, et al. A novel molecular interaction for the adhesion of follicular CD4 T cells to follicular DC. Eur J Immunol (2009) 39:695–703. doi: 10.1002/eji.200839116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reches A, Ophir Y, Stein N, Kol I, Isaacson B, Charpak Amikam Y, et al. Nectin4 is a novel TIGIT ligand which combines checkpoint inhibition and tumor specificity. J ImmunoTher Cancer (2020) 8:e000266. doi: 10.1136/jitc-2019-000266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lopez M, Aoubala M, Jordier F, Isnardon D, Gomez S, Dubreuil P, et al. The human poliovirus receptor related 2 protein is a new hematopoietic/endothelial homophilic adhesion molecule. Blood (1998) 92:4602–11. doi: 10.1182/blood.V92.12.4602 [DOI] [PubMed] [Google Scholar]
- 31. Sloan KE, Eustace BK, Stewart JK, Zehetmeier C, Torella C, Simeone M, et al. CD155/PVR plays a key role in cell motility during tumor cell invasion and migration. BMC Cancer (2004) 4:73. doi: 10.1186/1471-2407-4-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anderson AC, Joller N, Kuchroo VK. Lag-3, Tim-3, and TIGIT: Co-inhibitory receptors with specialized functions in immune regulation. Immunity (2016) 44:989–1004. doi: 10.1016/j.immuni.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, Carnemolla B, et al. Identification of PVR (CD155) and nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med (2003) 198:557–67. doi: 10.1084/jem.20030788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fuchs A, Cella M, Giurisato E, Shaw AS, Colonna M. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155). J Immunol (2004) 172:3994–8. doi: 10.4049/jimmunol.172.7.3994 [DOI] [PubMed] [Google Scholar]
- 35. Fittje P, Hoelzemer A, Garcia-Beltran WF, Vollmers S, Niehrs A, Hagemann K, et al. HIV-1 nef-mediated downregulation of CD155 results in viral restriction by KIR2DL5+ NK cells. PloS Pathog (2022) 18:e1010572. doi: 10.1371/journal.ppat.1010572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Husain B, Ramani SR, Chiang E, Lehoux I, Paduchuri S, Arena TA, et al. A platform for extracellular interactome discovery identifies novel functional binding partners for the immune receptors B7-H3/CD276 and PVR/CD155. Mol Cell Proteomics (2019) 18:2310–23. doi: 10.1074/mcp.TIR119.001433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gilfillan S, Chan CJ, Cella M, Haynes NM, Rapaport AS, Boles KS, et al. DNAM-1 promotes activation of cytotoxic lymphocytes by nonprofessional antigen-presenting cells and tumors. J Exp Med (2008) 205:2965–73. doi: 10.1084/jem.20081752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iguchi-Manaka A, Kai H, Yamashita Y, Shibata K, Tahara-Hanaoka S, Honda S, et al. Accelerated tumor growth in mice deficient in DNAM-1 receptor. J Exp Med (2008) 205:2959–64. doi: 10.1084/jem.20081611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nabekura T, Kanaya M, Shibuya A, Fu G, Gascoigne NR, Lanier LL, et al. Costimulatory molecule DNAM-1 is essential for optimal differentiation of memory natural killer cells during mouse cytomegalovirus infection. Immunity (2014) 40:225–34. doi: 10.1016/j.immuni.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun H, Huang Q, Huang M, Wen H, Lin R, Zheng M, et al. Human CD96 correlates to natural killer cell exhaustion and predicts the prognosis of human hepatocellular carcinoma. Hepatology (2019) 70:168–83. doi: 10.1002/hep.30347 [DOI] [PubMed] [Google Scholar]
- 41. Chan CJ, Martinet L, Gilfillan S, Souza-Fonseca-Guimaraes F, Chow MT, Town L, et al. The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat Immunol (2014) 15:431–8. doi: 10.1038/ni.2850 [DOI] [PubMed] [Google Scholar]
- 42. Bi J, Zheng X, Chen Y, Wei H, Sun R, Tian Z. TIGIT safeguards liver regeneration through regulating natural killer cell-hepatocyte crosstalk. Hepatology (2014) 60:1389–98. doi: 10.1002/hep.27245 [DOI] [PubMed] [Google Scholar]
- 43. Salzberger W, Martrus G, Bachmann K, Goebels H, Heß L, Koch M, et al. Tissue-resident NK cells differ in their expression profile of the nutrient transporters Glut1, CD98 and CD71. PloS One (2018) 13:e0201170. doi: 10.1371/journal.pone.0201170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Martrus G, Goebels H, Langeneckert AE, Kah J, Flomm F, Ziegler AE, et al. CD49a expression identifies a subset of intrahepatic macrophages in humans. Front Immunol (2019) 10:1247. doi: 10.3389/fimmu.2019.01247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Langeneckert AE, Lunemann S, Martrus G, Salzberger W, Hess LU, Ziegler AE, et al. CCL21-expression and accumulation of CCR7(+) NK cells in livers of patients with primary sclerosing cholangitis. Eur J Immunol (2019) 49:758–69. doi: 10.1002/eji.201847965 [DOI] [PubMed] [Google Scholar]
- 46. Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell (2015) 160:299–312. doi: 10.1016/j.cell.2014.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hagen SH, Henseling F, Hennesen J, Savel H, Delahaye S, Richert L, et al. Heterogeneous escape from X chromosome inactivation results in sex differences in type I IFN responses at the single human pDC level. Cell Rep (2020) 33:108485. doi: 10.1016/j.celrep.2020.108485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hagen SH, Hennesen J, Altfeld M. Assessment of escape from X chromosome inactivation and gene expression in single human immune cells. STAR Protoc (2021) 2:100641. doi: 10.1016/j.xpro.2021.100641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hipp N, Symington H, Pastoret C, Caron G, Monvoisin C, Tarte K, et al. IL-2 imprints human naive b cell fate towards plasma cell through ERK/ELK1-mediated BACH2 repression. Nat Commun (2017) 8:1443. doi: 10.1038/s41467-017-01475-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Livak KJ, Wills QF, Tipping AJ, Datta K, Mittal R, Goldson AJ, et al. Methods for qPCR gene expression profiling applied to 1440 lymphoblastoid single cells. Methods (2013) 59:71–9. doi: 10.1016/j.ymeth.2012.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol (2013) 31:545–52. doi: 10.1038/nbt.2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zambello R, Barilà G, Manni S, Piazza F, Semenzato G. NK cells and CD38: Implication for (Immuno)Therapy in plasma cell dyscrasias. Cells (2020) 9(3):768. doi: 10.3390/cells9030768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barrow AD, Martin CJ, Colonna M. The natural cytotoxicity receptors in health and disease. Front Immunol (2019) 10. doi: 10.3389/fimmu.2019.00909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Harmon C, Jameson G, Almuaili D, Houlihan DD, Hoti E, Geoghegan J, et al. Liver-derived TGF-β maintains the Eomes(hi)Tbet(lo) phenotype of liver resident natural killer cells. Front Immunol (2019) 10:1502. doi: 10.3389/fimmu.2019.01502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang Z, Gothe F, Pennamen P, James JR, McDonald D, Mata CP, et al. Human interleukin-2 receptor β mutations associated with defects in immunity and peripheral tolerance. J Exp Med (2019) 216:1311–27. doi: 10.1084/jem.20182304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Duhan V, Hamdan TA, Xu HC, Shinde P, Bhat H, Li F, et al. NK cell-intrinsic FcϵRIγ limits CD8+ T-cell expansion and thereby turns an acute into a chronic viral infection. PloS Pathog (2019) 15:e1007797. doi: 10.1371/journal.ppat.1007797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kamishikiryo J, Fukuhara H, Okabe Y, Kuroki K, Maenaka K. Molecular basis for LLT1 protein recognition by human CD161 protein (NKRP1A/KLRB1). J Biol Chem (2011) 286:23823–30. doi: 10.1074/jbc.M110.214254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morandi F, Horenstein AL, Chillemi A, Quarona V, Chiesa S, Imperatori A, et al. CD56brightCD16- NK cells produce adenosine through a CD38-mediated pathway and act as regulatory cells inhibiting autologous CD4+ T cell proliferation. J Immunol (2015) 195:965–72. doi: 10.4049/jimmunol.1500591 [DOI] [PubMed] [Google Scholar]
- 59. Nayyar G, Chu Y, Cairo MS. Overcoming resistance to natural killer cell based immunotherapies for solid tumors. Front Oncol (2019) 9:51. doi: 10.3389/fonc.2019.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Marquardt N, Kekäläinen E, Chen P, Lourda M, Wilson JN, Scharenberg M, et al. Unique transcriptional and protein-expression signature in human lung tissue-resident NK cells. Nat Commun (2019) 10:3841. doi: 10.1038/s41467-019-11632-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Adams NM, Lau CM, Fan X, Rapp M, Geary CD, Weizman OE, et al. Transcription factor IRF8 orchestrates the adaptive natural killer cell response. Immunity (2018) 48:1172–1182.e1176. doi: 10.1016/j.immuni.2018.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wendel M, Galani IE, Suri-Payer E, Cerwenka A. Natural killer cell accumulation in tumors is dependent on IFN-γ and CXCR3 ligands. Cancer Res (2008) 68:8437–45. doi: 10.1158/0008-5472.CAN-08-1440 [DOI] [PubMed] [Google Scholar]
- 63. Martín-Fontecha A, Thomsen LL, Brett S, Gerard C, Lipp M, Lanzavecchia A, et al. Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nat Immunol (2004) 5:1260–5. doi: 10.1038/ni1138 [DOI] [PubMed] [Google Scholar]
- 64. Hanna J, Bechtel P, Zhai Y, Youssef F, McLachlan K, Mandelboim O. Novel insights on human NK cells’ immunological modalities revealed by gene expression profiling. J Immunol (2004) 173:6547–63. doi: 10.4049/jimmunol.173.11.6547 [DOI] [PubMed] [Google Scholar]
- 65. Fu B, Wang F, Sun R, Ling B, Tian Z, Wei H. CD11b and CD27 reflect distinct population and functional specialization in human natural killer cells. Immunology (2011) 133:350–9. doi: 10.1111/j.1365-2567.2011.03446.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pappu R, Schwab SR, Cornelissen I, Pereira JP, Regard JB, Xu Y, et al. Promotion of lymphocyte egress into blood and lymph by distinct sources of sphingosine-1-phosphate. Science (2007) 316:295–8. doi: 10.1126/science.1139221 [DOI] [PubMed] [Google Scholar]
- 67. Melsen JE, Lugthart G, Vervat C, Kielbasa SM, van der Zeeuw SAJ, Buermans HPJ, et al. Human bone marrow-resident natural killer cells have a unique transcriptional profile and resemble resident memory CD8(+) T cells. Front Immunol (2018) 9:1829. doi: 10.3389/fimmu.2018.01829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang Q, Bi J, Zheng X, Chen Y, Wang H, Wu W, et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat Immunol (2018) 19:723–32. doi: 10.1038/s41590-018-0132-0 [DOI] [PubMed] [Google Scholar]
- 69. Chauvin J-M, Ka M, Pagliano O, Menna C, Ding Q, DeBlasio R, et al. IL15 stimulation with TIGIT blockade reverses CD155-mediated NK-cell dysfunction in melanoma. Clin Cancer Res (2020) 26:5520–33. doi: 10.1158/1078-0432.CCR-20-0575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. MacParland SA, Liu JC, Ma X-Z, Innes BT, Bartczak AM, Gage BK, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun (2018) 9:4383. doi: 10.1038/s41467-018-06318-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jameson G, Robinson MW. Insights into human intrahepatic NK cell function from single cell RNA sequencing datasets. Front Immunol (2021) 12:649311. doi: 10.3389/fimmu.2021.649311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Aizarani N, Saviano A, Sagar Mailly L, Durand S, Herman JS, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature (2019) 572:199–204. doi: 10.1038/s41586-019-1373-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature (2019) 575:512–8. doi: 10.1038/s41586-019-1631-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Meng F, Li L, Lu F, Yue J, Liu Z, Zhang W, et al. Overexpression of TIGIT in NK and T cells contributes to tumor immune escape in myelodysplastic syndromes. Front Oncol (2020) 10:1595. doi: 10.3389/fonc.2020.01595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Doyle EH, Aloman C, El-Shamy A, Eng F, Rahman A, Klepper AL, et al. A subset of liver resident natural killer cells is expanded in hepatitis c-infected patients with better liver function. Sci Rep (2021) 11:1551. doi: 10.1038/s41598-020-80819-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lima M, Leander M, Santos M, Santos AH, Lau C, Queiros ML, et al. Chemokine receptor expression on normal blood CD56(+) NK-cells elucidates cell partners that comigrate during the innate and adaptive immune responses and identifies a transitional NK-cell population. J Immunol Res (2015) 2015:839684. doi: 10.1155/2015/839684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Castriconi R, Carrega P, Dondero A, Bellora F, Casu B, Regis S, et al. Molecular mechanisms directing migration and retention of natural killer cells in human tissues. Front Immunol (2018) 9:2324. doi: 10.3389/fimmu.2018.02324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hamann I, Unterwalder N, Cardona AE, Meisel C, Zipp F, Ransohoff RM, et al. Analyses of phenotypic and functional characteristics of CX3CR1-expressing natural killer cells. Immunology (2011) 133:62–73. doi: 10.1111/j.1365-2567.2011.03409.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Carlsten M, Norell H, Bryceson YT, Poschke I, Schedvins K, Ljunggren H-G, et al. Primary human tumor cells expressing CD155 impair tumor targeting by down-regulating DNAM-1 on NK cells. J Immunol (2009) 183:4921–30. doi: 10.4049/jimmunol.0901226 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Storage of data is performed by the Leibniz Institute of Virology on an internal server. Raw data will be made available upon request and can be shared after confirming that data will be used within the scope of the originally provided informed consent. Requests to access these datasets should be directed to the corresponding author.