ABSTRACT
Shopping malls offer various niches for microbial populations, potentially serving as sources and reservoirs for the spread of microorganisms of public health concern. However, knowledge about the microbiome and the distribution of human pathogens in malls is largely unknown. Here, we examine the microbial community dynamics and genotypes of potential pathogens from floor and escalator surfaces in shopping malls and adjacent road dusts and greenbelt soils. The distribution pattern of microbial communities is driven primarily by habitats and seasons. A significant enrichment of human-associated microbiota in the indoor environment indicates that human interactions with surfaces might be another strong driver for mall microbiomes. Neutral community models suggest that the microbial community assembly is strongly driven by stochastic processes. Distinct performances of microbial taxonomic signatures for environmental classifications indicate the consistent differences of microbial communities of different seasons/habitats and the strong anthropogenic effect on homogenizing microbial communities of shopping malls. Indoor environments harbored higher concentrations of human pathogens than outdoor samples, also carrying a high proportion of antimicrobial resistance-associated multidrug efflux genes and virulence genes. These findings enhanced the understanding of the microbiome in the built environment and the interactions between humans and the built environment, providing a basis for tracking biothreats and communicable diseases and developing sophisticated early warning systems.
IMPORTANCE Shopping malls are distinct microbial environments which can facilitate a constant transmission of microorganisms of public health concern between humans and the built environment or between human and human. Despite extensive investigation of the natural environmental microbiome, no comprehensive profile of microbial ecology has been reported in malls. Characterizing microbial distribution, potential pathogens, and antimicrobial resistance will enhance our understanding of how these microbial communities are formed, maintained, and transferred and help establish a baseline for biosurveillance of potential public health threats in malls.
KEYWORDS: antimicrobial resistance, built environment, human pathogen, microbial assembly, microbial interactions
INTRODUCTION
The built environment is the collection of all manufactured structures, encompassing transportation systems, commercial facilities, and other human-constructed physical surroundings, and represents important human-modified ecosystems with unique microbial assemblages (1). Humans are exposed to colonized bacteria, fungi, and viruses, possibly altering the trajectory of human health. Physical surfaces of buildings are primary ecological sites for microbial adhesion and biofilm formation, and microbial communities are vastly different between various types of built environments and surfaces (2–4). Building occupants contribute significantly to the mall microbiome by releasing human-associated microbes, transmitting outdoor bioparticles, and resuspending microorganisms on the surfaces (4–7). Environmental factors like temperature, geography, meteorology, urbanization, and pets have been associated with the complexity of the microbiome in the built environment (8, 9). Management practices such as routine cleaning and disinfectant usage could lead to unselective removal and killing of microorganisms, thus markedly changing the adaptation strategies (e.g., antimicrobial resistance) of microorganisms and impacting the composition and assembly of microbial residents (10, 11). Advances in high-throughput sequencing have characterized the microbiome in various built environments, including sewer systems (12, 13), hospitals (8, 10), metro systems (14, 15), households (16, 17), and dormitory environments (7, 18). However, patterns of the distribution and dynamics of microbial communities in shopping malls have not yet been reported.
Shopping malls are large enclosed communal gathering places of urban societies with high occupant diversity, densities, and turnovers which people from all walks of life and with different cultures, ages, or physical conditions visit for entertainment, refreshment, and business. Microbial contamination of such a public area could render malls to be a source and reservoir of infections via close interactions between individuals and surfaces (e.g., handrails, floors, and buttons) (14, 15, 19). The interactions could be mediated by microbial transfer between shoes and floor, the exchange of microbes between skin and handles, and the release of gut-related microbes in washrooms (5, 20–22). Although there is a paucity of evidence to manifest direct transfer of microorganisms from built environments to humans, some investigations have observed the potential transmission of pathogens and antimicrobial resistance to humans through surfaces and equipments (7, 23). Several pioneering studies for the mall microbiome have detected high bacterial densities on tables, trays, and cleaning cloths in food courts (23, 24). Potential pathogens such as Staphylococcus aureus, Pseudomonas spp., and Gram-negative bacilli have been isolated from shopping malls, and more than 50% of samples were detected with bacterial contamination (25, 26). Multidrug-resistant pathogenic species, including Stenotrophomonas, Aeromonas, Acinetobacter, Pseudomonas, and Bacillus, were detected from escalator handrails, lift buttons, and shopping carts by culture-based methods (11, 27). Furthermore, recent advances on the coronavirus pandemic have confirmed that shopping malls were associated with locally transmitted cases of COVID-19 and were identified as one of the superspreading environments for COVID-19 (28–30). These findings suggest that exposure to the mall microbiome might contribute to health burdens of human infectious diseases (15, 31).
Antimicrobial resistance (AMR) is a global health threat that has resulted in high mortality and health care costs. Research on hospitals and metro systems has confirmed that contaminated surfaces or fomites contribute to the spread of bacterial infections and AMR (14, 29, 31, 32). Enterobacteriaceae comprise a number of pathogens, such as Klebsiella, Enterobacter, Escherichia coli, Salmonella, and Citrobacter, and are commonly used as an indicator of hygiene and contamination in environments (33). The emergence and spread of AMR in Enterobacteriaceae complicate the treatment of infectious diseases. The World Health Organization (WHO) has prioritized carbapenem-resistant Enterobacteriaceae as a top-priority pathogens for an urgent need to develop new antibiotics (34). Available evidence has demonstrated that exposure to disinfectants and cleaning agents in built environments poses a possible risk of AMR development of Enterobacteriaceae species (35–37), and the environmental exposures of built environments may contribute to the skin microbiome and resistome (29, 31). Therefore, deciphering AMR of Enterobacteriaceae species is crucial for better understanding the transmission and evolution of AMR and pathogens in built environments.
In the present study, microbial communities, together with human pathogens and their antimicrobial resistance, were characterized from 20 populated shopping malls as well as the surrounding road dust and greenbelt soil in spring and summer. We hypothesized that microbial communities exhibit distinct season and habitat specificity and that microbial community assembly is driven mainly by stochastic processes owing to regular cleaning and disinfection in malls. We further predicted that human and outdoor (road dust and greenbelt soil) microbiomes contribute largely to the mall microbiome via direct or indirect transmission, and thus mall surfaces harbor more potential human pathogens and AMR determinants. To test these hypotheses, bacterial 16S rRNA gene and fungal internal transcribed spacer (ITS) amplicon sequencing was conducted to characterize the microbial community structures and compositions. Neutral community models (NCM), SourceTracker models, and random forest models were employed to depict the microbial community assembly processes, identify the microbial signatures related to environmental preference, and track the potential microbial source of shopping malls. High-throughput quantitative PCR (HT-qPCR) assays for human pathogens and genomic analysis of Enterobacteriaceae isolates were further performed to determine the pathogen distribution and the AMR and virulence gene burdens.
RESULTS
Microbial community diversity and composition.
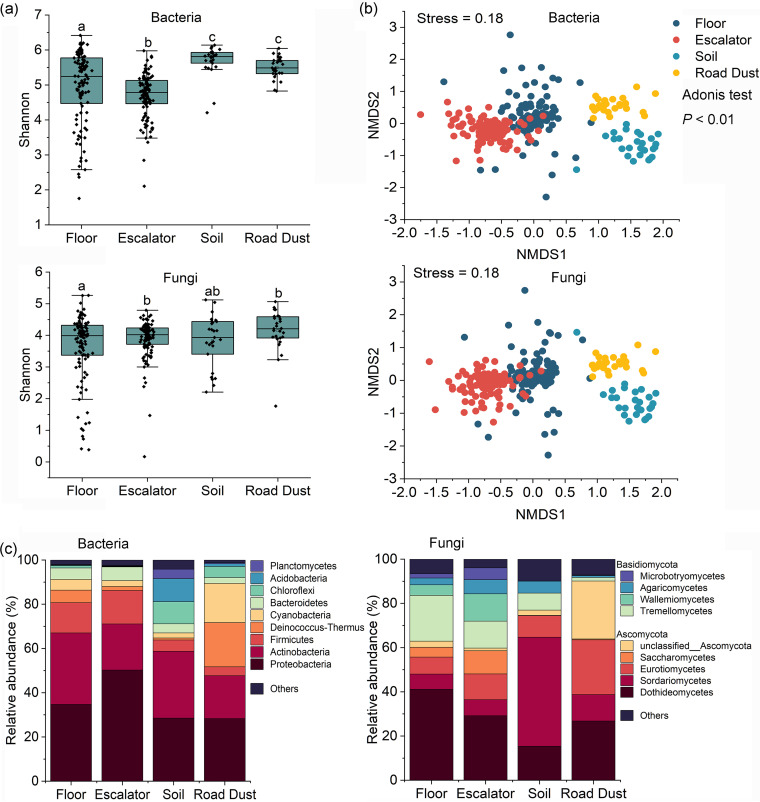
Microbial community diversity and composition were evaluated by amplicon sequence analysis of 274 DNA samples obtained from floor surfaces, escalator surfaces, road dusts, and greenbelt soils (see Table S1 in the supplemental material). In total, 25,806 bacterial amplicon sequence variants (ASVs) and 19,656 fungal ASVs were assigned, and floor surfaces had the highest number of ASVs, followed by escalators, road dusts, and greenbelt soils. There were 1,276 shared bacterial ASVs observed in four habitats, while shared fungal ASVs were not found in any of the four habitats. The bacterial community of floor surfaces had the highest species richness (Chao 1), followed by escalators, road dusts, and greenbelt soils (Fig. S1a). However, significantly higher diversities (Shannon and Simpson diversities) were found in the bacterial communities of greenbelt soils and road dusts than of floors and escalators (P < 0.05) (Fig. 1a; Fig. S1b). For the alpha-diversity of the fungal community, escalators had the lowest ASV number, and road dust harbored the highest number of ASVs (P < 0.05) (Fig. S1c). The fungal community of floor surfaces showed significantly lower Shannon and Simpson index values than the others (P < 0.05) (Fig. 1a; Fig. S1d). Significant seasonal variation in bacterial or fungal alpha-diversity was not observed (P > 0.05), while both bacterial and fungal communities were significantly grouped with habitats (Adonis test, P < 0.001) and seasons (Adonis test, P < 0.001) (Fig. 1b; Fig. S1e and f). Clustering of microbial communities from floors or escalators was not observed across malls (Fig. S2).
FIG 1.
Diversity and composition of the floor, escalator, greenbelt soil, and road dust microbial communities. (a) Shannon diversity of bacterial and fungal communities across different inhabitants. (b) Distribution patterns of microbial communities visualized using NMDS analysis based on Bray-Curtis distance. (c) Relative abundances of bacterial taxa (at the phylum level) and fungal taxa (at the class level).
Diversity and composition of floor, escalator, soil, and road dust microbial communities. (a and b) Chao 1 index and Simpson index of bacterial communities. (c and d) Chao 1 index and Simpson index of fungal communities. Different letters indicate significant differences among different samples (mean ± standard deviation [SD]; ANOVA, P < 0.05). NMDS analysis revealed significant seasonal differences in floor (e) and escalator (f) microbial communities (Adonis test, P < 0.01). (g and h) Bacterial (at the phylum level) and fungal (at the class level) community compositions varied with seasons (spring and summer). Download FIG S1, TIF file, 1.3 MB (1.4MB, tif) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Principle-coordinate analysis based on Bray-Curtis distance showed that the microbial community (bacterial community [a and b] and fungal community [c and d]) from the surfaces of floors or escalators was not grouped across shopping malls. Download FIG S2, TIF file, 0.6 MB (635.4KB, tif) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary for the collected samples. Download Table S1, DOCX file, 0.1 MB (82.8KB, docx) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proteobacteria and Actinobacteria were the predominant bacterial phyla across all samples, occupying 46.0% to 70.5% of bacterial communities (Fig. 1c; Fig. S1g). Firmicutes was the third most abundant phylum on floor and escalator surfaces. Greenbelt soils harbored more abundant Chloroflexi and Acidobacteria (P < 0.05) and road dusts harbored more Cyanobacteria and Deinococcus-Thermus (P < 0.05) than did the other habitats. A total of 14 fungal phyla were detected among all samples, and the most abundant phyla were Ascomycota and Basidiomycota (Fig. 1c), in which Dothideomycetes was the most abundant class of bacteria in floor, escalator, and road dust samples, representing 26.8% to 41.1% of the overall fungal communities, while Sordariomycetes was the dominant class in greenbelt soils. Seasonal variations in fungal community composition were also observed in all samples. Summer samples were significantly dominated by Eurotiomycetes, Saccharomycetes, and Wallemiomycetes, and spring samples mainly harbored Tremellomycetes, Agaricomycetes, Microbotryomycetes, and Dothideomycetes (one-way analysis of variance [ANOVA] tests, P < 0.05) (Fig. S1h).
Determination of core microbial communities.
We defined the core microbial communities as taxa detected in more than 80% of samples according to previous studies (12, 38). The core bacterial communities contained 36 bacterial ASVs, accounting for 3.9% to 19.9% of bacterial sequences. These core bacterial species were not uniformly abundant across all sample types. The most abundant core bacterial ASVs were ASV5031_g_Acinetobacter (0.9%), ASV4883_g_Enhydrobacter (0.84%), and ASV18928_g_Kocuria (0.82%), in which Acinetobacter baumannii and Kocuria kristinae were associated with infectious diseases. The core fungal community contained only one ASV (ITS_745_g_Cladosporium, commonly causing allergies and asthma), accounting for 5.6% of the total fungal sequences and occurring exclusively on escalators (8.7%) and floors (5.2%). The shared microbial communities presenting in all samples (100% of samples) of each habitat were also determined. A majority of the shared bacterial and fungal ASVs in soils were also observed in the shared microbial communities of road dusts. In contrast, none of shared microbial ASVs observed in soils or road dusts was found in escalator or floor microbial communities and vice versa.
Microbial interaction network and community assembly.
A maximum number of significant (P < 0.05) bacterial pairwise correlations was observed for floors (12,621), followed by those for escalators (9,837), road dusts (6,435), and soils (4,374) (Fig. S3). In the fungal community, floors harbored the maximum number of significant pairwise relationships (5,961), followed by soils (4,480), road dusts (4,480), and escalators (4,414). Among these microbial associations, the number of positive correlations was significantly higher than the negative ones (Table S2). Topological features of cooccurrence networks based on significant (P < 0.01) and robust (Pearson’s r = 1) microbial correlations showed more complex interactions in bacterial communities than fungal communities, and the networks were more dense in road dust and soil samples than in escalator and floor samples.
Correlation network analysis depicted the microbial interactions in different habitats. A connection represents a strong (Pearson’s r = 1) and significant (P < 0.01) correlation. Nodes indicate the taxonomic affiliation at the family level. Node size is proportional to the number of connections. Download FIG S3, TIF file, 2.5 MB (2.5MB, tif) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Topological features of cooccurrence networks for microbial communities. Download Table S2, DOCX file, 0.04 MB (40.8KB, docx) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
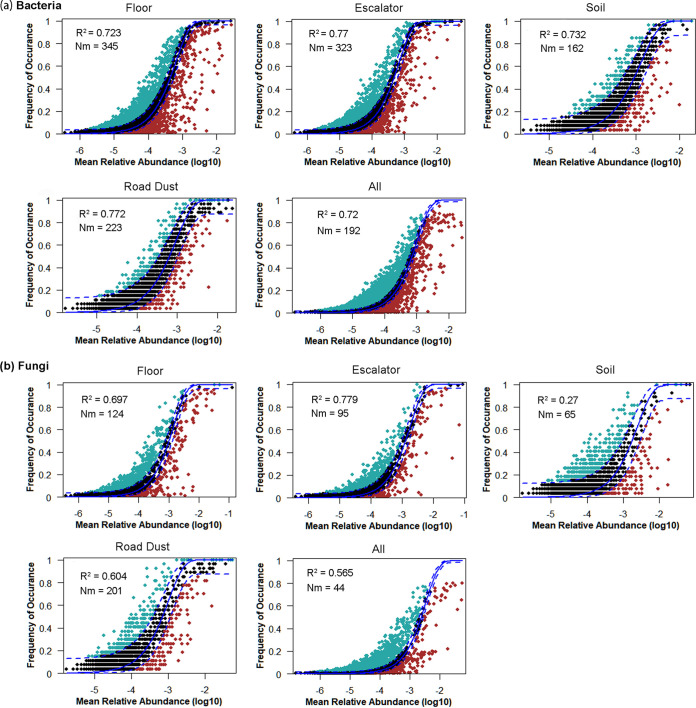
To further explore the community assembly process controlling microbial diversity patterns, we applied the NCMs to characterize the relationship between the predicted ASV occurrence frequencies and their relative abundances (Fig. 2). The best-fit neutral model revealed that the models explained 70% to 77.2% of bacterial community variances for each habitat and overall samples. The NCMs of fungal communities also explained large proportions of variation for floor (69.7%), escalator (77.9%), road dust (60.4%), and overall (56.5%) samples, while the NCM for the soil fungal community explained only 27% of the variance. The values of both Nm (estimate of dispersal between communities) and m (immigration rate) of the bacterial communities exceeded those of the fungal communities, where soil fungal communities were observed to have the lowest species dispersal (Nm = 65) and immigration rate (m = 0.14) (Fig. S4a). The normalized stochasticity ratio (NST) index was also calculated to evaluate the relative importance of deterministic (<50%) and stochastic (>50%) community assembly processes. NSTs of both bacterial and fungal communities were substantially above the 50% boundary (Wilcoxon test, P < 0.001), except that those of the floor bacterial community (56%) and soil fungal community (52.5%) were close to the margin, indicating the significant role of stochastic processes in microbial assembly (Fig. S4b).
FIG 2.
NCM of bacterial (a) and fungal (b) communities across floor, escalator, soil, and road dust. “All” represents microbial communities from all habitats. Dark dots indicate occurrence frequency within the 95% confidence interval (dashed blue lines). ASVs that occur more and less frequently than predicted by NCM are marked in green and red, respectively. The coefficient of determination (R2) is the goodness of fit of the neutral model, and it ranges from 0 (no fit) to 1 (perfect fit). Nm indicates the estimates of the metacommunity size times immigration rate. N represents the metacommunity size, and m is the immigration rate.
(a) The immigration rates at the community level were predicted by the neutral community model. Immigration rates indicate the probability that a dead individual is replaced by an immigrant. (b) The normalized stochasticity ratio (NST) index evaluated the relative importance of deterministic (<50%) and stochastic (>50%) community assembly processes. Download FIG S4, TIF file, 0.5 MB (575.2KB, tif) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Identification of microbial signatures.
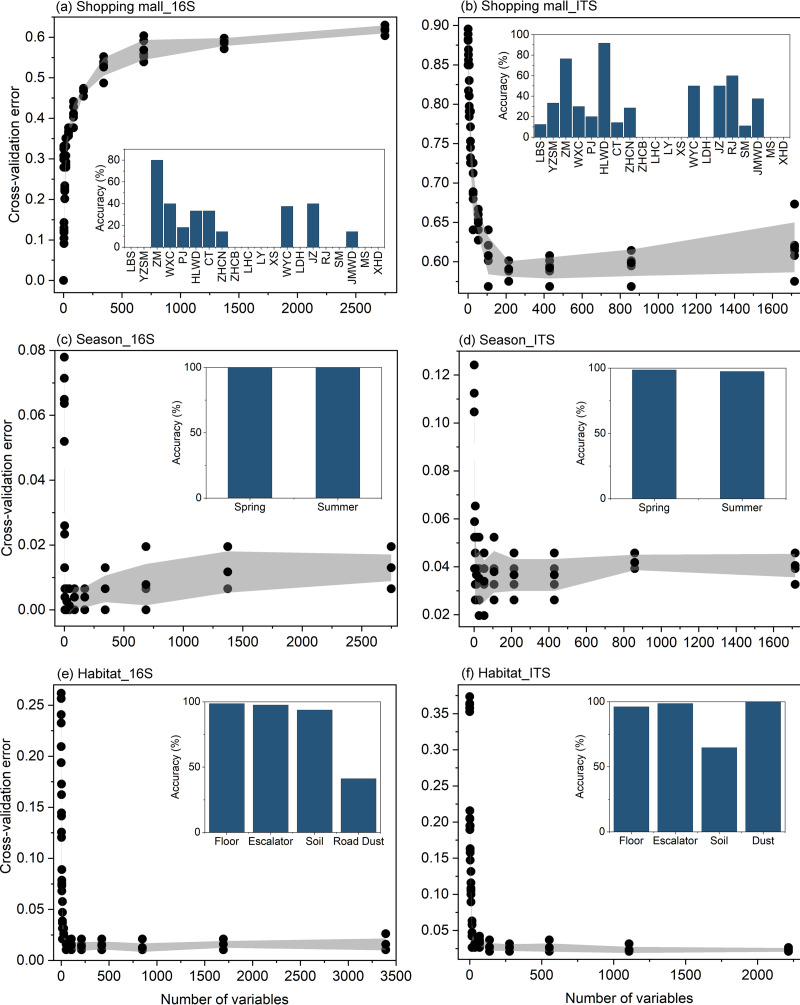
To determine the predictive potential of microbial fingerprints for shopping mall, season, and habitat discrimination, we trained the random forest classifiers (RFCs) for microbial communities and evaluated their performance in differentiating samples with their correct origins based on a 10-fold cross-validation framework (Fig. 3; Fig. S5). The trained RFCs were highly sensitive and specific for determining from which seasons or habitats a sample was taken, which indicates consistent differentiation of microbial communities from different seasons or habitats (Table S3). However, microbial community-based identification of individual malls showed high classification error ratios (68.83% of out-of-bag (OOB) error rate of bacterial community; 64.05% of out-of-bag error rate of fungal community). This is likely due to the presence of the core microbiomes in the floor and escalator microbial communities from shopping malls. Moreover, the sample size in each shopping mall also affected the accuracy of RFCs. Based on the MeanDecreaseAccuracy and MeanDecreaseGini metrics, the most predictive taxonomic signatures for season and habitat classifications were evaluated by the trained RF models. The top 10 ASVs/ITS sequences with the highest feature importance scores were mostly affiliated with the bacterial phyla Actinobacteria and Proteobacteria or the fungal phyla Ascomycota and Basidiomycota.
FIG 3.
Classification accuracy of the optimized random forest models for assigning samples to shopping malls, seasons, and habitats. The top 10 important bacterial and fungal signatures were selected as the optimal biomarker sets to optimize the random forest model based on the five cross-validation sets of trained samples. Embedded histograms revealed that the prediction performance of random forest models was evaluated using the testing set (validation set), measured as accuracy.
A random forest model indicates the predictor importance (percentage of increase in mean square error) of dominant ASVs (>274 in total number) as drivers for microbial community in the whole profile. The accuracy importance measure was computed for each tree and averaged over the forest (1,000 trees). Percentage increases in the mean squared error (MSE) of variables were used to estimate the importance of these predictors, and higher MSE% values imply more important predictors. The embedded charts showed that the true-positive percentage and the false-positive percentage achieved an area under the curve (AUC) value of 0.9024 with s 95% confidence interval (CI) of 0.8168 to 0.9669 between trained (n = 183) and tested (n = 91) samples in the validation cohort (P < 0.0001). Download FIG S5, TIF file, 2.0 MB (2MB, tif) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of predictive accuracy of random forest supervised learning models for habitat, season, and shopping mall classification. Download Table S3, DOCX file, 0.05 MB (48.8KB, docx) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Tracking human microbial footprint in built environments.
Since occupants and outdoor environments are the two main sources of microbes in built environments, we evaluated the potential contribution of their microbial communities to the indoor microbiomes (floors and escalators) of shopping malls by using Bayesian-based source tracking models (Fig. S6). SourceTracker analysis revealed that human palms were a major source of escalator microbiota, approximately 60% of which was associated with human palms. Bacterial taxa associated with air, road dust, and soil were predicted to have a minor influence on the escalator community, accounting for 1% to 2.5% of escalator microbes. Similarly, floor microbiota contained a high proportion of ASVs with human palm (~ 25%) and road dust (15%) sources, while air (5.5%) and soil (2%) contributed a small percentage of the microbial composition of floors. However, a considerable proportion of floor ASVs were predicted with unknown sources.
SourceTracker models predicted the proportions of bacterial taxa from floors and escalators that come from possible sink environments. The bacterial communities from soils, road dust, air, and human palms were used as sources. Download FIG S6, TIF file, 0.1 MB (132.8KB, tif) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Prevalence of potential human pathogens and antimicrobial resistance.
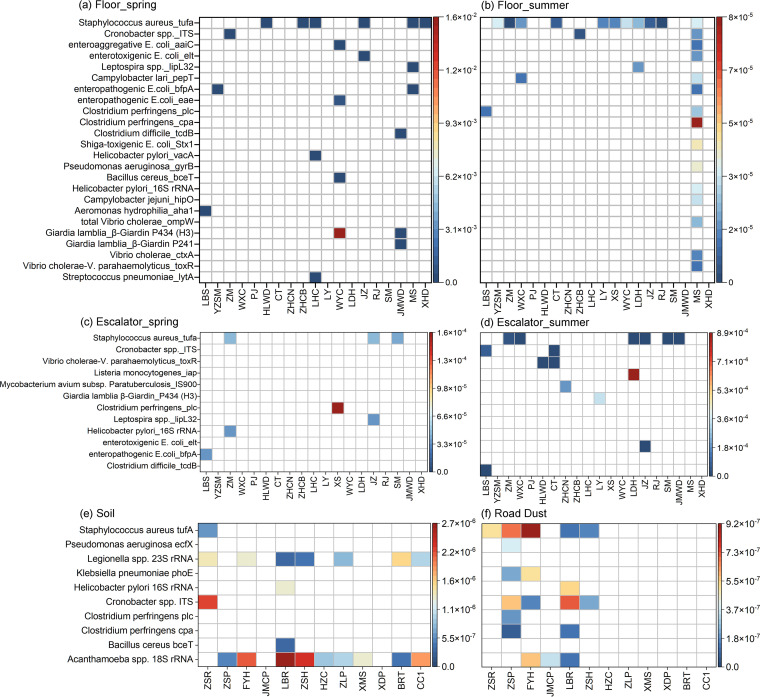
HT-qPCR assays showed that a highest relative abundance of pathogens was observed for floors (6.0 × 10−7 to 8.3 × 10−2 copies/copy of 16S rRNA), followed by escalators (3.9 × 10−4 to 4.5 × 10−3 copies/copy of 16S rRNA), soils (2.3 × 10−6 to 2.4 × 10−5 copies/copy of 16S rRNA), and road dusts (1.8 × 10−6 to 1.1 × 10−5 copies/copy of 16S rRNA) (Fig. 4). Seasonal variations in the distribution of marker genes for pathogens were observed. For examples, floors in the spring harbored more abundant marker genes than floors in the summer, and for escalators, marker genes were more abundant in summer than in spring. Staphylococcus aureus was the most prevalent pathogen, with varied detection frequencies for floors (75%), escalators (25%), and road dusts (42%). Acanthamoeba spp., which was the most frequently detected pathogen in soils, with 75% of sites positive, is commonly found in natural environments and responsible for a fatal encephalitis and keratitis in humans.
FIG 4.
Heat map showing the incidence and relative abundance (copies/copy of 16S rRNA gene) of marker genes for human pathogens detected from floors (a and b), escalators (c and d), soils (e), and road dusts (f) by using HT-qPCR assays.
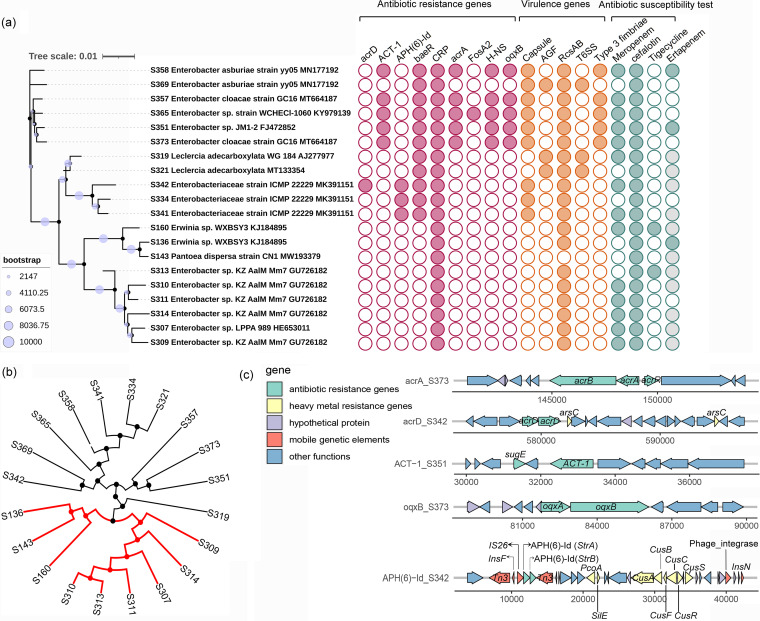
A total of 302 bacterial strains were isolated from CHROMagar Escherichia coli and coliforms plates, and 20 strains were affiliated with the family Enterobacteriaceae, all of which were retrieved from floors (Fig. 5a). Core genome single nucleotide polymorphism (SNP)-based phylogenetic analysis revealed a clustering profile similar to that of the 16S rRNA-based phylogenetic tree (Fig. 5b). A majority of strains were resistant to cephalothin (19/20) and meropenem (15/20), and a few of the isolates were resistant to tigecycline (2/20) and ertapenem (3/10) (Fig. 5a). All strains carried the CRP gene (ARO:3000518), which encodes a regulator associated with the expression of the MdtEF multidrug efflux pump and is responsible for fluoroquinolone, macrolide, and penam resistance. Strains S358 (Enterobacter asburiae), S357 (Enterobacter cloacae), S365 (Enterobacter sp.), S351 (Enterobacter sp.), and S373 (Enterobacter cloacae) harbored the same antibiotic resistance genes (ARGs), including ACT-1 (beta-lactamase conferring resistance to carbapenem, cephalosporin, cephamycin and penam; ARO:3001821), baeR (a regulator for the expression of MdtABC and AcrD efflux pump complexes encoding resistance to aminocoumarin and aminoglycoside; ARO:3000828), acrA (AcrAB-TolC multidrug efflux for triclosan, cephalosporin, and fluoroquinolone; ARO:3004042), the H-NS gene (a regulator for the resistance-nodulation-division [RND]-type multidrug exporters for cephalosporin, cephamycin, fluoroquinolone, and tetracycline; ARO:3000676), and oqxB (RND efflux pump conferring resistance to fluoroquinolone, glycylcycline and tetracycline, ARO: 3003923). Discrepancies between antimicrobial resistance phenotyping and genotyping were also observed in some isolates. For example, tigecycline resistance-related tet gene variants were not found, although the resistance phenotype was detected in isolates S160 and S313. We further analyzed the accessory genes of several ARGs and found that the ACT-1-carrying isolates possessed the same genomic backbone comprising a resistance gene cluster, sugE (quaternary ammonium compound-resistance protein)–orf–ACT-1 (Fig. 5c). The contigs carrying APH(6)-Id harbored complex genetic contexts, including transposon, ARGs, and copper resistance genes (Tn3-insF-IS26-strA-strB-Tn3 and cusA-cusB-cusC-cusR-orf-cusS-orf-orf-orf-orf-orf-phage_integrase-orf-orf-insN). The main resistance mechanisms of these detected ARGs were multidrug efflux systems and antibiotic target alteration. For virulence-associated genes (VAGs), rcsAB was frequently detected in these isolates (19/20, 95%), which is related to the regulation of VAGs. Other VAGs encoding capsule (antiphagocytosis), AGF (curli fibers/thin aggregative fimbriae), T6SS (secretion system), and type 3 fimbriae (mediating biofilm formation) were found with detection frequencies ranging from 15% to 45%.
FIG 5.
Genomic analysis and antimicrobial susceptibility of Enterobacteriaceae species (n = 20) retrieved from floors and escalators. (a) Phylogenetic tree (left) of all strains based on full-length 16S rRNA gene sequences (bootstrap = 10,000). A bubble plot (right) reflects the detection of antibiotic resistance genes (annotated by searching against CARD database; red) and virulence genes (annotated by searching against VFDB database; orange) from genome sequences and antimicrobial susceptibility test (green; strains marked in gray are the ones for which the ertapenem susceptibility test was not performed). (b) A rooted maximum-likelihood phylogenetic tree was constructed based on the alignment of the core genome single nucleotide polymorphisms. The strains in the red branches clustered together, which was consistent with the clustering of their full-length 16S rRNA genes. (c) Flanking regions of the detected antibiotic resistance genes in contigs. Arrows indicate the direction of gene transcription. Different genes are indicated by different colors. Genes with ≥98% amino acid identity and a query coverage of ≥99% were annotated by mapping sequences against the NCBI nr database. AGF, curli fibers/thin aggregative fimbriae.
DISCUSSION
Shopping malls represent the main source of microbial exposures for human occupants, providing a unique venue for microbial interactions and exerting direct or indirect effects on human living space and health. Our study characterized the microbial community profiles, community assembly mechanisms, and potential pathogens of the floors and escalators of shopping malls and their surrounding road dusts and greenbelt soils. These habitats represent distinct ecological niches with homogeneous dispersals (community evenness) of species, various genetic pools (species richness), and different community compositions. Homogeneous dispersal of bacterial species dominated within road dusts and greenbelt soils, while floors and escalators might have a large genetic pool of microorganisms. It was presumed that soil and dust legacy possibly affected community evenness, and the infiltration of outdoor air/dust or biological particles from cleaning processes being settled contributed to the microbial richness of mall surfaces. The fungal alpha-diversity in the built environments was in accordance with the values reported by previous studies (39), and significantly higher alpha-diversity was observed in road dust than in floor surfaces, suggesting higher species number and species evenness for road dust.
Microbial communities were significantly clustered with habitats, and the shared taxa for each habitat occupied a large proportion of microbial community exclusively in their respective habitat, indicating that microbial distribution patterns could be strongly driven by habitats. The disparities of environmental conditions in the investigated habitats, such as the availability of nutrients, humidity, air exchange rate, temperature fluctuations, and UV radiation contributed to the spatial heterogeneity of the microbiomes (40). Distinct seasonal distribution of microbial communities was also observed in malls. The significant increase in the number of shoppers in summer could be an important factor for the seasonal distribution of microbial communities. With the increased occupant number in summer, the exchange of microorganisms between humans and the environment would be more frequent, resulting in different microbial compositions. It also should be noted that samples in this study were collected in spring and summer owing to the climatic conditions of the target city, a subtropical maritime climate without typical winter weather. For those cities with four distinct seasons, analysis of samples collected from each season would provide a more comprehensive profile of the seasonal distribution of microbial communities. Previous studies of the transit microbiome suggested that the surface types were the major determinant of the variations in microbial composition, and human-surface interactions largely shaped the community composition (4, 14, 15, 41). We also observed a significant enrichment of the human skin-associated microbiome on shopping mall floors and escalator handrails, suggesting that human-environment interactions were another strong driver for community patterns of the mall microbiome. The low sensitivity and high error ratios of RF classifiers for differentiating malls indicated the classification confusion for distinguishing one sample as having come from a specific mall. The possible explanations were that the similar environmental conditions, operational modes, and human activities across malls exert a homogenizing effect on mall surface microbial communities, hindering the precise discrimination of specific shopping malls (18). The sample size in each shopping mall is another important factor affecting the accuracy of models for differentiating the individual malls. The small data set with many outliers, missing values, or skewed data would impact the score of each decision tree of the random forest classifier.
Periodic anthropogenic perturbations in built environments (e.g., regular cleaning, disinfection, hand contact, and walking) would create opportunities for microorganisms to adapt to or colonize the environment and for communities to reassemble (42). Microorganisms can establish a range of relationships that generate increased benefits for the community. Microbial interaction networks may elucidate more about the inner workings of a community and the processes governing the assembly of community. Our analysis revealed that both bacterial and fungal networks consisted mainly of positively synergistic interactions. Indoor environments (floors and escalators) harbored more significant pairwise relationships, whereas more complex, dense, and robust microbial interconnections were observed in outdoor environment communities (soils and road dusts). In the surface environment, most microorganisms might be concurrently transported to the habitats from the same source through cleaning materials or walking, thus resulting in many pairwise relationships. However, regular cleaning and disinfection are possibly an important modifying factor for regulating the interactions of the surface microbiome with decreased microbial biomass and viability on surfaces, hence causing reduced microbial complex interactions. Soil and road dust would provide more nutrients or relatively stable conditions, resulting in a profitable niche for microbial fitness and interactions (43, 44).
The built environment is considered a microbial wasteland, where microbes passively accumulate and the process of microbial colonization is intrinsically stochastic (3, 43). The combination of biotic factors, such as the interactions between newcomers and resident microorganisms, and abiotic factors, such as nutrient availability, may change the process of community assembly (40, 43). We determined the ecological processes (neutral versus selective) underpinning microbial community assembly in these built environment habitats. The composition of the overall microbial community in these built environments was consistent with neutral model predictions, suggesting that the stochastic balance between the loss and gain of microbes (such as stochastic growth, death, and immigration) shaped the variations in the microbial communities (44). The neutral model generally incorporates passive dispersal (for example, sampling individuals from a source pool of available species) to ecological drift (random births and deaths of individuals) as a neutral process (45). However, owing to the microbial flux between human skin and floor/escalator and/or seasonal fluctuation, community dispersal might be the more relevant neutral process for floors and escalators. For soil and road dust, it is difficult to infer the relative role of dispersal versus ecological drift due to a lack of temporal data. We also observed minimum values of the NCM parameters R2, Nm, and m in the soil fungal community. This could be attributed largely to plant genotype and development in greenbelt soils, which exerted strong effects on microbiome assembly (46). The NST index demonstrated that the importance of stochastic processes was not similar between bacterial and fungal communities in the same habitat. Bacterial community assemblies were more stochastic than fungi in soil and road dust. The main possible explanation is that fungal hyphae penetrate or become entangled with the soil or dust aggregates, and the dispersal of the fungal community can be both deterministic and stochastic (47, 48).
Shopping malls are the main environment where people share microbes, and many diseases associated with human activities may have their origins in malls (10). Previous studies have revealed that pathogens and AMR were frequently detected in built environments, e.g., nosocomial infectious pathogens in hospital (19) and VAG- or ARG-carried microbes in metro systems (14, 15, 31), and could spread to humans by close interactions between individuals and surface microbiota. Our observation showed that 0.82% to 8.7% of potential human pathogens were identified in the core microbiomes of the built environments. HT-qPCR assays also revealed that human pathogens were prevalent and that indoor environments harbored more abundant pathogens than outdoor samples, indicating that human exposure to these mall surfaces might represent a potential health risk. Skin-to-surface direct contact and/or shedding of biological particles possibly introduces and transmits the microbial contaminants indoor (20, 49–51); humidity, air temperature, and the occupant density and source of ventilation air can also influence the abundance and transmission of pathogenic microorganisms in indoor environments (2).
Enterobacteriaceae species are important pathogens in health care- and community-associated infections worldwide, and the emergence and spread of resistance among Enterobacteriaceae species are threatening antibiotic treatment efficacy. Genomic analysis showed that a high diversity of AMR genes (e.g., CRP, oqxB, acrA, the H-NS gene, and baeR) encoding the multidrug efflux pump for antibiotics, disinfectants, and detergents (e.g., triclosan, fluoroquinolones, and cephalosporin) was detected in Enterobacteriaceae isolates. Previous reports suggested that the high proportions of multidrug efflux genes were frequently found in microbe-controlled compartments, including hospital-associated surfaces, intensive care units, and cleanroom facilities (10), and the multidrug efflux systems were one of the most frequently reported resistance mechanisms in Enterobacteriaceae species (8). Increased confinement and cleaning (regular exposure to disinfectants and cleaning reagents) in indoor environments were reported to be associated with an overall high level of virulence and antimicrobial resistance of microbial populations (10). Although tigecycline-resistant tet variants were not detected, the gene oqxB encoding RND type efflux pumps associated with tigecycline susceptibility was detected in resistant strains. Thus, the prevalence of the efflux pumps possibly also explained the incompatibility between genotype and phenotype of tigecycline-resistant strains (50, 52). Plasmid was not assembled in the present study, but the flanking regions of ARGs [e.g., APH(6)-Id encoding beta-lactamase] carried by several potential pathogens contained transposon-related genes (e.g., Tn3, IS26, and insF), suggesting potential transposon-mediated horizontal transfer of ARGs between human pathogens and other environmental strains.
MATERIALS AND METHODS
Sample collection.
Surface samples from 20 shopping malls were collected in March (spring, average daytime temperature of 21.6°C and relative humidity of 72%) and July (summer, average daytime temperature of 35°C and relative humidity of 85%) of 2020 in Xiamen, China (see Table S1 in the supplemental material). Samples were not collected in autumn and winter. Since the target city, Xiamen, is a subtropical city near the Tropic of Cancer, without typical winter, the weather conditions are similar in spring and autumn. Air humidity in shopping malls was kept in the range of 50% to 60%, and the temperature was 22°C to 27°C (https://www.cma.gov.cn/). We noticed that the occupancy in malls dramatically increased in summer compared to spring owing to the mitigation of the COVID-19 pandemic after April 2020 in China (https://new.qq.com/rain/a/20210312A0572S00). For all malls, cleaning and sanitizing (commonly with sodium hypochlorite) were regularly performed each day before opening (7:00 a.m. to 9:00 a.m.). Discontinuous cleaning operations were also performed during daily business hours according to foot traffic. Sampling time was set to the preferred afternoon time (3:00 p.m. to 6:00 p.m.) of shoppers.
A sterile nylon-flocked swab with 1 mL of transport medium (liquid Amies elution swab 481C; Copan, Italy) was used for collecting surface samples according to a previous study by Afshinnekoo et al. (14). Briefly, two swabs from floor surfaces close to each entrance for one shopping mall were taken by swabbing an area of approximately 6 cm by 6 cm for 1 min. For the escalator surfaces, only the right handrails of both ascending and descending escalators were swabbed with a palm-size (~0.01-m2) surface area during one transfer. Swabs were immediately placed into collection tubes, with immersion in the transport medium. To detect background contamination caused by air biological materials during sample collection, a buffer-dampened empty swab was held in the air for 1 min in each mall as a negative control. Road dust was collected from the streets (approximately 50 to 100 m away from the malls) around 12 shopping malls by using 75% ethanol-sterilized brushes or vacuum cleaners. To achieve adequate dust, road dust samples from more than two sites on the same street were pooled, and the sampling area for each site depended on the amount of available dust. Approximately 50 g of greenbelt surface soil (~0 to 20 cm) in close proximity to the road dust sampling sites was also sampled using sterile scoops. A total of 474 samples were collected, including 222 floor swabs, 218 escalator swabs, 27 soil samples, and 27 road dust samples. The collected samples were immediately kept on ice and then transported to a −20°C freezer within 3 h.
DNA extraction and amplicon sequencing.
Surface samples in transport medium were thawed at room temperature, and the entire nylon fiber flocked swab tips were cut by sterile scissors and sterilized with 75% ethanol before passage through a flame. Then, the medium and the swab tip for one sample were transferred into a collection tube in a FastDNA spin kit for soil DNA extraction (MP Biomedicals, USA). To obtain adequate DNA, DNA from two swabs from the same entrance/escalator was pooled in 70 μL of elution buffer. For road dust and soils, 0.5-g samples were used for DNA extraction. Thus, 274 DNA samples were used for microbial community analysis, including 111 DNA samples from floors, 109 from escalators, 27 from road dust, and 27 from soils (Table S1). DNA concentrations were measured by a Quant-iT double-stranded DNA (dsDNA) high-sensitivity assay kit (Invitrogen) and a Qubit 3.0 fluorometer (Invitrogen) according to the protocols provided. DNA extraction and amplification for negative controls were performed using the same protocol.
Amplicons for bacterial 16S rRNA gene were generated using the barcoded primers 515F and 806R (53), and the fungal internal transcribed spacer (ITS) was amplified with the barcoded primers ITS1F and ITS2R (39). A DNA library was constructed and amplicon sequencing was performed on an Illumina MiSeq PE250 sequencing instrument (Shanghai Majorbio, China). Further details for amplicon sequencing can be found in Text S1 in the supplemental material. The DNA concentrations of negative controls were below the detection limit of a Qubit 3.0 fluorometer (Invitrogen), and amplification of the 16S rRNA gene and ITS returned negative results.
Detailed information for amplicon sequencing, HT-qPCR assays for human pathogens, identification of the Enterobacteriaceae isolates from malls, neutral community models, and random forest models. Download Text S1, DOCX file, 0.04 MB (44.3KB, docx) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
HT-qPCR assays for human pathogens.
TaqMan-based HT-qPCR analysis for human pathogens was performed by using a WaferGen SmartChip real-time PCR system (TaKaRa, Japan) according to our previous study (54). HT-qPCR assays simultaneously quantify 68 marker genes for 33 human pathogens that are commonly associated with respiratory infections, intestinal illness, keratitis, and other diseases. Data analysis for HT-qPCR was performed according to the methods in Text S1 in the supplemental material.
Isolation and antimicrobial susceptibility testing of Enterobacteriaceae species.
Coliforms were isolated from the floor and escalator transport samples by using CHROMagar ECC agar (France) in accordance with the manufacturer’s protocol. Taxonomic identification of the isolates was performed by sequencing the full-length 16S rRNA gene (Text S1). Antibiotic susceptibility to meropenem, cephalothin, tigecycline, and ertapenem (Oxoid, UK) was determined for the identified Enterobacteriaceae species using the disc diffusion method according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST, version 10.0; https://www.eucast.org/) and the Clinical and Laboratory Standards Institute (CLSI 2015, M100-S25; https://clsi.org/). Reference strain Escherichia coli DH5α served as the quality control strain.
Whole-genome sequencing of Enterobacteriaceae species.
Enterobacteriaceae isolate DNA was extracted using a Wizard genomic DNA purification kit (Promega, USA). Indexed DNA libraries were constructed with an insert size of 300 bp by using a NEBNext Ultra DNA library prep kit (New England Biolabs, USA), and sequencing was performed on an Illumina HiSeq × 10 system with a minimum of 100-fold coverage (Shanghai Majorbio, China).
Bioinformatics.
(i) Phylotype analysis for amplicons. Raw sequencing data of 16S rRNA/ITS gene amplicons were demultiplexed by tag sequences using QIIME python scripts (split_libraries_fastq.py) (55); the DADA2 pipeline based on amplicon sequence variants (ASVs) was applied for microbial diversity analysis. Briefly, the core sample inference algorithm was used for filtering and trimming demultiplexed sequences (maximum number of N [maxN] = 0, maximum number of expected errors allowed in a read [maxEE] = 2, and DADA2 truncate the read at the first nucleotide with a quality score of 2 [truncQ = 2]) (56). Subsequently, error rates were checked, deduplication was performed, and forward/reverse reads were merged. Chimeras were removed from the merged sequences, and a feature table of ASVs was generated. Singletons, chloroplasts, and mitochondrial sequences were removed from the final data set. The sequence variants were assigned to taxonomic lineages against the SILVA reference database (version 132) for bacteria and the UNITE ITS database for fungi by using a naive Bayesian classifier method (57).
(ii) Whole-genome sequencing analysis. Sequencing adapters were removed, and quality filtering of the acquired reads was conducted using FastQC. Draft genomes were de novo assembled using SOAPdenovo2 (58) and visualized by CLC Genomics Workbench 8 (CLC Bio). The putative coding sequences (CDS) were predicted using Glimmer v3.02 (59). Protein sequences were converted from CDS and annotated using Diamond against the NR, Swiss-Prot, Pfam, EggNOG, GO, and KEGG databases (E value ≤ 1e−5). ARGs were identified by matching the sequences to the Comprehensive Antibiotic Resistance Database (CARD, version 1.1.3) (60) and the ResFinder database (61). VAGs were annotated by searching the assembled contigs against the virulence factor database VFDB (62). Integron_Finder was applied to identify the integron by detecting the promoters and attI sites with the use of INFERNAL and HMMER profiles (43–65). These genes were screened with a strict cutoff value of amino acid identity of ≥98% and a query coverage of ≥99%. Phylogenetic trees based on the alignments of full-length 16S rRNA gene sequences and SNPs in the core genomes were constructed using PhyML based on a maximum likelihood approach (bootstrap = 10,000) and displayed using iTOL (66).
Statistical analysis.
Analysis for microbial alpha-diversity and beta-diversity was performed in the open-source R environment (v4.0.3) with the package vegan 2.2.0 (67). Three alpha-diversity indices, including Shannon, Simpson, and Chao 1 indexes, were calculated, and the beta-diversity distribution variation was evaluated using Bray-Curtis dissimilarity-based nonmetric multidimensional scaling (NMDS) or principal-coordinate analysis (PCoA). A permutational multivariate analysis of variance (PERMANOVA, Adonis test) was performed to assess the significance of dissimilarity in microbial beta-diversity distribution. To understand the interactions between microorganisms, correlation analysis of microbial taxa at the family level was conducted using a pairwise Pearson method, and ASV networks were visualized using Gephi. P values were adjusted for comparisons with the false discovery rate (FDR) algorithm to reduce the false-positive results (68). For comparative analysis between seasons/sample types, paired t tests and one-way ANOVA tests were performed using SPSS (IBM). All statistical tests were considered significant at a P of <0.05. To determine the potential sources of the indoor (floors and escalators) microbial communities, we downloaded the data sets of 16S rRNA gene amplicons from previous studies of the outdoor air microbiome (Xiamen, China) and the human skin (palm) microbiome (69, 70). The microbiota from road dust, greenbelt soils, outdoor air, and human palms were used as the sources, and microbial communities on floors and escalators were treated as the sinks. SourceTracker models were constructed using Bayesian SourceTracker in QIIME with default settings (http://qiime.org/tutorials/source_tracking.html) (71). We also assessed the stochasticity of community assembly using a neutral community model (NCM) and a normalized stochasticity ratio (NST) index. The relationships between the detection frequency of microbial taxa and their relative abundance across the wider metacommunity were predicted in the NCM models (72). Random forest (RF) supervised learning models were trained for illuminating the forensic potential of the microbiome and predicting shopping mall, season, and sample type classifications (73). More details about NCM and RF models can be found in Text S1.
Our study represents a comprehensive analysis of the mall microbiome and provides a better understanding of microbial distribution, assembly, pathogen distributions, and antimicrobial resistance burdens in shopping malls and their adjacent road dust and soils. It was demonstrated that these shopping mall-related habitats had distinct bacterial community compositions, and environmental factors (seasons and habitats) and human activities commonly affected the variations in microbial communities. The identification of microbial signatures indicated the predictive potential of microbial communities for shopping mall, season, and habitat discrimination. Stochastic processes largely contributed to the microbial community assembly. Additionally, more human pathogens were observed in indoor environments, carrying a high proportion of antimicrobial resistance-associated multidrug efflux genes and virulence genes. These data provide a background baseline for further study of the mall microbiome by integrating multidimensional factors from human population, time, environment, and geography. A dynamic surveillance of human pathogens and antimicrobial resistance in malls could also provide scientific data for public policymaking regarding environmentally mediated transmission of harmful microorganisms.
Data availability.
The raw amplicon sequences were submitted to the Sequence Read Archive (SRA) under accession no. PRJNA707496. All full-length sequences of 16S rRNA genes and the genome assemblies of Enterobacteriaceae species were deposited under GenBank no. MZ461606–MZ461903 and SRA accession no. PRJNA749475, respectively.
ACKNOWLEDGMENTS
This study was financially supported by the National Key Research and Development Plan (grant no. 2020YFC1806902) and the Natural Science Foundation of China (grant no. 81991535, 42021005, 21936006, and 32061143015).
Contributor Information
Jian-Qiang Su, Email: jqsu@iue.ac.cn.
Emily B. Graham, Pacific Northwest National Laboratory
REFERENCES
- 1.Gilbert JA, Stephens B. 2018. Microbiology of the built environment. Nat Rev Microbiol 16:661–670. doi: 10.1038/s41579-018-0065-5. [DOI] [PubMed] [Google Scholar]
- 2.Kembel SW, Meadow JF, O'Connor TK, Mhuireach G, Northcutt D, Kline J, Moriyama M, Brown GZ, Bohannan BJ, Green JL. 2014. Architectural design drives the biogeography of indoor bacterial communities. PLoS One 9:e87093. doi: 10.1371/journal.pone.0087093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibbons SM. 2016. The built environment is a microbial wasteland. mSystems 1:e00033-16. doi: 10.1128/mSystems.00033-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu T, Joice R, Vallarino J, Abu-Ali G, Hartmann EM, Shafquat A, DuLong C, Baranowski C, Gevers D, Green JL, Morgan XC, Spengler JD, Huttenhower C. 2016. Urban transit system microbial communities differ by surface type and interaction with humans and the environment. mSystems 1:e00018-16. doi: 10.1128/mSystems.00018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leung MH, Lee PKH. 2016. The roles of the outdoors and occupants in contributing to a potential pan-microbiome of the built environment: a review. Microbiome 4:21. doi: 10.1186/s40168-016-0165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shan YF, Wu WD, Fan W, Haahtela T, Zhang GC. 2019. House dust microbiome and human health risks. Int Microbiol 22:297–304. doi: 10.1007/s10123-019-00057-5. [DOI] [PubMed] [Google Scholar]
- 7.Sharma A, Richardson M, Cralle L, Stamper CE, Maestre JP, Stearns-Yoder KA, Postolache TT, Bates KL, Kinney KA, Brenner LA, Lowry CA, Gilbert JA, Hoisington AJ. 2019. Longitudinal homogenization of the microbiome between both occupants and the built environment in a cohort of United States Air Force cadets. Microbiome 7:70. doi: 10.1186/s40168-019-0686-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lax S, Sangwan N, Smith D, Larsen P, Handley KM, Richardson M, Guyton K, Krezalek M, Shogan BD, Defazio J, Flemming I, Shakhsheer B, Weber S, Landon E, Garcia-Houchins S, Siegel J, Alverdy J, Knight R, Stephens B, Gilbert JA. 2017. Bacterial colonization and succession in a newly opened hospital. Sci Transl Med 9:eaah6500. doi: 10.1126/scitranslmed.aah6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mora M, Wink L, Kögler I, Mahnert A, Rettberg P, Schwendner P, Demets R, Cockell C, Alekhova T, Klingl A, Krause R, Zolotariof A, Alexandrova A, Moissl-Eichinger C. 2019. Space Station conditions are selective but do not alter microbial characteristics relevant to human health. Nat Commun 10:3990. doi: 10.1038/s41467-019-11682-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahnert A, Moissl-Eichinger C, Zojer M, Bogumil D, Mizrahi I, Rattei T, Martinez JL, Berg G. 2019. Man-made microbial resistances in built environments. Nat Commun 10:968. doi: 10.1038/s41467-019-08864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masuwa K, Kalonda A, Malama S, Chimana M. 2020. Bacteriological analysis of escalator handrails and lift buttons of selected shopping malls in Lusaka Zambia: the public health risk implication. Int J Res Stud Microbiol Biotechnol 6:22–27. [Google Scholar]
- 12.Su JQ, An XL, Li B, Chen QL, Gillings MR, Chen H, Zhang T, Zhu YG. 2017. Metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China. Microbiome 5:84. doi: 10.1186/s40168-017-0298-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu L, Ning D, Zhang B, Li Y, Zhang P, Shan X, Zhang Q, Brown MR, Li Z, Van Nostrand JD, Ling F, Xiao N, Zhang Y, Vierheilig J, Wells GF, Yang Y, Deng Y, Tu Q, Wang A, Zhang T, He Z, Keller J, Nielsen PH, Alvarez PJJ, Criddle CS, Wagner M, Tiedje JM, He Q, Curtis TP, Stahl DA, Alvarez-Cohen L, Rittmann BE, Wen X, Zhou J, Global Water Microbiome Consortium . 2019. Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat Microbiol 4:1183–1195. doi: 10.1038/s41564-019-0617-0. [DOI] [PubMed] [Google Scholar]
- 14.Afshinnekoo E, Meydan C, Chowdhury S, Jaroudi D, Boyer C, Bernstein N, Maritz JM, Reeves D, Gandara J, Chhangawala S, Ahsanuddin S, Simmons A, Nessel T, Sundaresh B, Pereira E, Jorgensen E, Kolokotronis SO, Kirchberger N, Garcia I, Gandara D, Dhanraj S, Nawrin T, Saletore Y, Alexander N, Vijay P, Hénaff EM, Zumbo P, Walsh M, O'Mullan GD, Tighe S, Dudley JT, Dunaif A, Ennis S, O'Halloran E, Magalhaes TR, Boone B, Jones AL, Muth TR, Paolantonio KS, Alter E, Schadt EE, Garbarino J, Prill RJ, Carlton JM, Levy S, Mason CE. 2015. Geospatial resolution of human and bacterial diversity with city-scale metagenomics. Cell Syst 1:72–87. doi: 10.1016/j.cels.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danko D, Bezdan D, Afshin EE, Ahsanuddin S, Bhattacharya C, Butler DJ, Chng KR, Donnellan D, Hecht J, Jackson K, Kuchin K, Karasikov M, Lyons A, Mak L, Meleshko D, Mustafa H, Mutai B, Neches RY, Ng A, Nikolayeva O, Nikolayeva T, Png E, Ryon KA, Sanchez JL, Shaaban H, Sierra MA, Thomas D, Young B, Abudayyeh OO, Alicea J, Bhattacharyya M, Blekhman R, Castro-Nallar E, Cañas AM, Chatziefthimiou AD, Crawford RW, Filippis FD, Deng YP, Desnues C, Dias-Neto E, Dybwad M, Elhaik E, Ercolini D, Frolova A, Dennis G, Gootenberg JS, Graf AB, Green DC, Hajirasouliha I, Hastings JJ, International MetaSUB Consortium, et al. 2021. A global metagenomic map of urban microbiomes and antimicrobial resistance. Cell 184:3376–3393.e17. doi: 10.1016/j.cell.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, Gibbons SM, Larsen P, Shogan BD, Weiss S, Metcalf JL, Ursell LK, Vázquez-Baeza Y, Treuren WV, Hasan NA, Gibson MK, Colwell R, Dantas G, Knight R, Gilbert JA. 2014. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding LJ, Zhou XY, Zhu YG. 2020. Microbiome and antibiotic resistome in household dust from Beijing, China. Environ Int 139:105702. doi: 10.1016/j.envint.2020.105702. [DOI] [PubMed] [Google Scholar]
- 18.Richardson M, Gottel N, Gilbert JA, Lax S. 2019. Microbial similarity between students in a common dormitory environment reveals the forensic potential of individual microbial signatures. mBio 10:e01054-19. doi: 10.1128/mBio.01054-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lax S, Gilbert J. 2015. Hospital-associated microbiota and implications for nosocomial infections. Trends Mol Med 21:427–432. doi: 10.1016/j.molmed.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 20.Meadow JF, Altrichter AE, Kembel SW, Moriyama M, O'Connor TK, Womack AM, Brown GZ, Green JL, Bohannan BJM. 2014. Bacterial communities on classroom surfaces vary with human contact. Microbiome 2:7. doi: 10.1186/2049-2618-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood M, Gibbons SM, Lax S, Eshoo-Anton TW, Owens SM, Kennedy S, Gilbert JA, Hampton-Marcell JT. 2015. Athletic equipment microbiota are shaped by interactions with human skin. Microbiome 3:25. doi: 10.1186/s40168-015-0088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rai S, Singh DK, Kumar A. 2021. Microbial, environmental and anthropogenic factors influencing the indoor microbiome of the built environment. J Basic Microbiol 61:267–292. doi: 10.1002/jobm.202000575. [DOI] [PubMed] [Google Scholar]
- 23.Dingsdag S, Coleman NV. 2013. Bacterial communities on food court tables and cleaning equipment in a shopping mall. Epidemiol Infect 141:1647–1651. doi: 10.1017/S0950268812002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elsergany M, Moussa M, Ahsan A, Khalfan A, Eissa A. 2015. Exploratory study of bacterial contamination of different surfaces in four shopping malls in Sharjah, UAE. J Environ Occup Sci 4:101–105. doi: 10.5455/jeos.20150611030039. [DOI] [Google Scholar]
- 25.Al-Ghamdi AK, Abdelmalek SMA, Ashshi AM, Faidah H, Shukri H, Jiman-Fatani AA. 2011. Bacterial contamination of computer keyboards and mice, elevator buttons and shopping carts. Afr J Microbiol Res 5:3998–4003. doi: 10.5897/AJMR11.770. [DOI] [Google Scholar]
- 26.Gerba CP, Maxwell S. 2012. Bacterial contamination of shopping carts and approaches to control. Food Protection Trends 32:747–749. [Google Scholar]
- 27.Irshaid FI, Jacob JH, Khwaldh AS. 2014. Contamination of the handles and bases of shopping carts by pathogenic and multi-drug resistant bacteria. Eur Respir J 10:242–254. [Google Scholar]
- 28.Shen Y, Xu WJ, Li CW, Handel A, Martinez L, Ling F, Ebell M, Fu XF, Pan JR, Ren JP, Gu WL, Chen EF. 2020. A cluster of novel coronavirus disease 2019 infections indicating person-to-person transmission among casual contacts from social gatherings: an outbreak case-contact investigation. Open Forum Infect Dis 7:ofaa231. doi: 10.1093/ofid/ofaa231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dietz L, Horve PF, Coil DA, Fretz M, Eisen JA, Wymelenberg KVD. 2020. 2019 novel coronavirus (COVID-19) pandemic: built environment considerations to reduce transmission. mSystems 5:e00245-20. doi: 10.1128/mSystems.00375-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loo BPY, Tsoi KH, Wong PPY, Lai PC. 2021. Identification of superspreading environment under COVID-19 through human mobility data. Sci Rep 11:4699. doi: 10.1038/s41598-021-84089-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang K, Ni YQ, Li J, Imamovic L, Sarkar C, Kobler MD, Heshiki Y, Zheng T, Kumari S, Wong JCY, Archna A, Wong CWM, Dingle C, Denizen S, Baker DM, Sommer MOA, Webster CJ, Panagiotou G. 2018. The environmental exposures and inner- and intercity traffic flows of the metro system may contribute to the skin microbiome and resistome. Cell Rep 24:1190–1202.e5. doi: 10.1016/j.celrep.2018.06.109. [DOI] [PubMed] [Google Scholar]
- 32.Gu DX, Dong N, Zheng ZW, Lin D, Huang M, Wang LH, Chan EWC, Shu LB, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 33.Cave R, Cole J, Mkrtchyan HV. 2021. Surveillance and prevalence of antimicrobial resistant bacteria from public settings within urban built environments: challenges and opportunities for hygiene and infection control. Environ Int 157:106836. doi: 10.1016/j.envint.2021.106836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Health Organization. 2017. Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. https://www.who.int/publications/i/item/WHO-EMP-IAU-2017.12.
- 35.Donaghy JA, Jagadeesan B, Goodburn K, Grunwald L, Jensen ON, Jespers AD, Kanagachandran K, Lafforgue H, Seefelder W, Quentin MC. 2019. Relationship of sanitizers, disinfectants, and cleaning agents with antimicrobial resistance. J Food Prot 82:889–902. doi: 10.4315/0362-028X.JFP-18-373. [DOI] [PubMed] [Google Scholar]
- 36.Bouzada MLM, Silva VL, Moreira FAS, Silva GA, Diniz CG. 2010. Antimicrobial resistance and disinfectants susceptibility of persistent bacteria in a tertiary care hospital. J Microbiol Antimicrob 2:105–112. [Google Scholar]
- 37.Tandukar M, Oh S, Tezel U, Konstantinidis KT, Pavlostathis SG. 2013. Long-term exposure to benzalkonium chloride disinfectants results in change of microbial community structure and increased antimicrobial resistance. Environ Sci Technol 47:9730–9738. doi: 10.1021/es401507k. [DOI] [PubMed] [Google Scholar]
- 38.Gajardo K, Rodiles A, Kortner TM, Krogdahl Å, Bakke AM, Merrifield DL, Sørum H. 2016. A high-resolution map of the gut microbiota in Atlantic salmon (Salmo salar): a basis for comparative gut microbial research. Sci Rep 6:30893. doi: 10.1038/srep30893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Limon JJ, Skalski JH, Underhill DM. 2017. Commensal fungi in health and disease. Cell Host Microbe 22:156–165. doi: 10.1016/j.chom.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung MH, Wilkins D, Li EK, Kong FK, Lee PK. 2014. Indoor-air microbiome in an urban subway network: diversity and dynamics. Appl Environ Microbiol 80:6760–6770. doi: 10.1128/AEM.02244-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faust K, Raes J. 2012. Microbial interactions: from networks to models. Nat Rev Microbiol 10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 42.HilleRis LJ, Adler PB, Harpole WS, Levine JM, Mayfield MM. 2012. Rethinking community assembly through the lens of coexistence theory. Annu Rev Ecol Evol Syst 43:227–248. doi: 10.1146/annurev-ecolsys-110411-160411. [DOI] [Google Scholar]
- 43.Furman O, Shenhav L, Sasson G, Kokou F, Honig H, Jacoby S, Hertz T, Cordero OX, Halperin E, Mizrahi I. 2020. Stochasticity constrained by deterministic effects of diet and age drive rumen microbiome assembly dynamics. Nat Commun 11:1904. doi: 10.1038/s41467-020-15652-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan BJ. 2016. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J 10:655–664. doi: 10.1038/ismej.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venkataraman A, Bassis CM, Beck JM, Young VB, Curtis JL, Huffnagle GB, Schmidt TM. 2015. Application of a neutral community model to assess structuring of the human lung microbiome. mBio 6:e02284-14. doi: 10.1128/mBio.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao C, Montoya L, Xu L, Madera M, Hollingsworth J, Purdom E, Singan VR, Vogel J, Hutmacher R, Dahlberg J, Coleman-Derr D, Lemaux P, Taylor JW. 2020. Fungal community assembly in drought-stressed sorghum shows stochasticity, selection, and universal ecological dynamics. Nat Commun 11:34. doi: 10.1038/s41467-019-13913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lan GY, Quan F, Yang C, Sun R, Chen BQ, Zhang XC, Wu ZX. 2022. Driving factors for soil fungal and bacterial community assembly in topical forest of China. Appl Soil Ecol 177:104520. doi: 10.1016/j.apsoil.2022.104520. [DOI] [Google Scholar]
- 48.Zhang W, Zhao ZY, Lv FL, Yin YY, Wang ZH, Zhao ZY, Li ZY, Zhai BN. 2021. Fungal alpha diversity influences stochasticity of bacterial and fungal community assemblies in soil aggregates in an apple orchard. Appl Soil Ecol 162:103978. [Google Scholar]
- 49.Stephens B. 2016. What have we learned about the microbiomes of indoor environments? mSystems 1:e00083-16. doi: 10.1128/mSystems.00083-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rashid T, VonVille HM, Hasan I, Garey KW. 2016. Shoe soles as a potential vector for pathogen transmission: a systematic review. J Appl Microbiol 121:1223–1231. doi: 10.1111/jam.13250. [DOI] [PubMed] [Google Scholar]
- 51.Deshpande A, Cadnum JL, Fertelli D, Sitzlar B, Thota P, Mana TS, Jencson A, Alhmidi H, Koganti S, Donskey CJ. 2017. Are hospital floors an underappreciated reservoir for transmission of healthcare-associated pathogens? Am J Infect Control 45:336–338. doi: 10.1016/j.ajic.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 52.Osei SJ, Govinden U, Bester LA, Essack SY. 2016. Colistin and tigecycline resistance in carbapenemase-producing Gram-negative bacteria: emerging resistance mechanisms and detection methods. J Appl Microbiol 121:601–617. doi: 10.1111/jam.13169. [DOI] [PubMed] [Google Scholar]
- 53.Zhou J, Deng Y, Shen L, Wen C, Yan Q, Ning D, Qin YJ, Xue K, Wu LY, He ZL, Voordeckers JW, Nostrand JDV, Buzzard V, Michaletz ST, Enquist BJ, Weiser MD, Kaspari M, Waide R, Yang YF, Brown JH. 2016. Temperature mediates continental-scale diversity of microbes in forest soils. Nat Commun 7:12083. doi: 10.1038/ncomms12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.An XL, Wang JY, Pu Q, Li H, Pan T, Li HQ, Pan FX, Su JQ. 2020. High-throughput diagnosis of human pathogens and fecal contamination in marine recreational water. Environ Res 190:109982. doi: 10.1016/j.envres.2020.109982. [DOI] [PubMed] [Google Scholar]
- 55.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li LG, Dechesne A, He ZM, Madsen JS, Nesme J, Sørensen SJ, Smets BF. 2018. Estimating the transfer range of plasmids encoding antimicrobial resistance in a wastewater treatment plant microbial community. Environ Sci Technol Lett 5:260–265. doi: 10.1021/acs.estlett.8b00105. [DOI] [Google Scholar]
- 57.Delgado SL, Restrepo S, Bustos JR, Zambrano MM, Anzola JM. 2016. Mycofier: a new machine learning based classifier for fungal ITS sequences. BMC Res Notes 9:402. doi: 10.1186/s13104-016-2203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, Tang JB, Wu GX, Zhang H, Shi YJ, Liu Y, Yu C, Wang B, Lu Y, Han CL, Cheung DW, Yiu SM, Peng SL, Zhu XQ, Liu GM, Liao XK, Li YR, Yang HM, Wang J, Lam TW, Wang J. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guitor AK, Raphenya AR, Klunk J, Kuch M, Alcock BP, Surette M, McArthur A, Poinar H, Wright GD. 2019. Capturing the resistome: a robust and reliable targeted capture method for detecting antibiotic resistance determinants. Antimicrob Agents Chemother 64:e01324-19. doi: 10.1128/AAC.01324-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu B, Zheng DD, Jin Q, Chen LH, Yang J. 2019. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47:D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nawrocki EP, Eddy S. 2013. Infernal 11: 100-fold faster RNA homology searches. Bioinformatics 29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cury J, Jové T, Touchon M, Néron B, Rocha EP. 2016. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res 44:4539–4550. doi: 10.1093/nar/gkw319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 30. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 67.R Core Team. 2016. A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 68.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statist Soc B 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 69.Li H, Zhou XY, Yang XR, Zhu YG, Hong YW, Su JQ. 2019. Spatial and seasonal variation of the airborne microbiome in a rapidly developing city of China. Sci Total Environ 665:61–68. doi: 10.1016/j.scitotenv.2019.01.367. [DOI] [PubMed] [Google Scholar]
- 70.Leung MHY, Tong X, Wilkins D, Cheung HHL, Lee PKH. 2018. Individual and household attributes influence the dynamics of the personal skin microbiota and its association network. Microbiome 6:26. doi: 10.1186/s40168-018-0412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST. 2011. Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sloan WT, Lunn M, Woodcock S, Head IM, Nee S, Curtis TP. 2006. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol 8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- 73.Breiman L. 2001. Random forests. Mach Learn 45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Diversity and composition of floor, escalator, soil, and road dust microbial communities. (a and b) Chao 1 index and Simpson index of bacterial communities. (c and d) Chao 1 index and Simpson index of fungal communities. Different letters indicate significant differences among different samples (mean ± standard deviation [SD]; ANOVA, P < 0.05). NMDS analysis revealed significant seasonal differences in floor (e) and escalator (f) microbial communities (Adonis test, P < 0.01). (g and h) Bacterial (at the phylum level) and fungal (at the class level) community compositions varied with seasons (spring and summer). Download FIG S1, TIF file, 1.3 MB (1.4MB, tif) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Principle-coordinate analysis based on Bray-Curtis distance showed that the microbial community (bacterial community [a and b] and fungal community [c and d]) from the surfaces of floors or escalators was not grouped across shopping malls. Download FIG S2, TIF file, 0.6 MB (635.4KB, tif) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary for the collected samples. Download Table S1, DOCX file, 0.1 MB (82.8KB, docx) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Correlation network analysis depicted the microbial interactions in different habitats. A connection represents a strong (Pearson’s r = 1) and significant (P < 0.01) correlation. Nodes indicate the taxonomic affiliation at the family level. Node size is proportional to the number of connections. Download FIG S3, TIF file, 2.5 MB (2.5MB, tif) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Topological features of cooccurrence networks for microbial communities. Download Table S2, DOCX file, 0.04 MB (40.8KB, docx) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(a) The immigration rates at the community level were predicted by the neutral community model. Immigration rates indicate the probability that a dead individual is replaced by an immigrant. (b) The normalized stochasticity ratio (NST) index evaluated the relative importance of deterministic (<50%) and stochastic (>50%) community assembly processes. Download FIG S4, TIF file, 0.5 MB (575.2KB, tif) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A random forest model indicates the predictor importance (percentage of increase in mean square error) of dominant ASVs (>274 in total number) as drivers for microbial community in the whole profile. The accuracy importance measure was computed for each tree and averaged over the forest (1,000 trees). Percentage increases in the mean squared error (MSE) of variables were used to estimate the importance of these predictors, and higher MSE% values imply more important predictors. The embedded charts showed that the true-positive percentage and the false-positive percentage achieved an area under the curve (AUC) value of 0.9024 with s 95% confidence interval (CI) of 0.8168 to 0.9669 between trained (n = 183) and tested (n = 91) samples in the validation cohort (P < 0.0001). Download FIG S5, TIF file, 2.0 MB (2MB, tif) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of predictive accuracy of random forest supervised learning models for habitat, season, and shopping mall classification. Download Table S3, DOCX file, 0.05 MB (48.8KB, docx) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SourceTracker models predicted the proportions of bacterial taxa from floors and escalators that come from possible sink environments. The bacterial communities from soils, road dust, air, and human palms were used as sources. Download FIG S6, TIF file, 0.1 MB (132.8KB, tif) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed information for amplicon sequencing, HT-qPCR assays for human pathogens, identification of the Enterobacteriaceae isolates from malls, neutral community models, and random forest models. Download Text S1, DOCX file, 0.04 MB (44.3KB, docx) .
Copyright © 2022 An et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The raw amplicon sequences were submitted to the Sequence Read Archive (SRA) under accession no. PRJNA707496. All full-length sequences of 16S rRNA genes and the genome assemblies of Enterobacteriaceae species were deposited under GenBank no. MZ461606–MZ461903 and SRA accession no. PRJNA749475, respectively.