Abstract
Protein D has previously been demonstrated to be associated with Escherichia coli ribosomes by the radical-free and highly reducing method of two-dimensional polyacrylamide gel electrophoresis. In this study, we show that protein D is exclusively present in the 30S ribosomal subunit and that its gene is located at 33.6 min on the E. coli genetic map, between ompC and sfcA. The gene consists of 45 codons, coding for a protein of 5,096 Da. The copy number of protein D per ribosomal particle varied during growth and increased from 0.1 in the exponential phase to 0.4 in the stationary phase. For these reasons, protein D was named SRA (stationary-phase-induced ribosome-associated) protein and its gene was named sra. The amount of SRA protein within the cell was found to be controlled mainly at the transcriptional level: its transcription increased rapidly upon entry into the stationary phase and was partly dependent on an alternative sigma factor (sigma S). In addition, global regulators, such as factor inversion stimulation (FIS), integration host factor (IHF), cyclic AMP, and ppGpp, were found to play a role either directly or indirectly in the transcription of sra in the stationary phase.
The ribosomal proteins of Escherichia coli were systematically characterized in the beginning of the 1970s, and a unified nomenclature, S1 through S21 for the 30S subunit and L1 through L34 for the 50S subunit, was proposed on the basis of their behavior in two-dimensional gel electrophoresis (17, 44, 45, 46). Using a modified and improved method, termed radical-free and highly reducing method of two-dimensional polyacrylamide gel electrophoresis (RFHR 2-D PAGE), we discovered four additional proteins, proteins A, B, C, and D, that were associated with E. coli ribosomes (38, 39, 40).
The gene for protein A (40) was located between infC (initiation factor 3) and rplT (ribosomal protein L20) (26, 27, 31) at 38 min, suggesting that the three constitute a new ribosomal protein operon in E. coli. The gene for protein B (40) was found to be identical to the protein X gene of the spc operon (8). Proteins A and B were consequently named L35 and L36 and their genes were named rpmI and rpmJ, respectively, in accordance with the standard nomenclature for E. coli ribosomal proteins and their genes. Protein C (41) is likely to be the full-length product of the rpmE gene, coding for L31, because the N-terminal amino acid sequence completely matches that of L31 and the molecular mass of protein C is larger than that of L31 by about 1,000 Da. More recently, L31 was proved to arise through cleavage of protein C by protease VII, an outer membrane enzyme (A. Wada, unpublished data).
Protein D is a small basic protein that can be isolated from the 30S subunit after dissociation of the 70S ribosome. It migrates much faster than S21 and to a point below the L32 spot upon RFHR 2-D PAGE of purified 70S ribosomal proteins. Thus, it is the smallest protein component of the 30S subunit of E. coli. When the 70S ribosome was dissociated into the core particle and split proteins by CsCl density gradient centrifugation (34), protein D was found in the soluble split-protein fraction (39). Furthermore, in simultaneous reconstitution experiments with 50S and 30S subunits, protein D was exclusively associated with the 30S subunit (39).
In this study, we have identified the gene encoding protein D by determining its N-terminal amino acid sequence and have analyzed the mechanism of control of its expression in terms of growth phase and the transcriptional factors involved. Protein D was named SRA (stationary-phase-induced ribosome-associated) protein.
MATERIALS AND METHODS
Bacterial strains, plasmids, phage, and culture conditions.
E. coli K-12 strains, plasmids, and phage (Table 1) were grown in medium E (37) containing 2% Polypeptone and supplemented with 0.5% glucose (EP medium) at 37°C with shaking at 100 cycles/min. Cells were harvested in the middle exponential and/or stationary phase of cell growth and stored at −80°C until use.
TABLE 1.
Bacterial strains, phage, and plasmids
| Strain, phage, or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| Strains | ||
| W3110 | F−; prototroph | |
| W3350 | F−gal-1 gal-2 | |
| MC4100 | F− Δ(argF-lac)U169 araD139 rpsL150 relA1 thiA deoC1 ptsF25 flbB5301 | 7 |
| Q13 | Hfr met ma pnp tyr | 33 |
| KY1461 | F− Δ(ara-leu)697 Δ(lac-pro) thi trpA38(Oc) | 25 |
| MG1655 | F−; prototroph | 12 |
| UM122 | Hfr thi katF::tet | 22 |
| ME8465 (JB100) | Hfr mal-1(Tc) ompR::tet | 5 |
| CU284 | MC4100 Δhns::kan | 48 |
| YK4207 | YK1100 Δ3[hip]::cat Δ82[himA]::tet trp+ | 11 |
| YK1100 | W3110 trpC9941 | C. Yanofsky |
| AQ7996 | fis::kan | 4 |
| KI251 | F−cya-283 ilv::tet trpR trpA9605 his-29 | 1 |
| CF1678 | MG1655 ΔrelA251::kam ΔspoT209::cat | 47 |
| KY1471 | KY1461 katF::tet | This work |
| KY1472 | KY1461 ompR::tet | This work |
| KY1473 | KY1461 Δhns::kan | This work |
| KY1474 | KY1461 Δ3[hip]::cat Δ82[himA]::tet | This work |
| KY1475 | KY1461 fis::kan | This work |
| KY1476 | KY1461 cya-283 | This work |
| Phage | ||
| λ pF13 | imm21 | 15 |
| λ pF13(Psra-lacZ) | imm21 Psra-lacZ | This work |
| Plasmids | ||
| pKV7350 | pUC119-277L | This work |
| pKV7351 | pUC119-277L sra::kan | This work |
| pUC-4K | ||
| pBAD-αβγ | 51 | |
| pMS4342 | 15 | |
| pFF6 | 18 | |
| pMS4342(Psra-lacZ) | This work | |
| pFF6(Psra-lacZ) | This work |
Preparation of ribosomes, ribosomal subunits, and ribosomal proteins.
Crude and high-salt-washed ribosomes were prepared according to the methods of Noll et al. (29) and Horie et al. (16). Subunits were isolated from the high-salt-washed ribosome preparation by sucrose density gradient centrifugation as described previously (38). Proteins were prepared by the acetic acid method of Hardy et al. (13), dialyzed against 2% acetic acid, lyophilized, and stored at −80°C until use.
RFHR 2-D PAGE and determination of copy number of SRA (protein D).
Lyophilized ribosomal proteins (approximately 0.3 mg/gel) were analyzed by RFHR 2-D PAGE as described previously (38) with the following modifications: the volume of glacial acetic acid used in the sample charging buffer (50×) was 7.4 ml, and the gel thickness used was 2 mm. After the gels were stained with amido black 10B in 1% acetic acid and destained with 1% acetic acid, the protein spots on the gels were scanned with a PD 110 personal densitometer (Molecular Dynamics Co.). The copy number of SRA (protein D) was determined by calculating the optical density values obtained per unit of molecular weight and normalizing them with the average values for S18, S19, L32, and L33, each of which has one copy per ribosome (38). The standard deviations for these calculations were approximately 25%.
Amino acid sequence analysis of SRA.
After electrophoresis, the SRA protein was blotted from the gel to a polyvinylidene difluoride membrane. Sequence analysis was performed using a model 477A protein sequencer (Perkin-Elmer Applied Biosystems) as described previously (42).
Gene locus and nucleotide sequencing.
The amino acid sequence data obtained were used to design a degenerate probe: AAT/CCGT/CCAA/GGCT/C/ G/ACGT/CCAT/CATT/CCTGGG. The probe was used to screen Kohara λ clone library clones (19). Positive clones obtained were 1G9 (277), 9B8 (278), and 10C7 (279), which are located at 33 min. A chromosomal insert in clone 277 was subcloned into either pUC118 or pUC119, and the fragment containing the sra gene was sequenced by the dideoxy method (24).
Construction of λpF13(Psra-lacZ).
Five regions upstream of the ATG initiation codon of the sra gene as well as 39 bp downstream of it were amplified by PCR. The primers used contained a KpnI site to facilitate cloning into the pMS4342 plasmid vector for operon fusion. The inserts thus cloned were recombined in vivo into phage vector λpF13 (15, 50), and the resulting phage, λpF13(Psra-lacZ), was lysogenized into strain KY1461. The forward primers used and the distances (base pairs) from the ATG initiation codon (in parentheses) are as follows: 5′-CGGGTACCGGATATTCCGAATAAAATCCGG-3′ (595), 5′-GCGGTACCTCATGGTGTTAAAATATAAA-3′ (455), 5′-GCGGTACCGAATCACTAGTTTACTTAT-3′ (325), 5′-GCGGTACCCTGACCAAATGGGTTGAAGC-3′ (200), and 5′-GCGGTACCCACGTGTTTCTTCGCTACGG-3′ (62). The reverse primer, 5′-CGGTCGACCAGTCCAAGAATATGACGTGCC-3′, contained a SalI site. The DNA of plasmid pKV7350 was used as a template in all PCRs. The correctness of the sequences of the PCR products was confirmed by nucleotide sequencing. Unless otherwise stated, λpF13(Psra-lacZ) with the region 595 bp upstream of the ATG of sra was used throughout this work.
Measurement of the level of transcription of the sra gene.
The level of transcription was estimated by measuring the β-galactosidase activity of the lacZ reporter in each lysogen constructed as described above. For the integration host factor (IHF)- or cyclic AMP (cAMP)-defective mutant into which λpF13(Psra-lacZ) could not be lysogenized, plasmid pFF6(Psra-lacZ) (18), a mini-F plasmid derivative carrying an ara-trp-lac fusion identical to that carried by λpF13, was used instead.
Determination of β-galactosidase activity.
Aliquots (0.1, 0.2, or 0.5 ml) of a culture were mixed with Z buffer (0.9, 0.8, or 0.5 ml, respectively) in an ice bath. Cells were disrupted by the addition of chroroform and sodium dodecyl sulfate and were assayed for β-galactosidase activity essentially as described by Miller (28). The average of four to eight assays is reported.
Construction of deletions of the sra gene.
The regions flanking the sra gene were amplified by PCR using pKV7350 (pUC119-277L) as the template and 5′-TTTAACTCCTCAATCCTGTAGCTAG-3′ and 5′-TTCATAACCATCAGTCCTCAATGAC-3′ as the primers. The resultant PCR product was restricted with HincII and ligated into a kanamycin-resistant cassette with HincII sites at both ends (derived from plasmid pUC-4K). Whether plasmid pKV7351 (pUC119-277L-sra::kan) was correctly constructed or not was examined by nucleotide sequencing. The plasmid was then linearized with SphI and EcoRI and transformed into MG1655 cells carrying plasmid pBAD-αβγ, which increases recombination frequency (51). Kanamycin-resistant transformants were selected, and deletion of the sra gene in them was confirmed by PCR. Subsequently, P1 phage prepared on one of the transformants was used to transduce MG1655 cells, and kanamycin-resistant transductants were selected. The sra gene deletion was similarly confirmed by PCR, and the strain was designated MG1655Δsra::kan.
Primer extension.
MG1655, MG1655Δsra::kan, and MG1655 carrying pKY7350 (pUC119-277L) were grown at 37°C in 10 ml of Luria-Bertani medium to the stationary phase of growth (2 h after cessation of the turbidity increase). Cells in 1 ml of culture were harvested by centrifugation at 5,000 × g for 15 min, and RNA was extracted using an RNA-DNA extraction kit (Qiagen Inc.). All solutions were treated with diethylpyrocarbonate (Sigma) prior to use. The concentration of RNA was estimated from the absorbance at 260 nm, and the quality and degree of degradation of RNA were assessed by electrophoresis on a 1.5% agarose gel containing 1.8% formaldehyde. The primer used for extension was complementary to the coding region of sra and was 5′-TTATGGTCCAGTCCAAGAATATGACGTGCCTGACGGTTCG-3′. For the primer extension assays, 10 pmol of a 32P-end-labeled oligonucleotide primer and 40 μg of RNA template were mixed, heated at 65°C for 90 min, and annealed by allowing the mixture to cool slowly to room temperature. The extension reaction was performed according to the method described by Triezenberg (35). The reaction mixtures were boiled for 2 min and loaded onto a 5% denaturing polyacrylamide sequencing gel. The sequencing reaction was performed using a BcaBEST dideoxy sequencing kit (Takara) and 3.2 pmol of the same primer as that used for the primer extension reaction.
RESULTS
SRA is present exclusively in ribosomal particles.
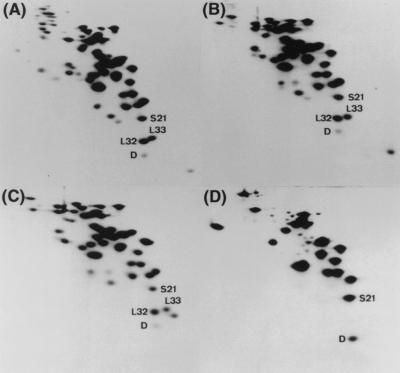
Strain W3110 was cultured and harvested at the stationary phase (after 24 h of growth), and cell extracts, crude ribosomes, high-salt-washed ribosomes, and the dissociated 30S subunit were prepared and analyzed by RFHR 2-D PAGE (Fig. 1). Subsequently, the quantity of SRA protein on each electropherogram was densitometrically determined as described in Materials and Methods. The copy number of the SRA protein was about 0.4 per ribosome in the cell extracts, and the level was maintained in the subsequent processes of ribosome preparation, including dissociation into subunits. These results indicate that SRA is strongly associated with 30S subunit particles not only in the exponential phase, as described previously (38), but also in the stationary phase. Free SRA protein was not detected in the soluble fraction after sedimentation of crude ribosomes by centrifugation at 165,000 × g for 90 min (data not shown). Therefore, we believe that SRA is an integral part of the ribosome, especially of the 30S subunit.
FIG. 1.
Association of SRA (protein D) with the 30S ribosomal subunit. Electropherograms from RFHR 2-D PAGE analysis of the ribosomal proteins in total cell extracts (A), crude ribosomes (B), high-salt-washed ribosomes (C), and 30S subunits (D) prepared from W3110 cells grown to the stationary phase. Protein D (SRA), S21, L32, and L33 spots are indicated.
The copy number of SRA is dependent upon the cellular growth phase.
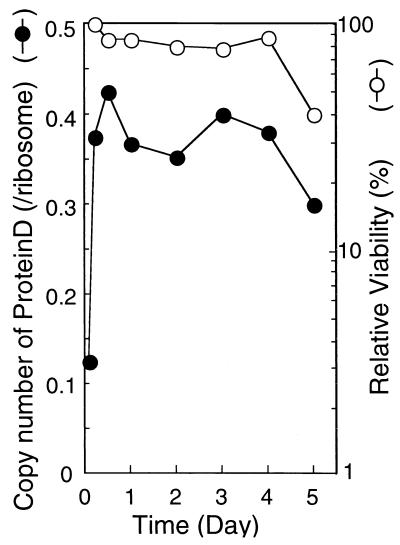
Cells of strain W3110 were harvested in the middle exponential to late stationary phase of growth, and the proteins prepared from crude ribosomes were analyzed by RFHR 2-D PAGE. The results shown in Fig. 2 demonstrate that the copy number of SRA increased from 0.1 in the middle exponential phase to 0.4 in the stationary phase. Cell viability remained constant at 70% or more during the 4 days of culturing (Fig. 2).
FIG. 2.
Cellular viability and copy number of SRA in the stationary phase. Cultures of W3110 cells were sampled at different times from the exponential phase through the stationary phase of growth. Optical density at 600 nm and colony-forming ability were determined, and the relative viability values obtained were normalized to that in the late exponential phase. Proteins extracted from harvested cells were analyzed by RFHR 2-D PAGE. The copy number of SRA was determined as described in Materials and Methods.
To test whether this stationary-phase induction of SRA occurs in various E. coli strains, copy numbers of SRA were measured and compared for five K-12 strains, W3110, W3350, MG1655, KY1461, and Q13, the last strain being an RNase I- and polynucleotide phosphorylase-defective mutant. In all five strains, the amounts of SRA in different growth phases were more or less the same (data not shown). This was also true for E. coli strain B/r and several natural isolates contained in the ECOR collections (data not shown). These results suggest that the stoichiometry of SRA changes depending upon the growth phase regardless of the E. coli strain analyzed.
Amino acid sequence analysis of SRA.
An RFHR 2-D PAGE gel of 70S ribosomal proteins was blotted onto a polyvinylidene difluoride membrane, and the SRA spot was subjected to protein sequence analysis. The sequence of the first 37 amino acid residues from the N terminus was MKSNRQARHILGLDHKISNQRKIVTEGDKSSVVNNPT. A protein with this sequence has not yet been identified among the ribosomal proteins or ribosome-associated proteins from E. coli or any other organism. We identified this protein in 1992 (unpublished data) when E. coli genome sequencing had not yet been completed.
Gene locus and DNA sequence of the sra gene.
A mixed DNA probe (26-mer) (described in Materials and Methods) was synthesized based on the amino acid sequence of the SRA protein from the fourth asparagine so as to perform screening of Kohara clones (19). Clones 1G9 (#277), 9B8 (#278), and 10C7 (#279) were found to hybridize with the probe. They are all located at 33.6 min on the E. coli genetic map. A total of 2,257 bp containing the gene (138 bp) for SRA (i.e., sra) was sequenced using the dideoxy method (reference 24 and data not shown).
The sra gene has 45 codons and is terminated with TAA. The nucleotide sequence was in complete agreement with the amino acid sequence of SRA described above. The molecular mass of the SRA protein was calculated to be 5,096 Da, a value distinctly smaller than the value of 5,900 Da estimated from its migration in RFHR 2-D PAGE (39). The pl was calculated to be 11.6. The absence of cysteine residues is consistent with results obtained by the iodoacetic acid method (39). The gene has a typical Shine-Dalgarno sequence and a rho-independent terminator forming a distinct hairpin loop. However, there is no clear consensus sequence for the −10 and −35 promoter regions (described in more detail below). A DNA fragment containing the gene was previously sequenced by Mahajan et al. (23) as part of the region upstream of the gene for a translational fusion protein (sfcA-recE). Both sequences were identical across the coding region boundary. The sequence was also consistent with the genomic sequencing data reported by both a Japanese group (http://ecoli.aist-nara.ac.jp/) and a Wisconsin group (6).
The nucleotide sequence data were registered at DDBJ/EMBL/GenBank in 1992 under accession number D13179. At that time, we assigned protein D and its gene as ribosome protein S22 and gene rpsV, respectively. However, as described above, we have changed the name of the protein to SRA and the name of its gene to sra, mainly because of the very low copy number of the protein in the 30S ribosomal subunit, especially in exponentially growing cells.
Construction and characterization of an sra deletion mutant.
An sra disruptant (MG1655Δsra::kan) was constructed as described in Materials and Methods, and its disruption was confirmed by PCR and Northern and Western blot analyses (data not shown) (see below for Northern analysis). To analyze the phenotype of the mutant, it was grown at 37°C for 6 to 7 days in either Luria broth or EP medium along with the parental wild-type strain (MG1655). Samples of each culture were withdrawn at various times from the exponential phase through the stationary phase of growth, and colony-forming ability was tested. Very few differences were observed between the strains (data not shown).
For E. coli, it is known that the translational activity of the ribosomes of cells in the stationary phase of growth is low and that 70S ribosomes associate through a ribosomal modification factor (RMF) induced in the stationary phase to form dimers (100S) that are inactive in translation (41, 43). Because the amount of the SRA protein increased during the stationary phase, we examined whether the sra deletion would affect the formation of 100S ribosomes in the stationary phase (24 h). However, no significant differences in the sucrose density gradient centrifugation patterns of 30S, 50S, 70S, or 100S ribosomes were observed between the wild type and the sra deletion mutant (data not shown).
The level of transcription of the sra gene is increased in the stationary growth phase.
As described above, the copy number of the SRA protein per ribosome increased during the stationary phase. To elucidate the control mechanism for sra gene expression, we analyzed the level of expression of sra during the transition from the exponential phase to the stationary phase of growth by measuring β-galactosidase activity with lacZ reporters.
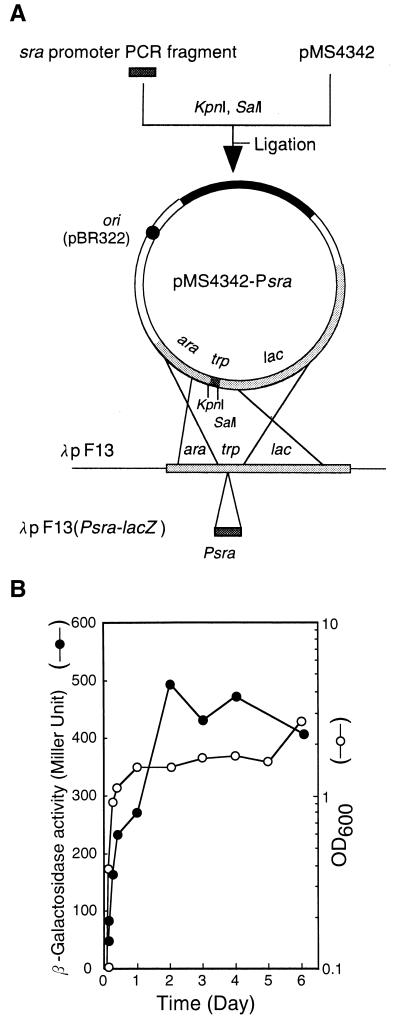
For this purpose, the λpF13(Psra-lacZ) (Fig. 3A) lysogen of strain MG1655 was grown in EP medium, and its β-galactosidase activity was monitored at different times during growth. The activity was low during the exponential phase but increased substantially when cells entered the stationary phase (Fig. 3B). The same results were obtained with the λpF13(Psra-lacZ) lysogen of strain KY1461. These results suggest that the increase in the amount of SRA resulted from an increased level of trancription of the sra gene.
FIG. 3.
Construction of phage λpF13(Psra-lacZ) and growth-phase-dependent induction from the sra promoter. (A) Construction of phage λpF13(Psra-lacZ). The sra promoter region was PCR amplified, digested, and cloned into plasmid pMS4342 digested with KpnI and SalI. The resultant sra-lacZ fusion was transferred to phage vector λpF13 as described in Materials and Methods (see the text for details). (B) Growth-phase-dependent induction from the sra promoter. MG1655 cells lysogenic for λpF13(Psra-lacZ) were grown at 37°C in EP medium for 6 days. Samples from cultures prepared in separate tubes were withdrawn at various times, and the optical density at 600 nm (OD600) and β-galactosidase activity in Miller units were determined.
Effects of sigma S and global regulators on the transcription of sra.
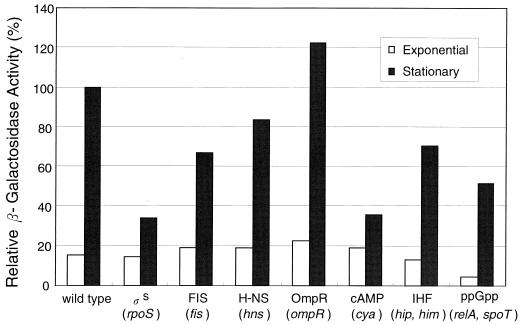
It is known that RNA polymerase with sigma S transcribes a large number of genes in the stationary phase. We examined whether the expression of the sra gene is dependent on sigma S. An rpoS mutant strain, KY1461 katF::tet (KY1471), as well as parental wild-type strain KY1461, carrying one copy of λpF13(Psra-lacZ), was grown at 37°C in EP medium. The level of expression of sra in both exponential- and stationary-phase cultures was estimated by measuring β-galactosidase activity. The level of expression of sra in the exponential phase was low in both the rpoS mutant strain and the wild-type strain. In contrast, when cultures entered the stationary phase, sra expression increased in the wild-type strain but not in the rpoS mutant strain, which showed only about 30% the sra expression of the wild-type strain (Fig. 4).
FIG. 4.
Effects of global regulators on the transcription of sra. Cells of KY1461 lysogenic for λpF13(Psra-lacZ) and containing either rpoS, fis, hns, ompR, or relA and spoT mutations and cells of KY1461 wild type or containing cya or hip and him mutations and carrying pFF6(Psra-lacZ) were grown at 37°C in EP medium. Cultures in either the exponential phase (optical density at 600 nm, 0.4) or the stationary phase (after 24 h) of growth were assayed for β-galactosidase activity, expressed relative to that of wild-type cells in the stationary phase.
The P1 promoter of the bolA gene is a typical sigma S-dependent promoter (20). We compared the β-galactosidase activities of a PbolA-lacZ operon fusion in the presence and absence of active sigma S. The β-galactosidase activity in the stationary phase of a λpF13(Psra-lacZ) lysogen of KY1461 was twice that of a λpF13(PbolA-lacZ) lysogen carrying an rpoS mutation (data not shown). As a further control, we assayed the level of expression of the rmf gene (49), which is sigma S independent. As anticipated, the β-galactosidase activities of a λpF13(Prmf-lacZ) lysogen were the same in the stationary phase regardless of the rpoS mutation (unpublished data). We interpret these results to indicate that the transcription of sra in the stationary phase is partly dependent on sigma S.
Many E. coli genes whose expression is enhanced in the stationary phase, regardless of whether the expression is dependent on rpoS, are also regulated either positively or negatively by other global regulators (for example, see reference 14). We examined the effects of some global regulators (factor inversion stimulation [FIS], H-NS, OmpR, cAMP, ppGpp, and IHF) on the expression of sra by using global regulator-defective mutants lysogenized with λpF13(Psra-lacZ) or carrying pFF6(Psra-lacZ) and by measuring β-galactosidase activity. In the exponential phase, only a few differences were observed between the wild-type strains and the mutants defective in FIS, H-NS, IHF, OmpR, cAMP, and ppGpp. However, the expression of sra during the stationary phase in the mutants defective in FIS, IHF, cAMP, and ppGpp was significantly lower than that in the corresponding wild-type strains, as shown in Fig. 4. The only exception was an ompR-defective mutant in which sra expression was slightly higher (Fig. 4). These results suggest that FIS, IHF, cAMP, and ppGpp are involved in the positive regulation of sra expression during the stationary growth phase.
Determination of the transcriptional start site of sra by primer extension.
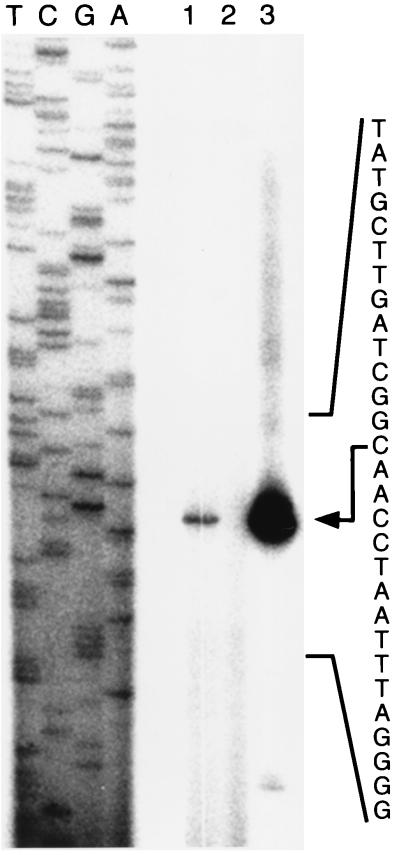
Based on the DNA sequence data alone, it was difficult to predict the promoter sequence of the sra gene. Therefore, its transcriptional start site was experimentally determined by primer extension using mRNA prepared from stationary-phase cells. The start site of the major transcript of sra was determined to be located at the 83rd C counted from the translational initiation codon (ATG) of sra (Fig. 5). The sra transcript was not observed in the stationary-phase culture of the sra deletion mutant strain. The −10 region for the sra promoter is likely to be 5′-TATGCT-3′, and the −35 region is likely to be 5′-AGCACC-3′. Since no other transcripts were observed for the region about 600 bp upstream of the translational initiation codon, the promoter identified here is the major one for the sra gene. By Northern analysis, only one sra transcript was detected in the stationary-phase culture, but no transcript was observed in the exponential-phase culture. These results are consistent with the β-galactosidase reporter assay results described above.
FIG. 5.
Identification of the transcriptional start site of sra. The transcriptional start site was determined by primer extension using the oligonucleotide primer described in Materials and Methods. Sequence ladders were generated by the dideoxy method with the same primer and pKV7350 as the template. RNA was extracted from cultures of MG1655 (lane 1), MG1655Δsra::kan (lane 2), and MG1655(pKV7350) (lane 3) grown at 37°C to the stationary phase. The nucleotide sequence of the sra promoter region is indicated to the right, and an arrow indicates the transcriptional start site of the major transcript.
In cells carrying a high copy number of the sra plasmid (pKV7350), minor transcripts were observed in addition to the major one (Fig. 5). One of the likely start site for these minor transcripts was the T at −55, although these transcripts were not analyzed further.
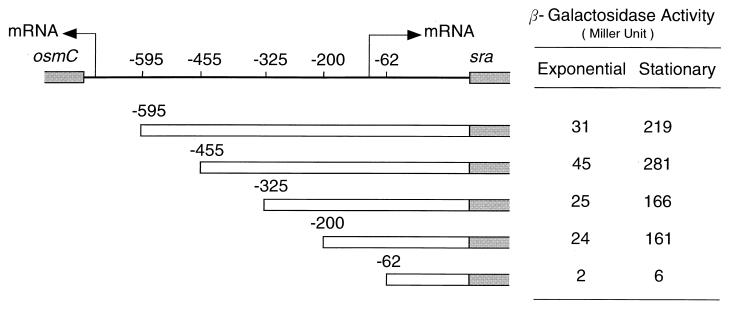
Effects of upstream cis elements on sra transcription.
Experimental determination of the transcriptional start site of the sra gene led to the prediction of the −10 and −35 regions of the sra promoter. Based on these data, we searched in the region upstream of the sra promoter elements for likely cis elements that might be recognized by global regulators of transcription. sra-lacZ fusion constructs containing regions 595, 455, 325, 200, or 62 bp upstream of the initiation (ATG) codon of sra (see Materials and Methods) were used for this purpose, and the β-galactosidase activity of cell samples taken at various times was assayed (Fig. 6). The sra −62 construct showed very low activity, most likely because it lacked some of the promoter elements. The sra −200 and sra −325 constructs, which contained the sra promoter, were both active, but the sra −455 construct showed about 75% higher β-galactosidase activity. These results suggested that the region from −455 to −325 bp might contain a cis element(s) that can be recognized by one or more of the global regulators that were shown to positively affect sra expression in the stationary phase (Fig. 4).
FIG. 6.
Search for cis elements in the region upstream of the sra promoter. KY1461 cells lysogenized with various λpF13(Psra-lacZ) constructs were grown at 37°C in EP medium. Cultures in either the exponential phase (optical density at 600 nm [OD600], 0.4) or the stationary phase (OD600, 1.5) were subjected to a β-galactosidase assay. The negative numbers indicate the construct endpoints measured from the A of the translation initiation codon ATG. The start sites and directions of transcription of sra and osmC are indicated by arrows.
Homologous genes in other bacteria.
To determine to what extent the sra gene is phylogenetically conserved in other organisms, nucleotide and protein databases were searched by BLAST. Homologous genes were identified in Salmonella enterica serovars Typhimurium, Typhi, and Paratyphi and Klebsiella pneumoniae (Fig. 7A). The genes in these bacteria code for proteins of 47, 47, 41, and 46 amino acid residues, respectively. The N-terminal domain of the proteins predicted from these genes was particularly well conserved in all five bacteria. Moreover, by aligning the nucleotide sequences of the upstream region of the gene, the Shine-Dalgarno sequence, mRNA start site, and −10 and −35 regions identified in E. coli were found to be perfectly conserved in all the bacteria except for K. pneumoniae, in which the degree of conservation was lower (Fig. 7B). These data suggest not only that each of the four bacteria harbors a gene identical to E. coli sra but that it is transcribed in a fashion identical to that found in E. coli.
FIG. 7.
Comparison of SRA and its homologs. (A) Alignment of amino acid sequences of SRA proteins from E. coli (ECO), Salmonella serovar Typhimurium (STM), Salmonella serovar Typhi (STY), Salmonella serovar Paratyphi (SPA), and K. pneumoniae (KPN). Dark boxes indicate complete sequence identity in all, and light boxes indicate identity in three or four organisms. (B) Alignment of the nucleotide sequence of the region upstream of the sra gene. Shaded boxes indicate identity in at least four organisms. The asterisk, −10 and −35, and SD show the transcriptional start site, RNA polymerase consensus sequences, and Shine-Dalgarno sequence of the E. coli sra gene, respectively.
DISCUSSION
In this paper, we have presented evidence indicating that the ribosome of E. coli contains a protein of unknown function, termed SRA, whose copy number increases in the stationary phase of growth. The relative copy number of the SRA protein remained constant throughout the ribosome purification steps, including washing with high concentrations of salts (Fig. 1). In addition, the protein bound exclusively to the 30S subunit in an in vitro simultaneous reconstitution experiment with both ribosomal subunits (39). Also, the protein was not detected in a free form in the supernatant after sedimentation of ribosomes by centrifugation. These data suggest that SRA is exclusively associated with ribosomes; hence, ribosomes must be the site of its function. Among the ribosomal proteins identified and characterized so far in various organisms, there are quite a few, including the S1 protein of E. coli, whose copy numbers are rather low. However, the copy number of SRA is even lower than that of S1, especially in the exponential phase of growth although, interestingly, it increases about fourfold during the transition to the stationary phase (Fig. 2). Taking these features into account, we adopted the name SRA.
The increase in the amount of the SRA protein is likely to reflect its unknown physiological role in the stationary phase. However, we were unable to discern any anomalies associated with the sra deletion mutation, not only during exponential growth but also upon prolonged culturing of the mutant. Thus, the precise function of the protein in relation to ribosomes remains to be investigated further.
RMF is highly expressed in the stationary phase and binds to the 50S subunits of 70S ribosomes. RMF plays a role in the dimerization of 70S ribosomes into translationally inactive 100S particles. It is believed that the loss of aminoacyl-tRNA binding is responsible for the reduced translational activity (43). The formation of 100S ribosomes and the levels of expression of RMF and SRA appear to be in parallel with each other over the entire growth cycle of E. coli. Nonetheless, an sra deletion mutant and its parental wild-type strain were indistinguishable from each other in terms of the expression of RMF and the formation of 100S ribosomes.
From the expression profile of sra, its transcription appears to be dependent on factors that are involved in the regulation of genes active in the stationary phase. Indeed, the β-galactosidase assay of the sra-lacZ operon fusion indicated that the level of transcription of sra in the rpoS mutant was only about 30% the wild-type level in the stationary phase (Fig. 4). In accordance with these results, we observed by Northern analysis that transcription from the sra promoter was lower in the rpoS mutant (data not shown). Therefore, the promoter of sra is partly dependent on sigma S. Furthermore, global transcriptional regulators, such as FIS, IHF, cAMP, and ppGpp, were found to positively regulate directly or indirectly the transcription of sra during the stationary phase, and the region between −455 and −325 bp is likely to be responsible for the action of the regulators (Fig. 6). The low level of transcription of sra observed in the exponential phase with the wild-type strain was also the case with mutants defective in the global regulators mentioned above (Fig. 4). Therefore, these global regulators do not interfere with sra expression in the exponential phase.
Various global regulators have been demonstrated to have both upregulatory and down-regulatory effects on genes in E. coli. For instance, the cAMP-cAMP receptor protein (CRP) complex has been implicated as an activator of carbon starvation-responsive genes (32). Various genes, including rpoS and several sigma S-dependent genes, such as bolA, are negatively controlled by cAMP-CRP (20, 21). Other global regulators, such as IHF, H-NS, and FIS, are histone-like DNA-binding proteins with a variety of functions. IHF stimulates the expression of dps in the stationary phase in a sigma S-dependent fashion (2), and the level of IHF increases approximately 10-fold during the transition to the stationary phase. Mutants of hns generally have increased levels of sigma S-controlled genes during exponential growth, and the expression of sigma S is stimulated by a posttranscriptional mechanism in hns mutants (3). Thus, it is known that H-NS is a pleiotropic exponential-phase inhibitor of the expression of many stationary-phase-responsive genes. It is an abundant protein in growing cells, and its concentration further increases upon entry into the stationary phase (9, 36). FIS is a relatively abundant DNA-binding protein found in exponentially growing cells. FIS binds sites upstream of promoters for rRNA and tRNA genes and strongly stimulates their promoter activity (30). The level of ppGpp in cells increases under stringent growth conditions. ppGpp has pleiotropic effects on various genes. In a relA spoT double mutant, which is essentially devoid of ppGpp, the cellular sigma S content is greatly decreased because ppGpp has a positive regulatory action on sigma S (10). Thus, many global regulators, including cAMP, H-NS, IHF, and ppGpp, influence the expression of sigma S, which in turn may regulate the transcription of sra in the stationary phase. However, details of the mechanisms of fine control of sra expression in the exponential phase as well as in the stationary phase by sigma S, OmpR, FIS, IHF, cAMP, and/or ppGpp remain to be investigated further.
A search for sra homologs in other organisms in the nucleotide databases revealed the presence of homologous genes in Salmonella serovar Typhimurium and three other enterobacteria (Fig. 7). However, no homologous genes could be found in gram-positive bacteria such as Bacillus subtilis, in spite of extensive searches with different search options (K. Isono and N. Ogasawara, personal communication). This result suggests that the sra gene is likely to be specific to gram-negative, most likely enteric, bacteria. Knowing that its copy number increases in the stationary phase, we are interested in establishing the function of SRA in association with ribosomes, particularly in the stationary phase of growth of E. coli and other enteric bacteria. Perhaps SRA plays a role in cells survival under unfavorable environmental conditions upon prolonged culturing.
ACKNOWLEDGMENTS
We are grateful to K. Isono for critical reading of the manuscript and helpful discussions. We also thank H. Inokuchi and A. Ishihama for helpful discussions; M. Ueta and M. Kanemori for laboratory supplies; and C. G. Kurland, A. F. Stewart, Y. Akiyama, Y. Kano, T. Katayama, K. Ikehara, and A. Nishimura for gifts of plasmids and bacterial strains.
This work was supported in part by grants awarded to A. Wada and C. Wada from the Ministry of Education, Science and Culture, Japan.
Footnotes
Corresponding author. Present address: Department of Physics, Osaka Medical College, 2-41, Sawaragi-Cho, Takatsuki 569-0084, Japan. Phone: 81-726-75-6905. Fax: 81-726-74-6033. E-mail: phy003@art.osaka-med.ac.jp.
REFERENCES
- 1.Akiyama Y, Ito K. The SecY membrane component of the bacterial protein export machinery: analysis by new electrophoretic methods for integral membrane proteins. EMBO J. 1985;4:3351–3356. doi: 10.1002/j.1460-2075.1985.tb04088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol Microbiol. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 3.Barth M, Marschall C, Muffler A, Fischer D, Hengge-Aronins R. Role for the histone-like protein H-NS in growth-phase-dependent and osmotic regulation of S and many S-dependent genes in Escherichia coli. J Bacteriol. 1995;177:3455–3464. doi: 10.1128/jb.177.12.3455-3464.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates D B, Boye E, Asai T, Kogoma T. The absence of effect of gid or mioC transcription on the initiation of chromosomal replication in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:12497–12502. doi: 10.1073/pnas.94.23.12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman M L, Enquist L W, Silhavy T J. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. [Google Scholar]
- 6.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 8.Cerretti D P, Dean D, Davis G R, Bedwell D M, Nomura M. The spc ribosomal protein operon of Escherichia coli: sequence and cotranscription of the ribosomal protein genes and a protein export gene. Nucleic Acids Res. 1983;11:2599–2616. doi: 10.1093/nar/11.9.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dersch P, Schmidt K, Bremer E. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth phase control and autoregulation. Mol Microbiol. 1993;8:875–889. doi: 10.1111/j.1365-2958.1993.tb01634.x. [DOI] [PubMed] [Google Scholar]
- 10.Gentry D R, Hernandez V J, Nguyen L H, Jensen D B, Cashel M. Synthesis of the stationary-phase sigma factor sigma S is positively regulated by ppGpp. J Bacteriol. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goshima N, Kano Y, Tanaka H, Kohno K, Iwaki T, Imamoto F. IHF suppresses the inhibitory effect of H-NS on HU function in the hin inversion system. Gene. 1994;141:17–23. doi: 10.1016/0378-1119(94)90122-8. [DOI] [PubMed] [Google Scholar]
- 12.Guyer M S, Reed R R, Steitz J A, Low K B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45:135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- 13.Hardy S J S, Kurland C G, Voynow P, Mora G. The ribosomal proteins of Escherichia coli. I. Purification of the 30S ribosomal proteins. Biochemistry. 1969;8:2897–2905. doi: 10.1021/bi00835a031. [DOI] [PubMed] [Google Scholar]
- 14.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM, Press; 1996. pp. 1497–1512. [Google Scholar]
- 15.Hirano M, Shigesada K, Imai M. Construction and characterization of plasmid and lambda phage vector systems for study of transcriptional control of Escherichia coli. Gene. 1987;57:89–99. doi: 10.1016/0378-1119(87)90180-6. [DOI] [PubMed] [Google Scholar]
- 16.Horie K, Wada A, Fukutome H. Conformational studies of Escherichia coli ribosomes with the use of acridine orange as a probe. J Biochem. 1981;60:449–461. doi: 10.1093/oxfordjournals.jbchem.a133492. [DOI] [PubMed] [Google Scholar]
- 17.Kaltschmidt E, Wittman H G. Ribosomal proteins. VII. Two-dimensional polyacrylamide gel electrophoresis for fingerprinting of ribosomal proteins. Anal Biochem. 1970;36:401–412. doi: 10.1016/0003-2697(70)90376-3. [DOI] [PubMed] [Google Scholar]
- 18.Kitagawa M, Wada C, Yoshioka S, Yura T. Expression of ClpB, an analog of the ATP-dependent protease regulatory subunit in Escherichia coli, is controlled by a heat shock sigma factor (sigma 32) J Bacteriol. 1991;173:4247–4253. doi: 10.1128/jb.173.14.4247-4253.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genome library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 20.Lange R, Hengge-Aronis R. Growth-phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor sigma S. J Bacteriol. 1991;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange R, Hengge-Aronis R. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 22.Loewen P C, Triggs B L. Genetic mapping of katF, a locus that with katE affects the synthesis of a second catalase species in Escherichia coli. J Bacteriol. 1984;160:668–675. doi: 10.1128/jb.160.2.668-675.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahajan S K, Chu C C, Willis D K, Templin A, Clark A J. Physical analysis of spontaneous and mutagen-induced mutants of Escherichia coli K-12 expressing DNA exonuclease VIII activity. Genetics. 1990;125:261–273. doi: 10.1093/genetics/125.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 25.Matsunaga F, Kawasaki Y, Ishiai M, Nishikawa K, Yura T, Wada C. DNA-binding domain of the RepE initiator protein of the mini-F plasmid: involvement of the carboxyl-terminal region. J Bacteriol. 1995;177:1994–2001. doi: 10.1128/jb.177.8.1994-2001.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayaux J-F, Fayat G, Fromant M, Springer M, Grunberg-Manago M, Blanquet S. Structural and transcriptional evidence for related thrS and infC expression. Proc Natl Acad Sci USA. 1983;80:6152–6156. doi: 10.1073/pnas.80.20.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller H I. Primary structure of the himA gene of Escherichia coli: homology with DNA-binding protein HU and association with the phenylalanyl-tRNA synthetase operon. Cold Spring Harbor Symp Quant Biol. 1984;49:691–698. doi: 10.1101/sqb.1984.049.01.078. [DOI] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 29.Noll M, Hapke B, Schreire M H, Noll H. Structural dynamics of bacterial ribosomes. I. Characterization of vacant couples and their relation to complexed ribosomes. J Mol Biol. 1973;75:281–294. doi: 10.1016/0022-2836(73)90021-1. [DOI] [PubMed] [Google Scholar]
- 30.Rao L, Ross W, Appleman J A, Gaal T, Leirmo S, Schlax P J, Record M T, Jr, Gourse R L. Factor independent activation of rrnB P1. An “extended” promoter with an upstream element that dramatically increases promoter strength. J Mol Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 31.Sacerdot C, Fayat G, Dessen P, Springer M, Blanquet S. Sequence of a 1.26-kb DNA fragment containing the structural gene for E. coli initiation factor IF3: presence of an AUU initiator codon. EMBO J. 1982;1:311–315. doi: 10.1002/j.1460-2075.1982.tb01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultz J E, Latter G I, Martin A. Differential regulation by cyclic AMP of starvation protein synthesis in Escherichia coli. J Bacteriol. 1988;170:3903–3909. doi: 10.1128/jb.170.9.3903-3909.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thang M N, Thang D C, Grunberg-Manago M. An altered polynucleotide phosphorylase in E. coli mutant Q13. Biochem Biophys Res Commun. 1967;28:374–379. doi: 10.1016/0006-291x(67)90320-8. [DOI] [PubMed] [Google Scholar]
- 34.Traub P, Nomura M. Structure and function of Escherichia coli ribosomes. I. Partial fractionation of the functionally active ribosomal proteins and reconstitution of artificial subribosomal particles. J Mol Biol. 1968;34:575–593. doi: 10.1016/0022-2836(68)90182-4. [DOI] [PubMed] [Google Scholar]
- 35.Triezenberg S J. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1992. Primer extension; pp. 4.8.1–4.8.5. [DOI] [PubMed] [Google Scholar]
- 36.Ueguchi C, Kakeda M, Mizuno T. Autoregulatory expression of the Escherichia coli hns gene encoding a nucleoid protein: H-NS functions as a repressor of its own transcription. Mol Gen Genet. 1993;236:171–178. doi: 10.1007/BF00277109. [DOI] [PubMed] [Google Scholar]
- 37.Vogel H J, Bonner D M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 38.Wada A. Analysis of Escherichia coli ribosomal proteins by an improved two dimensional gel electrophoresis. I. Detection of four new proteins. J Biochem. 1986;100:1583–1594. doi: 10.1093/oxfordjournals.jbchem.a121866. [DOI] [PubMed] [Google Scholar]
- 39.Wada A. Analysis of Escherichia coli ribosomal proteins by an improved two dimensional gel electrophoresis. II. Characterization of four new proteins. J Biochem. 1986;100:1595–1605. doi: 10.1093/oxfordjournals.jbchem.a121867. [DOI] [PubMed] [Google Scholar]
- 40.Wada A, Sako T. Primary structures of and genes for new ribosomal proteins A and B in Escherichia coli. J Biochem. 1987;101:817–820. doi: 10.1093/jb/101.3.817. [DOI] [PubMed] [Google Scholar]
- 41.Wada A, Yamazaki Y, Fujita N, Ishihama A. Structure and probable genetic location of a “ribosome modulation factor” associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc Natl Acad Sci USA. 1990;87:2657–2661. doi: 10.1073/pnas.87.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wada A, Koyama K, Maki Y, Shimoi Y, Tanaka A, Tsujii H. A 5 kDa protein (SCS23) from the 30S subunit of the spinach chloroplast ribosome. FEBS Lett. 1993;319:115–118. doi: 10.1016/0014-5793(93)80048-y. [DOI] [PubMed] [Google Scholar]
- 43.Wada A, Igarashi K, Yoshimura S, Aimoto S, Ishihama A. Ribosome modulation factor stationary growth phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem Biophys Res Commun. 1995;214:410–417. doi: 10.1006/bbrc.1995.2302. [DOI] [PubMed] [Google Scholar]
- 44.Wittmann H G. Two-dimensional polyacrylamide gel electrophoresis for separation of ribosomal proteins. Methods Enzymol. 1974;30:497–505. doi: 10.1016/0076-6879(74)30050-x. [DOI] [PubMed] [Google Scholar]
- 45.Wittmann H G, Stofflet G, Hindennach I, Kurland C G, Birge E A, Randall-Hazelbauer L, Nomura M, Kaltschmidt E, Mizushima S, Traut R R, Bickle T A. Correlation of 30S ribosomal proteins of Escherichia coli isolated in different laboratories. Mol Gen Genet. 1971;111:327–333. doi: 10.1007/BF00569784. [DOI] [PubMed] [Google Scholar]
- 46.Wittmann H G, Littlechild J A, Wittmann-Liebold B. Structure of ribosomal proteins. In: Chambliss G, Craven G R, Davies J, Davies K, Kahan L, Nomura M, editors. Ribosomes: structure, function and genetics. Baltimore, Md: University Park Press; 1980. pp. 51–88. [Google Scholar]
- 47.Xiao H, Kalma M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 48.Yamada H, Yoshida T, Tanaka K, Sasakawa C, Mizuno T. Molecular analysis of the Escherichia coli hns gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol Gen Genet. 1991;230:332–336. doi: 10.1007/BF00290685. [DOI] [PubMed] [Google Scholar]
- 49.Yamagishi M, Matsushima H, Wada A, Sakagami M, Fujita N, Ishihama A. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase- and growth rate-dependent control. EMBO J. 1993;12:625–630. doi: 10.1002/j.1460-2075.1993.tb05695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yano R, Imai M, Yura T. The use of operon fusions in studies of the heat-shock response: effects of altered sigma 32 on heat-shock promoter function in Escherichia coli. Mol Gen Genet. 1987;207:24–28. doi: 10.1007/BF00331486. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Buchholz F, Muyrers J P, Stewart A F. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]