Abstract
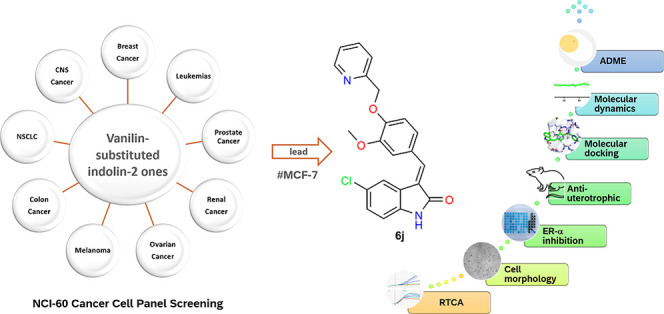
The structure-based design introduced indoles as an essential motif in designing new selective estrogen receptor modulators employed for treating breast cancer. Therefore, here, a series of synthesized vanillin-substituted indolin-2-ones were screened against the NCI-60 cancer cell panel followed by in vivo, in vitro, and in silico studies. Physicochemical parameters were evaluated with HPLC and SwissADME tools. The compounds demonstrated promising anti-cancer activity for the MCF-7 breast cancer cell line (GI = 6–63%). The compound with the highest activity (6j) was selective for the MCF-7 breast cancer cell line (IC50 = 17.01 μM) with no effect on the MCF-12A normal breast cell line supported by real-time cell analysis. A morphological examination of the used cell lines confirmed a cytostatic effect of compound 6j. It inhibited both in vivo and in vitro estrogenic activity, triggering a 38% reduction in uterine weight induced by estrogen in an immature rat model and hindering 62% of ER-α receptors in in vitro settings. In silico molecular docking and molecular dynamics simulation studies supported the stability of the ER-α and compound 6j protein–ligand complex. Herein, we report that indolin-2-one derivative 6j is a promising lead compound for further pharmaceutical formulations as a potential anti-breast cancer drug.
1. Introduction
With 2.3 million cases and 685 000 deaths in 2020, breast cancer has become the most widespread cancer worldwide. Breast cancer comprises various genetic and epigenetic factors with explicit clinical implications.1,2 Different types of breast cancer are usually described by their dependence on the estrogen receptor, ER, progesterone receptor, PR, and/or human epithelial receptor 2, HER2, with ER positive (ER+) cases accounting for 75% of all cases.3 Since ER receptors are dysregulated in cancer cells, they are involved in uncontrolled cell proliferation, metastasis, and cancer invasiveness.4 Consequently, antagonizing ER receptors is part of the first-line therapy for ER+ breast cancer cases. Tamoxifen was an ER antagonist initially adopted as a targeted therapy to prevent the estrogen-stimulated proliferation of breast tumor cells. Nevertheless, it was promptly elucidated that tamoxifen possesses tissue-selective agonist traits.5,6 This partial agonistic activity restrains antagonism, puts the therapeutic effectiveness of tamoxifen into question, and might explain some of tamoxifen’s adverse effects.7 These negative effects were mitigated by developing second- and third-generation ER antagonists, currently called selective estrogen receptor modulators (SERMs).8,9 SERMs share a potent ER antagonistic profile in breast tissue, protecting bone tissue without a uterotrophic profile.10,11
Different heterocycles were introduced during the development of the second and third generations of SERMs, such as the benzothiophene-based raloxifene12 and the indole-based bazedoxifene.13 The use of nitrogen-containing heterocycles may induce a polarized behavior that contributes to establishing an efficient interaction with ER-α receptors.14−16 The third generation of SERM, bazedoxifene, I, (Figure 1), is an indole-based modulator approved in 2013 to treat and prevent postmenopausal osteoporosis17 with several current trials for application in breast cancer18,19 and schizophrenia.20 It was designed by replacing the benzothiophene core of raloxifene with an indole ring.18,21 It showed tumor suppressor activity in ER+ breast cancer patients.21 It held the potential to counteract the acquired hormonal resistance observed with other SERMs in breast cancer cell lines.22 It even induced anti-proliferative activity in triple-negative breast cancer via decreasing the expression of p-STAT3 and inhibiting IL-6/GP130 pathways.23 Such effects contribute to anti-tumor effects observed in non-hormone sensitive cancer cell lines such as head and neck24 and gastric and pancreatic.23,25 Subsequently, bazedoxifene was used as a template for designing several potential anticancer agents with the ability to modulate estrogen activity for use in breast cancer cell lines as shown in Figure 1.26 Furthermore, indole using is not confined to SERMs but also widely goes to the design of several anticancer agents.27 It can be seen in several anticancer drugs, such as sunitinib, anlotinib, osimertinib, and other agents in clinical trials such as semaxinib.28,29 Additionally, indole and its derivatives, such as isatin, are functional motifs in the design of anti-cancer agents with diverse mechanisms.30 They can evoke an anti-cancer profile by inhibiting tubulin polymerization, some tyrosine kinases (such as Akt, EGFR, and ALK), the HDAC enzyme, and topoisomerase.30−32 Multiple indole derivatives were also designed to target breast cancer cell lines.33,34 Indolin-2-one was merged with a chalcone pharmacophore to produce a series of 3-(2-oxo-2-phenylethylidene)indolin-2-ones (6a–o, Figure 1) that considerably inhibited the proliferation of MDA-MB-231, MDA-MB-468, and MCF-7 breast cancer cells with IC50s’ of 8.54, 4.76, and 3.59 μM, respectively.35 Merging the pharmacophore of the SERMs bazedoxifene, I and pipendoxifene, II with the previously reported anti-cancer compound III, herein, we focus the biological activity of synthesized indole-2-one derivatives 6a–o for potential synergism of the anti-breast cancer activity observed in both compounds while retaining the inhibition of the ER-α receptor.
Figure 1.
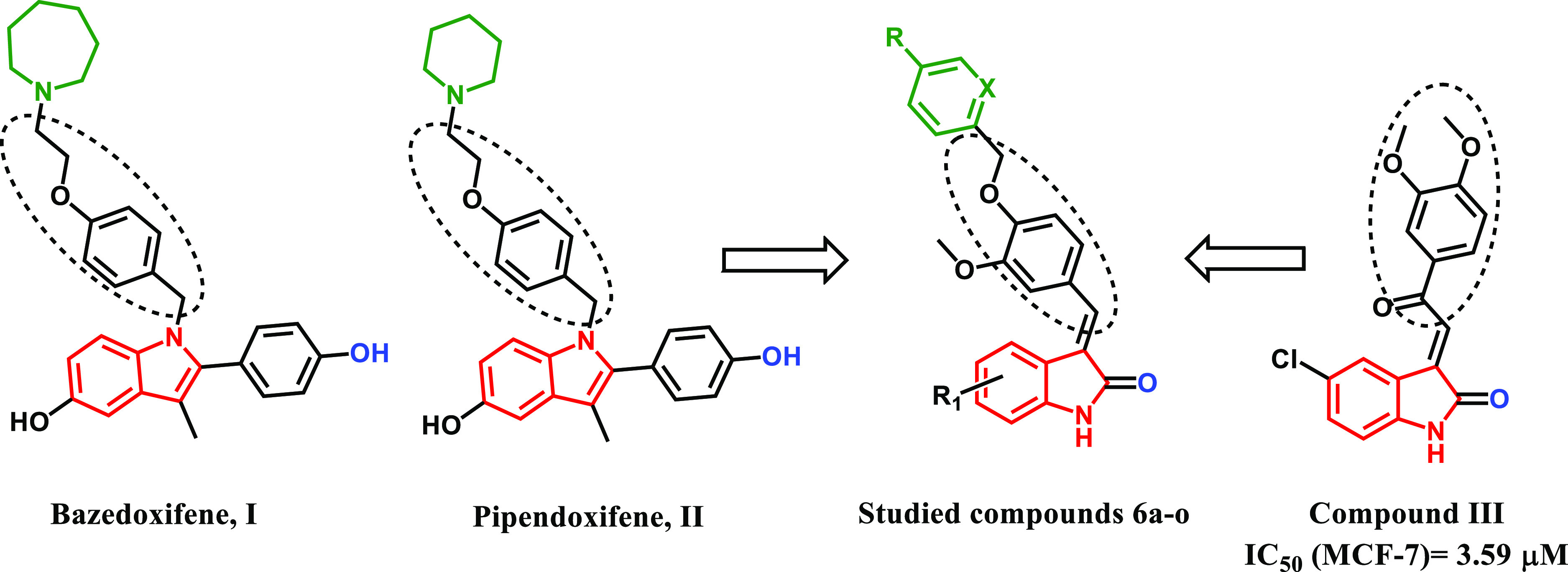
Structures of indole-based SERMs bazedoxifene I, pipendoxifene II, the previously reported anti-breast cancer indole derivative compound III, and the studied compounds 6a–o.
2. Results
2.1. Chemistry
The route for the synthesis of oxindoles 5a–c and 6a–o has been previously reported by our group and is summarized in Scheme 1.36 Vanillin or alkylated vanillin derivatives reacted with different oxindole derivatives, yielding compounds 5a–c and 6a–o, respectively. The resulted compounds were a mixture of E and Z isomers and used without separation as the previous literature reported that the E isomer is mainly the major isomer37−39 with the possibility of interconversion between the two isomers in methanol within 2 days.40,41 Compounds’ identities were confirmed by comparing mp and NMR data to those we had previously reported.36
Scheme 1. Compounds 5a–c and 6a–o, Which Have Been Previously Synthesized and Screened for Anti-Cancer Activity with the NCI-60 Cancer Cell Line Panel in This Study.
2.2. NCI-60 Cell Line One-Dose In Vitro Cytotoxicity Screening
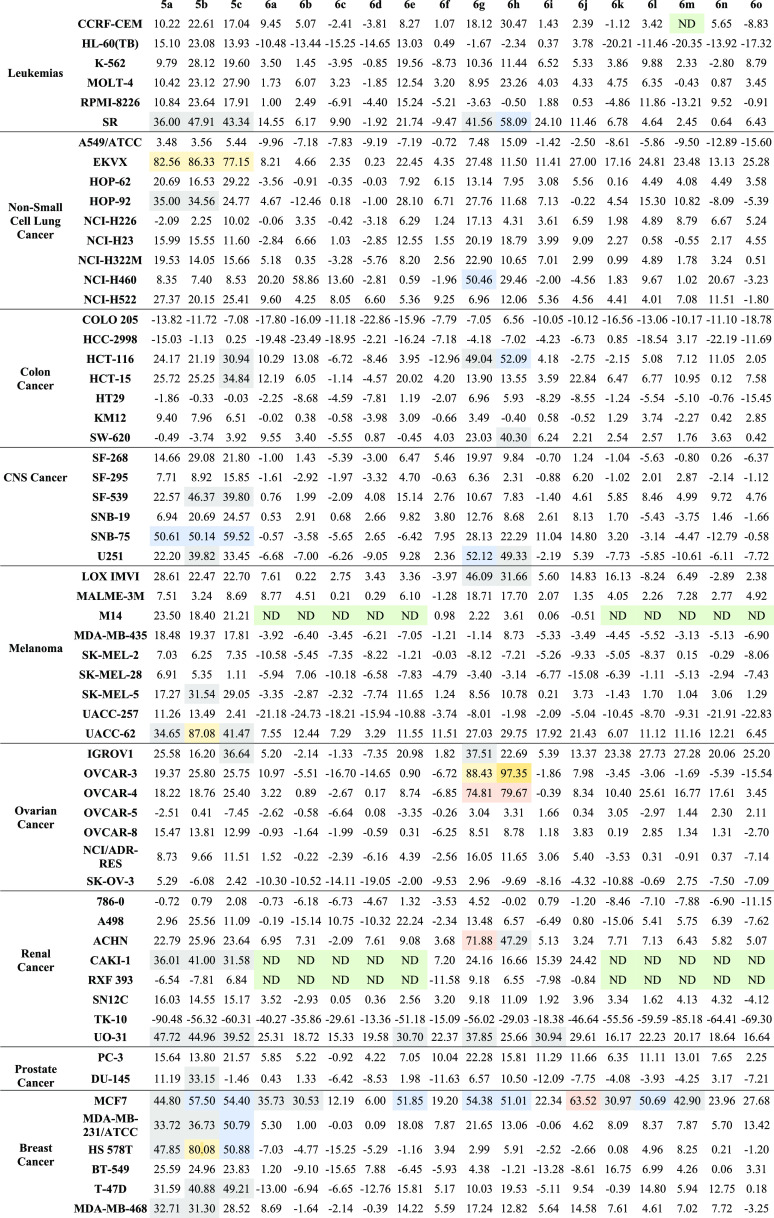
Compounds 5a–c and 6a–o were tested using standard NCI protocols for in vitro activity at the National Cancer Institute (NCI, Bethesda, Maryland, USA), wherein compounds were tested using one single concentration of 10 μM against 60 cell lines of nine different cancer types. The results are expressed as growth inhibition (GI, %) and listed in Table 1. Data showed a weak to moderate activity against leukemia, the central nervous system (CNS), melanoma, ovarian, renal, and prostate cancers. Excellent activity was observed against a single NSCLC cell line, EKVX, for compounds 5a–c (GIs = 77–86%, Table 1) with no observed activity for indoles with substituted vanillin 6a–o. Similarly, compounds 5a-o showed good activity against the SNB-75 CNS cancer cell line (GIs = 50–59%, Table 1) with a very weak activity for compounds 6a–o. The results also revealed excellent activity of compounds 6g and 6h against ovarian cancer cell lines OVCAR-3 and OVCAR-4. The GI observed was highest and ranged from 74 to 97%. All tested compounds exhibited a consistent inhibition against the MCF-7 breast cancer cell line with GIs of 6–63%. Compound 6j showed the highest activity with a GI of 63%, while compounds 5b–c, 6e, 6g–h, and 6l showed moderate activity with GIs of 50–65%. The MCF-7 cell line was selected for further testing since it demonstrated the only consistent activity.
Table 1. GI (%) Induced by 10 μM of Compounds 5a–c and 6a–o against the NCI-60 Cancer Cell Line Panela.
Cells shaded green = non determined activity; cells shaded gray for GI >30%; cells shaded blue for GI >50%; and cells shaded orange for GI >60%.
2.3. Real-Time Cellular Analysis against the MCF-7 Breast Cancer Cell Line and MCF-12A Normal Breast Cell Line
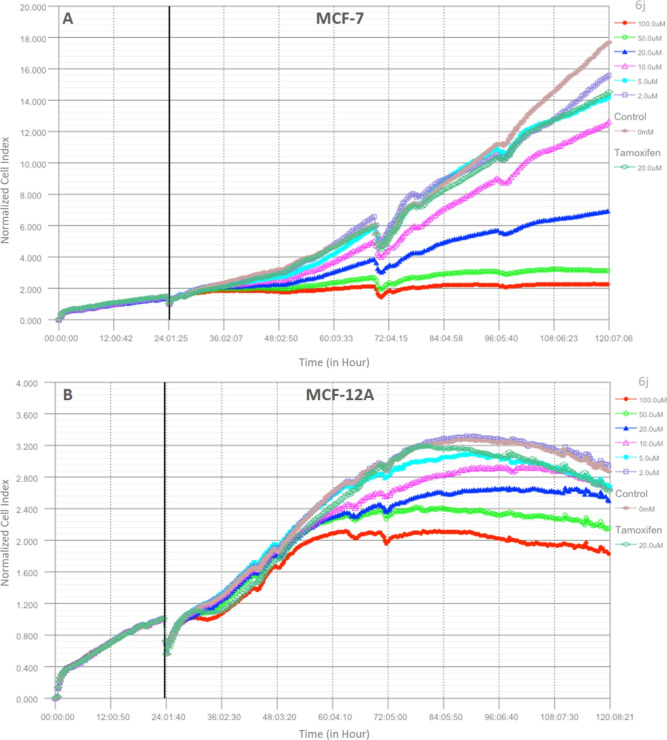
Since compound 6j displayed the most potent anti-breast cancer activity against MCF-7 cells in the NCI-60 panel, we aimed to further investigate the effects of compound 6j by using the iCELLigence real-time cell analysis system. Therefore, compound 6j was applied in serial doses (2, 5, 10, 20, 50, and 100 μM) to MCF-7 cells and parallel to MCF-12A cells to determine the selectivity and safety. The treatments were performed 24 h after seeding the cells on system-specific biosensor-based plates. A total of 120 h of analyses were monitored, with cell viability measurements taken every 15 min. Figure 2a,b shows the results as a normalized cell index graph. IC50 values at 24, 48, 72, and 96 h after compound treatments were calculated by the iCELLigence software and are given in Table 2. In addition, the viability percentage values for each dose in these periods were calculated and are summarized in Table 3 for MCF-7 and Table 4 for MCF-12A. As seen in Figure 2a, compound 6j completely inhibited the growth of MCF-7 cells at all time points at concentrations of 100 and 50 μM. In these treatments, cells were not killed dramatically after adding the highest two doses of 6j (100 and 50 μM); instead, they entered the stationary phase. 20 μM of 6j approximately inhibited the cell growth of MCF-7 cells at 50% in all time points, while 10 μM of 6j inhibited the growth of MCF-7 cells by 25%. The same GI curves were observed in cells treated with 5 μM 6j and 20 μM tamoxifen. Treatment with 2 μM of 6j was ineffective compared to the other doses, but it slightly reduced the cell proliferation compared to that of the control. IC50 values after the 6j treatments were calculated as 120.86 μM at 24 h, 16.19 μM at 48 h, 17.01 μM at 72 h, and 16.12 μM at 96 h.
Figure 2.
Dynamic monitoring of the effects of compound 6j on MCF-7 and MCF-12A cells with the iCELLigence real-time cell analysis system (A) MCF-7 and (B) MCF-12A cell lines.
Table 2. IC50 and R2 Values Obtained from Different Time Points Following Compound 6j Treatments to MCF-7 and MCF-12A Cell Lines.
| MCF-7 |
MCF-12A |
|||
|---|---|---|---|---|
| time points (h) | IC50 value (μM) | R2 | IC50 value (μM) | R2 |
| 24 | 120.86 | 0.9962 | 506.11 | 0.9298 |
| 48 | 16.19 | 0.9998 | 81.87 | 0.9828 |
| 72 | 17.01 | 0.9951 | 311.68 | 0.9960 |
| 96 | 16.12 | 0.9959 | 206.87 | 0.9831 |
Table 3. MCF-7 Cell Viability (%) at Different Time Points after Treatment with Different Concentrations of 6j or Tamoxifen (Relative to Control).
| cell viability (% ± SEM) |
||||||||
|---|---|---|---|---|---|---|---|---|
| compound 6j |
tamoxifen | |||||||
| time point (h) | control | 100 μM | 50 μM | 20 μM | 10 μM | 5 μM | 2 μM | 20 μM |
| 24 | 98.36 ± 0.00 | 57.46 ± 0.02 | 62.66 ± 0.00 | 70.25 ± 0.02 | 80.00 ± 0.01 | 84.83 ± 0.01 | 91.38 ± 0.01 | 86.84 ± 0.02 |
| 48 | 97.46 ± 0.02 | 33.25 ± 0.02 | 43.36 ± 0.00 | 64.29 ± 0.02 | 76.59 ± 0.01 | 83.01 ± 0.02 | 91.38 ± 0.01 | 86.84 ± 0.02 |
| 72 | 96.91 ± 0.03 | 20.25 ± 0.01 | 27.99 ± 0.00 | 51.20 ± 0.01 | 76.59 ± 0.01 | 83.01 ± 0.02 | 91.38 ± 0.01 | 85.59 ± 0.04 |
| 96 | 96.91 ± 0.03 | 13.43 ± 0.00 | 18.43 ± 0.00 | 40.49 ± 0.00 | 74.16 ± 0.00 | 82.54 ± 0.01 | 90.67 ± 0.01 | 80.31 ± 0.07 |
Table 4. MCF-12A Cell Viability (%) at Different Time Points after Treatment with Different Concentrations of 6j or Tamoxifen (Relative to Control).
| cell viability (% ± SEM) |
||||||||
|---|---|---|---|---|---|---|---|---|
| compound 6j |
tamoxifen | |||||||
| time point (h) | control | 100 μM | 50 μM | 20 μM | 10 μM | 5 μM | 2 μM | 20 μM |
| 24 | 99.43 ± 0.01 | 82.05 ± 0.00 | 90.25 ± 0.01 | 93.58 ± 0.00 | 97.22 ± 0.01 | 99.06 ± 0.01 | 98.70 ± 0.00 | 84.00 ± 0.01 |
| 48 | 98.78 ± 0.00 | 67.30 ± 0.00 | 77.81 ± 0.01 | 81.96 ± 0.01 | 88.48 ± 0.02 | 93.73 ± 0.02 | 97.43 ± 0.02 | 84.00 ± 0.01 |
| 72 | 98.78 ± 0.00 | 62.63 ± 0.01 | 72.70 ± 0.01 | 80.57 ± 0.01 | 87.93 ± 0.02 | 92.74 ± 0.01 | 97.43 ± 0.02 | 84.00 ± 0.01 |
| 96 | 98.78 ± 0.00 | 61.26 ± 0.00 | 72.54 ± 0.01 | 80.57 ± 0.01 | 87.93 ± 0.02 | 92.17 ± 0.00 | 97.43 ± 0.02 | 84.00 ± 0.01 |
In contrast to MCF-7 breast cancer cells, 6j displayed no effects on MCF-12A healthy breast cells during the first 24 h of treatment at any dose. It also exhibited a 5 times’ safer profile than that of MCF-7 at the 48th h. After 48 h, based on the IC50 values (506.11 μM at 24 h, 81.87 μM at 48 h, 311.68 μM at 72 h, and 206.87 μM at 96 h), the cells started to recover, and the safer profile continued afterward.
2.4. Morphological Assessment of 6j-Treated MCF-7 and MCF-12A Cell Lines
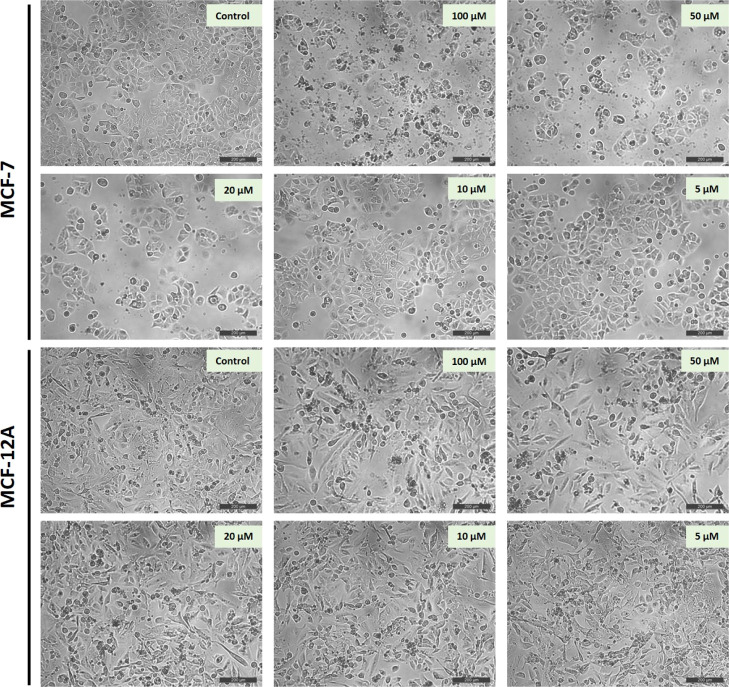
In addition to the viability analyses, morphological evaluations were performed after treating the cells with different doses of 6j (5, 10, 20, 50, and 100 μM) to better understand what was going on in the plate wells. The cells were photographed under an inverted microscope 48 h after 6j treatment. Figure 3 shows the effects of a 48 h treatment of 6j on MCF-7 and MCF-12A cells. Consistent with the iCELLigence GI curves, at 50 and 100 μM doses, MCF-7 cells remained stable by stopping cell division without being toxic, but MCF-12A cells continued to proliferate. 20 μM of 6j primarily inhibited the growth of the MCF-7 cells, while the cells displayed a healthy phenotype. The cell morphology has not deteriorated, and the membrane structures were preserved in a healthy way in the 6j-applied cells. Compared to the control group of MCF-7 cells, no decrease in cell number was observed in MCF-12A cells treated with 6j, especially at doses of 20, 50, and 100 μM.
Figure 3.
Effects of different concentrations of 6j on the MCF-7 and MCF-12A cell morphology photographed under an inverted microscope 48 h after 6j treatment. Scale bar represents 200 μm.
2.5. In Vivo Anti-Estrogenic (Anti-Uterotrophic) Activity of Compound 6j
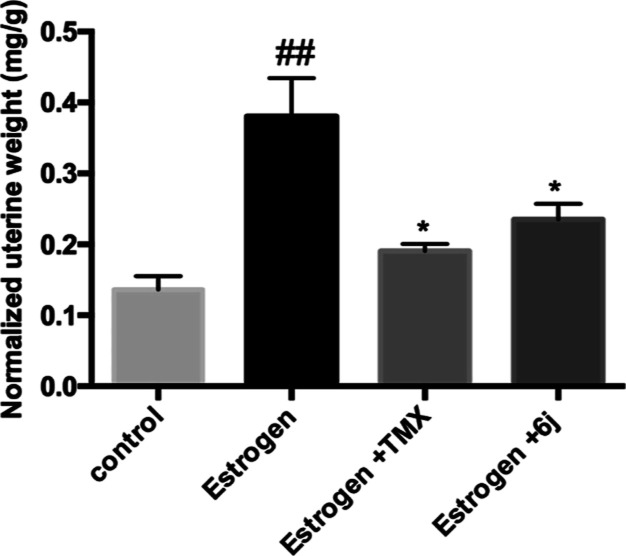
As shown in Figure 4, the anti-uterotrophic activities of tamoxifen and 6j are expressed as normalized uterine weight and were calculated upon orally treating the rats with each compound (20 mg/kg) over three independent experiments. Estrogen alone caused a significant increase in uterus weight compared to the control, while both tamoxifen and 6j significantly inhibited the estrogen-induced uterotrophic effect. The measured anti-uterotrophic activity of 6j was 38% compared to that of tamoxifen (50%).
Figure 4.
Bar chart showing the in vivo antiestrogenic activity of tamoxifen (TMX) and compound 6j. ## denotes a significant difference from the control group at p < 0.01 * denotes a significant difference from the estrogen group at p < 0.05.
2.6. In Vitro ER-α Inhibitory Activity of Compound 6j
The in vitro inhibitory activity of compound 6j against ER-α was measured via ELISA assay to confirm the observed anti-estrogenic activities of compound 6j. Compound 6j inhibited 62% of ER-α activity on MCF-7 cells compared to 71% for tamoxifen. The results are listed in Table 5.
Table 5. Concentration of the ER-α Receptor in MCF-7 Cells Treated with Compound 6j or Tamoxifen Compared to the Control.
| results |
||
|---|---|---|
| compound | ER-α pg/mL (mean ± SEM) | inhibition (%) |
| 6j | 424.9 ± 17.2 | 62.4 |
| tamoxifen | 327 ± 16.7 | 71.1 |
| control | 1131 ± 49.7 | 0 |
2.7. Evaluation of Physicochemical Parameters
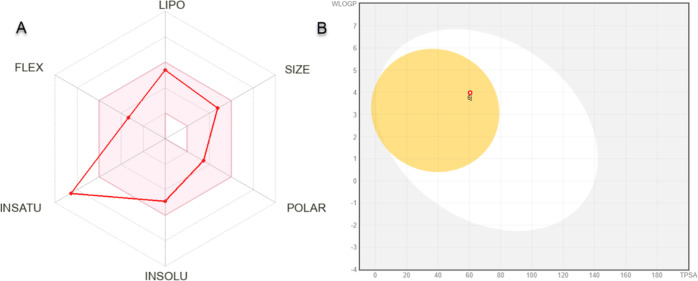
Studying drug solubility is a crucial part of the pre-formulation study. It is an integral phase that every drug has been through in any development process to determine its bioavailability and the best excipients used during formulation. The solubility of compound 6j was detected using HPLC in methanol, ethanol, and acetonitrile. Unfortunately, the method used did not detect any water solubility for 6j, while its solubility in organic solvents ranged from 55 to 58 mg/mL. In detail, HPLC detected the solubility for 6j to be 55.04 mg/mL in methanol, 57.33 mg/mL in ethanol, and 58.73 mg/mL in acetonitrile. With such results, compound 6j requires the addition of a surfactant to increase its solubility, especially in water. Different types of surfactants could be used to study their effect on increasing solubility in future plans. We see that this compound has the potential to go through the formulation study. Additionally, it is a fact that some drug molecule candidates are not approved as drugs, although they are active due to their poor absorption, distribution, metabolism, and excretion (ADME) properties. Estimating these properties of synthesized compounds as in silico is a useful approach in terms of medicinal chemistry.42,43 Accordingly, the active compound 6j was analyzed with SwissADME. The physicochemical properties of compound 6j, molecular weight (392.83 g/mol), fraction Csp3 (0.09), rotatable bonds (5), H-bond acceptors (4), H-bond donors (1), molar refractivity (112.41), and topological polar surface area (TPSA) (60.45 Å2) were measured. For lipophilicity, log Po/w (XLOGP3) (3.93), log Po/w (WLOGP) (3.98), log Po/w (MLOGP) (2.66), log Po/w (SILICOS-IT) (5.03) and consensus It was measured as log Po/w (3.75). For water solubility, log S (ESOL) is in the moderately soluble class with a value of −4.90. The ADME radar plot of 6j is shown in Figure 5A. The colored area in this plot indicates that the compounds are in the appropriate range for predicted oral bioavailability. In terms of pharmacokinetics, compound 6j has high gastrointestinal absorption, it is blood–brain barrier (BBB)-permeant, it is not a substrate of P-glycoprotein, and it is an inhibitor of CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4. In Figure 5B, the BOILED-Egg diagram obtained by comparing WLOGP and TPSA of 6j is shown. This diagram shows that 6j was passively permeable from the BBB, passively absorbed from the gastrointestinal tract, and was not effluated from the CNS by the P-glycoprotein if it was a red dot. The drug-likeness status of 6j was detected as suitable according to Lipinski, Ghose, Veber, Egan, and Muegge’s limited rules.44 Considering all these parameters and data, it is predicted that compound 6j will exhibit a favorable ADME profile.
Figure 5.
(A) Radar plot and (B) BOILED-Egg diagram obtained from the SwissADME server of compound 6j.
2.8. Molecular Docking Analysis
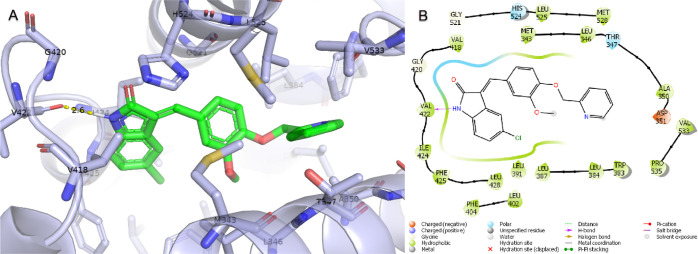
Molecular docking studies were performed to estimate the interaction pattern and binding energy of the active molecule compound 6j at the ER-α active site.45,46 For the control of molecular docking, the cocrystal ligand located in the estrogen receptor (PDB ID: 5W9C) crystal structure was self-docking and the root-mean-square deviation (rmsd) between its natural pose and docking pose was measured as 0.58 Å.47 The two compounds were almost completely superimposed by the rmsd value. After docking validation, compound 6j and standard compound tamoxifen were docked to the estrogen receptor active site. The Glide gscore value, which is the binding energy of compound 6j, was measured as −8.225 kcal/mol and tamoxifen’s as −9.694 kcal/mol. The binding poses and protein–ligand interactions of compound 6j were analyzed and are shown in Figure 6. Accordingly, compound 6j has a 2.6 Å long H bond with Val422, a polar interaction with Thr347 and His524, a negative charge with Phe425, and created hydrophobic interactions with Asp351, Met343, Leu346, Ala350, Trp383, Leu384, Leu387, Leu402, Phe404, Val418, Gly420, Val422, Ile424, Leu428, Gly521, Leu525, and Met528. Tamoxifen, on the other hand, formed both a face bridge and an H bond with Asp351, a polar interaction with Thr347, His524, and Asn532, a negative charge with Glu353 and Asp351, a positive charge with Arg394, and gave hydrophobic interactions with Met343, Ley346, Leu349, Ala350, Leu354, Trp383, Leu384, Leu387, Met388, Leu391, Phe404, Val418, Ile424, Gly521, Leu525, Val553, Val534, Pro535, and Leu539.
Figure 6.
Glide molecular docking interactions of ER-α with compound 6j. (A) Binding pose of 6j in an ER-α active site. (B) Protein–ligand schematic interaction diagram of the ER-α and 6j complex. (PDB ID: 5W9C).
2.9. Molecular Dynamics Simulations
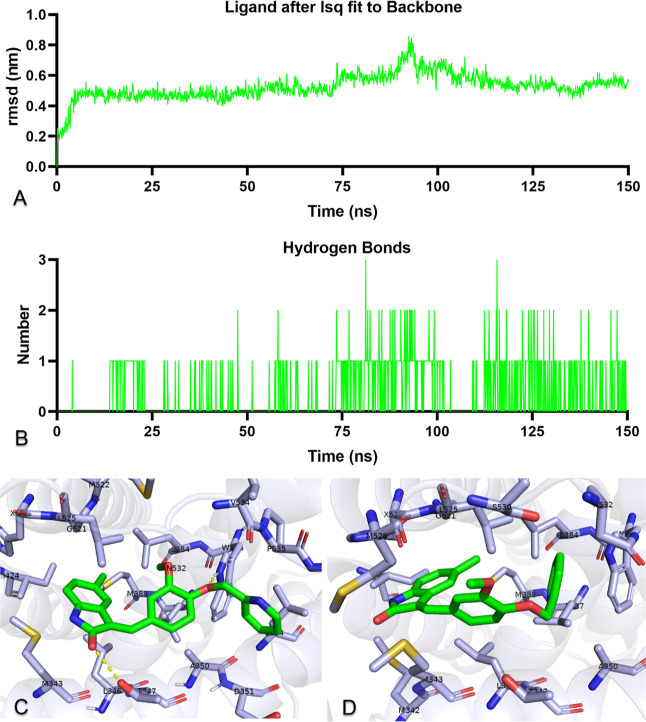
To investigate and prove in silico the stability of ER-α with compound 6j, 150 ns molecular dynamics simulations of the protein–ligand complex of ER-α and 6j were performed.48,49 It is a metric that numerically shows the difference between superimposed rmsd’s and is elegantly utilized in molecular dynamics simulations. Data on the rmsd measurement obtained by fitting compound 6j to the ER-α are shown in Figure 7A. Compound 6j after the first 10 ns of pre-simulation is below 0.6 nm and stable up to 75 ns, with a peak up to 0.8 nm around 95 ns, below 0.6 nm after 115 ns, and stable left. The other trajectory analysis is the H-bond analysis, which expresses the change with time, showing the number of H bonds between ER-α and compound 6j. As shown in Figure 7B, there was very sparse H bond formation in the first 15 ns, and after 15 ns, there was often one and sometimes two H bond formations.
Figure 7.
Molecular dynamics simulation trajectory analysis. (A) rmsd plot showing the stability of compound 6j with respect to the ER-α. (B) Number of H bonds formed between compound 6j and ER-α active site residues over 150 ns. (C,D) Binding poses of compound 6j with ER-α at 100 and 150 ns, respectively.
Binding poses at 100 and 150 ns were analyzed to analyze protein–ligand dynamic interactions and changes. In Figure 7C,D, the binding modes of 6j at 100 and 150 ns at the ER-α active site are shown. Accordingly, 6j and Trp383 yielded one H bond (1.98 Å), Asp351 yielded a negative charge, Thr347, Ser536, and Asn532 yielded a polar interaction, and Met 343, Leu346, Ala350, Leu354, Leu384, Met388, Leu387, Ile424, Gly521, Met522, His524, Leu525, Val534, Pro535, and Leu539 yielded hydrophobic interactions at 100 ns. Compound 6j had polar interactions with Thr347, Ser536, and Asn532 and hydrophobic interactions with Met342, Met343, Leu346, Ala350, Trp383, Leu384, Leu387, Met388, Ile424, Gkly521, His524, Leu525, and Met528 at 150 ns. In addition, an animation video was created from the molecular dynamics trajectory to monitor the protein–ligand interactions of ER-α and 6j at the active site for 150 ns and is presented in Video S1 of the Supporting Information. It was understood that 6j remained stable in the active site, although some interaction types and residues changed over time.
Finally, the binding free energy molecular mechanics Poisson–Boltzmann surface area (MMPBSA) formed between the protein and ligand for 150 ns was calculated from 1500 frames with the formula Δ: complex–receptor–ligand. The total binding energy MMPBSA value between ER-α and compound 6j was calculated as–30.47 ± 1.52 kcal/mol from the sum of van der Waals, electrostatic energy, electrostatic solvation free energy evaluated from the generalized Born equation, and the nonpolar component of the solvation energy, gas-phase energy, and solvation free energy. The standard deviation here was as low as 1.52 kcal/mol and an energy value of −30.47 kcal/mol was another factor indicating protein–ligand stability.
3. Discussion
The use of indole-containing compounds in the fight against breast cancer is extensively described in the literature.26,34,50 In addition to its tubulin polymerization inhibitory activity,51−53 indolin-2-one has been reported to possess anti-estrogenic activity,14,15,54 making it an effective tool in the design of medications against breast cancer. Although there was scattered cytotoxic activity of certain compounds such as 5a–c against some cell lines, the consistent activity of all test compounds 5a–c and 6a–o against the ER+ MCF-7 cell lines was similar to that of previous reports. A deeper look into the NCI in vitro anticancer screening revealed that an insignificant very weak activity was observed against ER– cells such as MDA-MB-231/ATCC with GI not exceeding 15%.
To confirm the antiproliferative activity observed against the MCF-7 cell line, the cell viability was assessed. Cell viability is regulated by biological pathways dependent on various intrinsic and extrinsic factors, and measuring the cell viability is adequately critical to the overall function and understanding of the physiology of cells. Cell viability can be measured by using several different techniques. Unlike traditional cell-based end-point assays, the xCELLigence system is a non-invasive, real-time cell analysis technology that can continuously monitor cellular dynamics, which provides more sensitive and consistent results. This technology uses electrical impedance measurement to detect cellular phenotypic changes and dynamically monitor cell proliferation via sensors.55−57 Also, these sensors allow the performance of a wide range of cell-based assays such as proliferation, cytotoxicity, migration, and invasion assays.57 Also, it distinguishes from other assays by allowing users to make the right decisions according to the current biological state of the cell before any manipulation. It eliminates the intensive steps of classical tests and risks such as being affected by some compounds due to the optical detection methods and affecting the consistency of the result.58
Viability results confirmed the data obtained from the NCI, and compound 6j was able to stop the proliferation of MCF-7 cells at different concentrations and time points. Interestingly, compound 6j showed double the activity as that observed with the use of tamoxifen at the same concentration (20 μM); 6j induced a 30% inhibition compared to that of the same dose of tamoxifen (Table 5). These results indicate that 6j shows cytostatic activity on MCF-7 cells without killing the cells in a toxic way as there is no significant increase in activity with increasing incubation time. Additionally, data from MCF-12A cells demonstrate the selective inhibition efficacy at all doses and periods of 6j, Table 4 and Figure 2. In summary, 6j shows cytostatic activity against MCF-7 ER-positive breast cancer cells, and it displays a safe profile by not showing any effect on healthy MCF-12A cells. The same conclusion was reached on examining the impact of 6j on the MCF-7 cell morphology, Figure 3. When all these results are taken together, it has been determined that 6j has a selective and safe cytostatic effect on MCF-7 breast cancer cells.
Investigations to further explore the mechanism of action of compound 6j suggested its ability to block estrogen receptors. This fact was supported by NCI data mentioned earlier wherein the observed antiproliferative activity was observed only with MCF-7, which is reported to express ER+ no significant activity was identified on ER– cell lines. This assumption was supported by the ability of 6j to antagonize the effects induced by estrogen on the rat uterus. The immature rat uterotrophic model is primarily employed to validate the impacts of estrogen agonists and antagonists on immature rats’ uteri. The model is inferred to determine the activity of a compound in the uterus quickly and accurately and can be utilized in either an agonist or antagonist mode. It depends on estrogen’s uterotrophic properties, which promote uterus development. Immature rats are employed for this test, and since they have not attained sexual maturity, endogenous estrogen has a negligible role in the estimation. After exposure to estrogen for the first time (estrone is given for 3 days), the uteri weight markedly increases as they develop quickly over these 3 days. This effect could be antagonized by co-administration of an estrogen antagonist, while estrogen agonists enhance such stimulation. Thus, the difference in uterine weight between the vehicle control and treated animals is taken as perceptive evidence of estrogen agonistic or antagonistic activities. This model successfully predicted these compounds’ clinical reactions in women.59,60 The results obtained by this model in the current study suggested an estrogen antagonistic activity attained by compound 6j as it caused a 38% reduction of the uteri weight induced by estrogen, Figure 4.
The estrogen receptors primarily mediate estrogen-induced physiological process subtypes ER-α and β. An in vitro assay against ER-α supported these data with a subtype predominant in the uterus and mammary glands.4 ER-α is the subtype usually correlated to the development of both hormone-dependent and hormone-independent cancers. It is closely associated with cancer formation, metastasis, drug resistance, and prognosis.61 Thus, the ability of compound 6j to antagonize estrogen, especially in cancer settings, was further assessed by an in vitro ER-α assay. The assay went on with the experiments mentioned above and confirmed the ability of 6j to counteract 62% of estrogen found in MCF-7 cell lines, Table 5.
Moreover, theoretical docking studies of 6j supported the experimental data and suggested a potential binding mode with the ER-α active site in a manner very similar to that of tamoxifen. According to in silico molecular docking and dynamic simulations, although compound 6j and tamoxifen show close interactions, it is understood that it inhibits ER-α by showing different binding poses and interactions.
4. Conclusions
The current study described a series of indolin-2-one derivatives (5a–c and 6a–o) as potential anti-breast cancer agents with anti-estrogenic activity. All the tested compounds exhibited weak to potent activity against the MCF-7 breast cancer cell line, where compound 6j showed the highest observed activity with a GI of 63%. The cell viability results confirmed the data obtained from the NCI. Compound 6j showed cytostatic activity against MCF-7 ER+ breast cancer cells. It displayed a safe profile without any significant effect on the healthy MCF-12A normal breast cell line. The results revealed that compound 6j has a selective and safe cytostatic effect on MCF-7 breast cancer cells. Moreover, the results of the immature rat uterotrophic model and in vitro ER-α assay suggested an estrogen antagonistic activity attained by compound 6j. Furthermore, molecular modelings are consistent with the experimental data. They predicted the potential binding patterns of the newly synthesized compound 6j with the ER-α active site in a manner close to that of tamoxifen. Collectively, these results suggested that the herein reported indolin-2-one derivative 6j is a promising lead compound for further optimization and development as a potentially efficient anti-breast cancer drug.
5. Materials and Methods
5.1. NCI-60 Cell Line One Dose In Vitro Cytotoxicity Screening
Anticancer activity was tested against 60 cancer cell lines at the NCI, Bethesda, USA. The screening process was done with a single dosage of 10 μM according to NCI protocols published on the NCI website https://dtp.cancer.gov/discovery_development/nci-60/methodology.htm.
5.2. Cell Lines and Culture Conditions
MCF-7 (Cat. no. HTB-22) (human estrogen receptor-positive breast cancer) and MCF-12A (Cat. no. CRL-10782) (human non-tumorigenic mammary epithelial) cell lines were obtained from the American Type Culture Collection (ATCC, Rockville, Maryland, USA). MCF-7 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Biological Industries, Haemek, Israel) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Biowest, Nuaillé, France), 2 mM l-glutamine (Biological Industries, Haemek, Israel), 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco, Waltham, MA, USA), and 2.5 μg/mL plasmocin (Invivogen, Toulouse, France) at 37 °C in a 5% CO2 humidified incubator. MCF-12A cells were cultured in a DMEM/F-12 Nutrient Mixture (Ham) (DMEM/F12 1:1 with HEPES and l-glutamine) (Gibco, Waltham, MA, USA) with 10% heat-inactivated FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 10 μg/mL insulin (Humulin R, Lilly, Indianapolis, USA), 20 ng/mL epidermal growth factor (Abcam, Cambridge, UK), 0.5 mg/mL hydrocortisone (Dekort, Deva Ilac, Istanbul, Turkey), and 2.5 μg/mL plasmocin in a humidified atmosphere of 5% CO2 at 37 °C. The cells were routinely cultured in cell culture flasks and checked regularly under an inverted microscope. Cells reaching 80% confluency were passaged by treatment with 0.25% trypsin–EDTA. Total cell numbers were counted by the trypan blue dye exclusion method using a hemocytometer prior to the experiments.
5.3. Monitoring the Cellular Activities with the iCELLigence Real-Time Cell Analysis System
The iCELLigence real-time cell analysis system was used to conduct a real-time and label-free examination of the activities of compound 6j on cells as we previously described.62 In brief, following a background measurement with 200 μL of complete medium on iCELLigence E-plate L8, 100 μL of MCF-7 or MCF-12A cells was seeded at a density of 5.0 × 103 per well. During the 120 h monitoring, the system took impedance measurements via biosensors every 15 min. At the 24th h of incubation, the cells were treated with increasing concentrations of the 6j compound (2, 5, 10, 20, 50, and 100 μM) in duplicate. For cell culture experiments, compound 6j was dissolved in DMSO (Sigma, St. Louis, USA) at a stock concentration of 20 mM. For treatments, dilutions were prepared from 20 mM stock with a cell growth medium, with a final DMSO concentration of 0.1%. The medium containing 0.1% DMSO was also used as a negative control. 20 μM tamoxifen (Tocris Bioscience, Bristol, UK) was included in the study set as a positive control. Data were recorded by the iCELLigence software for 120 h and analyzed at the end of the study. The IC50 values at 24, 48, 72, and 96 h after the treatments were calculated using the software using six different doses’ normalized cell index values.
5.4. Morphological Assessment of 6j-Treated MCF-7 and MCF-12A Cell Lines
Morphological studies were performed to observe the effects of 6j on MCF-7 and MCF-12A cells, as previously reported.63 In brief, 5 × 105 cells were seeded into six-well plates and incubated for 24 h. Afterward, increasing doses of compound 6j (5, 10, 20, 50, and 100 μM) were applied to the cells. The medium containing 0.1% DMSO was used as the untreated control. 48 h after treatments, the cells were photographed under an inverted microscope, Leica DM IL LED with a DFC-290 camera (Leica, Wetzlar, Germany).
5.5. In Vitro ER-α Inhibitory ELISA Assay of Compound 6j
An in vitro ER-α inhibitory ELISA assay was performed using a Human ER-α/Estrogen Receptor ELISA Kit (Sandwich ELISA) (Lifespan Biosciences, Seattle, Washington, USA) as previously described.64 The cells were plated at a density of 2000 cells/well in a 96-well plate. Treatment was done with 1 μg/mL of 6j or tamoxifen in triplicate, leaving three wells as the untreated control. After 24 h, the pellets of the cells were collected by centrifugation. The cells were washed three times with PBS and then lysed by ultrasonication, and the supernatant was collected for testing. The wells were loaded with 100 μL of either standards or samples and incubated for 90 min at 37 °C. The wells were washed with 1× wash buffer for removing any unbound sample and 100 μL 1× biotinylated detection antibody was next put in and incubated for 1 h at 37 °C. The wells were rewashed with 1× wash buffer, and 100 μL 1× HRP conjugate was then added and incubated for 30 min at 37 °C. A third wash with 1× wash buffer was done and 90 μL of the TMB substrate was added. The TMB substrate reacted with the HRP enzyme, ensuring a color development, and the reaction was terminated using 50 μL of a stop solution. Finally, the optical density (OD) of the well was measured at a wavelength of 450 nm ± 2 nm. The OD of an unknown sample was calculated by correlation with a standard curve generated by standards with known concentrations.
5.6. In Vivo Anti-Estrogenic (Anti-Uterotrophic) Activity of Compound 6j
The anti-estrogenic activity was assessed as previously described.65−67 All experiments were carried out in accordance with the recommendations of the International Animal Care and Use Committee. The experimental protocol was approved by “The Commission on the Ethics of Scientific Research”, Faculty of Pharmacy, Minia University (no. ES30/2021). 20-day-old Wistar immature female rats (40–50 g) from the animal care facility of Nahda University at Beni Suef (NUB) were allowed to acclimatize to lab conditions for 3 days before the experiment with free access to food and water. Estradiol was diluted in olive oil and subcutaneously injected on the loose dorsal skin in a dose of 10 μg/kg/day. Estradiol was diluted in olive oil and subcutaneously injected on the loose dorsal skin in a dose of 10 μg/kg/day. Tamoxifen was used in a dose of 20 mg/kg/day.66 Both compounds were dissolved in a mixture of DMSO, Tween 20, and saline (1:1:8, respectively) and orally administered. The rats were randomly assigned to three groups (n = 6) subjected to daily s.c. injections of estradiol, except for the control group. All rats receiving estradiol received an oral dose of tamoxifen or an equimolar dose of 6j daily for 3 consecutive days, except for the control group. On the 4th day, all rats were sacrificed by cervical dislocation, and the uteri were dissected free of fat and weighed immediately. The inhibition of uterine growth compared with the growth produced by estradiol alone was used to measure the anti-uterotrophic effect. The results were expressed as percent inhibition from the formula
Wv = mean uterine weights from animals treated with the vehicle, Ws = mean uterine weights from animals treated with estradiol and Ws + t = mean uterine weights from animals treated with a combination of estradiol and the test compound. It is noteworthy that doses of the tested compounds are calculated on a molar basis. One-way ANOVA followed by Tukey’s multiple comparisons test was performed using GraphPad Prism version 6.00 for Mac (GraphPad Software, La Jolla California, USA). Values are expressed as mean ± SEM.
5.7. Solubility Tests and Computational ADME
The solubility of compound 6j in various solvents (methanol, ethanol, and acetonitrile) was evaluated by adding an excess amount of the drug in a stoppered container with 0.5 mL aliquots of the used solvent. Continuous shaking was carried out in a water bath at 37 ± 1 °C for 48 h. Aliquots of the filtrate were adequately diluted with a suitable solvent and analyzed using HPLC as previously reported.68,69 The in silico ADME study of compound 6j was performed via the SwissADME server (http://www.swissadme.ch/), and some physicochemical properties, lipophilicity, water solubility, pharmacokinetics, and drug-likeness properties were calculated.44,70,71
5.8. Molecular Docking
A molecular docking study was performed with the Maestro GUI of Schrödinger v2022.2.72 For the estrogen receptor, PDB ID: 5W9C(73) from the RCSB Protein Data Bank was selected and prepared with the Protein Preparation Wizard module by choosing OPLS4 force fields.74 The missing residues in the 5W9C structure were replaced with the Prime module. The 3D structure of compound 6j and standard tamoxifen was prepared using the LigPrep module at pH = 7 ± 2 with OPLS4 force fields. Based on the cocrystal ligand in the 5W9C structure, the active site as x: 14.880, y: −11.277, z: −27.903, and 20*20*20 Å3 was created with the Receptor Grid Generation module. Molecular docking was performed with the Glide SP75,76 of the Ligand Docking module. 2D schematic interactions were created with the Maestro Ligand Interaction module, and the 3D binding pose was created by PyMOL Molecular Graphics System v2.4.1.
5.9. Molecular Dynamics Simulations
The stability of the compound 6j protein–ligand complex with the estrogen receptor obtained by Glide SP molecular docking was tested by molecular dynamics simulation using Gromacs v2021.2.77−79 The files required for molecular dynamics such as solvation of the protein–ligand complex and neutralization by adding 0.15 M KCl were created with the CHARMM-GUI server (https://charmm-gui.org/).80 Topology files of the protein and ligand were created using Amber FF99SB.81,82 Molecular dynamics simulation was carried out at 300 K and 1 atm pressure. A molecular dynamics simulation with a 150 ns duration was run. The rmsd and hydrogen bond analyses of the protein and ligand were performed with gmx rmsd and gmx hbond scripts. Biding free energy MMPBSA was calculated using gmx_MMPBSA83 tools from 1500 frames recorded between 0 and 150 ns.
Acknowledgments
The molecular dynamics numerical calculations reported in this paper were partially performed at TUBITAK ULAKBIM in TURKEY, High Performance and Grid Computing Center (TRUBA resources). Authors extend their appreciation to the Scientific Research Deanship Fund at the University of Hail, Saudi Arabia through project number RG-21 131.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c07793.
Molecular dynamics trajectory to monitor the protein–ligand interactions of ER-α and 6j (MP4)
This work was supported by the Scientific Research Deanship at the University of Hail, Saudi Arabia, through project number RG-21 131.
The authors declare no competing financial interest.
Supplementary Material
References
- Malik J. A.; Ahmed S.; Jan B.; Bender O.; Al Hagbani T.; Alqarni A.; Anwar S. Drugs Repurposed: An Advanced Step towards the Treatment of Breast Cancer and Associated Challenges. Biomed. Pharmacother. 2022, 145, 112375. 10.1016/j.biopha.2021.112375. [DOI] [PubMed] [Google Scholar]
- Bender O.; Atalay A.. Polyphenol Chlorogenic Acid, Antioxidant Profile, and Breast Cancer. In Cancer; Elsevier, 2021; pp 311–321. [Google Scholar]
- Dai X.; Cheng H.; Bai Z.; Li J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. 10.7150/jca.18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D.; Kumar S.; Narasimhan B. Estrogen Alpha Receptor Antagonists for the Treatment of Breast Cancer: A Review. Chem. Cent. J. 2018, 12, 107. 10.1186/s13065-018-0472-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher B.; Costantino J. P.; Wickerham D. L.; Redmond C. K.; Kavanah M.; Cronin W. M.; Vogel V.; Robidoux A.; Dimitrov N.; Atkins J.; Daly M.; Wieand S.; Tan-Chiu E.; Ford L.; Wolmark N.; Breast other N. S. A.; Investigators B. P. Tamoxifen for Prevention of Breast Cancer: Report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J. Natl. Cancer Inst. 1998, 90, 1371–1388. 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- Jordan V. C. Tamoxifen as the First Targeted Long-Term Adjuvant Therapy for Breast Cancer. Endocr. Relat. Cancer 2014, 21, R235–R246. 10.1530/erc-14-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottamal M.; Kang B.; Peng X.; Wang G. From Pure Antagonists to Pure Degraders of the Estrogen Receptor: Evolving Strategies for the Same Target. ACS Omega 2021, 6, 9334–9343. 10.1021/acsomega.0c06362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzdar A. U.; Come S. E.; Brodie A.; Ellis M.; Goss P. E.; Ingle J. N.; Johnston S. R.; Lee A. V.; Osborne C. K.; Vogel V. G.; Hart C. S. Proceedings of the First International Conference on Recent Advances and Future Directions in Endocrine Therapy for Breast Cancer: Summary Consensus Statement. Clin. Cancer Res. 2001, 7, 4335s–4337s. [PubMed] [Google Scholar]; ; discussion 4411s-4412s
- Serrano D.; Lazzeroni M.; Gandini S.; Macis D.; Johansson H.; Gjerde J.; Lien E.; Feroce I.; Pruneri G.; Sandri M.; Bassi F.; Brenelli F.; Luini A.; Cazzaniga M.; Varricchio C.; Guerrieri-Gonzaga A.; DeCensi A.; Bonanni B. A Randomized Phase II Presurgical Trial of Weekly Low-Dose Tamoxifen versus Raloxifene versus Placebo in Premenopausal Women with Estrogen Receptor-Positive Breast Cancer. Breast Cancer Res. 2013, 15, R47. 10.1186/bcr3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennari L.; Merlotti D.; Nuti R. Selective Estrogen Receptor Modulator (SERM) for the Treatment of Osteoporosis in Postmenopausal Women: Focus on Lasofoxifene. Clin. Interv. Aging 2010, 5, 19–29. 10.2147/cia.s6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B. Y.; Kim S. A.; Malla B.; Kim S. Y. The Effect of Selective Estrogen Receptor Modulators (SERMs) on the Tamoxifen Resistant Breast Cancer Cells. Toxicol. Res. 2011, 27, 85–93. 10.5487/tr.2011.27.2.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelhamid R.; Luo J.; VandeVrede L.; Kundu I.; Michalsen B.; Litosh V. A.; Schiefer I. T.; Gherezghiher T.; Yao P.; Qin Z.; Thatcher G. R. J. Benzothiophene Selective Estrogen Receptor Modulators Provide Neuroprotection by a Novel GPR30-Dependent Mechanism. ACS Chem. Neurosci. 2011, 2, 256–268. 10.1021/cn100106a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S. L. New Selective Estrogen Receptor Modulators (SERMs) in Development. Curr. Osteoporos. Rep. 2010, 8, 151–153. 10.1007/s11914-010-0025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla R.; Gupta K. B.; Upadhyay S.; Dhiman M.; Jaitak V. Design, Synthesis and Biological Evaluation of Novel Indole-Xanthendione Hybrids as Selective Estrogen Receptor Modulators. Bioorg. Med. Chem. 2018, 26, 266–277. 10.1016/j.bmc.2017.11.040. [DOI] [PubMed] [Google Scholar]
- Singla R.; Prakash K.; Bihari Gupta K.; Upadhyay S.; Dhiman M.; Jaitak V. Identification of Novel Indole Based Heterocycles as Selective Estrogen Receptor Modulator. Bioorg. Chem. 2018, 79, 72–88. 10.1016/j.bioorg.2018.04.002. [DOI] [PubMed] [Google Scholar]
- Singla R.; Jaitak V. Multitargeted Molecular Docking Study of Natural-Derived Alkaloids on Breast Cancer Pathway Components. Curr. Comput. Aided Drug Des. 2017, 13, 294–302. 10.2174/1573409913666170406144642. [DOI] [PubMed] [Google Scholar]
- Rossini M.; Lello S.; Sblendorio I.; Viapiana O.; Fracassi E.; Adami S.; Gatti D. Profile of Bazedoxifene/Conjugated Estrogens for the Treatment of Estrogen Deficiency Symptoms and Osteoporosis in Women at Risk of Fracture. Drug Des. Dev. Ther. 2013, 7, 601–610. 10.2147/dddt.s47807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickar J. H.; Komm B. S. Selective Estrogen Receptor Modulators and the Combination Therapy Conjugated Estrogens/Bazedoxifene: A Review of Effects on the Breast. Post Reprod. Heal. 2015, 21, 112–121. 10.1177/2053369115599090. [DOI] [PubMed] [Google Scholar]
- Fabian C. J.; Nye L.; Powers K. R.; Nydegger J. L.; Kreutzjans A. L.; Phillips T. A.; Metheny T.; Winblad O.; Zalles C. M.; Hagan C. R.; Goodman M. L.; Gajewski B. J.; Koestler D. C.; Chalise P.; Kimler B. F. Effect of Bazedoxifene and Conjugated Estrogen (Duavee) on Breast Cancer Risk Biomarkers in High-Risk Women: A Pilot Study. Cancer Prev. Res. 2019, 12, 711–720. 10.1158/1940-6207.capr-19-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni J.; Gavrilidis E.; Worsley R.; Van Rheenen T.; Hayes E. The Role of Estrogen in the Treatment of Men with Schizophrenia. Int. J. Endocrinol. Metabol. 2013, 11, 129. 10.5812/ijem.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis-Wambi J. S.; Kim H.; Curpan R.; Grigg R.; Sarker M. A.; Jordan V. C. The Selective Estrogen Receptor Modulator Bazedoxifene Inhibits Hormone-Independent Breast Cancer Cell Growth and Down-Regulates Estrogen Receptor α and Cyclin D1. Mol. Pharmacol. 2011, 80, 610–620. 10.1124/mol.111.072249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning S. W.; Jeselsohn R.; Dharmarajan V.; Mayne C. G.; Karimi M.; Buchwalter G.; Houtman R.; Toy W.; Fowler C. E.; Han R.; Lainé M.; Carlson K. E.; Martin T. A.; Nowak J.; Nwachukwu J. C.; Hosfield D. J.; Chandarlapaty S.; Tajkhorshid E.; Nettles K. W.; Griffin P. R.; Shen Y.; Katzenellenbogen J. A.; Brown M.; Greene G. L. The SERM/SERD Bazedoxifene Disrupts ESR1 Helix 12 to Overcome Acquired Hormone Resistance in Breast Cancer Cells. Elife 2018, 7, e37161 10.7554/eLife.37161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J.; Chen X.; Fu S.; Zhang R.; Pan L.; Cao Y.; Wu X.; Xiao H.; Lin H.-J.; Lo H.-W.; Zhang Y.; Lin J. Bazedoxifene Is a Novel IL-6/GP130 Inhibitor for Treating Triple-Negative Breast Cancer. Breast Cancer Res. Treat. 2019, 175, 553–566. 10.1007/s10549-019-05183-2. [DOI] [PubMed] [Google Scholar]
- Yadav A.; Kumar B.; Teknos T. N.; Kumar P. Bazedoxifene Enhances the Anti-Tumor Effects of Cisplatin and Radiation Treatment by Blocking IL-6 Signaling in Head and Neck Cancer. Oncotarget 2016, 8, 66912. 10.18632/oncotarget.11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thilakasiri P.; Huynh J.; Poh A. R.; Tan C. W.; Nero T. L.; Tran K.; Parslow A. C.; Afshar-Sterle S.; Baloyan D.; Hannan N. J.; Buchert M.; Scott A. M.; Griffin M. D.; Hollande F.; Parker M. W.; Putoczki T. L.; Ernst M.; Chand A. L. Repurposing the Selective Estrogen Receptor Modulator Bazedoxifene to Suppress Gastrointestinal Cancer Growth. EMBO Mol. Med. 2019, 11, e9539 10.15252/emmm.201809539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karadayi F. Z.; Yaman M.; Kisla M. M.; Keskus A. G.; Konu O.; Ates-Alagoz Z. Design, Synthesis and Anticancer/Antiestrogenic Activities of Novel Indole-Benzimidazoles. Bioorg. Chem. 2020, 100, 103929. 10.1016/j.bioorg.2020.103929. [DOI] [PubMed] [Google Scholar]
- Makar S.; Saha T.; Swetha R.; Gutti G.; Kumar A.; Singh S. K. Rational Approaches of Drug Design for the Development of Selective Estrogen Receptor Modulators (SERMs), Implicated in Breast Cancer. Bioorg. Chem. 2020, 94, 103380. 10.1016/j.bioorg.2019.103380. [DOI] [PubMed] [Google Scholar]
- Pandit B.; Sun Y.; Chen P.; Sackett D. L.; Hu Z.; Rich W.; Li C.; Lewis A.; Schaefer K.; Li P.-K. Structure-Activity-Relationship Studies of Conformationally Restricted Analogs of Combretastatin A-4 Derived from SU5416. Bioorg. Med. Chem. 2006, 14, 6492–6501. 10.1016/j.bmc.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Chow L. Q. M.; Eckhardt S. G. Sunitinib: From Rational Design to Clinical Efficacy. J. Clin. Oncol. 2007, 25, 884–896. 10.1200/jco.2006.06.3602. [DOI] [PubMed] [Google Scholar]
- Dadashpour S.; Emami S. Indole in the Target-Based Design of Anticancer Agents: A Versatile Scaffold with Diverse Mechanisms. Eur. J. Med. Chem. 2018, 150, 9–29. 10.1016/j.ejmech.2018.02.065. [DOI] [PubMed] [Google Scholar]
- Dhuguru J.; Skouta R. Role of Indole Scaffolds as Pharmacophores in the Development of Anti-Lung Cancer Agents. Molecules 2020, 25, 1615. 10.3390/molecules25071615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y.; Wen X.; Gong Y.; Wang X. Current Scenario of Indole Derivatives with Potential Anti-Drug-Resistant Cancer Activity. Eur. J. Med. Chem. 2020, 200, 112359. 10.1016/j.ejmech.2020.112359. [DOI] [PubMed] [Google Scholar]
- Liu J.; Ming B.; Gong G.-H.; Wang D.; Bao G.-L.; Yu L.-J. Current Research on Anti-Breast Cancer Synthetic Compounds. RSC Adv. 2018, 8, 4386–4416. 10.1039/c7ra12912b. [DOI] [Google Scholar]
- Kaur K.; Jaitak V. Recent Development in Indole Derivatives as Anticancer Agents for Breast Cancer. Anticancer Agents Med. Chem. 2019, 19, 962–983. 10.2174/1871520619666190312125602. [DOI] [PubMed] [Google Scholar]
- Karthikeyan C.; Solomon V. R.; Lee H.; Trivedi P. Design, Synthesis and Biological Evaluation of Some Isatin-Linked Chalcones as Novel Anti-Breast Cancer Agents: A Molecular Hybridization Approach. Biomed. Prev. Nutr. 2013, 3, 325–330. 10.1016/j.bionut.2013.04.001. [DOI] [Google Scholar]
- Bender O.; Shoman M. E.; Ali T. F. S.; Dogan R.; Celik I.; Mollica A.; Hamed M. I. A.; Aly O. M.; Alamri A.; Alanazi J.; Ahemad N.; Gan S. H.; Malik J. A.; Anwar S.; Atalay A.; Beshr E. A. M. Discovery of oxindole-based FLT3 inhibitors as a promising therapeutic lead for acute myeloid leukemia carrying the oncogenic ITD mutation. Arch. Pharm. 2022, e2200407 10.1002/ardp.202200407. [DOI] [PubMed] [Google Scholar]
- Lozinskaya N. A.; Babkov D. A.; Zaryanova E. V.; Bezsonova E. N.; Efremov A. M.; Tsymlyakov M. D.; Anikina L. V.; Zakharyascheva O. Y.; Borisov A. V.; Perfilova V. N.; Tyurenkov I. N.; Proskurnina M. V.; Spasov A. A. Synthesis and biological evaluation of 3-substituted 2-oxindole derivatives as new glycogen synthase kinase 3β inhibitors. Bioorg. Med. Chem. 2019, 27, 1804–1817. 10.1016/j.bmc.2019.03.028. [DOI] [PubMed] [Google Scholar]
- Fareed M. R.; Shoman M. E.; Hamed M. I. A.; Badr M.; Bogari H. A.; Elhady S. S.; Ibrahim T. S.; Abuo-Rahma G. E. D. A.; Ali T. F. S. New Multi-Targeted Antiproliferative Agents: Design and Synthesis of IC261-Based Oxindoles as Potential Tubulin, CK1 and EGFR Inhibitors. Pharmaceuticals 2021, 14, 1114. 10.3390/ph14111114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.; Xu J.; Zhao X.; Kang C. Synthesis of Novel 3-(Benzothiazol-2-Ylmethylene)Indolin-2-Ones. J. Chem. Res. 2017, 41, 537–540. 10.3184/174751917x15040891974776. [DOI] [Google Scholar]
- Amombo G. M. O.; Kramer T.; Lo Monte F.; Göring S.; Fach M.; Smith S.; Kolb S.; Schubenel R.; Baumann K.; Schmidt B. Modification of a promiscuous inhibitor shifts the inhibition from γ-secretase to FLT-3. Bioorg. Med. Chem. Lett. 2012, 22, 7634–7640. 10.1016/j.bmcl.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Andreani A.; Granaiola M.; Locatelli A.; Morigi R.; Rambaldi M.; Varoli L.; Vieceli Dalla Sega F.; Prata C.; Nguyen T. L.; Bai R.; Hamel E. Cytotoxic Activities of Substituted 3-(3,4,5-Trimethoxybenzylidene)-1,3-Dihydroindol-2-Ones and Studies on Their Mechanisms of Action. Eur. J. Med. Chem. 2013, 64, 603–612. 10.1016/j.ejmech.2013.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik I.; Ayhan-Kılcıgil G.; Karayel A.; Guven B.; Onay-Besikci A. Synthesis, Molecular Docking, in Silico ADME, and EGFR Kinase Inhibitor Activity Studies of Some New Benzimidazole Derivatives Bearing Thiosemicarbazide, Triazole, and Thiadiazole. J. Heterocycl. Chem. 2022, 59, 371–387. 10.1002/jhet.4431. [DOI] [Google Scholar]
- Doganc F.; Celik I.; Eren G.; Kaiser M.; Brun R.; Goker H. Synthesis, in Vitro Antiprotozoal Activity, Molecular Docking and Molecular Dynamics Studies of Some New Monocationic Guanidinobenzimidazoles. Eur. J. Med. Chem. 2021, 221, 113545. 10.1016/j.ejmech.2021.113545. [DOI] [PubMed] [Google Scholar]
- Daina A.; Michielin O.; Zoete V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibuh B. Z.; Khanna S.; Taneja P.; Sarkar P.; Taneja N. K. Molecular Docking, Synthesis and Anticancer Activity of Thiosemicarbazone Derivatives against MCF-7 Human Breast Cancer Cell Line. Life Sci. 2021, 273, 119305. 10.1016/j.lfs.2021.119305. [DOI] [PubMed] [Google Scholar]
- Canário C.; Matias M.; Brito V.; Santos A. O.; Falcão A.; Silvestre S.; Alves G. New Estrone Oxime Derivatives: Synthesis, Cytotoxic Evaluation and Docking Studies. Molecules 2021, 26, 2687. 10.3390/molecules26092687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küçükoğlu K.; Acar Çevik U.; Nadaroglu H.; Celik I.; Işık A.; Bostancı H. E.; Özkay Y.; Kaplancıklı Z. A. Design, Synthesis and Molecular Docking Studies of Novel Benzimidazole-1, 3, 4-Oxadiazole Hybrids for Their Carbonic Anhydrase Inhibitory and Antioxidant Effects. Med. Chem. Res. 2022, 31, 1771–1782. 10.1007/s00044-022-02943-6. [DOI] [Google Scholar]
- Shylaja R.; Loganathan C.; Kabilan S.; Vijayakumar T.; Meganathan C. Synthesis and evaluation of the antagonistic activity of 3-acetyl-2H-benzo[g]chromen-2-one against mutant Y537S estrogen receptor alpha via E-Pharmacophore modeling, molecular docking, molecular dynamics, and in-vitro cytotoxicity studies. J. Mol. Struct. 2021, 1224, 129289. 10.1016/j.molstruc.2020.129289. [DOI] [Google Scholar]
- Celik I.; Tallei T. E. A computational comparative analysis of the binding mechanism of molnupiravir’s active metabolite to RNA-dependent RNA polymerase of wild-type and Delta subvariant AY.4 of SARS-CoV-2. J. Cell. Biochem. 2022, 123, 807–818. 10.1002/jcb.30226. [DOI] [PubMed] [Google Scholar]
- Marconett C. N.; Sundar S. N.; Poindexter K. M.; Stueve T. R.; Bjeldanes L. F.; Firestone G. L. Indole-3-Carbinol Triggers Aryl Hydrocarbon Receptor-dependent Estrogen Receptor (ER)α Protein Degradation in Breast Cancer Cells Disrupting an ERα-GATA3 Transcriptional Cross-Regulatory Loop. Mol. Biol. Cell 2010, 21, 1166–1177. 10.1091/mbc.e09-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Yang J.; Niu L.; Hu D.; Li H.; Chen L.; Yu Y.; Chen Q. Structural Insights into the Design of Indole Derivatives as Tubulin Polymerization Inhibitors. FEBS Lett. 2020, 594, 199–204. 10.1002/1873-3468.13566. [DOI] [PubMed] [Google Scholar]
- Patil R.; Patil S. A.; Beaman K. D.; Patil S. A. Indole molecules as inhibitors of tubulin polymerization: potential new anticancer agents, an update (2013-2015). Future Med. Chem. 2016, 8, 1291–1316. 10.4155/fmc-2016-0047. [DOI] [PubMed] [Google Scholar]
- Kaur R.; Kaur G.; Gill R. K.; Soni R.; Bariwal J. Recent Developments in Tubulin Polymerization Inhibitors: An Overview. Eur. J. Med. Chem. 2014, 87, 89–124. 10.1016/j.ejmech.2014.09.051. [DOI] [PubMed] [Google Scholar]
- Hendy M. S.; Ali A. A.; Ahmed L.; Hossam R.; Mostafa A.; Elmazar M. M.; Naguib B. H.; Attia Y. M.; Ahmed M. S. Structure-based drug design, synthesis, In vitro, and In vivo biological evaluation of indole-based biomimetic analogs targeting estrogen receptor-α inhibition. Eur. J. Med. Chem. 2019, 166, 281–290. 10.1016/j.ejmech.2019.01.068. [DOI] [PubMed] [Google Scholar]
- Bender O.; Atalay A. Evaluation of Anti-Proliferative and Cytotoxic Effects of Chlorogenic Acid on Breast Cancer Cell Lines by Real-Time, Label-Free and High-Throughput Screening. Marmara Pharm. J. 2018, 22, 173. 10.12991/mpj.2018.54. [DOI] [Google Scholar]
- Bender O.; Gunduz M.; Cigdem S.; Hatipoglu O. F.; Acar M.; Kaya M.; Grenman R.; Gunduz E.; Ugur K. S. Functional analysis of ESM1 by siRNA knockdown in primary and metastatic head and neck cancer cells. J. Oral Pathol. Med. 2018, 47, 40–47. 10.1111/jop.12648. [DOI] [PubMed] [Google Scholar]
- Bird C.; Kirstein S. Real-Time, Label-Free Monitoring of Cellular Invasion and Migration with the XCELLigence System. Nat. Methods 2009, 6, v–vi. 10.1038/nmeth.f.263. [DOI] [Google Scholar]
- Lazarova I.; Zengin G.; Bender O.; Zheleva-Dimitrova D.; Uysal S.; Ceylan R.; Gevrenova R.; Aktumsek A.; Acar M.; Gunduz M. A comparative study of Bulgarian and Turkish Asphodeline lutea root extracts: HPLC-UV profiles, enzyme inhibitory potentials and anti-proliferative activities against MCF-7 and MCF-10A cell lines. J. Funct.Foods 2015, 15, 254–263. 10.1016/j.jff.2015.03.032. [DOI] [Google Scholar]
- Jain N.; Xu J.; Kanojia R. M.; Du F.; Jian-Zhong G.; Pacia E.; Lai M.-T.; Musto A.; Allan G.; Reuman M.; Li X.; Hahn D.; Cousineau M.; Peng S.; Ritchie D.; Russell R.; Lundeen S.; Sui Z. Identification and Structure–Activity Relationships of Chromene-Derived Selective Estrogen Receptor Modulators for Treatment of Postmenopausal Symptoms. J. Med. Chem. 2009, 52, 7544–7569. 10.1021/jm900146e. [DOI] [PubMed] [Google Scholar]
- Merchenthaler I. The Effect of Estrogens and Antiestrogens in Rat Models of Hot Flush. Drug Dev. Res. 2005, 66, 182–188. 10.1002/ddr.20057. [DOI] [Google Scholar]
- Liu Y.; Ma H.; Yao J. ERα, A Key Target for Cancer Therapy: A Review. OncoTargets Ther. 2020, 13, 2183–2191. 10.2147/ott.s236532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender O.; Llorent-Martínez E. J.; Zengin G.; Mollica A.; Ceylan R.; Molina-García L.; Luisa Fernández-de Córdova M.; Atalay A. Integration of in Vitro and in Silico Perspectives to Explain Chemical Characterization, Biological Potential and Anticancer Effects of Hypericum Salsugineum: A Pharmacologically Active Source for Functional Drug Formulations. PLoS One 2018, 13, e0197815 10.1371/journal.pone.0197815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahomoodally M. F.; Atalay A.; Nancy Picot M. C.; Bender O.; Celebi E.; Mollica A.; Zengin G. Chemical, Biological and Molecular Modelling Analyses to Probe into the Pharmacological Potential of Antidesma Madagascariense Lam.: A Multifunctional Agent for Developing Novel Therapeutic Formulations. J. Pharm. Biomed. Anal. 2018, 161, 425–435. 10.1016/j.jpba.2018.09.002. [DOI] [PubMed] [Google Scholar]
- Judson R. S.; Houck K.; Watt E.; Thomas R. On selecting a minimal set of in vitro assays to reliably determine estrogen agonist activity. Regul. Toxicol. Pharmacol. 2017, 91, 39. 10.1016/j.yrtph.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A.; Pakrasi P. Estrogenic and Antiestrogenic Properties of Clomiphene Citrate in Laboratory Mice. J. Biosci. 1995, 20, 665–673. 10.1007/bf02703306. [DOI] [Google Scholar]
- Ragab M. A.; Elagawany M.; Daabees H.; Ahmed A.-S. F.; Awad E. M.; Billon C.; Elgendy B.; Abouzid K. A. M.; Kassab S. E. Structure-Based Design and Synthesis of Conformationally Constrained Derivatives of Methyl-Piperidinopyrazole (MPP) with Estrogen Receptor (ER) Antagonist Activity. Bioorg. Chem. 2022, 119, 105554. 10.1016/j.bioorg.2021.105554. [DOI] [PubMed] [Google Scholar]
- Schweikart K. M.; Eldridge S. R.; Safgren S. L.; Parman T.; Reid J. M.; Ames M. M.; Goetz M. P.; Davis M. A. Comparative Uterotrophic Effects of Endoxifen and Tamoxifen in Ovariectomized Sprague-Dawley Rats. Toxicol. Pathol. 2014, 42, 1188–1196. 10.1177/0192623314525688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahadi H.; Emami S. Modification of 7-piperazinylquinolone antibacterials to promising anticancer lead compounds: Synthesis and in vitro studies. Eur. J. Med. Chem. 2020, 187, 111970. 10.1016/j.ejmech.2019.111970. [DOI] [PubMed] [Google Scholar]
- Abdel-Aa M. A. A.; Shaykoon S. A.; Mohamed M. S. A.; Abuo-Rahma M. F. A.; Abuo-Rahma G. E.-D. A. A. Antibacterial and Urease Inhibitory Activity of New Piperazinyl N-4 Functionalized Ciprofloxacin-Oxadiazoles. J. Mod. Res. 2019, 1, 1–7. 10.21608/jmr.2019.12650.1001. [DOI] [Google Scholar]
- Daina A.; Michielin O.; Zoete V. ILOGP: A Simple, Robust, and Efficient Description of n-Octanol/Water Partition Coefficient for Drug Design Using the GB/SA Approach. J. Chem. Inf. Model. 2014, 54, 3284–3301. 10.1021/ci500467k. [DOI] [PubMed] [Google Scholar]
- Daina A.; Zoete V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celi̇k I.; Onay-Besi̇kci̇ A.; Ayhan-Kilcigi̇l G. Approach to the Mechanism of Action of Hydroxychloroquine on SARS-CoV-2: A Molecular Docking Study. J. Biomol. Struct. Dyn. 2021, 39, 5792–5798. 10.1080/07391102.2020.1792993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximov P. Y.; Abderrahman B.; Fanning S. W.; Sengupta S.; Fan P.; Curpan R. F.; Rincon D. M. Q.; Greenland J. A.; Rajan S. S.; Greene G. L.; Jordan V. C. Endoxifen, 4-Hydroxytamoxifen and an Estrogenic Derivative Modulate Estrogen Receptor Complex Mediated Apoptosis in Breast Cancer. Mol. Pharmacol. 2018, 94, 812–822. 10.1124/mol.117.111385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C.; Wu C.; Ghoreishi D.; Chen W.; Wang L.; Damm W.; Ross G. A.; Dahlgren M. K.; Russell E.; Von Bargen C. D.; Abel R.; Friesner R. A.; Harder E. D. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. 10.1021/acs.jctc.1c00302. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Murphy R. B.; Repasky M. P.; Frye L. L.; Greenwood J. R.; Halgren T. A.; Sanschagrin P. C.; Mainz D. T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein–Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Banks J. L.; Murphy R. B.; Halgren T. A.; Klicic J. J.; Mainz D. T.; Repasky M. P.; Knoll E. H.; Shelley M.; Perry J. K.; Shaw D. E.; Francis P.; Shenkin P. S. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Celik I.; Sarıaltın S. Y.; Çoban T.; Kılcıgil G. Design, Synthesis, in Vitro and in Silico Studies of Benzimidazole-Linked Oxadiazole Derivatives as Anti-inflammatory Agents. ChemistrySelect 2022, 7, e202201548 10.1002/slct.202201548. [DOI] [Google Scholar]
- Abraham M. J.; Murtola T.; Schulz R.; Páll S. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1, 19. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Mellado M.; González C.; Mella J.; Aguilar L. F.; Celik I.; Borges F.; Uriarte E.; Delogu G.; Viña D.; Matos M. J. Coumarin-Resveratrol-Inspired Hybrids as Monoamine Oxidase B Inhibitors: 3-Phenylcoumarin versus Trans-6-Styrylcoumarin. Molecules 2022, 27, 928. 10.3390/molecules27030928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S.; Kim T.; Iyer V. G.; Im W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- Tian C.; Kasavajhala K.; Belfon K. A. A.; Raguette L.; Huang H.; Migues A. N.; Bickel J.; Wang Y.; Pincay J.; Wu Q.; Simmerling C. Ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theor. Comput. 2019, 16, 528–552. 10.1021/acs.jctc.9b00591. [DOI] [PubMed] [Google Scholar]
- Lee J.; Hitzenberger M.; Rieger M.; Kern N. R.; Zacharias M.; Im W. CHARMM-GUI Supports the Amber Force Fields. J. Chem. Phys. 2020, 153, 35103. 10.1063/5.0012280. [DOI] [PubMed] [Google Scholar]
- Valdés-Tresanco M. S.; Valdés-Tresanco M. E.; Valiente P. A.; Moreno E. Gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. 10.1021/acs.jctc.1c00645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.