ABSTRACT
Microbial communities can be structured by both deterministic and stochastic processes, but the relative importance of these processes remains unknown. The ambiguity partly arises from an inability to disentangle soil microbial processes from confounding factors, such as aboveground plant communities or anthropogenic disturbance. In this study, we characterized the relative contributions of determinism and stochasticity to assembly processes of soil bacterial communities across a large environmental gradient of undisturbed Antarctic soils. We hypothesized that harsh soils would impose a strong environmental selection on microbial communities, whereas communities in benign soils would be structured largely by dispersal. Contrary to our expectations, dispersal was the dominant assembly mechanism across the entire soil environmental gradient, including benign environments. The microbial community composition reflects slowly changing soil conditions and dispersal limitation of isolated sites. Thus, stochastic processes, as opposed to deterministic, are primary drivers of soil ecosystem assembly across space at our study site. This is especially surprising given the strong environmental constraints on soil microorganisms in one of the harshest environments on the planet, suggesting that dispersal could be a driving force in microbial community assembly in soils worldwide.
IMPORTANCE Because of their diversity and ubiquity, microbes provide an excellent means to tease apart how natural communities are structured. In general, ecologists believe that stochastic assembly processes, like random drift and dispersal, should dominate in benign environments while deterministic processes, like environmental filtering, should be prevalent in harsh environments. To help resolve this debate, we analyzed microbial community composition in pristine Antarctic soils devoid of human influence or plant communities for eons. Our results demonstrate that dispersal limitation is a surprisingly potent force of community limitation throughout all soil conditions. Thus, dispersal appears to be a driving force of microbial community assembly, even in the harshest of conditions.
KEYWORDS: community assembly, stochasticity, determinism, niche, dispersal
INTRODUCTION
Ever since Darwin published On the Origin of Species (1), ecologists have sought to understand the processes that control community composition. In general, communities can be assembled via either deterministic or stochastic processes (2, 3). Deterministic processes arise from niche theory, including both competitive exclusion (i.e., selection against overly similar species from occupying a community [4]) and environmental filtering (i.e., selection against overly dissimilar species when the environment is unsuitable [5]). Stochastic assembly, on the other hand, refers to probabilistic processes like dispersal, priority effects during colonization, and community drift via random births and deaths (2). There is as yet no clear trend as to whether determinism or stochasticity are more important to community structure. The relative importance of determinism and stochasticity varies among both studies and locations (3, 6–13). The balance between determinism and stochasticity is likely dictated by the abiotic environment but few studies explicitly link changes in community processes to changes in the environment using gradient studies (8, 10, 14). Identifying how community assembly mechanisms vary spatially has thus been described as one of the most important goals in ecology.
The ways in which community assembly mechanisms vary along environmental gradients remains contested due to a number of contrasting theories and results. On one hand, some have suggested that harsh environments should be stochastic and benign environments should be structured by deterministic competition (12). Indeed, competition among plant species is strongest in warm, resource-rich environments compared to cold and stressful environments (15). On the other hand, studies suggest that deterministic processes, such as environmental filtering, should also be strongest in harsh environments that impose severe constraints on occupancy and growth (16). For example, environmental filtering of plant communities was strongest in old-growth forests where light and nutrients were limiting (17). It is also possible that determinism is strongest in both harsh and resource-rich environments, while stochasticity dominates in the middle of the resource gradient, as implied by the stress-gradient hypothesis (18). Only a quantitative gradient study that allows determinism and stochasticity to vary in strength can test these hypotheses.
Soil microbial communities provide an excellent system to test assembly theory because microbial communities are incredibly speciose, omnipresent even in the harshest environments, and exhibit high spatial and temporal turnover (19). Microbes were historically thought to be uninhibited by dispersal limitation, as stated in the adage “everything is everywhere but the environment selects” (20). However, the Baas-Becking hypothesis has now been superseded by shifting emphasis toward resolving the “neutral versus niche” debate (21), including how stochastic and deterministic processes vary in strength with environmental conditions. Unfortunately, efforts to differentiate between determinism and stochasticity are complicated by two issues. First, a single mechanism can yield both deterministic and stochastic patterns. For example, dispersal constraints are typically considered stochastic, but can yield beta-diversity patterns that appear deterministic (5). Likewise, competition is generally thought to result in deterministic patterns of similarity; however, competition can also result in stochastic assembly even when niche processes are the driving force (22). Second, many null modeling methods partition total amounts of variation in beta-diversity into deterministic and stochastic components. Doing so forces determinism and stochasticity to trade off because an increase in the variability explained by one factor must be matched by a decrease in variation explained by the other. Yet there is no a priori reason to assume a zero-sum balance between determinism and stochasticity.
Here, we tested how community assembly mechanisms varied along an environmental gradient of soil conditions in the Shackleton Glacier region of Antarctica (Fig. 1). Specifically, we tested three hypotheses. Hypothesis 1: Both the environment and spatial patterns influence microbial community composition. This would indicate that community assembly is determined by a balance of both putative deterministic (i.e., environmental filtering) and stochastic (i.e., dispersal) mechanisms. Hypothesis 2: Deterministic assembly mechanisms are strongest in the harshest soils, but become less important as soil stress declines. Hypothesis 3: Tradeoffs between environmental filtering and dispersal are directly responsible for the observed changes in assembly mechanisms. We tested Hypotheses 2 and 3 by combining the power of traditional null modeling methods with a new model that explicitly accounts for different mechanisms and does not implicitly assume a trade-off between factors. By working with Antarctic soils, our study presents a novel ecosystem found nowhere else on the planet. First, the stability of Antarctic soils is unique compared to almost every other location on Earth (23), which have undergone extreme changes within the last 2 million years. Second, our study area has been devoid of aboveground plants since the Neogene (2.5 million years before present) and possibly the mid-Miocene (14 million years before present) (24, 25), such that we can test how changes in soil properties per se influence microbial assembly patterns. Finally, we used a new modeling approach to explicitly identify the relative importance of dispersal versus environment, providing a detailed assessment of assembly mechanisms. Our approach also expands on previous work by quantifying independent assembly mechanisms for colonization (i.e., presence/absence) and postcolonization success (i.e., relative abundance).
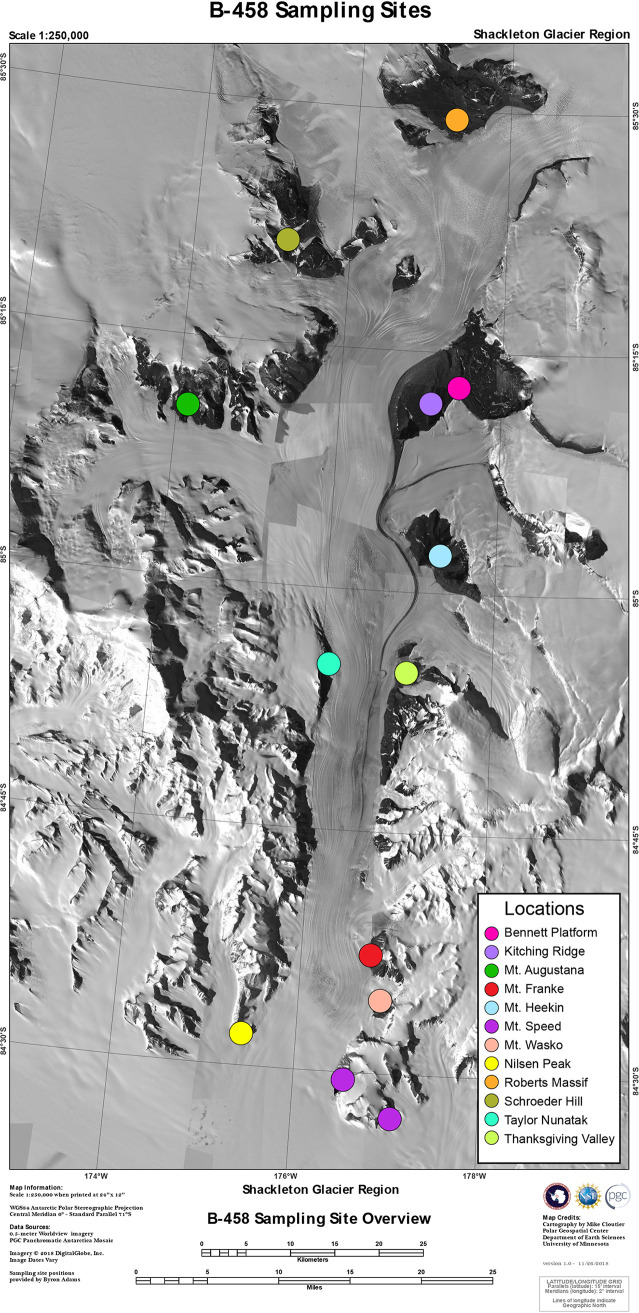
FIG 1.
Map of the Shackleton Glacier, with sampling sites denoted as colored point. Map by Mike Cloutier, Polar Geospatial Center (Imagery © 2021 Maxar; reproduced with permission).
RESULTS
Hypothesis 1: The environment and spatial arrangement control microbial community composition.
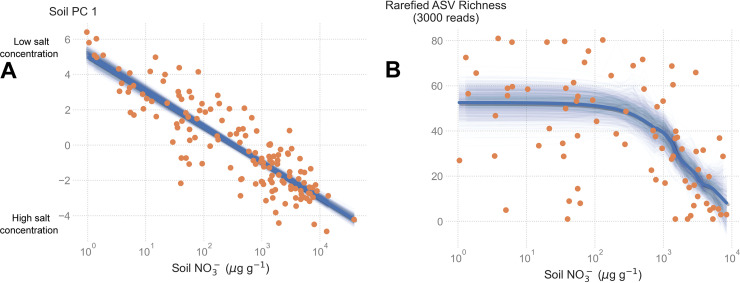
Both the environment and spatial distribution of sites controlled microbial community composition. Microbial species richness in particular was strongly regulated by the environment, but only in the harshest soils. Ordination of soil geochemistry yielded a single axis (Soil PC 1) that explained 56% of variation in geochemical and physical properties (Table S1). Soil PC 2 explained an additional 12% of geochemical variation among sites. Concentrations of Ca2+, Cl-, F-, K+, Mg2+, Na+, NH3, NO3-, PO43−, SiO2, SO42−, and Sr2+ all positively co-varied, clustering along Soil PC 1. Thus, Soil PC 1 was a salt concentration gradient that could be adequately represented by soil NO3- concentrations (pseudo-R2 = 0.82, Fig. 2A). As a result, we hereafter refer to soil NO3- concentrations to represent the soil salt gradient.
FIG 2.
Relationships between soil salt concentrations, exposure age, and microbial ASV richness. (A) The relationship between soil principal component (PC 1) and soil NO3- concentrations. The thick blue line shows the linear model fit to the observed data, and thin blue lines show 1,000 bootstrapped linear regressions. (B) Relationship between ASV richness and soil NO3- concentrations. Thick blue line shows the LOWESS fit to the observed data, while thin blue lines show 1,000 bootstrapped LOWESS estimates.
Variable loadings (distance calculation) of each variable on the first two principal component axes. Download Table S1, DOCX file, 0.01 MB (4.9KB, docx) .
Copyright © 2023 Lemoine et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The environment imposed a strong constraint on microbial community composition, as evidenced by the effect of soil NO3- on microbial diversity. Rarefied richness of amplicon sequence variants (R-ASV richness) appeared to be determined in large part by soil NO3- concentrations (i.e., the environment, Fig. 2B). Harsh soils with high NO3- concentrations had fewer ASVs present than benign environments (pseudo-R2 = 0.33, Fig. 2B). Mean R-ASV richness was stable (~53 ASVs) for soils with low NO3- concentrations until a threshold around 103 μg g−1 NO3-, beyond which R-ASV decline steeply (pseudo-R2 = 0.40, Fig. 2B).
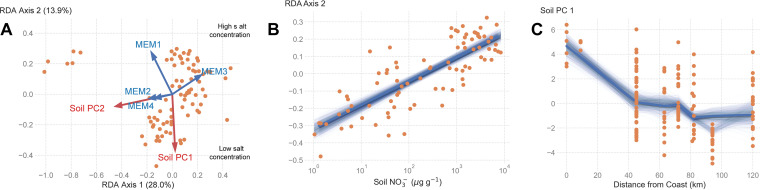
Redundancy analysis (RDA) demonstrated that microbial community composition was also constrained by both the environment and spatial site arrangement. RDA 1 represented both dispersal and environmental effects (Fig. 3A). In particular, RDA 1 separated the location closest to the polar plateau (Robert’s Massif) from all other sites (based on MEM3), whereas RDA 2 reflected the salt concentration gradient and characterized sites based on their distance from the coast (based on MEM1). Spatially, this means that inland sites further from the cost, particularly those on Robert’s Massif, had distinct microbial communities from low-elevation coastal sites. Environmentally, given that soil NO3- concentrations are a good proxy for soil saltiness (Fig. 2A), RDA 2 illustrated the degree to which soil salt concentrations influence microbial community composition (Fig. 3B). The similarity in soil salt concentrations among sites close to and distant from the coast suggests that the microbial uniqueness of the polar plateau sites stemmed from spatial isolation rather than changes in soil chemistry (Fig. 3C). Variance partitioning confirmed that both location and soil characteristics structure microbial communities. Environmental variables accounted for 16% of the variation in soil microbial communities, while spatial variables accounted for an additional 16% of variability. The covariation of soil characteristics along the spatial gradient explained another 9% of community variability.
FIG 3.
Redundancy analyses of microbial community composition. (A) Redundancy analysis (RDA) biplot of microbial community composition in relation to physical variables and spatial arrangement of sites. The cluster of points in the upper left corner are Robert’s Massif. (B) Relationship between RDA Axis 2 and soil NO3- concentrations. Thick blue line shows the linear model fit to all observed data, and thin blue lines show 1,000 bootstrapped linear regressions. (C) Relationship between Soil PC 1 and distance from the coast (km) for each site. Thick blue line shows the LOWESS fit to all observed data, and thin blue lines show 1,000 bootstrapped LOWESS fits.
Hypothesis 2: Deterministic processes should be strongest in the harshest soils.
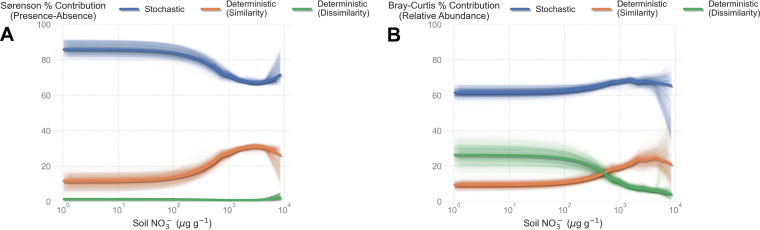
We tested the hypothesis that deterministic processes, like environmental filtering, should be strongest in the harshest soils using a null-modeling approach. We found that the relative contributions of stochastic and deterministic processes to community assembly changed across the soil salt concentration gradient, but not in the predicted way. In less stressful environments, stochastic processes contributed to the bulk of presence-absence composition (~90%), while deterministic similarity (i.e., environmental filtering) contributed the remaining variability (Fig. 4A). The relative importance of stochastic and deterministic assembly remained constant across the salt gradient up to 103 μg g−1 NO3- (Fig. 4A). In saltier soils, the contribution of stochastic processes declined, as expected, and also became more variable. However, stochastic processes still contributed 60% to 70% of the presence/absence variability. Deterministic environmental filtering, while important, was never the dominant process (Fig. 4). Results for relative taxon abundances were less certain due to the added complexities of predicting species abundance, but they exhibited patterns similar to the phylum-level analyses presented here (see supplemental information). In the most benign soils, stochastic processes contributed ~60% of the variation in microbial community turnover, while deterministic dissimilarity and similarity each averaged about 30% and 10% of the contribution, respectively (Fig. 4B). In the saltiest soils, the contribution of stochasticity increased to ~65%, while the importance of deterministic dissimilarity declined to negligible levels and was replaced entirely by deterministic similarity (Fig. 4B). These results suggest that less harsh environments are potentially regulated more via niche partitioning while stressful soils become increasingly structured via environmental filtering, but stochastic processes were the dominant structuring force across the entire soil gradient.
FIG 4.
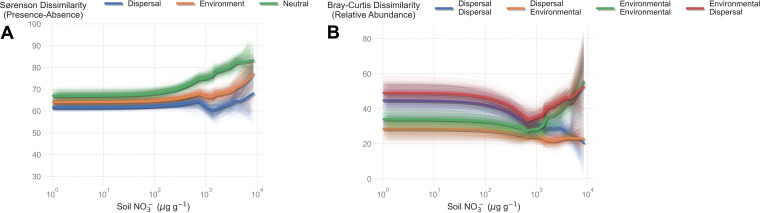
Microbial communities were largely structured by stochastic processes across all soil wetting ages. (A) Contribution of stochastic and deterministic processes to microbial community composition based on Sørenson dissimilarity, which records presence-absence dissimilarities. We calculated the average variance component for each site. Thick lines show a LOWESS regression against NO3- concentrations. Thin lines show 1,000 bootstrapped LOWESS regressions. (B) Contribution of stochastic and deterministic processes to microbial community composition based on Bray-Curtis dissimilarities, which record relative abundance dissimilarities. In all simulations, phylum richness was held fixed at the observed level for each site. Thick lines show a LOWESS regression against NO3- concentrations. Thin lines show 1,000 bootstrapped LOWESS regressions.
Hypothesis 3: Environmental filtering should be stronger than dispersal in the harshest soils.
We expected to find strong environmental filtering and weak dispersal limitation in most stressful soils with extremely high soil salt concentrations. However, our models did not support this hypothesis. With presence/absence data, the importance of dispersal increased with soil NO3- concentrations (Fig. 5A). Below 103 μg g−1 NO3-, neutral, environmental, and dispersal weightings all resulted in simulated communities that were 60% to 70% dissimilar from observed communities (Fig. 5A). Above 103 μg g−1 NO3-, neutral weighting and environmental weighting provided poorer fits to the data, through dispersal weighting maintained a 60% to 70% dissimilarity across all relative exposure ranges (Fig. 5A). This confirms that both environmental filtering and dispersal increase in importance as soils develop harsher environmental conditions, but dispersal remains the dominant factor for the presence/absence of microbial taxa in Antarctic soils.
FIG 5.
Dissimilarity of modeled communities to observed communities when modeled communities were structured by neutral, dispersal, or environmental processes. (A) Averaged Sørenson dissimilarity of simulated communities from observed communities. For each site, we took the average pairwise dissimilarities to the observed site from 1,000 model simulations. Thick lines show the LOWESS fit to the averaged dissimilarities as a function of soil NO3- concentrations. (B) For each site, we took the average pairwise dissimilarities to the observed site from 1,000 model simulations. Thick lines show the LOWESS fit to the average dissimilarities as a function of soil NO3- concentrations. Thin lines show 1,000 bootstrap simulations. In all simulations, phylum richness was fixed to the observed level for each site.
Simulations that included relative abundance based on Bray-Curtis dissimilarities yielded a slightly different interpretation. First, Bray-Curtis simulations fit the observed data much better than presence/absence data, with dissimilarities ranging from 20% to 60% (Fig. 5B). In other words, it mattered less whether taxa occurrence was dictated by dispersal or the environment, so long as abundance was determined by the environment. Above the 103 μg g−1 NO3- threshold, patterns reversed. The weightings for presence/absence became the most important factor, as both dispersal-weighted colonization models were ~40% more accurate than models with environmentally weighted colonization, regardless of abundance (Fig. 5B). Despite changes in the goodness of fit for some models, the best-fitting model remained consistent across the entire salt concentration gradient; dispersal-limited presence/absence and environmental-limited abundance consistently provided the best fits to observed community composition (Fig. 5B). From these results, we can conclude that, regardless of soil salt concentrations, dispersal is the primary factor determining which species appear in a community, while the environment determines how well those species perform once present.
DISCUSSION
Our data highlight the importance of extreme environmental gradients, as changes in soil microbial diversity only manifested in extremely harsh environments. Previous studies of community assembly demonstrated that deterministic and stochastic processes trade off, with stochastic processes dominating in benign soils and determinism increasing in harsher, low-nutrient soils (8, 10, 14). In Antarctic soils, salt concentrations form the basis of a stress gradient (Fig. 2), and thus, we expected environmental filtering to be the predominant force structuring microbial communities. Using diversity-based modeling approaches, we did indeed find evidence for strengthened deterministic processes in stressful soils, coupled with weakening stochastic effects (Fig. 4). Yet our modeling approach detected a consistently strong signal of dispersal limitation in soils salt concentration (Fig. 5). These lines of evidence are not contradictory, as stochastic processes consistently explained ~60% of the variation in community assembly, even in stressful soils, and the trade-off in determinism could in fact be due to a decline in other effects, such as drift (26).
Our modeling approach makes two key assumptions: that dispersal limitation can accurately be represented by geographic ranges, and that we measured all relevant environmental parameters. Regarding the assumption that bacterial dispersal is correlated with geographic distance, the degree of dispersal in bacterial communities is still an open question. Some studies have documented near-global dispersal of certain microbial taxa in the atmosphere (27), leading some researchers to propose that microbes are never limited by geographic barriers (28). Yet numerous experimental studies have documented dispersal limitation over very short scales (<1 km) for fungi (29, 30), bacteria (31), and archaea (32). The assumption that dispersal limitation could be represented as geographic separation therefore is reasonable. Our models imply that dispersal is an important process, because our models that included geographic dispersal limitation consistently outperformed other models. The second assumption underlying our support vector machine approach to model environmental niches is that we have measured all appropriate parameters. Our environmental parameters include soil water and salt concentrations, all of which are known to be strong determinants of Antarctic soil community composition (33–35). We are, therefore, confident that our support vector machine model can appropriately quantify relative habitat suitability for each taxon.
Our study represents a comprehensive examination of microbial community assembly, free from confounding factors due to the uniqueness of Antarctic soils remaining stable and free from anthropogenic disturbance for up to several million years. However, this stability is under threat. Antarctica is one of the most rapidly changing environments on the planet due to human activities (36), and soil communities are expected to respond rapidly to these changes (37). Studies conducted in pristine ecosystems relatively free from human disturbance are, therefore, crucial and becoming increasingly difficult to undertake. While previous work demonstrated that microbial community assembly processes rapidly become deterministic on short time scales (14), it is difficult to know whether strong determinism of microbial communities is driven by aboveground selection on microbial communities. Indeed, both plant and bacterial communities change rapidly during succession (38), and plant communities exert strong selection on microbial communities that can lead to a decline in richness and the appearance of environmental filtering in less than 100 years following deglaciation (39, 40). By examining a succession gradient that has been free of aboveground vegetation for millennia, our study suggests that the rapid inclination toward determinism in bacterial community succession is apparently an artifact of aboveground succession. In the absence of vegetation, deterministic processes did increase in importance. but never fully replaced stochastic processes as the strongest driver of microbial community assembly as soils aged. Thus, microbial communities per se might be more strongly dispersal limited than previously assumed.
MATERIALS AND METHODS
Sampling methods.
Soil samples were collected from the Shackleton Glacier region from December 2017 to January 2018. A total of 232 soil samples were collected from 12 sites running the length of the glacier. These sites represent a range of elevations (150 to 2221 m asl) across a 120-km north-south distance running from the Ross Ice Shelf to the Polar Plateau (Fig. 1). Between 4 to 26 soil samples were collected along transects located at each of the 12 sites to maximize variation in soil characteristics. Soils (0 to 10 cm depth) were collected in sterile polyethylene bags using clean hand trowels. GPS coordinates, photographs of the soil surface, elevation, and other metadata were recorded at the time of soil sample collection. All soils were transported to the field camp in insulated coolers, where they were frozen at –20°C and remained frozen until processed at the University of Colorado, Boulder, CO, USA.
Soil geochemistry.
Gravimetric soil moisture was measured by weighing 50 g of subsampled soil pre- and postoven drying at 105°C for 24 h. The water-soluble salt data interpreted in this study were reported by Diaz et al. (41, 42). Briefly, soils were leached at a 1:5 soil:DI water ratio for 24 h. The leachate was filtered through a 0.4 μm Nuclepore membrane filter and analyzed for major anions (F-, Cl-, Br-, SO42−) on a Dionex ICS-2100 ion chromatograph, and cations (K+, Na+, Ca2+, Mg2+) on a PerkinElmer Optima 8300 Inductively Coupled Plasma-Optical Emission Spectrometer (ICP-OES), and nutrients (NO3-, NO2-, PO43−, SiO2, NH3) on a Skalar San++ Automated Wet Chemistry analyzer (41–43). We focused on the subset of soil samples from Diaz et al. (41, 42) that had all biogeochemical measurements within instrumentation detection limits. We did not measure soil organic carbon because it is nearly absent from the Transantarctic Mountains (TAM), being below 0.5% and rarely above 2% (44). We also did not measure pH because it is relatively consistent (7.5 to 9.5) throughout the TAM (45, 46), and historical surveys found near-neutral soil pH at even the most isolated, harshest, saltiest site: Robert’s Massif (47). Further, soil pH and soil water content are nearly colinear in the TAM (45), such that including soil pH in analyses would likely not influence results. We also measured Sr2+ and PO4−3, but excluded these two variables from all analyses because they were below detection limit at most sites.
Microbial DNA sequencing and analyses.
DNA was extracted from each soil sample using the Qiagen Dneasy Powersoil HTP 96 kit (Qiagen, Germantown, MD, USA), following the manufacturer’s recommendations. Extracted DNA was PCR-amplified using a primer pair that targets the hypervariable V4 region of the archaeal and bacterial 16s rRNA gene (515F/806R), with appropriate Illumina adapters and unique barcode sequences. Amplified product was cleaned and normalized to equimolar concentrations using SequalPrep Normalization Plates (Thermo Fisher Scientific, Carlsbad, CA, USA) and sequences on an Illumina MiSeq using the V2 2 × 150 bp paired-end Illumina sequencing kits (Illumina, San Diego, CA, USA). 16s rRNA gene sequences were processed using the DADA2 pipeline (48) and were quality filtered and clustered into amplicon sequence variants (ASVs) with all sequences in a given ASV sharing 100% sequence similarity. Taxonomic information was assigned to ASVs using a Naive Bayesian classifier (49), which takes the set of ASVs generated and compares them to a training set of reference sequences from the 16s rRNA bacterial and archaeal SILVA database (50, 51). Samples that met a threshold of 586 reads per sample were included in downstream analyses. A total of 86 samples met this threshold and had the complete set of corresponding geochemical data, and these 86 samples were used in all downstream analyses.
Hypothesis 1: The environment and spatial arrangement control microbial community composition.
Our first hypothesis predicts that microbial community composition will be controlled by both the soil environment and the spatial arrangement of sites. We tested this hypothesis by analyzing the relationship between microbial species richness and soil properties, by analyzing the relationship between soil properties, spatial distribution, and microbial community composition, and finally by partitioning variance in microbial community composition between the environment and spatial site arrangement.
Principal-component analysis of soil geochemistry.
First, we collapsed multivariate metrics of soil characteristics into fewer dimensions using a principal-component analysis (PCA) of soil geochemistry and water content. The variables included salts (F- [ppm], Cl- [ppm], SO42− [ppm], Na+ [ppm], Mg2+ [ppm], K+ [ppm], Ca2+ [ppm], SiO2 [ppm], NO3- [ppm], NH3 [ppb]), and gravimetric soil moisture. All variables were log-transformed prior to PCA to improve linearity, and log-transformed variables were then all scaled to N(0, 1) distributions to remove the effects of different measurement scales. PCA proceeded via SVD decomposition of the standardized geochemistry matrix (52).
Rarefaction.
To analyze how whether the environment controlled ASV richness, we first rarefied ASV richness to a standard number of reads to account for differing sampling depths among sites. Individual-level ASV richness was rarefied using the asymptotic equivalent ΣiS(1 – [1 – Ni/N]N*), where S is the total number of ASVs, Ni is the number of reads for the ith ASV, N is the total number of reads in the sample, and N* is the rarefied number of reads (1 to 3,000) (53). We standardized ASV richness at all sites to 3,000 reads, the smallest number of total reads from the final 86 samples, to standardize ASV richness across all sites to the same, minimum number of reads. Standardizing to a higher number of reads would require extrapolation beyond the data for species-poor sites, and rarefaction curves showed that all sites reached an asymptote, such that 3,000 reads sufficiently captured species richness (Fig. S1). All remaining 86 samples (after removing unreliable samples with fewer than 586 reads) had at least 3,000 reads, and we calculated rarefied richness at the smallest number of reads so that no sample had to extrapolate beyond the observed number of reads.
Rarefaction curves for each site showed clear asymptotes by 3,000 reads, indicating that our sampling had sufficiently captured ASV richness. Red line shows 3,000 reads. Download FIG S1, PDF file, 0.03 MB (29.3KB, pdf) .
Copyright © 2023 Lemoine et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Redundancy analysis of microbial communities.
We then assessed how both the environment and the spatial arrangement of sites affected microbial community composition conducting RDA of Hellinger-transformed microbial communities that included both environmental and spatial predictors. Environmental predictors were the first two soil principal component axes. Spatial predictors were Moran’s Eigenvector Maps (MEMs). To construct MEMs, we first calculated pairwise distances (km) between all sites in a matrix D. Following Legendre and Legendre (52), we set the threshold value for distances at the maximum distance of a minimum spanning tree for D; any pairwise distance above this threshold (as well as the diagonal elements) were set to four times the threshold amount to generate a truncated matrix D*. We then conducted a principal coordinates analysis (PCoA) on D*. From the PCoA, we kept only the eigenvectors associated with positive eigenvalues. We visually inspected these 12 eigenvectors for interpretable spatial patterns by plotting a map of the sites and coloring the points according to eigenvector score. The first four MEMs resulted in interpretable patterns and were thus used in downstream analyses. The remaining eight MEMs did not describe spatial patterns that we could easily categorize or discern and were excluded from subsequent analyses. The RDA included predictors of an environmental matrix E containing the first two soil principal components and a spatial matrix S containing the first four MEMs. The RDA returned six axes that cumulatively explained 45% of the variation in fitted site scores. RDA was conducted at both the phylum and ASV levels (Fig. S3).
Following RDA, we used variance partitioning to describe the amount of variation in soil microbial communities accounted for by spatial and environmental variables. Variance partitioning proceeded by creating three RDA models; spatial only (S), environment only (E), and spatial + environment (S+E) predictors. We calculated the variance explained (R2) of each model, and used the following equations to isolate the variance due to each component:
Hypothesis 2: Deterministic processes should be strongest in the harshest soils.
We tested the hypothesis that deterministic assembly processes would be strongest in the harshest soils by partitioning variation in β-diversity into “deterministic” and “stochastic” variability for every site along the environmental gradient. First, we partitioned the variation in β-diversity using the method of Ning et al. (13) with the following procedure:
We calculated neutral regional frequency weights wf by calculating the proportion of sites occupied for each ASV. Proportions were then divided by the proportion sums across all ASVs to normalize into a probability weighting (i.e., Σwf = 1).
We calculated neutral regional abundance weights wa by summing the total number of reads for each ASV across all sites, then divided ASV totals by the total number of reads for all ASVs and sites.
For each site, we randomly drew species occurrences based on regional frequency weights wf. ASV richness was fixed at observed ASV richness so that differences in community composition did not reflect differences in richness.
Once null-model species had been chosen for each site, we calculated Dirichlet weighting as α = wa*/min(wa*), where wa* are the regional weights only for those species chosen in Step 3. Relative abundances for the null species were then drawn from a Dirichlet(α) distribution.
We then calculated pairwise distances among sites using either Sørensen or Bray-Curtis pairwise site dissimilarities. This procedure resulted in 1,000 estimates of neutral Sørensen and Bray-Curtis pairwise site dissimilarities. We next calculated pairwise dissimilarities based on the observed data O. To partition variation in stochastic (stoch), deterministic similarity (det-a), and deterministic dissimilarity (det-b), we estimated E(d) as the expected pairwise dissimilarity based on neutral simulations, effectively averaging dij across all 1,000 simulations. Then,
a. If :
b. If :
c.
These steps yielded one estimate of each component (stoch, det-a, det-b) for each site to all other sites. In other words, stoch was an 86 × 86 matrix that contained the contribution of stochasticity to dissimilarities among all sites. For graphics and analyses, we took the average contribution of stoch, det-a, det-b for every site to all other sites (i.e., averaged by columns) used LOWESS regression to identify the relationships between each component and soil nitrate (i.e., salt) concentrations.
Hypothesis 3: Environmental filtering should be stronger than dispersal in the harshest soils.
The above null models can only separate out “deterministic” versus “stochastic” assembly processes, and do not reflect the underlying processes. To directly assess how the strength of environmental filtering and dispersal vary along an environmental gradient, we simulated communities following different “rules,” with rules consisting of probability weights corresponding to the process in question. The two processes being tested were dispersal and environmental filtering; thus, we generated probability weights related to dispersal probability and niche suitability.
(i) Dispersal probabilities. Dispersal weights (wd) were taken as the inverse-distance weighted relative abundance for each ASV. That is, the dispersal weight for ASV k at site I (wd-ik) was equal to the average relative abundance of ASV k at all other sites, weighted by the inverse distance to each site: wd-ik = Σj≠1 (1/dij)xjk/Σj≠1 (1/dij), such that distant sites with high abundances were down-weighted, while nearby sites with high abundances were up-weighted, as the most likely correspond to “source” populations.
(ii) Environmental probabilities. Environmental weights (we) were calculated using a support vector machine learning algorithm (Python module scikit-learn). Prior to analyses, relative abundance data were converted to presence/absence data. We then used the environmental matrix consisting of all principal components to predict the probability of occurrence of each ASV at each site. The SVC function from scitkit-learn uses a radial basis function for the kernel. The probability predictions are generated using logistic regression against the model scores, and then trained with 5-fold cross validation.
Neutral simulations included four different rules: neutral dispersal wherein probabilities were relative regional frequencies wf described above, neutral abundance wherein probabilities were regional abundances wa described above, dispersal weights wd, and environmental weights we. We simulated communities in two steps: first, species were randomly filtered for occurrence according to one of the four rules. Next, for the species that passed the first filter, abundances of those species were randomly generated based on a second rule. The combination of rules reflects seven different assembly processes:
Completely neutral (wf → wa)
Environmental filtering for occurrence, neutral determination of abundance (we → wa)
Dispersal limitation for occurrence, neutral determination of abundance (wd → wa)
Environmental filtering for occurrence, environmental filtering for abundance (we → we)
Environmental filtering for occurrence, dispersal limitation of abundance (we → wd)
Dispersal limitation for occurrence, environmental filtering of abundance (wd → we)
Dispersal limitation for occurrence, dispersal limitation of abundance (wd → wd)
For each simulation, we calculated the Sørensen and Bray-Curtis dissimilarities of observed communities to simulation communities. We repeated 1,000 simulations and averaged the dissimilarities for each site across all simulations. The environmental and distance weights were only weakly correlated on the log-scale (Fig. S2).
All analyses were conducted at both the phylum and ASV level. We present phylum-level analyses in text, and ASV-level results are available as supplemental information (Fig. S4 and S5). All code and data used here are available on Figshare (10.6084/m9.figshare.21614226).
Scatterplots between dispersal and environmental weights on the natural and log-log scales. Download FIG S2, SVG file, 0.1 MB (104.1KB, svg) .
Copyright © 2023 Lemoine et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Redundancy analyses at the individual ASV level. Download FIG S3, SVG file, 0.8 MB (890KB, svg) .
Copyright © 2023 Lemoine et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Partitioning variance between stochastic and deterministic forces at the ASV level. Download FIG S4, SVG file, 1.2 MB (1.2MB, svg) .
Copyright © 2023 Lemoine et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Neutral models at the ASV level. Download FIG S5, SVG file, 3.0 MB (3.1MB, svg) .
Copyright © 2023 Lemoine et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data availability.
Microbial sequences and soil geochemistry available at the NCBI Sequence Read Archive under BioProject accession number (PRJNA699250).
ACKNOWLEDGMENTS
Conceptualization: B.J.A., D.H.W., W.B.L., N.F., I.D.H., N.P.L.; methodology: W.B.L., M.A.D., N.F., N.B.D., I.D.H., D.H.W., B.J.A., N.P.L.; investigation: W.B.L., M.A.D., N.F., N.B.D., B.J.A., D.H.W., N.P.L.; visualization: N.P.L.; supervision: B.J.A., D.H.W., W.B.L., N.F., I.D.H.; writing—original draft: N.P.L.; writing—review & editing: B.J.A., D.H.W., W.B.L., N.F., I.D.H., M.A.D., N.B.D., A.L.C.F.
Authors declare they have no competing interests. This work was supported by the following awards: National Science Foundation – Office of Polar Programs 1341631 (W.B.L.); National Science Foundation – Office of Polar Programs 1341648 (D.H.W.); National Science Foundation – Office of Polar Programs 1341629 (N.F.); National Science Foundation – Office of Polar Programs 1341736 (B.J.A.); National Science Foundation – Office of Polar Programs 1043681 (Polar Geospatial Center); National Science Foundation – Office of Polar Programs 1559691 (Polar Geospatial Center); National Science Foundation – Division of Environmental Biology 1941390 (N.P.L.); National Science Foundation – Graduate Research Fellowship 60041697 (M.A.D.).
Contributor Information
Nathan P. Lemoine, Email: nathan.lemoine@marquette.edu.
Miguel Lurgi, Swansea University.
REFERENCES
- 1.Darwin C. 1859. On the origin of species. W. Clowes and Sons, London, UK. [Google Scholar]
- 2.Vellend M. 2010. Conceptual synthesis in community ecology. Q Rev Biol 85:183–206. doi: 10.1086/652373. [DOI] [PubMed] [Google Scholar]
- 3.Chase JM, Myers JA. 2011. Disentangling the importance of ecological niches from stochastic processes across scales. Philos Trans R Soc Lond B Biol Sci 366:2351–2363. doi: 10.1098/rstb.2011.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacArthur RH. 1970. Species packing and competitive equilibrium for many species. Theor Popul Biol 1:1–11. doi: 10.1016/0040-5809(70)90039-0. [DOI] [PubMed] [Google Scholar]
- 5.Zhou J, Ning D. 2017. Stochastic community ecology: does it matter in microbial ecology? Microbiol Mol Biol Rev 81:1–32. doi: 10.1128/MMBR.00002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoener TW. 1983. Rate of species turnover decreases from lower to higher organisms: a review of the data. Oikos 41:372–377. doi: 10.2307/3544095. [DOI] [Google Scholar]
- 7.Langenheder S, Székely AJ. 2011. Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J 5:1086–1094. doi: 10.1038/ismej.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myers JA, Harms KE. 2011. Seed arrival and ecological filters interact to assemble high-diversity plant communities. Ecology 92:676–686. doi: 10.1890/10-1001.1. [DOI] [PubMed] [Google Scholar]
- 9.Ferrenberg S, O'Neill SP, Knelman JE, Todd B, Duggan S, Bradley D, Robinson T, Schmidt SK, Townsend AR, Williams MW, Cleveland CC, Melbourne BA, Jiang L, Nemergut DR. 2013. Changes in assembly processes in soil bacterial communities following a wildfire disturbance. ISME J 7:1102–1111. doi: 10.1038/ismej.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J, Deng Y, Zhang P, Xue K, Liang Y, Van Nostrand JD, Yang Y, He Z, Wu L, Stahl DA, Hazen TC, Tiedje JM, Arkin AP. 2014. Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc Natl Acad Sci USA 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Powell JR, Karunaratne S, Campbell CD, Yao H, Robinson L, Singh BK. 2015. Deterministic processes vary during community assembly for ecologically dissimilar taxa. Nat Commun 6:8444. doi: 10.1038/ncomms9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conradi T, Temperton VM, Kollmann J. 2017. Resource availability determines the importance of niche-based versus stochastic community assembly in grasslands. Oikos 126:1134–1141. doi: 10.1111/oik.03969. [DOI] [Google Scholar]
- 13.Ning D, Deng Y, Tiedje JM, Zhou J. 2019. A general framework for quantitatively assessing ecological stochasticity. Proc Natl Acad Sci USA 116:201904623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tripathi BM, Stegen JC, Kim M, Dong K, Adams JM, Lee YK. 2018. Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J 12:1072–1083. doi: 10.1038/s41396-018-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callaway RM, Brooker RW, Choler P, Kikvidze Z, Lortie CJ, Michalet R, Paolini L, Pugnaire FI, Newingham B, Aschehoug ET, Armas C, Kikodze D, Cook BJ. 2002. Positive interactions among alpine plants increase with stress. Nature 417:844–848. doi: 10.1038/nature00812. [DOI] [PubMed] [Google Scholar]
- 16.Spasojevic MJ, Suding KN. 2012. Inferring community assembly mechanisms from functional diversity patterns: the importance of multiple assembly processes. J Ecol 100:652–661. doi: 10.1111/j.1365-2745.2011.01945.x. [DOI] [Google Scholar]
- 17.Lemoine NP, Shue J, Verrico B, Erickson D, Kress WJ, Parker JD. 2015. Phylogenetic relatedness and leaf functional traits, not introduced status, influence community assembly. Ecology 96:2605–2612. doi: 10.1890/14-1883.1. [DOI] [PubMed] [Google Scholar]
- 18.Bertness MD, Leonard GH. 1997. The role of positive interactions in communities: lessons from intertidal habitats. Ecology 78:1976–1989. doi: 10.1890/0012-9658(1997)078[1976:TROPII]2.0.CO;2. [DOI] [Google Scholar]
- 19.Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baas-Becking L. 1934. Geobiologie of inleiding tot de milieukunde. W.P. Van Stockum & Zoon, The Hague, the Netherlands. [Google Scholar]
- 21.Wennekes PL, Rosindell J, Etienne RS. 2012. The neutral-niche debate: a philosophical perspective. Acta Biotheor 60:257–271. doi: 10.1007/s10441-012-9144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tilman D. 2004. Niche tradeoffs, neutrality, and community structure: a stochastic theory of resource competition, invasion, and community assembly. Proc Natl Acad Sci USA 101:10854–10861. doi: 10.1073/pnas.0403458101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett PJ. 2013. Resolving views on Antarctic Neogene glacial history - the Sirius debate. Earth Environ Sci Trans R Soc Edinburgh 104:31–53. doi: 10.1017/S175569101300008X. [DOI] [Google Scholar]
- 24.Lewis AR, Marchant DR, Ashworth AC, Hedenäs L, Hemming SR, Johnson JV, Leng MJ, Machlus ML, Newton AE, Raine JI, Willenbring JK, Williams M, Wolfe AP. 2008. Mid-Miocene cooling and the extinction of tundra in continental Antarctica. Proc Natl Acad Sci USA 105:10676–10680. doi: 10.1073/pnas.0802501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rees-Owen RL, Gill FL, Newton RJ, Ivanović RF, Francis JE, Riding JB, Vane CH, Lopes dos Santos RA. 2018. The last forests on Antarctica: reconstructing flora and temperature from the Neogene Sirius Group, Transantarctic Mountains. Org Geochem 118:4–14. doi: 10.1016/j.orggeochem.2018.01.001. [DOI] [Google Scholar]
- 26.Stegen JC, Lin X, Fredrickson JK, Chen X, Kennedy DW, Murray CJ, Rockhold ML, Konopka A. 2013. Quantifying community assembly processes and identifying features that impose them. ISME J 7:2069–2079. doi: 10.1038/ismej.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown JKM, Hovmøller MS. 2002. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297:537–541. doi: 10.1126/science.1072678. [DOI] [PubMed] [Google Scholar]
- 28.Finlay BJ. 2002. Global dispersal of free-living microbial eukaryote species. Science 296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 29.Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D. 2006. Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Philos Trans R Soc Lond B Biol Sci 361:1947–1963. doi: 10.1098/rstb.2006.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peay KG, Garbelotto M, Bruns TD. 2010. Evidence of dispersal limitation in soil microorganisms: isolation reduces species richness on mycorrhizal tree islands. Ecology 91:3631–3640. doi: 10.1890/09-2237.1. [DOI] [PubMed] [Google Scholar]
- 31.Kerr B, Riley MA, Feldman MW, Bohannan BJM. 2002. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 32.Whitaker RJ, Grogan DW, Taylor JW. 2003. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science (80-) 301:2002–2004. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- 33.Nkem JN, Virginia RA, Barrett JE, Wall DH, Li G. 2006. Salt tolerance and survival thresholds for two species of Antarctic soil nematodes. Polar Biol 29:643–651. doi: 10.1007/s00300-005-0101-6. [DOI] [Google Scholar]
- 34.Adams BJ, Wall DH, Gozel U, Dillman AR, Chaston JM, Hogg ID. 2007. The southernmost worm, Scottnema lindsayae (Nematoda): diversity, dispersal and ecological stability. Polar Biol 30:809–815. doi: 10.1007/s00300-006-0241-3. [DOI] [Google Scholar]
- 35.Poage MA, Barrett JE, Virginia RA, Wall DH. 2008. The influence of soil geochemistry on nematode distribution, McMurdo Dry Valleys, Antarctica. Arct Alp Res 40:119–128. doi: 10.1657/1523-0430(06-051)[POAGE]2.0.CO;2. [DOI] [Google Scholar]
- 36.Meredith MP, King JC. 2005. Rapid climate change in the ocean west of the Antarctic Peninsula during the second half of the 20th century. Geophys Res Lett 32:1–5. [Google Scholar]
- 37.Yergeau E, Bokhorst S, Kang S, Zhou J, Greer CW, Aerts R, Kowalchuk GA. 2012. Shifts in soil microorganisms in response to warming are consistent across a range of Antarctic environments. ISME J 6:692–702. doi: 10.1038/ismej.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Booth TH, Nix HA, Busby JR, Hutchinson MF. 2014. BIOCLIM: the first species distribution modelling package, its early applications and relevance to most current MaxEnt studies. Divers Distrib 20:1–9. doi: 10.1111/ddi.12144. [DOI] [Google Scholar]
- 39.Jangid K, Whitman WB, Condron LM, Turner BL, Williams MA. 2013. Soil bacterial community succession during long-term ecosystem development. Mol Ecol 22:3415–3424. doi: 10.1111/mec.12325. [DOI] [PubMed] [Google Scholar]
- 40.Williams MA, Jangid K, Shanmugam SG, Whitman WB. 2013. Bacterial communities in soil mimic patterns of vegetative succession and ecosystem climax but are resilient to change between seasons. Soil Biol Biochem 57:749–757. doi: 10.1016/j.soilbio.2012.08.023. [DOI] [Google Scholar]
- 41.Diaz MA, Corbett LB, Bierman PR, Adams BJ, Wall DH, Hogg ID, Fierer N, Lyons WB. 2021. Relative terrestrial exposure ages inferred from meteoric 10Be and NO3− concentrations in soils along the Shackleton Glacier, Antarctica. Earth Surf Dyn 9:1363–1380. doi: 10.5194/esurf-9-1363-2021. [DOI] [Google Scholar]
- 42.Diaz MA, Li J, Michalski G, Darrah TH, Adams BJ, Wall DH, Hogg ID, Fierer N, Welch SA, Gardner CB, Lyons WB. 2020. Stable isotopes of nitrate, sulfate, and carbonate in soils from the Transantarctic Mountains, Antarctica: a record of atmospheric deposition and chemical weathering. Front Earth Sci 8:341. doi: 10.3389/feart.2020.00341. [DOI] [Google Scholar]
- 43.Welch KA, Lyons WB, Whisner C, Gardner CB, Gooseff MN, McKnight DM, Priscu JC. 2010. Spatial variations in the geochemistry of glacial meltwater streams in the Taylor Valley, Antarctica. Antarct Sci 22:662–672. doi: 10.1017/S0954102010000702. [DOI] [Google Scholar]
- 44.Lyons WB, Deuerling K, Welch KA, Welch SA, Michalski SG, Walters WW, Nielsen U, Wall DH, Hogg ID, Adams BJ. 2016. The soil geochemistry in the Beardmore Glacier region, Antarctica: implications for terrestrial ecosystem history. Sci Rep 6:26189–26188. doi: 10.1038/srep26189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sokol ER, Herbold CW, Lee CK, Cary SC, Barrett JE. 2013. Local and regional influences over soil microbial metacommunities in the Transantarctic Mountains. Ecosphere 4:1–24. doi: 10.1890/ES13-00136.1. [DOI] [Google Scholar]
- 46.Scarrow JW, Balks MR, Almond PC. 2014. Three soil chronosequences in recessional glacial deposits near the polar plateau, in the Central Transantarctic Mountains, Antarctica. Antarct Sci 26:573–583. doi: 10.1017/S0954102014000078. [DOI] [Google Scholar]
- 47.Claridge GGC, Campbell IB. 1968. Soils of the Shackleton Glacier region, Queen Maud Range, Antarctica. New Zeal J Sci 11:171–218. [Google Scholar]
- 48.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glockner FO. 2014. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Legendre P, Legendre L. 1998. Numerical ecology, 2nd edition. Elsevier Science. [Google Scholar]
- 53.Colwell RK, Chao A, Gotelli NJ, Lin SY, Mao CX, Chazdon RL, Longino JT. 2012. Models and estimators linking individual-based and sample-based rarefaction, extrapolation and comparison of assemblages. J Plant Ecol 5:3–21. doi: 10.1093/jpe/rtr044. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Variable loadings (distance calculation) of each variable on the first two principal component axes. Download Table S1, DOCX file, 0.01 MB (4.9KB, docx) .
Copyright © 2023 Lemoine et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Rarefaction curves for each site showed clear asymptotes by 3,000 reads, indicating that our sampling had sufficiently captured ASV richness. Red line shows 3,000 reads. Download FIG S1, PDF file, 0.03 MB (29.3KB, pdf) .
Copyright © 2023 Lemoine et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Scatterplots between dispersal and environmental weights on the natural and log-log scales. Download FIG S2, SVG file, 0.1 MB (104.1KB, svg) .
Copyright © 2023 Lemoine et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Redundancy analyses at the individual ASV level. Download FIG S3, SVG file, 0.8 MB (890KB, svg) .
Copyright © 2023 Lemoine et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Partitioning variance between stochastic and deterministic forces at the ASV level. Download FIG S4, SVG file, 1.2 MB (1.2MB, svg) .
Copyright © 2023 Lemoine et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Neutral models at the ASV level. Download FIG S5, SVG file, 3.0 MB (3.1MB, svg) .
Copyright © 2023 Lemoine et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Microbial sequences and soil geochemistry available at the NCBI Sequence Read Archive under BioProject accession number (PRJNA699250).