Abstract
A series of (8-hydroxyquinoline) gallium(III) complexes (CP-1–4) was synthesized and characterized by single X-ray crystallography and density functional theory (DFT) calculation. The cytotoxicity of the four gallium complexes toward a human nonsmall cell lung cancer cell line (A549), human colon cancer cell line (HCT116), and human normal hepatocyte cell line (LO2) was evaluated using MTT assays. CP-4 exhibited excellent cytotoxicity against HCT116 cancer cells (IC50 = 1.2 ± 0.3 μM) and lower toxicity than cisplatin and oxaliplatin. We also evaluated the anticancer mechanism studies in cell uptake, reactive oxygen species analysis, cell cycle, wound-healing, and Western blotting assays. The results showed that CP-4 affected the expression of DNA-related proteins, which led to the apoptosis of cancer cells. Moreover, molecular docking tests of CP-4 were performed to predict other binding sites and to confirm its higher binding force to disulfide isomerase (PDI) proteins. The emissive properties of CP-4 suggest that this complex can be used for colon cancer diagnosis and treatment, as well as in vivo imaging. These results also provide a foundation for the development of gallium complexes as potent anticancer agents.
Introduction
Cancer is a challenging disease to treat; therefore, new and effective anticancer drugs are urgently needed. Many anticancer drugs, such as the chemotherapeutic drug cisplatin, cross-link with purine bases in DNA, interfere with DNA repair mechanisms, cause DNA damage, and induce apoptosis in cancer cells.1,2 However, the use of platinum drugs, including cisplatin, is limited by side effects, such as drug resistance, vomiting, and kidney toxicity.3−5 Therefore, the development of nonplatinum drugs with low toxicity and strong targeting ability has gained attention.6,7 The ion radius of gallium is similar to iron; gallium can also be absorbed through cellular iron transport systems and can be substituted into ferrous proteins.8,9 In contrast to iron, gallium levels cannot be restored under physiological conditions.10 Additionally, it inhibits the function of gallium complex proteins and the growth of cells. Thus, this element acts a “Trojan horse hypothesis” by destroying the protein structure that adds iron to bacteria and cancer cells.9,11 However, the toxicity of gallium hydroxide, which is formed by soluble water, severely limits its application. Recent studies have shown that gallium nitrate and other gallium compounds also have antitumor properties.12 A number of gallium complexes, including tris(8-quinolinolato) gallium(III), tris(maltolato) gallium(III), phenol-based gallium(III), and paminothiourea gallium(III) complexes, have been synthesized and evaluated in vitro and in vivo, and gallium(III) nitrate was shown to reduce the proliferation of malignant cells.13−16 Clinically, gallium(III) complexes are used to treat microbial, inflammatory, cancer, and bone metabolic diseases.17 The primary therapeutic mechanism of these effects involves elevated levels of reactive oxygen species (ROS) and mitochondrial dysfunction. An accumulation of ROS leads to disruption of the mitochondrial membrane potential, which results in increased mitochondrial membrane permeability; under this condition, apoptosis-related proteins can enter the cytoplasmic matrix to promote apoptosis.18,19
Quinoline ligands are an important pharmacodynamic group, and their derivatives show excellent pharmacological properties. Quinoline has several biological activities, including antimicrobial,20,21 antifungal,22 antileishmaniasis, antimalarial, antioxidant, and anticancer effects.23,24 The 8-hydroxyquinoline derivatives are effective targeted broad-spectrum drug molecule used to treat a wide range of life-threatening diseases, such as cancer, HIV infection, and neurodegenerative diseases,28−32 and exert in vitro antitumor activity.25−27 Various development and drug research centers are working to chemically or structurally modify their functional groups to construct broad-spectrum bioactive candidates.33,34
In this study, we developed gallium(III) compounds and tested their potential as anticancer agents. Four novel gallium(III) complexes {[Ga(HQL)3] (CP-1), [Ga(MQL)3] (CP-2), [Ga(ClQL)3] (CP-3), and [Ga(BrQL)4] (CP-4)} were synthesized by reacting different 8-hydroxyquinoline derivatives with gallium(III) metal salts (Scheme 1). Analysis of their anticancer activity showed that CP-3 and CP-4 were highly selective for HCT116 cells. In addition, CP-1, CP-3, and CP-4 displayed bioimaging properties. Preliminary examination of the anticancer mechanisms of CP-1, CP-3, and CP-4 revealed that these compounds induce the production of ROS and may differ from cisplatin in their anticancer mechanisms. CP-4 also inhibits protein expression and predicted other protein-binding sites through molecular docking.
Scheme 1. Structures of Four Gallium(III) Complexes.
2. Experimental Section
2.1. Materials and Characterization
Gallium nitrate, 8-hydroxyquinoline (L-1), 2-methyl-8-hydroxyquinoline (L-2), 5-chloro-8-hydroxyquinoline (L-3), and 5-bromo-8-hydroxyquinoline (L-4) were purchased from Sigma-Aldrich, and all reagents were not further purified. Fluorescence spectroscopy and lifetime analysis of CP-1–4 were performed on a Horiba Jobin Yvon Fluorolog-3. Fluorescence quantum yield was performed on a Edinburgh FLS1000 fluorescence spectrophotometer. The FT-IR absorption spectra for CP-1 and CP-2 were collected from KBr pellets using a Thermo Scientific FTIR-Nicolet iS10 spectrometer in the range 4000–400 cm–1. The UV–visible spectra of CP-1–4 and L-1–4 were obtained using a UV-240 IPC spectrophotometer. High-resolution mass spectrometry (HRMS) measurements of CP-1–4 were acquired by an LTQOrbitrapXL. 1H NMR spectra (Avance500, Bruker, Germany) were used to analyze the molecular structures of CP-1–4.
2.2. Synthesis of CP-1 and CP-2
8-Hydroxyquinoline (2.25 mmol, 326.55 mg) and 2-methyl-8-hydroxyquinoline (2.25 mmol, 358.16 mg) were each dissolved in 40 mL of absolute methanol, and Ga(NO3)3 (0.75 mmol, 191.85 mg) was dissolved in 6.5 mL of absolute ethanol. The prepared Ga(NO3)3 was gradually added to both the methanol solution of 8-hydroxyquinoline and 2-methyl-8-hydroxyquinoline, continuously stirred for 16 h at 80 °C, and then cooled to room temperature (20 °C). NH3·H2O (WNH3 = 25%–28%) was added until no yellow green precipitate was produced (pH ≈ 10). After filtering, the precipitate was washed with absolute ethanol and heated to 100 °C in an oven. When the pH became constant, the ammonia was removed. Yield: (318.5 mg, 84%) for L-1 and (322.5 mg, 79%) for L-2. To obtain single crystals of CP-1 and CP-2, 5 mg of solid CP-1 and CP-2 was dissolved in 1 mL of dichloromethane, 4 mL of anhydrous methanol was slowly added along the vessel wall to form layers, and the mixture was then incubated for approximately 5 days. 1H NMR (500 MHz, chloroform-d), CP-1: δ 8.88 (d, J = 15 Hz, 2H), 8.30 (m, J = 10 HZ, 3H), 7.51–7.36 (m, 6H), 7.16 (d, J = 10 Hz, 4H), 7.04 (d, J = 5 Hz, 3H). CP-2: δ 8.02 (d, J = 10 Hz, 3H), 7.39 (t, J = 5 Hz, 5H), 7.12 (d, J = 10 Hz, 1H), 7.08–6.99 (m, 10H), 2.57 (s, 9H), (Figures S1 and S2). HRMS-ESI (CP-1): m/z, calcd for C27H18GaN3O3 = 502.0677; found = 502.0679 [M + H]+. HRMS-ESI (CP-2): m/z, calcd for C30H24GaN3O3 = 566.0966; found = 566.0976 [M + Na]+, (Figures S5 and S6). FT-IR: CP-1 aromatic ring skeleton, 1577 cm–1, 1497 cm–1, 1461 cm–1; υ(C–O), 1289 cm–1, 1268 cm–1; υ(C–N), 1379 cm–1, 1326 cm–1; υ(Ga–N), 628 cm–1; υ(Ga–O), 526 cm–1. CP-2 aromatic ring skeleton, 1577 cm–1, 1528 cm–1,1508 cm–1; υ(C–O), 1289 cm–1, 1268 cm–1; υ(C–N),1384 cm–1; υ(−CH3), 2925 cm–1; υ(Ga–N), 658 cm–1; υ(Ga–O), 532 cm–1 (Figure S9).
2.3. Synthesis of CP-3 and CP-4
5-Chloro-8-hydroxyquinoline (2.25 mmol, 404.10 mg) and 5-bromo-8-hydroxyquinoline (2.25 mmol, 687.38 mg) were each dissolved in 50 mL of methanol. Ga(NO3)3 (0.75 mmol, 191.85 mg) was dissolved in 6.5 mL of absolute ethanol. The prepared Ga(NO3)3 was gradually added to both the methanol solution of 5-chloro-8-hydroxyquinoline and 5-bromo-8-hydroxyquinoline, continuously stirred for 80 °C at 24 h, and then cooled to room temperature. NH3·H2O (WNH3 = 25%–28%) was added, and a small amount of precipitation was observed. Yield: (413.3 mg, 91%) for L-3 and (664.9 mg, 88%) for L-4. To obtain single crystals of CP-3 and CP-4, 4 mg of solid CP-3 and CP-4 was dissolved in 1.5 mL of trichloromethane, 4.5 mL of anhydrous methanol was slowly added along the vessel wall to form layers, and the samples were incubated for approximately 1 week. 1H NMR (500 MHz, chloroform-d), CP-3: δ 8.81 (d, J = 25 Hz, 2H), 8.67 (m, J = 15 Hz, 3H), 7.57 (d, J = 5 Hz, 5H), 7.43 (d, J = 30 Hz, 2H), 7.06 (s, 3H). CP-4: δ 8.90 (d, J = 30 Hz, 2H), 8.65–8.55 (m, 3H), 7.75 (d, J = 10 Hz, 3H), 7.61–7.41 (m, 4H), 7.02 (s, 3H), (Figures S3 and S4). HRMS-ESI (CP-3): m/z, calcd for C27H15GaCl3N3O3 = 605.9499; found, 605.9500 [M + H]+. HRMS-ESI (CP-4): m/z, calcd for C27H15GaBr3N3O3 = 739.7963; found = 739.7972 [M + H]+, (Figures S7 and S8). FT-IR: CP-3 aromatic ring skeleton, 1574 cm–1, 1496 cm–1,1459 cm–1; υ(C–O), 1259 cm–1; υ(C–N), 1369 cm–1,1323 cm–1; υ(-Cl), 783 cm–1; υ(Ga–N), 677 cm–1; υ(Ga–O), 547 cm–1. CP-4 aromatic ring skeleton, 1573 cm–1, 1491 cm–1, 1455 cm–1; υ(C–O),: 1258 cm–1, 1222 cm–1; υ(C–N), 1364 cm–1,1320 cm–1; υ(-Br), 626 cm–1; υ(Ga–N), 640 cm–1; υ(Ga–O), 539 cm–1 (Figure S9).
According to the UV–visible spectra of the complexes and ligands (Figure S10), gallium ions were π–π conjugated with the quinoline ring to form a quinoline B ring (L-1, 312 nm → CP-1, 317 nm; L-2, 304 nm → CP-2, 313 nm; L-3 ,330 nm → CP-3, 340 nm; L-4, 329 nm → CP-4, 336 nm) red shift [CP-1, λmax (MeOH)/nm = 377 (ε/dm3 mol–1 cm–1 = 17 253); CP-2, λmax (MeOH)/nm = 356 (ε/dm3 mol–1 cm–1 = 7385); CP-3, λmax (MeOH)/nm = 395 (ε/dm3 mol–1 cm–1 = 13843); CP-4, λmax (MeOH)/nm = 387 (ε/dm3 mol–1 cm–1 = 10 065)].
2.4. X-ray Crystallography
A single crystal with suitable dimensions was mounted on a glass capillary for X-ray diffraction analyses. Crystallographic data were collected on a Bruker Smart AXS CCD diffractometer with graphite–monochromate Mo–Kα radiation (λ = 0.71073 Å) using the ω-scan technique. Empirical absorption corrections were applied using the SADABS program. The structure of each compound was solved using the SHELXL package and refined using the full matrix least-squares method on the basis of F2 using the SHELX-2014 program.35,36 All nonhydrogen atoms were in different Fourier syntheses and finally refined on the basis of anisotropic thermal parameters. CCDC: 2183374 for CP-1, 2183373 for CP-2, 2181298 for CP-3, and 2190520 for CP-4.
2.5. Fluorescence Characteristics
The fluorescence intensities of CP-1–4, prepared at 100 μM [dimethyl sulfoxide (DMSO)] and then gradually diluted to 50, 25, 12.5, 6.25, 3.125, and 1.5625 μM, as well as lifetimes (6.25, 12.5, 25, 50, 100 μM) and solid quantum yields, were evaluated. The fluorescence changes of CP-1–4 (100 μM) within 72 h were tested.
2.6. Cell Activity Screening
The cytotoxicity of CP-1–4 toward various cell types was measured for 72 h. A549, HCT116, and LO2 cells were incubated in 96-well plates (5 × 103 cells/plate) overnight. The original medium (A549 and HCT116 in RPMI-1640 medium, LO2 in DMEM medium) was removed, CP-1–4 (100, 50, 25, 12.5, 6.25, 3.125, 1.5625 μM) was added to each plate, and the cells were incubated for 72 h. Equal volume medium was used as the control group. The medium was removed, 20 μL of MTT solution (5 mg/mL) was added, and the cells were cultured for 4 h. The medium was removed, and 150 μL of DMSO was added. The optical density (OD) was measured using a microplate reader (490 nm). HeLa, HepG2, HCC1806, and HCT116 cells were next inoculated into 96-well plates (5 × 103 cells/plate) and incubated overnight; the cells were incubated for 48 h with CP-1, CP-3, and CP-4 (10 μM), after which cytotoxicity and fluorescence were measured. Cellular activity was measured using sulforhodamine B (SRB). The cell culture conditions were 37 °C and 5% CO2.
HCT116 cells in the logarithmic growth stage were collected, counted, inoculated at 3 × 105 cells/mL into 12-well plates, and incubated at 37 °C for 12 h. When the cells had adhered to the well wall, the medium was removed, and fresh medium containing the corresponding concentrations of gallium(III) complexes was added for an additional 24 h of incubation. After incubation, the cell status was observed; when apoptosis began, prediluted Hoechst 33342 dye (1 μg/mL) was added to the cell culture panel to fully cover the cell surface at 37 °C, and the culture was incubated in a constant temperature cabinet in the dark for 15 min. The cells were vacuumed with dye solution before onboard assays, washed three times with phosphate-buffered saline (PBS) (pH 7.2–7.4), and observed under a Leica DMI3000B inverted fluorescence microscope. Images were evaluated using LASV4.3 software.
2.7. Cellular Uptake and Distribution Experiments
Confocal laser scanning microscopy (CLSM) can be used to quantitatively analyze the uptake of various substance by cells. HCT116 cells (1 × 106 cells/mL) were seeded into a 35 mm glass dish for 24 h. After the cells were treated for 5 h with CP-1, CP-3, and CP-4, the cells were gently rinsed with PBS (pH 7.2–7.4) and immobilized at 37 °C using 4% polyformaldehyde for 20 min. After the cells were rinsed with PBS (pH 7.2–7.4), the cell nuclei were stained with 4,6-diamidine-2-phenylindole (DAPI) in the dark for 10 min. The cells were washed with PBS (pH 7.2–7.4), and excess DAPI was removed. The stained cells were observed using an FV10i confocal laser. DAPI was visualized with 359 nm excitation light, and the corresponding emission was observed at 461 nm. The excitation and emission wavelengths of the CPs were 428 and 536 nm, respectively.
2.8. Cell ROS Analysis
Thereafter, 3 × 105 HCT116 cells were inoculated into 12-well plates and incubated at 37 °C for 24 h. DCFH-DA fluorescence probes were diluted by 1:1000 with serum-free medium, which gave a final concentration of 10 μM. After 24 h, the cells were exposed to 10 μM gallium(III) compounds and incubated for 24 h. DCFH-DA was diluted to the appropriate volume to remove the compound-containing medium. The cells were cultured for 20 min at 37 °C, the dye was removed, and the cells were washed three times with serum-free medium to completely remove free DCFH-DA. Images were captured, and excitation wavelengths of 488 and 525 nm, respectively, were observed using an inverted fluorescence microscope and LAS V4.3.
2.9. Wound-Healing Assay
Cell migration and repair were analyzed in a wound-healing assay. A marked pen was used to mark the back of a 12-well plate at 0.5 cm intervals, across the wells, with three horizontal lines running across each well. HCT116 cells were inoculated and added to the plate. When the cells were affixed to the wall, cells in the logarithmic growth phase were plated at 3 × 105 cells/mL. The plates were placed in an incubator at 37 °C overnight, during which the cells affixed to the plate walls. Scratches were made according to the lines on the back of the plate using 200 μL sterile pipet tips, and cells in the middle of the scratch were discarded. Cells were treated with 2 μM (prepared in serum-free medium), and the scratches were photographed under a microscope. The cells were incubated at 37 °C with the corresponding concentration of fresh medium. After 24 h, the cell culture plate was removed, and the scratch position was photographed again.
2.10. Cell Cycle Analysis
HCT116 cells in the logarithmic growth stage were inoculated into a six-well plate at a density of 1 × 106 cells per well; 1 mL of medium was added to each well and was supplemented with another 2 mL. The cells were incubated at 37 °C for 24 h. The media was removed, and the cells were washed 2–3 times with PBS (pH 7.2–7.4); 2 mL of the fresh medium was added to each well, and 10 μM drug was added for 24 h incubation. The drug-containing medium was removed, the cells were washed three times with precooled PBS (pH 7.2–7.4), pancreatic enzymes were added for digestion, the enzymes were discarded, and the suspended cells were washed with PBS (pH 7.2–7.4). The cell suspensions were transferred into 1.5 mL tubes, centrifuged (2000 rpm, 5 min), and the supernatants were discarded. The resulting cell pellets were washed twice with precooled PBS (pH 7.2–7.4) and centrifuged again. Thereafter, 500 μL of 70% ethanol was added to the tube, shaken well to prevent cell aggregation, and placed at −20 °C for 48 h. After centrifugation (2000 rpm, 5 min), the purge solution was discarded; the cells were washed once with PBS (pH 7.2–7.4) and centrifuged (2000 rpm, 5 min), and the supernatant was discarded. PBS (pH 7.2–7.4, 250 μL) was added to the tube to resuspend the cell. RNase A (2.5 μL) was added and incubated at 37 °C in a water bath for 30 min, and then 12.5 μL of propidium iodide (PI) was added and incubated for 30 min at room temperature in the dark. A NovaCyte flow cytometer was used according to the manufacturer’s instructions, and Mod Fit analysis was used for the structural cell cycle fitting software.
2.11. Western Blot Analysis
HCT116, SW480, and SW620 cells were treated with different concentrations of CP-4 for 24 h. The medium was removed by vacuum, and radioimmunoprecipitation assay buffer (RIPA) containing protease and phosphatase inhibitors was used to lyse the cells for 30 min on ice. The protein concentration in the cell lysate was measured using a BCA kit. The proteins were separated using polyacrylamide gel electrophoresis (PAGE) and transferred onto a polyvinylidene fluoride (PVDF) membrane. After skim milk sealing, the primary antibody was added, the membrane was shaken overnight at 4 °C, and the corresponding secondary antibody was added and incubated at room temperature for 1 h. ImageQuant LAS4000 Biomolecular imager (GE Healthcare, Little Chalfont, UK) was used for visualization.
2.12. Computational Details
Density functional theory (DFT) calculations were performed using Gaussian 09 (revision E.01) packages.37 The ground-state geometry was optimized out at the ωB97X-D level using the def2-SVP basis set with an ultrafine grid.38,39 The implicit solvent effect of methanol media was taken into consideration using the self-consistent reaction field (SCRF) method via the solvent model based on density (SMD) model.40 We employed time-dependent DFT (TDDFT) theory at the ωB97X-D/def2-TZVP/SMD level to calculate the first 15 excited states. The frontier molecular orbitals, electron and hole distributions, and electrostatic potentials (ESP) analyses were performed using the Multiwfn packages.41
Molecular docking was performed using Pymol and Autodock-Vina software.42,43 Simple optimization of the ligand structure and structural correction, protonation, constraint, and minimization of the receptor protein were performed by the program. Finally, the active site of the protein was searched for via docking with the ligand.
3. Results and Discussion
3.1. Structure and Stability of CP-1–4
Synthetic CP-1–4 were validated through DFT and X-ray crystallography. As shown in Figure S11, CP-1–4 have the same coordination environment, and the atomic structure was obtained at the ωB97X-D(SMD = methanol)/Def2-TZVP energy level and experiments; it can be found that the structure obtained by DFT (Figure 1a) is basically consistent with the single crystal structure (Figure 1b). The crystal structures of CP-1–4 exhibited the same structural framework. Each complex consists of a gallium(III) ion metal center and three quinoline derivative ligands in zero-dimensional structure. The detailed crystal parameters are shown in the Supporting Information (Tables S1–S5). In order to explore the solubility of CP-1–4 in different solutions (Figure S12), we prepared a concentration of 150 μmol/L (greater than the maximum concentration of MTT assay); placed it in water, methanol, ethanol, DMSO, chloroform, acetonitrile, ethyl acetate, water + 5% PEG, water + 2.5% Tween (animal experiment), and cyclohexane; and then tested the ten solutions for 72 h (average temperature 18 °C). Organic polar solvents have excellent solubility for CP-1–4. In nonpolar solvents, CP-1–4 precipitates to varying degrees, which may be attributed to the principle of similitude compatibility.
Figure 1.
Comparison of (a) DFT theoretical structures and (b) single-crystal structures of four gallium complexes.
We then further evaluated the stability of the complexes in air by testing them with 1H NMR after 3 months. We found that their chemical shifts were highly consistent, thereby indicating that the four complexes are stable in air (storage temperature: 25 °C). 1H NMR (500 MHz, chloroform-d), CP-1: δ 8.88, 8.30, 7.51–7.36, 7.16, 7.04. CP-2: δ 8.02, 7.39, 7.12, 7.08–6.99, 2.57. CP-3: δ 8.81, 8.67, 7.57, 7.43, 7.06. CP-4: δ 8.90, 8.65–8.55, 7.75, 7.61–7.41, 7.02. Three months later, CP-1: δ 8.89, 8.29, 7.52–7.39, 7.16, 7.06. CP-2: δ 8.01, 7.39, 7.12, 7.07–6.98, 2.57. CP-3: δ 8.91, 8.66, 7.57, 7.42, 7.08. CP-4: δ 8.95, 8.61–8.52, 7.79, 7.60–7.40, 7.03.
Finally, the stability of CP-1–4 in water was further determined by testing the fluorescence of CP-1–4 within 72 h. From Figure S13, we can see that the fluorescence intensity of CP-1–4 is basically consistent, which indicates that the material has excellent stability.
3.2. Electrostatic Potentials and HOMO–LUMO
The electrostatic potentials of gallium(III), L-1–4, and CP-1–4 were calculated using DFT to further explore the binding mechanism. As shown in Figure 2, gallium(III) is positively charged (red), whereas both N and O on the ligands are negatively charged (blue). Gallium(III) binds to N and O on the ligands at positive and negative positions to form corresponding Ga–N and Ga–O bonds. The highest occupied molecular orbital (HOMO) and lowest occupied molecular orbital (LUMO) were evaluated. The electron donor is known as the HOMO, the electron receptor is known as the LUMO, and the energy gap between the HOMO and LUMO explains the charge transfer interaction within the molecule. A higher value indicates that a complex is more stable and vice versa. High stability indicates low chemical reactivity, whereas low stability indicates high chemical reactivity.44 Increasing the softness increases chemical activity, and increasing the hardness decreases the chemical activity.45,46 As determined from the HOMO–LUMO orbital and orbital energy gap (Tables S6 and S7, Figures S14 and S15), the electron cloud is concentrated on the pyridine ring of the ligand and is composed of the π → π* orbital of C and N. CP-1–4 showed high orbital energy differences of 6.9606, 6.0635, 6.7897, and 7.2436 eV in the HOMO–LUMO orbit, respectively. CP-4 showed the largest range of orbital energy, and thus, was considered highly stable.
Figure 2.
DFT calculation of electrostatic potentials of four ligands and gallium(III) complexes (positively charged red area, negatively charged blue area).
3.3. Fluorescence Properties
CP-1–4 were dissolved in DMSO solution, configured to 100 μM, and gradually diluted by half for fluorescence intensity testing, as shown in Figure 3a–d. CP-1–4 were fluorescent with ordered intensities of CP-1 > CP-3 ≈ CP-4 > CP-2 at the same concentration; these results may be applicable for in vitro tracking of drug distributions. Analysis of the DFT computational electron/cavity distribution of CP-1–4 (Figure 4) indicated that the electrons in CP-1–4 were transferred between ligands, as the electron–cavity distribution of a single ligand was evident. However, when corresponding complexes were formed, some ligands existed only in cavities without electrons. We hypothesized that the ligand-to-ligand charge-transfer (LLCT) transition occurred primarily. In addition, the fluorescence lifetimes of the four gallium(III) complexes were determined at different concentrations using third-order fitting (Figure S16) and were found to decrease gradually with increasing concentration. In addition, the solid quantum yields were also measured (Figure S17).
Figure 3.
Fluorescence intensities of four gallium(III) complexes (a) CP-1 (λex = 400 nm), (b) CP-2 (λex = 439 nm), (c) CP-3 (λex = 450 nm), and (d) CP-4 (λex = 421 nm) at different concentrations.
Figure 4.
Electron (green) and cavity (blue) distributions of four gallium(III) complexes calculated by DFT.
3.4. Cell Viability
The cytotoxicity of CP-1–4 at different concentrations was evaluated against A549 and HCT116 cancer cell lines and normal LO2 cells in an MTT assay. Cisplatin and oxaliplatin were used as reference control treatments (Table 1). As shown in Figure 5, CP-1–4 were cytotoxic toward all cancer cells and inhibited cell proliferation in a dose-dependent manner after 72 h of treatment. According to the 50% inhibitory concentration (IC50) (Table 1), CP-1 inhibited A549 and HCT116 cancer cells at a concentration of 6.25 μM, whereas CP-2 inhibited A549 cancer cells while being more toxic toward LO2 cells. In addition, CP-3 and CP-4 exhibited strong cytotoxicity values of 91.98% and 95.19%, respectively, against HCT116 cells at 25 μM. The IC50 values of CP-3 (2.0 μM for HCT116 cells) and CP-4 (1.2 μM for HCT116 cells) were lower than those of cisplatin (5.7 μM for HCT116 cells) and oxaliplatin (8.3 μM for HCT116 cells). In addition, CP-3 and CP-4 showed lower toxicity than cisplatin (0.7 μM) and oxaliplatin (2.3 μM) in LO2 cells, which may reduce the system toxicity of cisplatin and oxaliplatin. This may be because CP-3 and CP-4 possess halogen groups that influence the charge distribution of the gallium(III) complexes by increasing electrical binding to cell receptors and enhancing the antineoplastic efficacy.
Table 1. IC50 Values of Cisplatin, Oxaliplatin, and CP-1-4 after 72 h.
| IC50 (μM)a |
|||
|---|---|---|---|
| compounds | A549 cells | HCT116 cells | LO2 cells |
| cisplatin | 7.5 ± 0.7 | 5.7 ± 0.0 | 0.7 ± 0.0 |
| oxaliplatin | 3.2 ± 0.2 | 8.3 ± 0.4 | 2.3 ± 0.3 |
| CP-1 | 3.8 ± 1.1 | 1.2 ± 0.2 | 8.0 ± 1.0 |
| CP-2 | 6.1 ± 0.8 | 4.7 ± 0.7 | 3.3 ± 0.5 |
| CP-3 | 8.7 ± 0.8 | 2.0 ± 0.3 | 23.3 ± 5.3 |
| CP-4 | 8.7 ± 1.0 | 1.2 ± 0.3 | 30.0 ± 5.4 |
IC50, 50% inhibitory concentration.
Figure 5.
Cell viability of (a) A549, (b) HCT116, and (c) LO2 cell line treated with four gallium(III) complexes at different concentrations for 72 h.
On the basis of these results, sulforhodamine B (SRB) was used to further examine the cytotoxicity of 10 μM CP-1, CP-3, and CP-4 toward HeLa, HepG2, HCC1806, and HCT116 cancer cells. As shown in Figure 6, the three gallium(III) complexes exhibited varying degrees of inhibition for all four cancer cell lines. CP-4 showed the strongest inhibitory effects in HCC1806 and HCT116 cells with values of 91.36% and 83.81%, respectively. HCT116 cells were stained with Hoechst 33342 at 24 h after administration (10 μM). Compared with other treatment groups (cisplatin, oxaliplatin, CP-1 and CP-3), HCT116 cells treated with CP-4 showed brighter blue fluorescence, which suggests that CP-4 inhibited HCT116 cell growth and induced apoptosis (Figure 7), which is consistent with the MTT and SRB results.
Figure 6.

Comparative toxicity of CP-1, CP-3, and CP-4 against the tested different cell lines at 10 μM after 48 h of exposure.
Figure 7.

Hoechst 33342 staining detected morphological changes in HCT116 cells after treatment by varied compounds at 10 μM for 24 h. All scale bars = 10 μm.
3.5. Cellular Uptake and Distribution Analyses
Cell uptake distribution studies revealed that the CPs exhibited strong fluorescence in cells, which is consistent with the fluorescence characteristic observed at the same concentrations (10 μM) described above (Section 3.3). According to the merge maps (Figure 8a) and Pearson correlation coefficients (a value of 0.8–1 indicates a strong correlation) (Figure 8b), the CPs can enter the nucleus, thereby providing insight into the mechanisms by which these complexes bind to DNA (Figure S18) in the nucleus and lead to apoptosis (when CP-4 was mixed with CT-DNA, the fluorescence intensity of CP-4 decreased in varying degrees with the increase of CT-DNA concentration, which indicates that our material could bind to DNA), as well as their dual functions (anticancer activity and in vitro imaging). DAPI dye showed blue fluorescence and revealed the location of cell nuclei. The CPs showed green fluorescence and revealed their location in the cells.
Figure 8.
(a) CLSM of HCT116 cells interacted with CP-1, CP-3, and CP-4 at 10 μM for 5 h. (b) 2D intensity histogram of CPs’ fluorescence colocation and Pearson correlation coefficient (PCC). All scale bars = 50 μm.
3.6. ROS Detection
ROS accumulation is an important factor that leads to apoptosis, and increasing the levels of ROS is an effective anticancer strategy. To investigate the anticancer mechanisms of CP-1, CP-3, and CP-4, we assessed ROS levels in cancer cells at 24 h after treatment with these complexes (10 μM). Figure 9a shows that the three gallium(III) complexes and a control group led to increased ROS production in HCT116 cells. The ROS strength of CP-4 is stronger than that of cisplatin, oxaliplatin, and CP-1–3 (Figure 9b). The fluorescence intensity was proportional to the ROS content, which suggests that CP-4 more effectively induces increases in ROS levels and apoptosis than the effects of the other complexes.
Figure 9.

(a) Generation of reactive oxygen species induced by different compounds in HCT116 cells at 10 uM for 24 h. (b) Mean fluorescence intensity of different complexes. All scale bars = 10 μm.
3.7. Wound-Healing Assay
Cell migration indicates the ability of cells to metastasize and invade tissues. The effects of cisplatin, oxaliplatin, CP-1, CP-3, and CP-4 on HCT116 cell migration was analyzed using scratch assays. In the scratch experiment, the closer the scratch width is to 0 h, the lower the healing. The narrower the scratch distance after 24 h, the better the drug effect. As shown in Figure 10, all three gallium(III) compounds tested inhibited HCT116 cell migration to varying degrees and showed stronger effects than oxaliplatin and equivalent effects as cisplatin, which suggests that the compounds inhibit tumor cell proliferation.
Figure 10.
(a) Migration inhibition (wound-healing assay) of HCT116 untreated or treated with the tested compounds for 24 h at the 2 μM concentrations. (b) Different healing rates. All scale bars = 10 μm.
3.8. Cell Cycle Assay
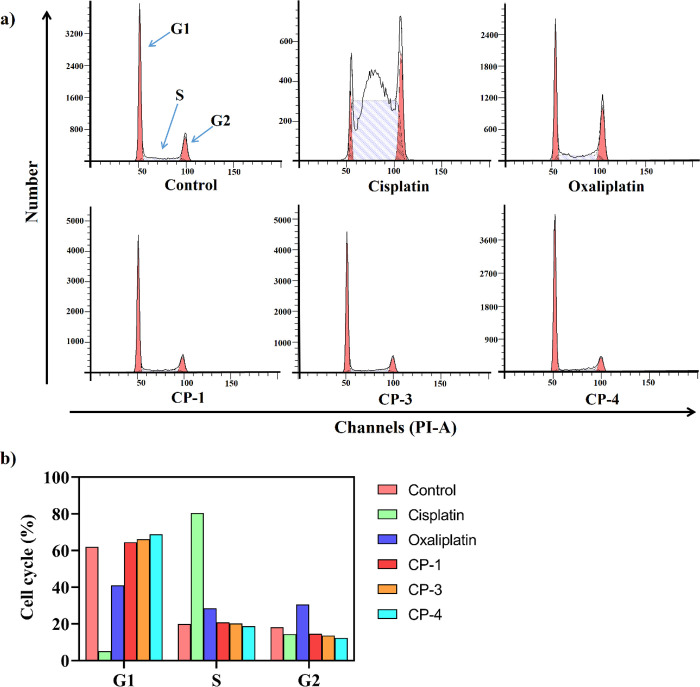
The cell cycle distribution was examined using flow cytometry to investigate the potential mechanism of action for CP-1, CP-3, and CP-4 in inducing apoptosis. Treatment of HCT116 cells with the three gallium(III) complexes for 24 h caused significant changes in the cell cycle. As shown in Figure 11, compared with control treatment with cisplatin, the three gallium(III) complexes showed a significant blocking effect in the G1 phase, whereas cisplatin mainly blocked the cell cycle in the S phase, and oxaliplatin blocked it in the G1/S/G2 phase with comparable effects; therefore, the anticancer mechanisms of the gallium(III) complexes may differ from those of cisplatin and oxaliplatin, and cancer cell proliferation may be inhibited by blocking DNA synthesis and cell cycle progression.
Figure 11.
(a) Cell cycle distribution was tested by flow cytometric analysis of DNA content after treatment with 10 μM CPs and platinum compounds for 24 h in HCT116 cells. (b) Different cell cycle ratios.
3.9. Western Blot Analysis
Considering the excellent therapeutic effect of CP-4 on colorectal cancer, we further studied the effect of CP-4 on related proteins (overexpression or low expression of these proteins in cancer) by combining different doses of oxaliplatin and CP-4 (2.5 and 5 μM) and treating human colon cancer cell lines (HCT116, SW480, and SW620) with these agents. The Western blotting results showed that CP-4 significantly induced the expression of cyclin D1, which is a cyclin progression protein, compared with the DMSO group and was more effective than oxaliplatin at both low (2.5 μM) and high (5 μM) concentrations. Furthermore, CP-4 inhibited the expression of the antiapoptotic proteins Mcl-1, induced cleavage of the apoptotic protein PARP, and was more pro-apoptotic than oxaliplatin (Figure 12). These results suggest that CP-4 promotes proliferation and induces apoptosis in colon cancer cell lines.
Figure 12.
Colon cancer cells were treated with CP-4 (2.5 and 5 μM) for 24 h, and the expression levels of related proteins were detected by WB method, with Tubulin used as an internal reference.
3.10. Molecular Docking
Disulfide isomerases (PDI) and glycogen synthase kinase-3 (GSK-3β) are involved in cell and tissue physiology and pathophysiology, as well as cell survival and proliferation in different cancers.47−49 Molecular docking simulation was performed to further predict the mechanism of action of CP-4, which showed the best anticancer activity. The synthesized gallium(III) complex and PDI proteases were examined at the molecular level through molecular docking of PDI (PDB ID: 2C0G) to determine the site of action (Figure 13a and Table 2). Metallic gallium(III) interacts with ASP1029, a negatively charged B-chain protein, whereas functional groups in the compound act as hydrogen bond donors to the A-chain protein and ASP1029. In contrast, GSK-3β (PDB ID: 6Y9S) was selected for molecular docking with CP-4 to explore their binding interaction. CP-4 and GSK-3β proteins interacted mainly between the benzene rings in the complex and Gly63 and Tyr140 to form π–π conjugates, as well as hydrogen bonds between the complex and IL-62 amino acids (Figure 13b and Table 3), thereby suggesting that CP-4 has higher affinity for PDI than for GSK-3β, and that CP-4 has a higher binding force to PDI.
Figure 13.
Interaction networks of CP-4 with (a) PDI and (b) GSK-3β in Docking Mod.
Table 2. Interaction Energies between CP-4 and Responsive Amino Acid Residues in Disulfide Isomerases (PDI).
| amino acid residues | interaction energy (kJ mol–1) |
|---|---|
| Cys 1024 | –0.38 |
| Cys 1027 | –0.06 |
| Asp 1029 | –7.31 |
| Ala 1052 | –0.14 |
| Ser 1053 | 0.16 |
| Pro 1054 | –0.40 |
| Thr 1080 | –1.36 |
| Gly 1082 | 2.76 |
| Glu 1090 | 0.30 |
Table 3. Interaction Energies between CP-4 and Responsive Amino Acid Residues in Glycogen Synthase Kinase-3 (GSK-3β).
| amino acid residues | interaction energy (kJ mol–1) |
|---|---|
| Ile 62 | –2.35 |
| Gly 63 | 1.45 |
| Asn 64 | –0.16 |
| Phe 67 | –1.19 |
| Val 70 | –0.73 |
| Lys 85 | –0.11 |
| Thr 138 | 0.77 |
| Thr 140 | 0.98 |
| Arg 141 | 1.65 |
| Lys 183 | 0.56 |
| Gln 185 | 0.45 |
| Asn 186 | –0.80 |
| Asp 200 | –0.28 |
In addition, CP-4 is the most potential therapeutic drug, and we conducted the oil partition coefficient experiment on it (Table S8).
4. Conclusions
Four gallium(III) complexes were designed and synthesized. CP-1, CP-3, and CP-4 showed high fluorescence intensity and antitumor activity, which is respectively helpful for monitoring drug distributions in vitro and for curing cancer. Analysis of their antineoplastic activity showed that the four gallium(III) complexes potently inhibited ROS-induced apoptosis in various cancer cells. In addition, CP-4 significantly inhibited the HCT116 cell line and was less toxic than cisplatin or oxaliplatin. The four gallium(III) complexes entered the nucleus and were imaged on the basis of their emissive properties. The gallium(III) complexes strongly blocked the G1 phase, in contrast to cisplatin, which blocks the S phase, which suggests that these compounds inhibit the proliferation of tumor cells by preventing DNA synthesis in the G1 phase. CP-4 inhibited the expression of Mcl-1 and cyclinD1 and induced the cleavage of the apoptotic protein PARP. CP-4 showed a high binding force to PDI proteins, which is useful for predicting anticancer targets. These results provide a foundation for developing gallium complexes as potent anticancer agents and for the exploration of target proteins of gallium(III) anticancer drugs.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (grant nos. 21967019, 21977080, 81830087, and U2102203), National Key R&D Program of China (2020YFA0112300), the Yunnan Fundamental Research Projects (202101AS070050) and Yunnan Province Young and middle-aged academic and technical leaders Program 2011CI002, and the Postgraduate Research and Innovation Foundation of Yunnan University (No. 2021Y385). The authors thank the Advanced Analysis and Measurement Center of Yunnan University for the sample testing service. The authors would like to thank Tong Qi from Shiyanjia Lab (www.shiyanjia.com) for the 1H NMR.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c07742.
CCDC 2183374 (CIF)
CCDC 2183373 (CIF)
CCDC 2181298 (CIF)
CCDC 2190520 (CIF)
Details of characterization data, including coordination environment, UV–visible spectra, FT-IR spectra, 1H NMR, HRMS, HOMO–LUMO orbitals, energy differences, crystal data, selected bond lengths and angles, and Fluorescence property.(PDF)
Author Present Address
⊥ Academy of Biomedical Engineering, Kunming Medical University, Kunming 650500, China
Author Contributions
# These authors contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Dasari S.; Bernard Tchounwou P. Cisplatin in Cancer Therapy: Molecular Mechanisms of Action. Eur. J. Pharmacol. 2014, 740, 364–378. 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palchaudhuri R.; Hergenrother P. J. DNA as a Target for Anticancer Compounds: Methods to Determine the Mode of Binding and the Mechanism of Action. Curr. Opin. Biotechnol. 2007, 18 (6), 497–503. 10.1016/j.copbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Fang L.; Qin X.; Zhao J.; Gou S. Construction of Dual Stimuli-Responsive Platinum(IV) Hybrids with NQO1 Targeting Ability and Overcoming Cisplatin Resistance. Inorg. Chem. 2019, 58 (3), 2191–2200. 10.1021/acs.inorgchem.8b03386. [DOI] [PubMed] [Google Scholar]
- Karumban K. S.; Muley A.; Raut R.; Gupta P.; Giri B.; Kumbhakar S.; Misra A.; Maji S. Mononuclear Co(Ii) Polypyridyl Complexes: Synthesis, Molecular Structure, DNA Binding/Cleavage, Radical Scavenging, Docking Studies and Anticancer Activities. Dalt. Trans. 2022, 51, 7084–7099. 10.1039/D1DT04144D. [DOI] [PubMed] [Google Scholar]
- Wang D.; Lippard S. J. Cellular Processing of Platinum Anticancer Drugs. Nat. Rev. Drug Discovery 2005, 4 (4), 307–320. 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- Angeles-Boza A. M.; Bradley P. M.; Fu P. K. L.; Wicke S. E.; Bacsa J.; Dunbar K. R.; Turro C. DNA Binding and Photocleavage in Vitro by New Dirhodium(II) Dppz Complexes: Correlation to Cytotoxicity and Photocytotoxicity. Inorg. Chem. 2004, 43 (26), 8510–8519. 10.1021/ic049091h. [DOI] [PubMed] [Google Scholar]
- Tanaka T.; Yukawa K.; Umesaki N. Combination Effects of Irradiation and Irinotecan on Cervical Squamous Cell Carcinoma Cells in Vitro. Oncol. Rep. 2005, 58 (4), 2286–2298. 10.3892/or.14.5.1365. [DOI] [PubMed] [Google Scholar]
- Chitambar C. R.; Narasimhan J. Targeting Iron-Dependent DNA Synthesis with Gallium and Transferrin-Gallium. Pathobiology 1991, 59 (1), 3–10. 10.1159/000163609. [DOI] [PubMed] [Google Scholar]
- Goss C. H.; Kaneko Y.; Khuu L.; Anderson G. D.; Ravishankar S.; Aitken M. L.; Lechtzin N.; Zhou G.; Czyz D. M.; McLean K.; Olakanmi O.; Shuman H. A.; Teresi M.; Wilhelm E.; Caldwell E.; Salipante S. J.; Hornick D. B.; Siehnel R. J.; Becker L.; Britigan B. E.; Singh P. K. Gallium Disrupts Bacterial Iron Metabolism and Has Therapeutic Effects in Mice and Humans with Lung Infections. Sci. Transl. Med. 2018, 10 (460), eaat7520. 10.1126/scitranslmed.aat7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apseloff G. Therapeutic Uses of Gallium Nitrate: Past, Present, and Future. American Journal of Therapeutics. 1999, 6, 327–339. 10.1097/00045391-199911000-00008. [DOI] [PubMed] [Google Scholar]
- Chitambar C. R. Medical Applications and Toxicities of Gallium Compounds. Int. J. Environ. Res. Public Health 2010, 7 (5), 2337–2361. 10.3390/ijerph7052337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitambar C. R.; Antholine W. E. Iron-Targeting Antitumor Activity of Gallium Compounds and Novel Insights into Triapine-Metal Complexes. Antioxidants Redox Signal. 2013, 18 (8), 956–972. 10.1089/ars.2012.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjos K. D.; Cawthray J. F.; Polishchuk E.; Abrams M. J.; Orvig C. Gallium(III) and Iron(III) Complexes of Quinolone Antimicrobials. Dalt. Trans. 2016, 45 (33), 13146–13160. 10.1039/C6DT01315E. [DOI] [PubMed] [Google Scholar]
- Chitambar C. R.; Al-Gizawiy M. M.; Alhajala H. S.; Pechman K. R.; Wereley J. P.; Wujek R.; Clark P. A.; Kuo J. S.; Antholine W. E.; Schmainda K. M. Gallium Maltolate Disrupts Tumor Iron Metabolism and Retards the Growth of Glioblastoma by Inhibiting Mitochondrial Function and Ribonucleotide Reductase. Mol. Cancer Ther. 2018, 17 (6), 1240–1250. 10.1158/1535-7163.MCT-17-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsen A.; Collery P.; Garnotel R.; Brassart B.; Etique N.; Mohamed Sabry G.; Elsherif Hassan R.; Jeannesson P.; Desmaële D.; Morjani H. A New Gallium Complex Inhibits Tumor Cell Invasion and Matrix Metalloproteinase MMP-14 Expression and Activity. Metallomics 2017, 9 (8), 1176–1184. 10.1039/C7MT00049A. [DOI] [PubMed] [Google Scholar]
- Enyedy É. A.; Dömötör O.; Bali K.; Hetényi A.; Tuccinardi T.; Keppler B. K. Interaction of the Anticancer Gallium(III) Complexes of 8-Hydroxyquinoline and Maltol with Human Serum Proteins. J. Biol. Inorg. Chem. 2015, 20 (1), 77–88. 10.1007/s00775-014-1211-9. [DOI] [PubMed] [Google Scholar]
- Farrell N. P.; Gorle A. K.; Peterson E. J.; Berners-Price S. J.. Metalloglycomics. In Metallo-Drugs: Development and Action of Anticancer Agents; Sigel A., Sigel H., Freisinger E., Sigel R. K.O., Eds; Metal Ions in Life Sciences, Vol. 18; De Gruyter, 2018; pp 109–140. [DOI] [PubMed] [Google Scholar]
- Bullock G. C.; Richardson C. L.; Schrott V.; Gunawardena N. D.; Cole T. N.; Corey C. G.; Shiva S. S. The Role of Mitochondrial Metabolism and Redox Signaling in Iron Deficiency Anemia. Blood 2015, 126 (23), 2145–2145. 10.1182/blood.V126.23.2145.2145. [DOI] [Google Scholar]
- Di Stefano A.; Frosali S.; Leonini A.; Ettorre A.; Priora R.; Di Simplicio F. C.; Di Simplicio P. GSH Depletion, Protein S-Glutathionylation and Mitochondrial Transmembrane Potential Hyperpolarization Are Early Events in Initiation of Cell Death Induced by a Mixture of Isothiazolinones in HL60 Cells. Biochim. Biophys. Acta - Mol. Cell Res. 2006, 1763 (2), 214–225. 10.1016/j.bbamcr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Jeon J. H.; Lee C. H.; Lee H. S. Antimicrobial Activities of 2-Methyl-8-Hydroxyquinoline and Its Derivatives against Human Intestinal Bacteria. J. Appl. Biol. Chem. 2009, 52 (2), 202–205. 10.3839/jksabc.2009.037. [DOI] [Google Scholar]
- Enquist P. A.; Gylfe Å.; Hägglund U.; Lindström P.; Norberg-Scherman H.; Sundin C.; Elofsson M. Derivatives of 8-Hydroxyquinoline - Antibacterial Agents That Target Intra- and Extracellular Gram-Negative Pathogens. Bioorg. Med. Chem. Lett. 2012, 22 (10), 3550–3553. 10.1016/j.bmcl.2012.03.096. [DOI] [PubMed] [Google Scholar]
- Musiol R.; Jampilek J.; Buchta V.; Silva L.; Niedbala H.; Podeszwa B.; Palka A.; Majerz-Maniecka K.; Oleksyn B.; Polanski J. Antifungal Properties of New Series of Quinoline Derivatives. Bioorg. Med. Chem. 2006, 14 (10), 3592–3598. 10.1016/j.bmc.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Cheghib N.; Abdaoui M.; Cresteil T.; Aubert G.; Kraus J. L. Synthesis and Anti-Proliferative Activity of Novel Quinolin-8-Ol Derivatives. Polycycl. Aromat. Compd. 2011, 31 (3), 143–153. 10.1080/10406638.2011.581261. [DOI] [Google Scholar]
- Chan S. H.; Chui C. H.; Chan S. W.; Kok S. H. L.; Chan D.; Tsoi M. Y. T.; Leung P. H. M.; Lam A. K. Y.; Chan A. S. C.; Lam K. H.; Tang J. C. O. Synthesis of 8-Hydroxyquinoline Derivatives as Novel Antitumor Agents. ACS Med. Chem. Lett. 2013, 4 (2), 170–174. 10.1021/ml300238z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Xu Y.; Liang M.; Ke Q.; Fang Y.; Xu H.; Jin X.; Huang C. Honeycomb-like Polysulphone/Polyurethane Nanofiber Filter for the Removal of Organic/Inorganic Species from Air Streams. J. Hazard. Mater. 2018, 347, 325–333. 10.1016/j.jhazmat.2018.01.012. [DOI] [PubMed] [Google Scholar]
- Sgarlata C.; Arena G.; Bonomo R. P.; Giuffrida A.; Tabbì G. Simple and Mixed Complexes of Copper(II) with 8-Hydroxyquinoline Derivatives and Amino Acids: Characterization in Solution and Potential Biological Implications. J. Inorg. Biochem. 2018, 180, 89–100. 10.1016/j.jinorgbio.2017.12.002. [DOI] [PubMed] [Google Scholar]
- Serrao E.; Debnath B.; Otake H.; Kuang Y.; Christ F.; Debyser Z.; Neamati N. Fragment-Based Discovery of 8-Hydroxyquinoline Inhibitors of the HIV-1 Integrase-Lens Epithelium-Derived Growth Factor/P75 (IN-LEDGF/P75) Interaction. J. Med. Chem. 2013, 56 (6), 2311–2322. 10.1021/jm301632e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwanjang W.; Prachayasittikul S.; Prachayasittikul V. Effect of 8-Hydroxyquinoline and Derivatives on Human Neuroblastoma SH-SY5Y Cells under High Glucose. PeerJ. 2016, 4, e2389. 10.7717/peerj.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q. P.; Chen Z. F.; Qin J. L.; He X. J.; Li Y. L.; Liu Y. C.; Huang K.-B.; Liang H. Studies on Antitumor Mechanism of Two Planar Platinum(II) Complexes with 8-Hydroxyquinoline: Synthesis, Characterization, Cytotoxicity, Cell Cycle and Apoptosis. Eur. J. Med. Chem. 2015, 92, 302–313. 10.1016/j.ejmech.2014.12.052. [DOI] [PubMed] [Google Scholar]
- Zou H. H.; Meng T.; Chen Q.; Zhang Y. Q.; Wang H. L.; Li B.; Wang K.; Chen Z. L.; Liang F. Bifunctional Mononuclear Dysprosium Complexes: Single-Ion Magnet Behaviors and Antitumor Activities. Inorg. Chem. 2019, 58 (4), 2286–2298. 10.1021/acs.inorgchem.8b02250. [DOI] [PubMed] [Google Scholar]
- Meng T.; Tang S. F.; Qin Q. P.; Liang Y. L.; Wu C. X.; Wang C. Y.; Yan H. T.; Dong J. X.; Liu Y. C. Evaluation of the Effect of Iodine Substitution of 8-Hydroxyquinoline on Its Platinum(II) Complex: Cytotoxicity, Cell Apoptosis and Telomerase Inhibition. Medchemcomm 2016, 7 (9), 1802–1811. 10.1039/C6MD00201C. [DOI] [Google Scholar]
- Oliveri V.; Viale M.; Aiello C.; Vecchio G. New 8-Hydroxyquinoline Galactosides. the Role of the Sugar in the Antiproliferative Activity of Copper(II) Ionophores. J. Inorg. Biochem. 2015, 142, 101–108. 10.1016/j.jinorgbio.2014.09.017. [DOI] [PubMed] [Google Scholar]
- Shamle N. J.; Tella A. C.; Obaleye J. A.; Balogun F. O.; Ashafa A. O. T.; Ajibade P. A. 11Synthesis, Crystal Structures, Electrochemistry and Biological Evaluation of Tris(Quinolin-8-Olato) Mn(III) and Co(III) Complexes Methanol Solvate. J. Mol. Struct. 2020, 1219, 128585. 10.1016/j.molstruc.2020.128585. [DOI] [Google Scholar]
- Gupta R.; Luxami V.; Paul K. Insights of 8-Hydroxyquinolines: A Novel Target in Medicinal Chemistry. Bioorg. Chem. 2021, 108, 104633. 10.1016/j.bioorg.2021.104633. [DOI] [PubMed] [Google Scholar]
- De Angelis Curtis S.; Kurdziel K.; Materazzi S.; Vecchio S. Crystal Structure and Thermoanalytical Study of Cobalt(II) and Nickel(II) Complexes with 2,2′-Bis-(4,5-Dimethylimidazole). Thermochim. Acta 2010, 510 (1–2), 75–81. 10.1016/j.tca.2010.06.025. [DOI] [Google Scholar]
- Papi F.; Potticary J.; Lanza A. E.; Hall S. R.; Gemmi M. Crystal Structure of a Peculiar Polycyclic Aromatic Hydrocarbon Determined by 3D Electron Diffraction. Cryst. Growth Des. 2021, 21 (11), 6341–6348. 10.1021/acs.cgd.1c00820. [DOI] [Google Scholar]
- Frisch M. J.; Trucks G. W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Mennucci B.; Petersson G. A.; Nakatsuji H.; Caricato M.; Li X.; Hratchian H. P.; Izmaylov A. F.; Bloino J.; Zheng G.; Sonnenberg J. L.; Hada M.; Ehara M.; Toyota K.; Fukuda R.; Hasegawa J.; Ishida M.; Nakajima T.; Honda Y.; Kitao O.; Nakai H.; Vreven T.; Montgomery J. A. Jr., Peralta J. E.; Ogliaro F.; Bearpark M.; Heyd J. J.; Brothers E.; Kudin K. N.; Staroverov V. N.; Keith T.; Kobayashi R.; Normand J.; Raghavachari K.; Rendell A.; Burant J. C.; Iyengar S. S.; Tomasi J.; Cossi M.; Rega N.; Millam J. M.; Klene M.; Knox J. E.; Cross J. B.; Bakken V.; Adamo C.; Jaramillo J.; Gomperts R.; Stratmann R. E.; Yazyev O.; Austin A. J.; Cammi R.; Pomelli C.; Ochterski J. W.; Martin R. L.; Morokuma K.; Zakrzewski V. G.; Voth G. A.; Salvador P.; Dannenberg J. J.; Dapprich S.; Daniels A. D.; Farkas O.; Foresman J. B.; Ortiz J. V.; Cioslowski J.; Fox D. J.. Gaussian 09, Revision E.01; Gaussian, Inc., Wallingford, CT, 2013. [Google Scholar]
- Chai J. Da; Head-Gordon M. Long-Range Corrected Hybrid Density Functionals with Damped Atom-Atom Dispersion Corrections. Phys. Chem. Chem. Phys. 2008, 10 (44), 6615–6620. 10.1039/b810189b. [DOI] [PubMed] [Google Scholar]
- Weigend F.; Ahlrichs R. Balanced Basis Sets of Split Valence, Triple Zeta Valence and Quadruple Zeta Valence Quality for H to Rn: Design and Assessment of Accuracy. Phys. Chem. Chem. Phys. 2005, 7 (18), 3297–3305. 10.1039/b508541a. [DOI] [PubMed] [Google Scholar]
- Marenich A. V.; Cramer C. J.; Truhlar D. G. Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113 (18), 6378–6396. 10.1021/jp810292n. [DOI] [PubMed] [Google Scholar]
- Lu T.; Chen F. Multiwfn: A Multifunctional Wavefunction Analyzer. J. Comput. Chem. 2012, 33 (5), 580–592. 10.1002/jcc.22885. [DOI] [PubMed] [Google Scholar]
- Yuan S.; Chan H. C. S.; Hu Z. Using PyMOL as a Platform for Computational Drug Design. WIREs Comput. Mol. Sci. 2017, 7, e1298. 10.1002/wcms.1298. [DOI] [Google Scholar]
- Ali M. R.; Sadoqi M.; Møller S. G.; Boutajangout A.; Mezei M. Assessing the Binding of Cholinesterase Inhibitors by Docking and Molecular Dynamics Studies. J. Mol. Graph. Model. 2017, 76, 36–42. 10.1016/j.jmgm.2017.06.027. [DOI] [PubMed] [Google Scholar]
- Depeursinge A.; Racoceanu D.; Iavindrasana J.; Cohen G.; Platon A.; Poletti P.-A.; Muller H. Fusing Visual and Clinical Information for Lung Tissue Classification in HRCT Data. Artif. Intell. Med. 2010, 50, 13–21. 10.1016/j.artmed.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Alizadeh M.; Mirjafary Z.; Saeidian H. Straightforward Synthesis, Spectroscopic Characterizations and Comprehensive DFT Calculations of Novel 1-Ester 4-Sulfonamide-1,2,3-Triazole Scaffolds. J. Mol. Struct. 2020, 1203, 127405. 10.1016/j.molstruc.2019.127405. [DOI] [Google Scholar]
- Elangovan N.; Thomas R.; Sowrirajan S. Synthesis of Schiff Base (E)-4-((2-Hydroxy-3,5-Diiodobenzylidene)Amino)-N-Thiazole-2-Yl)Benzenesulfonamide with Antimicrobial Potential, Structural Features, Experimental Biological Screening and Quantum Mechanical Studies. J. Mol. Struct. 2022, 1250, 131762. 10.1016/j.molstruc.2021.131762. [DOI] [Google Scholar]
- Badolato M.; Carullo G.; Aiello F.; Garofalo A. Synthesis and Experimental Validation of New PDI Inhibitors with Antiproliferative Activity. J. Chem. 2017, 2017, 2370359. 10.1155/2017/2370359. [DOI] [Google Scholar]
- Chandel S.; Singh R.; Gautam A.; Ravichandiran V. Screening of Azadirachta Indica Phytoconstituents as GSK-3β Inhibitor and Its Implication in Neuroblastoma: Molecular Docking, Molecular Dynamics, MM-PBSA Binding Energy, and in-Vitro Study. J. Biomol. Struct. Dyn. 2022, 40, 12827. 10.1080/07391102.2021.1977705. [DOI] [PubMed] [Google Scholar]
- Vergoten G.; Bailly C. Molecular Docking Study of GSK-3β Interaction with Nomilin, Kihadanin B, and Related Limonoids and Triterpenes with a Furyl-δ-Lactone Core. J. Biochem. Mol. Toxicol. 2022, 36 (9), e23130. 10.1002/jbt.23130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.