Abstract
Dephosphocoenzyme A (dephospho-CoA) kinase catalyzes the final step in coenzyme A biosynthesis, the phosphorylation of the 3′-hydroxy group of the ribose sugar moiety. Wild-type dephospho-CoA kinase from Corynebacterium ammoniagenes was purified to homogeneity and subjected to N-terminal sequence analysis. A BLAST search identified a gene from Escherichia coli previously designated yacE encoding a highly homologous protein. Amplification of the gene and overexpression yielded recombinant dephospho-CoA kinase as a 22.6-kDa monomer. Enzyme assay and nuclear magnetic resonance analyses of the product demonstrated that the recombinant enzyme is indeed dephospho-CoA kinase. The activities with adenosine, AMP, and adenosine phosphosulfate were 4 to 8% of the activity with dephospho-CoA. Homologues of the E. coli dephospho-CoA kinase were identified in a diverse range of organisms.
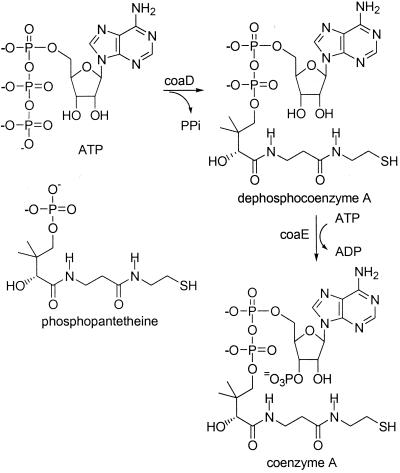
Coenzyme A (CoA) is an essential cofactor in a wide variety of biochemical pathways (1). It is estimated that about 4% of all enzymes use CoA or a thioester of CoA as a substrate (24). The final two steps in CoA biosynthesis are coupling of phosphopantetheine with ATP to form dephosphocoenzyme A (dephospho-CoA) and the subsequent phosphorylation of the 3′-hydroxyl group to form CoA (Fig. 1) (3). Genes coding for six of the seven enzymes involved in the biosynthesis of phosphopantetheine have been identified (10, 14, 20, 26). The final two reactions of CoA biosynthesis in mammalian cells were first attributed to a single bifunctional enzyme by Hoagland and Novelli (8). This was verified by Worrall and Tubbs, who purified a protein from pork liver that possessed both the phosphopantetheine adenylyltransferase and dephospho-CoA kinase activities (25). Subsequently, these two activities were shown to be part of a multifunctional enzyme complex in baker's yeast (5, 6). We first showed that the final two steps in CoA biosynthesis in Corynebacterium ammoniagenes (formerly Brevibacterium ammoniagenes) are catalyzed by distinct proteins that were readily separated by ion-exchange chromatography (12). The gene from Escherichia coli coding for phosphopantetheine adenylyltransferase was recently cloned, and the crystal structure of the enzyme was determined (7, 9). We report here purification of dephospho-CoA kinase from wild-type C. ammoniagenes and identification of the gene coding for a homologous protein in E. coli. This gene was cloned using PCR and overexpressed, and the resulting protein was purified. Enzyme assays and product characterization were used to show that the resulting recombinant protein is indeed dephospho-CoA kinase. The dephospho-CoA kinase gene is now designated coaE, based on the previous naming of the pantothenate kinase gene as coaA and of the phosphopantetheine adenylyltransferase gene as coaD (3, 7).
FIG. 1.
Final two steps of CoA biosynthesis (coaD, phosphopantetheine adenylyltransferase; coaE, dephospho-CoA kinase).
MATERIALS AND METHODS
Materials.
Dephospho-CoA, dilithium CoA, NADH, pyruvate kinase, lactic dehydrogenase, phosphoenolpyruvate, ATP, AMP, adenosine phosphosulfate (APS), DEAE Sepharose, and Q Sepharose were purchased from Sigma. A Sephacryl S-200 gel filtration column was purchased from Pharmacia. E. coli W3110 and C. ammoniagenes ATCC 6871 were obtained from the American Type Culture Collection. PCR primers were purchased from Integrated DNA Technologies, and the pEt 28b(+) vector was purchased from Novagen.
Enzyme and protein assays.
All enzyme assays were carried out at 25°C, and changes in absorbance at 340 nm were monitored. One unit of activity corresponded to the formation of 1 μmol of product/min using a molar absorbtivity for NADH of 6,220 M−1 cm−1. Dephospho-CoA kinase activity was measured as described previously (12). Each 1-ml assay mixture contained KCl (20 mM), MgCl2 (10 mM), ATP (10 mM), NADH (0.3 mM), phosphoenolpyruvate (0.4 mM), pyruvate kinase (10 U), and lactic dehydrogenase (4 U) in 50 mM Tris buffer (pH 8.5). After addition of the dephospho-CoA kinase sample, the reaction was initiated by adding dephospho-CoA (0.4 mM). Adenosine, APS, or AMP (0.4 mM) was substituted for dephospho-CoA in assays performed with these alternate substrates. To assay for CoA formation, each 1-ml assay mixture contained MgCl2 (10 mM), ATP (10 mM), acetyl phosphate (0.4 mM), phosphate acetyltransferase (2 U), and a dephospho-CoA kinase sample (0.02 U). The reaction was initiated by adding dephospho-CoA (0.4 mM), and thioester formation was observed at 240 nm (ɛ240 = 5.4 × 103 M−1 cm−1). Concentrations of dephospho-CoA solutions were measured by end point assays by using dephospho-CoA kinase. Kinetic parameters were determined by fitting the data to the equation for a sequential reaction mechanism by using Origin 50. For proof of product structure, a solution containing dephospho-CoA (2.2 mg, 3.0 μmol), ATP (0.1 μmol), MgCl2 (1.0 μmol), KCl (1.0 μmol), and phosphoenolpyruvate (3.5 μmol) in 5 ml of H2O was adjusted to pH 8.0, pyruvate kinase (5 U) and recombinant dephospho-CoA kinase (2 U) were added, and the solution was left at room temperature for 30 min. The CoA produced was purified by high-performance liquid chromatography on a preparative C18 column (21.4 mm by 25 cm; 5 min with 5% methanol in 10 mM phosphate buffer [pH 4.5], followed by a linear gradient to 40% methanol over 30 min) and had an elution time of 20 min. The product was lyophilized and redissolved in D2O for nuclear magnetic resonance (NMR) analysis. Protein determinations were carried out by the method of Bradford (4), using bovine serum albumin as the standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out with 12% polyacrylamide gels by using Tris-glycine buffer (11).
Purification of wild-type dephospho-CoA kinase.
C. ammoniagenes was grown at 26°C to an optical density at 650 nm of 1.7 in 2 liters of liquid medium containing (per liter) 10 g of glucose, 15 g of peptone, 3 g of dipotassium hydrogen phosphate, 2 of g of sodium chloride, 1 g of yeast extract, and 0.1 g of anhydrous magnesium sulfate. All subsequent steps were performed at 4°C. Cells were harvested by centrifugation, and the wet cell paste (16.9 g) was suspended in Tris buffer (50 mM Tris [pH 8.0], 5 mM dithiothreitol [DTT], 0.2 mM phenylmethylsulfonyl fluoride) to give a total volume of 60 ml. Cells were disrupted with a French press, and cellular debris was removed by centrifugation (10,000 × g, 20 min).
The supernatant (about 50 ml) was loaded onto a DEAE Sepharose column (2.5 by 10 cm) which had been preequilibrated with DEAE buffer (25 mM Tris [pH 8.0], 5 mM DTT). The column was washed with DEAE buffer containing NaCl (0.2 M). The column was eluted with a 300-ml linear NaCl gradient (0.2 to 0.5 M NaCl) in DEAE buffer. Dephospho-CoA kinase activity eluted at 0.37 M NaCl.
The active fractions were pooled (11 ml), and MgCl2 was added to a final concentration of 0.01 M. The resulting combined fraction was then applied to a Red A agarose column (1.5 by 4.5 cm) which had been equilibrated with Red A agarose buffer (0.02 M Tris [pH 8.0], 0.01 M MgCl2, 2 mM DTT). The enzyme was allowed to bind to the Red A agarose for 35 min, and the column was washed with Red A agarose buffer (20 ml). The column was washed with the same buffer (15 ml) containing ATP (2 mM), and then the column was allowed to stand for 1 h. Dephospho-CoA kinase was eluted from the column with Red A agarose buffer (15 ml) containing ATP (2 mM). The active fractions (6 ml) were combined and applied to a Q Sepharose column (1.5 by 2.5 cm) equilibrated with Q buffer (50 mM Tris [pH 7.5], 2 mM DTT). The column was washed with Q buffer (10 ml) and eluted with a gradient of the same buffer (100 ml) containing KCl (0 to 0.5 M). Dephospho-CoA kinase activity eluted at 0.32 M KCl. The active fractions were pooled and concentrated to 2 ml. A portion of the resulting sample was submitted for N-terminal sequence analysis.
Cloning and expression of the coaE gene.
The coaE gene was amplified by using E. coli W3110 genomic DNA as the template in a PCR. The forward primer included an Ala codon (GCC) inserted after the start codon to form an NcoI restriction site: GGA ACC ATG GCC AGG TAT ATA GTT GCC TTA. The reverse primer included an HindIII restriction site after the stop codon: GCG CAA GCT TAC GGT TTT TCC TGT GAG ACA. The amplification cycle was as follows: denaturation at 94°C for 45 s, annealing at 58°C for 45 s, and extension at 72°C for 2 min. Amplification was performed for 32 cycles and was followed by a final extension step at 72°C for 10 min. The resulting PCR product was subcloned into the NcoI-HindIII site of the pEt28b(+) expression plasmid. The resulting plasmid, designated pEt/coaE, was sequenced by using the T7 promoter primer. E. coli BL21(DE3) was transformed with pEt28b/coaE to obtain an overexpression strain. All steps were carried out by using standard molecular biology procedures (16).
Purification of recombinant dephospho-CoA kinase.
E. coli BL21(DE3)(pEt28b/coaE) was grown at 37°C in Luria-Bertani medium (500 ml) containing kanamycin (50 μg/ml). Isopropyl-β-d-thiogalactopyranoside (IPTG) (100 μM) was added when the optical density at 600 nm reached 0.6, and incubation was continued for 6 h at 37°C. Cells (4.5 g) were harvested by centrifugation and disrupted with a French press. Chromatography on DEAE Sepharose was performed as described above for the C. ammoniagenes enzyme with a 0.0 to 0.3 M NaCl gradient. Dephospho-CoA kinase activity eluted at 0.15 M NaCl. Active fractions were pooled and loaded onto a Q Sepharose column (1.5 by 10 cm) equilibrated with Q Sepharose buffer. The column was washed with Q Sepahrose buffer (30 ml) and eluted with a 150-ml linear KCl gradient (0.0 to 0.5 M KCl) in the same buffer. Dephospho-CoA kinase activity eluted at 0.20 M KCl.
RESULTS
Purification of dephospho-CoA kinase from C. ammoniagenes.
The initial steps for purification of dephospho-CoA kinase from C. ammoniagenes were performed as described previously (12). Dephospho-CoA kinase activity could not be measured in the crude cell lysate due to high background values but was readily measured in fractions resulting from chromatography of the crude lysate on DEAE Sepharose (Table 1). Active fractions from the DEAE column were then applied to a Red A agarose column. While most of the protein was not bound by the column, the dephospho-CoA kinase was bound after a few minutes of equilibration. The enzyme was selectively eluted by using a 2 mM solution of ATP, and almost 200-fold purification was obtained with this step. The enzyme fractions obtained in this way contained only minor impurities as determined by SDS-PAGE. Final purification was achieved by further chromatography on Q Sepharose. The purified enzyme produced one band at a molecular mass of about 25 kDa.
TABLE 1.
Purification of dephospho-CoA kinase from C. ammoniagenesa
| Purification step | Protein (mg) | Activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 1,600 | 3.6b | 0.0023 | ||
| DEAE Sepharose | 270 | 3.6 | 0.013 | 100 | 5.7 |
| Red A agarose | 1.2 | 3.0 | 2.5 | 83 | 1,100 |
| Q Sepharose | 0.2 | 1.3 | 6.5 | 36 | 2,800 |
The data were obtained with 16.9 g of wet cell paste.
Dephospho-CoA kinase activity could not be measured in the crude extract because of high background activity. The yield of DEAE Sepharose activity was thus assumed to be 100%.
Cloning, expression, and purification of recombinant dephospho-CoA kinase from E. coli.
The purified dephospho-CoA kinase obtained from C. ammoniagenes was subjected to 30 cycles of N-terminal sequencing, which resulted in the sequence MKIIG LIGGI GSGKT TVAQL FVDEG FPLVD. This sequence was used to search the nonredundant protein sequence databases using BLAST (2). The search revealed a homologous protein (60% identity, 74% positives) encoded by the yacE gene of E. coli (Fig. 2), as well as homologous proteins (up to 78% identical) from several other sources. The 206-amino-acid protein has a calculated molecular mass of 22,600 Da. The coaE gene from E. coli W3110 was amplified by PCR, and the resulting product was inserted into pEt28b(+). The resulting plasmid, designated pEt/coaE, was sequenced by using the T7 promoter primer. E. coli BL21(DE3) containing this plasmid expressed the dephospho-CoA kinase upon induction with IPTG. The recombinant dephospho-CoA kinase was initially purified by anion-exchange chromatography on DEAE Sepharose, like the wild-type C. ammoniagenes enzyme (Table 2). The resulting dephospho-CoA kinase activity was found to not bind to Red A agarose. The DEAE fractions were then purified further by anion-exchange chromatography on Q Sepharose. The recombinant dephospho-CoA kinase was found to be pure by SDS-PAGE. Twelve cycles of N-terminal sequencing of the recombinant protein gave the sequence ARYIVALTGGIG.
FIG. 2.
Alignment of protein sequences homologous to the E. coli dephospho-CoA kinase sequence. Six protein sequences exhibiting high homology to the E. coli dephospho-CoA kinase sequence identified from the WIT genomic database (15) were aligned by using ClustalW (21). Ec, E. coli (WIT database no. REC04326); Bs, Bacillus subtilis (RBS02900); Ss, Synechocystis sp. (RCY14763); Ce, Caenorhabditis elegans (RCE05784); Sc, Saccharomyces cerevisiae (RSC03337); Ta, Thermus aquaticus (sp/Q56416); Hs, Homo sapiens (RHS05130). Asterisks indicate identical residues, colons indicate highly conserved residues; and periods indicate less highly conserved residues. The kinase motif of the E. coli sequence is underlined.
TABLE 2.
Purification of recombinant dephospho-CoA kinase from E. coli BL21(DE3)(pEt28b/coaE)a
| Purification step | Protein (mg) | Activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 750 | 760 | 1.0 | 100 | |
| DEAE Sepharose | 63 | 390 | 6.2 | 51 | 6.2 |
| Q Sepharose | 13 | 290 | 22 | 38 | 22 |
The data were obtained with 4.5 g of wet cell paste.
Preliminary characterization of the recombinant dephospho-CoA kinase.
In addition to the standard assay for dephospho-CoA kinase activity based on ADP formation, an assay for CoA formation by the recombinant enzyme was performed by using phosphotransacetylase and acetyl phosphate with monitoring of thioester formation at 240 nm. This assay gave the same activity as the standard assay. A sample resulting from preparative phosphorylation of dephospho-CoA with the recombinant enzyme was subjected to 1H NMR analysis, and the spectrum obtained was identical to that of authentic CoA. The purified enzyme eluted from a calibrated Sephacryl S-100 column with an elution volume equivalent to a molecular mass of 22.2 ± 1.1 kDa, suggesting that the native enzyme exists as a monomer.
Kinetic constants for dephospho-CoA kinase were determined by measuring the initial rates obtained with different concentrations of ATP and dephospho-CoA. Km values of 0.74 and 0.14 mM were determined for dephospho-CoA and ATP, respectively. Activities were also observed when adenosine, APS, and AMP were used as substrates, and the activities observed were 4, 8, and 5% respectively, of the activity when 0.4 mM dephospho-CoA was used as the substrate. The recombinant dephospho-CoA kinase exhibited high activity over a broad pH range, and the maximum activity occurred at about pH 8.5.
DISCUSSION
C. ammoniagenes was chosen as a source of dephospho-CoA kinase for purification based on the higher activities of the enzymes of CoA biosynthesis in this bacterium than in E. coli and other microorganisms (18) and because of prior success in our lab with purification of phosphopantetheine adenylyltransferase and partial purification of dephospho-CoA kinase from this organism (12). Purification of the dephospho-CoA kinase turned out to be fairly simple, largely due to the high degree of purification achieved during the very selective elution of dephospho-CoA kinase from Red A agarose when ATP was used. A BLAST search using the N-terminal sequence of the purified enzyme suggested that dephospho-CoA kinase of E. coli was the protein encoded by yacE. The 30 amino acid residues determined for the Corynebacterium dephospho-CoA kinase were 60% identical to the predicted N-terminal amino acid sequence encoded by the E. coli gene (Fig. 2), and the molecular mass of the deduced yacE protein (22.6 kDa) was approximately the same as that of the purified dephospho-CoA kinase (about 25 kDa). While yacE and its homologues have been characterized as hypothetical kinases or ATP-binding proteins, no specific function of the yacE protein has been proposed previously. Cloning and expression of yacE, now renamed coaE, provided the recombinant dephospho-CoA kinase.
The enzyme APS kinase, which catalyzes phosphoryl transfer from ATP to the 3′-hydroxyl group of APS, was previously reported to accept dephospho-CoA as a substrate, although the specific activity was not reported (17). The dephospho-CoA kinase gene does not have significant homology to the APS kinase gene. APS and two other potential alternate substrates, adenosine and AMP, were tested as substrates for the recombinant dephospho-CoA kinase. All three of these compounds showed significant activity, although the activities were much less than that observed when dephospho-CoA was the substrate. These results and the NMR product analysis which showed that dephospho-CoA was phosphorylated on the 3′-hydroxyl group provide evidence that dephospho-CoA is the natural substrate for the recombinant enzyme. We are unaware of any enzymes other than dephospho-CoA kinase and APS kinase that catalyze 3′-phosphorylation of a nucleotide, although DNA and RNA polymerases catalyze 3′-nucleotidylation (24).
The Km determined for dephospho-CoA (0.74 mM) is much higher than the value reported for the dephospho-CoA kinase activity of the partially purified bifunctional enzyme from rat liver (0.01 mM) (19) and somewhat higher than the value previously measured for the C. ammoniagenes dephospho-CoA kinase (0.12 mM) (13). Like the rat liver enzyme, maximum activity is observed at a high pH. The combined subunit molecular weights of the E. coli phosphopantetheine adenylyltransferase (18,000) (7) and dephospho-CoA kinase (22,600) are somewhat less than the molecular weight of the bifunctional enzyme from pork liver (57,000) (25). There is no information concerning the structure of the pork liver enzyme or any other mammalian bifunctional enzyme that permits further analysis of similarities to or differences from the equivalent monofunctional enzymes from E. coli.
Surprisingly, coaE is not clustered with any genes related to CoA biosynthesis or metabolism in any of the microbial genomes sequenced. Notably, some chromosomal clustering is observed between coaE and some genes for NAD de novo biosynthesis. More than 60 homologues of the E. coli coaE gene product were identified by using the WIT system (15), and sequences from a range of organisms are shown in Fig. 2. The different sequences have several areas of high homology, including the Walker kinase motif (22, 23) (residues 9 to 17 of the E. coli enzyme). The homologues in Fig. 2 illustrate that the coaE gene is present in a wide range of organisms, including bacteria, fungi, and animals, although no plant species have been identified. Identification of a human homologue of the monofunctional dephospho-CoA kinase is somewhat surprising, given the evidence that there is a bifunctional phosphopantetheine adenylyltransferase–dephospho-CoA kinase in rat and pork livers (8, 25).
Identification of the dephospho-CoA kinase gene provides a major missing piece of information in the genetics of CoA biosynthesis. Of the nine enzymes required for CoA biosynthesis, only the gene for phosphopantothenoyl cysteine synthetase has not been identified yet (3). Identification of homologues of coaE should facilitate further studies of CoA biosynthesis in other organisms.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant GM45831. The NMR facility at Stony Brook is supported by National Science Foundation grant CHE9413510. The Center for Analysis and Synthesis of Macromolecules at Stony Brook is supported by National Institutes of Health grant RR02427 and the Center for Biotechnology.
We thank Zuzana Zachar, Stefan Tafrov, Ann Sutton, Rolf Sternglanz, and Peter Tonge for advice and technical assistance. We thank Chi-Huey Wong and coworkers for providing an APS kinase expression plasmid.
REFERENCES
- 1.Abiko Y. Metabolism of coenzyme A. In: Greenberg D M, editor. Metabolic pathways. 3rd ed. Vol. 7. New York, N.Y: Academic Press; 1975. pp. 1–25. [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begley T P, Kinsland C, Taylor S, Tandon M, Nicewonger R, Wu M, Chiu H-J, Kelleher N, Campobasso N, Zhang Y. Cofactor biosynthesis: a mechanistic perspective. Top Curr Chem. 1998;195:93–142. [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Bucovaz E T, Rhoades J L, Morrison J C, Fryer J E, Morrison W C, Whybrew W D. Characteristics of coenzyme A-synthesizing protein complex of bakers yeast. Fed Proc. 1977;36:876. [Google Scholar]
- 6.Bucovaz E T, Macleod R M, Morrison J C, Whybrew W D. The coenzyme A-synthesizing protein complex and its proposed role in CoA biosynthesis in bakers' yeast. Biochimie. 1997;79:787–798. doi: 10.1016/s0300-9084(97)86938-6. [DOI] [PubMed] [Google Scholar]
- 7.Geerlof A, Lewendon A, Shaw W V. Purification and characterization of phosphopantetheine adenylyltransferase from Escherichia coli. J Biol Chem. 1999;274:27105–27111. doi: 10.1074/jbc.274.38.27105. [DOI] [PubMed] [Google Scholar]
- 8.Hoagland M B, Novelli G D. Biosynthesis of coenzyme A from phosphopantetheine and of pantetheine from pantothenate. J Biol Chem. 1953;207:767–773. [PubMed] [Google Scholar]
- 9.Izard T, Geerlof A. The crystal structure of a novel bacterial adenylyltransferase reveals half of sites reactivity. EMBO J. 1999;18:2021–2030. doi: 10.1093/emboj/18.8.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kupke T, Uebele M, Schmid D, Jung G, Blaesse M, Steinbacher S. Molecular characterization of lantibiotic-synthesizing enzyme EpiD reveals a function for bacterial Dfp proteins in coenzyme A biosynthesis. J Biol Chem. 2000;275:31838–31846. doi: 10.1074/jbc.M004273200. [DOI] [PubMed] [Google Scholar]
- 11.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Martin D P, Drueckhammer D G. Separate enzymes catalyze the final two steps of coenzyme A biosynthesis in Brevibacterium ammoniagenes: purification of pantetheine phosphate adenylyltransferase. Biochem Biophys Res Commun. 1993;192:1155–1161. doi: 10.1006/bbrc.1993.1537. [DOI] [PubMed] [Google Scholar]
- 13.Martin D P, Bibart R T, Drueckhammer D G. Synthesis of novel analogs of acetyl coenzyme A: mimics of enzyme reaction intermediates. J Am Chem Soc. 1994;116:4660–4668. [Google Scholar]
- 14.Merkel W K, Nichols B P. Characterization and sequence of the Escherichia coli panBCD gene cluster. FEMS Microbiol Lett. 1996;143:247–252. doi: 10.1111/j.1574-6968.1996.tb08488.x. [DOI] [PubMed] [Google Scholar]
- 15.Overbeek R, Larsen N, Pusch G D, Dsouza M, Selkov E, Jr, Kyrpides N, Fonstein M, Maltsev N, Selkov E. WIT: integrated system for high-throughput genome sequence analysis and metabolic reconstruction. Nucleic Acids Res. 2000;28:123–125. doi: 10.1093/nar/28.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Satishchandran C, Taylor J C, Markham G D. The ORF1 of the gentamicin-resistance operon (aac) of Pseudomonas aeruginosa encodes adenosine 5-phosphosulfate kinase. Mol Microbiol. 1993;9:1223–1227. doi: 10.1111/j.1365-2958.1993.tb01251.x. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu S, Kubo K, Morioka H, Tani Y, Ogata K. Studies on metabolism of pantothenic acid in microorganisms. 9. Some aspects of enzyme activities involved in coenzyme A biosyntheis in various microorganisms. Agric Biol Chem. 1974;38:1015–1021. [Google Scholar]
- 19.Skrede S, Halvorsen O. Mitochondrial pantetheinephosphate adenylyltransferase and dephospho-CoA kinase. Eur J Biochem. 1983;131:57–63. doi: 10.1111/j.1432-1033.1983.tb07231.x. [DOI] [PubMed] [Google Scholar]
- 20.Song W J, Jackowski S. Cloning, sequencing, and expression of the pantothenate kinase (coaA) gene of Escherichia coli. J Bacteriol. 1992;174:6411–6417. doi: 10.1128/jb.174.20.6411-6417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson J D, Higins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Traut T W. The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide-binding sites. Eur J Biochem. 1994;222:9–19. doi: 10.1111/j.1432-1033.1994.tb18835.x. [DOI] [PubMed] [Google Scholar]
- 23.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webb E C. Enzyme nomenclature. San Diego, Calif: Academic Press; 1992. [Google Scholar]
- 25.Worrall D M, Tubbs P K. A bifunctional enzyme complex in coenzyme A biosynthesis: purification of pantetheine phosphate adenylyltransferase and dephospho-CoA kinase. Biochem J. 1983;215:153–157. doi: 10.1042/bj2150153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng R, Blanchard J S. Kinetic and mechanistic analysis of the E. coli panE-encoded ketopantoate reductase. Biochemistry. 2000;39:3708–3717. doi: 10.1021/bi992676g. [DOI] [PubMed] [Google Scholar]