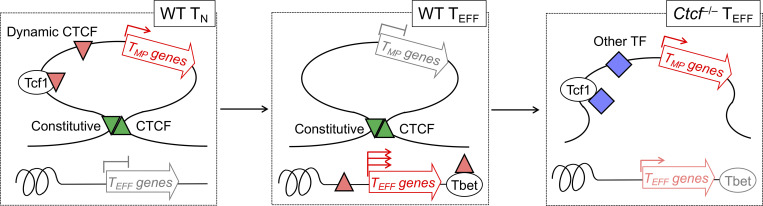
Xue, Peng, and colleagues employed multiomics approaches including Hi-C to demonstrate that CTCF promotes cytotoxic effector differentiation through dynamic redistribution, local chromatin accessibility control, and higher-order genomic reorganization, while enclosing memory precursor–associated genes within insulated neighborhoods to restrain their activation.
Abstract
Differentiation of effector CD8+ T cells is instructed by stably and dynamically expressed transcription regulators. Here we show that naive-to-effector differentiation was accompanied by dynamic CTCF redistribution and extensive chromatin architectural changes. Upon CD8+ T cell activation, CTCF acquired de novo binding sites and anchored novel chromatin interactions, and these changes were associated with increased chromatin accessibility and elevated expression of cytotoxic program genes including Tbx21, Ifng, and Klrg1. CTCF was also evicted from its ex-binding sites in naive state, with concomitantly reduced chromatin interactions in effector cells, as observed at memory precursor–associated genes including Il7r, Sell, and Tcf7. Genetic ablation of CTCF indeed diminished cytotoxic gene expression, but paradoxically elevated expression of memory precursor genes. Comparative Hi-C analysis revealed that key memory precursor genes were harbored within insulated neighborhoods demarcated by constitutive CTCF binding, and their induction was likely due to disrupted CTCF-dependent insulation. CTCF thus promotes cytotoxic effector differentiation by integrating local chromatin accessibility control and higher-order genomic reorganization.
Graphical Abstract
Introduction
CD8+ T lymphocytes are cytotoxic cells that lyse cells infected with intracellular pathogens and malignantly transformed cells (Chung et al., 2021; McLane et al., 2019). In response to acute viral or bacterial infections, antigen-specific naive CD8+ T (TN) cells are activated and undergo clonal expansion to generate effector CD8+ T cells that are equipped with cytotoxic molecules. The effector cells are heterogenous: The KLRG1loIL-7Rαhi or Tcf1hi subset shows increased potential to become memory CD8+ T cells and is considered as memory precursors (TMP), while cells with the opposite phenotype (KLRG1hiIL-7Rαlo, Tcf1lo) are fully differentiated cytotoxic effector (TEFF) cells, with reduced contribution to memory T cell pool (Gullicksrud et al., 2017; Herndler-Brandstetter et al., 2018; Joshi et al., 2007; Pais Ferreira et al., 2020). The CD8+ T cell differentiation requires instruction by transcription factors (TFs), which usually exhibit three distinct expression patterns: (1) induced expression after activation such as Tbet and Blimp1, (2) substantial repression, especially in TEFF cells, such as Tcf1 and Myb, and (3) relative stable expression such as Runx3, which is nonetheless essential for Blimp1 induction and Tcf1 repression (Gautam et al., 2019; Joshi et al., 2007; Kallies et al., 2009; Shan et al., 2017; Wang et al., 2018). Dynamic TF expression has been at the center of attention, and genome-wide TF occupancy is frequently interpreted as stochastic events. It remains less understood if redistribution of key TFs, even those stably expressed, contributes to fate decision and differentiation process of activated CD8+ T cells.
CCCTC binding factor (CTCF) was initially discovered as a transcriptional regulator, but is now best known for its ability to mediate long-range chromatin interaction and organize genome in three-dimensional space (Ohlsson et al., 2001; Pongubala and Murre, 2021; Zhao et al., 2022). Topologically associating domains (TADs) are recognized as physically and functionally isolated units in mammalian genome organization (Dixon et al., 2016). TADs consist of sub-TADs or insulated neighborhoods that are smaller in size and provide finer gene regulation (Hnisz et al., 2016). CTCF binding at the boundaries of TADs or insulated neighborhoods is strong and constitutive across different cell types, consistent with its insulator function that shields from external enhancer activity or heterochromatin spreading. CTCF binding is also prevalent within TADs or insulated neighborhoods, exhibits cell type specificity, and contributes to the formation of promoter-enhancer loops (Arzate-Mejia et al., 2018). During T cell development, CTCF cooperates with Bcl11b to facilitate the formation of chromatin loops specific to T cell lineage-committed cells (Hu et al., 2018). In naive CD8+ T cells, CTCF is recruited by Tcf1 and Lef1 at non-constitutive binding sites, and acquires novel binding sites in response to IL-7 and IL-15 stimulation, to promote homeostatic proliferation (Shan et al., 2022b). CTCF binding strength is also altered by IL-2 in T helper 1 CD4+ cells polarized in vitro (Chisolm et al., 2017). It remains unknown if and how the versatile functions of CTCF are utilized in CD8+ T cells responding to acute infections. Using an in vivo infection model, we found that CTCF exhibited dynamic redistribution in the CD8+ T cell genome in response to TCR stimulation, and both dynamic and constitutive CTCF binding acted in concert in spatial genome reorganization to promote TEFF differentiation.
Results
CTCF redistribution is associated with chromatin accessibility and transcriptomic changes in effector CD8+ T cells
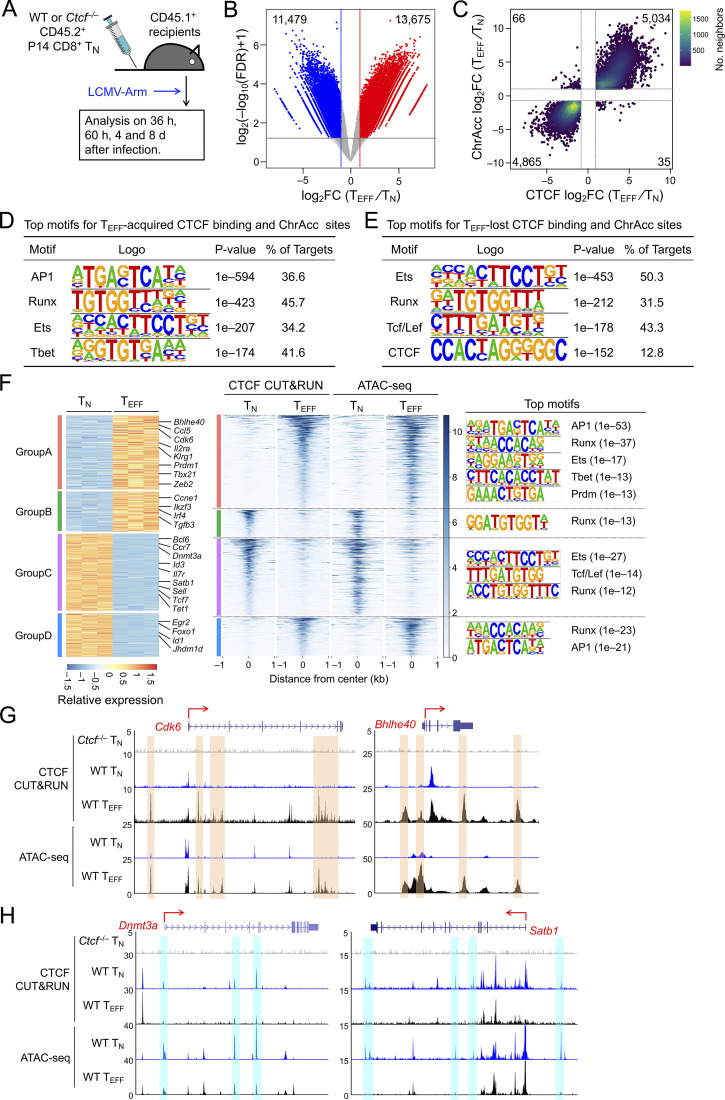
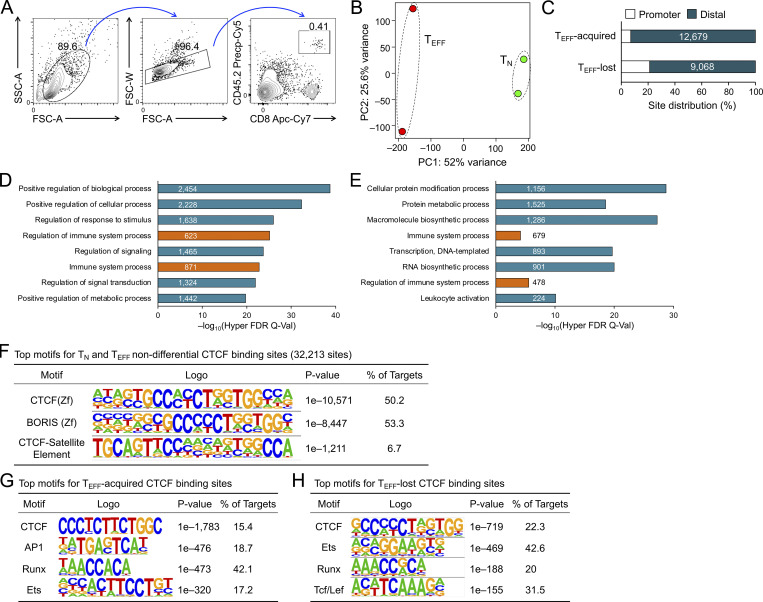
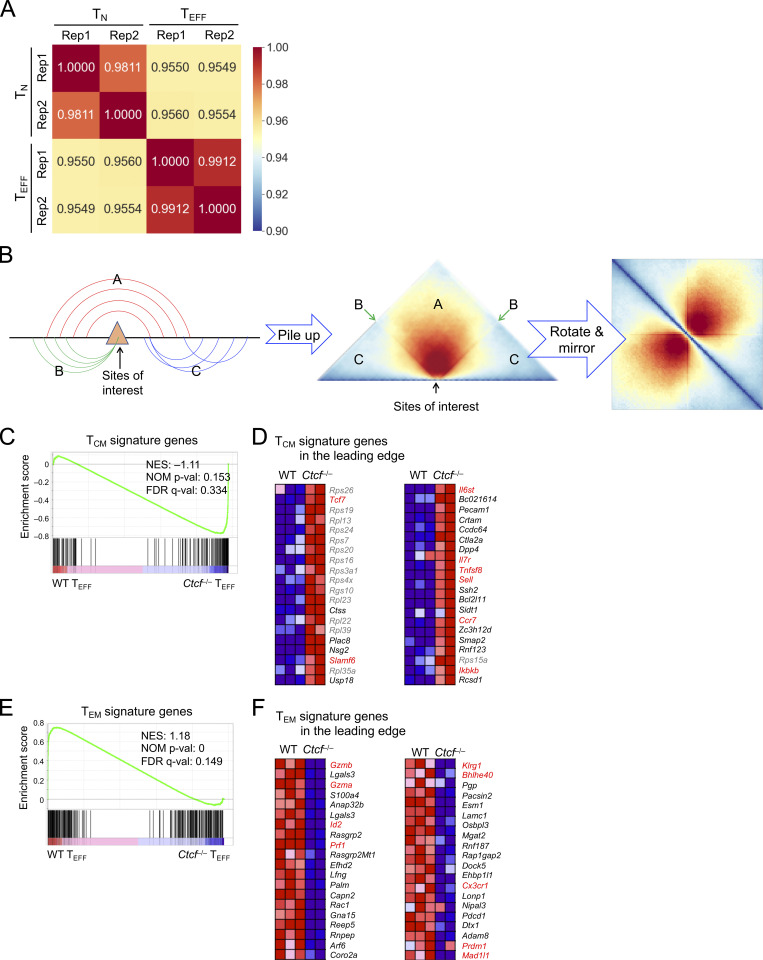
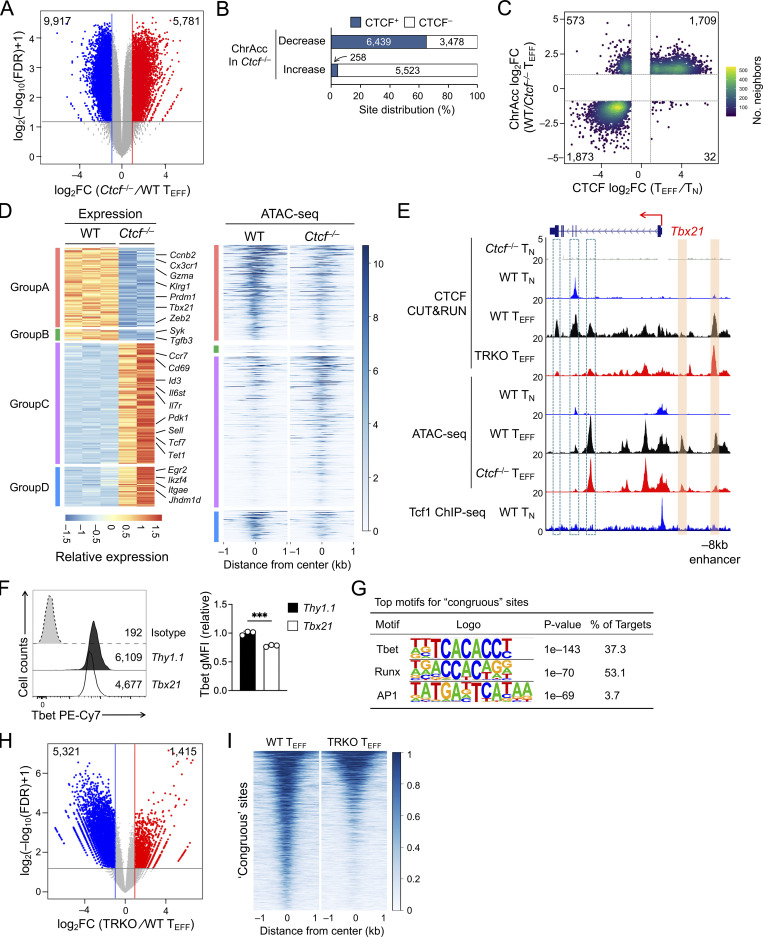
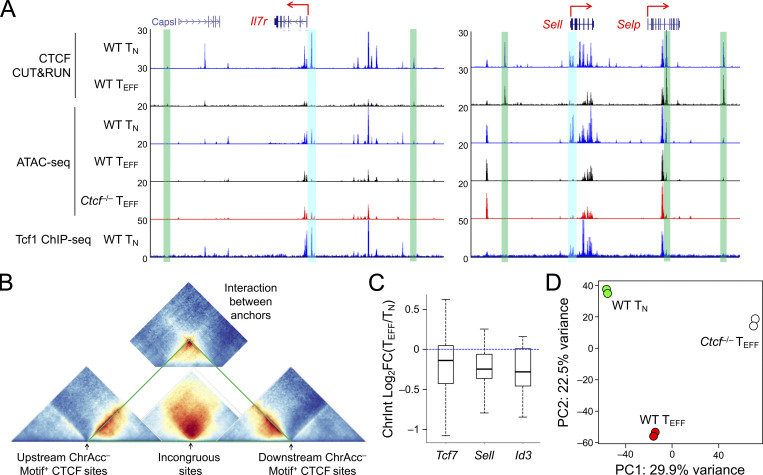
To determine CTCF occupancy in antigen-responding CD8+ T cells in vivo, we isolated CD45.2+ naive CD8+ T cells expressing the transgenic P14 TCR which is specific for the glycoprotein 33–41 epitope (GP33) of lymphocytic choriomeningitis virus (LCMV), and adoptively transferred into WT CD45.1+ recipients, followed by infection with LCMV Armstrong strain (LCMV-Arm) to elicit acute viral infection (Fig. 1 A and Fig. S1 A). P14 cells were sort-purified on day 4 post-infection (4 dpi) as early TEFF cells and were subjected to CTCF Cleavage Under Targets and Release Using Nuclease (CUT&RUN) analysis along with TN cells. With IgG CUT&RUN in WT TN and CTCF CUT&RUN in CTCF-deficient TN cells (see below) as negative controls, a total of 57,366 high-confidence CTCF binding sites in TN and TEFF cells were identified; among which, 13,675 sites showed evident CTCF binding in TEFF only or increased binding strength in TEFF over TN cells, while 11,479 sites showed negligible binding signals in TEFF or decreased binding strength in TEFF compared to TN cells (called TEFF-acquired and TEFF-lost CTCF sites, respectively), indicating dynamic redistribution of CTCF in the CD8+ T cell genome after in vivo activation (Fig. 1 B and Fig. S1 B). Over 80% of the “dynamic” CTCF sites were detected in distal regulatory regions and were linked to genes in “immune system process” as determined with the Genomic Regions Enrichment of Annotations Tool (GREAT) analysis (Fig. S1, C–E; and Table S1). Among the constitutive CTCF binding sites between TN and TEFF cells, CTCF motif was the most enriched and was detected in over 50% of the target sequences (Fig. S1 F). TEFF-acquired and TEFF-lost CTCF sites also had CTCF consensus sequence as the top motif, consistent with its ability to bind DNA directly, while these sites were also enriched in Ets and Runx motifs, suggesting that CTCF can be actively recruited to or evicted from Ets and Runx binding sites (Fig. S1, G and H).
Figure 1.
CTCF is redistributed and concordantly affects ChrAcc during TEFF cell differentiation. (A) Experimental design. Donor P14 cells were isolated from LNs, and the donor-derived effector cells were analyzed in the spleen of recipients throughout this study. (B) Volcano plot showing differential CTCF binding strength and statistical significance between WT TEFF and TN cells, with values denoting the numbers of differential CTCF binding sites. (C) Scatter plot showing distribution between differential CTCF binding strength and differential ChrAcc sites in comparisons between WT TEFF and TN cells, with values denoting site numbers. (D and E) Top motifs in TEFF-acquired and TEFF-lost CTCF + ChrAcc sites (quadrants i and iii in C, respectively) based on HOMER analysis. (F) Heatmaps showing DEGs and their associated differential CTCF binding and ChrAcc sites. DEGs linked to key GO terms (Fig. S2 E) were allocated into different groups based on the pattern of CTCF binding and ChrAcc changes, where select genes are marked and the color scale denotes relative strength of each molecular feature. Top motifs for CTCF + ChrAcc sites in each group are listed. (G and H) Tracks of CTCF CUT&RUN and ATAC-seq in WT TN and TEFF cells at select genes in groups A (G) or C (H), which are associated with concordantly acquired CTCF + ChrAcc sites (denoted with orange bars in G) or with concordantly lost CTCF + ChrAcc sites (denoted with blue bars in H). Tracks in gray denote CTCF CUT&RUN in Ctcf−/− TN cells as a negative control.
Figure S1.
Characterization of dynamic CTCF binding sites in early TEFF compared to TN cells. (A) Gating strategy to identify CD45.2+ donor-derived P14 CD8+ T cells in WT CD45.1+ recipients. (B) PCA of CTCF binding profile in WT TN and early TEFF cells, where normalized CTCF CUT&RUN signals on merged peaks were used as input data. (C) Distribution of dynamic CTCF binding sites in promoter (TSS+/−1 kb) and distal regulatory regions, where values in bars denote actual site numbers. (D and E) Functional annotation of TEFF-acquired (D) and TEFF-lost (E) CTCF binding sites based on GREAT analysis using the TSS+/−100 kb association rule. Non-redundant GO terms were selected from top 20 Hyper Rank for display, where values in bars denote site numbers and orange bars denote specific connection with immune functions. (F) Top three motifs in TN and TEFF non-differential CTCF binding sites based on HOMER analysis, with the known motif results shown. (G and H) Top motifs in TEFF-acquired (G) and TEFF-lost (H) CTCF binding sites based on HOMER analysis, with the de novo motif results shown.
Mapping chromatin accessibility (ChrAcc) with Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) showed extensive changes between TN and early TEFF cells, with over 15,000 sites becoming more “open” and another 14,000 sites becoming more “closed” in TEFF cells (Fig. S2, A and B). Stratifying these differential ChrAcc sites with dynamic CTCF binding sites showed highly concordant changes, that is, ∼1/3 of more open ChrAcc sites in TEFF cells were associated with TEFF-acquired CTCF sites, while about 1/3 of more closed ChrAcc sites in TEFF cells were associated with TEFF-lost CTCF sites (Fig. 1 C). Motif analysis of TEFF-acquired CTCF + ChrAcc sites showed significant enrichment of AP1 and Tbet motifs, besides Runx and Ets motifs (Fig. 1 D), suggesting that TCR-mobilized AP1 factors (e.g., BATF and Jun/Fos) and the induced Tbet contributed to CTCF recruitment for chromatin opening. On the other hand, the TEFF-lost CTCF + ChrAcc sites were enriched in Tcf/Lef motif, besides Ets and Runx (Fig. 1 E); because Tcf1 and Lef1 are downregulated upon CD8+ T cell activation (Zhao et al., 2010), the loss of CTCF binding and ChrAcc was at least partly a passive event, following partner TF expression changes in TEFF cells.
Figure S2.
Integration of dynamic CTCF binding with transcriptomic and ChrAcc changes during TN to TEFF cell differentiation. (A) PCA of the ChrAcc profiles of WT TN, WT and Ctcf−/− early TEFF cells, where normalized ATAC-seq signals on merged peaks were used as input data. (B) Volcano plot showing differential ChrAcc sites between WT TN and early TEFF cells, where values denote numbers of differential ChrAcc sites. FC, fold change. (C) PCA of transcriptomic profiles in WT TN, WT and Ctcf−/− early TEFF cells, where TPM for each gene in different samples were used as the input data. (D) Volcano plot showing DEGs between WT TN and early TEFF cells, where values denote numbers of DEGs. (E) Functional annotation of DEGs associated with concordant changes in CTCF binding and ChrAcc changes. DEGs from WT TN vs. early TEFF comparison were grouped into A–D based on association with concordantly acquired or lost CTCF + ChrAcc sites (as illustrated in Fig. 1 F), with gene and site numbers listed. DEGs in each group was analyzed with DAVID for functional annotation, and select GO terms of interest were displayed along with gene numbers and enrichment scores. All genes in the listed GO terms were visualized in gene expression heatmaps, and the gene-associated CTCF binding and ChrAcc changes are shown in heatmaps as parallel panels in Fig. 1 F. (F) Tracks of CTCF CUT&RUN and ATAC-seq in WT TN and TEFF cells at select genes in group B, which are associated with concordantly lost CTCF + ChrAcc sites (denoted with blue bars). Note that these CTCF binding sites may function as transcriptional silencers in TN cells, and their eviction in TEFF cells results in target gene induction. (G) Tracks of CTCF CUT&RUN and ATAC-seq in WT TN and TEFF cells at select genes in group D, which are associated with concordantly acquired CTCF + ChrAcc sites (denoted with orange bars). Note that these acquired CTCF binding sites may function as transcriptional silencers in TEFF cells.
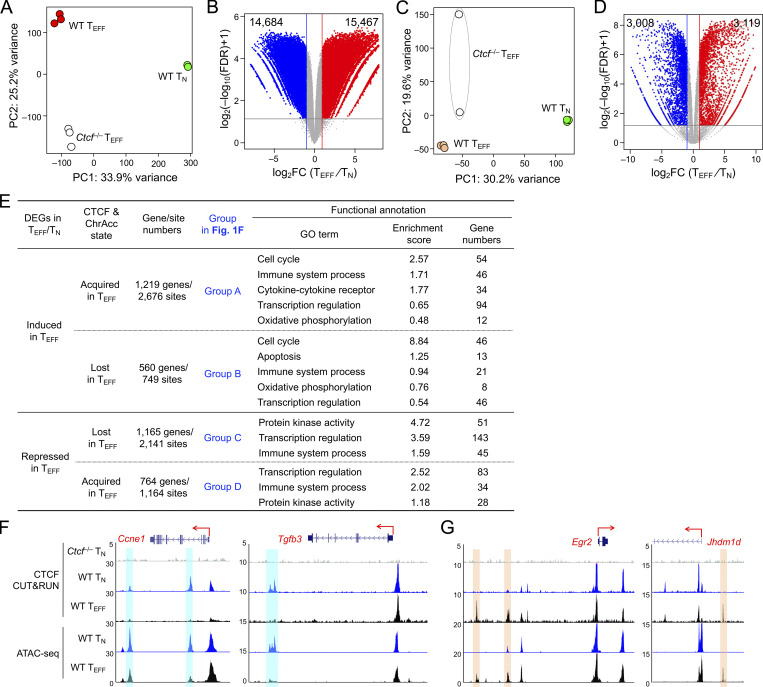
Transcriptomic analysis of TN and early TEFF cells with RNA sequencing (RNA-seq) identified 3,119 upregulated and 3,008 downregulated genes in TEFF compared to TN cells (Fig. S2, C and D; and Table S2). To specifically investigate the impact of dynamic CTCF redistribution, we focused on differentially expressed genes (DEGs) that harbored concordantly acquired or lost CTCF + ChrAcc sites (quadrants i and iii in Fig. 1 C, respectively) in the “−50 kb to +50 kb” genomic region flanking their transcription start sites (TSSs). Over 1,200 upregulated genes in TEFF cells were associated with concordantly acquired CTCF + ChrAcc sites at about 2.2 sites/gene, and included “cell cycle,” “immune system process,” and “transcription regulation” as top gene ontology (GO) terms (Fig. S2 E). These genes included cyclin-dependent kinases (such as Cdk6), surface proteins associated with activated CD8+ T cells (Il2ra and Klrg1), and key TFs including (Bhlhe40, Tbx21, Prdm1, and Zeb2; group A in Fig. 1, F and G), indicating the TEFF-acquired CTCF binding is directly associated with induction of chromatin opening and cytotoxic program in CD8+ T cells activated in vivo. On the other hand, 1,165 downregulated genes in TEFF cells were associated with concordantly lost CTCF + ChrAcc sites at about 1.8 sites/gene, and included “immune system process” and “transcription regulation” as top GO terms (Fig. S2 E). These genes included surface proteins associated with naive and central memory T cells (Sell, Ccr7, and Il7r), TFs (Tcf7, Id3, and Bcl6), epigenetic regulator (Dnmt3a) and genome organizer (Satb1; group C in Fig. 1, F and H). We also noted that upregulated genes in TEFF cells were associated with lost CTCF + ChrAcc sites and downregulated genes with acquired CTCF + ChrAcc sites (groups B and D in Fig. 1 F, respectively; Fig. S2, E–G). We recently observed that a Tcf1/Lef1-dependent ChrAcc site in the Prdm1 gene locus functions as a silencer to restrain Blimp1 expression in TN cells (Shan et al., 2021b). By interference, these CTCF sites in groups B and D may have engaged in silencer activity for target gene regulation. The concordantly acquired or lost CTCF + ChrAcc sites frequently shares similar motifs, but exhibited preferential usage of Runx motif when associated with silencer activity (groups B vs. C and D vs. A, Fig. 1 F right panels). Collectively, these observations suggested that dynamic CTCF distribution is associated with ChrAcc and transcriptomic changes following TEFF cell differentiation in vivo.
CTCF redistribution is associated with dynamic chromatin interaction changes in effector CD8+ T cells
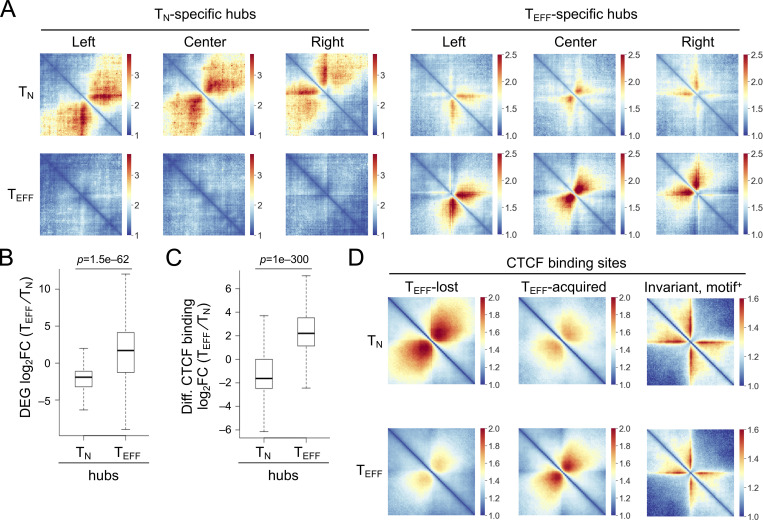
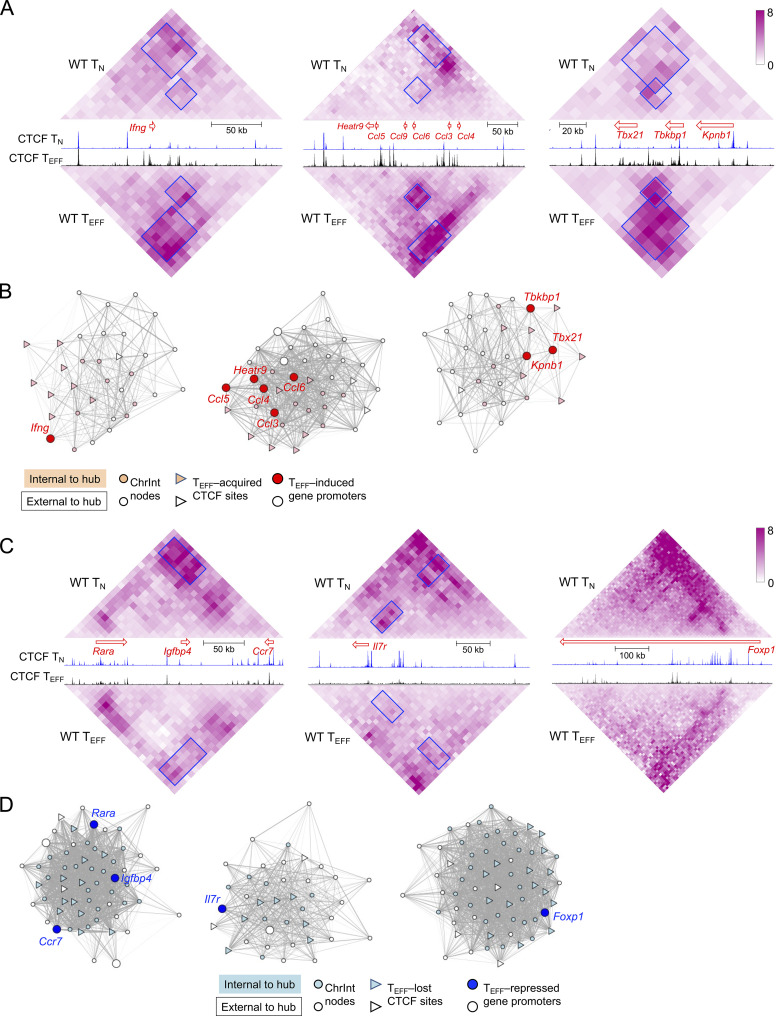
Given the important role of CTCF in regulating three-dimensional chromatin architecture, we performed in situ Hi-C on KLRG1+IL-7Rα− TEFF cells isolated on 8 dpi, and both replicates showed strong reproducibility (Fig. S3 A). In comparison with Hi-C data in TN cells (Shan et al., 2021b; Shan et al., 2022b), we applied the HiCHub algorithm (Li et al., 2022 Preprint), which identifies cell-type-specific chromatin interaction (ChrInt) hubs that contain collective unidirectional ChrInt changes in one cell type over the other, going beyond punctual ChrInt loops between two anchors. This analysis identified 775 TN-specific and 893 TEFF-specific hubs, which showed distinct ChrInt patterns as displayed in Hi-C pile-up graphs (Fig. 2 A). Cross-comparison with DEGs between TN and early TEFF cells showed that TEFF-specific DEGs were highly enriched in TEFF-specific hubs and vice versa (Fig. 2 B and Table S3), consistent with current view that increased ChrInt is largely associated with elevated gene transcription (Cuartero et al., 2022). Furthermore, the TEFF-acquired and TEFF-lost CTCF binding sites were highly enriched in TEFF- and TN-specific hubs, respectively (Fig. 2 C). We then performed Hi-C pile-up analysis centered on CTCF binding with different molecular characteristics (Fig. S3 B). For constitutive CTCF binding sites that were strictly invariant between TN and TEFF cells (≤1.1-fold differences in binding strength) and positive for its own motif, there was little interaction between their flanking regions in both cell types, suggesting that these sites had conserved insulation functions in TN and TEFF cells (Fig. 2 D, right column). For TEFF-acquired CTCF sites, their flanking regions had weak ChrInt in TN cells, and showed greatly strengthened ChrInt in TEFF cells (Fig. 2 D, middle column). As exemplified at the TEFF-induced Tbx21, Ifng, and Ccl gene loci, there was a highly concordant increase in multiple TEFF-acquired CTCF sites and extensively elevated ChrInt in TEFF over TN cells (Fig. 3 A), where the elevated ChrInt was observed among TEFF-acquired CTCF sites and between TEFF-acquired and constitutive CTCF sites. Visualization of the ChrInt hubs in the 3D space using network graphs showed that the TEFF-acquired CTCF sites were in architectural proximity with target gene promoters (Fig. 3 B), and multiple genes in the same hub showed concomitant induction in TEFF cells such as several Ccl genes, and Tbx21 together with Tbkbp1 and Kpnb1. On the other hand, the TEFF-lost CTCF sites were at the center of genomic regions with extensive ChrInt in TN cells, and the ChrInt was substantially attenuated in TEFF cells (Fig. 2 D, left column), as exemplified at the TEFF-downregulated Il7r, Ccr7, and Foxp1 genes (Fig. 3, C and D). These data suggest that dynamic CTCF redistribution is intimately linked to concordant genomic reorganization to promote effector differentiation.
Figure S3.
Hi-C data characterization and GSEA of TCM and TEM signature genes. (A) Correlation heatmap showing reproducibility of Hi-C libraries from TN and TEFF cells. Pearson correlation coefficient between Hi-C library pairs was determined using ChrInt scores at 10 kb resolution as the input data (see details in Materials and methods). (B) Diagram illustrating the summation approach underlying the Hi-C pile-up profile centered on specific genomic locations (sites of interest). The ChrInt between the regions flanking the sites of interest is aggregated in section A, while that within the region on either side of the sites is in sections C. The interaction of the sites with its flanking regions is projected on lines B. (C–F) RNA-seq data on TCM and TEM cells were retrieved and processed in-house. By requiring ≥1.5-fold changes, FDR < 0.05, and fragments per kilobase of exon per million mapped fragments at the higher expression condition ≥1, we identified 144 genes as TCM signature and 231 genes as TEM signature genes. The enrichment of these signature genes in WT and Ctcf−/− early TEFF transcriptomes was determined with GSEA. (C and D) TCM signature is enriched in Ctcf−/− early TEFF cells, with 49 TCM signature genes in the leading edge. C, enrichment plot; D, heatmaps of the top 40 genes in the leading edge, with genes of interest in red font. (E and F) TEM signature is enriched in WT early TEFF cells, with 44 TEM signature genes in the leading edge. E, enrichment plot; F, heatmaps of the top 40 genes in the leading edge, with genes of interest in red font. Marked in enrichment plot are NES (normalized enrichment score), NOM P val (nominal P values), and FDR q-val (FDR q values) as output from GSEA. Note that similar results were obtained with TCM and TEM signature gene sets defined in GSE147080 (Millner et al., 2020).
Figure 2.
TN and TEFF cells form distinct chromatin interaction hubs. (A) Heatmaps showing chromatin interactions in TN- and TEFF-specific hubs through Hi-C pile-up analysis centered on hub center and edges in both cell types. (B) Box plots summarizing expression changes of DEGs in TN- and TEFF-specific hubs. (C) Box plots summarizing binding strength changes of differential CTCF binding sites in TN- and TEFF-specific hubs. The box center lines denote the median, box edge denotes interquartile range (IQR), and whiskers denote the most extreme data points that are no more than 1.5 × IQR from the edge. P values were determined with one-sided Mann–Whitney U test. (D) Heatmaps showing ChrInt among +/−500 kb regions centered on the TEFF-lost, TEFF-acquired, and motif+ invariant (fold change ≤1.1) CTCF binding sites, based on pile-up analysis of Hi-C data in TN and TEFF cells, where color scale denotes ChrInt strength.
Figure 3.
CTCF redistribution is associated with genomic architectural changes in TEFF cells. (A and C) Diamond graphs showing ChrInt in TN (top) and TEFF cells (bottom) surrounding cytotoxic effector genes (Ifng, Ccl, and Tbx21 in A) or memory precursor–associated genes (Ccr7, Il7r, and Foxp1 in C) gene loci, as displayed on WashU epigenome browser. Blue boxes denoting ChrInt “patches” showing marked changes between TN and TEFF cells. Shown in the middle are gene size and transcription orientation along with CTCF CUT&RUN tracks in TN and TEFF cells. (B and D) Network view of TEFF-specific hubs harboring Ifng, Ccl, and Tbx21 genes (B) and TN-specific hubs harboring Ccr7, Il7r, and Foxp1 genes (D). Filled nodes represent 10 kb bins belonging to the network community underlying the hub, while open nodes belong to +/−100 kb extended genomic regions from the hub. Gray lines denote increased (B) or decreased (D) ChrInt in TEFF compared to TN cells, with line width representing log2 fold change of ChrInt between TEFF and TN cells.
CTCF promotes differentiation of effector CD8+ T cells
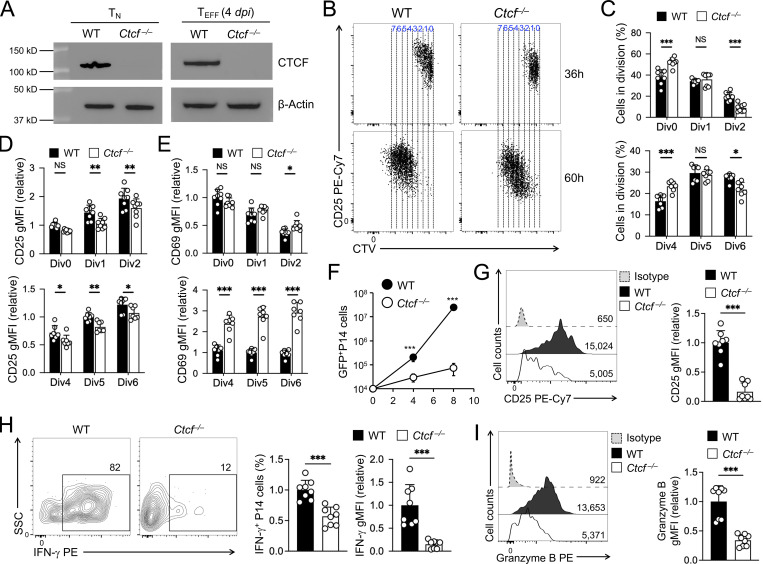
To determine the biological impact of CTCF on TEFF cell differentiation, we generated P14-Tg+hCD2-Cre+Rosa26GFPCtcf+/+ (WT) and P14-Tg+hCD2-Cre+Rosa26GFPCtcfFL/FL (Ctcf−/−) mice where the hCD2-Cre transgene ablated CTCF in mature T cells with high efficiency (Fig. 4 A), without affecting thymic development or causing aberrant T cell activation (Shan et al., 2022b). WT and Ctcf−/− naive P14 CD8+ T cells were adoptively transferred, and recipients infected with LCMV-Arm (Fig. 1 A). Within 36–60 h after infection, WT and Ctcf−/− CD8+ T cells were activated and initiated proliferation at the early response stage, where loss of CTCF moderately delayed cell division, reduced CD25 induction but elevated CD69 expression (Fig. 4, B–E). In contrast, Ctcf−/− TEFF cells failed to accumulate at 4 and 8 dpi, showing ∼7- and 330-fold reduction than WT cells, respectively (Fig. 4 F). On 4 dpi, Ctcf−/− early TEFF cells showed more pronounced reduction in CD25 expression (Fig. 4 G). These observations suggest that TEFF cells are progressively dependent on CTCF to complete effector differentiation. In fact, Ctcf−/− early TEFF cells showed profoundly impaired in IFN-γ production and greatly diminished granzyme B expression (Fig. 4, H and I), consistent with the notion that CTCF is required for activating and/or sustaining the cytotoxic program in TEFF cells.
Figure 4.
Ablating CTCF impairs TEFF cell accumulation and cytotoxic functions. (A) Immunoblotting for CTCF in sorted WT or Ctcf−/− GFP+ naive CD8+ T cells (before adoptive transfer, left) and early TEFF cells (4 dpi from infected recipients), with β-actin as loading control. Data are representative from two independent experiments. (B and C) Cell division of CTV-labeled WT or Ctcf−/− naive CD45.2+GFP+CD8+ T cells at 36 h (top) or 60 h (bottom) after transfer into CD45.1+ recipient mice followed by LCMV-Arm infection, with representative dot plots (B) and cumulative data on cell frequency in each cell division (C) shown. (D and E) Relative expression of CD25 (D) and CD69 (E) in WT or Ctcf−/− CD8+ T cells in individual cell divisions at 36 or 60 h after infection. gMFI, geometric mean fluorescence intensity. (F) Numbers of WT or Ctcf−/− effector CD8+ T cells on 4 and 8 dpi. (G–I) Detection of CD25 expression (G), IFN-γ production (H), and granzyme B expression (I) in WT or Ctcf−/− CD8+ T cells on 4 dpi. Data in B, H, and I are from two independent experiments. Data in C–F are means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not statistically significant by two-tailed Student’s t test. Source data are available for this figure: SourceData F4.
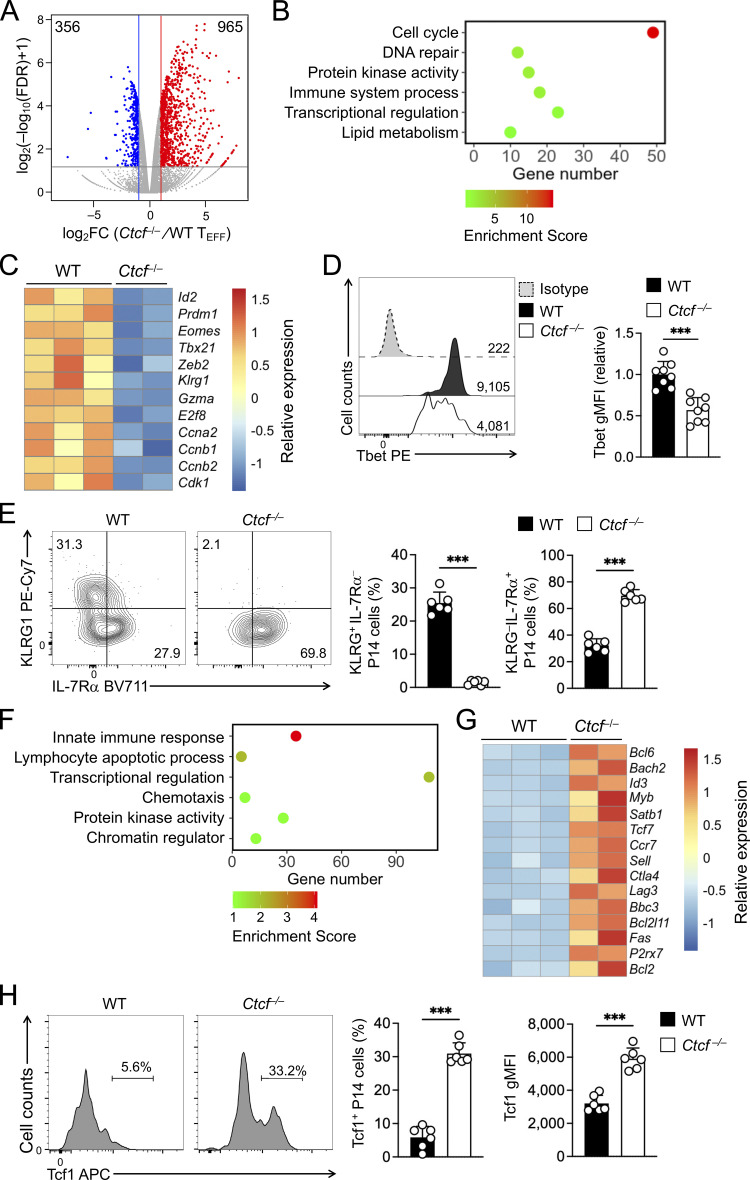
Because the few Ctcf−/− TEFF cells detected on 8 dpi contained substantial portion of undeleted cells, we focused molecular characterization on WT and Ctcf−/− early TEFF cells isolated on 4 dpi, where CTCF protein ablation remained complete (Fig. 4 A). RNA-seq analysis identified 356 genes that showed diminished expression in Ctcf−/− compared to WT TEFF cells (Fig. 5 A, Fig. S2 C, and Table S4). These downregulated genes were enriched in GO terms such as “cell cycle” (e.g., Ccnb1, Cdk1, and E2f8), “immune system process” (e.g., Gzma and Klrg1), and “transcription regulation” (including Tbx21, Prdm1, Id2, and Zeb2; Fig. 5, B and C). Consistent with known requirements for CTCF in cell proliferation in other cell types, CTCF-dependent regulation of cell cycle genes in TEFF cells may thus be a major cause for the progressive loss of Ctcf−/− TEFF cells at 4 and 8 dpi (Fig. 4 F). Furthermore, the expression of Tbet and KLRG1 proteins was substantially reduced in Ctcf−/− TEFF cells (Fig. 5, D and E). These CTCF-dependent TEFF functional aspects were strongly associated with the concordantly acquired CTCF binding and increased ChrAcc in WT TEFF cells (compare with Fig. 1), highlighting an essential role for redistributed CTCF in activating the cytotoxic program in TEFF cells, besides sustaining cell proliferation.
Figure 5.
Ablating CTCF impairs TEFF program but favors TMP program. (A) Volcano plot showing DEGs between WT and Ctcf−/− early TEFF cells isolated on 4 dpi, with values denoting gene numbers. (B and F) GO terms for genes downregulated (B) or upregulated (F) in Ctcf−/− compared to WT early TEFF cells, as determined with the DAVID Bioinformatics Resources, with dot color denoting enrichment scores. (C and G) Heatmaps of select downregulated (C) or upregulated genes (G) in Ctcf−/− compared to WT early TEFF cells. (D and H) Detection of Tbet (D) and Tcf1 (H) proteins by intranuclear staining in WT and Ctcf−/− early TEFF cells isolated on 4 dpi. Values in half-stacked histographs (D) denote gMFI, and those in separate histographs (H) denote percentage of Tcf1+ subset. (E) Detection of KLRG1 and IL-7Rα proteins by surface staining in WT and Ctcf−/− early TEFF cells isolated on 4 dpi, with values denoting percentages of KLRG1hiIL-7Rα− and KLRG1loIL-7Rα+ subsets. Data in D, E, and H are from two independent experiments, and cumulative data are means ± SD. ***, P < 0.001 by two-tailed Student’s t test. gMFI, geometric mean fluorescence intensity.
On the other hand, Ctcf−/− TEFF cells had 965 genes expressed at higher levels than WT TEFF cells (Fig. 5 A and Table S4). These upregulated genes included those in “lymphocyte apoptotic process” (including both pro-survival Bcl2 and proapoptotic genes Bbc3 and Bcl2l11, with the latter two encoding PUMA and BIM, respectively), surface proteins associated with coinhibitory pathways (e.g., Ctla4 and Lag3) or TCM cells (such as Ccr7, Sell, and Il7r, encoding CCR7, CD62L, and IL-7Rα, respectively), and key transcription regulators associated with TCM cells, including Tcf7 (encoding Tcf1), Id3, Bcl6, and Myb (Fig. 5, F and G). Gene set enrichment analysis (GSEA) further showed that TCM signature genes were strongly enriched in Ctcf−/− TEFF cells (Fig. S3, C and D), while TEM signature genes were depleted (Fig. S3, E and F), consistent with the impaired induction of effector cytotoxic program in the absence of CTCF. Indeed, increased portion of Ctcf−/− TEFF cells expressed IL-7Rα and Tcf1 proteins, and Tcf1 protein expression was elevated in Ctcf−/− over WT TEFF cells (Fig. 5, E and H). These data suggested that CTCF is necessary to prevent premature apoptosis and co-inhibition, and in addition, CTCF may also contribute to suppressing TMP fate and hence ensuring TEFF cell differentiation.
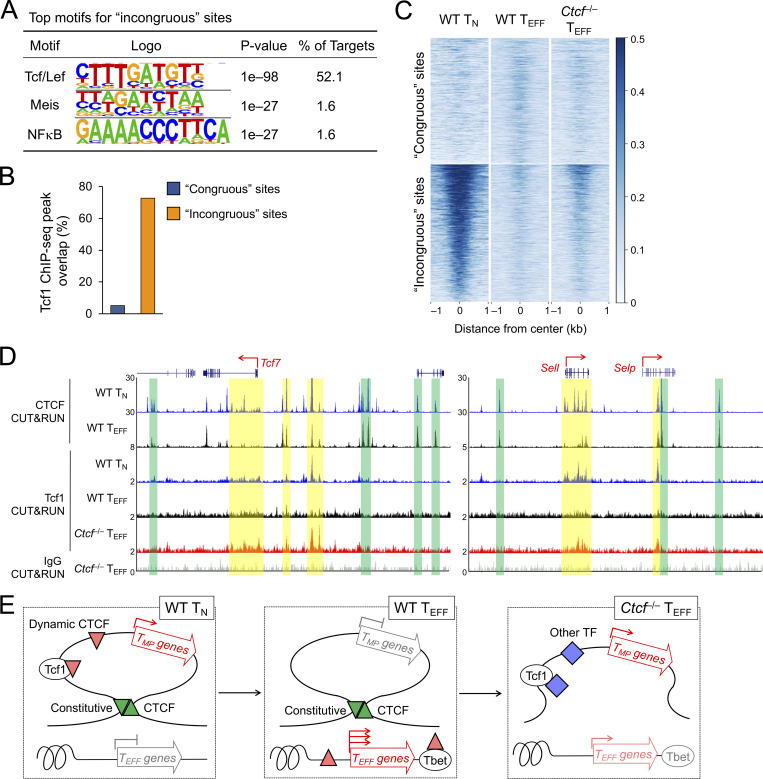
To further determine the direct contribution of CTCF to target gene regulation, ATAC-seq was performed on WT and Ctcf−/− early TEFF cells, which identified 9,917 sites showing decreased ChrAcc and 5,781 sites showing increased ChrAcc upon loss of CTCF (Fig. 6 A and Fig. S2 A). Stratifying the differential ChrAcc sites with all CTCF binding sites (including both dynamic and constitutive) detected in early TEFF cells showed that over 60% sites with decreased ChrAcc in Ctcf−/− cells were bound by CTCF, while <6% sites with increased ChrAcc in Ctcf−/− cells overlapped with CTCF binding (Fig. 6 B), indicating a predominant role of CTCF in establishing and/or maintaining open chromatin state in TEFF cells. To specifically assess the contribution of dynamically redistributed CTCF binding events, we stratified the differential ChrAcc sites between WT and Ctcf−/− TEFF cells with dynamic CTCF binding sites derived from the TEFF vs. TN comparison in Fig. 1. This analysis showed that the TEFF-acquired CTCF sites showed frequent overlap with decreased ChrAcc sites in Ctcf−/− over WT TEFF cells (quadrant i, Fig. 6 C). The 1,709 sites were in line with the expectation that TEFF-acquired CTCF binding directly establish and/or maintain chromatin open state in TEFF cells, and hence called “congruous” CTCF sites herein.
Figure 6.
TEFF-acquired CTCF binding activates effector transcriptional program. (A) Volcano plot showing differential ChrAcc sites between WT and Ctcf−/− early TEFF cells isolated on 4 dpi, with values denoting site numbers. (B) Distribution of CTCF binding sites in WT TEFF cells (both dynamic and constitutive) in differential ChrAcc sites between Ctcf−/− and WT early TEFF cells. (C) Scatterplot showing distribution between dynamic CTCF binding sites (in WT TEFF vs. TN comparison) and differential ChrAcc sites (in WT vs. Ctcf−/− TEFF comparison), where the 1,709 sites in quadrant i and the 1,873 sites in quadrant iii are defined as congruous and incongruous sites, respectively. (D) Heatmaps showing DEGs (in WT vs. Ctcf−/− early TEFF comparison) and associated differential ChrAcc sites (overlapping with congruous or incongruous CTCF binding) between WT and Ctcf−/− TEFF cells. The DEGs were grouped based on gene expression patterns defined in WT TEFF vs. TN comparison in Fig. 1 F, with select genes marked. (E) Tracks of CTCF CUT&RUN, ATAC-seq, and Tcf1 ChIP-seq at the Tbx21 gene locus. Orange bars denote TEFF-acquired CTCF binding sites linked to CTCF-dependent ChrAcc in TEFF cells, and open bars with dotted line denote CTCF binding sites that depended on Tbet and Runx3 (TRKO, Tbx21−/−Runx3−/−). (F) Tbet expression in TEFF cells in which sgRNAs targeting Tbx21 −8 kb enhancer (marked in E, Chr.11) or Thy1 constitutive CTCF binding site (Chr.9) were retrovirally delivered together with dCas9-KRAB-MeCP2. Values in half-stacked histographs (left) denote gMFI, and cumulative data are means ± SD (right) from two independent experiments. ***, P < 0.001 by two-tailed Student’s t test. gMFI, geometric mean fluorescence intensity. (G) Top motifs in the congruous CTCF sites (defined in quadrant i in C), with the incongruous sites as background based on HOMER analysis. (H) Volcano plot showing differential CTCF binding sites between WT and Tbx21−/−Runx3−/− (TRKO) early TEFF cells isolated on 4 dpi, with values denoting site numbers. (I) Heatmap showing CTCF binding strength at the congruous CTCF sites in WT and Tbx21−/−Runx3−/− (TRKO) early TEFF cells.
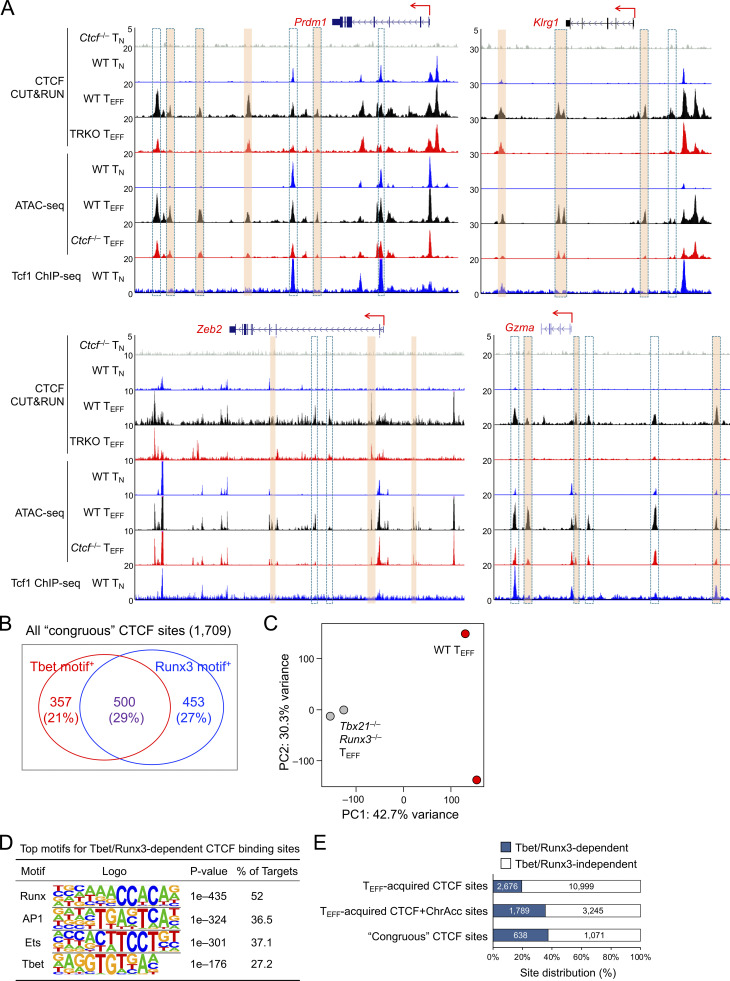
These congruous CTCF sites were strongly associated with downregulated genes in Ctcf−/− compared to WT TEFF cells (group A in Fig. 6 D), indicating that these sites functioned as enhancers for inducing cytotoxicity genes in TEFF cells, as observed in the Tbx21, Prdm1, Klrg1, Zeb2, and Gzma gene loci (Fig. 6 E and Fig. S4 A, compare tracks 6 and 7 at sites marked with orange bars). To further substantiate this notion, we employed the short guide RNA (sgRNA)–directed dCas9-KRAB-MeCP2 complex to block a Tbx21 −8 kb upstream congruous CTCF site, which contained a CTCF motif (Fig. 6 E). In the complex, the nuclease-dead Cas9 (dCas9) retains the DNA binding ability but does not generate double-strand DNA breaks, hence avoiding cellular toxicity; the fusion of dCas9 with repression domains from Krüppel-associated box (KRAB) and methyl-CpG binding protein 2 (MeCP2) has the capacity to antagonize promoter/enhancer activities in a site-directed manner (Thakore et al., 2015; Yeo et al., 2018). Compared with a negative control where a constitutive CTCF binding site in the Thy1 locus was used, targeting the −8 kb congruous CTCF site upstream of Tbx21 consistently diminished Tbet protein expression in TEFF cells (Fig. 6 F), validating the enhancer activity of the regulatory element that acquired CTCF binding and CTCF-dependent chromatin opening in TEFF cells. As noted above, the congruous sites could also function as transcriptional silencers, and some of these were linked to upregulated genes in Ctcf−/− TEFF cells (group D in Fig. 6 D).
Figure S4.
TEFF-acquired CTCF binding activates effector transcriptional program and is partly dependent on Tbet and Runx3. (A) Tracks of CTCF CUT&RUN, ATAC-seq, and Tcf1 ChIP-seq at the genes in the cytotoxic program. Orange bars denote TEFF-acquired CTCF binding sites linked to CTCF-dependent ChrAcc in TEFF cells, and bars with dotted lines denote CTCF binding sites that depended on Tbet and Runx3 (TRKO, Tbx21−/−Runx3−/−). (B) Detection of Tbet and Runx motifs in the congruous CTCF sites based on HOMER analysis. Values are motif+ site numbers and those in parentheses denote detection frequency. (C) PCA of the CTCF binding profiles of WT and Tbx21−/−Runx3−/− early TEFF cells, where normalized CTCF CUT&RUN signals on merged peaks were used as input data. (D) Top motifs in Tbet/Runx3-dependent CTCF sites (5,321 sites as defined in Fig. 6 H) based on HOMER analysis. (E) Distribution of Tbet/Runx3-dependent CTCF sites among TEFF-acquired CTCF binding sites (13,675 sites, defined in Fig. 1 B), TEFF-acquired concordant CTCF + ChrAcc sites (5,034 sites, defined in Fig. 1 C), and congruous CTCF sites (1,709 sites, defined in Fig. 6 C).
Motif analyses of the congruous CTCF sites showed enrichment of Tbet, Runx, and AP1 TF motifs (Fig. 6 G), suggesting that CTCF can be recruited by these TFs in addition to directly accessing target genes through its own DNA-binding capacity. In fact, >50% of the congruous CTCF sites contained either Tbet or Runx motif, and about 30% contained both motifs (Fig. S4 B). To substantiate this point, we generated P14-Tg+hCD2-Cre+ Rosa26GFP Tbx21FL/FL Runx3FL/FL (Tbx21−/−Runx3−/−) mice (Shan et al., 2017), used WT and Tbx21−/−Runx3−/− P14 cells in adoptive transfer and LCMV-Arm infection (as in Fig. 1 A), and then performed CTCF CUT&RUN on early TEFF cells isolated on 4 dpi. Over 5,000 CTCF binding sites showed diminished binding strength in Tbx21−/−Runx3−/− compared to WT early TEFF cells (Fig. 6 H and Fig. S4 C), and these sites were indeed enriched in Runx and Tbet motifs (Fig. S4 D). Among the Tbet/Runx3-dependent CTCF binding sites, ∼50% (2,676 sites) were acquired in TEFF over TN cells, constituting ∼20% of all TEFF-acquired CTCF sites (Fig. S4 E) and a greater portion of the congruous CTCF sites (Fig. 6 I and Fig. S4 E). Specifically, the TEFF-acquired CTCF sites at the Tbx21 intron regions, Prdm1, Klrg1, and Gzma loci were abrogated or greatly reduced in binding strength in the absence of Tbet and Runx3 (Fig. 6 E and Fig. S4 A, compare tracks 3 and 4, at sites marked with dotted lines). These observations corroborate the recruitment of CTCF by Tbet and/or Runx3 to promote activation of the cytotoxic transcriptional program. It is of interest to note that CTCF binding at the Tbx21 −8 kb enhancer was not affected by loss of Tbet and Runx3 (Fig. 6 E), indicating that CTCF directly activates this enhancer through its own motif in TEFF cells, and highlighting diverse mechanisms by which CTCF supports TEFF cell differentiation.
CTCF restrains memory precursor fate in activated CD8+ T cells
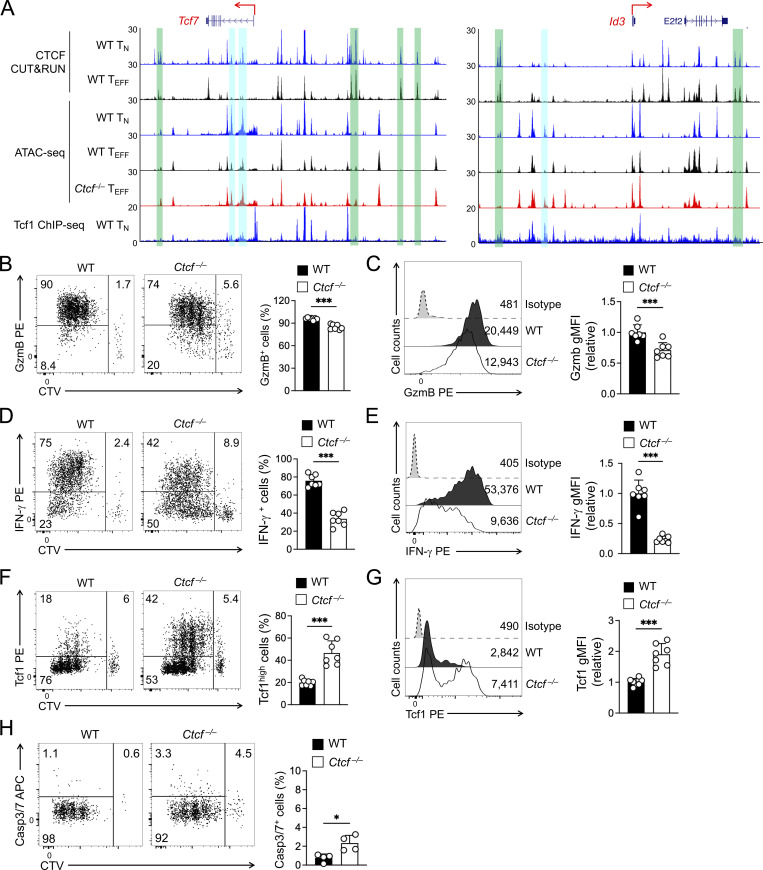
In addition to acquisition of novel CTCF binding sites, the WT TEFF cells lost or had attenuated CTCF binding at ∼12,000 sites, and about one third of these sites showed concordantly decreased ChrAcc compared to TN cells (Fig. 1, B and C). These concordant TEFF-lost CTCF + ChrAcc sites were weakened in such a way that in many cases they were not identified as CTCF binding or open chromatin sites in TEFF cells any longer, as exemplified at the Dnmt3a and Satb1 gene loci (Fig. 1 H). For a conventional TF in the context of unidirectional cellular differentiation, one may expect that genetic ablation of the TF would have similar effect as loss of the TF binding, and hence would not affect ChrAcc or associated gene expression. To our surprise, the TEFF-lost CTCF + ChrAcc sites were associated with more open ChrAcc sites in Ctcf−/− compared to WT early TEFF cells (quadrant iii, Fig. 6 C), and the 1,873 sites are hence called “incongruous” sites herein. The incongruous sites were predominantly associated with upregulated genes in Ctcf−/− TEFF cells (group C in Fig. 6 D), and these genes were associated with the TMP transcriptional program, as exemplified at the Tcf7, Sell, Il7r, and Id3 gene loci (Fig. 7 A and Fig. S5 A). These data demonstrated that a set of genes, which were actively transcribed in TN but destined for silencing in WT TEFF cells, resisted repression and at least partly maintained active transcription in CTCF-deficient TEFF cells.
Figure 7.
CTCF has an early impact on TEFF and TMP gene expression in antigen-experienced CD8+ T cells. (A) Tracks of CTCF CUT&RUN, ATAC-seq, and Tcf1 ChIP-seq at the Tcf7 and Id3 gene loci linked to the incongruous sites (denoted with blue bars). The green bars denote ChrAcc− constitutive CTCF binding sites that function as boundary anchors for insulated neighborhoods. (B–G) Detection of TEFF- and TMP-characteristic proteins in CTV-labeled WT or Ctcf− CD45.2+GFP+CD8+ T cells within 60 h after in vivo activation by LCMV-Arm infection. B and C, intracellular detection of granzyme B; D and E, intracellular detection of GP33-stimulated IFN-γ production; F and G, intranuclear detection of Tcf1. In B, D, and F, representative dot plots showing percentage of actively dividing cells that expressed proteins of interest; in C, E, and G, half-stacked histographs showing gMFI of the proteins. (H) Detection of active caspase3/7 in CTV-labeled WT or Ctcf−/− CD8+ T cells within 60 h after in vivo activation. Cumulative data in B–H are means ± SD (right) from at least two independent experiments. *, P < 0.05; ***, P < 0.001 by two-tailed Student’s t test. gMFI, geometric mean fluorescence intensity.
Figure S5.
TMP genes are harbored in insulated neighborhoods demarcated by constitutive CTCF binding and gain access to Tcf1 in CTCF-deficient TEFF cells. (A) Tracks of CTCF CUT&RUN, ATAC-seq, and Tcf1 ChIP-seq at the Il7r and Sell gene loci linked to the incongruous sites (denoted with blue bars). The green bars denote ChrAcc− constitutive CTCF binding sites that are positioned at the boundaries of insulated neighborhoods. (B) Diagram illustrating the relative positions of Hi-C–based ChrInt pile-up profile centering at sites of interest. The incongruous site-centered heatmap (from Fig. 8 A, without mirroring image) is in the middle, and flanked by the boundary anchor–centered heatmaps (from Fig. 8 B, without mirroring image) on each side. Summation of interactions between the boundary anchors (from Fig. 8 C) is positioned on the top, as insulating knots. (C) ChrInt changes between TEFF and TN cells in the insulated neighborhoods harboring Tcf7, Sell, and Id3 genes. The insulated neighborhoods are marked in triangles in solid green lines in Fig. 8 E, and changes in each ChrInt pair within the neighborhoods between TEFF and TN cells are summarized in boxplots, where center lines denote the median, box edge denotes IQR, and whiskers denote the most extreme data points that are no more than 1.5 × IQR from the edge. (D) PCA of Tcf1 binding profile in WT TN, WT and Ctcf−/− TEFF cells, where library size-normalized Tcf1 CUT&RUN signals on merged peaks were used as input data.
Based on RNA-seq and phenotypic/functional characterizations, Ctcf−/− early TEFF cells showed evident decrease in KLRG1+ but increase in IL-7Rα+ subset compared with WT cells (Fig. 5 E). Such subset ratio changes in Ctcf−/− cells may have intrinsic bias toward TMP-like cells, leading to the observed ChrAcc landscape changes showing impaired TEFF but enhanced TMP transcriptional programs. According to the asymmetrical cell division/signaling models, the TEFF vs. TMP fate decision is made during the first few divisions of activated CD8+ T cells (Arsenio et al., 2015; Nish et al., 2017). Retention of Tcf1 expression in early dividing cells is strongly associated with TMP fate (Gullicksrud et al., 2017; Lin et al., 2016; Pais Ferreira et al., 2020). Within 60 h after LCMV-Arm infection, WT P14 cells robustly induced granzyme B expression and IFN-γ production, while a small portion retained Tcf1 expression (Fig. 7, B–G), consistent with previous reports (Bird et al., 1998; Jenkins et al., 2008; Lin et al., 2016). Ctcf−/− P14 cells largely retained the ability of granzyme B induction but showed more pronounced reduction in IFN-γ production (Fig. 7, B–E), indicating that CTCF-deficient cells maintained the ability of inducing key cytotoxic genes during the initial cell division stage, albeit with decreased magnitude. On the other hand, a larger portion of Ctcf−/− P14 cells in initial divisions retained Tcf1 expression, with increased Tcf1 protein levels compared to WT cells (Fig. 7, F and G), suggesting an early bias toward TMP fate in CTCF-deficient cells. Transcriptomic analysis showed increased pro-apoptotic as well as pro-survival genes in Ctcf−/− early TEFF cells (Fig. 5 G), and we then measured cell apoptosis to directly measure the net impact of CTCF deficiency. Activation of caspase-3/7 was detected at a modestly increased frequency in Ctcf−/− compared to WT P14 cells in initial divisions (Fig. 7 H), but the frequency of apoptotic cells was so low that the differences may not constitute a major fate-deciding factor at the initial cell division stage. Collectively, these data support the notion that CTCF promotes TEFF but restrains TMP cell fate during the first few cell divisions of activated CD8+ T cells.
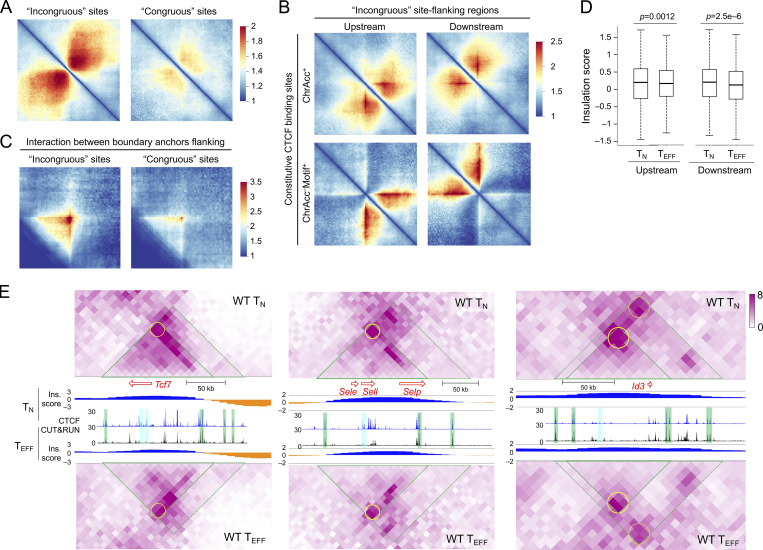
CTCF binding at the TMP-characteristic genes was lost in TEFF cells, yet ablating CTCF protein resulted in increased accessibility and expression of the TMP genes. How to reconcile these seemingly paradoxical observations? A key molecular distinction between WT and Ctcf−/− TEFF cells is that WT cells lost CTCF binding at the incongruous sites, while Ctcf−/− TEFF cells lost both dynamic and constitutive CTCF binding sites. CTCF is distinct from a conventional TF in its ability of establishing TADs and sub-TADs/insulated neighborhoods, besides mediating enhancer–promoter interactions (Dixon et al., 2016; Hnisz et al., 2016). We, therefore, deduced that constitutive CTCF binding sites and their associated chromatin architectural roles influenced the behavior of the incongruous sites. Utilizing in situ Hi-C data in TN cells (Shan et al., 2021b; Shan et al., 2022b), we found that the incongruous sites were embedded in highly connected genome regions with extensive chromatin interactions in TN cells; by contrast, the congruous sites were less connected with their neighboring regions (Fig. 8 A). We then examined the constitutive CTCF binding sites in the flanking regions of incongruous sites and analyzed two major groups based on their molecular characteristics. The first group included constitutive CTCF sites with robust ChrAcc signals (irrespective of presence of CTCF motifs); these ChrAcc+ constitutive CTCF sites could function as enhancers and were indeed in more connected genomic regions (Fig. 8 B, top panels). In contrast, the second group included constitutive CTCF sites that did not have robust ChrAcc but contained CTCF motifs; these ChrAcc− motif+ constitutive CTCF sites showed distinct features: Their neighboring regions proximal to the incongruous sites had substantially denser intra-region chromatin interactions than the regions distal to the incongruous sites, and significantly, the interactions between their proximal and distal regions were sparse (Fig. 8 B, bottom panels). These observations suggested that the ChrAcc− motif+ constitutive CTCF sites functioned as insulators shielding the incongruous sites. In addition, the interactions between ChrAcc− motif+ constitutive CTCF sites on the opposite sides of the incongruous sites was highly robust, while those between such sites flanking the congruous sites were much weaker (Fig. 8 C), corroborating the notion that the incongruous sites and their associated genes are enclosed in insulated neighborhoods in TN cells, with ChrAcc− motif+ constitutive CTCF sites demarcating the boundaries (Fig. S5 B). Because the incongruous and congruous sites were more frequently associated with TMP and TEFF genes, respectively (Fig. 6 D), these findings further suggested that TMP genes are more frequently flanked by boundary-forming CTCF binding sites than TEFF genes. Additionally, the insulation effect by the ChrAcc− motif+ constitutive CTCF sites was preserved, or even strengthened in TEFF cells (Fig. 8 D), as determined with insulation scores (Crane et al., 2015), where a lower score is indicative of stronger insulation effect.
Figure 8.
Constitutive CTCF binding forms insulated neighborhoods encompassing TMP genes. (A) Heatmaps showing ChrInt among +/−500 kb regions centered on the incongruous (left) and congruous sites (right), based on pile-up analysis of Hi-C data in TN cells. (B) Heatmaps showing ChrInt pile-up profile centered on ChrAcc+ (top) and ChrAcc− motif+ (bottom) constitutive CTCF binding sites that are located at 100–200 kb upstream or downstream of the incongruous sites in TN cells.. (C) Heatmap showing the interaction strength between upstream and downstream ChrAcc− constitutive CTCF binding sites flanking the incongruous sites in TN cells.based on aggregate peak analysis. In A–C, color scale denotes ChrInt strength. (D) Boxplots showing insulation scores at the upstream (left) and downstream (right) ChrAcc− motif+ constitutive CTCF binding sites flanking the incongruous sites in TN and TEFF cells. The P values are determined with one-sided Mann–Whitney U test. (E) Heatmaps showing ChrInt strength at the Tcf7, Sell, and Id3 gene loci and associated incongruous sites in TN and TEFF cells. The green-edged triangle marks an insulated neighborhood, and that with dotted lines represents extended regions where additional ChrAcc− constitutive CTCF binding sites may contribute to enforcing the boundary. The yellow circle represents the insulating knots formed through interactions between the boundary anchors. The end of red arrow represents TSS of genes of interest, and the length is proportional to the gene length. CTCF binding tracks in WT TN and TEFF cells are the same as Fig. 7 A (for Tcf7 and Id3) or Fig. S5 A (for Sell), shown as location reference for the constitutive (green) and dynamic (blue) CTCF binding sites. Insulation (Ins) scores of each 10 kb bin across each gene locus are displayed, with lower scores in orange representing stronger insulation effects.
At the TMP-associated gene loci specifically, Tcf7, Id3, and Sell genes were in fact located in genomic regions that had key features of insulated neighborhoods, that is, showing stronger intra-region chromatin interactions but sparse or weaker interactions with its neighbor regions (Fig. 8 E, compare ChrInt signals within to those outside the green triangles), and having ChrAcc− constitutive CTCF binding sites as “boundary anchors” on both ends of the regions (marked with green bars in Fig. 8 E). Importantly, the “insulating knots” formed through the interactions between these boundary anchors were largely sustained at these loci in TEFF cells (marked with yellow circles in Fig. 8 E), and the insulation effect was evidently observed at the boundary anchors and beyond, manifested as score values at or below zero (where the negative values were shown in orange in Fig. 8 E). These case studies further corroborated that the structure of insulated neighborhoods is preserved from naive to antigen-experienced CD8+ T cells at key TMP genes. On the other hand, ChrInt within the insulated neighborhoods encompassing Tcf7, Id3, and Sell genes showed a decreasing trend in TEFF compared to TN cells (Fig. 8 E and Fig. S5 C), concordant with evicted CTCF binding and downregulated TMP genes in TEFF cells. These findings suggested that regulation of TMP genes occurs “locally” within the insulated neighborhoods, being shielded from external interference beyond the boundary anchors. In Ctcf−/− TEFF cells, the loss of CTCF binding at the boundary anchors likely disrupted the integrity of the insulated neighborhoods (Nora et al., 2017), and hence exposing the incongruous sites to TFs other than CTCF.
In fact, the incongruous sites had Tcf/Lef as the top motif, which were found in >50% of the target sites (Fig. 9 A). Stratifying with previously reported Tcf1 chromatin immuoprecipitation using sequencing (ChIP-seq) peaks in TN cells (Shan et al., 2021b), the incongruous CTCF sites were highly enriched in Tcf1 binding compared with the congruous sites (Fig. 9 B). Specifically, Tcf1 ChIP-seq peaks overlapped with the incongruous sites in the Tcf7, Id3, Il7r, and Sell genes (blue bars in Fig. 7 A and Fig. S5 A), but rarely found at constitutive CTCF binding sites (green bars in Fig. 7 A and Fig. S5 A) or congruous sites in the Tbx21, Prdm1, Klrg1, Zeb2, and Gzma gene loci (Fig. 6 E and Fig. S4 A). These analyses suggest that the incongruous sites are intrinsically more accessible to Tcf1.
Figure 9.
Constitutive CTCF binding limits access to TMP genes by Tcf1. (A) Top motifs of the incongruous CTCF sites (defined in quadrant iii in Fig. 6 C), with the congruous sites as background based on HOMER analysis. (B) Distribution of Tcf1 ChIP-seq peaks in the congruous and incongruous sites. (C) Heatmaps show Tcf1 binding strength at the congruous (top) and incongruous (bottom) sites, as detected with CUT&RUN in WT TN, WT and Ctcf−/− TEFF cells. (D) Tracks of CTCF and Tcf1 CUT&RUN in TN or TEFF cells at the Tcf7 and Sell gene loci. The CTCF binding tracks in WT TN and TEFF cells are the same as Fig. 8 E as reference for the boundary anchors (denoted with green bars). Yellow bars highlight increased Tcf1 binding signals in Ctcf−/− over WT TEFF cells. Note that Tcf1 binding in Ctcf−/− TEFF cells remained weaker than that in WT TN cells, as displayed on different y-axis scales. (E) Diagram showing the proposed model on the cooperative actions of constitutive and dynamic CTCF binding in promoting TEFF cell differentiation. Genes in the TEFF program acquire novel CTCF binding and increased ChrAcc (upward red triangles), directly or indirectly by Tbet or other TFs (such as those in AP1, Runx, and Ets families, not depicted). On the other hand, genes in the TMP program are positioned in insulated neighborhoods, represented by a single loop demarcated with convergent, constitutive CTCF binding sites (green triangles). The structure of the insulated neighborhoods is preserved in TN and TEFF cells, and the downregulation of TMP genes is likely controlled locally, associated with Tcf7 gene silencing and eviction of CTCF binding (downward red triangle) within the insulated neighborhoods. Genetic ablation of CTCF impairs dynamic CTCF-mediated activation of TEFF genes and disrupts constitutive CTCF-organized insulated chromatin structure, resulting in access to TMP genes by activating TFs (blue diamonds) including Tcf1.
As demonstrated above, Ctcf−/− TEFF cells retained higher frequency and levels of Tcf1 expression at both initial cell division and early TEFF stages, compared to WT TEFF cells (Fig. 5 H; and Fig. 7, F and G). To directly test the accessibility of incongruous sites to Tcf1 after CD8+ T cell activation, we performed Tcf1 CUT&RUN in WT and Ctcf−/− TEFF cells, with that in TN cells as a positive control and IgG CUT&RUN in Ctcf−/− TEFF cells (which retained Tcf1 expression) as a negative control (Fig. S5 D). Consistent with analysis using Tcf1 ChIP-seq data (Fig. 9 B), Tcf1 binding signals detected with CUT&RUN in TN cells were robust at the incongruous sites but were close to the background level at the congruous sites (Fig. 9 C). In line with strong downregulation of Tcf1 after CD8+ T cell activation, Tcf1 binding at the incongruous sites was largely lost in WT TEFF cells, but partly retained in Ctcf−/− TEFF cells (Fig. 9 C). The retained Tcf1 peaks were found at the Tcf7 and Sell gene loci, within the boundary anchors (Fig. 9 D, compare tracks 4 and 5 at sites marked with yellow bars). In contrast, the congruous sites did not acquire Tcf1 binding in either WT or Ctcf−/− TEFF cells (Fig. 9 C), in spite of increased Tcf1 protein expression in the latter. These data corroborate the shielding effect by the boundary anchors and lend further support to the notion that CTCF restrains TMP fate by utilizing its constitutive binding to establish insulated neighborhoods that encompass TMP genes (Fig. 9 E).
Discussion
Activation of the cytotoxic transcriptional program is quintessential for CD8+ T cells to eliminate target cells infected with intracellular pathogens. Comparative analysis of in situ Hi-C between naive and effector CD8+ T cells revealed extensive genome reorganization during the effector cell differentiation process. De novo chromatin interaction hubs, manifested as unidirectionally increased chromatin loops in aggregation, formed around effector genes such IFN-γ and granzyme A, and key transcription regulators such as Tbet, Zeb2, and Bhlhe40. Underlying the formation of effector-specific hubs is at least partly ascribed to CTCF, which acquires novel binding sites in effector CD8+ T cells. As a TF itself, CTCF accesses its own binding motif to activate effector gene transcription in activated CD8+ T cells, as exemplified at the Tbx21 −8 kb enhancer. In addition, CTCF cooperates with T cell–lineage specific TFs, such as Tcf1 and Lef1, as a transcriptional coregulator to control identity and homeostatic proliferation of naive CD8+ T cells (Shan et al., 2022b). In this capacity, CTCF was indeed recruited to a substantial portion of novel binding sites in a Tbet/Runx3-dependent manner in effector cells. These findings indicate that dynamic redistribution of CTCF in CD8+ T cell genome represents a novel mechanism in effector CD8+ T cell differentiation in response to acute infections. In this context, CTCF not only acts through local regulatory elements but also nucleates extensive chromatin interactions encompassing critical effector genes. The resulting chromatin architectural changes likely stabilize and sustain effector gene transcription to ensure complete acquisition of cytotoxic capacity by activated CD8+ T cells.
CTCF-mediated genome organization functions in at least two distinct capacities, one is to bridge promoter-enhancer interactions to facilitate transcription activation, as discussed above, and the other is to form boundaries of TADs/insulated neighborhoods to shield influence from external enhancers or silencers. Consistent with reported observations that TAD structure is largely conserved during cell differentiation (Beagan and Phillips-Cremins, 2020; Yu and Ren, 2017), the constitutive CTCF binding sites between naive and effector CD8+ T cells, especially those with CTCF motifs, showed conserved insulation effects in both cell types. The insulation by CTCF applies not only to TADs in mega-million base pairs, but also to genomic structures on a smaller scale such as sub-TADs/insulated neighborhoods nested within TADs (Beagan and Phillips-Cremins, 2020; Yu and Ren, 2017). In naive CD8+ T cells, the CTCF-insulated neighborhoods were found at memory precursor–associated genes, such as those encoding Tcf1 and CD62L. Within such insulated neighborhoods, CTCF likely acts in the capacity of TF or cofactor to support gene transcription in naive CD8+ T cells, and its eviction from these sites after cell activation is highly concordant with downregulation of their associated genes and decreased chromatin interactions within the insulated neighborhoods. In contrast, the insulation effects by the boundary anchors demarcating the insulated neighborhoods remained robust in both naive and antigen-experienced CD8+ T cells. We posit that retention of the insulated chromatin structure in effector cells serves a critical biological purpose, that is, to limit re-expression of TMP genes and ensure completion of effector differentiation. In support of this view, genetic ablation of CTCF led to increased expression of memory precursor genes, likely resulting from disruption of the insulated neighborhoods and exposure of their encompassed memory precursor genes to external regulators including Tcf1. Under this scenario, the feed-forward regulation of Tcf7 gene by Tcf1 protein itself may thus forge a self-amplification loop that promotes TMP fate. Collectively, CTCF-mediated promoter/enhancer interaction and constitutive insulation act in concert to ensure effector differentiation while limiting memory potentials.
Accumulating evidence suggests that effector CD8+ T cell differentiation is a stepwise, gradual process, including initial cell division, early effector, and late effector stages. During the initial cell division stage, TCR-induced, early response transcription regulators such as BATF in the AP1 family and IRF4 may function as “pioneer factors” to establish ChrAcc and epigenetic landscape that are characteristic of effector cells (Kurachi et al., 2014; Man et al., 2017). BATF has been demonstrated to recruit CTCF for chromatin opening in CD4+ T cells activated ex vivo (Pham et al., 2019), and this is likely the case for CD8+ T cells where CTCF contributes more to IFN-γ production, albeit to a lesser extend to granzyme B induction and cell cycle progression. It is believed that through asymmetrical cell division and/or asymmetrical signaling, the TEFF and TMP fates are decided during the initial few cell divisions, and such heterogeneity persists to later stages (Arsenio et al., 2015; Nish et al., 2017). An important impact of CTCF ablation was increased retention of Tcf1 and hence a bias toward TMP fate in the early dividing cells, which is likely ascribed to disruption of CTCF-insulated chromatin structure, as discussed above.
DNA demethylation and erasure of repressive histone marks likely take extra time to allow corresponding regulatory elements to become accessible to activating factors, because inhibition of DNA methyltransferases and histone deacetylases accelerates cytokine-producing capability in activated T cells (Bird et al., 1998). In line with this view, in spite of rapid induction of many effector-associated genes at the initial cell division stage, many others, including key transcription regulators such as Blimp1, Tbet and Bhlhe40, are only evidently induced till the early effector stage (Best et al., 2013). Some of these gradually induced effector genes showed clear dependence on CTCF, with one example of particular interest as Tbet. The Tbx21 gene locus acquires several novel CTCF binding sites in early effectors, with an −8 kb upstream enhancer directly accessed by CTCF. Tbet per se contributes to CTCF recruitment in early effector cells, and in fact, the TEFF-acquired CTCF binding sites in Tbx21 introns depended on intact expression of Tbet and Runx3. Based on these findings, we posit that CTCF acts in a sequential manner, including initial induction of Tbet by itself, followed by cooperativity with Tbet to form a feed forward loop to enforce the effector fate and further differentiation. Tbet is unlikely the sole CTCF recruiting factor, and CTCF may thus have broadened regulatory roles through cooperating with both stably expressed and induced factors in early effectors. These multilayered actions may hence underlie the increased dependence on CTCF for the early effectors to sustain activation of the cytotoxic program, their proliferative capacity and survival. In summary, our findings highlight the critical requirements for CTCF to activate local enhancers and reorganize genomic architecture for target gene regulation, and the integrated actions of CTCF promote generation of cytotoxic effectors and anti-viral/tumor immunity.
Materials and methods
Mice
C57BL/6J (B6), B6.SJL, hCD2-Cre, Rosa26GFP, Runx3FL/FL (Naoe et al., 2007), and Tbx21FL/FL (Intlekofer et al., 2008) mice were from The Jackson Laboratory. CtcfFL/FL mice were provided by N. Galjart (Erasmus University Medical Center, Rotterdam, Netherlands) and A. Melnick (Weill Cornell Medicine, New York, NY, USA; Heath et al., 2008). All compound mouse strains used in this work were from in-house breeding at the animal care facility of Center for Discovery and Innovation, Hackensack University Medical Center. The mice were housed at 18–23°C with 40–60% humidity, with 12-h light/12-h dark cycles. All mice used in this study were 6–12 wk of age, and both sexes were used without randomization or blinding. All mouse experiments were performed under protocol approved by the Institutional Animal Use and Care Committees of Center for Discovery and Innovation, Hackensack University Medical Center.
Flow cytometry and immunoblotting
Single-cell suspensions were prepared from the spleen, LNs, and surface or intracellularly stained as described (Shan et al., 2022a). The fluorochrome-conjugated antibodies were as follows: anti-CD8 (53–6.7), anti-TCRβ (H57-597), anti-CD45.2 (104), anti-Granzyme B (GB12), anti-IFN-γ (XMG1.2), anti-Tbet (4B10), anti-CD62L (MEL-14), anti-KLRG1 (2F1), anti-CD25 (PC61.5), anti-CD69 (H1.2F3), and anti-CD44 (IM7) were from Thermo Fisher Scientific; anti-Tcf1 (C63D9) from Cell Signaling Technology; anti-IL-7Rα (A7R34) and anti-CD45.1 (A20) were from BioLegend. For detection of Tcf1 and Tbet proteins, surface-stained cells were fixed and permeabilized with the Foxp3/Transcription Factor Staining Buffer Set (eBiosciences, Thermo Fisher Scientific), followed by incubation with corresponding fluorochrome-conjugated antibodies. Apoptotic cells were detected with FLICA 660 Caspase-3/7 detection kit (Bio-Rad). Data were collected on FACSCelesta or FACSVerse (BD Biosciences) and were analyzed with FlowJo software V10.2 (TreeStar). For validation of CTCF deletion efficiency, cell lysates from sorted GFP+ naive or early effector CD8+ T cells were immunoblotted with anti-CTCF antibody (D31H2; Cell Signaling Technology) following standard protocols.
Cell labeling, adoptive transfer, and viral infection
WT or Ctcf−/− naive P14 CD8+ T cells were obtained from spleen and LNs of littermates. For detection of cell activation and initial cell division, the cells were labeled with 10 μM Cell Trace Violet (CTV, Invitrogen, Thermo Fisher Scientific), and 1 × 106 of CTV-labeled CD45.2+Vα2+CD8+ T cells were adoptively transferred into CD45.1+ B6.SJL mice through tail vein injection. For analysis of effector CD8+ T cell differentiation, CD45.2+Vα2+CD8+ T cells were transferred without CTV labeling at 2 × 104 cells/recipient mouse. On the following day, the recipients were i.v. infected with 2 × 105 PFU of LCMV-Arm, and the donor-derived P14 effector CD8+ cells were analyzed at 36 and 60 h, or 4 and 8 dpi.
dCas9-mediated repression of a Tbx21 enhancer
The dCas9-KRAB-MeCP2 plasmid in the pcDNA3.3-_TOPO backbone was obtained from Addgene (#110821; Yeo et al., 2018), and the cassette was cloned into the retroviral vector pMSCV-IRES-mCherry FP vector (#52114; Addgene). The sgRNA retroviral vector, which contains U6 promoter-driven cassette and PGK promoter-driven Ametrine in the LMPd backbone (Chen et al., 2014), was kindly provide by Dr. Hongbo Chi (St. Jude Children’s Research Hospital, Memphis, TN, USA; Wei et al., 2019). sgRNAs for a constitutive CTCF binding site at the Thy1 locus were 5′-TATCATTCAAACCCTCACGT-3′ and 5′-AGCCTCTCCCTAAACCTTCC-3′, and those for the −8 kb Tbx21 enhancer were 5′-CGGTGGAGCTGACGGGCCCG-3′ and 5′-ATAGAGTGTGTATCAACACA-3′. The dCas9-KRAB-MeCP2-mCherry and dual sgRNA-Ametrine retroviruses were packaged separately in 293T cells as previously described (Li et al., 2018). WT P14 CD8+ T cells were enriched with negative selection and primed in vitro using anti-CD3 and anti-CD28 followed by spinofection with both retroviruses for two consecutive days. The retrovirally transduced P14 cells were adoptively transferred into B6.SJL recipients, followed by LCMV-Arm infection on the next day. 5 d later, mCherry+Ametrine+ P14 cells were sort-purified and intracellular stained for Tbet expression.
RNA-seq and data analysis
Data generation
The RNA-seq data for WT naive CD8+ T cells were previously reported (Shan et al., 2021b) and deposited at the Gene Expression Omnibus (GEO; GSE164712) under the SuperSeries of GSE164713. WT or Ctcf−/− CD45.2+GFP+CD8+ T cells were sorted from recipient spleens on 4 dpi as early TEFF cells, total RNA extracted, and cDNA synthesis and amplification were performed using SMARTer Ultra Low Input RNA Kit (Clontech) following manufacturer’s instruction. The resulting libraries were sequenced on Illumina’s HiSeq2000 in paired-end mode with read length of 150 nucleotides. The new RNA-seq data were deposited at GEO under GSE208129 in the SuperSeries of GSE208130.
Data analysis
The sequencing quality of RNA-seq libraries were assessed by FastQC (v0.11.9; https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The reads were mapped to mouse genome mm9 using hisat2 (v2.2.1; Kim et al., 2019). Samtools (v1.7; Li et al., 2009) was used to transfer sam files to bam files and sort bam files. Mapped reads were then processed by htseq-count (v1.99.2; Anders et al., 2015) to estimate expression levels of all genes. The expression level of a gene was expressed as a gene-level transcripts per kilobase million (TPM) value. Gene raw counts were processed by edgeR (v3.32.1; Robinson et al., 2010) to identify DEGs between a pair of conditions (quasi-likelihood test, robust, fold-change > = 2 and false discovery rate [FDR] < 0.05). The reproducibility of RNA-seq data was evaluated by applying the principal component analysis (PCA) for all genes. UCSC genes from the iGenome mouse mm9 assembly (http://support.illumina.com/sequencing/sequencing_software/igenome.html) were used for gene annotation.
ATAC-seq and data analyses
Data generation
CD44lo-medCD62Lhi naive CD8+ cells were sorted from WT C57BL/6 mice, and WT or Ctcf−/− CD45.2+GFP+CD8+ early TEFF cells were sorted from recipient spleens on 4 dpi for preparation ATAC-seq libraries as previously described (Shan et al., 2021a). Briefly, the sorted cells were treated in lysis buffer for 3 min on ice, and the extracted nuclei were resuspended in transposition mix containing 2.5 μl Transposase (Illumina) and incubated at 37°C for 30 min. The products were purified with MinElute Reaction Cleanup Kit (Qiagen), and then amplified by PCR for 12 cycles with barcoded Nextera primers (Illumina). DNA fragments in the range of 150–1,000 bp were recovered from 2% E-Gel EX Agarose Gels (Invitrogen, Thermo Fisher Scientific). The libraries were quantified using a KAPA Library Quantification kit and sequenced on Illumina HiSeq2000 in paired read mode with the read length of 150 nucleotides at the Admera Health. The ATAC-seq data were deposited at the GEO under GSE208120 in the SuperSeries of GSE208130.
Data processing
The sequencing quality of ATAC-seq libraries was assessed by FastQC. Trim Galore (v0.6.7; https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) was used to trim low quality sequences and remove adapters. Bowtie2 (v2.3.4.1; Langmead and Salzberg, 2012) was used to align the sequencing reads to the mm9 mouse genome, and only uniquely mapped reads (mapping quality [MAPQ] > 10) were retained. Samtools (Li et al., 2009) was used to transfer sam files to bam files and sort bam files. Picard MarkDuplicates (v2.21.6; https://github.com/broadinstitute/picard) was used to remove duplicate reads in the bam files. MACS2 (v2.1.1; Zhang et al., 2008) was used for peak calling with a stringent criteria of ≥4 summit fold change and FDR < 0.05. For ATAC peaks in a given condition, the mapped reads from replicates were pooled for peak calling. For consistency, the ATAC-seq peaks are referred to as ChrAcc sites in this work.
Reproducibility analysis and identification of differential ChrAcc sites
Peaks called by MACS2 in three conditions were merged into union peaks. Raw reads were counted in each library on the union peaks resulting in a matrix with rows representing peaks and columns representing libraries. The raw-count matrices were then subjected to row-wise normalization by peak length per kilobase and then column-wise normalization by the column sum per million. The normalized matrix was subjected to PCA analysis with the z-score option. To identify differential ChrAcc sites for pairwise comparisons, the raw count matrix for two conditions was used as input for edgeR (quasi-likelihood test, robust, fold-change ≥2 and FDR < 0.05).
CUT&RUN and data analyses
Data generation
WT or Ctcf−/− TN, WT or Tbx21−/−Runx3−/− TEFF cells were sorted as above and used in CUT&RUN (Skene and Henikoff, 2017) assay to map CTCF binding sites as previously described (Shan et al., 2022b). In brief, FACS-sorted live cells (1 × 105 cells/reaction) were bound to Concanavalin A–coated magnetic beads (Bangs Laboratories), and permeabilized with 0.05% (wt/vol) digitonin, and then incubated with anti-CTCF antiserum (Active Motif, 1 μl/reaction) or IgG overnight. After removal of unbounded antibodies with proper washing, the nuclei were incubated with protein A/G-micrococcal nuclease (MNase) fusion protein (produced in-house with prokaryotic expression plasmid from Addgene, plasmid #123461) for 1 h at 4°C. CaCl2 was then added to activate MNase activity and incubated on ice for 30 min. The reaction was quenched with stopping buffer, and the DNA fragments were purified with MinElute Reaction Cleanup Kit (Qiagen), and then amplified by PCR for 10–14 cycles with barcoded Nextera primers (Illumina). DNA fragments in the range of 150–1,000 bp were recovered from 2% E-Gel EX Agarose Gels (Invitrogen, Thermo Fisher Scientific). The libraries were quantified using a KAPA Library Quantification kit and sequenced on Illumina HiSeq4000 in paired read mode with the read length of 150 nucleotides at the Admera Health. The CTCF CUT&RUN data were deposited at the GEO under GSE208128 and GSE220526 in the SuperSeries of GSE208130.
For Tcf1 CUT&RUN, WT TN, WT or Ctcf−/− TEFF cells were sort-purified (4–6 × 105 cells for each replicate) and fixed with 1% formaldehyde for 10 min at room temperature and then suspended in Radioimmunoprecipitation Assay buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 0.2% SDS, 0.1% wt/vol sodium deoxycholate, and 1% Triton X-100) for nuclei extraction. The nuclei were then incubated with 0.5 μl rabbit anti-Tcf1 polyclonal antibody (C46C7; Cell Signaling Technology) or rabbit IgG in Antibody-binding buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, and 1% Triton X-100) overnight with rotation. The unbound antibody was removed by washing the nucleus pellet with the Antibody-binding buffer, and the nuclei were incubated with protein A/G-micrococcal nuclease (MNase) fusion protein for 1 h at 4°C. The unbound MNase was removed by washing with Wash buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 400 mM NaCl, and 1% Triton X-100). After suspended in Resuspension buffer (20 mM Tris-HCl, pH 7.5, 10 mM NaCl, and 0.1% Triton X-100), the antibody-bound MNase was activated by addition of CaCl2 (final concentration 2 mM) followed by 30 min incubation at 0°C. The reaction was quenched with Stopping buffer (20 mM Tris-HCl, pH 8.0, 10 mM EGTA, 20 mM NaCl, 0.2% SDS, and 0.2 µg/μl proteinase K), and then incubated at 65°C for 2 h to reverse the crosslinking. The DNA fragments were purified with MinElute Reaction Cleanup kit (QIAGEN), end-repaired, adaptor added, and then amplified with PCR for 10–14 cycles with barcoded Nextera primers (Illumina). The amplified DNA fragments in the range of 150-1,000 bp were recovered, and the libraries were sequenced as described above. The Tcf1 CUT&RUN data were deposited at the GEO (GSE220527) under the SuperSeries of GSE208130.
Data processing
The sequencing quality of the libraries was assessed by FastQC. Trim Galore was used to trim low quality sequences and remove adapters. Bowtie2 (Langmead and Salzberg, 2012) was used to align the sequencing reads to the mm9 mouse genome, and only uniquely mapped reads (MAPQ > 10) were retained. Samtools (Li et al., 2009) was used to transfer the sam files to bam files and sort bam files. Picard MarkDuplicates was used to remove duplicate reads in the bam files. MACS2 (Zhang et al., 2008) was used for CTCF peak calling, with the IgG CUT&RUN library used a negative control, where stringent criteria of ≥4 summit fold change and FDR < 0.05 were used. CTCF binding sites in a cell type were called by applying MACS2 to bam reads from biological replicates pooled together. Tcf1 peaks were called using MACS2 with genome background as control, with parameters of ≥2 summit fold change and FDR < 0.05.
Reproducibility analysis and identification of differential and constitutive CTCF binding sites
Significant peaks called by MACS2 from naive and early effector CD8+ T cells were merged into union peaks. Raw counts in each library were mapped onto those union peaks, resulting in a matrix with rows representing the peaks and columns representing the libraries. The raw-count matrix was then subjected to normalization as follows: Each row, representing a peak region, was normalized by length of each peak region per kilobase, and each column, representing a library, was then normalized by the column sum per million. The normalized matrix was subjected to PCA analysis with the z-score option. The raw-count matrix was used as input for edgeR (quasi-likelihood test, robust, fold-change ≥2 and FDR < 0.05) to identify differential CTCF binding sites between naive and early TEFF CD8+ T cells and those between WT and Tbx21–/–Runx3–/– early TEFF cells. The same approach was used for assessment of Tcf1 CUT&RUN reproducibility. Constitutive CTCF binding sites were defined as non-differential CTCF binding sites between TN and TEFF cells, and further differentiated based on (1) presence of CTCF motifs and (2) overlap with robust ATAC-seq peaks in TN cells (i.e., log10[q-val] < −50).
Visualization of sequencing tracks and peak heatmaps
We adopted the following normalization method to enable quantitative comparison of signal levels among different cell types/states. For the sequencing tracks of ATAC-seq and CTCF CUT&RUN, replicates were merged, and raw-count BigWig files were normalized separately in each molecular feature by the total number of reads on peaks (called by merged bam files in each condition with MACS2 threshold and controls as before). For the Tcf1 and IgG CUT&RUN track, the raw-count BigWig file was normalized by the total reads per million. Deeptools (Ramírez et al., 2016) was used to plot the peak heatmaps.
Hi-C data analyses
Data generation
P14 donor-derived KLRG1+IL-7Rα– CD8+ cells were sorted as TEFF cells on day 8 after LCMV-Arm infection. Hi-C was performed on the TEFF cells (in two replicates, 4 × 106 cells/replicate) following the same protocol as previously described (Shan et al., 2022b) except that Mbo I was used in lieu of three restriction enzymes. The Hi-C data in TEFF cells were deposited at the GEO (GSE220528) under the SuperSeries of GSE208130.
Data processing
Hi-C data in TEFF cells were processed together with that in naive CD8+ T cells (GSE164710; Shan et al., 2021b) for consistency. Hi-C FASTQ sequencing files were mapped to the mm9 mouse genome using distiller-nf mapping pipeline (https://github.com/mirnylab/distiller-nf) with default parameters. Read pairs on the same chromosome and with mapq> = 10 were retained, and cool files with 10 kb resolution were generated. Cool files were converted to text files using cooler (https://github.com/open2c/cooler) and then to hic files using juicer_tools.jar (v1.21.01; Durand et al., 2016) pre command for downstream analyses.
Hi-C replicates reproducibility
hic-straw (https://pypi.org/project/hic-straw/) was used to extract interaction scores from hic files with Knight-Ruiz (KR) normalization (Knight and Ruiz, 2013) and the observed/expected (o/e) option. Each chromosome was partitioned into 10 kb bins. For every 10 kb bin on each chromosome, row sum (sum of interaction scores with its flanking 50 bins, i.e., 500 kb on each side) was calculated. The Pearson correlation of the row sum values of bins were calculated for each pair of Hi-C libraries. The heatmap of Pearson correlation values was plotted to assess reproducibility. After validating reproducibility, the raw read counts from replicates in each cell type were pooled together for downstream analyses to enhance sensitivity.
HiCHub analysis
HiCHub uses a network approach for comparing chromatin interactions between two cell types/states (Li et al., 2022 Preprint). In brief, KR-normalized Hi-C matrices in two conditions were used as input before applying the LOESS normalization. Candidate hubs with P value < 1 × 10−5 was considered as cell-type-specific hubs. The promoters of DEGs from the two cell types were then stratified against cell-type-specific hubs to identify genes whose expression was evidently modulated by changes in the chromatin interaction network. The code for HiCHub is available at https://github.com/WeiqunPengLab/HiCHub.
Insulation score
The insulation score was calculated following a previously defined approach (Crane et al., 2015), using FAN-C package (Kruse et al., 2020) with the insulation command. In brief, a sum of all chromatin contacts in a sliding square window (300 kb for each side) was calculated for each bin on a chromosome along the Hi-C matrix diagonal. The sum was then divided by mean value of all bins on the chromosome and log2 transformed as insulation score, where a lower score indicates stronger insulation effect.
Hi-C pile-up profile and data visualization
The KR o/e normalized contact matrix extracted by hic-straw was used in Hi-C pile-up analysis. From the contact matrix, aggregation of submatrices centered on a peak-set (e.g., dynamic or constitutive CTCF binding sites) with an extension of +/−50 bins or aggregation of submatrices centered on pixels (interactions between anchors) with an extension of +/−30 bins were plotted. The KR o/e normalized contact matrix was also used to display chromatin interactions at specific gene loci in heatmaps.
Statistical analysis
For comparison between two experimental groups, two-tailed Student’s t test was used. The statistical significance for the multiomics analyses was determined using the processing algorithms, i.e., EdgeR for RNA-seq, MACS2 for ATAC-seq and CTCF CUT&RUN, HOMER for motif analysis, HiCHub and FAN-C for Hi-C analysis, GSEA, DAVID, and GREAT for gene pathway and ontology analyses.
Supplementary Material
shows dynamic CTCF binding sites associated with genes in “immune system process.”
shows DEGs between WT naive CD8 and WT TEFF cells on 4 dpi.
shows DEGs associated with concordant chromatin interaction hubs in TN and TEFF cells.
shows DEGs between WT and CTCF-deficient TEFF cells on 4 dpi.
contains original blots for Fig. 4.
Acknowledgments
We thank the Flow Cytometry Core facility at the Center for Discovery and Innovation (M. Poulus and W. Tsao) for cell sorting. We thank N. Galjart (Erasmus Medical Center, Rotterdam, Netherlands) for the permission of using Ctcf-floxed mouse strain, and A.M. Melnick and M.A. Rivas (Weill Cornell Medical College, New York, NY, USA) for providing the mice.
This study is supported in part by grants from the National Institutes of Health (AI112579 to H.-H. Xue, AI121080 and AI139874 to H.-H. Xue and W. Peng) and Veterans Affairs (BX005771 to H.-H. Xue).
Author contributions: J. Liu performed the experiments and analyzed the data, with assistance from W. Hu, X. Zhao, and Q. Shan; S. Zhu analyzed the high throughput sequencing data; W. Peng and H.-H. Xue conceived the project, supervised the study, and wrote the paper.
Data availability
The RNA-seq on WT and Ctcf−/− early TEFF cells, ATAC-seq on WT TN, WT and Ctcf−/− early TEFF cells, CTCF CUT&RUN data on WT and Ctcf−/− TN cells, CTCF CUT&RUN data on WT and Tbx21−/− Runx3−/− early TEFF cells, Tcf1 CUT&RUN data on WT TN cells, WT and Ctcf−/− early TEFF cells, and Hi-C data on TEFF cells were deposited at the GEO under GSE208130.
References
- Anders, S., Pyl P.T., and Huber W.. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenio, J., Metz P.J., and Chang J.T.. 2015. Asymmetric cell division in T lymphocyte fate diversification. Trends Immunol. 36:670–683. 10.1016/j.it.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzate-Mejía, R.G., Recillas-Targa F., and Corces V.G.. 2018. Developing in 3D: The role of CTCF in cell differentiation. Development. 145:145. 10.1242/dev.137729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagan, J.A., and Phillips-Cremins J.E.. 2020. On the existence and functionality of topologically associating domains. Nat. Genet. 52:8–16. 10.1038/s41588-019-0561-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best, J.A., Blair D.A., Knell J., Yang E., Mayya V., Doedens A., Dustin M.L., Goldrath A.W., and Immunological Genome Project Consortium . 2013. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat. Immunol. 14:404–412. 10.1038/ni.2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, J.J., Brown D.R., Mullen A.C., Moskowitz N.H., Mahowald M.A., Sider J.R., Gajewski T.F., Wang C.R., and Reiner S.L.. 1998. Helper T cell differentiation is controlled by the cell cycle. Immunity. 9:229–237. 10.1016/S1074-7613(00)80605-6 [DOI] [PubMed] [Google Scholar]
- Chen, R., Bélanger S., Frederick M.A., Li B., Johnston R.J., Xiao N., Liu Y.C., Sharma S., Peters B., Rao A., et al. 2014. In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity. 41:325–338. 10.1016/j.immuni.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisolm, D.A., Savic D., Moore A.J., Ballesteros-Tato A., León B., Crossman D.K., Murre C., Myers R.M., and Weinmann A.S.. 2017. CCCTC-binding factor translates interleukin 2- and α-ketoglutarate-sensitive metabolic changes in T cells into context-dependent gene programs. Immunity. 47:251–267.e7. 10.1016/j.immuni.2017.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, H.K., McDonald B., and Kaech S.M.. 2021. The architectural design of CD8+ T cell responses in acute and chronic infection: Parallel structures with divergent fates. J. Exp. Med. 218:218. 10.1084/jem.20201730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane, E., Bian Q., McCord R.P., Lajoie B.R., Wheeler B.S., Ralston E.J., Uzawa S., Dekker J., and Meyer B.J.. 2015. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 523:240–244. 10.1038/nature14450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuartero, S., Stik G., and Stadhouders R.. 2022. Three-dimensional genome organization in immune cell fate and function. Nat. Rev. Immunol. 10.1038/s41577-022-00774-5 [DOI] [PubMed] [Google Scholar]
- Dixon, J.R., Gorkin D.U., and Ren B.. 2016. Chromatin domains: The unit of chromosome organization. Mol. Cell. 62:668–680. 10.1016/j.molcel.2016.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand, N.C., Shamim M.S., Machol I., Rao S.S., Huntley M.H., Lander E.S., and Aiden E.L.. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3:95–98. 10.1016/j.cels.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam, S., Fioravanti J., Zhu W., Le Gall J.B., Brohawn P., Lacey N.E., Hu J., Hocker J.D., Hawk N.V., Kapoor V., et al. 2019. The transcription factor c-Myb regulates CD8+ T cell stemness and antitumor immunity. Nat. Immunol. 20:337–349. 10.1038/s41590-018-0311-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullicksrud, J.A., Li F., Xing S., Zeng Z., Peng W., Badovinac V.P., Harty J.T., and Xue H.H.. 2017. Differential requirements for Tcf1 long isoforms in CD8+ and CD4+ T cell responses to acute viral infection. J. Immunol. 199:911–919. 10.4049/jimmunol.1700595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, H., Ribeiro de Almeida C., Sleutels F., Dingjan G., van de Nobelen S., Jonkers I., Ling K.W., Gribnau J., Renkawitz R., Grosveld F., et al. 2008. CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J. 27:2839–2850. 10.1038/emboj.2008.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndler-Brandstetter, D., Ishigame H., Shinnakasu R., Plajer V., Stecher C., Zhao J., Lietzenmayer M., Kroehling L., Takumi A., Kometani K., et al. 2018. KLRG1+ effector CD8+ T cells lose KLRG1, differentiate into all memory T cell lineages, and convey enhanced protective immunity. Immunity. 48:716–729.e8. 10.1016/j.immuni.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz, D., Day D.S., and Young R.A.. 2016. Insulated neighborhoods: Structural and functional units of mammalian gene control. Cell. 167:1188–1200. 10.1016/j.cell.2016.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, G., Cui K., Fang D., Hirose S., Wang X., Wangsa D., Jin W., Ried T., Liu P., Zhu J., et al. 2018. Transformation of accessible chromatin and 3D nucleome underlies lineage commitment of early T cells. Immunity. 48:227–242.e8. 10.1016/j.immuni.2018.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer, A.M., Banerjee A., Takemoto N., Gordon S.M., Dejong C.S., Shin H., Hunter C.A., Wherry E.J., Lindsten T., and Reiner S.L.. 2008. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 321:408–411. 10.1126/science.1159806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, M.R., Mintern J., La Gruta N.L., Kedzierska K., Doherty P.C., and Turner S.J.. 2008. Cell cycle-related acquisition of cytotoxic mediators defines the progressive differentiation to effector status for virus-specific CD8+ T cells. J. Immunol. 181:3818–3822. 10.4049/jimmunol.181.6.3818 [DOI] [PubMed] [Google Scholar]
- Joshi, N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., and Kaech S.M.. 2007. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 27:281–295. 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies, A., Xin A., Belz G.T., and Nutt S.L.. 2009. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 31:283–295. 10.1016/j.immuni.2009.06.021 [DOI] [PubMed] [Google Scholar]
- Kim, D., Paggi J.M., Park C., Bennett C., and Salzberg S.L.. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37:907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, P.A., and Ruiz D.. 2013. A fast algorithm for matrix balancing. IMA J. Numer. Anal. 33:1029–1047. 10.1093/imanum/drs019 [DOI] [Google Scholar]
- Kruse, K., Hug C.B., and Vaquerizas J.M.. 2020. FAN-C: A feature-rich framework for the analysis and visualisation of chromosome conformation capture data. Genome Biol. 21:303. 10.1186/s13059-020-02215-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurachi, M., Barnitz R.A., Yosef N., Odorizzi P.M., DiIorio M.A., Lemieux M.E., Yates K., Godec J., Klatt M.G., Regev A., et al. 2014. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat. Immunol. 15:373–383. 10.1038/ni.2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F., Zeng Z., Xing S., Gullicksrud J.A., Shan Q., Choi J., Badovinac V.P., Crotty S., Peng W., and Xue H.H.. 2018. Ezh2 programs TFH differentiation by integrating phosphorylation-dependent activation of Bcl6 and polycomb-dependent repression of p19Arf. Nat. Commun. 9:5452. 10.1038/s41467-018-07853-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Yuan S., Zhu S., Xue H.-H., and Peng W.. 2022. HiCHub: A network-based approach to identify domains of differential interactions from 3D genome data. bioRxiv. (Preprint posted April 16, 2022). 10.1101/2022.04.16.488566 [DOI] [Google Scholar]
- Lin, W.W., Nish S.A., Yen B., Chen Y.H., Adams W.C., Kratchmarov R., Rothman N.J., Bhandoola A., Xue H.H., and Reiner S.L.. 2016. CD8+ T lymphocyte self-renewal during effector cell determination. Cell Rep. 17:1773–1782. 10.1016/j.celrep.2016.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man, K., Gabriel S.S., Liao Y., Gloury R., Preston S., Henstridge D.C., Pellegrini M., Zehn D., Berberich-Siebelt F., Febbraio M.A., et al. 2017. Transcription factor IRF4 promotes CD8+ T cell exhaustion and limits the development of memory-like T cells during chronic infection. Infect. Immun. 47:1129–1141.e5. 10.1016/j.immuni.2017.11.021 [DOI] [PubMed] [Google Scholar]
- McLane, L.M., Abdel-Hakeem M.S., and Wherry E.J.. 2019. CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 37:457–495. 10.1146/annurev-immunol-041015-055318 [DOI] [PubMed] [Google Scholar]
- Millner, J.J., Toma, C., He, Z., Millner, J.J., Kurd, N.S., Nguyen, Q.P., McDonald, B., Quezada, L., Widjaja, C.E., Witherden, D.A.,et al. 2020. Immunity. 52:808. 10.1016/j.immuni.2020.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoe, Y., Setoguchi R., Akiyama K., Muroi S., Kuroda M., Hatam F., Littman D.R., and Taniuchi I.. 2007. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J. Exp. Med. 204:1749–1755. 10.1084/jem.20062456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nish, S.A., Lin W.W., and Reiner S.L.. 2017. Lymphocyte fate and metabolism: A clonal balancing act. Trends Cell Biol. 27:946–954. 10.1016/j.tcb.2017.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora, E.P., Goloborodko A., Valton A.L., Gibcus J.H., Uebersohn A., Abdennur N., Dekker J., Mirny L.A., and Bruneau B.G.. 2017. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 169:930–944.e22. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson, R., Renkawitz R., and Lobanenkov V.. 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 17:520–527. 10.1016/S0168-9525(01)02366-6 [DOI] [PubMed] [Google Scholar]
- Pais Ferreira, D., Silva J.G., Wyss T., Fuertes Marraco S.A., Scarpellino L., Charmoy M., Maas R., Siddiqui I., Tang L., Joyce J.A., et al. 2020. Central memory CD8+ T cells derive from stem-like Tcf7hi effector cells in the absence of cytotoxic differentiation. Immunity. 53:985–1000.e11. 10.1016/j.immuni.2020.09.005 [DOI] [PubMed] [Google Scholar]
- Pham, D., Moseley C.E., Gao M., Savic D., Winstead C.J., Sun M., Kee B.L., Myers R.M., Weaver C.T., and Hatton R.D.. 2019. Batf pioneers the reorganization of chromatin in developing effector T cells via ets1-dependent recruitment of Ctcf. Cell Rep. 29:1203–1220.e7. 10.1016/j.celrep.2019.09.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongubala, J.M.R., and Murre C.. 2021. Spatial organization of chromatin: Transcriptional control of adaptive immune cell development. Front. Immunol. 12:633825. 10.3389/fimmu.2021.633825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez, F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., and Manke T.. 2016. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44:W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, Q., Hu S., Chen X., Danahy D.B., Badovinac V.P., Zang C., and Xue H.H.. 2021a. Ectopic Tcf1 expression instills a stem-like program in exhausted CD8+ T cells to enhance viral and tumor immunity. Cell. Mol. Immunol. 18:1262–1277. 10.1038/s41423-020-0436-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, Q., Hu S.S., Zhu S., Chen X., Badovinac V.P., Peng W., Zang C., and Xue H.H.. 2022a. Tcf1 preprograms the mobilization of glycolysis in central memory CD8+ T cells during recall responses. Nat. Immunol. 23:386–398. 10.1038/s41590-022-01131-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, Q., Li X., Chen X., Zeng Z., Zhu S., Gai K., Peng W., and Xue H.H.. 2021b. Tcf1 and Lef1 provide constant supervision to mature CD8+ T cell identity and function by organizing genomic architecture. Nat. Commun. 12:5863. 10.1038/s41467-021-26159-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, Q., Zeng Z., Xing S., Li F., Hartwig S.M., Gullicksrud J.A., Kurup S.P., Van Braeckel-Budimir N., Su Y., Martin M.D., et al. 2017. The transcription factor Runx3 guards cytotoxic CD8+ effector T cells against deviation towards follicular helper T cell lineage. Nat. Immunol. 18:931–939. 10.1038/ni.3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, Q., Zhu S., Chen X., Liu J., Yuan S., Li X., Peng W., and Xue H.H.. 2022b. Tcf1-CTCF cooperativity shapes genomic architecture to promote CD8+ T cell homeostasis. Nat. Immunol. 23:1222–1235. 10.1038/s41590-022-01263-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene, P.J., and Henikoff S.. 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife. 6:e21856. 10.7554/eLife.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore, P.I., D’Ippolito A.M., Song L., Safi A., Shivakumar N.K., Kabadi A.M., Reddy T.E., Crawford G.E., and Gersbach C.A.. 2015. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods. 12:1143–1149. 10.1038/nmeth.3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., Diao H., Getzler A.J., Rogal W., Frederick M.A., Milner J., Yu B., Crotty S., Goldrath A.W., and Pipkin M.E.. 2018. The transcription factor Runx3 establishes chromatin accessibility of cis-regulatory landscapes that drive memory cytotoxic T lymphocyte formation. Immunity. 48:659–674.e6. 10.1016/j.immuni.2018.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, J., Long L., Zheng W., Dhungana Y., Lim S.A., Guy C., Wang Y., Wang Y.D., Qian C., Xu B., et al. 2019. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature. 576:471–476. 10.1038/s41586-019-1821-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, N.C., Chavez A., Lance-Byrne A., Chan Y., Menn D., Milanova D., Kuo C.C., Guo X., Sharma S., Tung A., et al. 2018. An enhanced CRISPR repressor for targeted mammalian gene regulation. Nat. Methods. 15:611–616. 10.1038/s41592-018-0048-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M., and Ren B.. 2017. The three-dimensional organization of mammalian genomes. Annu. Rev. Cell Dev. Biol. 33:265–289. 10.1146/annurev-cellbio-100616-060531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D.M., Yu S., Zhou X., Haring J.S., Held W., Badovinac V.P., Harty J.T., and Xue H.H.. 2010. Constitutive activation of Wnt signaling favors generation of memory CD8 T cells. J. Immunol. 184:1191–1199. 10.4049/jimmunol.0901199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Zhu S., Peng W., and Xue H.H.. 2022. The interplay of transcription and genome topology programs T cell development and differentiation. J. Immunol. 209:2269–2278. 10.4049/jimmunol.2200625 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows dynamic CTCF binding sites associated with genes in “immune system process.”
shows DEGs between WT naive CD8 and WT TEFF cells on 4 dpi.
shows DEGs associated with concordant chromatin interaction hubs in TN and TEFF cells.
shows DEGs between WT and CTCF-deficient TEFF cells on 4 dpi.
contains original blots for Fig. 4.
Data Availability Statement
The RNA-seq on WT and Ctcf−/− early TEFF cells, ATAC-seq on WT TN, WT and Ctcf−/− early TEFF cells, CTCF CUT&RUN data on WT and Ctcf−/− TN cells, CTCF CUT&RUN data on WT and Tbx21−/− Runx3−/− early TEFF cells, Tcf1 CUT&RUN data on WT TN cells, WT and Ctcf−/− early TEFF cells, and Hi-C data on TEFF cells were deposited at the GEO under GSE208130.