Abstract
Plesiomonas shigelloides is a gram-negative pathogen which can utilize heme as an iron source. In previous work, P. shigelloides genes which permitted heme iron utilization in a laboratory strain of Escherichia coli were isolated. In the present study, the cloned P. shigelloides sequences were found to encode ten potential heme utilization proteins: HugA, the putative heme receptor; TonB and ExbBD; HugB, the putative periplasmic binding protein; HugCD, the putative inner membrane permease; and the proteins HugW, HugX, and HugZ. Three of the genes, hugA, hugZ, and tonB, contain a Fur box in their putative promoters, indicating that the genes may be iron regulated. When the P. shigelloides genes were tested in E. coli K-12 or in a heme iron utilization mutant of P. shigelloides, hugA, the TonB system genes, and hugW, hugX, or hugZ were required for heme iron utilization. When the genes were tested in a hemA entB mutant of E. coli, hugWXZ were not required for utilization of heme as a porphyrin source, but their absence resulted in heme toxicity when the strains were grown in media containing heme as an iron source. hugA could replace the Vibrio cholerae hutA in a heme iron utilization assay, and V. cholerae hutA could complement a P. shigelloides heme utilization mutant, suggesting that HugA is the heme receptor. Our analyses of the TonB system of P. shigelloides indicated that it could function in tonB mutants of both E. coli and V. cholerae and that it was similar to the V. cholerae TonB1 system in the amino acid sequence of the proteins and in the ability of the system to function in high-salt medium.
Plesiomonas shigelloides is a gram-negative bacterium associated with diarrheal disease in humans (4). The organism has been reported to cause several types of gastroenteritis, including acute secretory gastroenteritis (33), an invasive shigellosis-like disease (35), and a cholera-like illness (55). Extraintestinal infections, such as meningitis, bacteremia (2), and pseudoappendicitis (13), are also associated with P. shigelloides infection.
Many bacterial pathogens have iron transport systems that play a critical role in allowing the organism to establish an infection (for reviews, see references 31 and 41). Several bacterial pathogens, including P. shigelloides (9), obtain iron from heme or heme-containing compounds. Heme iron utilization systems have been examined in many gram-negative pathogens, including Vibrio cholerae (20, 21, 38), Vibrio vulnificus (30), Shigella dysenteriae (37, 60), Escherichia coli O157:H7 (54), Serratia marcescens (16, 26, 27), yersiniae (22, 50, 51, 53), Haemophilus (8, 11, 24, 32), neisseriae (7, 29, 52, 64), Pseudomonas (23, 28, 39), and Porphyromonas gingivalis (47). Many heme iron utilization systems studied to date require an outer membrane receptor which binds heme from the environment (for a review, see reference 58). TonB, with the aid of ExbBD, is thought to interact with the receptor to allow movement of heme into the periplasm. A periplasmic binding protein then moves the heme across the periplasm, and inner membrane permeases transport the heme into the cytoplasm.
A question yet to be resolved is what happens to the heme once it enters the cytoplasm of the cell. It is clear that heme iron is utilized as an iron source, but it is not clear how the heme is broken down. Recently, genes have been identified in the pathogenic neisseriae (64) and in the gram-positive pathogen Corynebacterium diphtheriae (45) that encode heme oxygenases which may break down the heme, releasing the iron into the cell. The heme oxygenase genes are required for heme iron utilization in both organisms. Whether other heme iron-utilizing bacteria use a similar mechanism to remove the iron from heme has not been determined.
In a previous study, P. shigelloides heme iron utilization genes were isolated (9). The goal of the present study was to further characterize the P. shigelloides system by examining the genes required for heme iron utilization and determining the functions of the proteins they encode. Our results indicate that the heme utilization system of P. shigelloides is most similar to that of V. cholerae in terms of the amino acid sequence and function of many of the proteins, the regulation of some of the genes, and in the linkage of TonB system genes to the heme iron utilization locus. In addition, we have identified a set of genes, one or more of which is required for heme iron utilization but not utilization of heme as a porphyrin source.
MATERIALS AND METHODS
Strains.
Bacterial strains, plasmids, and their sources are listed in Table 1. DPH-2 was a spontaneous heme utilization mutant isolated from P. shigelloides 9 after nalidixic acid enrichment as previously described (20). DHE-1, a hemA entB mutant of E. coli, was created by P1 transduction of a lysate from the entB mutant E. coli AB1515.24 (48) into the hemA mutant RK1065L.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| P. shigelloides | ||
| P. shigelloides 9 | Type strain | 9 |
| DPH-2 | Spontaneous heme iron utilization mutant of P. shigelloides 9 | This study |
| V. cholerae | ||
| CA401 | Wild type; classical strain | 15 |
| CA40130 | Vibriobactin synthesis mutant of CA401 | 17 |
| DHH-11 | TonB− mutant of CA40130 | 20 |
| E. coli | ||
| 1017 | entf::Tn5 mutant of HB101; F− Δ(gpt-proA)62 leuB6 glnV44 ara-14 galK2 lacY1 Δ(mcrC-mrr) rpsL20 Strrxyl-5 mtl-1 recA13 | S. M. Payne |
| RK1065L | hemA | R. Kadner |
| DHE-1 | RK1065L entB | This study |
| DH5α | F−endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA)U169 (φ80ΔlacZ M15) λ− | 18 |
| KP1032 | tonB::Kan mutant of W3110 F− IN(rrnD-rrnE)1 | 25 |
| Plasmids | ||
| pHPS1 | Ampr; 30-kb (partial) Sau3A fragment of P. shigelloides 9 DNA cloned into pJB8; carries hugAWXZ, tonB, exbBD | 9 |
| pHPS6 | Ampr; 48-kb (partial) Sau3A fragment of P. shigelloides 9 DNA cloned into pJB8; hugBCD | P. Daskaleros |
| pHUG1 | Cmr; pHUG2 without the 1.2-kb EcoRV-HindIII fragment; contains P. shigelloides tonB | This study |
| pHUG2 | Cmr; contains 2.7-kb AseI-HindIII fragment of pHUG10 cloned into the EcoRV-HindIII site of pACYC184; contains P. shigelloides tonB and exbBD | This study |
| pHUG2.1 | Cmr, Knr; pHUG2 with 1.2 kb-PstI fragment containing kanamycin cassette from pUC4K cloned into PstI site of pHUG2; disrupts P. shigelloides tonB | This study |
| pHUG3 | Tetr; 2.5-kb PstI fragment from pHUG10 cloned into PstI site of pAT153; contains hugA | This study |
| pHUG3.1 | Cbr; 2.5-kb PstI fragment from pHUG3 cloned into PstI site of pWSK29 | This study |
| pHUG3.2 | Tetr; 2.5-kb PstI fragment from pHUG3 cloned into PstI site of pLAFR3 | This study |
| pHUG4 | Ampr; 7.5-kb HindIII-PshAI fragment of pHUG10 cloned into HindIII-SmaI site of pWSK29; contains hugA, hugWXZ, tonB, exbBD | This study |
| pHUG7 | Ampr; 7-kb HindIII-NheI fragment from pHUG10 cloned into HindIII-XbaI site of pWSK29; contains hugA and hugWXZ | This study |
| pHUG8 | Cmr; pHUG10 without the 2.4-kb BsiWI fragment; contains hugA, tonbB, exbBD | This study |
| pHUG10 | Cmr; 10-kb HindIII fragment of pHPS1 cloned into pACYC184; contains hugA, hugWXZ, tonB, exbBD | This study |
| pHUG10.1 | Tetr; 10-kB HindIII fragment of pHUG10 cloned into pLAFR3 | This study |
| pHUG16 | Tetr; 6-kB EcoRI fragment of pHPS6 cloned into pACYC184; contains exbBD, hugBCD | This study |
| pHUGlac1 | Ampr; 2.3-kb BamHI fragment from pHUG10 cloned into pQF50; contains putative hugAW promoter in front of lacZ | This study |
| pTONlac1 | Ampr; 2.3-kb BamHI fragment from pHUG10 cloned into pQF50 in opposite orientation as in pHUGlac1; contains putative tonB promoter in front of lacZ | This study |
| pTEE1 | Cmr; 2.1-kb PCR fragment containing V. cholerae, tonB1, exbB1D1 cloned into pACYC184 | 38 |
| pCOS3 | Tetr; 30-kb partial Sau3A1 fragment of V. cholerae DNA cloned into pLAFR3; contains tonB2, exbB2D2 | 38 |
| pHUT3 | Ampr; 3-kb SalI-HindIII fragment of V. cholerae DNA cloned into pAT153; contains hutA | 20 |
| pHUT10 | Cmr; 10.3-kb HindIII fragment of V. cholerae DNA cloned into pACYC184; contains tonB1, exbB1D1, hutBCD and hutWXZ | 20 |
| pAC1 | Cmr; pACYC184 without the BamHI-SalI fragment containing the tetracycline resistance gene; used to provide antibiotic resistance to DPH-2 for triparental matings | This study |
| pETONBX | Ampr; PCR fragment containing E. coli tonB cloned into pWKS30 | A. Mey |
| pACYC184 | Cmr, Tetr; plasmid vector | 6 |
| pAT153 | Ampr, Tetr; plasmid vector | 56 |
| pWSK29 | Ampr; low-copy-number plasmid vector | 59 |
| pQF50 | Ampr; plasmid vector for construction of promoter lac fusions | 12 |
| pUC4K | Kanr, Ampr; vector containing kanamycin cassette | Pharmacia |
| pLAFR3 | Tetr; mobilizable cosmid cloning vector | 49 |
Media, chemicals, and enzymes.
Bacterial strains were routinely grown at 37°C in Luria (L) broth or on L agar. Ethylenediamine-di-(o-hydroxyphenyl acetic acid) (EDDA), deferrated as described by Rogers (44), was added to L broth or L agar to chelate nonheme iron. Antibiotics and supplements were used in the following concentrations: carbenicillin, 25 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 30 μg/ml (for E. coli) and 10 μg/ml (for P. shigelloides); tetracycline, 12.5 μg/ml (for E. coli) and 4.2 μg/ml (for P. shigelloides); δ-aminolevulinic acid (ALA), 80 μg/ml; and hemin, 7.6 μM.
β-Galactosidase assays.
β-Galactosidase assays were performed on mid-log-phase cultures as described by Miller (36) on E. coli DH5α that had been transformed with pHUGlac1 or pTONlac1. Overnight L broth cultures were washed twice in M9 medium (36) and diluted 1:25 into M9 medium containing 0.3% Casamino Acids deferrated as described by Pugsley and Reeves (43). Low-iron medium contained 300 μg of EDDA per ml, and high-iron medium contained 40 μM FeSO4. Independent experiments were conducted on each strain grown under the same conditions on three separate occasions.
Growth assays.
To detect the utilization of heme as an iron source, overnight cultures of E. coli 1017 containing the indicated plasmid(s) were diluted 1 to 1,000 into L broth or L broth containing 200 μg of EDDA per ml with or without hemin. Absorbance at 600 nm was measured after 15 to 18 h of growth. An absorbance of 1.4 or above was considered a positive result and 0.5 or below a negative result. In bioassays to detect utilization of heme as a porphyrin source, mid-log-phase cultures of DHE-1 were washed with saline and seeded into L agar with kanamycin at 5 × 104 cells/ml. Five microliters of 20-mg/ml ALA or 95 μM heme was spotted onto the media, and zones of growth were measured after 15 h. Bioassays to detect utilization of heme as an iron source in DHE-1 were performed in the same manner, except the cells were seeded into L agar containing ALA and EDDA (300 μg/ml). Other heme iron utilization bioassays were performed in manners similar to those of the three assays described above, with specific details being provided in the appropriate table.
Electroporation and triparental matings.
The electroporation of P. shigelloides was performed as previously described (38). Triparental mating with the mobilizing strain MM294/pRK2013 also was used to transfer recombinant plasmids into P. shigelloides.
DNA sequencing and analysis.
The DNA sequence of both strands of the insert in pHUG10 and other recombinant plasmids was determined with an ABI Prism 377 DNA sequencer from Applied Biosystems and was analyzed with the DNA Strider program (34). The BLAST program of the National Center for Biotechnology Information (1) was used to determine homologies of the deduced amino acid sequences, and MacVector ClustalW was used to determine protein identity and similarity.
Nucleotide sequence accession number.
The nucleotide and amino acid sequences corresponding to this region can be found under GenBank/EMBL accession no. AY008342.
RESULTS
Nucleotide sequence analysis of P. shigelloides heme iron utilization genes.
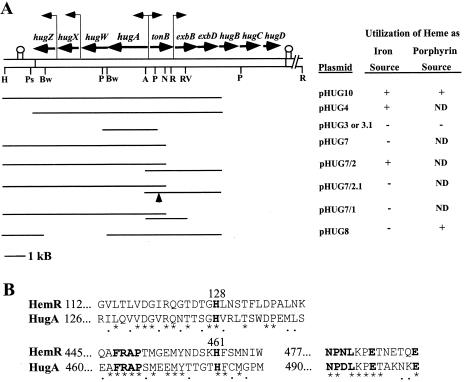
The genetic organization of the P. shigelloides heme iron utilization locus was determined by DNA sequence analysis of pHUG10, a plasmid which enables E. coli 1017 to utilize heme as an iron source, and of pHUG16, which contains hugBCD. Our analyses indicated the presence of 10 open reading frames (ORFs) which spanned approximately 10,000 nucleotides. Figure 1A shows the genetic organization of the region. The calculated molecular weights and pIs for these proteins and the percent identity and similarity of the predicted proteins to selected proteins in other organisms are shown in Table 2.
FIG. 1.
(A) Genetic and restriction enzyme map of cloned DNA from P. shigelloides heme iron utilization locus and identification of sequences required for utilization of heme as an iron source or as a porphyrin source. Genes carried by pHUG10 and pHUG16 are shown with horizontal arrows indicating the direction of transcription. Putative promoters are indicated with vertical bent arrows. Downstream of hugZ and hugD are regions of dyad symmetry indicated with two vertical lines with a circle at the top. The restriction enzyme sites shown are as follows: H, HindIII; Ps, PshA1; Bw, BsiWI; P, PstI; A, AseI; N, NheI; R, EcoRI; RV, EcoRV. Not all of the PstI, EcoRI, EcoRV, and PshAI sites are shown. The heme iron and heme porphyrin assays are described in Materials and Methods. ND, not determined. (B) Amino acid comparison of highly conserved heme receptor sequences in Y. enterocolitica HemR and P. shigelloides HugA. The numbers to the left of the sequences indicate the positions of the first amino acids. An asterisk below the sequence indicates identical amino acid residues and a period indicates conservative substitutions. The bold-faced sequences are those found by Bracken et al. (3) to be present in most heme receptors. Histidines 128 and 461 in HemR are important for the proper function of the protein (3).
TABLE 2.
Proteins with homology to products of P. shigelloides heme utilization locus
| P. shigelloides protein (MW)a (pI)b | Homologue (reference) | Amino acid identity (%) | Amino acid similarity (%) |
|---|---|---|---|
| HugA (71,900) (5.23) (mature) | H. influenzae HxuC (8) | 36 | 50 |
| Y. enterocolitica HemR (50) | 28 | 42 | |
| S. dysenteriae ShuA (37) | 28 | 41 | |
| E. coli O157:H7 ChuA (54) | 28 | 41 | |
| Yersinia pestis HmuR (53) | 28 | 41 | |
| V. cholerae HutA (21) | 25 | 41 | |
| HugW (50,200) (6.50) | V. cholerae HutW (19)c | 51 | 68 |
| V. parahaemolyticus PhuW (40) | 51 | 66 | |
| S. enterica serovar Typhimurium HemN (62) | 21 | 36 | |
| HugX (17,500) (7.36) | Y. pestis hypothetical protein Xd | 38 | 54 |
| V. cholerae HutX (19)e | 37 | 53 | |
| S. dysenteriae ShuX (60) | 34 | 50 | |
| HugZ (21,000) (5.11) | V. cholerae HutZ (19)f | 60 | 76 |
| H. influenzae hypothetical protein HI0854 (14) | 21 | 39 | |
| TonB (30,200) (10.18) | V. cholerae TonB1 (38) | 32 | 48 |
| V. parahaemolyticus TonB (40) | 31 | 42 | |
| P. aeruginosa TonB2 (63) | 26 | 39 | |
| E. coli TonB (42) | 24 | 34 | |
| V. cholerae TonB2 (38) | 20 | 29 | |
| ExbB (29,700) (10.64) | V. cholerae ExbB1 (38) | 41 | 56 |
| V. parahaemolyticus ExbB (40) | 40 | 54 | |
| V. cholerae ExbB2 (38) | 18 | 30 | |
| ExbD (15,700) (4.52) | V. cholerae ExbD1 (38) | 41 | 63 |
| Pseudomonas putida ToIRg | 22 | 46 | |
| V. cholerae ExbD2 (38) | 21 | 31 | |
| HugB (32,800) (8.99) | V. cholerae HutB (38) | 34 | 51 |
| Y. enterocolitica HemT (51) | 30 | 50 | |
| Y. pestis HmuT (53) | 30 | 49 | |
| HugC (36,800) (10.71) | V. cholerae HutC (38) | 52 | 70 |
| P. aeruginosa PhuUh | 50 | 66 | |
| Y. enterocolitica HemU (51) | 45 | 65 | |
| HugD (28,800) (8.45) | V. cholerae HutD (38) | 48 | 68 |
| Y. pestis HmuV (53) | 41 | 60 | |
| Y. enterocolitica HemV (51) | 39 | 60 |
The P. shigelloides hugA gene encodes a protein with a predicted leader sequence of 41 amino acids. HugA has homology with a number of outer membrane heme receptors (Table 2). In addition, HugA contains sequences found by Bracken et al. (3) to be present in most heme receptors but absent in outer membrane receptors not involved in heme uptake. Figure 1B shows a comparision of some of these sequences in Yersinia enterocolitica HemR and P. shigelloides HugA. Two histidines (residues 128 and 461 in HemR) (Fig. 1B) required for HemR receptor function (3) are conserved in HugA. HugA also contains the FRAP and NPNL boxes and two conserved glutamic acids on the carboxy-terminal side of the NPNL box (Fig. 1B). A putative TonB box with the sequence NEVLVTA is located 17 amino acids from the predicted amino terminus of the mature HugA. This sequence is similar to those of the putative TonB boxes in the V. cholerae heme receptor, HutA (DEVVVST) (21), and in the V. vulnificus heme receptor, HupA (DEVVVSA) (30). Among the three TonB boxes, there is identity in four of seven amino acids.
Downstream of hugA are three genes (hugWXZ) which are transcribed in the same direction as hugA (Fig. 1A). The first of these genes, hugW, which overlaps the stop codon of hugA, encodes a protein which shares homology to ORFs in other heme transport loci (Table 2) and has weaker homology with the Salmonella enterica serovar Typhimurium protein HemN, an oxygen-independent form of coproporphyrinogen oxidase. Coproporphyrinogen oxidases convert coproporphyrinogen III into protoporphyrin IX, a late step in heme biosynthesis.
The gene for HugX is 224 nucleotides downstream of hugW. HugX has homology to ORFs linked to heme transport systems in other organisms, including the V. cholerae HutX and the S. dysenteriae ShuX (Table 2), but no homology with ORFs of known function.
HugZ is 165 nucleotides downstream of hugX. HugZ shares homology to HutZ, an ORF linked to the V. cholerae heme utilization locus, and to Haemophilus influenzae hypothetical protein HI0854 (Table 2), neither of which has a known function. A potential stem-loop structure is located 143 nucleotides downstream of hugZ and may serve as a transcriptional terminator.
The other six ORFs are transcribed in the opposite direction of hugAWXZ (Fig. 1A). P. shigelloides TonB shares homology to TonB proteins in other organisms (Table 2). The exbB gene is 67 nucleotides downstream of tonB, and exbD overlaps exbB by 155 nucleotides. P. shigelloides ExbBD proteins share homology with ExbBD proteins in several organisms (Table 2). The start codon for the P. shigelloides hugB overlaps the stop codon for exbD. HugB is similar in amino acid sequence to periplasmic binding proteins found in several bacteria (Table 2). The P. shigelloides hugC begins 13 nucleotides downstream from hugB, and hugD begins 11 nucleotides downstream from hugC. HugCD share homology with a number of inner membrane permeases in other organisms (Table 2), and HugD contains Walker motif A, GPNGTGKS (amino acids 31 to 38), and motif B, LLMLDEPT (amino acids 168 to 175) (57), suggesting that it is the ATPase component of the complex. A potential stem-loop structure is located 56 nucleotides downstream of hugD.
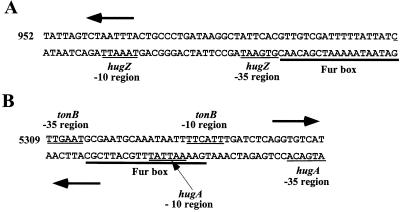
Predicted promoters for heme utilization genes.
Each of the genes in the P. shigelloides heme utilization locus except hugW, exbD, and hugBCD appears to contain its own promoter (Fig. 1A). The predicted promoter for hugZ contains a potential Fur box upstream of the putative −35 region (Fig. 2A). The putative promoters for hugAW and tonB are divergent overlapping promoters and also contain a sequence that resembles a Fur box (Fig. 2B). The putative Fur box, which overlaps the −10 region of each promoter, shares 12 of 19 nucleotides with the consensus sequence of the E. coli Fur box (10) and 13 of 19 nucleotides with the V. cholerae viuA downstream Fur box (5) (data not shown). To assess regulation of the predicted hugAW-tonB promoters, the hugA-tonB intergenic region was cloned in both orientations upstream of the promoterless lacZ gene in pQF50 to create pHUGlac1 and pTONlac1. β-Galactosidase assays were performed on E. coli DH5α containing the indicated plasmids following growth in high- or low-iron medium. When either transformed strain was grown in high-iron medium, the level of β-galactosidase activity was approximately 30 U. When the strains were grown under low-iron conditions, activity increased approximately 19-fold for both strains (577 in DH5α/pHUGlac1 and 558 in DH5α/pTONlac1). These data indicate that promoters in the hugA-tonB intergenic region are iron regulated.
FIG. 2.
Predicted promoters for hugZ, hugAW, and tonB. The DNA sequences of the putative hugZ promoter (A) and of the putative hugAW-tonB promoters (B) are shown. The numbers to the left of the sequences indicate the positions of the first nucleotide in the sequences. The heavy horizontal arrows indicate the direction of transcription. The predicted −10 and −35 regions are underlined and labeled. The putative Fur boxes are indicated with a heavy black line underneath the sequence.
Reconstitution of the P. shigelloides heme iron utilization system in E. coli 1017.
To determine which genes are necessary for heme iron utilization, the Ent− E. coli strain 1017 containing subclones of pHUG10 was tested for growth in L broth containing heme as the sole iron source (Fig. 1A). E. coli 1017/pHUG10 grew well in this medium, indicating that hugBCD are not required for heme iron utilization. E. coli 1017/pHUG4 also grew in media with heme as the iron source, indicating that sequences downstream of hugZ are not required for heme iron utilization. E. coli 1017/pHUG3, containing only hugA, or 1017/pHUG7, containing hugAWXZ, did not utilize heme as an iron source, indicating that the P. shigelloides TonB system genes are required. This was confirmed when 1017 containing both pHUG7 and pHUG2 was able to utilize heme. Heme iron utilization did not occur when 1017/pHUG7 was transformed with pHUG2.1, which contains a kanamycin cassette that disrupts tonB, or when the exbBD genes from pHUG2 were removed (pHUG1). These data indicate that P. shigelloides TonB requires ExbBD to function and that E. coli ExbBD cannot substitute for the P. shigelloides counterparts. To determine if the genes downstream of hugA are required for heme iron utilization, E. coli 1017 transformed with pHUG8, which contains hugA, tonB, and exbBD but is missing hugWXZ, was tested, and the strain failed to utilize heme.
Genes required for utilization of heme as a porphyrin source.
We wanted to determine if the genes needed for utilization of heme as an iron source are also required for utilization of heme that is incorporated in intact form into heme proteins. Bioassays were conducted with DHE-1, a hemA entB mutant of E. coli, which cannot synthesize heme in the absence of the precursor ALA and cannot transport heme to meet the porphyrin requirement. In the assay, the strains were seeded into L agar, and ALA or heme was spotted onto the plates.
DHE-1/pHUG10 grew around the heme spot, indicating that pHUG10 contained the necessary genes for heme to be utilized as a porphyrin source (Fig. 1A). DHE-1 containing pHUG8, which has only hugA, tonB, and exbBD, also could utilize heme as a porphyrin source, suggesting that hugWXZ are not required for utilization of heme as a porphyrin source. The strain transformed with pHUG3.1, which carries hugA, could not utilize heme as a porphyrin source. These data indicate that the heme receptor gene and the TonB system genes are required for the utilization of heme as a porphyrin source.
hugW, -X, or -Z is needed to prevent heme toxicity.
We also seeded the strains into iron-restricted medium containing ALA and supplied heme as an iron source. As expected, E. coli DHE-1/pHUG10 grew well in the assay, exhibiting a zone of growth of approximately 12 mm. In contrast, DHE-1/pHUG8 grew poorly and appeared to be inhibited by the higher concentration of heme present in the center of the spot. The strain exhibited a 14-mm zone of inhibition surrounded by a very faint 3-mm zone of growth. This suggested that the high level of heme was toxic to the cells.
We also determined the effect of several different concentrations of heme on the growth of DHE-1/pHUG8 and found that at higher heme concentrations, the zone of inhibition increased in diameter (data not shown). Because it is possible that growing the strain in media containing both ALA and heme resulted in the accumulation of toxic intermediates in the heme biosynthetic pathway that would not be present when either heme or ALA were present alone, the heme iron assay was performed with no ALA in the L agar. Growth around the heme spots was essentially the same as that observed when ALA was supplied in the L agar (data not shown). Growth inhibition in the presence of heme was also observed in a HemA+ strain (E. coli 1017), indicating that the inhibition is unlikely to be associated with the hemA mutation. These data suggest that hugW, -X, or -Z is necessary to prevent heme toxicity when heme is supplied as an iron source. It is not clear why heme supplied as an iron source, but not as a porphyrin source, is toxic to the cells. This could be due to the fact that when heme was supplied as an iron source, the cells are grown under low-iron conditions, which increases the expression of heme utilization proteins and which may allow higher levels of heme to enter the cytoplasm.
Determination that HugA is a heme receptor.
Our sequencing data indicated that HugA shares homology with a number of heme receptors, including V. cholerae HutA. Thus, we determined if HugA could function as a heme receptor by testing its ability to substitute for V. cholerae HutA. The V. cholerae heme utilization system can be reconstituted in E. coli 1017 by transforming the strain with recombinant plasmids pHUT3, which carries hutA, and pHUT10, which carries tonB1, exbB1D1, hutBCD, and hutWXZ. When E. coli 1017/pHUT3/pHUT10 was tested for growth in L broth with EDDA and heme, the strain utilized heme as an iron source, whereas E. coli 1017/pHUG10 could not, confirming previously published results (20) (Table 3). To determine if HugA can substitute for HutA, we moved pHUG3, which carries the P. shigelloides gene hugA, into 1017/pHUT10 to create a chimeric heme iron utilization system. This strain grew as well as 1017/pHUT3/pHUT10 in L broth with EDDA and heme, indicating that hugA encodes a protein that can replace the function of the V. cholerae HutA.
TABLE 3.
P. shigelloides HugA functions as a heme receptor in E. coli
| E. coli 1017 transformed with plasmid: | Absorbance of
cultures grown ina:
|
||
|---|---|---|---|
| L broth | L broth with EDDA | L broth with EDDA plus heme | |
| pHUT10/pHUT3 (V. cholerae system) | 1.20 | 0.05 | 0.95 |
| pHUT10/pHUG3 (hybrid system with P. shigelloides hugA) | 1.20 | 0.06 | 1.25 |
| pHUT10 (no receptor gene) | 1.02 | 0.05 | 0.08 |
Absorbance at 650 nm was taken after 18 h of growth.
To confirm these results, a heme iron utilization mutant of P. shigelloides 9 was isolated as described in Materials and Methods. The mutant DPH-2 failed to grow with heme or hemoglobin as an iron source (Table 4). DPH-2 was complemented with pHUG10.1, which contains all the P. shigelloides heme iron utilization genes characterized in this paper except hugBCD, or with pHUG3.2, which contains only hugA. The mutant also was complemented with pHUT2, which contains V. cholerae hutA. However, pHUT4, which contains V. cholerae tonB1, exbB1D1, hutBCD, and hutWXZ but not hutA could not utilize heme (Table 4). These data further support our contention that hugA encodes the P. shigelloides heme receptor. The results also indicate that hugA can be added to the list of P. shigelloides genes required for heme iron utilization.
TABLE 4.
Characterization of a P. shigelloides heme utilization mutant
| P. shigelloides strain | Growth with the iron
sourcea:
|
||
|---|---|---|---|
| Heme | Hemoglobin | FeSO4 | |
| P. shigelloides 9 (wild type) | +b | + | + |
| DPH-2 (heme utilization mutant) | −c | − | + |
| DPH-2/pHUG10.1 (contains hugAWXZ, tonB, exbBD) | + | + | + |
| DPH-2/pHUG3.2 (contains hugA) | + | + | + |
| DPH-2/pHUT2 (contains V. cholerae hutA) | + | + | + |
| DPH-2/pHUT4 (contains V. cholerae tonB1, exbB1D1, hutBCD, hutWXZ) | − | − | + |
Strains were seeded into L agar containing 50 μg of EDDA per ml at 105 cells/ml. Five microliters of 40 μM heme, 10 μM hemoglobin, or 10 mM FeSO4 was spotted onto the plates, and the plates were incubated for 15 h.
A plus represents a zone of growth of at least 8 mm around the spot of FeSO4 and 10 mm around the spots of heme and hemoglobin.
A minus indicates no detectable growth.
Complementation of tonB mutants with P. shigelloides TonB system genes.
Because P. shigelloides TonB and ExbBD resemble proteins found in TonB systems from other organisms, we wanted to determine if these proteins function as a TonB system. We moved the P. shigelloides TonB system on pHUG2 or E. coli tonB on pETONBX into KP1032, an E. coli tonB mutant, and performed complementation assays on the strains. The strains were grown in low-iron medium, which tests for the ability to acquire iron via the siderophore enterobactin, a TonB-dependent process. E. coli KP1032/pHUG2 grew as well in low-iron medium as the positive control, KP1032/pETONBX (Table 5), indicating that the P. shigelloides TonB system complements an E. coli tonB mutant. However, when the P. shigelloides tonB gene without exbBD was provided on pHUG1, the strain failed to grow. The fact that the P. shigelloides TonB by itself could not restore function but the entire TonB system could suggests that P. shigelloides TonB cannot function with the E. coli ExbBD proteins but that it can function with the enterobactin receptor in E. coli.
TABLE 5.
Complementation of an E. coli tonB mutant with the P. shigelloides TonB system genes
| E. coli KP1032 transformed with plasmid: | Absorbance of cultures grown
ina:
|
|
|---|---|---|
| L broth | L broth with EDDA (enterobactin utilization) | |
| No plasmid (TonB−) | 1.49 | 0.23 |
| pETONBX (E. coli tonB) | 1.48 | 1.51 |
| pHUG2 (P. shigelloides TonB system) | 1.55 | 1.34 |
| pHUG1 (P. shigelloides tonB) | 1.50 | 0.03 |
Absorbance was taken at 600 nm after 17 h of growth.
We also tested pHUG2 for complementation of DHH-11, a TonB− strain of V. cholerae. DHH-11/pHUG2 could acquire iron from heme and the siderophore vibriobactin, which are TonB-dependent processes, whereas the untransformed strain could not (data not shown).
Salt sensitivity of P. shigelloides TonB.
Seliger et al. (46) reported that high levels of salt diminish the function of V. cholerae TonB2 but have no impact on TonB1 function. This is thought to occur because TonB1 is longer than TonB2 (244 amino acids versus 206 amino acids). Thus, TonB1 can extend across the periplasm and interact with the receptor under high-salt conditions when the periplasm may become larger due to possible shrinkage of the cytoplasm. To determine if P. shigelloides TonB, which is 288 amino acids in length, can also function under high-salt conditions, we moved plasmids containing various TonB systems into E. coli 1017/pHUG7 (contains hugAWXZ). The following TonB systems were moved into the strain: the P. shigelloides TonB system on pHUG2, the V. cholerae TonB1 system on pTEE1, and the V. cholerae TonB2 system on pCos3. All the strains grew well in high-salt L broth, which showed that high levels of salt in iron-rich media did not inhibit growth (Table 6). The strains also grew in low-salt medium with heme as the iron source, which indicated that each of the three TonB proteins could interact with P. shigelloides HugA to allow heme to enter the cell (Table 6). When the strains were tested for their ability to utilize heme as an iron source in high-salt media, substantial growth was observed with E. coli 1017/pHUG7 containing the P. shigelloides TonB system or the V. cholerae TonB1 system but not in the strain containing the V. cholerae TonB2 system. These data suggest that P. shigelloides TonB is like V. cholerae TonB1 in its ability to function under high-salt conditions.
TABLE 6.
Ability of P. shigelloides TonB and V. cholerae TonB1 and TonB2 to function in high-salt medium
| E. coli 1017 transformed with plasmid: | Growth in media
containinga:
|
||||
|---|---|---|---|---|---|
| L broth
|
L broth with EDDA (low salt) | L broth with
EDDA plus heme
|
|||
| Low salt | High salt | Low salt | High salt | ||
| pHUG7/pHUG2 (P. shigelloides TonB) | 1.94 | 1.56 | 0.02 | 1.77 | 1.37 |
| pHUG7/pTEE1 (V. cholerae TonB1) | 2.20 | 1.77 | 0.00 | 1.90 | 1.40 |
| pHUG7/pCos3 (V. cholerae TonB2) | 2.03 | 1.76 | 0.05 | 1.30 | 0.40 |
Strains were grown in L broth containing 170 mM NaCl (low salt) or 625 mM NaCl (high salt) containing no EDDA or containing 200 μg of EDDA per ml. with or without hemin. Absorbance at 600 nm was taken after 18 h of growth.
DISCUSSION
Our analyses of the P. shigelloides heme iron utilization system indicates it contains an outer membrane receptor, HugA, which shares significant homology to other heme iron receptors. HugA also contains several motifs that are present in most heme receptors but are absent in receptors for other ligands. Our data indicate that HugA functions as a heme receptor in that it is interchangeable with the V. cholerae heme receptor in both a hugA mutant of P. shigelloides and in an E. coli strain that contains a hybrid heme iron utilization system consisting of hugA and V. cholerae tonB1, exbB1D1, hutBCD, and hutWXZ. The ability of these two heme receptors to function interchangeably may be due to the similar amino acid sequences of their TonB boxes (NEVLVTA in HugA and DEVVVST in HutA). The TonB box is the region of an outer membrane receptor that is thought to physically interact with the TonB protein. Other regions that the two proteins share in common also may play a role in allowing HugA and HutA to be interchangeable, as the two proteins share significant homology overall (Table 2).
P. shigelloides also has a TonB system which is required for heme iron utilization. The TonB system genes are arranged in the same order as those in the V. cholerae heme iron utilization system (tonB, exbB, exbD) and are linked to other genes specifically required for heme iron utilization. Linkage of a ligand transport system to tonB is unusual. P. shigelloides, V. cholerae (38), and possibly Vibrio parahaemolyticus (40) are the only organisms characterized to date which share this feature. The fact that these three organisms have TonB system genes linked to other heme iron utilization genes suggests that the genes were acquired simultaneously by horizontal gene transfer, and that at least initially these TonB systems were specific for heme iron utilization.
The P. shigelloides TonB and ExbBD sequences most closely resemble their counterparts in the V. cholerae TonB1 system. P. shigelloides TonB also is similar in size to V. cholerae TonB1 and like TonB1 can function in high-salt conditions. However, the P. shigelloides TonB system differs from the V. cholerae TonB1 system in that it can complement an E. coli TonB mutant, whereas the V. cholerae TonB1 system cannot (38). V. cholerae has a second TonB system (38) which, like the P. shigelloides system, complements an E. coli tonB mutant. Thus, it appears that the P. shigelloides TonB system shares features with both TonB systems in V. cholerae.
O'Malley et al. (40) suggested that V. parahaemolyticus and Vibrio alginolyticus have two tonB genes similar to those identified in V. cholerae, and two tonB genes have been identified in Pseudomonas aeruginosa (63). It is not clear if P. shigelloides also has two different tonB genes. No signal was detected in Southern blots of P. shigelloides chromosomal DNA probed with V. cholerae tonB2. This suggests that P. shigelloides does not have a tonB2 homologue but does not rule out the possibility that a second tonB is present that cannot be detected with the V. cholerae tonB2 probe at low stringency.
Many heme iron utilization systems characterized to date have genes encoding periplasmic and inner membrane permeases, which move the heme across the periplasm and into the cytoplasm. In V. cholerae these genes (hutBCD) are located downstream of exbD1. P. shigelloides also has homologues to these genes (hugBCD) which are located downstream of exbD. However, hugBCD are not needed to reconstitute the P. shigelloides heme iron utilization system in E. coli 1017, whereas V. cholerae hutBCD are needed to reconstitute the V. cholerae system in the same E. coli strain (38).
The role of the proteins encoded by P. shigelloides hugWXZ is not clear. Several bacterial heme iron utilization systems contain homologues of one or more of these proteins (Table 2), but functions have yet to be assigned to these proteins. Our results indicate that these three genes are not required for utilization of heme as a porphyrin source but are needed when heme is used as an iron source. When the P. shigelloides heme utilization system is reconstituted in E. coli in the absence of hugWXZ, heme supplied as an iron source is toxic to the cells. Stojiljkovic and Hantke (51) observed a similar result when they reconstituted the Y. enterocolitica heme iron utilization system in E. coli. In those experiments, hemS was required for utilization of heme as an iron source but not for utilization of heme as a porphyrin source, and heme toxicity was observed in the absence of hemS. The researchers hypothesized, but have not confirmed, that HemS breaks down the heme and releases iron into the cytoplasm. One or more of the proteins encoded by hugWXZ could serve a similar function in P. shigelloides, although none were found to share homology with HemS. It is also possible that these proteins may not be involved in breaking down the heme but rather in controlling the level of heme that enters the cytoplasm of the cell. The proteins could also be involved in heme storage, and their absence could result in accumulation of toxic levels of heme in the cytoplasm.
While P. shigelloides and V. cholerae have heme iron utilization systems that are similar to one another, the two organisms appear to be very different in the number of strategies each possesses for acquiring iron in the host. V. cholerae has several methods of acquiring iron from the host. V. cholerae utilizes the siderophores vibriobactin (17), ferrichrome (17), schizokinen (46), and enterobactin (61). P. shigelloides may not exhibit the same versatility. Daskaleros et al. (9) demonstrated that P. shigelloides does not synthesize a siderophore and that it cannot utilize vibriobactin or enterobactin. We conducted additional tests and found that it could not utilize ferrichrome or schizokinen or be crossfed by V. parahaemolyticus, which produces vibrioferrin, or by V. alginolyticus, which produces a yet-to-be-characterized siderophore (data not shown). Thus, the organism can use heme but not siderophores as an iron source. One could speculate that the absence of siderophore uptake systems in P. shigelloides may have a negative impact on its ability to survive in the environment and to infect potential hosts. This may explain why P. shigelloides appears to be less successful than V. cholerae in causing disease in a large number of humans.
ACKNOWLEDGMENTS
This study was supported by Grant Development funds from the University of Texas of the Permian Basin and Alliance for Minority Participation funds from the University of Texas System.
We thank Shelley Payne for her guidance and critical reading of the manuscript, Donald Allen and Douglas Hale for their strong support, and Alexandra Mey for kindly providing recombinant plasmids.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Billiet J, Kuypers S, Van Lierde S, Verhaegen J. Plesiomonas shigelloidesmeningitis and septicaemia in a neonate: report of a case and review of the literature. J Infect. 1989;19:267–271. doi: 10.1016/s0163-4453(89)90809-8. [DOI] [PubMed] [Google Scholar]
- 3.Bracken C S, Baer M T, Abdur-Rashid A, Helms W, Stojiljkovic I. Use of heme-protein complexes by the Yersinia enterocoliticaHemR receptor: histidine residues are essential for receptor function. J Bacteriol. 1999;181:6063–6072. doi: 10.1128/jb.181.19.6063-6072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenden R A, Miller M A, Janda J M. Clinical disease spectrum and pathogenic factors associated with Plesiomonas shigelloidesinfections in humans. Rev Infect Dis. 1988;10:303–316. doi: 10.1093/clinids/10.2.303. [DOI] [PubMed] [Google Scholar]
- 5.Butterton J R, Stoebner J A, Payne S M, Calderwood S B. Cloning, sequencing, and transcriptional regulation of viuA, the gene encoding the ferric vibriobactin receptor of Vibrio cholerae. J Bacteriol. 1992;174:3729–3738. doi: 10.1128/jb.174.11.3729-3738.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C J, Sparling P F, Lewis L A, Dyer D W, Elkins C. Identification and purification of a hemoblogin-binding outer membrane protein from Neisseria gonorrhoeae. Infect Immun. 1996;64:5008–5014. doi: 10.1128/iai.64.12.5008-5014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cope L D, Yogev R, Muller-Eberhard U, Hansen E J. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzaetype b. J Bacteriol. 1995;177:2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daskaleros P A, Stoebner J A, Payne S M. Iron uptake in Plesiomonas shigelloides: cloning of the genes for heme-iron uptake system. Infect Immun. 1991;59:2706–2711. doi: 10.1128/iai.59.8.2706-2711.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elkins C, Chen C-J, Thomas C E. Characterization of the hgbA locus encoding a hemoglobin receptor from Haemophilus ducreyi. Infect Immun. 1995;63:2194–2200. doi: 10.1128/iai.63.6.2194-2200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farinha M A, Kropinski A M. Construction of broad-host- range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol. 1990;172:3496–3499. doi: 10.1128/jb.172.6.3496-3499.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer K, Chakraborty T, Hof H, Kirchner T, Wamsler O. Pseudoappendicitis caused by Plesiomonas shigelloides. J Clin Microbiol. 1988;26:2675–2677. doi: 10.1128/jcm.26.12.2675-2677.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, et al. Whole-genome random sequencing and assembly of Haemophilus influenzaeRd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 15.Gardner E W, Lyles S T, Lankford C E. A comparison of virulence in Vibrio choleraestrains for embryonated egg. J Infect Dis. 1964;114:412. doi: 10.1093/infdis/114.5.412. [DOI] [PubMed] [Google Scholar]
- 16.Ghigo J M, Letoffe S, Wandersman C. A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J Bacteriol. 1997;179:3572–3579. doi: 10.1128/jb.179.11.3572-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths G L, Sigel S P, Payne S M, Neilands J B. Vibriobactin, a siderophore from Vibrio cholerae. J Biol Chem. 1984;259:383–385. [PubMed] [Google Scholar]
- 18.Hanahan D. Studies on transformation of Escherichia coliwith plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 19.Heidelberg J F, Elsen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson D P, Payne S M. Cloning and characterization of the Vibrio choleraegenes encoding the utilization of iron from hemin and hemoglobin. Mol Microbiol. 1993;7:461–469. doi: 10.1111/j.1365-2958.1993.tb01137.x. [DOI] [PubMed] [Google Scholar]
- 21.Henderson D P, Payne S M. Characterization of the Vibrio choleraeouter membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J Bacteriol. 1994;176:3269–3277. doi: 10.1128/jb.176.11.3269-3277.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hornung J M, Jones H A, Perry R D. The hmu locus of Yersinia pestisis essential for utilization of free hemin and heme-protein complexes as iron sources. Mol Microbiol. 1996;20:725–739. doi: 10.1111/j.1365-2958.1996.tb02512.x. [DOI] [PubMed] [Google Scholar]
- 23.Idei A, Kawai E, Akatsuka H, Omori K. Cloning and characterization of the Pseudomonas fluorescensATP-binding cassette exporter, HasDEF, for the heme acquisition protein HasA. J Bacteriol. 2000;181:7545–7551. doi: 10.1128/jb.181.24.7545-7551.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin H, Ren Z, Pozsgay J M, Elkins C, Whitby P W, Morton D J, Stull T L. Cloning of a DNA fragment encoding a heme-repressible hemoglobin-binding outer membrane protein from Haemophilus influenzae. Infect Immun. 1996;64:3134–3141. doi: 10.1128/iai.64.8.3134-3141.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larson R A, Thomas M G, Wood G E, Postle K. Partial suppression of an Escherichia coliTonB transmembrane domain mutation (delta V17) by a missense mutation in ExbB. Mol Microbiol. 1994;13:627–640. doi: 10.1111/j.1365-2958.1994.tb00457.x. [DOI] [PubMed] [Google Scholar]
- 26.Letoffe S, Ghigo J M, Wandersman C. Iron acquisition from heme and hemoglobin by a Serratia marcescensextracellular protein. Proc Natl Acad Sci USA. 1994;91:9876–9880. doi: 10.1073/pnas.91.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letoffe S, Nato F, Goldberg M E, Wandersman C. Interactions of HasA, a bacterial haemophore, with haemoglobin and with its outer membrane receptor HasR. Mol Microbiol. 1999;33:546–555. doi: 10.1046/j.1365-2958.1999.01499.x. [DOI] [PubMed] [Google Scholar]
- 28.Letoffe S, Redeker V, Wandersman C. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescensHasA haemophore. Mol Microbiol. 1998;6:1223–1234. doi: 10.1046/j.1365-2958.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 29.Lewis L A, Gray E, Wang Y-P, Roe B A, Dyer D W. Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon in Neisseria meningitidis. Mol Microbiol. 1997;23:737–749. doi: 10.1046/j.1365-2958.1997.2501619.x. [DOI] [PubMed] [Google Scholar]
- 30.Litwin C M, Byrne B L. Cloning and characterization of an outer membrane protein of Vibrio vulnificusrequired for heme utilization: regulation of expression and determination of the gene sequence. Infect Immun. 1998;66:3134–3141. doi: 10.1128/iai.66.7.3134-3141.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Litwin C M, Calderwood S B. Role of iron in regulation of virulence genes. Clin Microbiol Rev. 1993;6:137–149. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacIver I, Latimer J L, Liem H H, Muller-Eberhard U, Hrkal S, Hansen E J. Identification of an outer membrane protein involved in utilization of hemoglobin-haptoglobin complexes by nontypable Haemophilus influenzae. Infect Immun. 1996;64:3703–3712. doi: 10.1128/iai.64.9.3703-3712.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandal B K, Whale K, Morrison B C. Acute colitis due to Plesiomonas shigelloides. Br Med J. 1982;285:1539–1540. doi: 10.1136/bmj.285.6354.1539-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marck C. ‘DNA Strider’: a ‘C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNeeley D, Ivy P, Craft J C, Cohen I. Plesiomonas: biology of the organism and diseases in children. Pediatr Infect Dis J. 1984;3:176–181. [PubMed] [Google Scholar]
- 36.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 37.Mills M, Payne S M. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coliO157:H7. J Bacteriol. 1995;177:3004–3009. doi: 10.1128/jb.177.11.3004-3009.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Occhino D A, Wyckoff E E, Henderson D P, Wrona T J, Payne S M. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, and exbDgenes. Mol Microbiol. 1998;29:1493–1507. doi: 10.1046/j.1365-2958.1998.01034.x. [DOI] [PubMed] [Google Scholar]
- 39.Ochsner U A, Johnson Z, Vasil M L. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology. 2000;146:185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- 40.O'Malley S M, Mouton S L, Occhino D A, Deanda M T, Rashidi J R, Fusion K L, Rashidi C E, Mora M Y, Payne S M, Henderson D P. Comparison of the heme iron utilization systems of pathogenic vibrios. J Bacteriol. 1999;181:3594–3598. doi: 10.1128/jb.181.11.3594-3598.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Payne S M. Iron and virulence in the family Enterobacteriaceae. Crit Rev Microbiol. 1988;16:81–111. doi: 10.3109/10408418809104468. [DOI] [PubMed] [Google Scholar]
- 42.Postle K, Good R F. DNA sequence of the Escherichia coli tonBgene. Proc Natl Acad Sci USA. 1983;80:5235–5239. doi: 10.1073/pnas.80.17.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pugsley A P, Reeves P. Characterization of group B colicin-resistant mutants of Eshcerichia coliK-12: colicin resistance and the role of enterochelin. J Bacteriol. 1976;127:218–228. doi: 10.1128/jb.127.1.218-228.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogers H J. Iron-binding catechols and virulence in Escherichia coli. Infect Immun. 1973;7:438–444. doi: 10.1128/iai.7.3.438-444.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitt M P. Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J Bacteriol. 1997;179:838–845. doi: 10.1128/jb.179.3.838-845.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seliger, S. S., A. R. Mey, A.-M. Valle, and S. M. Payne. The two TonB systems in Vibrio cholerae: redundant and specific functions. Mol. Microbiol. 39:801–812. [DOI] [PubMed]
- 47.Shi Y, Ratnayake D B, Okamoto K, Abe N, Yamamoto K, Nakayama K. Genetic analysis of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J Biol Chem. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 48.Staab J F, Earhart C F. EntG activity of Escherichia colienterobactin synthetase. J Bacteriol. 1990;11:6403–6410. doi: 10.1128/jb.172.11.6403-6410.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stojiljkovic I, Hantke K. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 1992;11:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stojiljkovic I, Hantke K. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol Microbiol. 1994;13:719–732. doi: 10.1111/j.1365-2958.1994.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 52.Stojiljkovic I, Hwa V, de Martin S L, Nassif X, Heffron F, So M. The Neisseria meningitidishaemoglobin receptor: its role in iron utilization and virulence. Mol Microbiol. 1995;15:531–542. doi: 10.1111/j.1365-2958.1995.tb02266.x. [DOI] [PubMed] [Google Scholar]
- 53.Thompson J M, Jones H A, Perry R D. Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmumutants for hemin and hemoprotein utilization. Infect Immun. 1999;67:3879–3892. doi: 10.1128/iai.67.8.3879-3892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torres A G, Payne S M. Haem iron-transport system in enterohaemorrhagic Escherichia coliO157:H7. Mol Microbiol. 1997;23:825–833. doi: 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]
- 55.Tsukamoto T, Kinoshita Y, Shimada T, Sakazaki R. Two epidemics of diarrhoeal disease possibly caused by Plesiomonas shigelloides. J Hyg. 1978;80:275–280. doi: 10.1017/s0022172400053638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Twigg A J, Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980;283:216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- 57.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;8:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wandersman C, Stojiljkovic I. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr Opin Microbiol. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]
- 59.Wang R F, Kushner S R. Construction of versatile low-copy number vectors for cloning, sequencing, and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 60.Wyckoff E E, Duncan D, Torres A G, Mills M, Maase K, Payne S M. Structure of the Shigella dysenteriaehaem transport locus and its phylogenetic distribution in enteric bacteria. Mol Microbiol. 1998;28:1139–1152. doi: 10.1046/j.1365-2958.1998.00873.x. [DOI] [PubMed] [Google Scholar]
- 61.Wyckoff E E, Valle A-M, Smith S L, Payne S M. A multifunctional ATP-binding cassette transporter system from Vibrio choleraetransports vibriobactin and enterobactin. J Bacteriol. 1999;181:7588–7596. doi: 10.1128/jb.181.24.7588-7596.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu K, Elliott T. Cloning, DNA sequence, and complementation analysis of the Salmonella typhimurium hemNgene encoding a putative oxygen-independent coproporphyrinogen III oxidase. J Bacteriol. 1994;176:3196–3203. doi: 10.1128/jb.176.11.3196-3203.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao Q, Poole K. A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbDgenes. FEMS Microbiol Lett. 2000;184:127–132. doi: 10.1111/j.1574-6968.2000.tb09002.x. [DOI] [PubMed] [Google Scholar]
- 64.Zhu W, Hunt D J, Richardson A R, Stojiljkovic I. Use of heme compounds as iron sources by pathogenic neisseriae requires the product of the hemOgene. J Bacteriol. 2000;182:439–447. doi: 10.1128/jb.182.2.439-447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]