Highlights
-
•
NPAS4 is differentially expressed in the DLPFC following opioid overdose.
-
•
Gene expression in response to overdose may not reflect long-term opioid use.
-
•
Human postmortem brain tissue is central to clarifying the impact of opioid use.
Abstract
Background
Although preclinical models reveal the neurobiological pathways altered through opioid abuse, comprehensive assessments of gene expression in human brain samples are needed. Moreover, less is known about gene expression in response to fatal overdose. The primary goal of the present study was to compare gene expression in the dorsolateral prefrontal cortex (DLPFC) between brain samples of individuals who died of acute opioid intoxication and group-matched controls.
Methods
Postmortem tissue samples of the DLPFC from 153 deceased individuals (Mage = 35.4; 62% male; 77% European ancestry). Study groups included 72 brain samples from individuals who died of acute opioid intoxication, 53 psychiatric controls, and 28 normal controls. Whole transcriptome RNA-sequencing was used to generate exon counts, and differential expression was tested using limma-voom. Analyses were adjusted for relevant sociodemographic characteristics, technical covariates, and cryptic relatedness using quality surrogate variables. Weighted correlation network analysis and gene set enrichment analyses also were conducted.
Results
Two genes were differentially expressed in opioid samples compared to control samples. The top gene, NPAS4, was downregulated in opioid samples (log2FC = -2.47, adj. p = .049) and has been implicated in opioid, cocaine, and methamphetamine use. Weighted correlation network analysis revealed 15 gene modules associated with opioid overdose, though no intramodular hub genes were related to opioid overdose, nor were pathways related to opioid overdose enriched for differential expression.
Conclusions
Results provide preliminary evidence that NPAS4 is implicated in opioid overdose, and more research is needed to understand its role in opioid abuse and associated outcomes.
1. Introduction
According to the Centers for Disease Control and Prevention, more than 80% of drug overdose deaths in the United States involve opioids (O'donnell et al., 2020). Moreover, the use of illicitly manufactured fentanyl in the United States is rising, with two-thirds of opioid-related overdose deaths involving synthetic opioids (Scholl et al., 2019). In addition to the loss of life, opioid misuse and abuse is estimated to cost Americans as much as $78.5 billion per year (Florence et al., 2016). The individual and societal costs of opioid abuse underscore the need to clarify the molecular underpinnings of opioid abuse and its consequences, which will aid in the development of therapeutic targets and subsequent interventions to effectively treat opioid use disorder. Although preclinical models are an essential first step in understanding the neurobiological mechanisms underlying opioid abuse, examination of gene expression in human brain tissue is critical to accurately characterize neurobiological differences between opioid users and non-users (Egervari et al., 2019). The goal of the present study was to compare transcriptome-wide patterns of gene expression in the dorsolateral prefrontal cortex (DLPFC) of postmortem human adult brains among individuals who died of acute opioid intoxication and group-matched controls. The DLPFC was selected based on preclinical and clinical evidence demonstrating its role in addiction (Volkow and Morales, 2015; Kalivas et al., 2005; Kruyer and Chioma, 2020).
1.1. Neurobiology of opioid use
It is well established that persistent drug use contributes to alterations in the functioning of reward-processing networks in the brain, namely the mesocorticolimbic dopamine system (Hyman et al., 2006). This system consists of the mesocortical and mesolimbic pathways, which span key brain structures involved in addiction, such as the ventral tegmental area (VTA), nucleus accumbens (NAc) and prefrontal cortex (PFC). Dopamine neurons that originate in the VTA and terminate in the NAc contribute to the acute rewarding effects of drug use. Opioids, specifically, impact these reward pathways through the activation of various opioid receptors, primarily µ-opioid receptors. M-opioid receptors are widely expressed in the brain, including structures within the dopamine reward system (e.g., VTA). Prior work has shown that activation of µ-opioid receptors reduces the release of GABA neurons in the VTA, rostromedial tegmental nucleus (RMTg), and ventral pallidum (Galaj and Xi, 2021). By reducing the release of GABA (an inhibitory neuron), dopamine neurons in the VTA are disinhibited, resulting in a surge of dopamine in the NAc, which contributes to the acute rewarding effects of opioids (Ellis et al., 2021).
Prolonged changes in functioning of dopaminergic and glutamatergic transmission in the mesocorticolimbic system are associated with drug cravings (Jones et al., 2016), deficits in learning, memory, and attention (Arias et al., 2016), and increased impulsivity, which can last for years after abstinence (Biernacki et al., 2016). Moreover, evidence demonstrates that the PFC and NAc interact in a manner such that dopamine-related adaptations in the PFC influence behavioral responses to drug-related stimuli, while simultaneous glutamate-related adaptations in the NAc contribute to compulsive drug-seeking behavior (Kalivas et al., 2005). These findings underscore the interdependence of these brains regions and value in exploring changes in gene expression within the PFC.
1.2. Gene expression and opioid use
To date, several studies have assessed gene expression in the human brain in response to long-term opioid abuse. Alberston and colleagues studied changes in gene expression within the NAc of long-term heroin users (n = 7) compared to controls (n = 7), finding decreased expression of several genes encoding proteins involved in presynaptic release of neurotransmitters among heroin users (Albertson et al., 2006). Sillivan and colleagues examined gene expression in the striatum of heroin users (n = 22) compared to controls (n = 27), finding decreased expression of µ-opioid receptors, in addition to dysregulation of ERK signaling pathways, specifically, in heroin users compared to controls (Sillivan et al., 2013).
More recently, Saad and colleagues examined transcriptome-wide differences in gene expression in the midbrain between opioid users (n = 30) and controls (n = 20) (Saad et al., 2019). The authors found 545 differentially expressed genes in the midbrain of deceased opioid users, most of which were protein coding genes that were upregulated. In addition, weighted correlation network analysis revealed a gene cluster associated with inflammation. Lastly, Seney and colleagues examined transcriptome-wide differences in gene expression within the DLPFC and NAc between individuals diagnosed with an opioid use disorder (n = 20) and controls (n = 20) (Seney et al., 2021). The authors found numerous differentially expressed genes among cases compared to controls, and many differentially expressed genes overlapped across these two brain regions. Moreover, differential expression was enriched in pathways related to inflammation, lending support to the finding by Saad et al. that prolonged opioid use alters inflammatory processes in the body.
Although the evidence, to date, supports differential gene expression across multiple brain regions in response to long-term opioid use, changes in gene expression associated with overdose are less well characterized. Moreover, existing studies are limited in their sample sizes and available information on drug use history, which are common challenges in working with postmortem brain tissue. Although differential gene expression in response to an overdose may not reflect long-term opioid abuse, it can provide valuable insight into the molecular effects of acute intoxication at high relative doses of opioids. Thus, the goal of the present study was to examine differential gene expression in the DLPFC between human postmortem brain samples of individuals who died of an opioid overdose compared to controls.
2. Materials and methods
2.1. Sample collection
Postmortem brain samples (n = 160) were donated to the Lieber Institute for Brain Development from the Offices of the Chief Medical Examiner of the State of Maryland (MDH protocol #12–24) and of Western Michigan University Homer Stryker School of Medicine, Department of Pathology (WIRB protocol #1,126,332); One brain sample was acquired through a material transfer agreement from the National Institute of Mental Health (NIMH; donated through the Office of the Chief Medical Examiner of the District of Columbia: protocol NIMH#90-M-0142), all with the informed consent of legal next-of-kin at the time of autopsy. Subsequent analysis of DNA or RNA derived from human postmortem brain tissue is exempt from institutional ethics committees as research conducted on postmortem tissue is not considered Human Subjects Research by the Department of Health and Human Services.
At the time of donation, a 36-item next-of-kin informant telephone screening was conducted to obtain medical, social, demographic, and psychiatric history. Macroscopic and microscopic neuropathological examinations were conducted on every case by a board-certified neuropathologist to exclude for neurological problems, neuritic pathology, or cerebrovascular accidents. A retrospective clinical diagnostic review was conducted on every brain donor, consisting of the telephone screening, macroscopic and microscopic neuropathological examinations, autopsy and forensic investigative data, forensic toxicology data, extensive psychiatric treatment, substance abuse treatment, and/or medical record reviews, and whenever possible, family informant interviews. All data were compiled into a comprehensive psychiatric narrative summary that was reviewed by two board-certified psychiatrists to arrive at lifetime DSM-V psychiatric diagnoses (including substance use disorders/intoxication) and medical diagnoses.
The present analysis consisted of 153 postmortem human brain samples (Mage of death = 35.4; 62% male; 77% European ancestry). These samples consist of 72 opioid cases, 53 group-matched psychiatric controls, and 28 group-matched normal controls Table 1. contains demographic information on the study samples, split by group status. Non-psychiatric healthy controls were free from psychiatric and substance use diagnoses, and their toxicological data were negative for drugs of abuse. Psychiatric controls may have had a lifetime DSM-V psychiatric diagnosis (including substance use disorders/intoxication) and their toxicological data may have been positive for drugs of abuse, but an overdose involving opioids was not the cause of death. In a previous publication using this same cohort (Shu et al., 2021), we described the primary psychiatric diagnoses among the sample groups. Here, we provide additional data on any lifetime diagnoses of specific substance use disorders (e.g., opioid use disorder), based on the psychiatric narrative summaries. Of the original 160 brain samples, five psychiatric controls were removed for the cause of death involving opioids, and two addition samples (one opioid sample, one psychiatric control) were excluded due to mismatching genotyped and reported sex, resulting in the final sample of 153 brain specimens. Every brain donor had forensic toxicological analysis, which typically covered ethanol and volatiles, opiates, cocaine/metabolites, amphetamines, and benzodiazepines. Some donors also received supplemental directed toxicological analysis using National Medical Services, Inc., including nicotine/cotinine testing, cannabis testing, and the expanded forensic panel in postmortem blood (or, in rare cases, in postmortem cerebellar tissue) to cover any substances not tested. The following substances were considered opioids: codeine, morphine, oxycodone, hydrocodone, oxymorphone, hydromorphone, methadone, fentanyl, 6-monoacetylmorphine, and tramadol.
Table 1.
Demographic characteristics by study group.
| Analytic Sample (n = 153) |
Opioid Samples (n = 72) |
Psychiatric Controls (n = 53) |
Normal Controls (n = 28) |
|||||
|---|---|---|---|---|---|---|---|---|
| Demographic Categories | n (%) | Mean (SD) | n (%) | Mean (SD) | n (%) | Mean (SD) | n (%) | Mean (SD) |
| Sex | ||||||||
| Female | 58 (38%) | – | 22 (31%) | – | 21 (40%) | – | 15 (54%) | – |
| Male | 95 (62%) | – | 50 (69%) | – | 32 (60%) | – | 13 (46%) | – |
| Race | ||||||||
| African American | 35 (23%) | – | 10 (14%) | – | 10 (19%) | – | 13 (46%) | – |
| European American | 118 (77%) | – | 62 (86%) | – | 43 (81%) | – | 15 (54%) | – |
| Age of Death | – | 35.42 years (9.42) | – | 33.49 (8.86) | – | 36.22 (9.37) | – | 38.87 (10.03) |
| Postmortem Interval | – | 27.42 h (9.60) | – | 27.12 (8.95) | – | 27.67 (10.45) | – | 27.73 (9.87) |
| RNA Integrity Number | – | 7.79 (1.17) | – | 7.40 (1.35) | – | 8.18 (0.78) | – | 8.07 (0.96) |
2.2. Sample extraction and RNA measurement and preprocessing
Brains were hemisected and cut into 1.0–1.5-cm-thick coronal slabs, flash frozen, and stored at −80 °C. DLPFC gray matter approximating BA46/9 was dissected using a dental drill. Dissected DLPFC was pulverized and stored at −80 °C. Total RNA was extracted from ∼100 mg of pulverized tissue using the QIAGEN AllPrep DNA/RNA Mini Kit with on-column DNase treatment according to the manufacturer's protocol. The RNA quality was assessed with high-resolution capillary electrophoresis on an Agilent Bioanalyzer 2100 (Agilent Technologies). Paired-end strand-specific sequencing libraries were prepared from 300 ng total RNA using the TruSeq Stranded Total RNA Library Preparation kit with Ribo-Zero Gold ribosomal RNA (rRNA) depletion. Resulting RNA-seq libraries were sequenced on an Illumina HiSeq 3000 at the Lieber Institute for Brain Development Sequencing Core. Average sequencing depth was approximately 99.4 million reads per sample. RNA quality was adequate, with an average RNA integrity number (RIN) of 7.79 across all brain samples. Raw sequencing reads were processed into gene counts using a previously described pipeline (Collado-Torres et al., 2019). Briefly, reads were aligned to the genome using HISAT2 (Kim et al., 2019) and the 58,037 Gencode v25 genes were quantified from resulting alignments using featureCounts using paired-end stranded counting (Liao et al., 2014). We calculated the reads per kilobase of transcript, per million mapped reads (RPKM) from these counts, and genes with an average RPKM < 0.20 were excluded from downstream analyses. This resulted in 25,644 genes used in the differential expression analysis.
We also calculated principal components (PCs) and examined correlations between the top 10 PCs and phenotypic and technical covariates. Technical covariates included: RIN, rRNA assignment rate, exonic assignment rate, mitochondrial mapping rate (i.e., chrM Map Rate), concordant mapping rate (for paired-end sequencing), overall mapping rate, and External RNA Controls Consortium (ERCC) spike-in control transcripts. The top 10 PCs accounted for 49.6% of the variance in the data. The first two PCs were significantly correlated with all technical covariates, and the first PC was significantly correlated with observed sex. Thus, these variables were included among the list of covariates in the differential expression analysis. Lastly, because cell composition can confound the association between gene expression and outcomes of interest, we estimated nine cell types using Bisque (Jew et al., 2020) (see Section 2.3.1) for inclusion in our differential expression analysis.
2.3. Statistical methods
2.3.1. Estimation of cell composition
Cell composition in bulk DLPFC tissue was estimated using Bisque (Jew et al., 2020). Briefly, post-processed bulk tissue RNA-seq data was decomposed using a reference sample (n = 3) of single-nucleus RNA-seq data Tran et al. (2021). Estimates were derived for nine cell types: astrocytes, excitatory neurons, inhibitory neurons, macrophages, microglia, mural cells, oligodendrocytes, oligodendrocyte progenitor cells (OPC), and T cells. Cell type proportions were inspected and plotted using stacked bar charts (see Fig. S1). Given the small proportion of certain cell types (i.e., < 5% of total cells) across all samples, only the following four cell types were carried forward as potential covariates for the differential expression analysis: astrocytes, excitatory neurons, inhibitory neurons, and oligodendrocytes. However, when included in the model with the quality surrogate variables (qSVs), each cell type was strongly correlated with the fourth qSV (all p’s < 1e-5). In an effort to not over-fit the differential expression model, these cell type estimates were excluded from the final analytic model.
2.3.2. Differential expression analysis
Group-wise differences in gene expression between opioid-positive samples and controls were tested in R version 4.1 (R Core Team, 2021) using limma-voom (Ritchie et al., 2015). Specifically, a linear model was fit where gene expression was regressed on opioid overdose status (yes/no) across each of 25,644 expressed genes. Prior to the analysis, we collapsed the psychiatric and healthy control samples. We also identified 21 opioid-positive samples that, although cause of death was an opioid overdose, tested positive for cocaine or one of its metabolites at the toxicology screen. Thus, we added a covariate to the model to account for this. The analytic model is presented below, where Yij reflects average gene expression for sample i at gene j. The model adjusted for the following covariates: age of death, sex, race (Black/White), positive for cocaine (yes/no), postmortem interval (PMI), seven technical covariates, and 10 surrogate variables identified through quality surrogate variable analysis (qSVA) (Jaffe et al., 2017). Briefly, qSVA is an extension of surrogate variable analysis (Leek and Storey, 2007) and accounts for confounding due to brain degradation. Scalar variables, representing unmeasured sources of confounding, are identified through qSVA and can be included as covariates in an analytic model. Multiple testing correction was applied using the false discovery rate (FDR) procedure (Benjamini and Hochberg, 1995). This method controls the expected proportion of false positives among all statistical tests. It is less conservative than the commonly used Bonferroni correction, which controls for any expected false positive, but benefits from increased statistical power. Statistical significance was determined by an FDR corrected p-value < 0.10 (Benjamini and Hochberg, 1995).
2.3.3. Weighted gene co-expression network analysis
Weighted gene co-expression network analysis (WGCNA) was conducted using the WGCNA package in R (Langfelder and Horvath, 2008). Also known as weighted correlation network analysis, WGCNA is a systems biology method for describing correlation patterns among genes based on their expression profiles. Researchers can then examine correlation patterns of genes within these gene clusters, or modules, and test correlations between the modules and phenotypic traits of interest.
Prior to WGCNA, counts were transformed to log2-counts per million (logCPM) using voom, and the influence of covariates was regressed out. Next, we examined the data to ensure that neither genes nor samples had excessive missing values, and that there were no sample outliers. No samples or genes were excluded during this step. The “pickSoftThreshold()” function was then used to identify the soft thresholding power, β, to which co-expression similarity is raised to calculate adjacency (i.e., connectivity). The function returns various indices, namely a graph indicating the lowest power for which the scale-free topology fit index curve flattens (i.e., when the curve of R2 values flattens). Lastly, the WGCNA function, “blockwiseModules()”, is used to create correlation matrices based on similarity in the expression profiles of genes in the dataset. These correlation matrices are raised to the soft-thresholding power selected in the previous step, and hierarchical clustering is used to create modules and merge modules that are highly similar. In the present study, the “blockwiseModules()” function was implemented with the following parameters: power = 10, TOMType = “unsigned”, minModuleSize = 30, mergeCutHeight = 0.25, maxBlockSize = 26,000, corType = “bicor.”
Once the number of modules was determined, module eigengenes (i.e., the first principal component of a given module) were used to test correlations between each module and variables included in the analytic model, excluding quality surrogate variables. These results were then visualized using a heatmap. Modules that were significantly correlated with opioid overdose status were then selected and examined for hub genes. Lastly, modules correlated with opioid overdose status were used in gene-set enrichment analyses.
2.3.4. Hub gene identification and enrichment analysis
If a module from the WGCNA was significantly correlated with opioid overdose status (i.e., p < .05), these modules were further inspected for intramodular hub genes (Langfelder and Horvath, 2008). Intramodular hub genes were identified as genes with high gene-module memberships (i.e., > 0.80) and gene-trait significance (i.e., absolute value > 0.30). Lastly, the hub genes were used in gene ontology (GO) enrichment analyses using GOrilla (Eden et al., 2009). Specifically, hub genes in each module were entered as an unranked target list, separately, along with all genes in the study (n = 25,644) as the background list. We tested for enrichment within biological process, molecular function, and cellular component pathways. Significant enrichment was determined using an FDR adjusted p < .10.
3. Results
3.1. Drug use history
Every case in the study was assessed for a lifetime history of alcohol and substance use disorders. Among the opioid samples, 66 had a history of an opioid use disorder; two had a history of a cocaine use disorder; and four did not meet criteria for a lifetime substance use disorder despite dying of an opioid overdose. Average age of death was approximately 33 years (M = 33.49, SD = 8.86); and manner of death (i.e., the ruling by the medical examiner as to the determination of how the acute opioid intoxication death occurred) was either accidental (n = 19) or undetermined (n = 53). Age of onset for substance use was obtained for 58 of the 72 opioids samples, with the average age of onset being approximately 18 years old (M = 17.97, SD = 6.35; min = 10, max = 37). Among 24 opioid samples with data on age of first drug detox treatment, the average age was approximately 25 years old (M = 25.08, SD = 7.21; min = 15, max = 44).
3.2. Differential gene expression
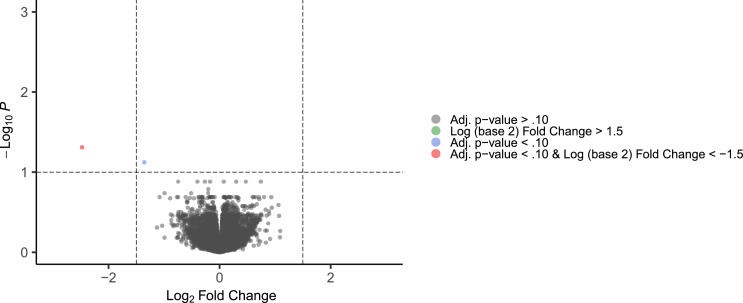
To ascertain differences in gene expression between opioid-positive samples compared to controls, we performed RNA-sequencing on postmortem brain specimens from those who died of acute opioid intoxication (n = 72) and group-matched controls (n = 81), as described in the Methods. Differential expression analysis revealed two genes that survived multiple testing correction (i.e., adj. p < .10; see Fig. 1), both of which were downregulated. The complete list of differentially expressed genes can be found in the supplementary material (Table S1). Notably, the top differentially expressed gene was Neuronal PAS Domain Protein 4 (NPAS4; log2 fold change [log2FC] = −2.47, adj. p = .049), which is an immediate early gene (IEG) that has been implicated in opioid, cocaine, and methamphetamine use (Taniguchi et al., 2017; Martin et al., 2012). The other significantly downregulated gene was RP11–667K14.3, a long intergenic non-protein coding gene. Other notable genes that approached our significance threshold were Regulator of G Protein Signaling 2 (RGS2; log2FC = −0.51, adj. p = .18), and another IEG, Early Growth Response 4 (EGR4, log2FC = −0.76, adj. p= .20).
Fig. 1.
Volcano plot of differentially expressed genes.
3.3. Weighted gene co-expression network analysis
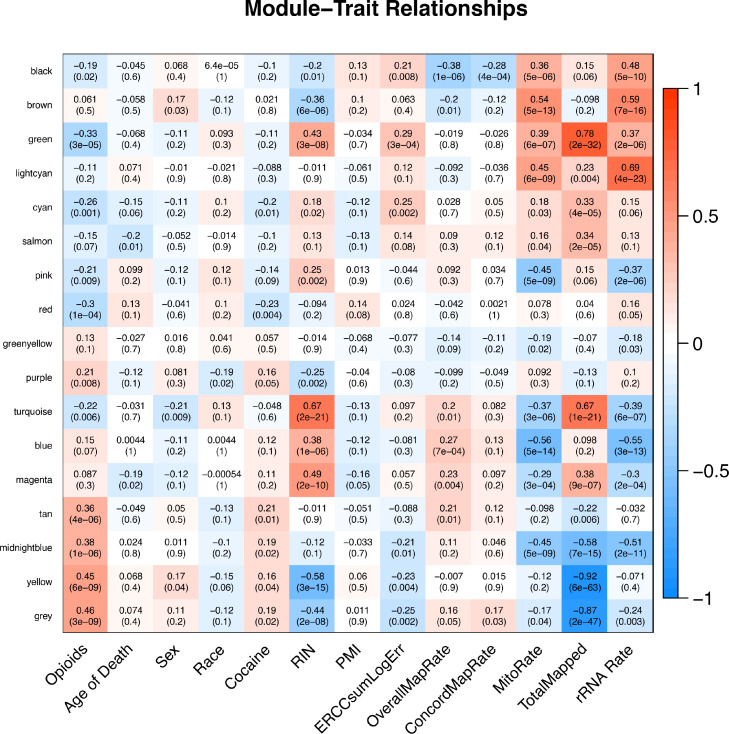
Next, we conducted WGCNA to cluster genes into modules based on correlations of expression patterns. The cluster and eigengene dendrograms from this analysis can be found in Figs. S2 and S3. Sixteen modules (labeled as colors; see Fig. 2) were identified, with the smallest module (light cyan) containing 41 genes and the largest module (turquoise) containing 3810 genes. It is important to note that the grey module in Fig. 2 containing 9017 genes is not a distinct module, but rather a list of genes that were not assigned to a specific module. So, although it is significantly correlated with opioid overdose status, we did not include it in downstream enrichment analyses given the limited conclusions that could be drawn from this subset of unassigned genes.
Fig. 2.
Heatmap of gene modules from WGCNA and study variables. Opioids = opioid overdose status (yes/no); Cocaine = cocaine or cocaine metabolite present during toxicology screen (yes/no); RIN = RNA integrity number; PMI = postmortem interval; ERCCsumLogErr = External RNA Controls Consortium spike-in control transcripts; overallMapRate = overall mapping rate; concordMapRate = concordant mapping rate; mitoRate = mitochondrial mapping rate; totalAssignedGene = exonic assignment rate; rRNA_rate = rRNA assignment rate.
As can be seen in Fig. 2, 10 modules were significantly correlated with opioid overdose status. Four modules were positively correlated with opioid status: purple (r = 0.008; 235 genes), tan (r = 0.36; 192 genes), midnight blue (r = 0.38; 94 genes), and yellow (r = 0.45; 2434 genes); and seven modules were negatively correlated with opioid overdose status: black (r = −0.19; 632 genes), green (r = −0.33; 1387 genes), cyan (r = −0.26; 106 genes), pink (r = −0.21; 282 genes), red (r = −0.30; 1326 genes), and turquoise (r = −0.22; 3810 genes). Surprisingly, NPAS4 was among the unassigned gene list within the grey module.
3.4. Hub gene identification and enrichment analysis
We then selected the two modules with the strongest positive (yellow) and negative (green) correlation with opioid overdose status and inspected for intramodular hub genes, which had a gene-module membership >0.80 and gene-trait significance > +/−0.30. The yellow module contained 88 hub genes and the green module contained 79 hub genes. The top 10 hub genes from each module are presented in Table 2, and the complete list of hub genes for the yellow and green modules can be found in Table S2 and Table S3, respectively. None of the hub genes we identified are clearly linked to opioid or substance use behaviors.
Table 2.
Top hub genes from WGCNA.
| Yellow Module | Green Module |
|---|---|
| DANT2 | CFL1 |
| XKR6 | CUEDC2 |
| LINC00632 | DRAP1 |
| HECW1-IT1 | CUTA |
| TNRC6A | TIMM50 |
| GJA6P | FXR2 |
| SETD5 | FKBP2 |
| RPS3AP29 | GAPDH |
| GPATCH8 | COX4I1 |
| PCDHB13 | MICOS13 |
| MBD5 | PSMB6 |
Note. Hub genes were identified as genes with a gene-module membership > 0.80 and gene-trait significance greater than an absolute value of 0.30.
Lastly, we conducted gene-set enrichment analyses for the hub genes in these two modules using GOrilla (Eden et al., 2009). There was no evidence of significant enrichment of differential expression among the yellow hub genes. We did detect significant enrichment among numerous process, molecular function, and cellular component pathways among the green module hub genes; however, none of these GO terms were relevant to opioid use (see File S1). Given the lack of information from the yellow hub genes, we conducted similar enrichment analyses of hub genes from the tan and midnight blue modules, which were positively associated with opioid overdose status. We did not detect enrichment in any pathways of differential expression for the midnight blue module; however, among genes in the tan module, there was significant enrichment of differential expression in genes that lie in pathways related to glutamatergic synapse function (GO:0,098,978). Otherwise, the results were similarly nondescript.
4. Discussion
The primary goal of this study was to examine transcriptome-wide differences in gene expression within the human DLPFC following acute opioid intoxication. Although there is evidence to support differential expression of numerous genes within the midbrain, NAc, and DLPFC in response to long-term use of opioids, to our knowledge, no human studies have exclusively focused on fatal overdose. We detected two FDR-significant differentially expressed genes between opioid-positive and control samples. The most notable of these genes was NPAS4, which has been identified in preclinical work related to cocaine and methamphetamine use (Taniguchi et al., 2017; Martin et al., 2012). Several other genes (RGS2, EGR4) previously implicated in opioid and cocaine use (Senese et al., 2020; Bisagno and Cadet, 2019) were also among the top-ranking genes in our analysis, although they did not meet our stringent significance threshold. Weighted correlation network analysis revealed several gene modules associated with opioid overdose status; however, enrichment analyses did not reveal meaningful enrichment in pathways related to opioid use. Nonetheless, the present findings advance our understanding of gene expression in relation to an opioid overdose, and point toward a gene of interest that may be relevant to both opioid abuse and acute intoxication.
The results support acute opioid intoxication influencing expression of at least one gene directly linked to substance use behaviors. The primary finding from this study was that NPAS4 was significantly under-expressed in opioid-positive samples compared to controls. NPAS4 is a protein coding gene that aids in the strengthening of neural connectivity and influences expression of brain-derived neurotrophic factor (BNDF) (Lin et al., 2008). As an IEG, NPAS4 is transcribed quickly in response to extracellular stimuli and serves as a transcription factor that regulates downstream gene expression (Sun and Lin, 2016). For example, NPAS4 has been shown to control GABAergic synapse development in an activity-dependent manner, and GABAergic deficits have been implicated in psychiatric disorders such as substance use disorders and major depressive disorder (Luscher et al., 2011). Recent work examining the NAc of Sprague Dawley rats showed that cocaine inhibits HDAC5 – a histone deacetylase enzyme – from entering the cell nucleus and limits production of Npas4; subsequent up-regulation of Npas4 is associated with increased drug-environment associations (Taniguchi et al., 2017). Interestingly, NPAS4 was downregulated in the present analysis. Preclinical evidence suggests that acute opioid use is associated with upregulation of NPAS4 (Bisagno and Cadet, 2019; Piechota et al., 2010). However, preclinical studies of methamphetamine use suggest that Npas4 is significantly downregulated hours after initial upregulation (Martin et al., 2012). The reasons for the discrepancy in findings related to NPAS4 make interpretation of this finding difficult. Based on next-of-kin interviews and psychiatric narratives, the opioid-positive samples likely were long-term users, though polysubstance use could affect patterns of gene expression.
It is also interesting to note several other genes implicated in substance use that approached significance, specifically, RGS2 and EGR4. RGS2 is particularly relevant given other known G-protein coupled receptors, namely µ-opioid receptors, that are affected by opioid use (Sutton et al., 2016) Sutton et al. (2016). demonstrated that, in response to morphine administration, deletion of Rgs7 increased reward and analgesia, while heightening withdrawal, in mice. EGR4 is interesting given the role of IEGs. As previously noted, IEGs are responsible for encoding transcription factors, which can interact with gene promoters to effect changes in gene expression at downstream genes (Bisagno and Cadet, 2019). Preclinical evidence suggests that Egr1, which is in the same gene family as EGR4, is differentially expressed in the forebrain of mice after morphine administration (ZióŁkowska et al., 2015). Further examination of these genes and their potential role in opioid abuse and overdose may elucidate alternative mechanisms through which opioid dependence, cravings, and other drug-related behaviors develop and persist.
Lastly, results from WGCNA and pathway analyses did not provide evidence of enrichment of differential expression along pathways relevant to opioid use. Moreover, there was no evidence of hub genes relevant to opioid use or overdose status. These results could be explained, in part, due to the limited samples size, or opioid overdose may indeed involve a unique biological response that does not overlap with long-term abuse. Given these null findings, in addition to the differential expression of NPAS4, replication is needed in long-term abuse and overdose samples to disentangle these findings.
4.1. Strengths and limitations
The primary strength of this study is the sample size. To date, the largest study to examine transcriptome-wide differences in gene expression within the DLPFC was carried out by Seney et al., who examined differential expression in 20 cases and 20 controls. In comparison, we had access to 81 control samples and 72 opioid-positive samples. As donation of postmortem brains and access to this tissue increases, it will be imperative to attempt to replicate extant studies to detect true signals. This study also benefitted from access to drug use history data. Posthumous psychiatric information is often difficult to obtain on postmortem samples. Although there is potential bias in next-of-kin interviews regarding drug use history, this information is valuable when attempting to interpret study findings. Lastly, our focus on opioid overdose sheds light on the biological response in the context of acute intoxication of a high level of opioids. Our finding regarding NPAS4 may or may not reflect both long-term and acute opioid use but provides an avenue for future work given extant evidence for NPAS4 in relation to use of several substances, including opioids.
This study also has several limitations to consider when interpreting the findings. First, although opioid users died of acute opioid intoxication, clearly defining a homogeneous sample of those with opioid use disorder, and no other substance use disorders, using postmortem next of kind interviews is difficult. This presents a challenge in teasing out effects due to chronic versus acute use, and polysubstance use. Examination of tissue within a single brain region also presents a limitation in interpretation of results. Although the PFC is implicated in addiction, the NAc is often the focus in studies of addiction (Volkow and Morales, 2015; Kalivas et al., 2005). Although Seney et al. demonstrated a high degree of overlap in differential gene expression between the DLPFC and NAc in opioid users, unique differences exist, and future studies should aim to examine multiple brain regions simultaneously. Lastly, future studies will benefit from more racially and ethnically diverse samples, as well as samples including individuals across various developmental stages. The latter may elucidate differential effects of opioid abuse depending on brain maturation and aging.
4.2. Conclusions
This study represents the first transcriptome-wide analysis of gene expression in the human DLPFC in response to acute opioid intoxication resulting in fatal overdose. Evidence suggests that opioid overdose is associated with differential expression of NPAS4, which is implicated in drug cravings and drug-environment cues in the contexts of cocaine and methamphetamine use. Additional genes implicated in substance abuse (RGS2, EGR4) provide promising avenues for continued research. Future studies examining chronic versus acute opioid use and polysubstance use, and those with larger, more racially and developmentally diverse samples will clarify these associations and aid in determining the value of differentially expressed genes as therapeutic targets for substance use disorders.
Disclosures
The authors declare no competing interests.
Data availability
Raw data that support the findings of this study are publicly available through the Sequence Read Archive (SUB9455518).
Supporting information
Fig. S1. Proportion of Estimated Cell Types by Brain Sample. Astro = astrocyte; Excit = excitatory neurons; Inhib = inhibitory neurons; Micro = microglia; Oligo = oligodendrocytes; OPC = oligodendrocyte progenitor cells
Fig. S2. Cluster Dendrogram from WGCNA.
Fig. S3. Eigengene Dendrogram from WGCNA.
Fig._S2.pdf
Fig._S3.pdf
File_S1.xlsx
Table_S1.txt
Table_S2.txt
Table_S3.txt
CRediT authorship contribution statement
David W. Sosnowski: Visualization, Formal analysis, Writing – original draft, Writing – review & editing. Andrew E. Jaffe: Visualization, Formal analysis, Writing – review & editing. Ran Tao: Data curation, Writing – review & editing, Investigation. Amy Deep-Soboslay: Data curation, Writing – review & editing, Investigation. Chang Shu: Writing – review & editing, Formal analysis. Sarven Sabunciyan: Writing – review & editing. Joel E. Kleinman: Data curation, Writing – review & editing, Investigation. Thomas M. Hyde: Data curation, Writing – review & editing, Investigation. Brion S. Maher: Conceptualization, Funding acquisition, Visualization, Formal analysis, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by the National Institute on Drug Abuse (R01 DA039408; PI: Brion S. Maher). David W. Sosnowski was supported by the National Institute on Drug Abuse’s Epidemiology Training Program (T32 DA007292–27; PI: Brion S. Maher). The authors would like to express their gratitude to our colleagues whose efforts have led to the donation of postmortem tissue to advance these studies, including at the Office of the Chief Medical Examiner of the State of Maryland and the Office of the Chief Medical Examiner of Kalamazoo County, Michigan. We also would like to acknowledge the contributions of Dr. Llewelyn Bigelow for his diagnostic expertise. Finally, we are indebted to the generosity of the families of the decedents, who donated the brain tissue used in these studies.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dadr.2022.100040.
Appendix. Supplementary materials
References
- O'donnell J., Gladden Matt R., Mattson C.L., Hunter C.T., Davis N.L. Morbidity and mortality weekly report vital signs: characteristics of drug overdose deaths involving opioids and stimulants-24 states and the district of Columbia. 2019 https://www.cdc.gov/drugoverdose/od2a/index.html. (accessed 17 Sep 2020). [DOI] [PMC free article] [PubMed]
- Florence C.S., Zhou C., Luo F., Xu L. The economic burden of prescription opioid overdose, abuse, and dependence in the United States, 2013. Med. Care. 2016;54:901–906. doi: 10.1097/MLR.0000000000000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egervari G., Kozlenkov A., Dracheva S., Hurd Y.L. Molecular windows into the human brain for psychiatric disorders. Mol. Psychiatry. 2019;24:653–673. doi: 10.1038/s41380-018-0125-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow N.D., Morales M. The brain on drugs: from reward to addiction. Cell. 2015;162:712–725. doi: 10.1016/j.cell.2015.07.046. [DOI] [PubMed] [Google Scholar]
- Kalivas P.W., Volkow N., Seamans J. Unmanageable motivation in addiction: a pathology in prefrontal-accumbens glutamate transmission. Neuron. 2005;45:647–650. doi: 10.1016/j.neuron.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Kruyer A., Chioma V.C., Kalivas P.W. The opioid-addicted tetrapartite synapse. Biol. Psychiatry. 2020;87:34–43. doi: 10.1016/j.biopsych.2019.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman S.E., Malenka R.C., Nestler E.J. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu. Rev. Neurosci. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- Galaj E., Xi Z.X. Progress in opioid reward research: from a canonical two-neuron hypothesis to two neural circuits. Pharmacol. Biochem. Behav. 2021;200 doi: 10.1016/j.pbb.2020.173072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R.J., Rahman T., Sherman J., Hurd Y.L. SnapShot: neurobiology of opioid use disorder. Cell. 2021;184:1648–1648.e1. doi: 10.1016/j.cell.2021.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Vadhan N.P., Luba R.R., Comer S.D. The effects of heroin administration and drug cues on impulsivity. J. Clin. Exp. Neuropsychol. 2016;38:709–720. doi: 10.1080/13803395.2016.1156652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias F., Arnsten J.H., Cunningham C.O., Coulehan K., Batchelder A., Brisbane M., et al. Neurocognitive, psychiatric, and substance use characteristics in opioid dependent adults. Addict. Behav. 2016;60:137–143. doi: 10.1016/j.addbeh.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernacki K., McLennan S.N., Terrett G., Labuschagne I., Rendell P.G. Decision-making ability in current and past users of opiates: a meta-analysis. Neurosci. Biobehav. Rev. 2016;71:342–351. doi: 10.1016/j.neubiorev.2016.09.011. [DOI] [PubMed] [Google Scholar]
- Albertson D.N., Schmidt C.J., Kapatos G., Bannon M.J. Distinctive profiles of gene expression in the human nucleus accumbens associated with cocaine and heroin abuse. Neuropsychopharmacology. 2006;31:2304–2312. doi: 10.1038/sj.npp.1301089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillivan S.E., Whittard J.D., Jacobs M.M., Ren Y., Mazloom A.R., Caputi F.F., et al. ELK1 transcription factor linked to dysregulated striatal mu opioid receptor signaling network and oprm1 polymorphism in human heroin abusers. Biol. Psychiatry. 2013;74:511–519. doi: 10.1016/j.biopsych.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad M.H., Rumschlag M., Guerra M.H., Savonen C.L., Jaster A.M., Olson P.D., et al. Differentially expressed gene networks, biomarkers, long noncoding RNAs, and shared responses with cocaine identified in the midbrains of human opioid abusers. Sci. Rep. 2019;9:1–9. doi: 10.1038/s41598-018-38209-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seney M.L., Kim S.M., Glausier J.R., Hildebrand M.A., Xue X., Zong W., et al. Transcriptional alterations in dorsolateral prefrontal cortex and nucleus accumbens implicate neuroinflammation and synaptic remodeling in opioid use disorder. Biol. Psychiatry. 2021;90:550–562. doi: 10.1016/j.biopsych.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu C., Sosnowski D.W., Tao R., Deep-Soboslay A., Kleinman J.E., Hyde T.M., et al. Epigenome-wide study of brain DNA methylation following acute opioid intoxication. Drug Alcohol Depend. 2021;221 doi: 10.1016/j.drugalcdep.2021.108658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado-Torres L., Burke E.E., Peterson A., Shin J.H., Straub R.E., Rajpurohit A., et al. Regional heterogeneity in gene expression, regulation, and coherence in the frontal cortex and hippocampus across development and schizophrenia. Neuron. 2019;103:203–216.e8. doi: 10.1016/j.neuron.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Jew B., Alvarez M., Rahmani E., Miao Z., Ko A., Garske K.M., et al. Accurate estimation of cell composition in bulk expression through robust integration of single-cell information. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-15816-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran M.N., Maynard K.R., Spangler A., Huuki L.A., Montgomery K.D., Sadashivaiah V., et al. Single-nucleus transcriptome analysis reveals cell-type-specific molecular signatures across reward circuitry in the human brain. Neuron. 2021;109:3088–3103.e5. doi: 10.1016/j.neuron.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl L., Seth P., Kariisa M., Wilson N., Baldwin G. Morbidity and mortality weekly report drug and opioid-involved overdose deaths-United States. 2019 https://www.cdc.gov/nchs/data/nvsr/nvsr61/(accessed 14 Sep 2020). [DOI] [PMC free article] [PubMed]
- R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2021. https://www.R-project.org/ [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A.E., Tao R., Norris A.L., Kealhofer M., Nellore A., Shin J.H., et al. QSVA framework for RNA quality correction in differential expression analysis. Proc. Natl. Acad. Sci. USA. 2017;114:7130–7135. doi: 10.1073/pnas.1617384114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., Storey J.D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M., Carreira M.B., Cooper Y.A., Bobadilla A.C., Heinsbroek J.A., Koike N., et al. HDAC5 and its target gene, NPAS4, function in the nucleus accumbens to regulate cocaine-conditioned behaviors. Neuron. 2017;96:130–144.e6. doi: 10.1016/j.neuron.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T.A., Jayanthi S., McCoy M.T., Brannock C., Ladenheim B., Garrett T., et al. Methamphetamine causes differential alterations in gene expression and patterns of histone acetylation/hypoacetylation in the rat nucleus accumbens. PLoS ONE. 2012;7:e34236. doi: 10.1371/journal.pone.0034236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senese N.B., Kandasamy R., Kochan K.E., Traynor J.R. Regulator of G-Protein Signaling (RGS) protein modulation of opioid receptor signaling as a potential target for pain management. Front. Mol. Neurosci. 2020;13:5. doi: 10.3389/fnmol.2020.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisagno V., Cadet J.L. Expression of immediate early genes in brain reward circuitries: differential regulation by psychostimulant and opioid drugs. Neurochem. Int. 2019;124:10–18. doi: 10.1016/j.neuint.2018.12.004. [DOI] [PubMed] [Google Scholar]
- Lin Y., Bloodgood B.L., Hauser J.L., Lapan A.D., Koon A.C., Kim T.K., et al. Activity-dependent regulation of inhibitory synapse development by NPAS4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Lin Y. NPAS4: linking neuronal activity to memory. Trends Neurosci. 2016;39:264–275. doi: 10.1016/j.tins.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B., Shen Q., Sahir N. The GABAergic deficit hypothesis of major depressive disorder. Mol. Psychiatry. 2011;16:383–406. doi: 10.1038/mp.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechota M., Korostynski M., Solecki W., Gieryk A., Slezak M., Bilecki W., et al. The dissection of transcriptional modules regulated by various drugs of abuse in the mouse striatum. Genome Biol. 2010;11:48. doi: 10.1186/gb-2010-11-5-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton L.P., Ostrovskaya O., Dao M., Xie K., Orlandi C., Smith R., et al. Regulator of G-protein signaling 7 regulates reward behavior by controlling opioid signaling in the striatum. Biol. Psychiatry. 2016;80:235–245. doi: 10.1016/j.biopsych.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZióŁkowska B., Gieryk A., Solecki W., PrzewŁocki R. Temporal and anatomic patterns of immediate-early gene expression in the forebrain of C57BL/6 and DBA/2 mice after morphine administration. Neuroscience. 2015;284:107–124. doi: 10.1016/j.neuroscience.2014.09.069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data that support the findings of this study are publicly available through the Sequence Read Archive (SUB9455518).