Abstract
This research investigated the mechanism of ozone treatment on sweet cherry (Prunus avium L.) by Lable-free quantification proteomics and physiological traits. The results showed that 4557 master proteins were identified in all the samples, and 3149 proteins were common to all groups. Mfuzz analyses revealed 3149 candidate proteins. KEGG annotation and enrichment analysis showed proteins related to carbohydrate and energy metabolism, protein, amino acids, and nucleotide sugar biosynthesis and degradation, and fruit parameters were characterized and quantified. The conclusions were supported by the fact that the qRT-PCR results agreed with the proteomics results. For the first time, this study reveals the mechanism of cherry in response to ozone treatment at a proteome level.
Keywords: sweet cherry (Prunus avium L.), ozone, label-free quantification proteomics, preservation, mfuzz
Introduction
Sweet cherry (Prunus avium L.) is an economically important horticultural crop cultivated in north China, which is thought to be a non-climacteric fruit. Sweet cherries are a significant and valuable fruit that are a fantastic source of many phytochemicals with health-promoting qualities (anthocyanins, vitamins, phenolics etc.), and are widely adored by consumers (Zhang et al., 2007; Calle et al., 2021; Clayton-Cuch et al., 2021; Gutierrez et al., 2021). However, several physiological or environmental conditions will have a significant impact on cherry fruits during storage, which will reduce their post-harvest quality and market value. Postharvest loss is a serious issue that affects the entire world, each year, more than one-third of fruits and vegetables are lost in the field and the subsequent supply chain (Romanazzi et al., 2016).
Because it safely and spontaneously breaks down into oxygen and free radicals quickly without leaving any lasting effects, ozone has gained acceptance as a food sanitizer, particularly in organic farming (Monaco et al., 2016). In 2001, ozone was approved by the FDA for the treatment of fresh fruit (FDA, 2001). A thorough study is currently being conducted on the ozonation method, which increases the shelf life of stored raw fruits and vegetables. For instance, numerous attempts have been made to use ozone therapy to lengthen the shelf life of carrot, lettuce, peach and berries (Tabakoglu and Karaca, 2018). Many papers have proved that gaseous ozone is also use to reduce formation of volatile chemicals, such as propene or acetylene, which induce unfavorable early fruit ripening. Initial research has mainly concentrated on the study of ozone to destroy hazardous microbes. Studies have shown that changes in the content of antioxidant components, such as flavonoids and other phenolic compounds, are positively impacted by gaseous ozone. Ozone can, however, damage nutrients, change the appearance of food, and impair food flavor when used improperly (Sachadyn-Kró et al., 2016). However, it is unknown how this potent oxidant may affect the food’s quality and safety for consumption.
The development of omics techniques has made it feasible to obtain understanding of the intricate processes that take place during cherry storage at the protein level. Proteome profiling techniques that reveal changes in protein content during fruits storage are currently uncommon and tend to concentrate mature or germination-stage seeds. In the current study, the dynamic change of proteins in cherry-treated ozone during cold stress stage was investigated using a label-free proteomics and bioinformatics analysis. This study, as far as we are aware, was the first to look at the molecular mechanism of ozone in cherry preservation. The findings provide vital information for cherry preservation as well as information on cherry molecular responses to ozone.
Materials and methods
Sample collection and treatment
Fresh “red lantern” sweet cherries that were undamaged, and commercially mature were acquired on June 1, 2020, in Ninghe District of Tianjin, China, and delivered to the National Engineering Technology Research Center for Agricultural Products Preservation (Tianjin). The samples were separated into two groups (ozone treatment group, TP and control check group, CK) and stored for 35 days at a temperature of 0 ± 0.5°C with a relative humidity of 90 ± 5%. One ozone treatment group (TP) was housed in 18 boxes with 1 kilogram of cherries each, each inside a huge tent. The cherry ozone treatment approach is based on the work of Gao et al. (2020). The ozone sensor (MIC-03, Shenzhen) is fixed in the huge tent with two fans of 0.4 m·s-1 generating a breeze of 0.2 m·s-1. Every 7 days, the TP sample was fumigated with 3 ppm ozone for two hours. Only the other control check group (CK) was boxed. At 0, 7, 14, 21, 28 and 35 days, physiological indicators and samples were collected for storage ( Figure 1 ). After being instantly frozen in liquid nitrogen, the cherry was kept at −80°C until protein and RNA extraction.
Figure 1.
The visual appearance of different treatment cherry fruit stored for 28 days at 0°C.
Physical and physiological analysis
Soluble solids, titratabole acidity, and firmness
Cherry samples were crushed in a mortar and passed through gauze to determine of the soluble solid content. Utilizing a PLA-1 digital hand-held pocket refractometer, the produced juice was examined (Atago Co., Ltd., Tokyo, Japan). Three times were used to repeat each measurement. The results are given as percentages (%) (Yang SB et al., 2021). Titratable acidity content was determined by titration. Three times were used to repeat each measurement. The results are shown as percentages (%) (Yang SB et al., 2021). Randomly select 10 sweet cherries, and the firmness of the sample were measured with TA.XT.Plus physical property analyzer along the two ends of the sweet cherry equator. The measurement parameters are: probe P/2 (Φ2 mm), puncture preparation speed is 2.00 mm/s, puncture speed is 2.00 mm/s, and puncture depth is 5mm. The result is indicated by “N” (Hu et al., 2020).
Measurement of total flavonoids, anthocycadins, total phenol, and ascorbic acid (Vc) content
The method of Zhao et al. (2021) was slightly modified to determine the total flavonoids, anthocycadins, total phenol, and ascorbic acid (Vc) content. The total phenol results were expressed as gram gallic acid equivalents per kilogram fresh weight (mg GA kg-1). Ascorbic acid results were expressed as milligram of ascorbic acid for kilograms (mg 100g-1 FW).
Measurement of key antioxidant enzyme activity
Activities of CAT, APX, POD, and SOD were determined as described by Porcher et al. (2020) and Mekontso et al. (2021), with slight alterations. All enzyme activity units were reported as U g−1.
Protein extraction, digestion and analysis
Protein was extracted from sweet cherry using the method that we previously established in studies that have been published (Huang and Hou, 2021). The powdered cherry samples were treated with trichloroacetic acid (TCA)-acetone extraction, and the crude proteins were lysated by urea extraction buffer (8 M urea, 0.1 M Tris-HCl, pH 8.5) and sonicated for 5 min (pulse 2 s on, 3 s off, 50% amplitude) in ice-water bath (BILON-650Y Ultrasonic cell pulverizer). Then, the protein is enzymatically hydrolyzed by FASP method, briefly, total protein (100μg) was loaded into the ultrafiltration centrifuge tubes (10 kD Microcon, Millipore), which then were centrifuged at 14,000×g for 15 min. After centrifugation, concentrates were reduced using 100 μL of 100 mM DTT-50mM NH4HCO3 (50°C, 30 min). Then, 100μL of 100 mM iodoacetamide (IAA) in urea extraction buffer was added, and incubated for 40 min in darkness. The filters were washed with 100μL of urea extraction buffer three times, and followed by 100μL of 25 mM NH4HCO3 buffer. The concentrate was then digested at 37°C for 18 hours with 200μL of trypsin (Promega), and the resulting peptides were collected as the filtrate.
The peptides were examined using an online Ultimate 3000 nanoflow liquid chromatography tandem Q-Extractive HF mass spectrometer (Thermo Scientific, Waltham, MA, USA). Briefly, the peptide mixture was loaded onto a reversed-phase trap column (Thermo Scientific Acclaim PepMap 100, 100μm×2cm, nanoViper C18) connected to a C18 reversed-phase analytical column (Thermo Scientific Acclaim PepMap 100, 15 cm long, 75 μm inner diameter, 3μm resin). MS data were obtained using a top 20 data-dependent acquirement (DDA) dynamically choosing the most abundant precursor ions from the survey scan (m/z150~2000) for higher-energy collisional dissociation (HCD) fragmentation. The following settings were made to the instrument: automatic gain control target was 3e6; dynamic exclusion duration, 30 s; resolution for survey scans, 120,000 at m/z 200; resolution for HCD spectra, 30,000 at m/z 200, and isolation width was m/z 2. The normalized collision energy was 27, and the underfill ratio was 0.1%. Each sample was analyzed in three technical triplicates (Huang and Hou, 2021).
Protein identification
Proteome Discoverer software (ver. 2.2, Thermo Scientific, Bremen, Germany) was utilized to evaluate the raw data, searches against UniProtKB/Swiss-Prot cherry database (release 2020_12, 35660 total entries, downloaded 12/26/20) were performed. Search parameters were: precursor error tolerance 10 ppm, fragment ion tolerance 0.5 Da, trypsin full specificity, maximum number of missed cleavages 2. Methionine oxidation and protein N-term acetylation were set as variable modifications and cysteine carbamidomethylation was designated as fixed modification. False discovery rate (FDR) <0.01. Razor and unique peptides were used for protein quantification (Huang and Hou, 2021).
Database searching and protein quantification
To assess the quality of the proteomic data, the coefficient of variation (CV) distribution for each sample was less than 10%. Unsupervised principal component analysis (PCA) was accomplished using R package “prcomp” within R language (www.r-project.org) and the data were scaled to zero-mean and unit variance before PCA (Li QQ et al., 2022; Li Y et al., 2022). Differential expression between TP/CK groups was analyzed with the limma R package (1.10.1). The resulting P values were adjusted according to the Benjamini and Hochberg approach for controlling the false discovery rate. Differentially expressed proteins were those determined by R package limma with an adjusted P-value < 0.05, |log2 (fold change)| > 1, false discovery rate < 0.01. (Yu et al., 2019).
Soft clustering analysis
Soft clustering can assign a gene to many clusters using the fuzzy c-means algorithm with time-course data on the gene expression (Kumar and Futschik, 2007). To analyze the expression trend of proteins with time, R software packages “Mfuzz” was used to soft clustering, which generates the membership values of proteins in a cluster to reflect the degree of a gene’s association with a cluster (Yuan and Cai, 2017). The similarity of protein expression vectors to each other can be determined by membership value (Yang CH et al., 2021).
Bioinformatics analysis
The KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis is useful to map unigenes onto known signaling pathways (Yu et al., 2019). The “clusterProfiler” package in R was used to conduct the functional analysis for DEPs in CK and TP groups, respectively. A hypergeometric distribution test was used to obtain the P-value of enriched pathways, Subsequently, the P-value was revised using BH (Benjamini and Hochberg) approach, and the adjusted P-value < 0.05 was served as the cut-off criterion (Yang CH et al., 2021).
Quantitative real-time PCR analysis
Eight randomly selected proteins were used to analyze the transcription level using qRT-PCR with GAPDH (GenBank No. LOC107418185) as the reference gene (Yang CH et al., 2021). Total RNA extraction, primer design, and PCR detection were described in the literature (Yang CH et al., 2021; Dahn et al., 2021). The information of the primers was shown in Table 1 .
Table 1.
Sequence of forward and reverse qRT-PCR primers by gene (feature ID).
| Feature ID | Forward (5’-3’) | Reverse (5’-3’) |
|---|---|---|
| LOC110759612 | AGTCGCTCAATCTCTCATCG | TACCCATTAAGCCTTGACAT |
| LOC110750545 | GAAACACCTCGTAAACCCTA | TTTGAGGGGAATGAATCTGG |
| LOC110753532 | TTTTCTTGAGACAGAGAGCC | TATCACTGCCATCCAGAGTA |
| LOC110770218 | GACACATTAGCAAAAACCCT | TTGTTCATGGAGCAACTACA |
| LOC110762082 | TCTTCTATATCGCGAAACCC | AGAATTCATTCAATGCCCCT |
| LOC110756609 | AAACAACGTTTGAAGGTTCC | ACAGTAGAAAGAAATGCAAAGG |
| LOC110760963 | TCAACCAGGAACTGAAAAG | AAAACACATCATCAATGGCG |
| LOC110766523 | ATCGGTCTCTTCTTCTACT | TAGCCAGAGAAGAAGATGA |
Results
Physical parameters
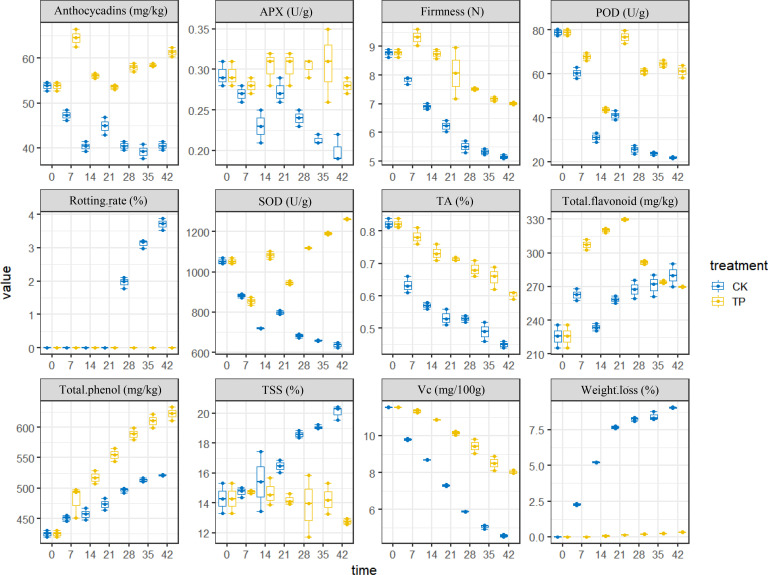
The physiological indicators of cherries were detected ( Table S1 ). For an intuitive view of the distribution of the different indicators between CK and TP groups, the boxplot was used for analysis. The box plot in Figure 2 shows the relative levels and changes of the 12 compounds (color, firmness, weight loss, pH, TSS content, and Vc content etc.) in sweet cherry. The level of weight loss, rotting rate and TSS was highest in the CK group and approximately twofold higher than the ozone treatment group. However, the other indicators of SOD, POD, firmness were higher in the ozone treatment, this result is consistent with previous literature reports (Wang et al., 2022). These results suggest a relationship between ozone treatment and traits, ozone can significantly affect the parameters of cherry.
Figure 2.
The 12 physiological characteristics of sweet cherry.
Firmness is the primary external quality factor affecting the marketability. Additional, the firmness of cherry play a crucial role in estimating how long it will preserve. Figure 2 showed that the firmness of cherry decreased with storage time in all groups, and the CK group was more significant, since it depends on the progress of the conversion of insoluble pectin to soluble pectin or even pectin acid. The firmness of cherry treated ozone with decreased by 26.48%, which is similar to the result reported by Zhang et al. (2021). As a result, the ozone can efficiently retards the softening of sweet cherry. Weight loss and rotting rate increased gradually throughout the storage time. The application of ozone treatment reduced the weight loss within the range of 0 to 0.32% compared with the control (range of 0-9.01%) from 0 to 42 days at 0°C ( Figure 2 ). Compared to the control group, the ozone treatment enhanced SOD, POD, and APX activities in sweet cherry. APX and POD activities in treated fruit were increased on 28 days at 0°C, reaching a peak value, respectively. However, SOD activity was higher in treated fruit than that in control during the periods of storage ( Figure 2 ).
Protein profiling from proteomics analysis
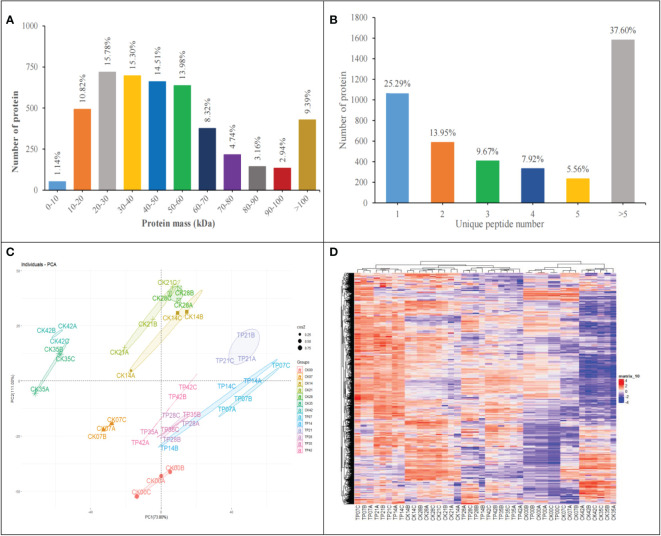
1335877 spectra were acquired using Label-free proteome analysis. After data filtering to eliminate low-scoring spectra, 624157 PSMs identified spectra were matched to 31155 peptides. Finally, total 4557 master proteins were identified with FDR of peptides was less than 0.01, 74.71% proteins were covered with 2 peptides or more ( Figure 3B ).
Figure 3.
Molecular weight distribution plot of all identified proteins (A). Distribution of proteins containing the different number of identified unique peptides (B). Principal component analysis of protein expression patterns (C). Heatmap of the 4457 identified proteins, different columns represent independent biological replicates (D).
The number of quantified proteins reached 90.69% of the total identified proteins, accounted for most molecular weight 0-10 kDa (1.14%), 10-20 kDa (10.82%), 20-30 kDa (15.78%), 30-40 kDa (15.30%), 40-50 kDa (14.51%), 50-60 kDa (13.98%), 60-70 kDa (8.32%), 70-80 kDa (4.74%), 80-90 kDa (3.16%) and 90-100 kDa (2.94%) ( Figure 3A ).
Figures 3C, D display PCA and HCA based on the 4557 master proteins. As indicated in Figure 3C , three replicates of each treatment were flocked together and each sample group was separated well at 13 sampling time points, demonstrating the reliabiliy of the sample collection and analysis; PCA revealed that the first principal component (PC1) reached 73.80% of the variance, and the second one (PC2) reached 11.00% of the variance, the contribution rate of PC1 and PC2 exceeds 80%. These results showed that the cherry fruits were affected significantly in the overall proteome dynamics, providing a suitable experimental dataset. HCA ( Figure 3D ) showed the association among samples (pearson correlation coefficient) according to the overall proteome profile, indicating that three replicates in the same stages had a high correlation ( Figure 3D ).
Expression cluster pattern of DEPs based on time course
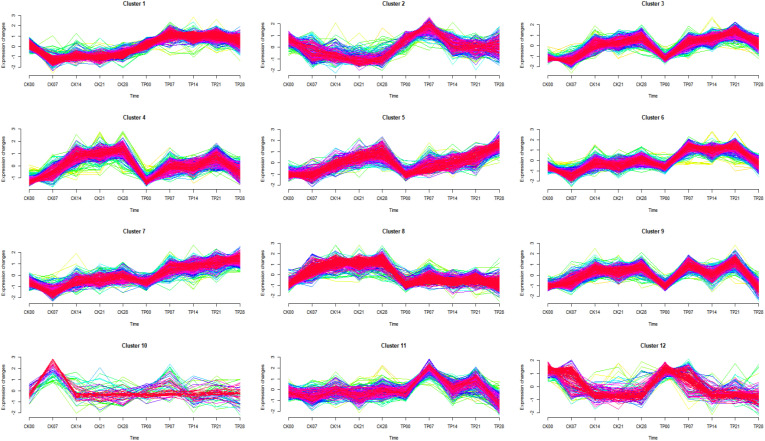
As seen in Figure S1 and Table S2 , 3149 proteins were common to the 13 samples. 3149 proteins used for Mfuzz analysis, which were selected by maximum {log2(Abundance + 1)} > 1 and the standard deviation of log2(Abundance + 1) > 1.
Soft clustering analysis (Mfuzz) was used to observe the trend of differentially expressed proteins with time series, and 12 protein clusters showing an appropriate separation were found ( Figure 4 ) (Meng et al., 2018). Mfuzz results showed the 12 proteins clusters, cluster 1 (293 proteins), cluster 2 (407 proteins), cluster 3 (154 proteins), cluster 4 (252 proteins), cluster 5 (284 proteins), cluster 6 (331 proteins), cluster 7 (183 proteins), cluster 8 (294 proteins), cluster 9 (264 proteins), cluster 10 (81 proteins), cluster 11 (455 proteins) and cluster 12 (151 proteins).
Figure 4.
Results of the Mfuzz clustering of 3149 differentially expressed proteins identified by Label-free proteomics based on their expression patterns.
Different clusters are merged into 4 groups, according to their trends ( Table 2 ). The group 1 from cluster 1, cluster 5, and cluster 7 gradually continuous increased with a time-dependent manner, and the protein content in CK was lower than that in TP at the later stage. The group 2 from cluster 2, and cluster 12 gradually continuous decrease decreased with a time-dependent manner, and the protein content in CK was lower than that in TP at the later stage. The group 3 from cluster 3, cluster 4, cluster 6, cluster 9, and cluster 11 gradually increase first then decreased with a time-dependent manner, and the protein content in CK was lower than that in TP at the later stage. The group 4 from cluster 8 and cluster 10 stable gradually with a time-dependent manner, and protein abundance in CK is higher than that in TP at the later stage. The proteins in the four groups were subjected to KEGG annotation and enrichment analysis ( Figure S2 and Table 2 ). As the biochemcial parameters of cherries gradually decreases with time during storage, 1456 proteins from group 3 with gradually down-regulation in serial passages were selected for further study.
Table 2.
subcluster of proteins of differential expression protein based on Mfuzz cluster analysis.
| Group | clusters | description | Numbers of Proteins | KEGG pathway |
|---|---|---|---|---|
| Group1 | 1,5,7 | Continuous increase | 760 |

|
| Group2 | 2,12 | Continuous decrease | 558 |

|
| Group3 | 3,4,6,9,11 | Increase first then decrease | 1456 |

|
| Group4 | 8,10 | Protein abundance in CK is higher than that in TP | 375 |

|
DEPs screening
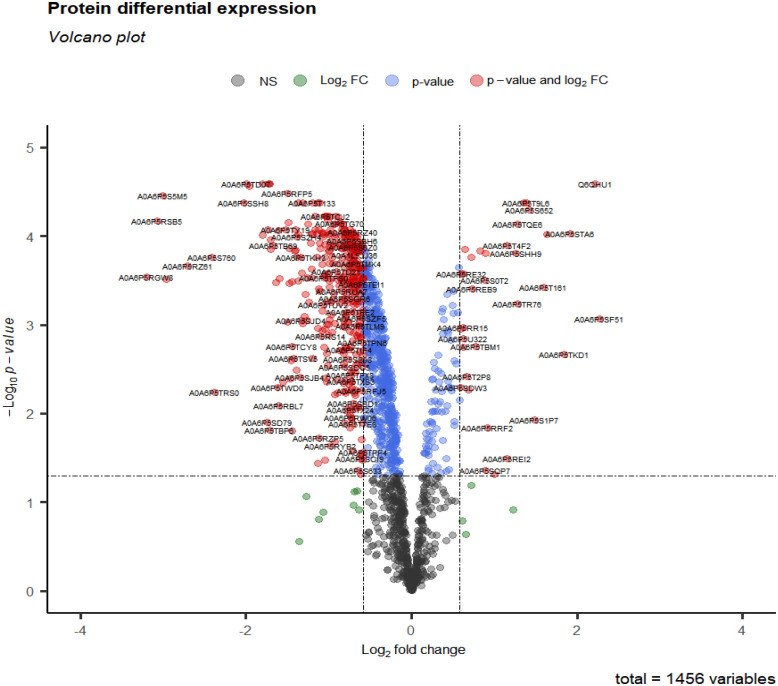
After data preprocessing, 1456 proteins were obtained from group 3 (cluster 3, 4, 6, 9 and 11) at 28 days. Under the threshold of |log2FC| ≥ 0.58, adj.p-value < 0.05, total 411 DEPs were selected for subsequent analysis. Compared with CK28 group, 32 up-regulated and 379 down-regulated proteins ( Figure 5 and Table S3 ) (Liu et al., 2016).
Figure 5.
Volcano plot of differential expression protein in 28 days at 0°C.
Identified protein KEGG functional annotation and enrichment analysis
The functions of proteins of group 3 were annotated based on the KEGG database. 411 uniprot IDs were converted to 409 ensemble gene IDs. 196 genes had KEGG annotations, a proportion of 47.68% of all proteins.
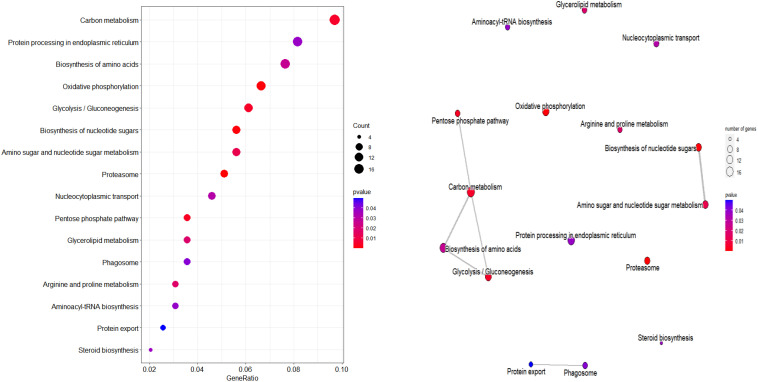
According to KEGG database, 196 proteins were enriched into 16 biological pathways (p-value<0.05) ( Figure 6 ). The pathways belonged to energy metabolism, genetic information processing, environmental information processing, and cellular processes. The most abundant proteins were involved in energy metabolism (4 pathways) which were belonging to the metabolism, followed by the translation (2 pathways) and folding, sorting and degradation (2 pathways) which belonged to genetic information processing, and thirdly, signal transduction (1 pathway) which belonging to environmental information processing, in addition, transport and catabolism (1 pathway) which belonging to cellular processes ( Table 3 ).
Figure 6.
KEGG pathway plot and Emapplot of DEPs in group 3. According to the KEGG database: “GeneRatio” means the ratio of the number of differentially expressed genes under this pathway term to the number of annotated genes under the pathway term. The larger the value is, the greater the enrichment degree is in kegg and emapplot.
Table 3.
KEGG pathway of group 3 ( Figure 6 ).
| KEGG name/ID | Gene name | Uniprot | Protein name/description | logFC |
|---|---|---|---|---|
| Pentose phosphate pathway (pavi00030) | LOC110759612 | A0A6P5ST42 | 6-phosphogluconate dehydrogenase, decarboxylating 3 | -0.69 |
| LOC110749634 | A0A6P5RR27 | probable 6-phosphogluconolactonase 4, chloroplastic | -0.70 | |
| LOC110768684 | A0A6P5TMK4 | uncharacterized protein LOC110768684 | -0.67 | |
| LOC110753532 | A0A6P5S8M0 | fructose-bisphosphate aldolase 1, cytoplasmic | -0.69 | |
| LOC110758049 | A0A6P5SHP0 | pyrophosphate–fructose 6-phosphate 1-phosphotransferase subunit alpha | -0.69 | |
| LOC110750379 | A0A6P5RYZ7 | probable 6-phosphogluconolactonase 1 | -0.69 | |
| LOC110767662 | A0A6P5THS0 | phosphoglucomutase, chloroplastic | -0.63 | |
| Glycolysis/Gluconeogenesis (pavi00010) | LOC110759242 | A0A6P5STH3 | phosphoglycerate kinase 3, cytosolic | -0.65 |
| LOC110750545 | A0A6P5RXW6 | aldehyde dehydrogenase family 3 member H1 isoform X2 | -0.61 | |
| LOC110760963 | A0A6P5SZC9 | LOW QUALITY PROTEIN: pyruvate kinase isozyme A, chloroplastic-like | -0.78 | |
| LOC110749720 | A0A6P5RWU3 | multiple inositol polyphosphate phosphatase 1-like isoform X1 | -0.71 | |
| LOC110753532 | A0A6P5S8M0 | fructose-bisphosphate aldolase 1, cytoplasmic | -0.69 | |
| LOC110762488 | A0A6P5SVE9 | cytosolic enolase 3 | -0.70 | |
| LOC110766537 | A0A6P5TEI1 | aldo-keto reductase family 4 member C10-like | -0.61 | |
| LOC110758049 | A0A6P5SHP0 | pyrophosphate–fructose 6-phosphate 1-phosphotransferase subunit alpha | -0.69 | |
| LOC110759031 | A0A6P5SSU7 | pyruvate kinase, cytosolic isozyme | -0.67 | |
| LOC110752150 | A0A6P5S4L3 | glyceraldehyde-3-phosphate dehydrogenase, cytosolic-like | -0.66 | |
| LOC110767662 | A0A6P5THS0 | phosphoglucomutase, chloroplastic | -0.63 | |
| LOC110770067 | A0A6P5TS82 | aldehyde dehydrogenase family 3 member H1-like | -0.80 | |
| Oxidative phosphorylation (pavi00190) | LOC110758582 | A0A6P5SGU2 | V-type proton ATPase subunit H | -1.03 |
| LOC110770218 | A0A6P5TSZ9 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial | -1.17 | |
| LOC110766344 | A0A6P5TDA1 | V-type proton ATPase subunit F | -0.79 | |
| LOC110744901 | A0A6P5RFK6 | ATP synthase subunit O, mitochondrial | -0.74 | |
| LOC110766094 | A0A6P5TD27 | cytochrome c1-2, heme protein, mitochondrial | -0.79 | |
| LOC110750913 | A0A6P5RZ61 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-A | -2.71 | |
| LOC110753052 | A0A6P5RXN7 | cytochrome b-c1 complex subunit 9-like | -0.69 | |
| LOC110764394 | A0A6P5T7A6 | V-type proton ATPase subunit C | -0.59 | |
| LOC110764920 | A0A6P5T963 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6 | -0.77 | |
| LOC110757961 | A0A6P5SNV3 | external alternative NAD(P)H-ubiquinone oxidoreductase B3, mitochondrial | -0.79 | |
| LOC110769121 | A0A6P5TM74 | cytochrome b-c1 complex subunit 7-2 | -1.46 | |
| LOC110768696 | A0A6P5TMD9 | cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like | -0.68 | |
| LOC110771972 | A0A6P5TXY8 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1 | -0.71 | |
| Glycerolipid metabolism (pavi00561) | LOC110759292 | A0A6P5SS33 | digalactosyldiacylglycerol synthase 2, chloroplastic isoform X1 | -0.92 |
| LOC110750545 | A0A6P5RXW6 | aldehyde dehydrogenase family 3 member H1 isoform X2 | -0.61 | |
| LOC110753288 | A0A6P5RYK4 | lysophospholipid acyltransferase 1-like | -1.26 | |
| LOC110766537 | A0A6P5TEI1 | aldo-keto reductase family 4 member C10-like | -0.61 | |
| LOC110770067 | A0A6P5TS82 | aldehyde dehydrogenase family 3 member H1-like | -0.80 | |
| LOC110771484 | A0A6P5TWD0 | 1-acyl-sn-glycerol-3-phosphate acyltransferase 2 | -1.61 | |
| LOC110760091 | A0A6P5SUX0 | D-glycerate 3-kinase, chloroplastic | -0.73 | |
| Amino sugar and nucleotide sugar metabolism (pavi00520) |
LOC110771554 | A0A6P5TXP5 | glucose-1-phosphate adenylyltransferase large subunit, chloroplastic/amyloplastic-like | -1.96 |
| LOC110762082 | A0A6P5T133 | UDP-glucuronic acid decarboxylase 6 isoform X1 | -1.20 | |
| LOC110756609 | A0A6P5SHI4 | UDP-glucuronic acid decarboxylase 5-like | -1.07 | |
| LOC110763904 | A0A6P5T5R8 | putative GDP-L-fucose synthase 2 | -0.80 | |
| LOC110747696 | A0A6P5RR53 | bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase | -0.65 | |
| LOC110753272 | A0A6P5S5E6 | bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase 1 | -0.66 | |
| LOC110769416 | A0A6P5TPT2 | NADH–cytochrome b5 reductase 1 | -0.87 | |
| LOC110757714 | A0A6P5SGI4 | GDP-mannose 4,6 dehydratase 1 | -0.72 | |
| LOC110753405 | A0A6P5S300 | alpha-1,4-glucan-protein synthase [UDP-forming] 2 | -0.64 | |
| LOC110767344 | A0A6P5THH5 | UDP-N-acetylglucosamine diphosphorylase 1 | -1.08 | |
| LOC110767662 | A0A6P5THS0 | phosphoglucomutase, chloroplastic | -0.63 | |
| Biosynthesis of nucleotide sugars (pavi01250) | LOC110771554 | A0A6P5TXP5 | glucose-1-phosphate adenylyltransferase large subunit, chloroplastic/amyloplastic-like | -1.96 |
| LOC110762082 | A0A6P5T133 | UDP-glucuronic acid decarboxylase 6 isoform X1 | -1.20 | |
| LOC110756609 | A0A6P5SHI4 | UDP-glucuronic acid decarboxylase 5-like | -1.07 | |
| LOC110763904 | A0A6P5T5R8 | putative GDP-L-fucose synthase 2 | -0.80 | |
| LOC110747696 | A0A6P5RR53 | bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase | -0.65 | |
| LOC110753272 | A0A6P5S5E6 | bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase 1 | -0.66 | |
| LOC110755885 | A0A6P5SAH1 | 2-dehydro-3-deoxyphosphooctonate aldolase 1-like | -0.59 | |
| LOC110757714 | A0A6P5SGI4 | GDP-mannose 4,6 dehydratase 1 | -0.72 | |
| LOC110753405 | A0A6P5S300 | alpha-1,4-glucan-protein synthase [UDP-forming] 2 | -0.64 | |
| LOC110767344 | A0A6P5THH5 | UDP-N-acetylglucosamine diphosphorylase 1 | -1.08 | |
| LOC110767662 | A0A6P5THS0 | phosphoglucomutase, chloroplastic | -0.63 | |
| Arginine and proline metabolism (pavi00330) | LOC110771241 | A0A6P5TVM3 | arginase 1, mitochondrial | -0.76 |
| LOC110750545 | A0A6P5RXW6 | aldehyde dehydrogenase family 3 member H1 isoform X2 | -0.61 | |
| LOC110757265 | A0A6P5SLA2 | proline iminopeptidase isoform X1 | -0.83 | |
| LOC110752072 | A0A6P5RYV2 | delta-1-pyrroline-5-carboxylate dehydrogenase 12A1, mitochondrial | -0.69 | |
| LOC110765925 | A0A6P5TCE0 | probable polyamine oxidase 2 | -1.59 | |
| LOC110770067 | A0A6P5TS82 | aldehyde dehydrogenase family 3 member H1-like | -0.80 | |
| Biosynthesis of amino acids (pavi01230) | LOC110771241 | A0A6P5TVM3 | arginase 1, mitochondrial | -0.76 |
| LOC110759242 | A0A6P5STH3 | phosphoglycerate kinase 3, cytosolic | -0.65 | |
| LOC110768015 | A0A6P5TJN5 | acetylornithine deacetylase | -0.73 | |
| LOC110763488 | A0A6P5T3N7 | tryptophan synthase alpha chain | -1.22 | |
| LOC110760963 | A0A6P5SZC9 | LOW QUALITY PROTEIN: pyruvate kinase isozyme A, chloroplastic-like | -0.78 | |
| LOC110773697 | A0A6P5U464 | serine acetyltransferase 1, chloroplastic-like | -0.65 | |
| LOC110751150 | A0A6P5S1I7 | serine acetyltransferase 5 | 0.89 | |
| LOC110768684 | A0A6P5TMK4 | uncharacterized protein LOC110768684 | -0.67 | |
| LOC110753532 | A0A6P5S8M0 | fructose-bisphosphate aldolase 1, cytoplasmic | -0.69 | |
| LOC110762488 | A0A6P5SVE9 | cytosolic enolase 3 | -0.70 | |
| LOC110758653 | A0A6P5SRD7 | 3-dehydroquinate synthase, chloroplastic | -0.61 | |
| LOC110759031 | A0A6P5SSU7 | pyruvate kinase, cytosolic isozyme | -0.67 | |
| LOC110752150 | A0A6P5S4L3 | glyceraldehyde-3-phosphate dehydrogenase, cytosolic-like | -0.66 | |
| LOC110768524 | A0A6P5TLM9 | diaminopimelate decarboxylase 1, chloroplastic-like | -0.61 | |
| LOC110754219 | A0A6P5SBM9 | threonine dehydratase biosynthetic, chloroplastic | -0.66 | |
| Nucleocytoplasmic transport (pavi03013) | LOC110771294 | A0A6P5TU18 | elongation factor 1-alpha | -1.73 |
| LOC110749570 | A0A6P5RQU6 | importin subunit beta-1 | -0.74 | |
| LOC110754597 | A0A6P5SCU9 | eukaryotic initiation factor 4A-3 | -0.73 | |
| LOC110763739 | A0A6P5T551 | importin subunit alpha-2 | -0.96 | |
| LOC110766679 | A0A6P5TEI0 | importin subunit beta-1-like | ||
| LOC110765688 | A0A6P5TC31 | importin-4 | -1.00 | |
| LOC110767294 | A0A6P5THD6 | importin subunit beta-1-like | -1.39 | |
| LOC110750711 | A0A6P5RW06 | importin subunit alpha-9 | -0.73 | |
| LOC110770330 | A0A6P5TTE8 | LOW QUALITY PROTEIN: RAN GTPase-activating protein 2 | -0.71 | |
| Steroid biosynthesis (pavi00100) | LOC110768162 | A0A6P5TKH3 | 24-methylenesterol C-methyltransferase 2 | -1.34 |
| LOC110750884 | A0A6P5S0I9 | cycloartenol synthase 2 | -1.38 | |
| LOC110766031 | A0A6P5TCY8 | probable 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | -1.44 | |
| LOC110768705 | A0A6P5TME5 | sterol 14-demethylase | -0.77 | |
| Protein processing in endoplasmic reticulum (pavi04141) | LOC110751012 | A0A6P5RQI0 | dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit STT3A | -1.11897 |
| LOC110762860 | A0A6P5SY54 | probable protein disulfide-isomerase A6 | -0.69 | |
| LOC110749749 | A0A6P5RV81 | eukaryotic translation initiation factor 2 subunit alpha homolog | -0.73 | |
| LOC110764580 | A0A6P5T7Q6 | SEC12-like protein 2 | -0.74 | |
| LOC110749381 | A0A6P5RTW9 | GTP-binding protein SAR1A-like | -0.79 | |
| LOC110754946 | A0A6P5S452 | protein transport protein Sec61 subunit gamma-like | -1.06 | |
| LOC110745511 | A0A6P5RHB1 | phospholipase A-2-activating protein | -0.80 | |
| LOC110745667 | A0A6P5RKT8 | ERAD-associated E3 ubiquitin-protein ligase component HRD3A | -0.66 | |
| LOC110758163 | A0A6P5SPT7 | ubiquitin thioesterase OTU1 | -0.87 | |
| LOC110757706 | A0A6P5SMW9 | heat shock protein 90-5, chloroplastic | -0.76 | |
| LOC110763918 | A0A6P5T5S9 | GTP-binding protein SAR1A-like | -0.78 | |
| LOC110766523 | A0A6P5TEM1 | hsp70 nucleotide exchange factor fes1-like | -0.59 | |
| LOC110744987 | A0A6P5R736 | protein transport protein Sec61 subunit alpha-like | -0.65 | |
| LOC110767398 | A0A6P5THX5 | dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit DAD1 | -0.67 | |
| LOC110758594 | A0A6P5SPJ9 | ERAD-associated E3 ubiquitin-protein ligase HRD1B-like | -1.02 | |
| LOC110750725 | A0A6P5RW23 | dnaJ protein P58IPK homolog | 0.69 | |
| Proteasome (pavi03050) |
LOC110752154 | A0A6P5RZ40 | 26S proteasome non-ATPase regulatory subunit 6 homolog | -0.71 |
| LOC110751327 | A0A6P5RY55 | 26S proteasome non-ATPase regulatory subunit 11 homolog | -1.00 | |
| LOC110768516 | A0A6P5TKB5 | 26S proteasome regulatory subunit S10B homolog B isoform X1 | -0.85 | |
| LOC110758254 | A0A6P5SIC0 | proteasome subunit alpha type-7 | -0.75 | |
| LOC110761611 | A0A6P5T0R9 | 26S proteasome non-ATPase regulatory subunit 2 homolog A | -0.68 | |
| LOC110759896 | A0A6P5SVN7 | 26S proteasome regulatory subunit 8 homolog A | -0.62 | |
| LOC110761950 | A0A6P5STQ4 | 26S proteasome non-ATPase regulatory subunit 14 homolog | -0.69 | |
| LOC110750779 | A0A6P5RPM8 | 26S proteasome regulatory subunit 6B homolog | -0.61 | |
| LOC110752287 | A0A6P5S1R5 | 26S proteasome non-ATPase regulatory subunit 13 homolog B | -0.72 | |
| LOC110764158 | A0A6P5T6N7 | 26S proteasome non-ATPase regulatory subunit 12 homolog A-like | -0.79 | |
| Aminoacyl-tRNA biosynthesis (pavi00970) | LOC110760913 | A0A6P5SQ42 | LOW QUALITY PROTEIN: tyrosine–tRNA ligase 1, cytoplasmic-like | -1.21 |
| LOC110767207 | A0A6P5TH00 | tryptophan–tRNA ligase, cytoplasmic | -0.63 | |
| LOC110759739 | A0A6P5SN62 | serine–tRNA ligase | -0.81 | |
| LOC110772028 | A0A6P5TYJ2 | asparagine–tRNA ligase, cytoplasmic 1-like isoform X1 | -0.87 | |
| LOC110764100 | A0A6P5T667 | cysteine–tRNA ligase 2, cytoplasmic | -0.82 | |
| LOC110765644 | A0A6P5TBP6 | tyrosine–tRNA ligase, chloroplastic/mitochondrial | -1.71 | |
| Phagosome (pavi04145) | LOC110758582 | A0A6P5SGU2 | V-type proton ATPase subunit H | -1.03 |
| LOC110766344 | A0A6P5TDA1 | V-type proton ATPase subunit F | -0.79 | |
| LOC110754946 | A0A6P5S452 | protein transport protein Sec61 subunit gamma-like | -1.06 | |
| LOC110764394 | A0A6P5T7A6 | V-type proton ATPase subunit C | -0.59 | |
| LOC110764808 | A0A6P5T8I7 | 25.3 kDa vesicle transport protein | -1.03 | |
| LOC110773403 | A0A6P5U2R9 | tubulin alpha-3 chain | -0.82 | |
| LOC110744987 | A0A6P5R736 | protein transport protein Sec61 subunit alpha-like | -0.65 | |
| Protein export (pavi03060) | LOC110748656 | A0A6P5RMW7 | signal recognition particle receptor subunit beta-like | -0.79 |
| LOC110754946 | A0A6P5S452 | protein transport protein Sec61 subunit gamma-like | -1.06 | |
| LOC110770381 | A0A6P5TSV5 | signal peptidase complex catalytic subunit SEC11A | -1.42 | |
| LOC110744987 | A0A6P5R736 | protein transport protein Sec61 subunit alpha-like | -0.65 | |
| LOC110757068 | A0A6P5SKK6 | signal recognition particle subunit SRP68 | -1.13 | |
| Carbon metabolism (pavi01200) | LOC110753798 | A0A6P5S0B5 | glycerate dehydrogenase | -1.19 |
| LOC110759612 | A0A6P5ST42 | 6-phosphogluconate dehydrogenase, decarboxylating 3 | -0.69 | |
| LOC110759242 | A0A6P5STH3 | phosphoglycerate kinase 3, cytosolic | -0.65 | |
| LOC110760963 | A0A6P5SZC9 | LOW QUALITY PROTEIN: pyruvate kinase isozyme A, chloroplastic-like | -0.78 | |
| LOC110749634 | A0A6P5RR27 | probable 6-phosphogluconolactonase 4, chloroplastic | -0.70 | |
| LOC110773146 | A0A6P5U2L1 | S-formylglutathione hydrolase | -0.61 | |
| LOC110773697 | A0A6P5U464 | serine acetyltransferase 1, chloroplastic-like | -0.65 | |
| LOC110751150 | A0A6P5S1I7 | serine acetyltransferase 5 | 0.89 | |
| LOC110768684 | A0A6P5TMK4 | uncharacterized protein LOC110768684 | -0.67 | |
| LOC110753532 | A0A6P5S8M0 | fructose-bisphosphate aldolase 1, cytoplasmic | -0.69 | |
| LOC110762488 | A0A6P5SVE9 | cytosolic enolase 3 | -0.70 | |
| LOC110752818 | A0A6P5S118 | dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like | -0.64 | |
| LOC110745262 | A0A6P5RGN9 | malate dehydrogenase, glyoxysomal-like | -1.11 | |
| LOC110759031 | A0A6P5SSU7 | pyruvate kinase, cytosolic isozyme | -0.67 | |
| LOC110752150 | A0A6P5S4L3 | glyceraldehyde-3-phosphate dehydrogenase, cytosolic-like | -0.66 | |
| LOC110753434 | A0A6P5S955 | ribulose bisphosphate carboxylase small chain clone 512-like | -0.66 | |
| LOC110750379 | A0A6P5RYZ7 | probable 6-phosphogluconolactonase 1 | -0.69 | |
| LOC110754219 | A0A6P5SBM9 | threonine dehydratase biosynthetic, chloroplastic | -0.66 | |
| LOC110760091 | A0A6P5SUX0 | D-glycerate 3-kinase, chloroplastic | -0.73 |
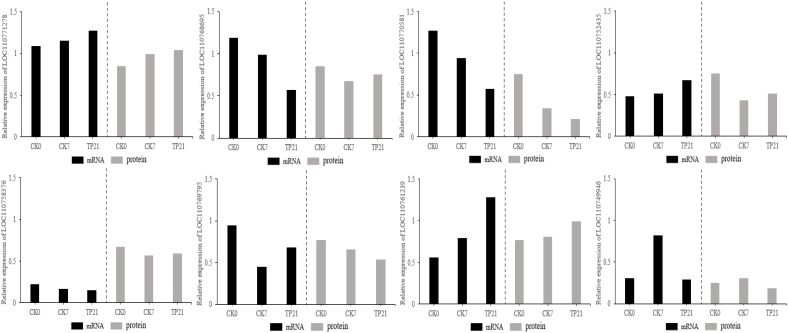
DEPs verification
To determine whether the levels of transcription and translation were correlated, the proteome data were checked at the transcription level. Results from qRT-PCR revealed that the expressions at the transcription level for most of the genes were similar to the results of cherry protein abundance among the eight randomly selected proteins ( Figure 7 ).
Figure 7.
Verification of genes of protein using qRT-PCR.
Discussion
Cherry fruit undergoes a rapid decay and disintegration process that involves several differential expression proteins during storage. In the study, CK disintegrated rapidly at 28 days of storage, while TP was more stable at TP vs CK, and the differential proteins were mainly related to energy metabolism, protein synthesis and modification, amino acid and cell wall metabolism.
Proteins associated with energy metabolism
The DEPs related to energy in TP vs CK group were involved in pentose phosphate pathway, glycolysis/gluconeogenesis, oxidative phosphorylation. Glycolysis refers to the process in which glucose is decomposed to produce pyruvate and provide energy under anaerobic conditions (Serina et al., 2019). Gluconeogenesis refers to the process in which non sugar substances such as pyruvate, pyruvic acid etc are used as precursors to synthesize glucose. There is a close relationship between gluconeogenesis and glycolysis. If glycolysis is active, gluconeogenesis is limited, conversely, if the key enzyme of glycolysis is inhibited, the acting enzyme of gluconeogenesis will be promoted (Quiroga et al., 2022). Glycolysis plays an important role in carbohydrate metabolism, and in this study, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, pyruvate kinase, and fructose-bisphosphate aldolase screened were a key enzyme in the process of glycolysis and were up-regulated in CK group. The up-regulated expression of hexokinase gene can promote the weakening of gluconeogenesis pathway, leading to the enhancement of pentose phosphate pathway and glycolysis pathway. NADH dehydrogenase, ATP synthase, cytochrome, V-type proteon ATPase in oxidative phosphorylation were up-regualted expression, this indicate that the more energy was produced in CK group, and the subsequent metabolic process should be at a high level.
Proteins associated with translation and folding, sorting and degradation
Proteins, an important part of the enzymes in many plants, providing nutrition and energy for plant storage, participating in the regulation of various plant metabolic processes, are closely related to plant metabolism, and stress resistance. In this study, This pathways (arginine and proline metabolism, aminoacyl-tRNA biosynthesis, protein processing in endoplasmic reticulum, protein export, nucleocytoplasmic transport, proteasome) related to DEPs belong to animo acid biosynthesis, translation, and folding, sorting and degradation. The final product of biological function is protein, and amino acids are a class of crucial natural molecules for building a variety of diverse peptides and proteins, which can also take part in the formation of metabolites in organisms (Huang et al., 2020; Huang et al., 2021). When plants are in a stress environment, in order to better adapt to this environment, amino acids in cells will respond to a certain extent. In the study, the results showed that the synthesis of amino acids of cherry was almost down-regulated after ozone treatment, indicating that the lower synthesis of amino acids may be a common response under the action of ozone, while the synthesis and expression of most amino acids in CK group were up-regulated, which further indicated that the CK group was more sensitive in responding to the process of ozone. Sufficient amino acids were provided for metabolic pathways, aminoacyl-tRNA biosynthesis and protein processing in endoplasmic reticulum are important metabolic pathways for protein formation. Aminoacylation of tRNAs was the critical step of protein biosynthesis, amino acids were first activated and then transfered to the tRNA (Cramer et al., 1991). In protein biosynthesis, aminoacyl-tRNA plays a crucial role in the process of transferring amino acids to the carboxyl end of peptide chain. Meanwhile, in our work, proteasome, protein processing in endoplasmic reticulum was associated with ozone-treated cherry. It shows that ozone treatment affected the expression of some proteins.
Proteins associated with cell wall metabolism
In our stuty, these DEPs were also enriched in several pathways such as biosynthesis of nucleotide sugars, amino sugar and nucleotide sugar metabolism ( Figure S3 ). The most common forms of amino sugars are glucosamine and galactosamine, and amino sugar metabolism is closely related to plant growth and development and environmental adaptability. Nucleotide sugar is an activated form of carbohydrate synthesis or mutual conversion. Ozone treatment may affect the expression of various metabolic enzymes of amino sugar and nucleotide sugar in cherry, and play a role in regulating the quality of cherry, especially firmness. Some enzymes in the amino sugar and nucleotide sugar metabolism pathway also participate in cell wall metabolism, such as chitinase, endogenous chitinase, mannose-1-phosphate guanyltransferase (MPG) and UDP glucuronic acid decarboxylase (UXS). Therefore, with the fruit at the ozone treatment, the cherry fruit degradation slowed down and attained a stable metabolism situation.
Proteins associated with lipid metabolism
These DEPs were also enriched in several pathways such as glycerolipid metabolism, steroid biosynthesis ( Figure S3 ). A specific fruit can be recognized by the combination of volatile organic chemicals that give sweet cherries their distinctive flavor. Volatiles are derived from metabolites like fatty acids and other compounds. Glycerolipids are the most abundant lipids in higher plants (Qin et al., 2020). Plant lipids include lipids, membrane lipids, signal molecules, photosynthetic pigments, essential oils, plant hormones and plant surface protective substances. They play an important role in plant growth, storage and stress response, and are widely involved in different biological processes. In addition to being an important structural material of organisms, lipids include the lipid bilayer of cell membrane and protective lipids on the surface of plant tissues and organs. Lipids are also very important physiological active substances and signal molecules in plants. For example, phosphatidic acid, brassinolide and phosphatidylinositol derivatives are widely involved in various biological processes in plants (Zheng and Li, 2017; Qin et al., 2020). Lipid is also an efficient energy storage material in plants. The energy stored by lipid per unit weight is much higher than that of carbohydrate and protein. Lipid storage, such as triglyceride, can provide sufficient energy and carbon source for biological metabolism. Membrane is sensitive to environmental changes, and abiotic stress directly affects membrane performance. In addition, glycerides are the main components of the membrane. Adjusting the composition, unsaturation and acyl chain length of the membrane also enables plants to maintain the integrity and fluidity of the membrane under environmental stress. Sequential action by acyl-releasing lipases and acyltransferases can replace the acyl groups of the bulk membrane glycerolipids while modifying the biophysical characterisitics of each lipid molecules to maintain the membrane bilayer structure. Free fatty acids (FAs) and a less lipophilic molecule, such as a lysophospholipid, are produced when acyl-releasing glycerolipid lipases hydrolyze the acyl groups from a glycerolipid molecule (Wang et al., 2019; Kim, 2020). Meanwhile, fatty acids can improve the tolerance of plants and reduce the damage of abiotic stress to plants.
Conclusion
In this study, 4557 master proteins were identified, and 3149 proteins were common to all groups. Mfuzz analyses revealed 3149 candidate proteins. KEGG annotation and enrichment analysis showed proteins related to carbohydrate and energy metabolism, protein, amino acids, and nucleotide sugar biosynthesis and degradation. The expressions on the transcription level of eight selected proteins by qRT-PCR validation were similar to the results of the protein level. Further study of the molecular structures of these proteins and their associated biological roles in cherry during storage.
Data availability statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) with the dataset identifier PXD037578.
Author contributions
YZ was responsible for the design of the experiments, and interpretation of data. ZH and YZ performed the experiments. YZ wrote the manuscript. NZ and HJ collected relevant materials. CD determined compounds and improved the results. XC assist in raw data uploading. JY improve the discussion. CC contributed to funding acquisition. HG contributed to reviewing and editing the manuscript.. All authors contributed to the article and approved the submitted version.
Funding Statement
The present study was supported by Graduate Scientific Research Innovation Project of Tianjin University of Science and Technology in 2021(KYS202157), the National Key R&D Program of China(2018YFF0213605), the Innovation Team of the Tianjin Forestry & Pomology Research System (ITTHRS2021000), Innovative research and experimental projects for young researchers (202009, 2021001); the Major Science and Technology Projects of Shandong Province (2019JZZY20617), Key Laboratory of Storage of Agricultural Products, Ministry of Agriculture and Rural Affairs (kf2020001, Kf2020003, Kf2020005, Kf2020006, kf2021001), The Project Program of Key Laboratory of Tianjin Key Laboratory of Food Quality and Health, China (TJS202101) and Major Innovation Pilot Project of Integration of Science, Education and Industry of Qilu University of Technology (Shandong Academy of Science) (No. 2022JBZ01-08), College Students’ Innovation and Entrepreneurship Training Program of Shandong Province (202210431008).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1065465/full#supplementary-material
References
- Calle A., Serradilla M. J., Wunsch A. (2021). QTL mapping of phenolic compounds and fruit colour in sweet cherry using a 6+9K SNP array genetic map. Sci. Hortic. 280, 109900. doi: 10.1016/j.scienta.2021.109900 [DOI] [Google Scholar]
- Clayton-Cuch D., Yu L., Shirley N., David Bradley D., Bulone V., Böttcher C. (2021). Auxin treatment enhances anthocyanin production in the non-climacteric sweet cherry (Prunus avium l.). Int. J. Mol. Sci. 22 (19), 10760. doi: 10.3390/ijms221910760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer F., Englisch U., Freist W., Stembach H. (1991). Aminoacylation of tRNAs as critical step of protein biosynthesis. Biochimie 73, 1027–1035. doi: 10.1016/0300-9084(91)90144-P [DOI] [PubMed] [Google Scholar]
- Dahn M. L., Dean C. A., Jo D. B., Coyle K. M., Marcato P. (2021). Human-specific GAPDH qRT-PCR is an accurate and sensitive method of xenograft metastasis quantification. Mol. Ther.: Methods Clin. Dev. 20, 398–408. doi: 10.1016/j.omtm.2020.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA (2001). Secondary direct food additives permitted in food for human consumption, final rule. Fed. Regist. 66, 33829–33820. [Google Scholar]
- Gao C. C., Lin Q., Dong C. H., Ji H. P., Yu J. Z., Chen C. K., et al. (2020). Effects of ozone concentration on the postharvest quality and microbial diversity of Muscat Hamburg grapes. RSC Adv. 10 (15), 9037–9045. doi: 10.1039/C9RA10479H [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez G., Figueroa C. R., Turner A., Munne-Bosch S., Schreiber L., Zeisler V., et al. (2021). Abscisic acid applied to sweet cherry at fruit set increases amounts of cell wall and cuticular wax components at the ripe stage. Sci. Hortic. 283, 110097. doi: 10.1016/j.scienta.2021.110097 [DOI] [Google Scholar]
- Huang X. L., Hou Z. H. (2021). Label-free quantitative proteomics analysis of jujube (Ziziphus jujuba mill.) during different growth stages. RSC Adv. 11 (36), 22106–22119. doi: 10.1039/d1ra02989d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. Y., Wang S. S., Song D., Cao X. F., Huang W. B., Ke S. Y. (2020). Discovery of gamma-lactam alkaloid derivatives as potential fungicidal agents targeting steroid biosynthesis. J. Agric. Food Chem. 68 (49), 14438–14451. doi: 10.1021/acs.jafc.0c05823 [DOI] [PubMed] [Google Scholar]
- Huang X. L., Zhao Y. H., Hou Z. H. (2021). Purification of ethyl linoleate from foxtail millet (Setaria italica) bran oil via urea complexation and molecular distillation. Foods 10 (8), 1925–1933. doi: 10.3390/foods10081925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y. N., Han Z. Y., Sun Y. Q., Wang S., Wang T., Wang Y., et al. (2020). ERF4 affects fruit firmness through TPL4 by reducing ethylene production. Plant J. 103, 937–950. doi: 10.1111/tpj.14884 [DOI] [PubMed] [Google Scholar]
- Kim H. U. (2020). Lipid metabolism in plants. Plants 9, 871. doi: 10.3390/plants9070871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L., Futschik M. (2007). Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2 (1), 5–7. doi: 10.6026/97320630002005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Jing L., Tu X. L. (2016). Weighted gene co-expression network analysis identifies specific modules and hub genes related to coronary artery disease. BCM Cardiovasc. Disord. 16 (1), 54–62. doi: 10.1186/s12872-016-0217-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang J. H., Wang K. T., Lyu S. H., Ren L. Y., Huang C. Y., et al. (2022). Comparison analysis of widely-targeted metabolomics revealed the variation of potential astringent ingredients and their dynamic accumulation in the seed coats of both carya cathayensis and carya illinoinensis. Food Chem. 374, 131688. doi: 10.1016/j.foodchem.2021.131688 [DOI] [PubMed] [Google Scholar]
- Li Q. Q., Yang S. P., Zhang R., Liu S. Y. (2022). Characterization of honey peach (Prunus persica (L.) batsch) aroma variation and unraveling the potential aroma metabolism mechanism through proteomics analysis under abiotic stress. Food Chem. 386, 132720. doi: 10.1016/j.foodchem.2022.132720 [DOI] [PubMed] [Google Scholar]
- Mekontso F. N., Duan W. H., Cisse E. H. M., Chen T. Y., Xu L. B. (2021). Alleviation of Postharvest chilling injury of carambola fruit by gamma-aminobutyric acid: physiological, biochemical, and structural characterization. Frontiers in Nutrition 8, 752583. doi: 10.3389/fnut.2021.752583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q., Gao J., Zhu H. W., He H., Lu Z., Hong M. H., et al. (2018). The proteomic study of serially passaged human skin fibroblast cells uncovers down-regulation of the chromosome condensin complex proteins involved in replicative senescence. Biochem. Biophys. Res. Commun. 505 (4), 1112–1120. doi: 10.1016/j.bbrc.2018.10.065 [DOI] [PubMed] [Google Scholar]
- Monaco K. A., Costa S. M., Minatel I. O., Correa C. R., Calero F. A., Vianello F., et al. (2016). Influence of ozonated water sanitation on postharvest quality of conventionally and organically cultivated mangoes after postharvest storage. Postharv. Biol. Technol. 120, 69–75. doi: 10.1016/j.postharvbio.2016.05.003 [DOI] [Google Scholar]
- Porcher A., Guérin V., Montrichard F., Lebrec A., Lothier J., Vian A. (2020). Ascorbate glutathione-dependent H2O2 scavenging is an important process in axillary bud outgrowth in rosebush. Ann. Bot. 126, 1049–1062. doi: 10.1093/aob/mcaa130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F., Lin L., Jia Y. X., Weiqi Li W. Q., Yu B. Z. (2020). Quantitative profiling of arabidopsis polar glycerolipids under two types of heat stress. Plants 9, 693. doi: 10.3390/plants9060693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroga J., Alarcon P., Manosalva C., Teuber S., Carretta M. D., Burgos R. A. (2022). D-lactate-triggered extracellular trap formation in cattle polymorphonuclear leucocytes is glucose metabolism dependent. Dev. Comp. Immunol. 135, 104492. doi: 10.1016/j.dci.2022.104492 [DOI] [PubMed] [Google Scholar]
- Romanazzi G., Simlanick J. L., Feliziani E., Droby S. (2016). Integrated management of postharvest gray mold on fruit crops. Postharv. Biol. Technol. 113, 69–76. doi: 10.1016/j.postharvbio.2015.11.003 [DOI] [Google Scholar]
- Sachadyn-Kró M., Materska M., Chilczuk B., Karas M., Jakubczyk A., Perucka I., et al. (2016). Ozone-induced changes in the content of bioactive compounds and enzyme activity during storage of pepper fruits. Food Chem. 211, 59–67. doi: 10.1016/j.foodchem.2016.05.023 [DOI] [PubMed] [Google Scholar]
- Serina J., Fernandes M. X., Castilho P. C. (2019). Effects of hydroxycinnamic acids on the glycolysis pathway. South Afr. J. Bot. 120, 219–229. doi: 10.1016/j.sajb.2018.06.016 [DOI] [Google Scholar]
- Tabakoglu N., Karaca H. (2018). Effects of ozone-enriched storage atmosphere on postharvest quality of black mulberry fruits (Morus nigra l.). LWT-Food Sci. Technol. 92, 276–281. doi: 10.1016/j.lwt.2018.02.044 [DOI] [Google Scholar]
- Wang K., Durrett T. P., Benning C. (2019). Functional diversity of glycerolipid acylhydrolases in plant metabolism and physiology. Prog. Lipid Res. 75, 100987. doi: 10.1016/j.plipres.2019.100987 [DOI] [PubMed] [Google Scholar]
- Wang S. Q., Li W. N., Zhang X. F., Li G., Li X. D., Chang H., et al. (2022). Metabolomics study of different germplasm resources for three ploygonatum species using UPLC-Q-TOF-MS/MS. Front. Plant Sci. 13, 826902. doi: 10.3389/fpls.2022.826902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. H., Ding M. J., Shao G. Q., Jia S. J., Yin X., Cui Y. H., et al. (2021). Kcnk3, Ggta1, and Gpr84 are involved in hyperbaric oxygenation preconditioning protection on cerebral ischemia–reperfusion injury. Exp. Brain Res. 239, 3601–3613. doi: 10.1007/s00221-021-06220-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. B., Meng Z. P., Li Y. N., Chen R. X., Yang Y. Z., Zhao Z. Y. (2021). Evaluation of physiological characteristics, soluble sugars, organic acids and volatile compounds in ‘Orin’ apples (Malus domestica) at different ripening stages. Molecules 26, 807. doi: 10.3390/molecules26040807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C., Cai J. F. (2017). Time-series expressiong profile analysis of fracture healing in young and old mice. Mol. Med. Rep. 16, 4529–4536. doi: 10.3892/mmr.2017.7198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. J., Wang H. X., Yue X., Liu B. Z. (2019). Dynamic immune and metabolism response of clam meretrix petechialis to vibrio challenge revealed by a time series of transcriptome analysis. Fish Shellf. Immunol. 94, 17–26. doi: 10.1016/j.fsi.2019.08.057 [DOI] [PubMed] [Google Scholar]
- Zhang X., Jiang Y. M., Peng F. T., He N. B., Li Y. J., Zhao D. C. (2007). Changes of aroma components in hongdeng sweet cherry during fruit development. Agric. Sci. China 6 (11), 1376–1382. doi: 10.1016/S1671-2927(07)60186-2 [DOI] [Google Scholar]
- Zhao H. D., Fu M. R., Du Y. M., Sun F., Chen Q. M., Jin T., et al. (2021). Combination treatment of 1-MCP plus ClO2 improves post-harvest quality of sweet cherry fruit. Sci. Hortic. 277, 109806. doi: 10.1016/j.scienta.2020.109806 [DOI] [Google Scholar]
- Zheng G. W., Li W. Q. (2017). Profiling membrane glycerolipids during gamma-ray-induced membrane injury. BMC Plant Biol. 17, 203–216. doi: 10.1186/s12870-017-1153-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) with the dataset identifier PXD037578.