Abstract
Background
An increasing number of trials indicate that treatment outcomes in cancer patients with metastatic disease are improved when targeted treatments are matched with druggable genomic alterations in individual patients (pts). An estimated 30–80% of advanced solid tumors harbor actionable genomic alterations. However, the efficacy of personalized cancer treatment is still scarcely investigated in larger, controlled trials due to the low frequency and heterogenous distribution of druggable alterations among different histologic tumor types. Therefore, the overall effect of targeted cancer treatment on clinical outcomes still needs investigation.
Study design/methods
ProTarget is a national, non-randomized, multi-drug, open-label, pan-cancer phase 2 trial aiming to investigate the anti-tumor activity and toxicity of currently 13 commercially available, EMA-approved targeted therapies outside the labeled indication for treatment of advanced malignant diseases, harboring specific actionable genomic alterations. The trial involves the Danish National Molecular Tumor Board for confirmation of drug-variant matches. Key inclusion criteria include a) measurable disease (RECIST v.1.1), b) ECOG performance status 0–2, and c) an actionable genomic alteration matching one of the study drugs. Key exclusion criteria include a) cancer type within the EMA-approved label of the selected drug, and b) genomic alterations known to confer drug resistance. Initial drug dose, schedule and dose modifications are according to the EMA-approved label. The primary endpoint is objective response or stable disease at 16 weeks. Pts are assigned to cohorts defined by the selected drug, genomic alteration, and tumor histology type. Cohorts are monitored according to a Simon’s two-stage-based design. Response is assessed every 8 weeks for the first 24 weeks, then every 12 weeks. The trial is designed similar to the Dutch DRUP and the ASCO TAPUR trials and is a partner in the Nordic Precision Cancer Medicine Trial Network. In ProTarget, serial fresh tumor and liquid biopsies are mandatory and collected for extensive translational research including whole genome sequencing, array analysis, and RNA sequencing.
Discussion
The ProTarget trial will identify new predictive biomarkers for targeted treatments and provide new data and essential insights in molecular pathways involved in e.g., resistance mechanisms and thereby potentially evolve and expand the personalized cancer treatment strategy.
Protocol version: 16, 09-MAY-2022.
Trial registration
ClinicalTrials.gov Identifier: NCT04341181.
Secondary Identifying No: ML41742.
EudraCT No: 2019–004771-40.
Keywords: Cancer genetics, Targeted therapies, Clinical trials, Cancer immunotherapy, Precision oncology, Tumor-agnostic therapy
Background
Personalized cancer care is rapidly evolving, as the biological understanding of the individual’s cancer disease increases. A deeper understanding of disease and host at the genomic level, coupled with accessible and affordable multiplex analysis of the transcriptome, proteome, and other aspects of the cancer, is leading the way towards new paradigms in the treatment of cancer, relying not solely on the tissue of origin, but on molecular tumor profiling.
Although actionable molecular targets are frequent in cancers, the heterogenous distribution across tumor types makes traditional randomized phase 3 clinical trials in precision medicine rare, especially in less frequently encountered molecular targets. However, evidence is mounting through small clinical trials, case series, and case reports that patient outcomes are improved when a targeted treatment is matched to a tumor harboring the molecular target [1–4]. Estimates are that 30–80% of advanced solid tumors harbor actionable genomic alterations [5–8].
Several challenges exist for identifying and providing the relevant treatment to the patients at need. Many oncologists and pathologists have sparse access to comprehensive genomic profiling for screening purposes and experts for interpretation of genomic test reports to guide scientifically informed decisions about the optimal use of targeted agents [9]. The relevant drug will in many cases be an already marketed drug to be prescribed outside the labeled indication or an investigational drug accessible only in a clinical trial. Marketed drugs may not be available to the treating physician due to reimbursement issues for patients treated in lieu of health insurances, or the use of the drug may not be approved for use in publicly funded health care systems due to high costs and/or sparse scientific evidence in the tumor type treated. As access to drugs may be limited and data acquisition and reporting may be sporadic in patients treated with molecular matched therapies in the off-label setting, the overall knowledge of clinical outcomes in this setting is limited. Currently, there are more than 30 marketed drugs targeting molecular pathways frequently aberrant in human tumors, e.g., EGFR, BRAF, MET and KIT, with several more in development (list of abbreviations, Table 1).
Table 1.
List of abbreviations
| Abbreviation | Definition |
|---|---|
| BRAF | B-Raf proto-oncogene, serine/threonine kinase |
| CA-125 | Cancer antigene 125 |
| CI | Confidence interval |
| CR | Complete response |
| CTCAE | Common Terminology Criteria for Adverse Events |
| ctDNA | circulating tumor DNA |
| EGFR | Epidermal growth factor receptor |
| EMA | European Medicines Agency |
| ERBB2 | erb-b2 receptor tyrosine kinase 2 |
| FDA | Food and Drug Administration |
| FFPE | Formalin fixed paraffin embedded |
| GCIG | Gynecological Cancer InterGroup |
| GCP | Good Clinical Practice |
| GOF | Gain of function |
| IMP | Investigational medical product |
| IMWG | International Myeloma Working Group |
| IV | Intravenous |
| KIT | Mast/stem cell growth factor receptor Kitq |
| LVEF | Left ventricle ejection fraction |
| MET | Hepatocyte growth factor receptor |
| DN-MTB | Danish National Molecular Tumor Board |
| OS | Overall survival |
| PCWG3 | Prostate Cancer Working Group 3 |
| PD | Progressive disease |
| PFS | Progression free survival |
| PR | Partial response |
| PSA | Prostate specific antigene |
| RANO | Response Assessment in Neuro-oncology Criteria |
| RECIST | Response Evaluation Criteria in Solid Tumours |
| SD | Stable disease |
| TIA | Transient ischemic attack |
| TMB | Tumor Mutational Burden |
| WES | Whole exome sequencing |
| WGS | Whole genome sequencing |
Matching molecular targets to relevant drugs may improve outcomes in cancer patients although large scale prospective, randomized, controlled trials are yet to be concluded. The randomized phase II trial SHIVA by Tourneau with matched molecular target treatment vs. physicians choice failed to demonstrate any significant different in progression free survival (PFS) between the two arms [10]. However, the meta-analysis by Schwaederle et al. of 570 phase II trials comparing patients receiving molecularly matched treatment to non-matched treatment demonstrated more favorable outcomes for patients receiving matched therapies [11]. Several individual, non-randomized phase 1 and phase 2 trials comparing molecularly matched treatment to non-matched treatment also demonstrated improved outcomes for the matched treatment groups. The Initiative for Molecular Profiling and Advanced Cancer Therapy (IMPACT) study (NCT00851032) by Tsimberidou et al. [7] analyzed 1144 patients of whom 40.2% had one or more genomic aberrations. Patients receiving a targeted therapy matched to a genomic aberration had median PFS and overall survival (OS) of 4.1 months and 10.2 months, respectively, compared to 2.4 and 8.2 months for non-matched patients. The MOSCATO-01 (Molecular Screening for Cancer Treatment Optimization, NCT01566019) [12] trial by Massard et al. included 1035 adult patients. An actionable molecular alteration was identified in 411 of 843 patients with a molecular profile and 199 patients were treated with a targeted therapy matched to a genomic alteration. The PFS from molecularly matched therapy was compared to the PFS for the most recent therapy on which the patient had disease progression (PFS2/PFS1 ratio). The PFS2/PFS1 ratio was > 1.3 in 33% of the patients (63/193). Objective responses were observed in 22 of 194 patients (11%; 95% CI, 7–17%), and median overall survival was 11.9 months (95% CI, 9.5–14.3 months).
The Danish study, CoPPO (Copenhagen Prospective Personalised Oncology) included 500 patients undergoing biopsy followed by whole exome sequencing (WES) and RNA sequencing. One hundred one patients (20%) received matched treatment based on either pathogenic variants or RNA expression levels of targets available in early clinical trials or off-label treatment. Objective response according to RECIST v1.1 was observed in 15 of 101 patients (0% complete response, 15% partial response), with a median PFS of 12 weeks (95% confidence interval, 9.9–14.4) [13].
The growing number of U.S. Food and Drug Administration (FDA) and/or European Medicines Agency (EMA) approved drugs for specific molecular targets in distinct histologies may lead to increased off-label use. Multiple prospective trials have recently been initiated to gather data on safety and outcomes, as well as extensive molecular data. These studies rely on extensive molecular profiling and decision making at molecular tumor boards [14] for matching patients with targeted therapies in a phase 2 open-label, prospective, non-randomized design. The American TAPUR (Testing the Use of FDA Approved Drugs That Target a Specific Abnormality in a Tumor Gene in People With Advanced Stage Cancer, NCT02693535) [15], the Dutch DRUP (Drug Rediscovery Protocol; NCT 02925234 [16], and Canadian CAPTUR (Canadian Profiling and Targeted Agent Utilization Trial NCT03297606) [17] are ongoing trials with purpose and design similar to the ProTarget trial (NCT04341181).
The primary aim of this trial is to investigate the anti-tumor activity and toxicity of commercially available, EMA-approved targeted therapies outside the labeled indications in the treatment of advanced malignant diseases harboring specific actionable genomic alterations. This protocol has been prepared according to the Standard Protocol Items: Recommendations for International Trials (SPIRIT) guidelines [18].
Design and methods
Study design
ProTarget is a Danish nationwide, interventional, multi-drug, open-label, pan-cancer, non-randomized, prospective phase 2 basket trial which aims to investigate the efficacy and safety of targeted anticancer drugs when used off-label in patients with a malignant disease harboring an actionable genomic alteration. Patients are recruited from eight different investigator sites at oncological centers. The trial aims to include 100 pts. annually.
Patients are identified through local genomic testing at each investigator site (Fig. 1). The proposed drug-variant-match (including full tumor genomic profile, tumor type and brief anonymous clinical case summary) must be submitted to and confirmed by the Danish National Molecular Tumor Board (DN-MTB) before informed consent can be obtained. The patient must meet all general and drug-specific criteria (Tables 2 and 3) before dosing in the trial. Fresh tumor biopsies are mandatory and obtained pre-treatment, during cycle one, and at progressive disease (PD). Breast or prostate cancer pts. with bone-only-disease, or patients with primary brain tumors, are eligible based on results from liquid biopsies only (e.g., circulating tumor DNA (ctDNA)). Liquid biopsies are collected from all patients at day 1 in each treatment cycle prior to dosing. Response assessment is performed every 8 weeks during the first 24 weeks, and then every 12 weeks.
Fig. 1.

ProTarget Study Schema. Potentially eligible pts. are identified by local tumor genomic testing at each site. The investigator presents the genomic profile and clinical history and status of the patient and proposes the drug-variant match. If confirmed by the DN-MTB, the pt. can sign drug-specific informed consent and enter screening. Patients failing screening may be re-assessed for another drug if relevant. Patients meeting all eligibility criteria will start treatment. Fresh biopsies are taken at screening, on-treatment and at PD. Liquid biopsies are taken at screening and at CXD1 before dosing. Treatment is continued until PD or unmanageable toxicity. The pt. may be re-assessed for another drug if a matching alteration exists. *MTB may include as treatment options: A) Confirmation of ProTarget drug-variant-match, B) Treatment of an alternate ProTarget genomic alteration, C) Treatment of non-ProTarget variant on/off protocol or off-label, D) No treatment/protocol available. ** Every 8 weeks for 24 weeks, then every 12 weeks. Abbreviations: DN-MTB: Danish National Molecular Tumor Board, ctDNA: circulating tumor DNA, C1D1: cycle one day one, CXD1: any cycle day one, CR: complete response, PR: partial response, SD: stable disease, PD: progressive disease
Table 2.
General inclusion criteria
| General inclusion criteria (abbreviated) | |
|---|---|
| 1) Patients ≥18 years of age with a histologically proven locally advanced or metastatic malignant disease for whom no standard treatment is available or indicated. | |
| 2) Ability to understand and the willingness to sign a written informed consent/assent document. | |
| 3) ECOG performance status 0–2. | |
| 4) Acceptable organ function. | |
| 5) Measurable or evaluable disease (e.g., RECIST v1.1 for solid tumors), defined as at least one lesion that can be accurately measured in at least one dimension. Patients who have assessable disease by physical or radiographic examination but do not meet these definitions of measurable disease are eligible and will be considered to have evaluable disease. | |
| 6) Patients must have one of the actionable alterations listed in Table 4: ProTarget Drugs and Acceptable Molecular Alterations | |
| 7) For oral IMPs, patients must be able to swallow and tolerate oral medication and must have no known malabsorption syndrome. | |
| 8) Women of child-bearing potential and men must agree to use highly effective contraception. |
Table 3.
General exclusion criteria
| General exclusion criteria (abbreviated) | |
|---|---|
| 1) Ongoing toxicity > CTCAE grade 2. Patients with ongoing peripheral neuropathy of ≥ CTCAE grade 3. | |
| 2) Prior treatment with the selected study drug. | |
| 3) Genomic alterations known to confer drug resistance. | |
| 4) Current treatment with other anti-cancer therapy (cytotoxic, biologic, radiation, or hormonal other than for replacement). Medications prescribed for supportive care that may potentially have an anti-cancer effect (e.g., megestrol acetate, bisphosphonates) or ongoing castration-intent therapy for prostate cancer, are accepted if they have been started ≥1 month prior to enrollment. | |
| 5) Female patients who are pregnant or nursing. Male and female patients who refuse to practice highly effective contraception methods. | |
| 6) Patients with known progressive brain metastases determined by serial imaging or declining neurologic function in the opinion of the treating physician. Patients with previously treated brain metastases must be clinically stable for at least 1 month after completion of treatment and off steroid treatment for one month prior to study enrollment. | |
| 7) Patients with preexisting uncontrolled cardiac conditions, LVEF known to be < 40%, stroke (including TIA) or acute myocardial infarction within 4 months before the first dose. | |
| 8) Patients with acute gastrointestinal bleeding within 1 month of start of treatment. | |
| 9) Patients with any other clinically significant medical condition which, in the opinion of the treating physician, makes it undesirable for the patient to participate in the study e.g., active infection, significant uncontrolled hypertension, severe psychiatric illness situations, or anticipated or planned anti-cancer treatment or surgery. | |
| 10) Patients who do not meet drug-specific eligibility requirements for the drug selected. | |
| 11) Patients whose disease is not measurable or assessable by radiographic imaging or physical examination (e.g., elevated serum tumor marker only) with the exception of ovarian cancer (CA-125) and prostate cancer (PSA). | |
| 12) Patients with known allergy/hypersensitivity to the study drug. |
Patient recruitment began 24-Aug-2020 and is ongoing. Last patient, last visit is undefined, and cohorts will open and reach completion successively depending on variant identification in individual pts.
Study objectives
The primary objective of the study is to evaluate the anti-tumor activity and toxicity of commercially available, EMA-approved targeted anti-cancer drugs used off-label to treat patients with advanced malignant disease harboring a known or predicted targetable genomic alteration.
The secondary objectives are 1) To perform refined biomarker analyses (e.g., WGS) on serial fresh tumor samples and liquid biopsies, and 2) to study mechanisms of resistance using serial fresh tumor and liquid biopsies.
Study endpoints
Primary study endpoints are:
Anti-tumor activity, defined as objective response, at 16 weeks assessed by disease specific response criteria
Stable disease at 16 weeks
Treatment-related and serious adverse events
Secondary study endpoints are:
Duration of response, progression-free survival and overall survival
Duration of study treatment (time on drug)
Percentage of screened patients treated based on their molecular tumor profile
Exploratory study endpoints:
Description of concordance between genomic tumor profile of pre-treatment tumor biopsies and genomic tumor profile according to tumor profiling tests that were used to enroll patients
Identification of patterns of resistance based on serial tumor biopsies and liquid biopsies
Study population and eligibility criteria
Patients with metastatic or advanced malignant disease with exhausted treatment options, or for whom no standard treatment exist, are eligible. The tumor must harbor a potentially actionable genomic alteration targetable by a drug accessible in the ProTarget Protocol and must be a cancer histology outside the FDA/EMA-labelled indication. Patients with a genomic alteration known to confer resistance to a specific drug (such as solvent front or gatekeeper mutations or traits causing redundant signaling) are not eligible to receive that agent. Additional inclusion and exclusion criteria may apply to specific drugs or drug-tumor type-variant matches (Table 4). In these cases, drug-specific eligibility criteria must be met after general eligibility criteria have been met.
Table 4.
ProTarget Drugs and Acceptable Molecular Alterations
| Drug | Acceptable Genomic Alterationsab | Excluded Genomic Alterationsc |
|---|---|---|
| Alectinib | EML4-ALK fusions or mutations, ROS1 fusions | None |
| Atezolizumab | MSI high | None |
|
POLE mutations: R150X, P286R, P286H, S297F, Y298fs, F367S, V411L, L424V, P436R, V437M, S459F, R573L, E597K, R665W, L698fs, R762W, R793C, K1008N, T1052M, R1111Q, L1235I, V1368M, R1519C, P1547S, R1826W, R1879C, Y1889C, S1892N, A1967V, A2213V, A2243T | ||
|
POLD1 mutations: W79L, P112fs, A930fs, N247I, R352C, Q461H, S478N, A864T, E1105D | ||
| TMB ≥10 mut/mb | ||
| Avelumab | MSI high | None |
|
POLE mutations: R150X, P286R, P286H, S297F, Y298fs, F367S, V411L, L424V, P436R, V437M, S459F, R573L, E597K, R665W, L698fs, R762W, R793C, K1008N, T1052M, R1111Q, L1235I, V1368M, R1519C, P1547S, R1826W, R1879C, Y1889C, S1892N, A1967V, A2213V, A2243T | ||
|
POLD1 mutations: W79L, P112fs, A930fs, N247I, R352C, Q461H, S478N, A864T, E1105D | ||
| TMB ≥10 mut/mb | ||
| Axitinib | VEGFR1 (FLT1), VEGFR2 (KDR), VEGFR3 (FLT-4) GOF mutations, amplification, or overexpression | None |
| Erlotinib |
EGFR exon 19 deletions in the region E746-E759 EGFR mutations: E709A/G/K, E884K, G719A/C/S, S768I, L858R, L861Q, L833V |
Any of the following EGFR mutations: L747S, T790M, or T854A Exon 20 insertions |
| Niraparib |
Germline or somatic BRCA1/BRCA2 inactivating mutations ATM/ATR mutations or deletions HRD positive d |
None |
| Pemigatinib | Mutations in PDGFRA, PDGFRB, or PCM1-JAK2 fusions. FGF/FGFR amplifications, mutation and fusions | None |
| Trastuzumab plus Pertuzumab |
ERBB2 amplification, overexpression, or mutations: G309A, G309E, S310F, D769H, D769Y, L755S, V777L, V842I, E321G, R896C ERBB2 P780insertions ERBB2 deletions in the region L755–T759 |
None |
| Trastuzumab emtansine | ERBB2 amplification, or overexpression, or presence of any activating ERBB2 mutations | None |
| Vemurafenib plus Cobimetinib | BRAF V600E/D/K/R mutations | Any mutations in MAP 2 K1, MAP 2 K2, MEK1, MEK2, NRAS |
| Vismodegib | PTCH1 deletion or inactivating mutations |
SMO mutations: D473G/H/Y, W535L GLI2 amplification |
Patients are eligible to receive one of the listed drugs if they have a non-indicated cancer harbouring a molecular alteration matching the drug
MSI: micro satellite instability; TMB: tumor mutational burden; GOF: gain of function; HRD: homologous recombination deficiency
aSource is FDA approved drug label, manufacturer data, [19–21]; Illumina Basespace Knowledge Network, QCI Precision Insight
bFor any of the genes listed, alterations such as point mutations, insertions, deletions, translocations and amplifications or overexpression may be acceptable to match a drug to that gene. If a proposed drug-variant match is not accepted by the automated matching rules process, consider requesting case review by the Molecular Tumor Board
cDetection of any of the alterations in this column will exclude the patient from receiving the matched drug treatment as these alterations are associated with drug resistance
dHRD is evaluated from cytoscan HD (ThermoFisher) SNP array where an HRD score is calculated based on the sum of loss of heterozygosity (LOH), telomeric allelic imbalance (TAI) and large-scale transitions (LST) [22]. When WGS data are available, HRD status can be supported by mutational signatures from Cosmic [23] and CO-Regulation Database (CORD [24])
A patient must meet all of the following criteria to be eligible to participate in this study:
Potential participants who meet any of the following criteria will be excluded.
Study procedures
Patients, who meet all eligibility criteria will be included. General study procedures are described in the following.
Actionable genomic alterations and drug selection
Potentially eligible patients must have at least one of the actionable genomic alterations (somatic or germline) listed in Table 4: ProTarget Drugs and Acceptable Molecular Alterations.
identified in their tumor and no variants conferring resistance to the relevant targeted anticancer therapy. The genomic alteration may be identified by any tumor genomic test or immunohistochemistry test performed on any type of tumor specimen (fresh frozen, RNAlater-preserved, formalin fixed paraffin embedded (FFPE)) or on ctDNA obtained from plasma (liquid biopsy) in a laboratory accredited by the competent local regulatory authority. Central confirmation of the actionable genomic alteration is performed retrospectively but is not a requisite for initiation of treatment. If more than one actionable genomic alteration is identified, the drug with the higher level of evidence supporting its use is preferred [19]. If several drugs with similar mode of action are available (i.e., PD-1 or PD-L1 inhibitors) randomization is performed by the trial coordinating team using Research Electronic Data Capture (REDCap v. 10.6.18, Vanderbilt University). Stratification factors include investigator site and performance status. REDCap is a secure, web-based software application building managing data for research studies [25, 26]. It is hosted at the Capital Region, Denmark.
Study drugs, treatment assignment and plan.
As of August 2022, 13 drugs are available in ProTarget and administered as monotherapy, unless otherwise indicated. Alectinib (Alecensa®), atezolizumab (Tecentriq®), erlotinib (Tarceva®), cobimetinib (Cotellic®) and vemurafenib (Zelboraf®) in combination, trastuzumab (Herceptin®) and pertuzumab (Perjeta®) in combination, trastuzumab emtansine (Kadcyla®), and vismodegib (Erivedge®) are supplied by Roche. Avelumab (Bavencio®) is supplied by Pfizer, as part of an alliance between Pfizer and Merck (CrossRef Funder ID: 10.13039/100009945), and axitinib (Inlyta®) is supplied by Pfizer. Niraparib (Zejula®) is supplied by GSK. Pemigatinib (Pemazyre®) is supplied by Incyte.
The study drugs are administered in cycles of 21–28 days. Patients are followed according to standard of care, unless otherwise specified in the drug-specific study manual. Initial drug dose and schedule, dose modifications, and management of treatment-related toxicities are performed according to the FDA and/or EMA approved label. All patients are followed for protocol-specified toxicity and efficacy outcomes including tumor response, progression-free survival and overall survival as well as duration of treatment. Treatment-related adverse events are graded according to Common Terminology Criteria for Adverse Events (CTCAE) version 5.0 and followed up until 1 month after the last administration of study drug. For all orally formulated drugs pts. complete dosing diaries and drug accountability is performed after each cycle. All pts. are treated free-of-charge, costs for transportation and accommodation are covered by the Danish healthcare system.
Response assessment and treatment duration
Response evaluation is performed every 8 weeks for the first 24 weeks and subsequently every 12 weeks until disease progression or treatment discontinuation. For patients with solid tumors other than glioblastoma response will be evaluated using the revised Response Evaluation Criteria in Solid Tumors (RECIST) guideline v 1.1 [27] and/or GCIG criteria [28] in case of CA125-based evaluation of patients with ovarian cancer and/or PCWG3 criteria for prostate cancer patients [29]. Bone-only breast cancer pts. will be evaluated using the MDA criteria [30, 31]. Being a non-randomized trial with objective response or non-progression as primary endpoint, confirmation of PR and CR is required ≥30 days after the first documentation of PR or CR, and documentation of non-progression per relevant diagnostic criteria for ≥16 weeks is required ≥2x and ≥ 28 days apart. For patients with multiple myeloma or B cell non-Hodgkin lymphoma, IMWG response criteria [32, 33] and CHESON/Lugano guidelines [34, 35] will be used, respectively. For glioblastoma patients, Response Assessment in Neuro-Oncology (RANO) criteria will be used [36]. Response is assessed by the local investigator. Study treatment will continue until unacceptable toxicity, PD, death, pregnancy, consent withdrawal or withdrawal at the discretion of the investigator. For patients treated with immunotherapy, treatment beyond radiographic progression is permitted provided the patient experiences clinical benefit, assessed by the local investigator.
Cohort definition and design
Each cohort is defined by the chosen study drug, relevant genomic alteration, and histologic tumor type (e.g., atezolizumab/TMB-high/prostate cancer). The ‘genomic alteration’ category is defined at gene level i.e., mutation, deletion, or amplification, e.g., ERBB2-mutation. Each cohort is monitored using a Simon-like two-stage ‘admissible’ monitoring plan to identify cohorts with evidence of activity [37, 38]. In short, eight participants are enrolled in stage one. If ≥1 patient achieves response on treatment (defined as ‘response’ per applicable criteria, or as stable disease for at least 16 weeks measured ≥2x and ≥ 28 days apart), an additional 16 participants are included in stage two, otherwise the cohort is permanently closed. If ≥5 out of 24 participants in stage two achieves response, further investigation of the drug-variant-tumor type combination is warranted. Response among ≤4 out of 24 participants will indicate lack of effect and the cohort is permanently closed. Patients are evaluable if they have received at least 1 cycle of oral drug or 2 administrations of IV drug, and if response is radiologically or clinically evaluable. Non-evaluable patients will be replaced.
Clinical data
Data are captured on electronic Case Report Forms (eCRFs) using REDCap with capture of adverse events pr. CTCAE v. 5.0, vital signs and physical examination at the beginning of every treatment cycle and at end of treatment.
Biological samples
Fresh tumor biopsies are preferably either 18G core-needle biopsies (3 samples, specimen length 22 mm) or surgical resection samples. Two samples are stored in RNAlater (Life Technologies) to determine DNA aberrations and changes in RNA expression during treatment, and one sample is formalin-fixed and paraffin-embedded (FFPE) for histopathologic analyses (including standard biomarker analyses where applicable). 7 mL EDTA whole blood is collected during screening to determine background DNA variation and the presence of germline variants. Liquid biopsies (ctDNA) are collected in every cycle as peripheral blood in BCT tubes (Streck Laboratories, Omaha, NE, USA) as previously described [39]. Biopsies and blood samples will be stored in a research biobank for up to 5 years after the end of the ProTarget study and then destroyed.
Sample analysis
Fresh tumor biopsies are analyzed by whole genome sequencing (WGS), RNA sequencing, and CNV-analysis (Illumina PCR-free, minimum 60x coverage, and Cytoscan) to identify genomic and phenotypic changes in the cancer cells during treatment as previously described [40, 41].
Statistical considerations
Each cohort is monitored using a Simon-like two-stage ‘admissible’ monitoring plan. Admissible designs lie between MiniMax and Optimal designs and have good characteristics of both (i.e., small maximum sample size, and low expected sample size under the null hypothesis of low activity). A true response rate (defined as CR/PR or SD at 16 weeks) of less than 10% will be considered of no clinical interest. A response rate of 30%, although not comprehensively reflecting efficacy [42] or more will be considered of sufficient interest to warrant further study in a confirmatory trial, as outlined in DRUP and TAPUR trials. This monitoring plan has 85% power and an alpha error rate of 7.8%. These operating characteristics were selected to represent a reasonable compromise between high power, low false positive rates, and desire for small sample sizes, especially in stage one.
Ethic considerations and dissemination
The study is conducted according to the international standards of ICH/Good Clinical Practice, monitored by the independent Danish GCP Units, and in full conformance with the “Declaration of Helsinki” and the Danish laws and regulations. The Protocol is approved by the Danish Ethics Committee (H-19089780; date of approval: 19-JUN-2020), the Danish Data Protection Agency (P-2020-210; 03-MAR-2020) and the Danish Medicines Agency (EudraCT 2019–004771-40; 17-FEB-2020).
All patients are informed about genetic findings revealed by genomic analysis and their potential consequences. In case of incidental findings with potential serious consequences for either the patient or the patient’s family, the patient will be offered referral for genetic counseling. Furthermore, the patients are informed that personal study-related data will be used by the Sponsor in accordance with the General Data Protection Regulation, the Data Protection Act and the Health Act. All patients are assigned a unique study ID to maintain patient confidentiality, if/when data is pooled with data from collaborating studies. In accordance with Danish law, research subjects are covered by Danish health care liability insurance.
Protocol amendments and modifications will be submitted for approval to the competent authorities and all relevant collaborators (e.g., sites, pharmacies, monitors, and funders) will be informed by the trial coordinating team. Results will be published in international and peer-reviewed scientific journals and presented at international conferences. Positive, negative as well as inconclusive results will be published. Designation of authorships will be based on the criteria of the Vancouver Convention (ICMJE).
Upon the completion of a cohort, individual and/or pooled cohort results will be published. General study results (such as overall submission, accrual, toxicity and efficacy analyses, as well as concordance between historic and pre-treatment genomic tumor profiles) will be published when appropriate. Individual case reports will only be published if clinically relevant. Publications will be prepared according to the Reporting Recommendations for Tumour Marker Prognostic Studies (REMARK [43]) and Standards for Reporting of Diagnostic Accuracy Studies (STARD [44]) guidelines.
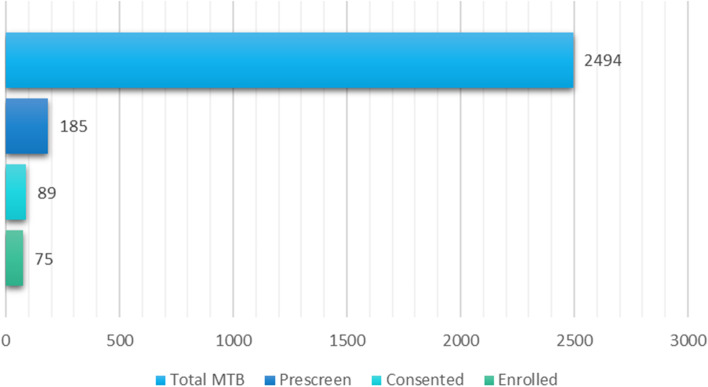
Study status
From study start 24-AUG-2020 to cut-off date 01-JUL-2022, 185 pts. of 2.494 pts. evaluated at DN-MTB have been pre-screened and found potentially eligible for ProTarget (Fig. 2). Of these 185 potentially eligible pts., 89 pts. have signed informed consent to participate, 75 pts. have been enrolled in treatment while 14 pts. failed screening. Fifty-four cohorts have been opened.
Fig. 2.
Patient enrollment status in ProTarget. The figure illustrates the number of molecular tumor profiles (Total MTB) assessed at the DN-MTB, and for ProTarget: the number of pre-screened pts. (Prescreen), number of pts. with informed consent (Consented), and number of pts. enrolled in treatment from study start 24-AUG-2020 to cut-off date 01-JUL-2022
Collaboration
The present trial design will result in a number of cohorts consisting of rare combinations of genomic alterations and tumor types which will be difficult to complete. To accommodate this challenge and ensure that all cohorts will provide conclusive data, the protocol has been developed with a similar design as the DRUP (NCT02925234) and TAPUR (NCT02693535) trials and the Nordic Precision Cancer Medicine Trial Network [45] has been established. The Nordic Network is established between DRUP, and the Nordic trials: ProTarget (NCT04341181), IMPRESS-Norway (NCT04817956), MEGALiT (Sweden, NCT04185831) and FINPROVE (Finland, NCT05159245) with the aim of merging data for specific cohorts in common. The network is focusing on further aligning objectives, endpoints and eCRFs to facilitate data aggregation, which will be based on generally accepted principles and involve relevant pseudonymized data and clinical outcomes. Data sharing will comply with applicable legislation, ethical approvals as well as Data Sharing Agreements and scientific publications based on joint cohorts will be discussed for each cohort and coordinated by the Data Sharing Committee.
Discussion and potential limitations
The ProTarget trial matches patients with non-curable malignant disease, harboring actionable genomic alterations, with relevant targeted drugs. The trial aims to evaluate the efficacy and safety of currently 13 EMA-approved targeted drugs, and study mechanisms of resistance using paired and serial tissue and liquid biopsies.
The ProTarget trial builds on the experiences and designs from matched therapy trials like DRUP and TAPUR and will provide data for the growing network of similar trials. Unique for the ProTarget trial is the extensive genomic profiling provided by the repeat biopsy design. By analyzing the extensive molecular data pre-, on-, and post-treatment, new insights can be gained in molecular pathways involved in e.g., intrinsic and acquired resistance to targeted anti-cancer therapies, clonal evolution, and predictive factors.
The DN-MTB plays a pivotal role in ProTarget; this national, multidisciplinary collaboration is attended by oncologists, molecular biologists, bioinformaticians, pathologists, and clinical geneticists from eight centers across Denmark covering 5.7 million inhabitants. It provides an opportunity for multidisciplinary evaluation and discussion of each case with regards to actionable genomic alterations, strong (dominant) onco-drivers, and potential resistance mutations, combined with the clinical history, histopathology, and patient status. The DN-MTB reviews approximately 1200 genomic profiles annually, mainly WGS/WES and large NGS panels. Thus, the DN-MTB ensures thorough and multidisciplinary pre-screening of each candidate before inclusion in the trial.
Patients are identified by local testing by any method in any type of tissue or blood sample for rapid, broad pre-screening of potential candidates. However, if data are derived from small NGS panels or IHC testing, treatment decisions may be made on potentially incomplete data. Furthermore, the tumor may have developed new oncogenic drivers or resistance mechanisms after the initial testing. To address these issues, fresh tumor biopsies are taken at baseline, analyzed by WGS, and presented at the DN-MTB to ensure that the genomic alteration is still present and relevant for targeted treatment. Treatment initiation and continuation decisions are not per se dependent on these protocol specific tests but may help guide treatment decisions for the individual patient and improve understanding of drug efficacy.
Each cohort is defined by study drug, the actionable genomic alteration, and the histologic tumor type; a design that eventually will give rise to cohorts of rare variant/tumor type-combinations difficult to accrue the required initial 8 subjects. To accommodate this issue, the protocol has been designed in similar to the DRUP and TAPUR trials with a European data sharing agreement for sharing such cohorts.
Tissue and liquid biopsies are collected for translational research for deeper understanding of targeted therapy resistance when applying large scale molecular profiling. The large amount of molecular data combined with the prospective clinical data will enable future research projects in a variety of fields including providing data for inductive hypothesis generation. Liquid biopsies are still in development as a tool for monitoring tumor progression, treatment response and in screening for therapeutic resistance in the individual patient. By collecting paired and serial tissue and liquid biopsies, this trial aims to investigate this modality further and combine findings with the solid tissue WGS and prospectively acquired clinical data. Monitoring of mean variant allele frequencies of selected variants across tumor types during treatment as a surrogate for PFS or response rate is currently being investigated in early clinical trials [46]. Large scale prospective studies comparing ctDNA and tumor tissue DNA before and during targeted treatment are few [47] but hold potential for using ctDNA for diagnostic and prognostic purposes.
In conclusion, data from this trial can potentially identify new matches between targeted treatments and actionable tumor genomic alterations, provide evidence for non-efficacy for other tumor type/genomic alterations and identify potential safety issues in marketed targeted therapies. New insights can be provided using whole genome data for exploratory analysis of determinants of efficacy and resistance.
Acknowledgements
We would like to thank patients and their relatives for participating in the trial. We thank our collaborators in DRUP and TAPUR for sharing their protocol and valuable experience, and for their contributions on trial- and cohort-design. Roche, Pfizer and Merck (CrossRef Funder ID: 10.13039/100009945), GSK, and Incyte reviewed this manuscript for medical accuracy only before journal submission. The authors are fully responsible for the content of this manuscript, and the views and opinions described in the publication reflect solely those of the authors.
Authors’ contributions
Study design: UL, KR, BEL, ML, JG, RLE, KHH, LHJ. Patient inclusion and follow-up: UL, KR, MH, IS, BEL, ML, CAH, JG, LS, RLE, KHH, ARK, LHJ, MIH, THO. Data management, analysis, and statistics: LH, TK, CWY, LBA, LB. Trial coordinating centre, Rigshospitalet: UL, TK, LH. Drafting of manuscript: TK and MH. All coauthors read and approved the final manuscript.
Funding
The ProTarget trial is funded by the Danish Cancer Society (R270-A15507). Drugs and funding for data handling are supplied by Roche, Pfizer (for avelumab, as part of an alliance between Pfizer and Merck [CrossRef Funder ID: 10.13039/100009945]), GSK, and Incyte. ProTarget is an Investigator-Initiated Trial; none of the funders participated in study design or conduct, but eligible molecular alterations as defined in Table 4 were agreed upon between sponsor and each pharmaceutical company.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
The Protocol is approved by the Danish Ethics Committee (H-19089780; date of first approval: 19-JUN-2020) and is conducted in full conformance with the “Declaration of Helsinki” and the Danish laws and regulations. Written informed consent was obtained from all the patients.
Consent for publication
Not applicable.
Competing interests
CAH: Within the last two years CAH has received honoraria for lectures from GSK and BMS.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311(19):1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagle N, Grabiner BC, Van Allen EM, Hodis E, Jacobus S, Supko JG, et al. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov. 2014;4(5):546–553. doi: 10.1158/2159-8290.CD-13-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peters S, Michielin O, Zimmermann S. Dramatic response induced by vemurafenib in a BRAF V600E-mutated lung adenocarcinoma. J Clin Oncol Off J Am Soc Clin Oncol. 2013;31(20):e341–e344. doi: 10.1200/JCO.2012.47.6143. [DOI] [PubMed] [Google Scholar]
- 4.Munoz J, Schlette E, Kurzrock R. Rapid response to vemurafenib in a heavily pretreated patient with hairy cell leukemia and a BRAF mutation. J Clin Oncol Off J Am Soc Clin Oncol. 2013;31(20):e351–e352. doi: 10.1200/JCO.2012.45.7739. [DOI] [PubMed] [Google Scholar]
- 5.Iyer G, Al-Ahmadie H, Schultz N, Hanrahan AJ, Ostrovnaya I, Balar AV, et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2013;31(25):3133–3140. doi: 10.1200/JCO.2012.46.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobain EF, Robinson DR, Wu Y-M, Worden FP, Smith DC, Schuetze S, et al. Clinical impact of high-throughput sequencing in patients with advanced cancer: lessons learned from the Michigan oncology sequencing center. J Clin Oncol. 2015;33(15_suppl):11057. doi: 10.1200/jco.2015.33.15_suppl.11057. [DOI] [Google Scholar]
- 7.Tsimberidou AM, Iskander NG, Hong DS, Wheler JJ, Falchook GS, Fu S, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res. 2012;18(22):6373–6383. doi: 10.1158/1078-0432.CCR-12-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macconaill LE, Garcia E, Shivdasani P, Ducar M, Adusumilli R, Breneiser M, et al. Prospective enterprise-level molecular genotyping of a cohort of cancer patients. J Mol Diagnostics. 2014;16(6):660–672. doi: 10.1016/j.jmoldx.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsimberidou A-M, Wen S, Hong DS, Wheler JJ, Falchook GS, Fu S, et al. Personalized medicine for patients with advanced cancer in the phase I program at MD Anderson: validation and landmark analyses. Clin Cancer Res an Off J Am Assoc Cancer Res. 2014;20(18):4827–4836. doi: 10.1158/1078-0432.CCR-14-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Tourneau C, Delord JP, Gonçalves A, Gavoille C, Dubot C, Isambert N, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16(13):1324–1334. doi: 10.1016/S1470-2045(15)00188-6. [DOI] [PubMed] [Google Scholar]
- 11.Schwaederle M, Zhao M, Lee JJ, Eggermont AM, Schilsky RL, Mendelsohn J, et al. Impact of precision medicine in diverse cancers: a meta-analysis of phase II clinical trials. J Clin Oncol. 2015;33(32):3817–25. [DOI] [PMC free article] [PubMed]
- 12.Massard C, Michiels S, Ferté C, Le Deley MC, Lacroix L, Hollebecque A, et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 2017;7(6):586–595. doi: 10.1158/2159-8290.CD-16-1396. [DOI] [PubMed] [Google Scholar]
- 13.Tuxen IV, Rohrberg KS, Oestrup O, Ahlborn LB, Schmidt AY, Spanggaard I, et al. Copenhagen prospective personalized oncology (COPPO)—clinical utility of using molecular profiling to select patients to phase I trials. Clin Cancer Res. 2019;25(4):1239–1247. doi: 10.1158/1078-0432.CCR-18-1780. [DOI] [PubMed] [Google Scholar]
- 14.Tamborero D, Dienstmann R, Rachid MH, Boekel J, Baird R, Braña I, et al. Support systems to guide clinical decision-making in precision oncology: the Cancer Core Europe molecular tumor board portal. Nat Med. 2020;26:992–4. [DOI] [PubMed]
- 15.Mangat PK, Halabi S, Bruinooge SS, Garrett-Mayer E, Alva A, Janeway KA, et al. Rationale and Design of the Targeted Agent and Profiling Utilization Registry Study. JCO Precis Oncol. 2018;2:1–14. doi: 10.1200/PO.18.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voest EE, van der Velden DL, Hoes L, Van Der Wijngaart H, Van Berge HM, Van Werkhoven E, et al. Drug rediscovery protocol: expanded use of existing anticancer drugs. Ann Oncol. 2019;30:v864–v865. doi: 10.1093/annonc/mdz394.016. [DOI] [PubMed] [Google Scholar]
- 17.Skamene T, Siu LL, Renouf DJ, Laskin JJ, Bedard PL, Jones SJM, et al. Canadian profiling and targeted agent utilization trial (CAPTUR/PM.1): a phase II basket precision medicine trial. J Clin Oncol. 2018;36(15_suppl).
- 18.Chan A-W, Tetzlaff JM, Altman DG, Laupacis A, Gøtzsche PC, Krleža-Jerić K, et al. SPIRIT 2013 statement: defining standard protocol items for clinical trials. Ann Intern Med. 2013;158(3):200–207. doi: 10.7326/0003-4819-158-3-201302050-00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meric-Bernstam F, Johnson A, Holla V, Bailey AM, Brusco L, Chen K, et al. A decision support framework for genomically informed investigational cancer therapy. J Natl Cancer Inst. 2015;107(7). [DOI] [PMC free article] [PubMed]
- 20.Jürgensmeier JM, Eder JP, Herbst RS. New strategies in personalized medicine for solid tumors: molecular markers and clinical trial designs. Clin Cancer Res an Off J Am Assoc Cancer Res. 2014;20(17):4425–4435. doi: 10.1158/1078-0432.CCR-13-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MD Anderson Cancer Center. Personalized Cancer Therapy - Knowledge Base for Precision Oncology. Available from: https://pct.mdanderson.org/#/home. Cited 2022 Jul 26.
- 22.Melinda LT, Kirsten MT, Julia R, Bryan H, Gordon BM, Kristin CJ, et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin Cancer Res. 2016;22(15):3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fahrenbach JP, Andrade J, McNally EM. The CO-Regulation Database (CORD): A Tool to Identify Coordinately Expressed Genes. PLoS One. 2014;9(3):e90408. doi: 10.1371/journal.pone.0090408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)-a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O’Neal L, et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform. 2019;95 Available from: https://pubmed.ncbi.nlm.nih.gov/31078660/. Cited 2022 Aug 24. [DOI] [PMC free article] [PubMed]
- 27.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 28.Rustin GJS, Vergote I, Eisenhauer E, Pujade-Lauraine E, Quinn M, Thigpen T, et al. Definitions for response and progression in ovarian cancer clinical trials incorporating recist 1.1 and CA 125 agreed by the gynecological cancer intergroup (GCIG) Int J Gynecol Cancer. 2011;21(2):419–423. doi: 10.1097/IGC.0b013e3182070f17. [DOI] [PubMed] [Google Scholar]
- 29.Scher HI, Morris MJ, Stadler WM, Higano C, Basch E, Fizazi K, et al. Trial design and objectives for castration-resistant prostate cancer: updated recommendations from the prostate cancer clinical trials working group 3. J Clin Oncol. 2016;34(12):1402–1418. doi: 10.1200/JCO.2015.64.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costelloe CM, Chuang HH, Madewell JE, Ueno NT. Cancer response criteria and bone metastases: RECIST 1.1, MDA and PERCIST. J Cancer. 2010;1(1):80–92. doi: 10.7150/jca.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamaoka T, Madewell JE, Podoloff DA, Hortobagyi GN, Ueno NT. Bone imaging in metastatic breast cancer. J Clin Oncol. 2004;22(14):2942–2953. doi: 10.1200/JCO.2004.08.181. [DOI] [PubMed] [Google Scholar]
- 32.Cheson BD, Fisher RI, Barrington SF, Cavalli F, Schwartz LH, Zucca E, et al. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J Clin Oncol Off J Am Soc Clin Oncol. 2014;32(27):3059–3068. doi: 10.1200/JCO.2013.54.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrington SF, Mikhaeel NG, Kostakoglu L, Meignan M, Hutchings M, Müeller SP, et al. Role of imaging in the staging and response assessment of lymphoma: consensus of the international conference on malignant lymphomas imaging working group. J Clin Oncol Off J Am Soc Clin Oncol. 2014;32(27):3048–3058. doi: 10.1200/JCO.2013.53.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durie BGM, Harousseau J-L, Miguel JS, Bladé J, Barlogie B, Anderson K, et al. International uniform response criteria for multiple myeloma. Leukemia. 2006;20(9):1467–1473. doi: 10.1038/sj.leu.2404284. [DOI] [PubMed] [Google Scholar]
- 35.Durie BGM, Harousseau J-L, Miguel JS, Bladé J, Barlogie B, Anderson K, et al. Erratum: international uniform response criteria for multiple myeloma. Leukemia. 2007;21(5):1134. doi: 10.1038/sj.leu.2404582. [DOI] [PubMed] [Google Scholar]
- 36.Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol Off J Am Soc Clin Oncol. 2010;28(11):1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 37.Simon R. Optimal two-stage designs for phase II clinical trials. Control Clin Trials. 1989;10(1):1–10. doi: 10.1016/0197-2456(89)90015-9. [DOI] [PubMed] [Google Scholar]
- 38.van der Velden DL, Hoes LR, van der Wijngaart H, van Berge Henegouwen JM, van Werkhoven E, Roepman P, et al. The drug rediscovery protocol facilitates the expanded use of existing anticancer drugs. Nature. 2019;574(7776):127–131. doi: 10.1038/s41586-019-1600-x. [DOI] [PubMed] [Google Scholar]
- 39.Ahlborn LB, Tuxen IV, Mouliere F, Kinalis S, Schmidt AY, Rohrberg KS, et al. Circulating tumor DNA as a marker of treatment response in BRAF V600E mutated non-melanoma solid tumors. Oncotarget. 2018;9(66):32570–32579. doi: 10.18632/oncotarget.25948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossing M, Sørensen CS, Ejlertsen B, Nielsen FC. Whole genome sequencing of breast cancer. Apmis. 2019;127(5):303. doi: 10.1111/apm.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabrielaite M, Torp MH, Rasmussen MS, Andreu-Sánchez S, Vieira FG, Pedersen CB, et al. A Comparison of Tools for Copy-Number Variation Detection in Germline Whole Exome and Whole Genome Sequencing Data. Cancers (Basel). 202;13(24) Available from: https://pubmed.ncbi.nlm.nih.gov/34944901/. Cited 2022 May 22. [DOI] [PMC free article] [PubMed]
- 42.Cherny NI, Dafni U, Bogaerts J, Latino NJ, Pentheroudakis G, Douillard JY, et al. ESMO-magnitude of clinical benefit scale version 1.1. Ann Oncol. 2017;28(10):2340–2366. doi: 10.1093/annonc/mdx310. [DOI] [PubMed] [Google Scholar]
- 43.Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): Explanation and Elaboration. PLOS Med. 2012;9(5):e1001216. doi: 10.1371/journal.pmed.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen JF, Korevaar DA, Altman DG, Bruns DE, Gatsonis CA, Hooft L, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open. 2016;6(11):e012799. doi: 10.1136/bmjopen-2016-012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dutch-Nordic Alliance for Precision Cancer Medicine launched | Netherlands Cancer Institute. Available from: https://www.nki.nl/news-events/news/dutch-nordic-alliance-for-precision-cancer-medicine-launched/. Cited 2022 Jul 12.
- 46.Rosen E, Silverman IM, Fontana E, Lee EK, Spigel DR, Højgaard M, et al. Circulating tumor DNA (ctDNA) determinants of improved outcomes in patients (pts) with advanced solid tumors receiving the ataxia telangiectasia and Rad3-related inhibitor (ATRi), RP-3500, in the phase 1/2a TRESR trial (NCT04497116). J Clin Oncol. 2022;40(16_suppl):3082. 10.1200/JCO.2022.40.16_suppl.3082.
- 47.Rothwell DG, Ayub M, Cook N, Thistlethwaite F, Carter L, Dean E, et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. 2019;25(5):738–743. doi: 10.1038/s41591-019-0380-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.