Abstract
Purpose
In image-guided adaptive brachytherapy (IGABT) a quantitative evaluation of the dosimetric changes between fractions due to anatomical variations, can be implemented via rigid registration of images from subsequent fractions based on the applicator as a reference structure. With available treatment planning systems (TPS), this is a manual and time-consuming process. The aim of this retrospective study was to automate this process. A neural network (NN) was trained to predict the applicator structure from MR images. The resulting segmentation was used to automatically register MR-volumes.
Material and Methods
DICOM images and plans of 56 patients treated for cervical cancer with high dose-rate (HDR) brachytherapy were used in the study. A 2D and a 3D NN were trained to segment applicator structures on clinical T2-weighted MRI datasets. Different rigid registration algorithms were investigated and compared. To evaluate a fully automatic registration workflow, the NN-predicted applicator segmentations (AS) were used for rigid image registration with the best performing algorithm. The DICE coefficient and mean distance error between dwell positions (MDE) were used to evaluate segmentation and registration performance.
Results
The mean DICE coefficient for the predicted AS was 0.70 ± 0.07 and 0.58 ± 0.04 for the 3D NN and 2D NN, respectively. Registration algorithms achieved MDE errors from 8.1 ± 3.7 mm (worst) to 0.7 ± 0.5 mm (best), using ground-truth AS. Using the predicted AS from the 3D NN together with the best registration algorithm, an MDE of 2.7 ± 1.4 mm was achieved.
Conclusion
Using a combination of deep learning models and state of the art image registration techniques has been demonstrated to be a promising solution for automatic image registration in IGABT. In combination with auto-contouring of organs at risk, the auto-registration workflow from this study could become part of an online-dosimetric interfraction evaluation workflow in the future.
Keywords: Brachytherapy, Image registration, Deep Learning, Auto Segmentation
1. Introduction
The standard of care for locally advanced cervical cancer (LACC) consists of external beam radiotherapy (EBRT) with concomitant chemotherapy followed by brachytherapy (BT) [1], [2]. Magnetic Resonance Imaging (MRI) based image-guided adaptive brachytherapy (IGABT) is considered an essential part of this gold-standard, resulting in excellent local control and reduced toxicity [2]. It uses three-dimensional MR-images of the patient with the applicator in-situ to create an individualized treatment plan, considering the 3D dose distribution and patient anatomy. The dose is prescribed to dose-volume parameters and optimized towards evidence-based dose limits according to international recommendations [1], [3]. Needles can be used in combination with the intracavitary applicator to further shape the dose distribution which results in improved target coverage [4].
A common schedule is to deliver BT in 4 fractions with two implants. Treatment planning, including imaging, applicator reconstruction and segmentation of targets and organs-at-risk (OAR), is usually performed on fraction 1 and fraction 3. A verification MRI is acquired prior to fraction 2 and 4 to detect major variations in implant geometry or anatomy. This can trigger various interventions (repositioning, adjustment of organ filling or replanning). Similar workflows have often been reported in the multicenter EMBRACE-I study [2], where 52% of the patients receiving MRI-guided adaptive BT in up to four high dose-rate (HDR) fractions were treated more than once with the same plan (EMBRACE study office, personal communication, December 1, 2021).
For IGABT planning or re-planning before another fraction is treated, an essential step is the applicator reconstruction [5]. Following imaging with the applicator in-situ, the source path needs to be accurately defined within the patient anatomy. For this purpose, modern treatment planning systems (TPS) offer a library of virtual 3D surface models of different applicators which are directly linked to corresponding source paths. In the TPS these models are manually aligned with the applicator depicted in the imaging volume. This is a critical step as geometrical reconstruction errors can lead to significant dose deviations in targets and OAR [6], [7].
State of the art IGABT treatment planning consists of: (i) applicator reconstruction, (ii) target and OAR delineation and (iii) dose optimization [1]. Depending on case complexity, the process takes a trained team of physician and physicist upwards of one hour to complete. Studies have shown that while systematic dosimetric uncertainties due to anatomical variations between subsequent fractions are low, random intra- and interfraction organ motion can have a significant dosimetric impact [8].
However due to time-constraints the scans are usually only visually inspected. A quantitative evaluation of the dosimetric changes due to anatomical variations currently requires the following steps within the TPS:
-
(i)
Reconstruction of the exact applicator position on planning and control MRI
-
(ii)
Definition of a set of reproducible applicator landmark points in planning and control MRI
-
(iii)
Landmark-based rigid registration of planning and control MRI
-
(iv)
Propagation and/or adaptation of OAR contours
-
(v)
Propagation of dose distribution
Two reasonable assumptions are made. First, the applicator is a rigid structure. MRI-compatible applicators are made from hardened plastic and thus deformations of the structure between subsequent fractions due to anatomical stresses are minimal. Second, as the applicator is the dose delivery device, the dose distribution is fixed to the applicator, and the target is fixed to the applicator by firm vaginal gauze packing. Therefore the target volume experiences no relative motion between fractions [9].
These assumptions make brachytherapy unique among other treatment techniques in radiation oncology. They allow the use of rigid registration based on the applicator for dose propagation.
Nevertheless, until now registering two BT MR-image series based on the applicator is a time-consuming process with no fully automatic commercial solutions available. While MRI offers superior soft-tissue contrast and is regarded as the gold standard for IGABT, it comes at the price of inferior applicator and needle representations as compared to CT. This increases the complexity of applicator reconstruction and poses an additional challenge for automatic image registration algorithms based on mutual information.
The required time investment limits the quantitative evaluation of inter and intra-fraction motions of OAR in clinical routine. An automatic solution for quantifying the dosimetric impact of organ motion could potentially lead to reduced differences between reported and delivered dose and improve outcome/-modelling for patients [10]. This study aims at developing a solution for the first part of an automated workflow for routine monitoring of interfraction variations.
In recent years, neural networks (NN) and related architectures have demonstrated success in various applications involving medical imaging [11], [12]. They achieve excellent segmentation performance [13] and are driving a new wave of research in automated treatment planning in radiation oncology [14].
In this study we i) compare different applicator-based rigid image registration algorithms for MR-IGABT, and ii) train a NN to predict the applicator structure to automate the registration process.
We hypothesize that by using the applicator structure as a reference, it is possible to register two MR-image series of subsequent BT fractions accurately and automatically for different applicator and needle geometries.
2. Materials and Methods
2.1. Patient Cohort
A cohort of 56 patients was available for this study. The retrospective use of clinical data was approved by the institutional ethics board (1400/2018).
Patients were treated at our institution between 2016 and 2019 for LACC. Patients received treatment according to the EMBRACEII protocol [15], which includes 25 × 1.8 Gy EBRT and four fractions of HDR BT to reach a High-Risk CTV D90 planning aim dose of 90-95 Gy EQD210Gy (Biologically equi-effective dose in 2 Gy fractions, α/β = 10 Gy).
The T2-weighted MR-images were acquired for treatment planning after applicator insertion using an open MR scanner (Siemens Magnetom C!, 0.35T, TSE, TR: 3290 ms, TE: 100 ms). Three image series, in para-transversal, sagittal and coronal orientation were recorded sequentially for clinical use. For this study, the para-transversal image series was used. All images had an in-plane resolution of 256 × 256, and voxel dimensions of 1.17 × 1.17 × 5 mm.
All patients were treated with a combination of intracavitary BT applicators with/without interstitial needles. Two different applicator models were used: the Vienna Ring Applicator [4], [16], and Venezia Applicator (Elekta, Venendaal). Different ring diameter and tandem length combinations were used. An overview of the configurations can be found in the Appendix Table A.1.
For each patient in our cohort, we exported two brachytherapy fraction treatment plans from the treatment planning system. By convention we refer to the first applied fraction as “fixed fraction”, and the second as “moving fraction”. All patients were treated with the same applicator for both fractions. All data was anonymized prior to export in DICOM format, using the TPS anonymization functionality.
2.2. Data Preprocessing
Two additional pre-processing steps were performed on the DICOM data after export. First, the contours of the applicator, which are usually not available in the TPS (Oncentra® Brachy – Elekta, Venendaal), were generated with an Elekta Applicator Slicer research plugin (Elekta, Venendaal) and treated as ground-truth masks. The plugin used the MRI and treatment plan with reconstructed applicator model to generate a standard radiotherapy (RT) structure file for the intracavitary applicator geometry. The resulting applicator mask was further processed with a hole filling algorithm to create continuous applicator geometries [17].
It is important to note that, even if the plugin was commercially available, the described method to extract these ground-truth segmentations would not be feasible for an automatic workflow, as it requires prior manual placement of an applicator library model.
Second, a python script was written to read and export all available dwell positions of the applicator from the treatment plan for the purpose of evaluating registration performance. Dwell positions were exported as x,y,z-coordiantes in the patient coordinate system. Only clinically relevant dwell positions were considered for the calculation of the registration error. (Fig. 1, Appendix Table A1).
Fig. 1.
3D visualization of the exported applicator dwell positions in the patient coordinate system.
2.3. Study Design
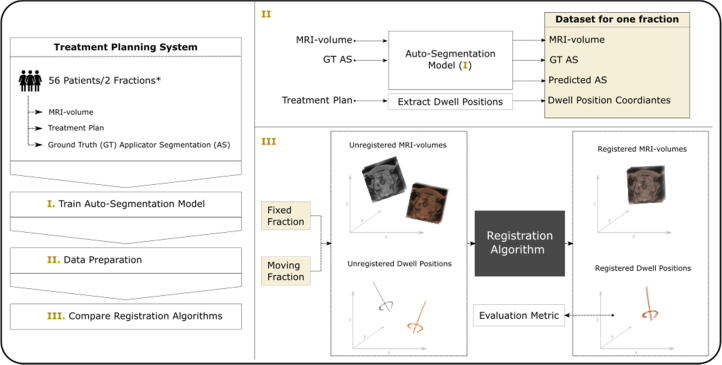
A summary of the major steps in this study is shown in Fig. 2. A NN was trained to predict the applicator segmentation, which was then used for rigid registration of MR-volumes from subsequent BT fractions. Different registration methods were compared. In a first step, ground-truth applicator masks were used to identify the most accurate registration approach. The method with the best results was then repeated with the predicted segmentations from the NN.
Fig. 2.
Summary of major steps in this study. For each patient in our cohort, two brachytherapy fraction treatment plans, and corresponding MRI-volumes and applicator segmentations were exported from the treatment planning system. An auto-segmentation neural network was trained to predict the applicator structure in unseen MR-volumes. Finally, different applicator-based rigid image registration algorithms were compared, initially with ground truth (GT), and eventually with predicted applicator segmentations (AS). The registration accuracy was evaluated by using the distance between dwell positions as a metric. *) For some patients only one fraction was available.
To get an unbiased measure of segmentation and registration performance, 10 randomly selected patients were split from the overall cohort and used as a test set. The remaining patients were used to train the NN. A description of the relevant cohorts’ parameters can be found in Table 1.
Table 1.
Summary of patient cohorts. BT: Brachytherapy; IC: Intercavitary; ICIS: Intercavitary/interstitial
| Cohort | n patients | n BT fractions | Technique |
|---|---|---|---|
| Train | 46 | 78 | IC: 15ICIS: 31 |
| Test | 10 | 20 | IC: 5ICIS: 5 |
For seven patients only one fraction was available. While these were used to train the neural network, they were not considered for the test set as no moving fraction was available (Table 1).
2.4. Image registration
The goal of the image registration was to align the fixed and the moving MR-volume as accurately as possible, with respect to the rigid applicator. Registration scripts were written using the python implementation of the Insight Toolkit (ITK) (Kitware, Clifton Park, NY), and the ITK implementation of elastix [18], [19], using default parameters.
Many uncertainties are still rooted in the use of deformable image registration (DIR) for brachytherapy [20]. Large anatomical deformations caused by organ movement, and the presence of the rigid applicator in the region of interest affect the registration performance. Applicator-based rigid registration however was found to be suitable for various tasks in clinical workflows for BT [20], and was therefore chosen for this work. Hence only rotation and translation were used as options for spatial transformations.
Five different rigid image registration algorithms were investigated:
“Default Rigid” (DR); used the full MRI-volumes and no applicator mask. “Applicator ROI” (AROI); used applicator masks to define valid sampling regions in MRI-volumes. “Applicator Mask” (AM); used no MRI and registered binary applicator masks directly. “Distance Map” (DM); used no MRI and registered distance maps generated from the applicator masks [21]. “Prediction” (DM*); Same as DM but used predicted applicator masks from the NN.
Two loss functions were used among the algorithms. Mutual Information (MI); a probabilistic loss using voxel information, and Kappa statistic (KS); a loss function specifically designed to register binary images (masks) [22].
Table 2 provides an overview of all tested configurations. Registration performance was evaluated on the test set.
Table 2.
Overview of the different investigated registration algorithms.
| “Default Rigid” (DR) | “Applicator ROI” (AROI) | “Applicator Mask” (AM) | “Distance Map” (DM) | “Prediction” (DM*) | |
|---|---|---|---|---|---|
| Implementation | Python/ITK | ||||
| Loss function | Mutual Information (MI) | MI | Kappa Statistic (KS) | MI | MI |
| Transform | 3D Translation + Rotation | ||||
| Image domain | MRI | MRI | Binary applicator mask | Euclidian distance map | Euclidian distance map |
| Description | Use full MRI volumes, and default settings. An out-of-the box solution that serves as a benchmark. | Use applicator masks to define valid sampling regions in MRIs. | Register binary applicator masks directly. | Transform binary mask to Euclidian distance maps and register the two distance maps. | Same as “Distance Map” but using the predicted applicator masks from the neural network. |
2.5. Automatic Applicator Segmentation
Two NN were trained to predict the applicator segmentation in MR-volumes. An often-used network architecture for medical image segmentation is U-NET [23], a CNN consisting of an encoder that learns global feature representations, followed by a decoder that upsamples these representations for pixel-wise classification. Influenced by recent advances in deep learning, a derived architecture called UNET-Transformer (UNETR) demonstrated superior performance for volumetric organ segmentation in medical images [24].
Because of the limited amount of available training data, we trained a 2D U-Net and 3D UNETR to compare which architecture delivers the best results. Both models use a Loss-function that computes both DICE Loss [25] and Cross Entropy Loss, and returns the sum of the two losses [26].
Data preparation involved: interpolation of voxel spacing to 1 × 1 × 3 mm3, and voxel intensity range min-max-scaling from 0 and 2000 to 0 and 1 for MRI. Data augmentation was used to artificially enlarge the dataset and improve model performance, including random flip, and rotation with a probability of 20%, along all axes (3D UNETR) or in the transversal plane (2D U-Net).
The 2D U-Net was trained using the para-transversal MR-slices as input, and outputs a single-channel label map of the same size. To boost model performance, a pretrained ResNet34 [27] was used in the encoder path. The model was trained using the fastai library [28]. Training hyperparameters were learning rate: 0.001, weight decay: 0.01, Adam optimizer, and a batch size of 12.
The 3D UNETR model used MRI volumes as input, and outputs a single-channel label map of the same size. Data augmentation further included random cropping of 192 × 192 × 32 voxel volumes. Prior to training, a hyperparameter search was executed [29], using the DICE coefficient as a performance metric (Equation (2)). The best parameters were found to be learning rate: 0.001, feature size: 16, weight decay: 1e-5, dimension of hidden layer: 256 and with residual blocks, the Adam optimizer, and a batch size of 1.
The final model was trained using 5-fold cross validation (CV). Predictions were made by averaging over the outputs of the 5-fold CV models. The network was trained using PyTorch [30]. Model architecture and loss function used implementations of the Medical Open Network for AI [31].
For both 2D U-Net and 3D UNETR, final model performance was evaluated on the test set using the DICE metric (Equation (2)).
2.6. Evaluation Metrics
The dwell positions of the reconstructed applicator were used to evaluate the registration accuracy. Clinically used treatment plans were available for both the fixed and moving fraction. The dwell positions derived from these applicator reconstructions were interpreted as the ground-truth.
Hence for each set of dwell positions in the fixed fraction, there existed a corresponding set of dwell positions in the moving fraction. To achieve a precise and interpretable metric of registration accuracy, the Euclidean distance between each pair of points was calculated and averaged over all positions, which resulted in a mean distance error in mm (MDE).
| (1) |
Where is the number of dwell positions, and and the coordinate vectors of the dwell position in fixed and moving fraction, respectively. To transform the dwell positions from the moving to the fixed domain, the resulting transformation matrix from the image registration was used.
To assess segmentation performance the DICE coefficient was used.
| (2) |
Where X and Y are the predicted and ground truth mask of the applicator. The coefficient measures the overlap between the two structures and returns a value between 0 and 1, with the latter representing a perfect match.
3. Results
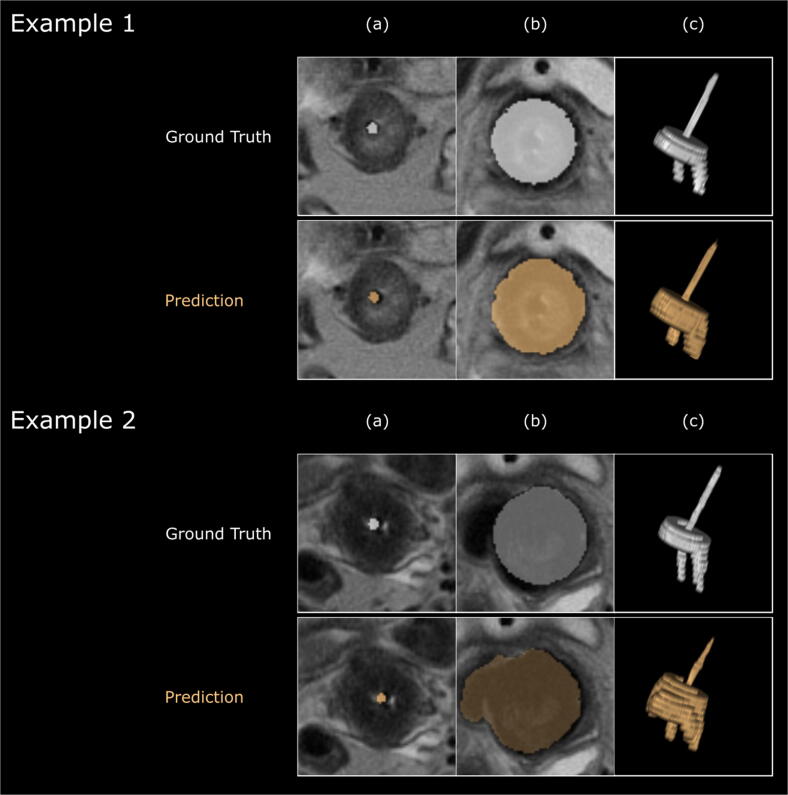
The mean DICE coefficient for the predicted applicator segmentations on the test set using UNETR and 2D U-Net was 0.70 ± 0.07 and 0.58 ± 0.04, respectively. Due to the superior performance of the UNETR, the predictions of this model were used for all further experiments. Fig. 3 shows the output of UNETR for two example patients in comparison with the ground truth.
Fig. 3.
Results of the auto-segmentation network from two example patients, and comparison with ground truth. Columns show (a) para-transversal slice through the tandem, (b) para-transversal slice through the ring of the applicator and (c) 3D rendering of the applicator structure. Example 2 represents a case where the network misclassified parts the applicator. Images and 3D rendering were generated with the MICE Toolkit (2020 NONPI Medical AB, Umeå, Sweden).
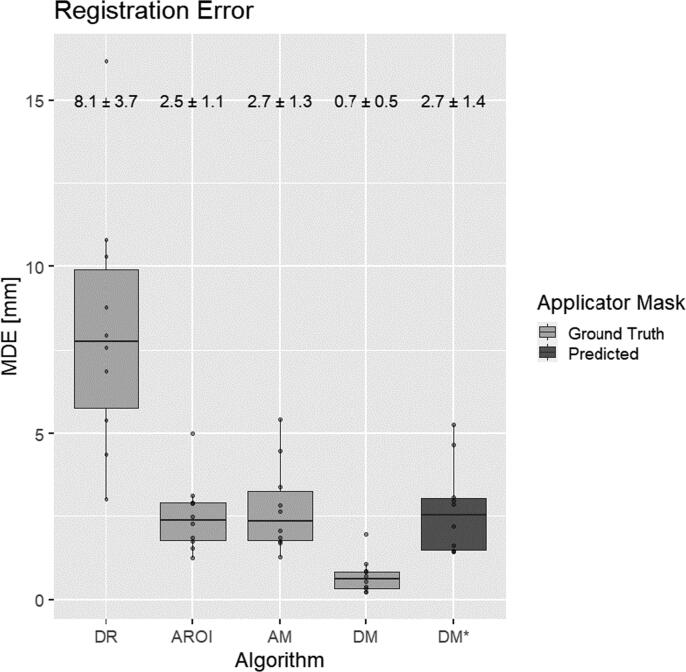
The results of the different registration algorithms, evaluated on the 10 patients in the test set, are summarized in Fig 4. DR, DROI and AM registration algorithms achieved MDE errors of 8.1 ± 3.7 mm, 2.5 ± 1.1 mm, and 2.7 ± 1.3 mm, respectively.
Fig. 4.
Registration error for different algorithms. Algorithm definitions according to Table 2 - Overview of the different investigated registration algorithms. Each box shows the distribution of the Mean Distance Error (MDE - average distance between dwell positions in mm). The lower and upper ends of the box correspond to the 25th and 75th quartiles respectively. Whiskers extend up to 1.5*IQR (inter-quartile range). Horizontal lines in the middle of each box represent the median value. Mean error and standard deviation are provided above.
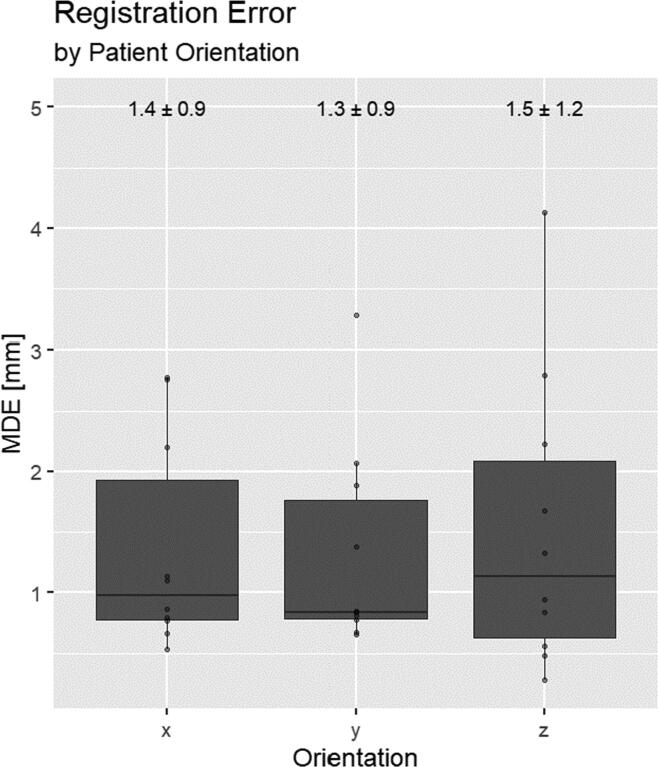
The best result was achieved by registering the distance maps generated from the ground truth applicator structures, resulting in an error of 0.7 ± 0.5 mm. Using the predicted applicator structures instead, the error was 2.7 ± 1.4 mm. For the latter, contributions of the different patient orientation components can be seen in Appendix Fig. A2. Fig. 5 shows an example of the registration result using the predicted applicator mask.
Fig. A2.
Registration error using predicted applicator mask (see DM* in Fig.4) stratified by patient orientation components. X: right to left. Y: Anterior to Posterior. Z: Caudal to Cranial.
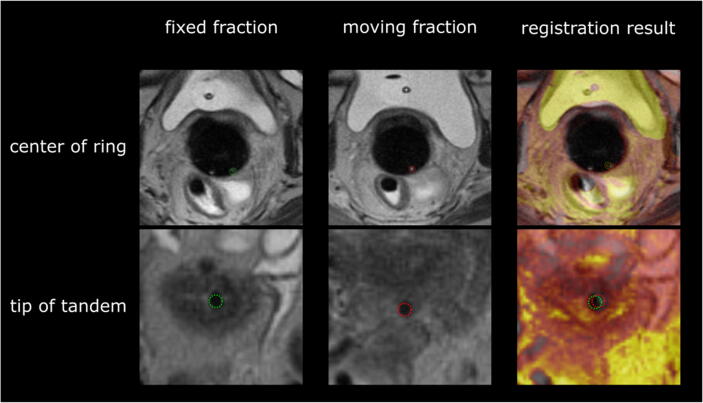
Fig. 5.
Example of registration result using the predicted applicator mask. Top row shows para-transversal slices through the center of the ring. Bottom row shows para-transversal slice through the tip of the tandem. Distinct landmarks of the applicator are marked with green circles (left, fixed fraction), and red circles (middle, moving fraction). The column on the right shows the registered and overlayed MRI-volumes, including landmarks, of both fractions. Moving fraction colored in orange.
4. Discussion
4.1. Previous work
Automating steps of the treatment workflow is a current and future research topic of interest in radiation oncology [12], [32], [33]. Although work has been published on auto-segmentation and applicator reconstruction using deep-learning methods, there is limited information about automatic image registration for BT, and no directly comparable work was found. Zaffino et al. trained a Convolutional Neural Network (CNN) to automatically segment needles from MR-images in gynecological BT with an average error of 2 ± 3 mm [34]. Hrinivich et al. used a 3D model to image approach for automatic applicator reconstruction on MRI [35], and reported dwell position errors within 2.1 mm. Zhang et al. trained a CT-based UNET to reconstruct channel paths with a Hausdorff distance below 1 mm [36].
4.2. MRI challenges
The advantages in terms of patient related outcome of using MRI instead of CT in IGABT have been demonstrated in the literature [2]. Investigating automatic image processing routines for MR-guided BT is therefore an important step towards future applications. However, MR-images for gynecological BT pose several distinct challenges for automatic contouring algorithms.
The applicators used in this study consist of two parts: the ring and intrauterine tandem. Both produce a void signal on T2-weighted MR-images.
In para-transversal slices the tandem can only be identified as a small circular structure. Similarly, the needles of interstitial implants have a similar shape, and close geometric proximity. In addition, there are other objects like the Foley catheter, blood vessels or the partially air-filled rectum that could potentially confuse auto-segmentation algorithms.
Furthermore, the rings in this study contain several holes that are used to guide interstitial needles. If holes fill with fluid during insertion, they can provide marker-style landmarks on MRI. These “markers” can help medical physicists when reconstructing the applicator manually. However, it is an inconsistent image feature, as it is subject to needle use and other anatomical factors and can thus not be assumed to be present beforehand.
Finally, the vaginal packing which holds the implant in place can further obscure the representation of the ring. Hence in a worst-case scenario it can be very challenging to identify the applicator ring on MRI.
A distinct challenge for this work was the use of clinical images from a low field (0.35 T), open MR scanner. The results are based on the retrospectively used clinical image data, with its limitations (non-isotropic, large slice thickness and no additional sequences within an acceptable scan time). Other MR scanners used in radiation oncology offer a higher field strength and thus superior resolution [37]. It is likely that with higher quality images the performance of image-processing algorithms and registration methods used in this work, would increase. As image series in sagittal and coronal direction were also recorded for clinical use, a possible extension of this work would be to include information from these volumes as well.
Finally, it should be noted that the use of MRI for every brachytherapy fraction is expensive and economically or logistically not feasible in many countries [38]. Studies have shown that using an applicator-based registration workflow, it would be possible to use a combination of MRI for the first fraction and subsequent CT based planning [9]. The workflow presented in this study could be adapted to accommodate these scenarios as well.
4.3. Segmentation Results
While the achieved mean DICE similarity of 0.70 is not particularly high, it indicates that the concept of using a neural network to automatically detect the applicator is feasible. Our network demonstrated that it was able to segment applicator structures of different configurations on MR-images, with and without needles. In cases with interstitial needles, it showed the ability to identify the tandem, and correctly distinguish between catheters or potentially confounding anatomical structures correctly (see Fig. 3, Example 1).
Overall, we see that the neural network can predict the applicator structure well. However, in some cases the auto-segmentation fails to capture parts of the geometry, leading to larger registration errors. This may originate from a limitation of this study, which is the comparatively small amount of training data for the neural network [12], [39]. This aspect could be mitigated by switching to a 2D-model architecture which uses MR-slices instead of volumes. However, our results show a considerable difference between 2D and 3D model performance. Considering that our 3D model was trained from scratch while the 2D model uses pretrained weights, indicates that volumetric context is an important factor to applicator autosegmentation.
To learn more details about the modes of failure and potential pitfalls, we analyzed the predicted segmentations. Predictions with high errors originate from MRI volumes where the network failed to accurately define the borders of the ring, while detecting the tandem proved to be less error prone. As can be seen in Fig. 3 - Example 2, the packing surrounding the ring can be a confounding factor for the network.
4.4. Registration Results
The comparatively low performance of the default rigid (DR) registration imply that the deformation of patient anatomy between BT fractions cannot accurately be captured by rigid registration techniques, using only MRI information.
Out of all tested rigid registration algorithms, the conversion of the applicator mask to a distance map and subsequent registration (DM) yielded the best results. The registration accuracy falls within reported uncertainties of manual applicator reconstruction in gynecological BT [6], [40]. A distance of up to 2 mm is reported as acceptable for image registrations of this kind [20].
The remaining error most likely stems from the fact that extracted segmentations from the TPS, even for the same applicator, are subject to some variability.
A closer look at the registration error stratified by patient orientation is shown in Appendix Fig. A2. Although not significant, the error in cranio-caudal direction is more pronounced. This is plausible, given that the MR-voxels have their largest extent in this dimension. Studies have found that the resulting dosimetric error from reconstruction uncertainties is greatest in anterior-posterior direction [7]. Hence in terms of dosimetric accuracy, a systematically larger error in cranio-caudal direction would be more manageable.
Re-running the registration with the predicted masks results in lower performance as compared to the ground-truth baseline. This result could be expected as the neural network is not able to fully replicate the ground-truth masks. It shows that the registration algorithm relies on accurate delineations of the applicator.
Our study shows that a registration error below 1 mm, which is well below the reported desired accuracy of 2 mm [20], is achievable with our technique. Thus, using an improved version of the auto-segmentation network or similar approaches, would enable a fully automatic registration workflow.
Importantly, each inference on the model and the registration algorithm, takes less than 30 s on a standard computer. This represents a significant time improvement from current manual practices, which based on clinical experience takes approximately 20 min. Furthermore, the quality of registration is independent of the experience of the operator. An implementation of such a workflow would therefore be critical towards fully automated online dosimetric evaluation of intra- and interfraction motion in IGABT.
4.5. Workflow
In clinical practice, the goal would be to automatically assess dosimetric changes from one BT fraction to the next. We assume a scenario where the patient completed the first fraction and thus MRI and treatment plan are available, and the same implant is used for the next fraction. An automated workflow using our method would proceed with the following steps: (1) Acquisition of control MRI. (2) Export of initial MRI, treatment plan and control MRI. (3) Execution of our algorithm to (i) predict applicator segmentations and (ii) register fixed and moving fraction. (4) Transfer of initial dose distribution and structures to the resulting MRI. (5) Adaptation of OAR contours. (6) Evaluation of dosimetric changes (7) Informed decision if treatment plan needs to be adapted.
Solutions for steps 4 and 6 already exist in the open-source domain or could be done in a TPS that allows the execution of custom scripts. NN have demonstrated considerable success in predicting OAR structures on MRI and could therefore be used for this purpose [13], [39]. Alternatively, OAR contours could be adapted by a physician in the TPS, which would make the process semi-automatic, but still more efficient than a fully manual workflow.
5. Conclusion
The results of this study show that automatic applicator-based image registration for MR-IGABT could be achieved by combining classical image registration algorithms with modern deep learning methods. It was demonstrated that our automatic workflow achieved the desired accuracy in a fraction of the time. The presented model would be attractive as it could be used for different applicator and needle configurations. Such automation would represent an important step towards future applications for routine monitoring of organ motion during treatment and could help to reduce dosimetric uncertainties.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: [Department of Radiation Oncology at Medical University of Vienna receives financial and/or equipment support for research and educational purposes from Elekta AB. CK, AS and NN received travel support and honoraria for educational activities from Elekta AB. YN is an employee of Elekta AB.]
Acknowledgements
We gratefully acknowledge the support of Elekta by provision of the Applicator Slicer research plugin used in this research. This research was funded by the Austrian Science Fund (FWF, project number KLI695-B33).
Appendix.
Table A1.
All used tandem and ring configurations, and their respective number of available dwell positions.
| Model | Applicator configuration: r(ing diameter) [mm] i(ntrauterine length) [mm] | n | Total number of dwell positions |
|---|---|---|---|
| Vienna | r26i60 | 25 | 60 |
| Vienna | r30i60 | 24 | 64 |
| Vienna | r30i40 | 16 | 64 |
| Vienna | r34i60 | 16 | 69 |
| Vienna | r26i40 | 9 | 50 |
| Venezia | r26i60 | 3 | 60 |
| Venezia | r30i50 | 2 | 64 |
| Venezia | r30i60 | 2 | 64 |
| Venezia | r26i50 | 1 | 60 |
References
- 1.Europe G. ICRU Report 89, Prescribing, Recording, and Reporting Brachytherapy for Cancer of the Cervix. J ICRU. 2013;13(1–2):1–10. doi: 10.1093/jicru/ndw027. [DOI] [PubMed] [Google Scholar]
- 2.Pötter R., et al. MRI-guided adaptive brachytherapy in locally advanced cervical cancer (EMBRACE-I): a multicentre prospective cohort study. Lancet Oncol. 2021;22(4):538–547. doi: 10.1016/S1470-2045(20)30753-1. [DOI] [PubMed] [Google Scholar]
- 3.Tanderup K., Nesvacil N., Kirchheiner K., Serban M., Spampinato S., Jensen N.B.K., Schmid M., Smet S., Westerveld H., Ecker S., Mahantshetty U., Swamidas J., Chopra S., Nout R., Tan L.T., Fokdal L., Sturdza A., Jürgenliemk-Schulz I., de Leeuw A., Lindegaard J.C., Kirisits C., Pötter R. Evidence-Based Dose Planning Aims and Dose Prescription in Image-Guided Brachytherapy Combined With Radiochemotherapy in Locally Advanced Cervical Cancer. Seminars in Radiation Oncology. 2020;30:311–327. doi: 10.1016/j.semradonc.2020.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Mahantshetty U., et al. Vienna-II ring applicator for distal parametrial/pelvic wall disease in cervical cancer brachytherapy: An experience from two institutions: Clinical feasibility and outcome. Radiother Oncol. 2019;141:123–129. doi: 10.1016/j.radonc.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Berger D., Dimopoulos J., Pötter R., Kirisits C. Direct reconstruction of the Vienna applicator on MR images. Radiother Oncol. 2009;93(2):347–351. doi: 10.1016/j.radonc.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Hellebust T.P., et al. Recommendations from Gynaecological (GYN) GEC-ESTRO working group: Considerations and pitfalls in commissioning and applicator reconstruction in 3D image-based treatment planning of cervix cancer brachytherapy. Radiother Oncol. 2010;96(2):153–160. doi: 10.1016/j.radonc.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Tanderup K., et al. Consequences of random and systematic reconstruction uncertainties in 3D image based brachytherapy in cervical cancer. Radiother Oncol. 2008;89(2):156–163. doi: 10.1016/j.radonc.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Nesvacil N., et al. A multicentre comparison of the dosimetric impact of inter- and intra-fractional anatomical variations in fractionated cervix cancer brachytherapy. Radiother Oncol. 2013;107(1):20–25. doi: 10.1016/j.radonc.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nesvacil N., Pötter R., Sturdza A., Hegazy N., Federico M., Kirisits C. Adaptive image guided brachytherapy for cervical cancer: A combined MRI-/CT-planning technique with MRI only at first fraction. Radiother Oncol. 2013;107(1):75–81. doi: 10.1016/j.radonc.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nesvacil N., Tanderup K., Lindegaard J.C., Pötter R., Kirisits C. Can reduction of uncertainties in cervix cancer brachytherapy potentially improve clinical outcome? Radiother Oncol. 2016;120(3):390–396. doi: 10.1016/j.radonc.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Boldrini L., Bibault J.E., Masciocchi C., Shen Y., Bittner M.I. Deep Learning: A Review for the Radiation Oncologist. Front Oncol. 2019;9(October) doi: 10.3389/fonc.2019.00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lundervold A.S., Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys. 2019;29(2):102–127. doi: 10.1016/j.zemedi.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Zhou T., Ruan S., Canu S. A review: Deep learning for medical image segmentation using multi-modality fusion. Array. 2019;3–4(July):100004. [Google Scholar]
- 14.Hesamian M.H., Jia W., He X., Kennedy P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J Digit Imaging. 2019;32(4):582–596. doi: 10.1007/s10278-019-00227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pötter R., et al. The EMBRACE II study: The outcome and prospect of two decades of evolution within the GEC-ESTRO GYN working group and the EMBRACE studies. Clin Transl Radiat Oncol. 2018;9:48–60. doi: 10.1016/j.ctro.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirisits C., Lang S., Dimopoulos J., Berger D., Georg D., Pötter R. The Vienna applicator for combined intracavitary and interstitial brachytherapy of cervical cancer: Design, application, treatment planning, and dosimetric results. Int J Radiat Oncol Biol Phys. 2006;65(2):624–630. doi: 10.1016/j.ijrobp.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 17.Soille P. 2nd ed. Springer; 2003. Morphological Image Analysis: Principles and Applications. [Google Scholar]
- 18.Klein S., Staring M., Murphy K., Viergever M.A., Pluim J.P.W. <emphasis emphasistype=“mono”>elastix</emphasis>: A Toolbox for Intensity-Based Medical Image Registration. IEEE Trans Med Imaging. 2010;29(1):196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 19.Shamonin D., Bron E., Lelieveldt B., Smits M., Klein S., Staring M. Fast Parallel Image Registration on CPU and GPU for Diagnostic Classification of Alzheimer’s Disease. Front Neuroinform. 2014;7:50. doi: 10.3389/fninf.2013.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swamidas J., Kirisits C., De Brabandere M., Hellebust T.P., Siebert F.-A., Tanderup K. Image registration, contour propagation and dose accumulation of external beam and brachytherapy in gynecological radiotherapy. Radiother Oncol. Feb. 2020;143:1–11. doi: 10.1016/j.radonc.2019.08.023. [DOI] [PubMed] [Google Scholar]
- 21.Maurer C., Qi R., Raghavan V. A Linear Time Algorithm for Computing Exact Euclidean Distance Transforms of Binary Images in Arbitrary Dimensions. IEEE Trans Pattern Anal Mach Intell. 2003;25:265–270. [Google Scholar]
- 22.Klein S, Staring M. Elastix: The Manual. October, 2015. p. 1–42.
- 23.Olaf Ronneberger T.B., Fischer P. UNet: Convolutional Networks for Biomedical Image Segmentation. IEEE Access. 2021;9:16591–16603. [Google Scholar]
- 24.Hatamizadeh A., et al. UNETR: Transformers for 3D. Medical Image Segmentation. 2021 [Google Scholar]
- 25.Milletari F., Navab N., Ahmadi S.-A. in 2016 Fourth International Conference on 3D Vision (3DV) 2016. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation; pp. 565–571. [Google Scholar]
- 26.Jadon S. A survey of loss functions for semantic segmentation. 2020 IEEE Conf Comput Intell Bioinforma Comput Biol CIBCB 2020; 2020.
- 27.He K., Zhang X., Ren S., Sun J. Deep Residual Learning for Image Recognition. CoRR. 2015;vol. abs/1512.0 [Google Scholar]
- 28.Howard J, Gugger S. fastai: A Layered API for Deep Learning; 2020. p. 1–27.
- 29.L. Biewald, “Experiment Tracking with Weights and Biases.” [Online]. Available: http://wandb.com/.
- 30.Paszke A. Curran Associates, Inc.; 2019. PyTorch: An Imperative Style, High-Performance Deep Learning Library; pp. 8024–8035. [Google Scholar]
- 31.M. Consortium. MONAI: Medical Open Network for AI; 2021.
- 32.Vandewinckele L., et al. Overview of artificial intelligence-based applications in radiotherapy: Recommendations for implementation and quality assurance. Radiother Oncol. 2020;153:55–66. doi: 10.1016/j.radonc.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Breedveld S., Storchi P.R.M., Voet P.W.J., Heijmen B.J.M. iCycle: Integrated, multicriterial beam angle, and profile optimization for generation of coplanar and noncoplanar IMRT plans. Med Phys. 2012;39(2):951–963. doi: 10.1118/1.3676689. [DOI] [PubMed] [Google Scholar]
- 34.Zaffino P., et al. Fully automatic catheter segmentation in MRI with 3D convolutional neural networks: Application to MRI-guided gynecologic brachytherapy. Phys Med Biol. 2019;64(16) doi: 10.1088/1361-6560/ab2f47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hrinivich W.T., Morcos M., Viswanathan A., Lee J. Automatic tandem and ring reconstruction using MRI for cervical cancer brachytherapy. Med Phys. 2019;46(10):4324–4332. doi: 10.1002/mp.13730. [DOI] [PubMed] [Google Scholar]
- 36.Zhang D., Yang Z., Jiang S., Zhou Z., Meng M., Wang W. Automatic segmentation and applicator reconstruction for CT-based brachytherapy of cervical cancer using 3D convolutional neural networks. J Appl Clin Med Phys. 2020;21(10):158–169. doi: 10.1002/acm2.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollard J.M., Wen Z., Sadagopan R., Wang J., Ibbott G.S. The future of image-guided radiotherapy will be MR guided. Br J Radiol. 2017;90(1073):20160667. doi: 10.1259/bjr.20160667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahantshetty U., et al. IBS-GEC ESTRO-ABS recommendations for CT based contouring in image guided adaptive brachytherapy for cervical cancer. Radiother Oncol. 2021;160:273–284. doi: 10.1016/j.radonc.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cardenas C.E., Yang J., Anderson B.M., Court L.E., Brock K.B. Advances in Auto-Segmentation. Semin Radiat Oncol. 2019;29(3):185–197. doi: 10.1016/j.semradonc.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Kirisits C., et al. Review of clinical brachytherapy uncertainties: Analysis guidelines of GEC-ESTRO and the AAPM. Radiother Oncol. 2014;110(1):199–212. doi: 10.1016/j.radonc.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]