Abstract
Insulin-mTOR signaling drives anabolic growth during organismal development, while its late-life dysregulation may detrimentally contribute to aging and limit lifespans. Age-related regulatory mechanisms and functional consequences of insulin-mTOR remain incompletely understood. Here we identify LPD-3 as a megaprotein that orchestrates the tempo of insulin-mTOR signaling during C. elegans aging. We find that an agonist insulin INS-7 is drastically over-produced in early life and shortens lifespan in lpd-3 mutants, a C. elegans model of human Alkuraya-Kučinskas syndrome. LPD-3 forms a bridge-like tunnel megaprotein to facilitate phospholipid trafficking to plasma membranes. Lipidomic profiling reveals increased abundance of hexaceramide species in lpd-3 mutants, accompanied by up-regulation of hexaceramide biosynthetic enzymes, including HYL-1 (Homolog of Yeast Longevity). Reducing HYL-1 activity decreases INS-7 levels and rescues the lifespan of lpd-3 mutants through insulin receptor/DAF-2 and mTOR/LET-363. LPD3 antagonizes SINH-1, a key mTORC2 component, and decreases expression with age in wild type animals. We propose that LPD-3 acts as a megaprotein brake for aging and its age-dependent decline restricts lifespan through the sphingolipid-hexaceramide and insulin-mTOR pathways.
What causes aging remains a contentious unresolved question. Major theories of aging, including the molecular damage and free radical theory, the antagonistic pleiotropy and hyperfunction theory of aging, have stimulated many studies to test predictions from each1–8. A new era in aging research was initiated by the discovery of exceptionally long-lived mutants in C. elegans, a genetically tractable model organism particularly suited for aging studies9–12. Reduced abundance or activity in components of the insulin pathway (the insulin receptor DAF-2 or PI-3 kinase AGE-1) extends longevity through mechanisms that have been extensively studied, and involve, at least in part, activation of geroprotective transcription factors (DAF-16, HSF-1 and SKN-1)13–15. While such loss-of-function models have been highly valuable and support critical roles of insulin-mTOR in aging from diverse organisms, whether and how insulin-mTOR hyperfunction may actively drive aging and age-associated pathological phenotypes remain underexplored. Key regulators that can prevent insulin-mTOR hyperfunction and orchestrate the tempo of aging remain unidentified.
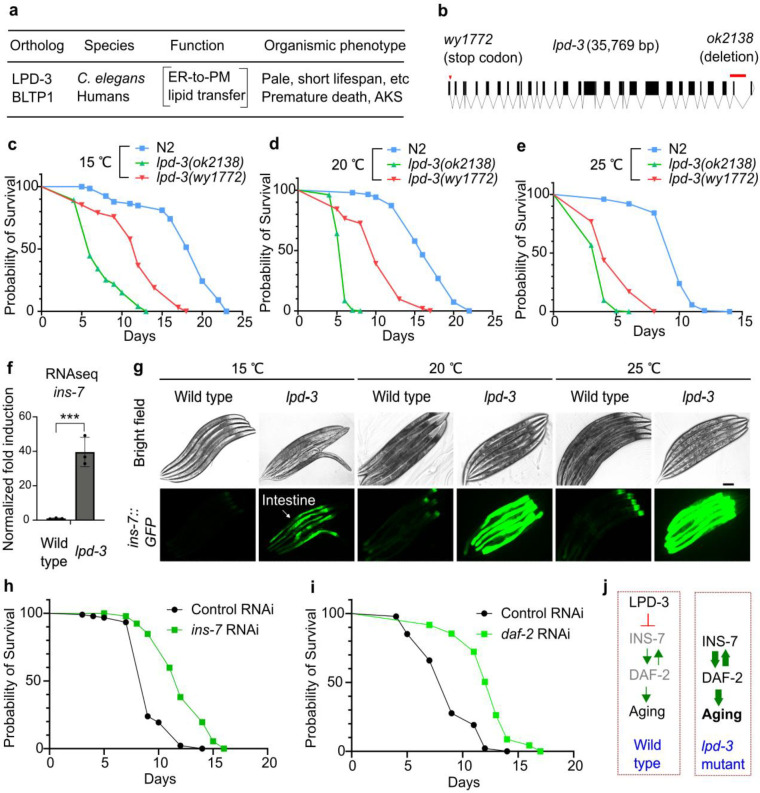
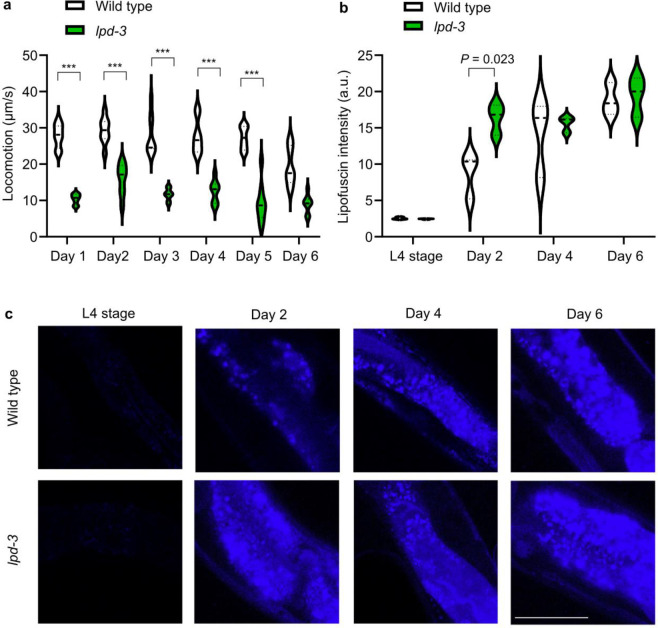
In recent studies, we identified lpd-3 in a mutagenesis screen and discovered that the 452 kDa megaprotein LPD-3 acts as a bridge-like tunnel protein (BLTP) with evolutionarily conserved roles in phospholipid trafficking from the endoplasmic reticulum (ER) to plasma membranes (PM) (Fig. 1a)16. Mutations in BLTP1, the human orthologue of lpd-3, cause a rare autosomal recessive genetic disorder, Alkuraya-Kucinskas syndrome (AKS)17–19. Since most AKS patients die prematurely of unknown etiology, we sought to determine the lifespan of lpd-3 mutants and explore how LPD-3 deficiency causes pathological phenotypes in C. elegans. We generated two backcrossed strains carrying a deletion allele ok2138 or CRISPR-generated stop-codon allele wy1772, respectively (Fig. 1b). We found that both strains showed strikingly shortened lifespans at three common temperature cultivation conditions tested (Fig. 1c–e). In addition, we found that lpd-3 mutants exhibited various common aging phenotypes, including age pigment and reduced behavioral locomotion that occurred earlier than in the wild type (Extended Data Fig. 1).
Fig. 1. Hyper-activation of ins-7 causes shortened lifespan in lpd-3 mutants.
a, Cellular function and organismic phenotypes for LPD-3 and the human homologue BLTP1. b, Gene diagram showing two alleles of lpd-3 examined for lifespan phenotypes. c, Lifespan analysis of wild type versus lpd-3 mutants with two different alleles (ok2138 and wy1772) grown at 15 °C. d, Lifespan analysis of wild type versus lpd-3 mutants with two different alleles (ok2138 and wy1772) grown at 20 °C. e, Lifespan analysis of wild type versus lpd-3 mutants with two different alleles (ok2138 and wy1772) grown at 25 °C. f, RNA-seq results showing increased ins-7 expression in lpd-3 mutants. Values are means ± S.D. *** indicates P < 0.001 (N = 3 biological replicates). g, Representative bright-field and epifluorescence images showing drastically increased ins-7p::ins-7::GFP abundance in lpd-3 mutants. Scale bar, 100 μm. h, Lifespan analysis of lpd-3 mutants with control or ins-7 RNAi at 20 °C, showing shortened lifespan rescued (P < 0.0001, log-rank test). i, Lifespan analysis of lpd-3 mutants with control or daf-2 RNAi at 20 °C, showing shortened lifespan rescued (P < 0.0001, log-rank test). j, Schematic diagrams illustrating a model of how LPD-3 regulates aging via INS-7 and DAF-2 in wild type and lpd-3 mutants.
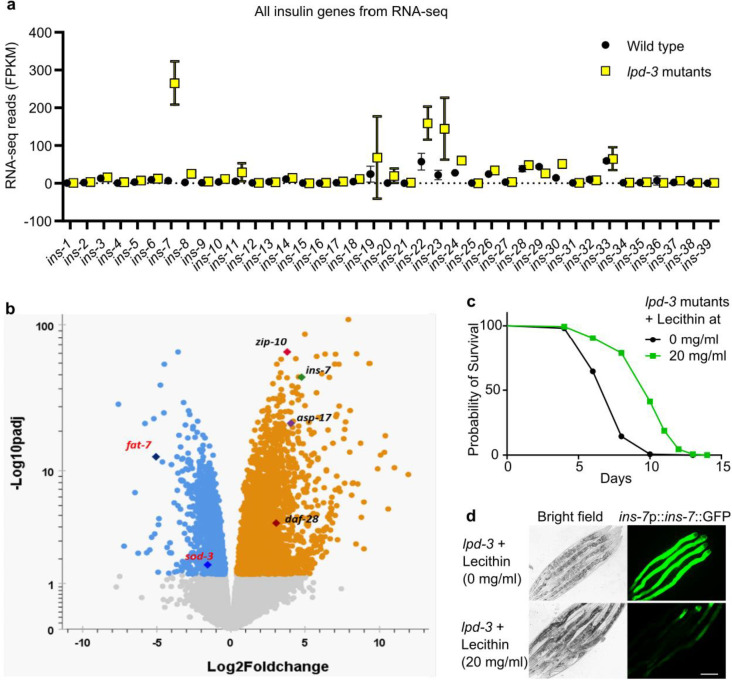
The drastically shortened lifespan of lpd-3 mutants prompted us to identify the underlying mechanisms. By analyzing RNA-seq datasets for lpd-3 mutants compared with wild type16, we discovered that LPD-3 deficiency led to a nearly 40-fold increase in the expression level of ins-7, which represents one of the most highly lpd-3 up-regulated genes and encodes an agonist insulin with previously reported effects on aging and lifespan in C. elegans20–22 (Fig. 1f). Expression of other insulin-encoding genes was much less regulated or largely unaltered (Extended Data Fig. 2a, b). To determine the spatiotemporal site of ins-7 regulation, we crossed an integrated ins-7p::INS-7::GFP translational reporter strain23 with the lpd-3 deletion mutant. In the wild type, ins-7p::INS-7::GFP is expressed at baseline levels that are detectable only in the anterior part of intestine at 15 °C starting from young adult stages (24 hrs post-L4), and increased at 20 °C and 25 °C (Fig. 1g). By contrast, ins-7p::INS-7::GFP in lpd-3 mutants showed drastically increased INS-7::GFP abundance and fluorescence throughout the intestine, and particularly under higher temperatures (20 or 25 °C). We showed previously that lpd-3 mutants are defective in ER-to-PM phospholipid trafficking, and exhibit sensitivity to thermal stress that can be normalized by Lecithin16. We confirmed in this study that Lecithin supplementation in the culture medium rescued both the shortened lifespan and ins-7 up-regulation phenotypes (Extended Data Fig. 2c, d).
To determine the causal role of ins-7 in the shortened lifespan phenotype of lpd-3 mutants, we used RNAi to knock down ins-7 expression in lpd-3 mutants. We found that treatment with RNAi against ins-7 led to a markedly extended lifespan of lpd-3 mutants, comparable to that in the wild type (Fig. 1h). We also observed such rescue of shortened lifespan phenotype of lpd-3 mutants by RNAi against daf-2 (Fig. 1i), which encodes the only known receptor of insulin in C. elegans. We note that control RNAi also led to a slight increase of lifespan in lpd-3 mutants (Fig. 1d, h), likely reflecting different lipid compositions of bacterial strains between OP50 and HT115 strains24,25 used in routine C. elegans culture and RNAi experiments, respectively. Reducing ins-7 expression has been previously shown to increase the lifespan of wild-type animals, implicating INS-7’s role in tissue entrainment by feedback regulation in C. elegans20–22. Our results support these early findings and reveal that overproduction of INS-7 is causally responsible for shortening lifespan in lpd-3 mutants, in a manner that requires the sole insulin receptor DAF-2 (Fig. 1j).
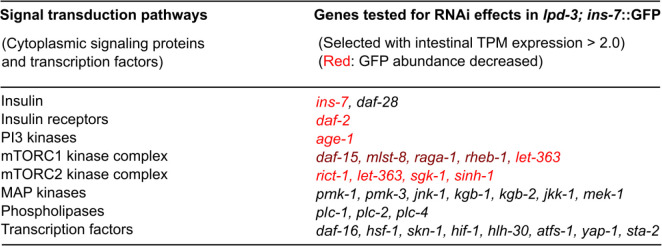
We next determined the regulatory mechanisms leading to ins-7 up-regulation and shortened lifespan in lpd-3 mutants. Taking advantage of the drastically up-regulated ins-7p:: INS-7::GFP reporter in lpd-3 mutants as a live fluorescent readout, we performed targeted RNAi screens to identify genes required for ins-7p:: INS-7::GFP up-regulation by lpd-3. We performed RNAi for those genes with reported adequate expression in the intestine (transcript per million, TPM > 2) and encoding mediators of major signal transduction pathways known to be involved in insulin signaling (the Ras/MAPK, phospholipase and PI3 kinase/mTORC1/mTORC2 pathways)26–28 (Extended Data Fig. 3). As positive controls, we verified strong effects of ins-7 or daf-2 RNAi in suppressing ins-7p::INS-7::GFP expression in lpd-3 mutants (Fig. 2a, b). From such screens, we found that RNAi against several genes encoding components of the mTORC2 complex in C. elegans, including let-363, rict-1, and sinh-1, led to a marked reduction of ins-7p::INS-7::GFP in lpd-3 mutants (Fig. 2a, b). RNAi against genes involved in the mTORC1 complex, except let-363 shared by mTORC1 and mTORC2, exhibited weaker effects (Fig. 2a).
Fig. 2. LPD-3 regulates INS-7 via the sphingolipid-ceramide-mTORC2 axis.
a, Table summarizing effects of RNAi against genes in the insulin/mTORC1/mTORC2 pathway on ins-7p::ins-7::GFP levels in lpd-3 mutants. * indicates GFP intensity observed under microscope. b, Representative bright-field and epifluorescence images showing ins-7p::ins-7::GFP up-regulation in lpd-3 mutants can be suppressed by RNAi against daf-2, rict-1 or hyl-1. Scale bar, 100 μm. c, Lipidomic quantification of hexaceramide species in wild type and lpd-3 mutants. Values are means ± S.D. (N = 3 biological replicates). d, Schematic showing biosynthetic pathways of hexaceramide, including sphingosine conversion to ceramide by HYL-1 and ceramide to hexaceramide by CGT-1/2/3. e, RNA-seq results showing increased hyl-1 and cgt-1/2/3 expression in lpd-3 mutants. Values are means ± S.D. ***P < 0.001 (N = 3 biological replicates). f, Lifespan analysis of lpd-3 mutants with control or rict-1 RNAi at 20 °C, showing rescued lifespan (P < 0.0001, log-rank test). g, Lifespan analysis of lpd-3 mutants with control or hyl-1 RNAi at 20 °C, showing rescued lifespan (P< 0.0001, log-rank test). h, Lifespan analysis of lpd-3 mutants with control or sgk-1 RNAi at 20 °C, showing rescued lifespan (P < 0.0001, log-rank test).
In parallel to such RNAi screens, we also performed lipidomic profiling of lpd-3 mutants compared with wild type. Among all major lipid species examined by LC-MS/MS, including phospholipids, glycerolipids and sphingolipids (Supplementary Table 1), the hexaceramide-type of sphingolipids showed marked up-regulation in lpd-3 mutants (Fig. 2c). Consistently, RNA-seq results revealed that genes encoding hexaceramide biosynthetic enzymes (Fig. 2d), including HYL-1 and CGT-1/2/3, were markedly up-regulated in lpd-3 mutants (Fig. 2e). As reduced LPD-3 activity results in impaired ER-to-PM phospholipid trafficking, a lipid homeostatic mechanism is likely triggered to up-regulate sphingolipids in lpd-3 mutants16. Given reported sphingolipid modulation of mTOR signaling and longevity29–31, we next explored the link of hexaceramide biosynthetic enzyme HYL-1 to ins-7. We found that hyl-1 RNAi led to strong suppression of ins-7p::INS-7::GFP in lpd-3 mutants (Fig. 2b). Functionally, lifespan analysis revealed that RNAi against rict-1 or hyl-1 led to the marked rescue of the shortened lifespan in lpd-3 mutants (Fig. 2f, g). RNAi against sgk-1, which encodes a major kinase32–34 mediating effects of mTORC2, showed similar rescuing effects (Fig. 2h). Taken together, these results indicate that the sphingolipid-mTOR pathway drives ins-7 over-expression, leading to the shortened lifespan in lpd-3 mutants.
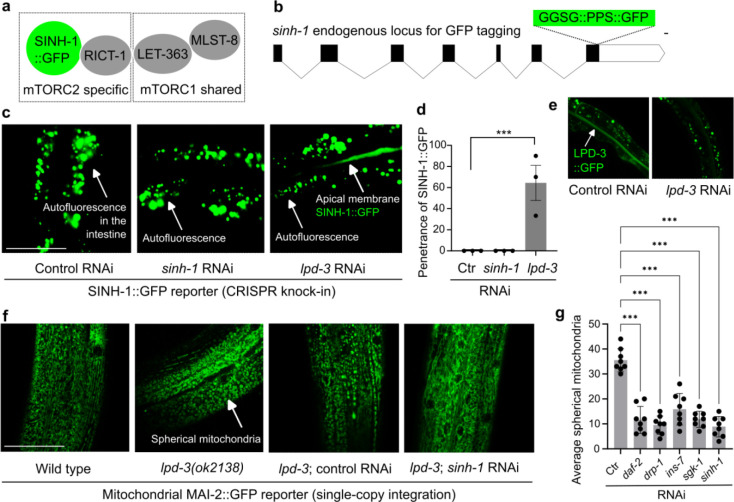
We next explored cellular mechanisms by which LPD-3 regulates mTORC2. The mTORC2 complex in C. elegans consists of the core subunit LET-363 shared by mTORC1, the mTORC2-specific subunits RICT-1 and SINH-1 (Fig. 3a). As the mammalian homologs of SINH-1 bind to PIP3 at the PM and mediate signaling from insulin receptors to the mTORC2 complex35–37, we tagged endogenous SINH-1 with GFP by CRISPR (Fig. 3b) and monitored how LPD-3 may affect the intracellular abundance and localization of SINH-1::GFP. In the wild type, we did not observe apparent SINH-1::GFP fluorescence throughout animals at the young adult stage (24 hrs post L4). By contrast, RNAi against lpd-3 or its mutations led to the marked increase of SINH-1::GFP fluorescence signals that were particularly enriched at loci in close proximity to PM in the intestine (Fig. 3c, d). As expected, RNAi against lpd-3 also effectively decreased the abundance of LPD-3 endogenously labeled with GFP by CRISPR (Fig. 3e, and see below). Since loss of LPD-3 causes PIP2/3 to accumulate in the ER membrane near the ER-PM junction16, increased SINH-1::GFP in lpd-3 mutants likely reflects its ectopic intracellular activity. In addition to the ins-7 phenotype (Fig. 2), we found that lpd-3 mutants exhibited morphologically spherical mitochondria with decreased branch lengths, whereas RNAi against genes encoding the mitochondrial fission machinery (DRP-1) or insulin-mTOR pathway components (INS-7, DAF-2, SINH-1, SGK-1) suppressed such phenotype (Fig. 3f, g). These results indicate that LPD-3 normally functions to restrict SINH-1 accumulation and thereby antagonize ectopic mTOR activity.
Fig. 3. Loss of LPD-3 dysregulates SINH-1/mTORC2 and mitochondria.
a, Schematic showing key components of the mTORC2 complex in the SINH-1::GFP tagged C. elegans. b, Schematic gene structure of sinh-1 showing the CRISPR-mediated knock-in allele encoding the endogenous SINH-1 tagged with a linker (GGGS), PreScission Protease site (PPS) and GFP. Scale bar: 100 bp. c, Representative confocal fluorescence images showing undetectable SINH-1::GFP signals in animals treated with control or sinh-1 RNAi but increased SINH-1::GFP enrichment along the apical intestinal membrane in animals treated with lpd-3 RNAi (non-specific gut autofluorescence not affected by sinh-1 or lpd-3 RNAi also indicated). d, Quantification of the penetrance of SINH-1::GFP enrichment at the apical intestinal membrane in animals with indicated RNAi treatment. Values are means ± S.E.M. ***P < 0.001 (N = 3 independent experiments, n > 10 in each trial). e, Representative confocal fluorescence images showing decreased LPD-3::GFP by lpd-3 RNAi. f, Representative confocal fluorescence images showing increased spherical MAI-2::GFP marked mitochondria in lpd-3 mutants (day 1) that can be normalized by sinh-1 but not control RNAi. g, Quantification showing rescued mitochondrial morphological defects in lpd-3 mutants by RNAi against genes encoding the mitochondrial fission machinery (drp-1) or insulin-mTOR pathway components (e.g. sinh-1). Values are means ± S.E.M. ***P < 0.001 (N = 8 biological replicates).
Are endogenous LPD-3 abundance and activity subject to regulation by normal aging in wild type animals? To address this question, we tagged endogenous LPD-3 with seven copies of the split GFP (GFP11) by CRISPR38–40, crossed the allele into a strain expressing GFP10 specifically in the intestine by the ges-1 promoter (Fig. 4a). We monitored how aging affects the intracellular abundance and localization of LPD-3::GFP based on the complementation of GFP1–10 and 7xGFP11 in the intestine, site of which we showed previously is critical for LPD-3 functions in phospholipid trafficking16. Confocal microscopy revealed that LPD-3::GFP is enriched along the apical membrane of the intestine at the L4 and young adult stages, with an age-dependent progressive decline in abundance at the stages of day 3, 5 and 9 (Fig. 4b, c). Functionally, we showed previously that LPD-3 facilitates ER-to-PM trafficking of phospholipids, including phosphatidylcholine and phosphatidylinositol. Using Akt-PH::GFP, which binds to the phosphatidylinositol species PIP2/PIP3, we found that aging caused a progressive decline of Akt-PH::GFP signals at the intestinal apical membrane while lpd-3 loss-of-function mutations exacerbated such age-dependent decline (Fig. 4d, e). By contrast, ins-7p::INS-7::GFP exhibited age-dependent increase in abundance while such age-dependent change was accelerated in lpd-3 mutants (Fig. 4f, g). These results demonstrate the age-dependent decline in both the abundance and functional activity of endogenous LPD-3 during normal C. elegans aging.
Fig. 4. LPD-3 abundance and functions decline with age in wild type animals.
a, Schematic gene structure of lpd-3 showing the CRISPR-mediated knock-in allele encoding the endogenous LPD-3 tagged with a split GFP1–10 that is complemented by an intestine-expressed GFP11. Scale bar: 500 bp. b, Representative confocal fluorescence images showing age-dependent decrease in the abundance of LPD-3::GFP signals in the intestinal apical membrane of animals at the stages of L4, day 1, 3, 5 and 9 post-L4. c, Quantification of the fluorescence intensities of LPD-3::GFP in animals with indicated ages. Values are means ± S.E.M. ***P < 0.001 (N = 6 biological replicates). d, Representative confocal fluorescence images showing age-dependent decrease in the abundance of Akt-PH::GFP signals that label PIP2/3 in the intestinal apical membrane of animals at the stages of L4, day 1, 3, 5 and 9 post-L4, in both wild type and lpd-3 mutants. Scale bar: 50 μm. e, Quantification of the fluorescence intensities of Akt-PH::GFP at apical intestinal membranes in animals with indicated ages. Values are means ± S.E.M. ***P < 0.001 (N = 8 biological replicates in each stage, N = 32 in each genotype). f, Representative confocal fluorescence images showing age-dependent increase in the abundance of ins-7p::INS-7::GFP signals in animals at the indicated stages in both wild type and lpd-3 mutants. Scale bar: 50 μm. e, Quantification of the fluorescence intensities of ins-7p::INS-7::GFP in animals with indicated ages. Values are means ± S.E.M., with ** P < 0.01 and ***P < 0.001 (N = 7 biological replicates).
Identification and subsequent mechanistic studies of exceptionally long-lived C. elegans mutants in the insulin-mTOR pathway have provided crucial insights into how the loss of insulin-mTOR function extends longevity. Activation of key transcription factors including DAF-16, HSF-1, and SKN-1 mediates geroprotective effects in these mutants by promoting somatic maintenance programs that attenuate molecular and cellular damages. Our studies provide a novel and complementary model in which we propose insulin-mTOR hyperfunction can actively drive aging downstream of LPD-3 and the sphingolipid-ceramide pathway (Extended Data Fig. 4). Our findings support the notion that somatic maintenance programs and molecular/cellular damaging agents (e.g. free radicals and/or cell death signals) from dysregulated mitochondria, both of which act downstream of insulin-mTOR signaling, may converge to control the tempo of aging progression. Based on new insights from C. elegans models, this view may imply to reconcile and unify two prevalent mechanistic theories of aging (molecular damage vs TOR hyperfunction)1–8.
Various sphingolipids have been shown to contribute to organismal aging and age-associated pathologies41–43. We identify LPD-3 as a critical megaprotein regulator of the phospholipid-sphingolipid rheostat, dysfunction of which can profoundly affect aging via insulin-mTOR hyperfunction. LPD-3 defines a member of a recently discovered and highly evolutionarily conserved protein family that controls lipid homeostasis in eukaryotic cell membranes16,44–46; it is currently unknown whether LPD-3 homologs may play similar roles in regulating insulin-mTOR and aging in other organisms. Detailed mechanisms linking LPD-3 to sphingolipid regulation, insulin over-production and aging also await further studies. Based on our findings and prior studies supporting emerging roles of sphingolipid/ceramides and conserved insulin-mTOR signaling from many species30,42,43,47,48, we propose that LPD-3 may act as a cellular brake that slows organismal aging by orchestrating its tempo and regulating the sphingolipid-insulin-mTOR axis likely in phylogenetically diverse organisms.
Methods
C. elegans culture and maintenance
All the C. elegans strains used in the current research were maintained in accordance with the standard laboratory procedures unless otherwise stated. Genotypes of strains used are as follows: N2 Bristol strain (wild type), lpd-3(ok2138), lpd-3(wy1772), yxIs13[ins-7p::ins-7::gfp; unc-122p::dsred], pwIs890[Pvha-6::AKT(PH)::GFP], xmSi[mai-2::GFP(single copy integration)], sinh-1(syb7471[GGSG::PPS::GFP]), lpd-3(wy1770[7xGFP11]); wyEx10609 [ges-1p:: GFP(110); odr-1p::GFP; ges-1p:: ERSP::mScarlet::MAPPER].
Lifespan assessment
The lifespan assay was conducted according to the standard protocol49. Briefly, wild type N2 strain was grown for two generations without starvation, and then embryos were synchronized through standard sodium hypochlorite treatment. These embryos were grown on OP50-seeded plates or transferred to RNAi-seeded NGM plates. Thereafter, 50–100 late-L4 larvae or young adults (24 hrs post-L4) were transferred to OP50-seeded plates or RNAi-seeded NGM plates supplemented with 50 μM of 5-fluoro-2-deoxyuridine (FUDR, Sigma) to prevent progeny growth. Assay populations were transferred to fresh OP50-seeded plate every 3–4 days. Live/dead/missing worms were scored on alternate days until the last surviving worm. The live worms were detected through touch provoke responses. The lifespan assays were repeated for at least three independent trials.
Epifluorescence and confocal microscopic imaging
The epifluorescence compound and confocal microscopes (Leica) were used to capture fluorescence images. Animals of various genotypes or treated with different RNAi were randomly picked and treated with 10 mM sodium azide in M9 solution (Sigma-Aldrich), symmetrically aligned on 2% agar pads on slides for imaging. The control and treatment groups were imaged under identical settings and conditions. The age-dependent decline in LPD-3::GFP strains was scored by calculating the mean fluorescence value from Z-stack of confocal planes in each worm at different time points near the apical membrane using the ImageJ software. Mitochondrial morphology was scored as the number of spherical mitochondria in each image counted manually. The penetrance of SINH-1::GFP in strains of different genotypes under different conditions was scored as a percentage of animals that exhibit detectable fluorescence with confocal microscopy.
Behavioral and age pigment/lipofuscin measurements during aging
The locomotion of worms was measured as previously described51. Briefly, the speed (total track length/time) of age-synchronized worms was recorded using WormLab System (MBF Bioscience) based on the midpoint positions of the worms at different time points. Each experiment was repeated three times with more than 10 animals per group. The age pigment/lipofuscin accumulation with age progression was measured as previously described52,53 by using ultraviolet light excitation coupled with detection through a blue/cyan filter and quantified by calculating the mean fluorescence value in each worm using the ImageJ software.
Lipidomic analysis
Wild type N2 strain and lpd-3(ok2138) mutants were grown under non-starved conditions for four generations at 20 °C, followed by bleach synchronization and harvest by M9 buffer at young adult stages. Frozen worm pellets (50 μl per sample, three independent biological replicates per genotype) are stored until lipidomic extraction and analysis. All solvents for lipidomic extraction and analysis used were either HPLC or LC/MS grade and purchased from Sigma-Aldrich (St Louis, MO, USA). Splash Lipidomix standards were purchased from Avanti (Alabaster, AL, USA). All lipid extractions were performed in 16×100mm glass tubes with PTFE-lined caps (Fisher Scientific, Pittsburg, PA, USA). Glass Pasteur pipettes and solvent-resistant plasticware pipette tips (Mettler-Toledo, Columbus, OH, USA) were used to minimize leaching of polymers and plasticizers. Samples were transferred to fresh glass tubes, and 1 mL of methanol, 1mL of water and 2mL of methyl tert-butyl ether (mTBE) were added for liquid-liquid extraction. The mixture was vortexed and centrifuged at 2,671 g for 5 min, resulting in two distinct liquid phases. The organic phase (upper phase) was transferred to a fresh tube with a Pasteur pipette and spiked with 20uL of a 1:5 diluted Splash Lipidomix standard mixture. The samples were dried under N2 and resuspended in 400 μL of hexane. Lipids were analyzed by LC-MS/MS using a SCIEX QTRAP 6500+ (SCIEX, Framingham, MA) equipped with a Shimadzu LC-30AD (Shimadzu, Columbia, MD) high-performance liquid chromatography (HPLC) system and a 150×2.1 mm, 5 μm Supelco Ascentis silica column (Supelco, Bellefonte, PA). Samples were injected at a flow rate of 0.3 ml/min at 2.5% solvent B (methyl tert-butyl ether) and 97.5% Solvent A (hexane). Solvent B was increased to 5% over 3 min and then to 60% over 6 min. Solvent B was decreased to 0% during 30 sec while Solvent C (90:10 (v/v) isopropanol-water) was set at 20% and increased to 40% during the following 11 min. Solvent C is increased to 44% over 6 min and then to 60% over 50 sec. The system was held at 60% solvent C for 1 min prior to re-equilibration at 2.5% of solvent B for 5 min at a 1.2 mL/min flow rate. Solvent D [95:5 (v/v) acetonitrile-water with 10 mM Ammonium acetate] was infused post-column at 0.03 ml/min. Column oven temperature was 25°C. Data was acquired in positive and negative ionization mode using multiple reaction monitoring (MRM). The LC-MS/MS data was analyzed using MultiQuant software (SCIEX). The identified lipid species were normalized to its corresponding internal standard.
Statistics and reproducibility
All the data in the current manuscript were analyzed using GraphPad Prism 9.2.0 Software (Graphpad, San Diego, CA) and presented as means ± S.E.M. unless otherwise specified, with significance P values calculated by unpaired two-sided t-tests (comparisons between two groups), one-way or two-way ANOVA (comparisons across more than two groups) and adjusted with Bonferroni’s multiple comparisons tests. Lifespan assay was quantified using Kaplan–Meier lifespan analysis and P values were calculated using the log-rank test.
Extended Data
Extended Data Fig. 1. Lipofuscin increase and behavioral decline of lpd-3 mutants early in aging.
a, Quantification of locomotion speed for wild type and lpd-3 mutants showing a rapid decline in lpd-3 mutants starting at day 1 (24 hrs post-L4 stage). *** indicates P < 0.001 (N = 30–50 animals per group). b, Lipofuscin/age pigment fluorescence intensities for wild type and lpd-3 mutants showing a more rapid decline in lpd-3 mutants at day 2 (48 hrs post-L4 stage). c, Representative lipofuscin/age pigment fluorescence images for wild type and lpd-3 mutants showing a more rapid decline in lpd-3 mutants at day 2 (48 hrs post-L4 stage). Scale bar, 100 μm.
Extended Data Fig. 2. Specific up-regulation of ins-7 by loss of LPD-3 and rescue by Lecithin.
a, RNA-seq reveals drastic and specific ins-7 up-regulation among all ins genes examined. Results were derived and analyzed from RNA-seq datasets we published earlier16. FPKM, Fragments Per Kilobase of transcript per Million mapped reads. b, Volcano plot showing significantly LPD-3 regulated genes, including fat-7 (downstream of SBP-1 and NHR-49), sod-3 (downstream of DAF-16) and zip-10 (downstream of ISY-1), asp-17 (downstream of ISY-1), and ins-7. c, Lifespan analysis of lpd-3 mutants with control or Lecithin treatment at 20 °C (P < 0.0001, log-rank test). d, Representative bright-field and epifluorescence images showing that ins-7p::ins-7::GFP up-regulation in lpd-3 mutants can be suppressed by exogenous Lecithin (20 mg/ml) treatment supplemented in culture media. Scale bar, 100 μm.
Extended Data Fig. 3. RNAi screens for genes affecting ins-7 in lpd-3 mutants.
Genes were selected for RNAi testing given their adequate expression values in intestine (TPM or transcript per million > 2.0) and putative roles in mediating insulin-mTOR signaling.
Extended Data Fig. 4. Model of how LPD-3 acts as a brake for aging through sphingolipid and insulin-mTOR regulatory pathways.

In the wild type, LPD-3 mediates phospholipid trafficking from ER to PM, and in turn, suppresses sphingolipid levels. Insulin and mTOR signaling are activated at moderate levels to promote early-life growth and late-life aging. In the lpd-3 mutants, reduced LPD-3 function leads to sphingolipid (hexaceramide type) up-regulation and hyperactivation of insulin and mTOR signaling. INS-7/DAF-2/mTORC2 forms a proposed vicious cycle to sustain insulin and mTOR activation through a positive feedback loop, culminating in hastened late-life aging and shortened lifespan. Red indicates inhibition, while green indicates activation. Solid arrows indicate proposed action based on this study, while dashed arrows indicate regulation inferred from the literature.
Supplementary Material
Supplementary Table 1 Lipidomic profiling of phospholipids, glycerolipids and sphingolipids by LC-MS/MS.
272 mass pairs were monitored by multiple reaction monitoring (MRM) mode. Data were background corrected and normalized with internal standards. Each group had three replicates (n=3) and their respective average (ave), standard deviation (sd), and relative standard deviation (RSD (%)) were calculated. The data file is organized by lipid class/subclass and when applied, their respective fatty acid composition (FA); Triacylglycerides (TAG), hexaceramides (Hexcer), phosphatidylcholine (PC), phosphatidylglycerol (PG), phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylinositol (PI), lyso-phosphatidylethanolamine (LPE).
Acknowledgments
Some strains were provided by Dr. Yun Zhang’s group at Harvard University, Dr. R. E. Navarro’s group at Universidad Nacional Autónoma de México, and by Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). The work was financially supported by the NIH/NIGMS grant 1R35GM139618, UCSF PBBR New Frontier Research, and UCSF BARI Investigator Award (D.K.M).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Blagosklonny M. V. Aging and immortality: quasi-programmed senescence and its pharmacologic inhibition. Cell Cycle 5, 2087–2102 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Blagosklonny M. V. The hyperfunction theory of aging: three common misconceptions. Oncoscience 8, 103–107 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gems D. The hyperfunction theory: An emerging paradigm for the biology of aging. Ageing Res Rev 74, 101557 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gladyshev V. N. Aging: progressive decline in fitness due to the rising deleteriome adjusted by genetic, environmental, and stochastic processes. Aging Cell 15, 594–602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longo V. D., Mitteldorf J. & Skulachev V. P. Programmed and altruistic ageing. Nat Rev Genet 6, 866–872 (2005). [DOI] [PubMed] [Google Scholar]
- 6.de Magalhães J. P. & Church G. M. Genomes optimize reproduction: aging as a consequence of the developmental program. Physiology (Bethesda) 20, 252–259 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Gladyshev V. N. The Free Radical Theory of Aging Is Dead. Long Live the Damage Theory! Antioxid Redox Signal 20, 727–731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sohal R. S. & Weindruch R. Oxidative stress, caloric restriction, and aging. Science 273, 59–63 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kenyon C. J. The genetics of ageing. Nature 464, 504–512 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Klass M. R. A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results. Mech Ageing Dev 22, 279–286 (1983). [DOI] [PubMed] [Google Scholar]
- 11.Kenyon C., Chang J., Gensch E., Rudner A. & Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464 (1993). [DOI] [PubMed] [Google Scholar]
- 12.Finch C. E. & Ruvkun G. The genetics of aging. Annu Rev Genomics Hum Genet 2, 435–462 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Tullet J. M. A. et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132, 1025–1038 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogg S. et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994–999 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Hsu A.-L., Murphy C. T. & Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300, 1142–1145 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Wang C. et al. A conserved megaprotein-based molecular bridge critical for lipid trafficking and cold resilience. Nat Commun 13, 6805 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kane M. S. et al. Endosomal trafficking defects in patient cells with KIAA1109 biallelic variants. Genes Dis 6, 56–67 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gueneau L. et al. KIAA1109 Variants Are Associated with a Severe Disorder of Brain Development and Arthrogryposis. Am J Hum Genet 102, 116–132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meszarosova A. U. et al. Two novel pathogenic variants in KIAA1109 causing Alkuraya-Kučinskas syndrome in two Czech Roma brothers. Clin Dysmorphol 29, 197–201 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Murphy C. T., Lee S.-J. & Kenyon C. Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc Natl Acad Sci U S A 104, 19046–19050 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy C. T. et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277–283 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Lee Y. et al. Reduced insulin/IGF1 signaling prevents immune aging via ZIP-10/bZIP-mediated feedforward loop. J Cell Biol 220, e202006174 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z. et al. Two insulin-like peptides antagonistically regulate aversive olfactory learning in C. elegans. Neuron 77, 572–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neve I. A. A. et al. Escherichia coli Metabolite Profiling Leads to the Development of an RNA Interference Strain for Caenorhabditis elegans. G3 (Bethesda) 10, 189–198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brooks K. K., Liang B. & Watts J. L. The Influence of Bacterial Diet on Fat Storage in C. elegans. PLoS One 4, e7545 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor S. R. et al. Molecular topography of an entire nervous system. Cell 184, 4329–4347.e23 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saltiel A. R. Insulin signaling in health and disease. J Clin Invest 131, e142241, 142241 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blackwell T. K., Sewell A. K., Wu Z. & Han M. TOR Signaling in Caenorhabditis elegans Development, Metabolism, and Aging. Genetics 213, 329–360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorner J. TOR complex 2 is a master regulator of plasma membrane homeostasis. Biochem J 479, 1917–1940 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu H., Shen H., Sewell A. K., Kniazeva M. & Han M. A novel sphingolipid-TORC1 pathway critically promotes postembryonic development in Caenorhabditis elegans. Elife 2, e00429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taniguchi M. et al. Regulation of autophagy and its associated cell death by ‘sphingolipid rheostat’: reciprocal role of ceramide and sphingosine 1-phosphate in the mammalian target of rapamycin pathway. J Biol Chem 287, 39898–39910 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soukas A. A., Kane E. A., Carr C. E., Melo J. A. & Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev 23, 496–511 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sohrabi S., Cota V. & Murphy C. T. Ce Lab, a Microfluidic Platform for the Study of Life History Traits, reveals Metformin and SGK-1 regulation of Longevity and Reproductive Span. bioRxiv 2023.01.09.523184 (2023) doi: 10.1101/2023.01.09.523184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones K. T., Greer E. R., Pearce D. & Ashrafi K. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol 7, e60 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu P. et al. PtdIns(3,4,5)P3-dependent Activation of the mTORC2 Kinase Complex. Cancer Discov 5, 1194–1209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zoncu R., Efeyan A. & Sabatini D. M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12, 21–35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calejman C. M., Doxsey W. G., Fazakerley D. J. & Guertin D. A. Integrating adipocyte insulin signaling and metabolism in the multi-omics era. Trends Biochem Sci 47, 531–546 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goudeau J. et al. Split-wrmScarlet and split-sfGFP: tools for faster, easier fluorescent labeling of endogenous proteins in Caenorhabditis elegans. Genetics 217, iyab014 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dickinson D. J. & Goldstein B. CRISPR-Based Methods for Caenorhabditis elegans Genome Engineering. Genetics 202, 885–901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jinek M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang H., Huang X. & Pang S. Regulation of the lysosome by sphingolipids: Potential role in aging. J Biol Chem 298, 102118 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trayssac M., Hannun Y. A. & Obeid L. M. Role of sphingolipids in senescence: implication in aging and age-related diseases. J Clin Invest 128, 2702–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laurila P.-P. et al. Sphingolipids accumulate in aged muscle, and their reduction counteracts sarcopenia. Nat Aging 2, 1159–1175 (2022). [DOI] [PubMed] [Google Scholar]
- 44.Braschi B., Bruford E. A., Cavanagh A. T., Neuman S. D. & Bashirullah A. The bridge-like lipid transfer protein (BLTP) gene group: introducing new nomenclature based on structural homology indicating shared function. Hum Genomics 16, 66 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neuman S. D., Levine T. P. & Bashirullah A. A novel superfamily of bridge-like lipid transfer proteins. Trends Cell Biol S0962–8924(22)00083–6 (2022) doi: 10.1016/j.tcb.2022.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levine T. P. Sequence Analysis and Structural Predictions of Lipid Transfer Bridges in the Repeating Beta Groove (RBG) Superfamily Reveal Past and Present Domain Variations Affecting Form, Function and Interactions of VPS13, ATG2, SHIP164, Hobbit and Tweek. Contact (Thousand Oaks) 5, 251525642211343 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cutler R. G., Thompson K. W., Camandola S., Mack K. T. & Mattson M. P. Sphingolipid metabolism regulates development and lifespan in Caenorhabditis elegans. Mech Ageing Dev 0, 9–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pandey T. & Ma D. K. Stress-Induced Phenoptosis: Mechanistic Insights and Evolutionary Implications. Biochemistry (Mosc) 87, 1504–1511 (2022). [DOI] [PubMed] [Google Scholar]
- 49.Sutphin G. L. & Kaeberlein M. Measuring Caenorhabditis elegans life span on solid media. J Vis Exp 1152 (2009) doi: 10.3791/1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valente A. J., Maddalena L. A., Robb E. L., Moradi F. & Stuart J. A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem 119, 315–326 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Vozdek R., Long Y. & Ma D. K. The receptor tyrosine kinase HIR-1 coordinates HIF-independent responses to hypoxia and extracellular matrix injury. Sci Signal 11, (2018). [DOI] [PubMed] [Google Scholar]
- 52.Pandey T. et al. Anti-ageing and anti-Parkinsonian effects of natural flavonol, tambulin from Zanthoxyllum aramatum promotes longevity in Caenorhabditis elegans. Exp Gerontol 120, 50–61 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Gerstbrein B., Stamatas G., Kollias N. & Driscoll M. In vivo spectrofluorimetry reveals endogenous biomarkers that report healthspan and dietary restriction in Caenorhabditis elegans. Aging Cell 4, 127–137 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 Lipidomic profiling of phospholipids, glycerolipids and sphingolipids by LC-MS/MS.
272 mass pairs were monitored by multiple reaction monitoring (MRM) mode. Data were background corrected and normalized with internal standards. Each group had three replicates (n=3) and their respective average (ave), standard deviation (sd), and relative standard deviation (RSD (%)) were calculated. The data file is organized by lipid class/subclass and when applied, their respective fatty acid composition (FA); Triacylglycerides (TAG), hexaceramides (Hexcer), phosphatidylcholine (PC), phosphatidylglycerol (PG), phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylinositol (PI), lyso-phosphatidylethanolamine (LPE).