This study characterizes the plasma cell and monoclonal antibody repertoire throughout the human colon at high resolution using matched single-cell V(D)J- and RNA-seq profiling, as well as antibody characterization of biopsies from patients with ulcerative colitis and healthy controls.
Abstract
Plasma cells (PCs) constitute a significant fraction of colonic mucosal cells and contribute to inflammatory infiltrates in ulcerative colitis (UC). While gut PCs secrete bacteria-targeting IgA antibodies, their role in UC pathogenesis is unknown. We performed single-cell V(D)J- and RNA-seq on sorted B cells from the colon of healthy individuals and patients with UC. A large fraction of B cell clones is shared between different colon regions, but inflammation in UC broadly disrupts this landscape, causing transcriptomic changes characterized by an increase in the unfolded protein response (UPR) and antigen presentation genes, clonal expansion, and isotype skewing from IgA1 and IgA2 to IgG1. We also directly expressed and assessed the specificity of 152 mAbs from expanded PC clones. These mAbs show low polyreactivity and autoreactivity and instead target both shared bacterial antigens and specific bacterial strains. Altogether, our results characterize the microbiome-specific colon PC response and how its disruption might contribute to inflammation in UC.
Introduction
Despite advances in ulcerative colitis (UC) treatment, many patients with poorly controlled disease still have to undergo colectomy (Tsai et al., 2020), and a better understanding of the disease is needed to identify new treatment targets. In particular, the role of microbiome-directed adaptive immunity in UC remains elusive (Neurath, 2019). Plasma cells (PCs) constitute a major fraction of lymphocytes in UC-associated inflammatory infiltrates, and their expansion is linked to the risk of disease recurrence (Bessissow et al., 2012). In addition, an Fc receptor variant (FcgR2) with decreased affinity to IgG1 is protective against UC in genome-wide association studies (GWAS) suggesting a role for IgG mAbs in the pathogenesis of UC (Bessissow et al., 2012; Castro-Dopico et al., 2019). Furthermore, recent RNA sequencing (RNA-seq) profiling has confirmed a significant expansion of IgG+ PCs in active UC compared with healthy controls (HCs) and suggested further B cell perturbations in UC such as the expansion of IFN-imprinted naive B cells (Uzzan et al., 2022). A striking feature of UC is the frequent coexistence of inflamed and unaffected mucosa within one patient (Smillie et al., 2019; Uzzan et al., 2022). Yet, the intraindividual differences in PC populations between these different disease states and colon areas within patients remain unknown. PCs in the human gut secrete grams of mAbs daily into the intestinal lumen (Spencer and Sollid, 2016) and some of these are thought to target colitogenic bacteria (Palm et al., 2014). However, the microbial targets, poly- and autoreactivity, of the PC repertoire throughout the human colon in health and UC also remains elusive.
Here, we used single-cell V(D)J- and RNA-seq profiling (sc(VDJ+RNA)-Seq) of HCs, UC patients in remission, and those with active inflammation to conduct an in-depth analysis of the mAb repertoire and matched PC expression states throughout different sites of the human colon. We show that areas of inflammation are associated with a shift from IgA to IgG, disruption of PC clonal architecture, upregulation of the unfolded protein response (UPR), and antigen presentation genes in PCs, and that IgG1 mAbs from inflamed colon PCs specifically target pathogenic bacterial strains.
Results
Single-cell profiling and V(D)J characterization of PCs throughout the colon in UC
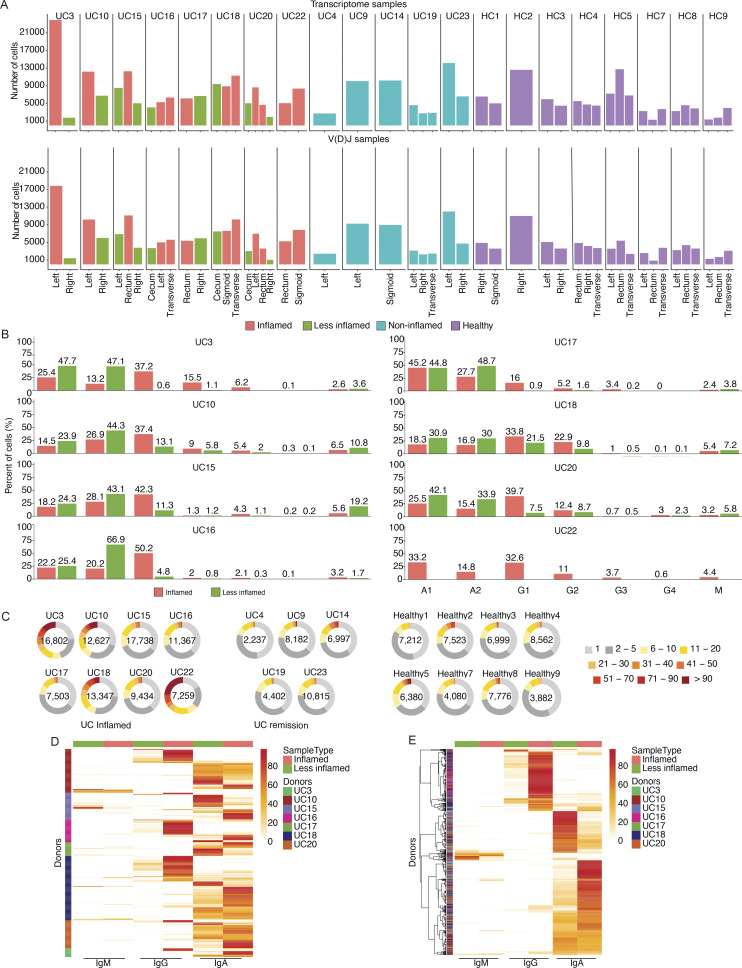
To understand the effect of UC on the colonic PC repertoire, we profiled by sc(VDJ+RNA)-Seq 181,124 PCs from 49 colon biopsies or mucosal resection samples from eight UC patients with at least one area of mucosal inflammation (12 inflamed and 9 matched less-inflamed samples from the same subjects), 5 UC patients in endoscopic and histologic remission (8 non-inflamed samples), and 8 HCs (20 healthy samples; Fig. 1 A and Fig. 2 A; Materials and methods). The 21 subjects had equal representation of gender (10 women, 11 men) and ranged in age from 28 to 75 yr; UC patients were diagnosed 1–20 yr prior to enrollment and were on different therapeutic regimens (Table S1). We digested colon samples to single-cell suspensions, performed FACS of CD27+CD38+ cells (5–20% of all cells; Fig. 1 B), and profiled sorted cells by 5′ directed single-cell RNA-seq (scRNA-seq) for both mRNA and paired V(D)J profiling (sc(VDJ+RNA)-Seq), recovering matched single-cell V(D)J and expression profiles, and yielding 2,237–16,802 high-quality cells per donor (Fig. 1 B and Fig. 2 A; Materials and methods).
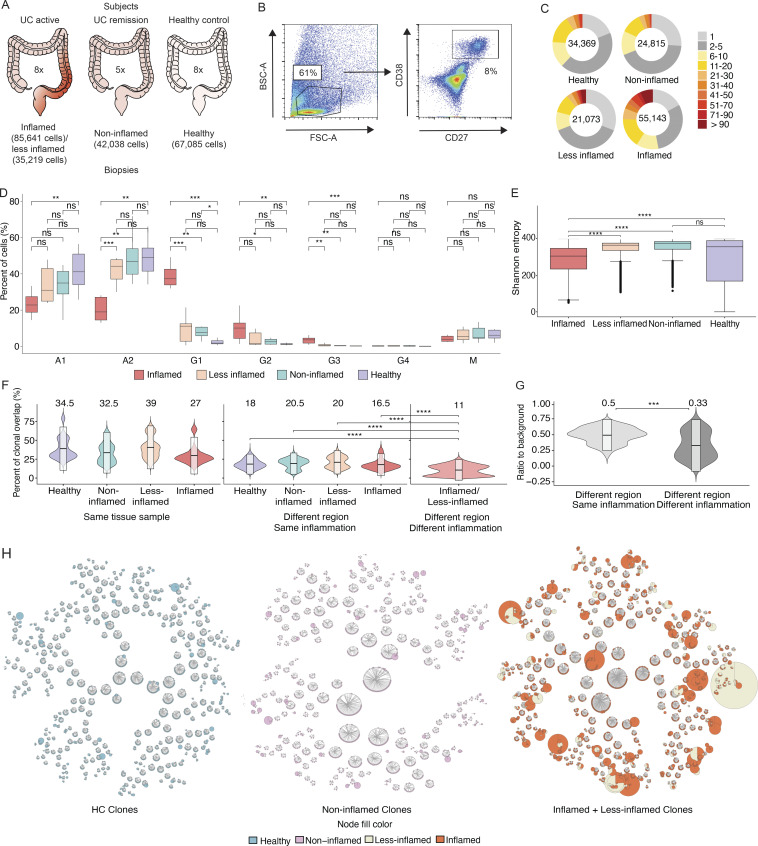
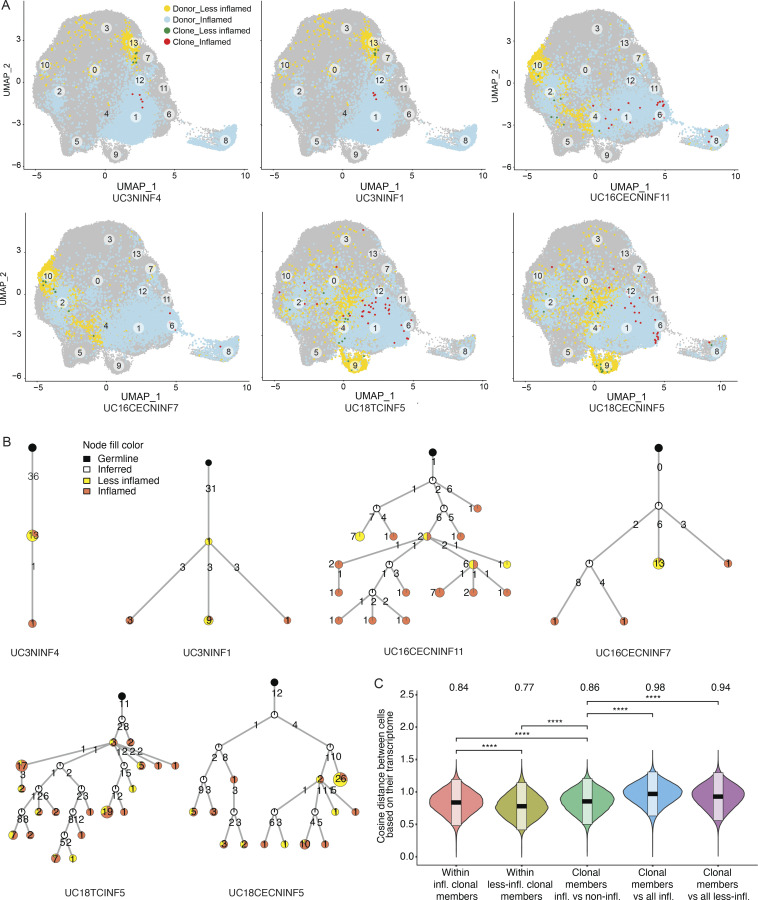
Figure 1.
Patient selection, PC sorting, isotype analysis, and clonal landscape. (A) Three patient categories recruited in this study with the number of subjects indicated in the center, respectively. Left: UC patients with inflammation; center: UC patients in remission; and right: HCs. The number of cells included in transcriptome analysis is indicated for each sample type. (B) Representative FACS plots with the gating strategy for the sorting of colon PCs. Cells isolated from digestion of colon biopsies and resection samples were gated on live cells based on their appearance in side scatter (BSC) and forward scatter (FSC). Of these, CD38-FITC and CD27-PE double-positive cells were selected for sorting and sequencing. (C) Pie charts show the expansion of differently sized PC clones for all samples grouped based on their inflammation status. Numbers in the center of the pie charts stand for the total number of cells analyzed in that particular plot. (D) Box plots displaying the distribution of the percentage of immunoglobulin isotypes (y axis) across samples grouped by their inflammation status (xaxis). Brackets indicate statistical significance using a one-sided non-parametric Wilcoxon test with *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, and ns indicating no statistical significance. Different samples taken from the same donor and with the same disease status are merged and represented as one data point in the distribution. (E) Box plots displaying the distribution of the Shannon entropy values of the PCs stratified by their inflammation status as indicated. Shannon entropy is a measure of population diversity, which is reversely related to the clonal expansion (Materials and methods). Brackets indicate statistical significance using a one-sided t test with ****P ≤ 0.0001, and ns indicating no statistical significance. (F) Violin plots displaying the distribution of the percentage of the shared clones between randomly sampled sets of PCs (n = 100; Materials and methods). Here, the two random samples of a specific donor that are to be evaluated for clonal overlap can belong to (i) same tissue sample (left), (ii) different colon regions with the same inflammation status (center), and (iii) different colon region with different inflammation status (right). The overlap between random samples that belong to different colon regions with the same inflammation status is significantly smaller (P value ≤ 0.0001, one-sided t test) than the random samples that belong to the same colon regions for all healthy, non-inflamed, less-inflamed, and inflamed groups. The overlap reduces significantly (P value ≤ 0.0001, one-sided t test) when the compared samples differ in their inflammation status. The boxes represent −2 SD, mean, and +2 SD. The values above each violin plot represent the median values of the distribution. (G) Distribution of the clonal overlap between randomly sampled PCs (n = 100) that are collected from different colon regions corrected for the background overlap percentages of the parent regions (Materials and methods). Bracket indicates statistical significance using a one-sided t test with ***P ≤ 0.001. The boxes represent −2 SD, mean, and +2 SD. The values above each violin plot represent the median values of the distribution. (H) Schematic representation of the B cell clones in healthy, non-inflamed, and inflamed samples, as indicated, show the clonal expansion and the isotype change in UC patients. Up to 50 of the largest clones from each donor are displayed. Each ring shows one distinct clone where the cells of the clone are depicted as individual nodes lined at the border of the clone ring and connected to each other by lines. The sizes of the nodes correspond to the number of the cells with the identical VDJ heavy chain sequence. The fill colors of the nodes indicate the inflammation status of the samples these particular cells belong to.
Figure 2.
Isotype analysis and clonal expansion. (A) Bar plots displaying the number of cells used in the downstream analysis (y axis) from various colon regions (x axis) of each subject. The upper graph shows the number of cells for which transcriptome profiling information was obtained and the lower graph shows the number of cells for which productive V(D)J sequence were obtained (Materials and methods). (B) Bar plots displaying the median percentage of each immunoglobulin isotype within the samples (y axis) of each donor with at least one area of inflammation stratified by the sample inflammation status as indicated (x axis). (C) Pie charts show the expansion of differently sized PC clones for each donor among UC patients with inflammation (left), UC patients in remission (middle), and HCs (right). Numbers in the center of the pie charts represent the total number of cells analyzed in that particular plot. (D and E) Heatmaps showing isotypes of expanded clones that are shared between inflamed and less-inflamed colon areas. The heatmaps display the percentage of each immunoglobulin isotype–inflammation status pair within each expanded clone with more than nine cells. The threshold of 10 cells was arbitrarily chosen to capture larger clones. Each row sums up to 100% and represents one expanded clone. Each column stands for the isotype–inflammation status pair. Heatmaps are organized based on donor (D) or enrichment in isotype–inflammation status pair (E).
A shift from IgA1/A2 to IgG1 PCs in inflammation
Consistent with early immunohistochemistry analyses (Baklien and Brandtzaeg, 1975) and prior single-cell atlases of the colon (Smillie et al., 2019; Uzzan et al., 2022), a significant skewing from IgA1 and IgA2 toward IgG1 occurred in inflamed samples, and to a smaller degree in less-inflamed and non-inflamed samples, when compared with HCs (Fig. 1 D; Materials and methods; tested with both multivariate test accounting for compositional dependencies and one-sided non-parametric Wilcoxon rank-sum test; P value <10−4). IgG1 was the most frequent isotype in PCs from inflamed samples, but IgG2 and IgG3 were also significantly elevated when compared with PCs from HCs (Fig. 1 D and Fig. 2 B; Materials and methods; tested with both multivariate test accounting for compositional dependencies and one-sided non-parametric Wilcoxon rank-sum test; P value <10−4). Thus, PCs in UC are significantly skewed toward IgG1 when compared with HCs, and this difference is most pronounced in inflamed tissue samples.
Clonal expansion, disruption of clonal architecture, and shift from Igk to Igl light chain usage in PCs from inflamed colon
Most (56–86%) B cells from the 21 subjects belonged to clones of three or more cells based on their V(D)J transcripts (Fig. 2 C). As an indication of significant clonal expansion, Shannon entropy and Simpson index decreased in PCs from inflamed biopsies compared with all other disease states (HCs, uninflamed biopsies from remission, and less-inflamed biopsies from subjects with active disease; Fig. 1 E and Fig. 2 C; inverse Simpson index not shown). Most of the 560 larger expanded clones (clone size >9 cells) that were present in both inflamed and matched less-inflamed colon areas from the same patient showed either IgG (166 of 560 clones) or IgA (375 of 560) dominance in inflamed areas or IgA (409 out of 560) dominance in the less-inflamed colon areas, and significantly maintained their dominant isotype across regions with different inflammation status with limited intraclonal crossover from IgG to IgA or vice versa (Spearman’s ρ = 0.4, P value < 2.2 × 10−16; Fig. 2, D and E; Data S1; Materials and methods).
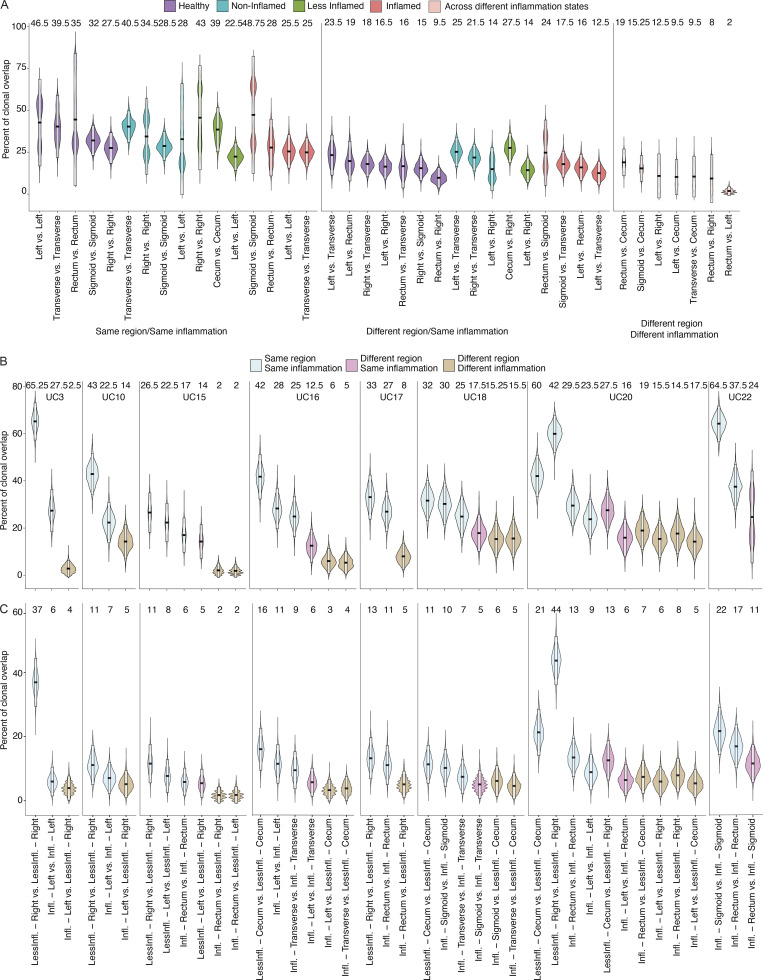
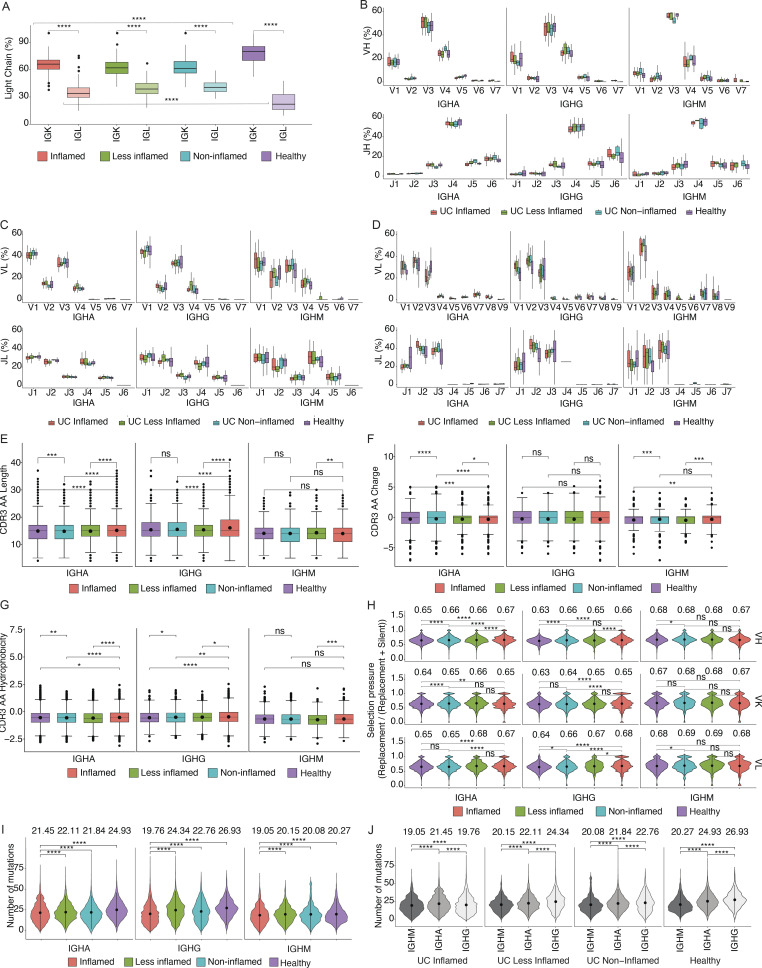
Estimating clonal overlap between randomly selected sets of 100 cells from the same sample or different colon regions within a patient (Materials and methods), the largest overlap was found among PCs within the same sample, followed by separate colon regions within a subject with the same inflammation state, where overlap was significantly higher than for regions within a subject with different inflammation states (Fig. 1, F and G; and Fig. S1, A and B; Materials and methods, one tailed t test, P value ≤ 10−3). This held true when expanded clones were collapsed to one cell to correct for different levels of clonal expansion (Fig. S1 C; Materials and methods). Identical clonal members (defined as a clonal member carrying the same heavy chain nucleotide sequence) were also significantly expanded in inflamed samples (Fig. 1 H). Although PCs expressing Igk light chains were more prevalent than Igl light chains in all samples, there was a significant shift from Igk to Igl usage in UC samples (inflamed, less-inflamed, or non-inflamed) compared with HCs (Fig. 3 A, one-sided non-parametric Wilcoxon rank-sum test, P value <10−4). VH and VL gene repertoires were consistent across antibody isotypes and disease states (Fig. 3, B–D; Materials and methods). Other antibody features such as CDRH3 length, charge and hydrophobicity, or inferred selection pressure (Materials and methods) differed across antibody isotypes and disease states, some reaching statistical significance (Fig. 3, E–H; Materials and methods). Consistent with prior studies (Uzzan et al., 2022), the level of somatic hypermutations was lower in PCs isolated from inflamed regions, and this difference was most pronounced among IgG+ cells (Fig. 3, I and J). Taken together, inflammation in UC is associated with increased clonal PC expansion including expansion of identical clonal members and disruption of the shared clonal landscape throughout different colon regions found in healthy subjects.
Figure S1.
Clonal overlap between PCs from different colon regions. (A and B) Violin plots display the distribution of the percentage of the shared clones between randomly sampled sets of PCs (n = 100; Materials and methods). In A, the two random samples of a specific donor that are to be evaluated for clonal overlap can belong to (i) same colon region (left), (ii) different colon regions with the same inflammation status (center), and (iii) different colon regions with different inflammation status (right) as indicated. In contrast to Fig. 1 F, each colon region is analyzed separately. Purple violin plots represent HC samples, turquoise plots represent non-inflamed samples from subjects in remission, green plots represent less-inflamed samples, and red plots represent inflamed samples in a UC patient with inflammation as indicated. Pink violins indicate samples that are being compared across different inflammation states. In B, samples are separated by the donor with blue violin plots representing overlap within the same colon region, pink different regions with the same inflammation status, and beige different regions with different inflammation status. For x-axis labeling, see C. (C) Violin plots displaying the distribution of the percentage of the shared clones between randomly sampled sets of 100 PCs (Materials and methods). As opposed to Fig. S1 B, expanded clones in each sample are collapsed into one cell. The two random samples of a specific donor that are to be evaluated for clonal overlap can belong to the same tissue sample (blue), different colon regions with same inflammation status (pink), and different colon regions with different inflammation statuses (beige). The boxes represent −2 SD (lower portion), mean (black line), and +2 SD (upper portion). The values above each violin plot represent the median values of the distribution.
Figure 3.
Colon PC antibody repertoire. Statistically significant differences are indicated with brackets. (A) Box plots display antibody light chain usage with kappa light chains shown in dark and lambda light chains in light colors. Antibodies are stratified by the disease state of the source tissue as indicated. Brackets indicate statistical significance using a one-sided non-parametric Wilcoxon rank-sum test, ****P ≤ 0.0001. (B–D) Box plots showing the V- and J-gene usage of antibody heavy chains (B), kappa light chains (C), and lambda light chains (D) as indicated. Red plots represent antibodies derived from PCs in inflamed tissue, green plots represent antibodies from less-inflamed tissue of UC patients with inflammation, turquoise plots represent antibodies from non-inflamed tissue in UC patients in remission, and purple plots represent antibodies from HCs. (E–G) Box plots displaying the CDRH3 amino acid length (E), CDRH3 amino acid charge (F), and CDRH3 amino acid hydrophobicity (G) of antibody heavy chains from PCs isolated from HCs (purple), non-inflamed tissue from UC patients in remission (turquoise), less-inflamed tissue from UC patients with inflammation (green), and inflamed tissue (red). Sequences were stratified based on antibody isotype. Brackets indicate statistical significance using a two-sided t test with **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, and ns, non-significant. (H) Violin plots showing selection pressure (nucleotide replacement mutation/nucleotide replacement + silent mutation) in antibody heavy chain V-genes (top row), kappa light chain V-genes (middle row), and lambda light chain V-genes (bottom row). Data are stratified based on the disease state of the source tissue and antibody isotype as indicated. Brackets indicate statistical significance using a one-sided Wilcoxon rank-sum test with *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001, and ns, non-significant. (I and J) Violin plots showing the distribution of the number of mutations in IgM, IgA, and IgG isotypes, stratified by the disease states. Black dots and lines display the mean values which are also written as text and ±1 SDs, respectively. Brackets display the P values obtained by two-sided Wilcoxon rank-sum test with ***P ≤ 0.001 and ****P ≤ 0.0001.
Cell-intrinsic expression changes in colon PCs during inflammation
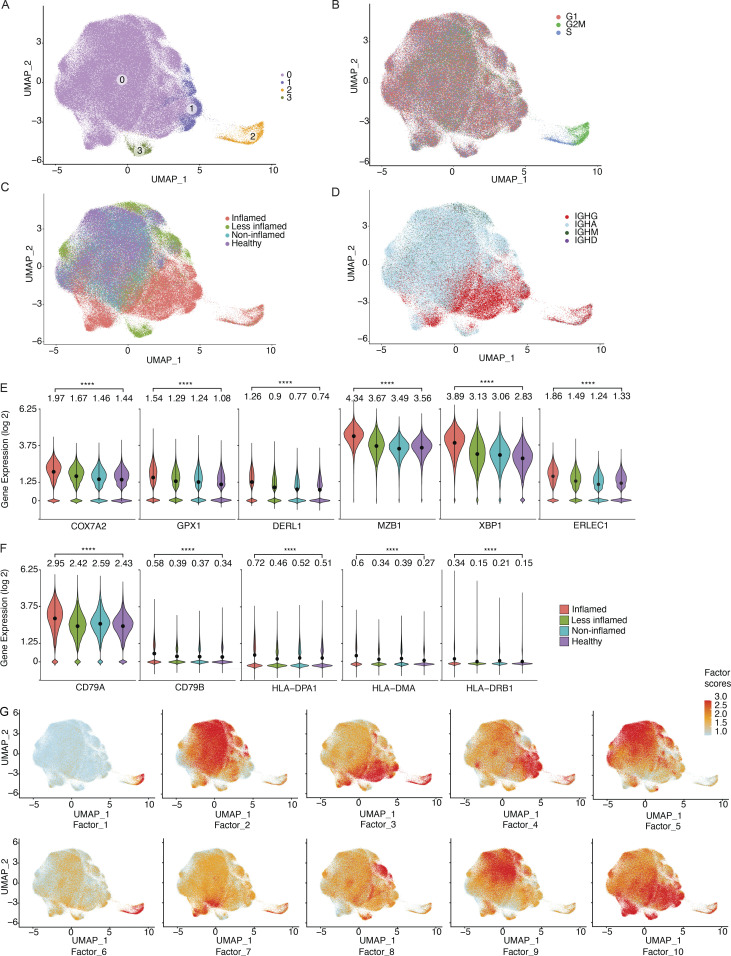
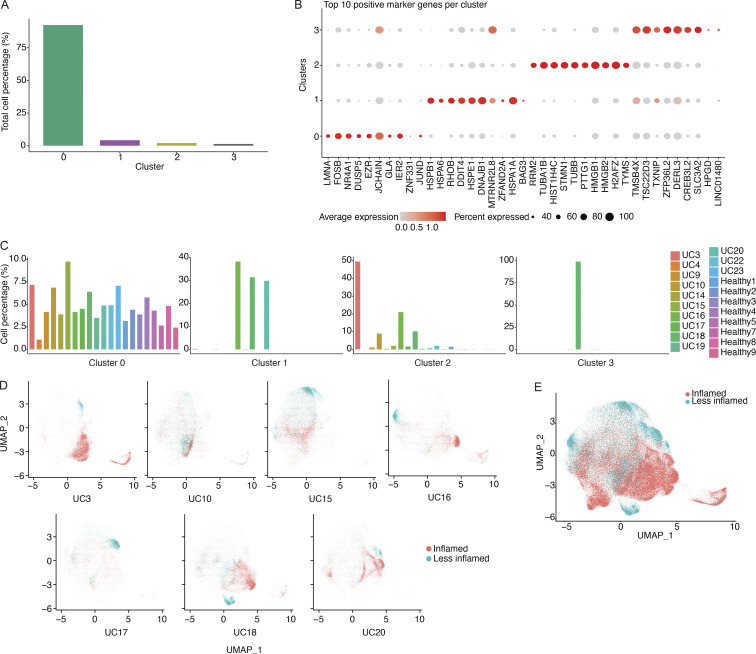
The scRNA-seq profiles of the 181,124 sorted PCs were partitioned into four main clusters (TCs), with one cluster (cluster 0) shared across all subjects (Fig. 4 A and Fig. S2, A–C). Marker genes for this cluster include NR4A1 and EZR, both of which are involved in the response to B cell receptor–antigen interactions (Tan et al., 2020; Treanor et al., 2011; Fig. S2 B). Cluster 1, with marker genes HSPA1A and HSPB1 that are upregulated in cellular stress (Garrido et al., 2006), and cluster 3, with marker genes DERL3 and SLC3A2 that are involved in the UPR (Hiller et al., 1996; Liu et al., 2018), were specific to a small number of subjects (Fig. S2 C); cluster 2 was enriched in cells in cell cycle phases G2 and S (Fig. 4 B). We observed a visually apparent separation between cells from inflamed vs. all other samples (Fig. 4 C), even when restricting only to cells from subjects with matched inflamed and less-inflamed samples (Fig. S2, D and E). Consistent with our PC isotype analysis above, IgG was enriched in clusters associated with inflammation (Fig. 4 D).
Figure 4.
Transcriptional characteristics of PCs from inflamed colon areas. (A) UMAP plot showing the cell embeddings based on the transcriptome. PCs are grouped into four clusters based on their transcriptomes. (B) Same UMAP as in A, but cells are colored based on their cell cycle phase G1 (red), G2M (green), or S (blue), as predicted by the cellular expression levels of the cell cycle genes (Butler et al., 2018; Stuart et al., 2019). (C and D) UMAP plots showing the cell embeddings colored by the (C) inflammation status of the tissue the cells were isolated from and (D) the antibody isotype of the cells. (E and F) Selected example genes involved in oxidative stress and UPR pathway (E) and antigen presentation (F) that are significantly differentially expressed (pseudobulk DE analysis, FDR < 0.05) between UC-inflamed and healthy samples. The violin plots display the distribution of the gene expression values per cell. Stars indicate the statistical significance of the non-parametric Wilcoxon rank sum test (****P ≤ 0.0001). The values above each violin plot and dots in each violin plot indicate the mean value of the distribution. (G) Latent factors that generate the main sources of variation in the transcriptome. We identified 10 latent factors that generate the variation in the transcriptome of the investigated colon PCs (Fig. S4). The 10 UMAP plots display the cells colored by the factor scores (Materials and methods) for each of the 10 factors.
Figure S2.
Transcriptional clusters of colon PCs. (A) Bar diagram displaying the fraction of all cells (in %) represented in each of the four transcriptional clusters. (B) Dot plot showing the relative expression of the top marker genes of each cluster (x axis) across all clusters (y axis). Each dot encodes both the detection rate and average gene expression in detected cells for a gene in a cluster. Darker color indicates higher average gene expression from the cells in which the gene was detected, and larger dot diameter indicates that the gene was detected in a greater proportion of cells from the cluster. (C) Bar diagram showing the representation of each of the four transcriptional clusters among all study subjects (x axis) as indicated. Each plot represents the indicated cluster and the fractions add up to 100% in each plot. (D) UMAP plots showing the cell embeddings colored by the inflammation status of the tissue the cells were isolated from for all subjects that had both inflamed and less-inflamed tissue. (E) UMAP plot showing all the cell embeddings shown in D but merged into one plot.
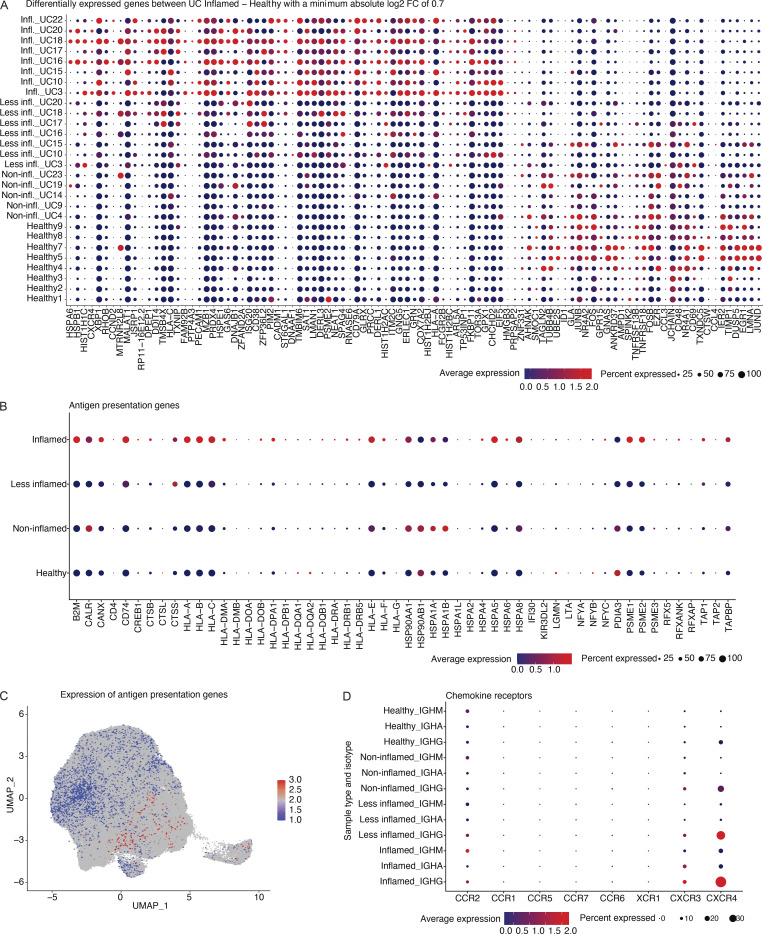
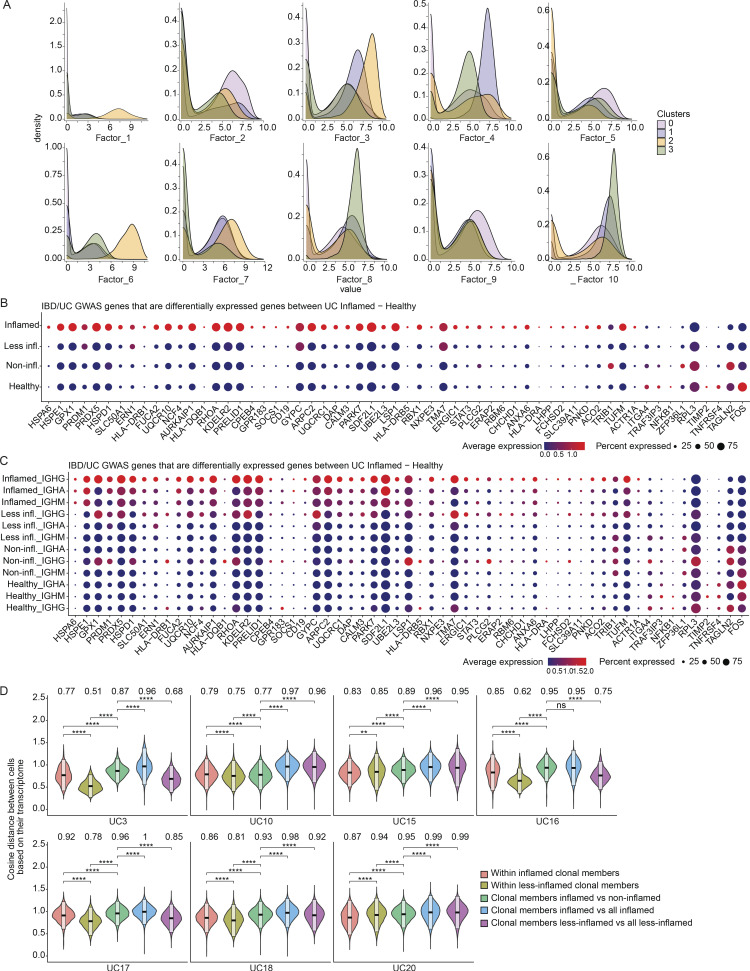
Genes differentially upregulated in PCs from inflamed compared with healthy colon areas across all cells (Fig. S3 A; Materials and methods; pseudobulk differential expression [DE] analysis, false discovery rate [FDR] < 0.05) included: COX7A2, upregulated in antibody-secreting PCs undergoing oxidative metabolism (Price et al., 2018); GPX1, encoding an intracellular antioxidant enzyme upregulated in response to oxidative stress (Bonifati et al., 2003; Dubuisson et al., 2004; Lubos et al., 2011); and ER stress-related genes DERL1, MZB1, XBP1, ERLEC1, and TMBIM6 (Cyster and Allen, 2019; Gaudette et al., 2020; Lisak et al., 2015; Minnich et al., 2016; Misiewicz et al., 2013), consistent with increased ER expansion to meet the demands of immunoglobulin secretion in PCs from inflamed areas (Fig. 4 E; pseudobulk DE analysis, FDR < 0.05; Fig. S3 A and Table S2). Several genes involved in antigen uptake and presentation (CD79A, CD79B, and several MHCII genes) were upregulated in PCs from inflamed areas (Fig. 4 F; pseudobulk DE analysis, FDR < 0.05; Fig. S3, B and C), possibly mirroring the role of PCs in celiac disease as antigen-presenting cells (Høydahl et al., 2019). CXCR4, encoding an important chemokine receptor for PC homing and survival (Cyster, 2003), was also differentially expressed only in IgG+ PCs from inflamed regions, consistent with prior findings in gut PCs from HIV and Crohn’s disease (Buckner et al., 2014; Fig. S3, A and D). Non-negative matrix factorization (NMF) of the scRNA-seq profiles of all 14 donors identified 10 programs (latent factors) underlying the variation across the cells (Materials and methods), which we annotated based on high-loading genes (Fig. S4 A; and Tables S3 and S4). The programs for cell division (Program 1), cellular biosynthetic processes and oxidative stress (Program 3), UPR response (Program 4), and MHC-II-expression (Program 7) increased in cells from inflamed regions (Fig. 4 G, Fig. 5 A, and Fig. S4 B). Multiple genes associated with inflammatory bowel disease (IBD) or UC through GWAS (Buckner et al., 2014; Huang et al., 2017; de Lange et al., 2017; Liu et al., 2015) are highly expressed in IgG+ PCs from UC-inflamed tissue, including genes related to oxidative stress (GPX1, PRDX5, and PARK7; Dubuisson et al., 2004; Lubos et al., 2011) and ER processes (KDELR2, SDF2L1; Sasako et al., 2019; Trychta et al., 2018; Fig. 5, B and C; Materials and methods; pseudobulk DE analysis, FDR < 0.05). In particular, SDF2L1 is upregulated during the acute UPR and functions as an ER chaperone to facilitate protein cargo secretion (You et al., 2021), and a coding variant in SDF2L1 was associated with Crohn’s disease risk in a recent large-scale whole exome sequencing study in Crohn’s disease patients (Sazonovs et al., 2021). Overall, genes associated with elevated antibody secretion, surface BCR expression, antigen presentation, and cell division are upregulated in PCs from inflamed colon areas.
Figure S3.
Genes that are higher expressed among PCs from inflamed tissue. (A) Dot plot showing the relative expression of the significantly differentially expressed genes when comparing PCs derived from inflamed tissue with PCs from HCs (x axis) across all subjects and disease states (y axis; pseudobulk DE analysis, FDR < 0.05). Each dot encodes both the detection rate and average gene expression in detected cells for a gene in a cluster. As indicated, dark red color indicates higher average gene expression from the cells in which the gene was detected, and larger dot diameter indicates that the gene was detected in a greater proportion of cells from the cluster. (B) Dot plot showing the relative expression of the antigen presentation genes (x axis) in PCs derived from tissue in different disease states as indicated (y axis). Color and size coding as in A. (C) UMAP plot showing the cell embeddings based on the transcriptome. Cells are colored based on their antigen presentation genes’ expression scores (Materials and methods), where PCs highlighted in dark red show the highest levels of expression. (D) Dot plot showing the relative expression of chemokine receptor genes (x axis) in PCs derived from tissue in different isotypes and disease states as indicated (y axis). Color and size coding as in A and B.
Figure S4.
Latent factor analysis. (A) Dot plots showing the top 15 gene ontology biological processes with the highest gene ratios in each of the 10 latent factors that were identified (Materials and methods). The size of the dots represents the number of genes in the significant gene list associated with the gene ontology term and the color of the dots represent the P-adjusted values. (B) Violin plots comparing the factor loadings between PCs from inflamed, less-inflamed, non-inflamed, and HC samples for 10 latent factors. ***P ≤ 0.001 and ****P ≤ 0.0001 using a one-sided t test.
Figure 5.
Expression levels of genes that are associated with IBD or UC through GWAS. (A) Stacked density plots displaying the distribution of cell factor scores stratified by the expression clusters. (B and C) Dot plots showing the relative expression of the selected genes (Buckner et al., 2014; Huang et al., 2017; de Lange et al., 2017; Liu et al., 2015) when comparing PCs derived from inflamed tissue with PCs from less-inflamed, non-inflamed, and HCs (y axis). In C, each disease state is stratified by antibody isotype. Each dot encodes both the detection rate and average gene expression in detected cells for a gene in a cluster. As indicated, the dark red color indicates higher average gene expression from the cells in which the gene was detected, and a larger dot diameter indicates that the gene was detected in a greater proportion of cells from the cluster. (D) Comparison of the pairwise cosine distances between PCs based on their transcriptome for each subject with inflamed and less-inflamed samples. Groups from left to right display the PC pair distance distributions between (i) inflamed clone members, (ii) less-inflamed clone members, (iii) inflamed and less-inflamed clone members, (iv) inflamed clone members and 100 randomly selected PCs of the donor that are inflamed and are not clone members, (v) clone members of less-inflamed samples and 100 randomly selected PCs of the donor that are less-inflamed and are not clone members. Brackets indicate statistical significance using a one-tailed t test with **P ≤ 0.01, **P ≤ 0.001, ****P ≤ 0.0001, and ns, non-significant.
PC expression profiles follow inflammation status rather than clonal membership
Examining the expression profiles of all cells from six representative PC clones from three donors with both inflamed and less-inflamed colon areas (Fig. 6, A and B; and Table S5), we found that PCs of the same clone separated mostly based on the inflammation status of their corresponding sample type. In all subjects with inflamed/less-inflamed samples, the pairwise cosine distances of PC expression profiles were significantly larger between clonal members across inflammation states compared with clonal members within inflamed or less-inflamed regions (Fig. 6 C; one tailed t test, P value ≤ 10−4; Materials and methods). With one exception (UC10), this was the case for each individual subject with matched inflamed and less-inflamed samples (Fig. 5 D; one tailed t test, P ≤ 10−4; Materials and methods). Thus, expression differences between PCs from inflamed and less-inflamed areas of the colon are apparent even for members of the same PC clone, suggesting that the local tissue environment is a dominant factor impacting their expression profiles.
Figure 6.
Transcriptomics of selected PC clones. (A) UMAP plots highlighting clonal members of six selected PC clones from three different subjects as indicated (Table S5). PCs from inflamed and less-inflamed colon areas from the respective subject are highlighted in blue and yellow, respectively. Red dots indicate clonal members belonging to the selected clone from inflamed colon areas and green dots from less-inflamed colon areas. (B) Phylogenetic trees summarize the clonal relationship of all members within the selected clones. Trees are rooted on a theoretical germline member (black node), uncolored nodes indicate inferred intermediates, and yellow and orange node colors indicate clonal members from less-inflamed or inflamed colon areas, respectively. Numbers on the connecting lines indicate the number of heavy chain mutations separating two nodes. (C) Comparison of the pairwise cosine distances between PCs based on their transcriptome. Groups from left to right display the PC pair distance distributions between (i) inflamed clone members, (ii) less-inflamed clone members, (iii) inflamed and less-inflamed clone members, (iv) inflamed clone members and 100 randomly selected PCs of the donor that are inflamed and are not clone members, (v) clone members of less-inflamed samples and 100 randomly selected PCs of the donor that are less-inflamed and are not clone members. There is a significant difference between transcriptional distances between clone members based on their inflammation type. Brackets indicate statistical significance using a one-tailed t test with ****P ≤ 0.0001.
Antibodies from PCs in inflamed colon regions are not polyreactive
Polyreactivity, defined as non-specific binding to unrelated antigens, is selected against throughout B cell development (Wardemann et al., 2003), but can arise during antibody affinity maturation (Tiller et al., 2007). The level of antibody polyreactivity in the human colon PC repertoire is unknown. Mouse-derived antibodies from small intestine PCs reportedly have high levels of polyreactivity (Bunker et al., 2017), whereas antibodies isolated from PCs in the small intestine from both HCs and inflamed regions in Crohn’s disease patients display lower levels of polyreactivity (Benckert et al., 2011; Kabbert et al., 2020). Similarly, autoreactivity is selected against during B cell development but can be generated during affinity maturation in germinal center reactions (Tiller et al., 2007) and is increased in peripheral B cell compartments of systemic lupus erythematosus patients (Wardemann et al., 2003).
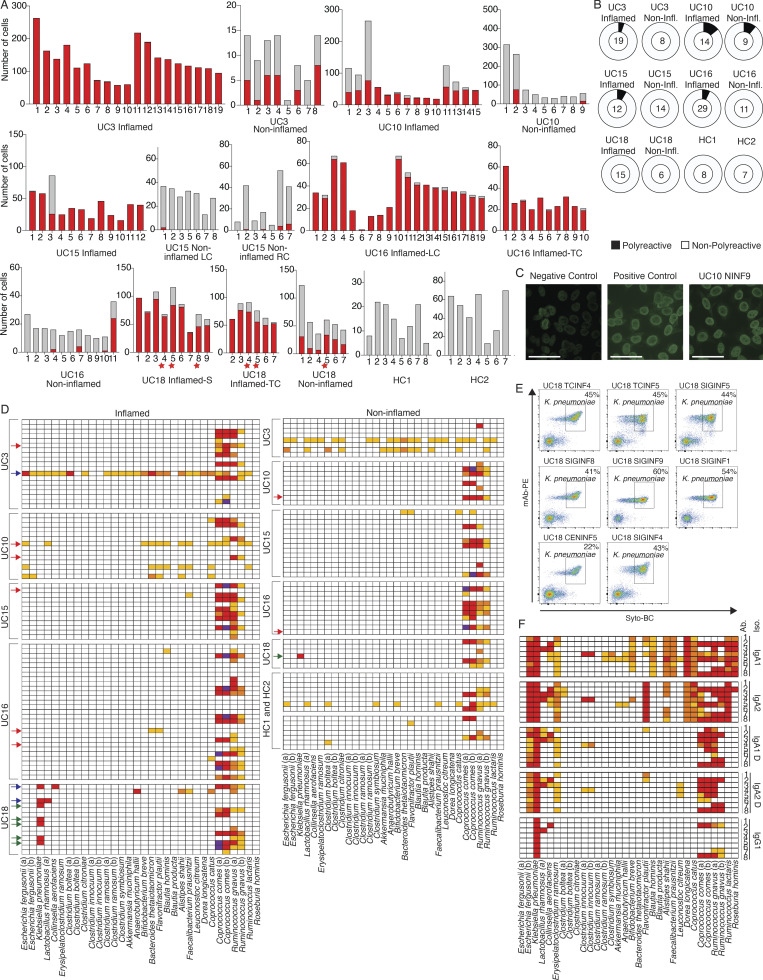
To evaluate polyreactivity and autoreactivity in mAbs from colon PCs in UC patients, we selected 152 mAbs from expanded clones from five UC patients and two HCs (Fig. 7 A and Table S5) and produced them as IgG1 mAbs irrespective of their original isotype to directly compare Fab reactivity. We defined polyreactivity as reactivity against at least two of the following antigens: single-stranded DNA, double-stranded DNA, LPS, insulin, or cardiolipin (Kabbert et al., 2020; Tiller et al., 2007; Fig. 7 B). We defined autoreactivity as binding against HEP-2 cells (Fig. 7 C; Tiller et al., 2007). We detected polyreactivity only in 6 of 89 (6.7%) antibodies from inflamed colon areas, 2 of 39 (5%) antibodies from less-inflamed colon areas, and 0 of 15 (0%) antibodies from HCs; the frequency of polyreactive antibodies was not significantly different between PCs from inflamed colon compared with HCs (P = 0.3, χ2 test; Fig. 7 B and Table S6). Polyreactivity rates were lower than those in mAbs isolated from HC human memory B cells (Tiller et al., 2007; overall 22.7%, P < 0.005, χ2 test) or in mouse colon-derived antibodies (Bunker et al., 2017; 29%, P < 0.005, χ2 test). Consistent with the low polyreactivity, only 1 of 152 mAbs (UC10NINF9) showed reactivity against HEP-2 cells (Fig. 7 C). Only 2 of 152 mAbs (UC16INF4 and UC16TNINF8), both originating from the same subject, showed reactivity against the recently described UC-associated autoantigen αVβ6 (Table S6; Kuwada et al., 2021; Uzzan et al., 2022), suggesting that, although frequently detected in the serum of UC patients (Kuwada et al., 2021), antibodies targeting this antigen are rarely found among local colon PCs in UC.
Figure 7.
Functional testing of selected mAbs. (A) Bar plots showing the clone size (number of cells, y axis) of each selected and tested mAb (x axis, Table S5) as well as its expansion in inflamed (red bar) or less-inflamed (gray bar) colon areas. mAbs are grouped based on the donor and colon area they were isolated from. The numbers below each clone correspond to the antibody names in Table S5. LC, left colon; RC, right colon; TC, transverse colon; S, sigmoid colon. Red stars indicate clones from which K. pneumoniae–binding mAbs were isolated (see below). (B) Pie charts summarizing the polyreactivity of all selected and tested mAbs. The number in the center of each pie indicates the number of mAbs tested and mAbs are grouped based on the donor and if they were isolated from an inflamed or less-inflamed colon area (Table S6). Polyreactivity was assessed by ELISA (Materials and methods) and experiments were repeated twice. (C) Representative HEP-2 cell IFA staining patterns of positive control and negative control serum as well as reactive mAb UC10NINF9 are shown. Scale bars represent 20 μm. (D) Heatmap showing the binding of all 152 selected mAbs against a panel of 32 bacterial strains in FACS. Each row represents one mAb and mAbs are sorted based on the donor and inflammation status of the colon region they were isolated from as indicated. White, <1% reactivity; light orange, 1–5% binding; dark orange, 5.1–10% binding; red, 10.1–50% binding; and purple, >50% binding. Red arrows indicate polyreactive mAbs (see above), blue arrows indicate mAbs with strong binding (>10%) to at least two different bacterial strains, and green arrows indicate mAbs only binding to K. pneumoniae. FACs experiments were repeated twice. (E) Representative FACS plots show the binding of reactive mAbs against bacterial strains used in D. Antibody binding was detected using a mouse anti-human IgG antibody coupled to PE, and bacteria were stained with SYTO-BC to exclude dead bacteria (Materials and methods). (F) Heatmap showing the binding of mAbs UC18CECNINF5 (1), UC18SIGINF1 (2), UC18SIGINF4 (3), UC18SIGINF5 (4), UC18SIGINF8 (5), UC18SIGINF9 (6), UC18TCINF4 (7), and UC18TCINF5 (8) in their IgG1, IgA1, and IgA2 forms as indicated against a panel of 32 bacterial strains in FACS. Color coding as in D. FACs experiments were repeated twice.
Colon PCs produce mAbs with strong binding to some bacterial strains
Sequencing of IgA-covered microbiota revealed preferential targeting of colitogenic bacteria (Palm et al., 2014), and PCs isolated from the small intestine of healthy subjects and Crohn’s disease patients show both specific and cross-species reactivity (Kabbert et al., 2020). However, evaluating specific antibody–microbiota interactions in the context of a complex patient-derived community is technically challenging because of the differences between stool samples and non-specific interactions, such as polyreactivity and binding to B cell superantigens (Bunker et al., 2019).
We, therefore, tested each of the 152 mAbs for their binding to single strains of a 32-strain bacterial panel that we assembled based on common members of the human microbiome and strains enriched in the microbiome of IBD patients (Franzosa et al., 2019; Ha et al., 2020; Hall et al., 2017; Lewis et al., 2015; Lloyd-Price et al., 2019; Shaw et al., 2016; Fig. 7 D). Consistent with prior reports of VH3-encoded heavy chains binding to a superantigen on Coprococcus comes as well as Ruminococcus gnavus (Bunker et al., 2019), 54 of 57 VH3-encoded antibodies showed strong binding against at least one of these strains (Fig. 7 D), and 54 of 60 antibodies with strong binding carried a VH3 heavy chain. Cross-strain reactivity with strong binding (>10% binding in FACS) to two or more bacterial strains (not counting non-specific superantigen binding as described above) was only detected in 3 of 152 antibodies (Fig. 7 D), none of which were polyreactive. Aside from R. gnavus and C. comes binding, six of 152 antibodies showed strong binding to a single bacterial strain, Klebsiella pneumoniae (Fig. 7, D and E). All K. pneumoniae–binding mAbs were isolated from one UC patient (UC18; Fig. 7, D and E; and Table S1), and each of these mAbs represents an expanded PC clone that spans multiple colon regions with preferential expansion in inflamed regions (Fig. 7 A; red stars).
To test if our 152 mAbs bind to stool bacteria, we grouped them into 12 mAb pools, including pools of VH3 and non-VH3 mAbs from inflamed, non-inflamed, and HC colon-derived PCs, as well as one mAb pool with all polyreactive mAbs (Table S7), and tested for binding against stool from C57BL/6J and RAG-1–deficient mice (to avoid crossdetection of IgG-bound bacteria in human stool samples). mAb-binding frequencies were low (0.3–4.1%, Fig. S5 A), consistent with the observed low level of polyreactivity and cross-strain reactivity (above).
Figure S5.
Binding of mAbs and mAb mixes to stool from RAG1-deficient and C57BL/6 mice and bacterial proteins and extracts. (A) FACS plots display SYTO-BC (x axis) and mAb mix staining (y axis) of stool from female and male RAG1-deficient and C57BL/6 mice with the double-positive population indicated through gating. Antibody binding was detected through a mouse anti-human IgG antibody coupled to PE (Materials and methods). The composition of each antibody mix used is summarized in Table S5 and bacterial staining was conducted so that each mAb was present at a concentration of 10 μg/ml. FACs experiments were repeated twice. (B) Table summarizing the fluorescence intensity values as measured on a GenePix 4000B imager (Axion) for the mAbs that showed binding in a screen of all 152 mAbs for binding to 50 bacterial lysates and antigens (Table S7 and Materials and methods). Values above 1,000 are considered positive and highlighted in red. (C) FACS plots display SYTO-BC (x axis) and mAb staining (y axis) of stool from RAG1-deficient mice with the double-positive population indicated through gating. K. pneumoniae–binding mAbs UC18CECNINF5, UC18SIGINF1, and UC18SIGINF5 in IgG1, IgA1, and IgA2 forms were used for staining as indicated and their binding was detected with mouse anti-human IgG or mouse anti-human IgA antibody coupled to PE (Materials and methods). FACs experiments were repeated twice.
Further testing of the 152 mAbs against an array of 50 bacterial lysates (Christmann et al., 2015; Materials and methods; Table S8) showed that six of eight mAbs binding to K. pneumoniae in FACS also bound K. pneumoniae lysates, two of them with at least eightfold lower crossreactivity against Enterococcus faecalis extract, Bacteroides thetaiotaomicron extract, and Bacteroides caccae extract (Fig. S5 B and Table S8). Another antibody from UC-inflamed tissue bound B. caccae extract, two bound elongation factor Tu from Bacteroides fragilis, and one bound Prevotella intermedia extract (Fig. S5 B and Table S8). None of these binding mAbs were polyreactive (Table S6 and Fig. 6 B). Finally, none of the 152 antibodies strongly bound common enteric viruses, including CMV, EBV, and rotavirus in ELISA (Table S6). We conclude that mAb–microbiota interactions from local PCs include specific interactions with K. pneumoniae as well as non-specific superantigen interactions with C. comes and R. gnavus.
IgA binds non-specifically to several microbiota
A significant fraction of microbiota in stool from mice and humans is bound by endogenously produced IgA (Palm et al., 2014), but how much of that binding activity reflects canonical Fab interactions with microbial surface antigens vs. non-canonical binding is unknown (Pabst and Slack, 2020).
To test if isotype switching changes the binding activity of antibodies to microbial targets, we chose three of our mAbs with known specificity to K. pneumoniae (see above) and tested their IgG1, IgA1, or IgA2 monomeric and dimeric forms for binding to stool from RAG-1–deficient mice (Fig. S5 C). While IgG1 forms of all three mAbs showed no binding, the same antibodies expressed as IgA showed increased binding to the stool samples tested, ranging from 1 to 5.9% (Fig. S5 C). To test if antibody isotype changes from either IgG1 to IgA1 or IgA2 lead to increased bacterial reactivity in mAbs isolated from human colon PCs, we expressed all eight K. pneumoniae binders from our collection (see above) as IgG1, IgA1, and IgA2 monomers and dimers and compared binding to our 32 bacterial strain panel (Fig. 7 F). While binding to K. pneumoniae was maintained after switching from IgG1 to both IgA1 and IgA2, several bacterial isolates bound to all mAbs in monomeric IgA1 or IgA2 or both isotypes, suggesting non-specific binding unrelated to the antigen-binding site. Some binding activity was maintained in dimeric IgA1 and IgA2 forms of the same mAbs (Fig. 7 F). We conclude that a significant fraction of microbiota shows non-specific binding to IgA1 or IgA2 molecules and that those interactions likely involve residues outside of the antigen-binding site.
Discussion
Immune responses to the microbiome play an important role in UC (Neurath, 2019), and the protective role of FCGR2A (Bessissow et al., 2012; Castro-Dopico et al., 2019), as well as disruption of both circulating and intestinal B cells (Uzzan et al., 2022) place PCs in the center of investigating novel therapeutic avenues in UC. Our results suggest that the clonal landscape in the healthy colon is dominated by IgA1/IgA2 PCs, which overlap significantly between different colon areas within subjects, and that UC disrupts this landscape, mostly through IgG1-dominant clones with lower levels of somatic hypermutations and limited overlap with IgA1/IgA2 clones, which could be a result of limited class switching within germinal center reactions (Roco et al., 2019). IgG1-dominant clones in inflamed regions also frequently contain multiple identical clonal members suggestive of a memory B cell origin in the setting of a secondary immune response. Interestingly, even UC patients in remission show an expansion, albeit less pronounced, of IgG1 PC clones that might constitute responses that remain dormant in remission but may readily expand upon recurrence of inflammation. Future longitudinal studies of UC patients will help elucidate the role of these clones in UC flares and disease progression. In addition, similar studies in subjects with other inflammatory conditions in the colon, such as Crohn’s disease or microscopic colitis, will help clarify how specific to UC these changes are.
Polyreactivity and autoreactivity are not significantly enriched in mAbs from inflamed colon areas and are overall rare in all disease states, a finding consistent with the highly specific mAb interactions with selected bacterial strains we identified for some mAbs. The expansive reactivity of multiple PC clones with K. pneumoniae in the inflamed samples from one subject raises the question if certain pathogens are preferentially targeted in inflamed colon areas. However, only a small number of mAb targets was identified, and further studies are required to map mAb reactivities to additional bacterial strains, evaluate cross-strain reactivity beyond our panel of 32 bacterial strains, and measure reactivity to fungi and other viruses. Beyond specific interactions, we also found significant non-specific VH-3 binding to R. gnavus and C. comes and demonstrated that an isotype switch from IgG1 to IgA1 and IgA2 mAbs leads to nonspecific binding against select bacterial strains. One potential limitation of this finding is that the IgA dimers tested in this study did not contain the secretory component which is present in most secreted forms of IgA. However, these findings underscore the multitude of mAb–microbiome interactions, which are captured with conventional IgA-seq of the microbiome, and the importance of investigations at the monoclonal level to understand the role of specific B cell responses in health and in inflammatory bowel diseases, such as UC. Certainly, the upregulation of MHCII-related genes in PCs from inflamed areas suggests that such specific B cell responses could also directly impact inflammatory T cell responses. Further studies of these interactions, the antigens involved, and how our findings apply to other inflammatory conditions in the colon are needed.
Materials and methods
Human subjects
All work with human samples was performed in accordance with approved Institutional Review Board (IRB) protocols which were reviewed by the Mass General Brigham IRB. HCs as well as UC patients were recruited through a patient cohort that has been created at Massachusetts General Hospital under IRB protocol 2004P001067, “Prospective Registry in Inflammatory Bowel Disease Study at Massachusetts General Hospital,” and all participants signed an informed written consent form. HCs were overall healthy individuals without any history of IBD, other autoimmune diseases, infectious colitis, or colon cancer. UC patients were included based on carrying a clinical diagnosis of UC. UC patients were determined to either have active disease or to be in remission based on macroscopic and histopathologic evaluation of the mucosa in the setting of endoscopic evaluation or of a resection sample (Table S1). None of the subjects had any known history of infectious colitis. Resection samples were obtained in the setting of total colectomies.
Single-cell dissociation from fresh colon mucosa samples
Biopsy bites or mucosal resection samples were immediately processed, and mucosal samples were placed into cryovials containing Advanced DMEM F-12 media (Thermo Fisher Scientific) and placed on ice for transport. Single-cell suspensions from collected mucosal samples were obtained using a modified version of a previously published protocol (Persson et al., 2013) as detailed below. Mucosal samples were first rinsed in 30 ml of ice-cold PBS. Each individual sample was then transferred to 5 ml of enzymatic digestion mix (base: RPMI1640, 100 U/ml penicillin [Thermo Fisher Scientific], 100 μg/ml streptomycin [Thermo Fisher Scientific], 10 mM HEPES [Thermo Fisher Scientific], 2% FCS [Thermo Fisher Scientific], and 50 μg/ml gentamicin [Thermo Fisher Scientific]), freshly supplemented immediately before with 100 mg/ml of Liberase TM (Roche) and 100 μg/ml of DNase I (Roche), and incubated at 37°C with 120 rpm rotation for 30 min. After 30 min, the enzymatic dissociation of the lamina propria was quenched by the addition of 1 ml of 100% FCS (Thermo Fisher Scientific) and 80 μl of 0.5 M EDTA and placed on ice for 5 min. Samples were typically fully dissociated at this step, and after gentle trituration with a P1000 pipette, filtered through a 40-mM cell strainer into a new 50 ml conical tube and rinsed with PBS to 30 ml total volume. This tube was spun down at 700 g for 10 min and resuspended in 500 μl of PBS with 5% FBS. Cell suspensions were then stained with anti-human CD27 PE (BD), CD38 FITC (BD), and CD19 APC (BD) and incubated for 20 min at 4°C before they were washed and resuspended in PBS with 5% FBS. PCs were then sorted using a Sony MA900 cell sorter by gating on live cells in the forward scatter and side scatter and on CD38-FITC and CD27-PE double-positive cells (Fig. 1 B). After sorting, cells were washed and counted using a hemocytometer and microscopy, before resuspending up to 10,000 cells in a volume of 32 μl for 5′ scRNA-seq (see below).
5′ scRNA-seq library generation
Cells were separated into droplet emulsions using the Chromium Next GEM Single-cell 5′ Solution (v1.1) and the 10x Chromium Controller. 5,000–10,000 cells were loaded per channel of the Chromium Next GEM single-cell 5′ (v1.1) Chip G. Following cell lysis, barcoded mRNA reverse transcription, and cDNA amplification, a 0.6× SPRI cleanup was performed, and the supernatant was set aside for Feature Barcoding library construction as instructed by the Chromium NextGEM single-cell V(D)J v1.1 protocol. A final elution of 45 μl was saved for further library construction. Using the saved supernatant of the 0.6× cDNA cleanup, Feature Barcoding libraries were completed according to the 5′ Next GEM (v1.1) Feature Barcoding library construction methods provided by 10x Genomics. Gene expression and V(D)J libraries were created according to the manufacturer’s instruction (10x Genomics), which includes enzymatic fragmentation, adaptor ligation, and sample index barcoding steps. The V(D)J libraries were created from the original following the Chromium NextGEM single-cell V(D)J v1.1 protocol.
scRNA-seq library sequencing
Gene expression, feature barcoding libraries, and BCR-enriched V(D)J libraries were sequenced either on a Nextseq500 (Illumina) using a high output 150 cycle flowcell, with the read configuration read 1: 28 cycles, read 2: 96 cycles, index read 1: 8 cycles, or on a HiSeq X (Illumina), using a 150-cycle flowcell with the read configuration: read 1: 28 cycles, read 2: 96 cycles, and index read 1: 8 cycles. Feature barcoding libraries were spiked into the gene expression libraries at 10–20% of the sample pool prior to sequencing. All BCR-enriched V(D)J libraries were pooled together and sequenced on a NextSeq500 (Illumina) using the same parameters as previously mentioned.
mAb production
mAb VDJ heavy chain and VJ light chain sequences of selected mAbs were produced as minigenes and cloned into IgG1, IgA1, or IgA2 heavy chain and kappa or lambda light chain expression vectors as previously described (Scheid et al., 2021; Wardemann et al., 2003). Matched mAb IgG1 heavy and light chain plasmids or matched mAb IgA1 or IgA2 heavy, light, and additional joining (J) chain (for IgA dimer production only) were cotransfected into Expi293F cells following the manufacturer’s instructions. Briefly, heavy and light chain plasmid DNAs were diluted in 1.5 ml Opti-Plex Complexation Buffer (Invitrogen) and mixed with 80 μl ExpiFectamine 293 Reagent (Invitrogen) diluted in 1.4 ml Opti-Plex Complexation Buffer (Invitrogen) and incubated at RT for 15 min. Complexes were slowly transferred (drop-wise) to a 125-ml flask containing 25 ml of Expi293F cells at a density of 3.0 × 106 viable cells/ml. Cells were incubated for 7 d at 37°C according to the manufacturer’s instructions. ExpiFectamine 293 Transfection Enhancer 1 and 2 were added 18 h after transfection. mAb containing supernatants were transferred to 50 ml Falcon tubes (Thermo Fisher Scientific) and harvested by centrifugation at 3,000 g for 20 min. IgG1 monomers were purified from transfection supernatants via protein G–coupled agarose beads (Cytiva) according to the manufacturer’s instructions. IgA1 and IgA2 monomers and dimers were purified from transfection supernatants using peptide M–coupled agarose beads (Invivogen) according to the manufacturer’s instructions. IgA dimers were further purified via 120 ml HiLoad 16/600 Superdex 200 pg (Cytiva) column on an ÄKTA Pure FPLC system operating at 4°C. Briefly, after equilibration of the column with PBS, each IgA preparation was loaded via 10 ml superloop at a flow rate of 1.25 ml/min, and 1.5 ml fractions were collected. Isolated peaks consistent with IgA multimers, IgA dimers, and IgA monomers were detected at 0.3–0.4, 0.4–0.5, and 0.5–0.6 column volumes, respectively. Fractions spanning all three peaks were collected and evaluated individually by running Criterion TGX precast gels (Bio-Rad) under non-reducing conditions with a Precision Plus Protein Kaleidoscope Prestained Protein Standard (Bio-Rad).
mAb ELISA testing
All ELISA testing was performed at a mAb concentration of 1 µg/ml, measured via A280 or IgG-specific ELISA as previously described (Tiller et al., 2008). mAb reactivities to EBV, CMV, and rotavirus were determined using IgG detection kits against the respective viruses: EBV (Abcam), CMV (Abcam), and rotavirus (amsbio). Polyreactivity ELISAs against single-stranded DNA, double-stranded DNA, LPS, and insulin and ELISA against cardiolipin (Sigma-Aldrich) were performed as previously described (Mouquet et al., 2010; Tiller et al., 2008). Similar to prior studies, polyreactivity was defined as reactivity to two or more of the aforementioned antigens (Kabbert et al., 2020; Tiller et al., 2008). As previously described, mAbs ED38, JB40, and mGO53 (Wardemann et al., 2003) were used as strongly polyreactive, moderately polyreactive, and non-polyreactive controls, respectively, at a concentration of 1–0.016 µg/ml to produce a standard curve. ELISAs against human integrin αVβ6 were performed as previously described (Uzzan et al., 2022) with 1 mM CaCl2 and MgCl2 supplemented to incubation buffers to stabilize the αVβ6 integrin heterodimer (Kuwada et al., 2021). Mouse anti-human αVβ6 MAB2077Z (Millipore) was used as a positive control mAb in ELISAs against human integrin αVβ6 at a concentration of 1–0.016 µg/ml to produce a standard curve.
Protein microarray and detection
Bacterial proteins and extracts were arrayed in quadruplicate on 16-pad nitrocellulose slides (Maine Manufacturing) at a concentration of 0.2 mg/ml using a Spotbot Personal Microarrayer (Arrayit). Recombinant proteins and bacterial extracts were diluted from stocks to 10 mM Tris (pH 7.4), 20% glycerol, and 0.1% SDS. To probe the microarray, pads were blocked for 1 h in SuperBlock (Thermo Fisher Scientific) and then probed with human mAbs diluted 1 to 250 (final concentration 1.2 μg/ml) in SuperBlock for 1 h. The pads were washed three times in PBS with 0.05% Tween 20; then, anti-human IgG secondary antibody labeled with Dylight 650 (Invitrogen) was applied at 1 to 1,000 (∼0.5 μg/ml) for 1 h in SuperBlock. The pads were then washed three times with PBS-Tween, distilled water, air-dried, and scanned using a GenePix 4000B imager (Axion).
Bacterial isolation and verification via 16S sequencing
All bacterial strains used in this study are detailed in Fig. 7 D. Bacterial isolation was carried out under anaerobic conditions. Briefly, bacteria were struck out from frozen stocks and streaked onto Cullen-Haiser Gut or yeast casitone fatty acid agar with carbohydrate media plates and incubated at 37°C for 24–72 h until isolated colonies were present. Single, isolated colonies were inoculated into new 15-ml Falcon tubes (Corning) containing Cullen-Haiser Gut or yeast casitone fatty acid agar with carbohydrate liquid medium and incubated at 37°C for 24–72 h until 600 nm optical density (OD600) of 0.2 or higher was achieved. Liquid cultures were mixed with a 50% glycerol, 1× PBS solution at a 1:1 ratio, aliquoted into cryovials, and stored at −80°C. To verify the identity of individual bacterial strains, 16S ribosomal RNA (rRNA) V1-V9 sequencing was carried out. Briefly, 5 μl of liquid bacterial cultures and 50 μl of hot-shot lysis buffer were combined in corresponding wells in a sterile 96-well PCR plate (Thermo Fisher Scientific). Plates were sealed and heated to 95°C for 10 min via PCR thermocycler (Bio-Rad). Plates were unsealed and 50 μl of neutralization buffer was added to each well. Template DNA was then added to premixed PCR reagents (1.5 μl 10 mM 16S rRNA forward prime [IDT], 1.5 μl 10 mM 16S rRNA reverse primer [IDT], 12.5 μl OneTaq 2× MasterMix [NEB], and 5 μl nuclease-free water) in a new 96-well plate. Three wells of PCR reagents without templated DNA were added as reagent controls. Plates were sealed and placed in a PCR thermocycler for the following protocol: step 1, 94°C for 30 s; step 2, 94°C for 30 s; step 3, 55°C for 60 s; step 4, 68°C, 90 s (cycle steps 2–4, 30 times); step 5, 68°C for 5 min; and step 6, 12°C, hold. Plates were submitted to Genewiz Inc. for enzymatic purification and 16S V1-V9 Sanger sequencing, with special instructions to use 2.5 μl of 10 mM 16S rRNA forward primer for sequencing. The taxonomic identity of each strain was verified by importing 16S rRNA data into Geneious Prime 2021.2.2 and performing standard nucleotide BLAST via blastn suite software (National Institutes of Health, National Library of Medicine, https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Bacterial prep for flow cytometry
Isolated strains verified by 16S rRNA sequencing were thawed on ice, resuspended, and filtered through sterile 50 µm CellTrics filters (Sysmex) into new 1.5-ml tubes. Tubes were centrifuged at 5,000 g for 8 min. Supernatants were removed by aspiration and pellets were resuspended in 1.5 ml cold PBS 0.25% BSA (Sigma-Aldrich). Tubes were centrifuged at 5,000 g for 8 min. Supernatants were removed by aspiration and pellets were resuspended in 1.0 ml cold PBS 0.25% BSA. Washed bacteria were diluted to OD600 of 0.2 in cold PBS 0.25% BSA and 50 μl was added per well to a Nunc 96-well, V-bottom, polypropylene plate (Thermo Fisher Scientific). mAbs were diluted in cold PBS 0.25% BSA and added to each well at a final concentration of 1.0 µg/ml. mAb testing was performed for binding to bacteria as described below.
Mouse stool sample collection and homogenization
Fresh fecal pellets were collected from adult WT C57BL/6J (Stock #000664; Jackson Laboratory) and Rag1tm1Mom (Stock #002216; Jackson Laboratory) mice, pooled by genotype, and stored at −80°C. Mouse fecal pellets were thawed and resuspended in 10 ml of BIOME-preserve anaerobic medium, transferred to a gentleMACS C tube (Miltenyi), and homogenized via gentleMACS Dissociator (Miltenyi) for three cycles of 61 s on the “intestine” setting. All stool homogenates were aliquoted into 2.0-ml cryovials (VWR) and stored at −80°C.
Stool sample preparation for flow cytometry
Homogenized stool samples were thawed on ice, resuspended, and centrifuged at 50 g for 15 min to remove large debris. Supernatants were filtered through sterile 50-µm filters into new 1.5-ml tubes. Tubes were centrifuged at 5,000 g for 8 min. Supernatants were removed by aspiration and pellets were resuspended in 1.5 ml cold PBS 0.25% BSA. Tubes were centrifuged at 5,000 g for 8 min. Supernatants were removed by aspiration and pellets were resuspended in 1.0 ml cold PBS 0.25% BSA. Washed stool samples were diluted to OD600 of 0.2 in cold PBS 0.25% BSA and 50 μl was added per well to a Nunc 96-well, V-bottom, polypropylene plate. mAbs were diluted in cold PBS 0.25% BSA and added to each well at a final concentration of 10.0 µg/ml. mAb testing was performed for binding to stool samples as described below.
mAb testing for binding to bacteria and stool samples
Following aforementioned bacteria or stool sample prep, plates were covered with foil Microseal (Bio-Rad) and incubated on ice for 30 min. Plates were centrifuged at 5,000 g for 8 min, plates were uncovered, and supernatants were decanted with force. Pellets were resuspended in 100 μl/well PE-conjugated mouse anti-human IgG (BD) diluted 1:100 in cold PBS 0.25% BSA or PE-conjugated goat anti-human IgA (Southern Biotech) diluted 1:1,000 in cold PBS 0.25% BSA. Plates were covered and incubated on ice for 20 min. Plates were centrifuged at 500 g for 1 min to remove potential condensate from foil covers. Plates were uncovered and stained bacteria were mixed with 100 μl/well of SYTO BC nucleic acid stain (Thermo Fisher Scientific) diluted 1:500 in cold PBS 0.25% BSA. Plates were covered and incubated on ice for 5 min. Plates were centrifuged at 5,000 g for 8 min, plates were uncovered, and supernatants were decanted with force. Pellets were resuspended in 250 μl cold PBS 0.25% BSA. Plates were centrifuged at 5,000 g for 8 min, plates were uncovered, and supernatants were decanted with force. Pellets were resuspended in 100 μl cold PBS 0.25% BSA. Stained bacteria or stool samples were analyzed via CytoFLEX S flow cytometers (Beckman Coulter), visualizing intact bacteria by gating on FSC+SSC+FITC+ cells and visualizing mAb-bound bacteria by gating on intact bacteria PE+ cells.
Preprocessing of scRNA-seq and V(D)J readouts
mRNA and V(D)J sequence reads were mapped to the reference human genome GRCh38-3.0.0 with the cloud-based Cumulus workflows (Li et al., 2020) using the CellRanger 3.0.2 software pipeline.
scRNA-seq analysis
For the mRNA data integration, count normalization, dimensionality reduction, clustering, cell scoring, and cluster marker genes detection, the Seurat R package (Butler et al., 2018; Stuart et al., 2019) was employed.
Preprocessing and batch correction
Cells that either do not have 10× standard high-quality heavy and light chain V(D)J sequences or that have more than 10% of their transcriptome reads coming from mitochondrial genes were filtered out before the downstream transcriptome analysis. For the UMI count normalization step, gene expression counts for each cell were divided by the total counts for that cell and multiplied by 106, which was then log-transformed using log1p. The top 2,000 variable genes were identified with the utilization of variance-stabilizing transformation, where a local polynomial regression (loess) model was fit to model the relationship of log(variance) and log(mean), and gene values were standardized using the observed mean and the expected variance given by the fitted line. Gene variance was then calculated on the standardized values after clipping to a maximum, which was defined as the square root of the number of cells. Batch effects, defined as the batch of the sequencing run, were regressed out from the normalized count values with the ComBat algorithm (Johnson et al., 2007) implemented in SVA R Package version 3.38.0. Finally, the resulting residuals were scaled and centered.
Dimensionality reduction, graph clustering, and uniform manifold approximation and projection (UMAP) visualization
Dimensionality reduction was done with PCA identifying the first 50 principal components. For clustering of the cells into expression clusters, a k-nearest neighbor graph of the cells was constructed (k = 20) using the 50 principal components. Next, this k-nearest neighbor graph was used to generate the shared nearest neighbor graph by calculating a Jaccard index between every cell and its k nearest neighbors. Then the Leiden algorithm (Traag et al., 2019) was used to find the clusters of the cells based on the generated shared nearest neighbor graph with a resolution of 0.024 decided based on the identifiability of the marker genes. Expression levels of immunoglobulin genes were discarded during the clustering step. A UMAP (McInnes et al., 2018) algorithm was run on the first 50 principal factors to obtain the 2D projections of the cells.
Scoring gene signatures
Module scores of the cell cycle genes and the antigen presentation genes were calculated by randomly selecting 100 genes as control genes for each gene in the module gene set.
DE analysis
Differentially expressed genes between UC-inflamed and healthy samples were identified with pseudobulk DE analysis using the DESeq2 R package (Love et al., 2014), where counts of each gene were aggregated at the sample level. Multiple testing correction was performed with the Benjamini–Hochberg procedure. Genes that had FDR < 0.05 were accepted as significantly differentially expressed between UC-inflamed and healthy samples.
NMF
NMF of the integrated single cell count matrix was done by utilizing the consensus NMF (cNMF; Kotliar et al., 2019) Python package. The number of high variance genes that were used for running the factorization was set to 3,000 and the loss function for NMF was frobenius. The optimal number of latent factors was decided based on the identifiability of the independent pathways verified by the gene set enrichment analysis. The genes with significantly high loadings per factor were defined to have loadings greater than three interquartile ranges higher than the 75th percentile. Gene set enrichment of the selected genes was performed with the clusterProfiler R package (Yu et al., 2012).
Antibody repertoire analysis
Preprocessing and B cell clone calling
The V(D)J contig assembly algorithm from 10x Genomics (https://support.10xgenomics.com/single-cell-vdj/software/pipelines/latest/algorithms/assembly) takes many forms of noise specific to scRNA-seq data into account while generating the assembled V(D)J sequences. Nevertheless, only the cells with high-quality heavy and light chain V(D)J contig sequences were selected, and V(D)J gene annotations were assigned by using IGBLAST (version 1.14.0) software with the Change-O R package (Gupta et al., 2015). Cells with more than one high-quality heavy or light chain sequence (i.e., double expressors) were excluded from the downstream repertoire analysis. Donor-specific B cell clones were identified by utilization of the Change-O R package on the combined cell population of all collected biopsies of the donor. The appropriate threshold for trimming the hierarchical clustering into B cell clones was found by inspecting the bimodal distribution of the distance between each sequence in the data and its nearest neighbor.
Comparison of isotype percentage between samples of different disease status
For isotype distribution comparison analysis for each biopsy sample, one representative member of each clone with unique heavy and light chain sequences was selected. Next, the percentage of the IgA1, IgA2, IgG1, IgG2, IgG3, IgG4, and IgM isotypes within the selected cells was calculated for each sample. Statistical significance of the differences between the distributions of the isotype percentages between samples with different disease statuses was tested with both non-parametric Wilcoxon rank sum test and Dirichlet-multinomial regression, implemented as DirichReg function in DirichletReg R package (Maier, 2014), to account for the fact that the percentage values of all isotypes within a sample sum up to 100.
Mutational load analysis
Replacement and silent mutation inference based on the scRNA-seq V(D)J sequences of the donor cells (Rubelt et al., 2012) was performed by the Shazam R Package (Yaari et al., 2012), where the region definition parameter was set to be “IMGT_V_BY_SEGMENTS,” which provides no subdivisions and treats the entire V segment as a single region.
CDR3 region length and amino acid physicochemical property analysis
CDRH3 length was defined based on IMGT definition (Lefranc et al., 2015) with the addition of two conserved amino acid residues to assist in clonal analysis (Nouri and Kleinstein, 2018). CDRH3 amino acid charges were calculated by the Alakazam R package (Gupta et al., 2015) using the method of Moore (Moore, 1985), excluding the N-terminus and C-terminus charges, and normalizing by the number of informative positions. Hydrophobicity scores were calculated with the Alakazam R package using the method of Kyte and Doolittle (Kyte and Doolittle, 1982).
Shannon entropy values were calculated using the Alakazam R package. For each donor, the transcriptome cluster-specific Hill diversity index (Hill, 1973), improved by Chao et al. (2014) and Chao et al. (2015), was calculated by setting the diversity order equal to 1 with the Alakazam R package. For each run, the number of bootstrap realizations was set to 1,000 and the minimum number of observations to sample was set to 20.
Clonal overlap analysis
To compare the clonal repertoire between two samples, 100 cells were randomly sampled from each sample 1,000 times, and for each pair of random selections, we calculated the percentage of cells that belong to clones that have members in each of the two random samples. The distribution of these 1,000 clonal overlaps is taken as a measure of repertoire similarity between the two compared samples. The same testing schema is repeated with one clonal representative cell per sample to control for the bias that might have been introduced due to the greater number of cells in the expanded clones.
Analysis of isotype conservation within clones across regions with different inflammation status
Dominant isotype conservation across inflammation status in the expanded clones (n > 9) that span both inflamed and less-inflamed regions of the donor was tested by calculating the Spearman’s correlation coefficient between the clonal percentage differences of the IgG and IgA cells coming from the inflamed samples (column 4 minus column 6 of Fig. 2 D) versus the clonal percentage differences of the IgG and IgA cells from the less-inflamed samples (column 3 minus column 5 of Fig. 2 D).
Comparison of transcriptional distance between cells based on the clonal membership
To test the partitioning of the clones on the expression landscape, for each clone, which has members from both inflamed and less-inflamed regions, the pairwise cosine distance was calculated between (i) members of the clone from samples with the same inflammation status, (ii) members of the clone from samples with a different inflammation status, (iii) 200 randomly selected cells of the donor that are not members of the clone from samples with the same inflammation status, (iv) 200 randomly selected cells of the donor that are not members of the clone from samples with different inflammation status. The significance of the difference between the distributions of these cosine distances was tested with a one-tailed t test.
Online supplemental material
Fig. S1 shows the clonal overlap between PCs from different colon regions. Fig. S2 shows the transcriptional clusters of colon PCs. Fig. S3 shows genes that are higher expressed among PCs from inflamed tissue. Fig. S4 shows latent factor analysis. Fig. S5 shows the binding of mAbs and mAb mixes to stool from RAG1-deficient and C57BL/6 mice and bacterial proteins and extracts. Table S1 shows patient characteristics. Table S2 displays differentially expressed genes in UC-inflamed compared with healthy samples. Table S3 displays the scores of each cell for the 10 latent factors identified by the non-negative factorization of the scRNA-seq count matrix. Table S4 displays the gene loadings per each of the 10 latent factors identified by the non-negative factorization of the scRNA-seq count matrix. Table S5 summarizes all mAbs cloned and produced from human colon tissue in this study. Table S6 summarizes mAb reactivities in ELISA. Table S7 summarizes the composition of all 13 mAb mixes used in Fig. S5 and the characteristics of the mAbs in each mix as indicated. Table S8 summarizes all antigens used in the protein and bacterial extract microarray. Data S1 shows the heatmap and phylogenetic trees of expanded clones with IgG and IgA isotypes.
Supplementary Material
shows patient characteristics.
shows differentially expressed genes in UC-inflamed compared to healthy samples.
shows the scores of each cell for the 10 latent factors identified by the non-negative factorization of the scRNA-seq count matrix.
shows the gene loadings per each of the 10 latent factors identified by the non-negative factorization of the scRNA-seq count matrix.
shows all mAbs cloned and produced from human colon tissue in this study.
shows mAb reactivities in ELISA.
shows the composition of all 13 mAb mixes used in Fig. S5 and the characteristics of the mAbs in each mix as indicated.
shows all antigens used in the protein and bacterial extract microarray.
shows heatmap and phylogenetic trees of expanded clones with IgG and IgA isotypes. Heatmap displaying the percentage of IGHA, IGHG, and IGHM isotype usage (columns) within expanded clones (n >9). Lineage trees of the expanded clones (n >9) which have both IgG and IgA cells. Nodes are colored based on the isotype of the majority of cells within each given node. Node size corresponds to the number of identical cells written in the center of the node.
Acknowledgments
We thank Heather Kang for editorial assistance with the manuscript and figures. We thank all study participants who devoted time to our research and the clinical staff. We thank Patricia Rogers and Natan Pirete (Broad Institute) for support with cell sorting. Cell sorting was performed at the Flow Cytometry Facility of the Broad Institute, and sequencing of preconstructed DNA libraries was performed at the Genomics Platform (Broad Institute).
This work was supported by the National Institutes of Health (RC2 DK114784 and P30 DK043351 to R.J. Xavier), the Crohn's and Colitis Foundation, and the Klarman Cell Observatory.
Author contributions: J.F. Scheid, B. Eraslan, D.R. Plichta, D.B. Graham, A. Regev and R.J. Xavier conceived the study, designed experiments, and wrote the manuscript with contributions from J. Deguine and other authors. J.F. Scheid, A.N. Ananthakrishnan, and A.T. Jess established and orchestrated the patient cohort. J.F. Scheid established and performed cell staining and sorting. B. Eraslan performed computational analysis with guidance from A. Regev. B. Eraslan and J.F. Scheid analyzed repertoire and transcriptome data with guidance from A. Regev. J.F. Scheid and A. Hudak performed mAb cloning, production, purification, and binding characterization. E.M. Brown, D. Sergio, and H. Vlamakis grew bacterial stocks and assisted in mAb bacterial binding experiments. L.W. Duck and C.O. Elson developed and performed mAb-binding assay to bacterial lysates. T.M. Delorey, D. Phillips, A. Lefkovith, and A. Regev developed and performed scRNA-seq library preparation and coordinated scRNA-seq.
Data availability
Antibody sequences and scRNA-seq count matrices will be deposited in the Single Cell Portal of the Broad Institute. Raw scRNA-seq files will be deposited on the platform DUOS. Source codes of the scRNA-seq and Ab repertoire analyses are deposited on GitHub (https://github.com/EraslanBas/UCPlasmaCellRepertoire). Antibody sequences and scRNA-seq count matrices are available under Single Cell Portal accession no. SCP1690 (https://singlecell.broadinstitute.org/single_cell/study/SCP1690/remodeling-of-human-colon-plasma-cell-repertoire-in-ulcerative-colitis). Raw FASTQ files are available under controlled access at DUOS accession no. DUOS-000148 (https://duos.broadinstitute.org/dataset_catalog).
References
- Baklien, K., and Brandtzaeg P.. 1975. Comparative mapping of the local distribution of immunoglobulin-containing cells in ulcerative colitis and Crohn’s disease of the colon. Clin. Exp. Immunol. 22:197–209 [PMC free article] [PubMed] [Google Scholar]
- Benckert, J., Schmolka N., Kreschel C., Zoller M.J., Sturm A., Wiedenmann B., and Wardemann H.. 2011. The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen-specific. J. Clin. Invest. 121:1946–1955. 10.1172/JCI44447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessissow, T., Lemmens B., Ferrante M., Bisschops R., Van Steen K., Geboes K., Van Assche G., Vermeire S., Rutgeerts P., and De Hertogh G.. 2012. Prognostic value of serologic and histologic markers on clinical relapse in ulcerative colitis patients with mucosal healing. Am. J. Gastroenterol. 107:1684–1692. 10.1038/ajg.2012.301 [DOI] [PubMed] [Google Scholar]
- Bonifati, V., Rizzu P., Squitieri F., Krieger E., Vanacore N., van Swieten J.C., Brice A., van Duijn C.M., Oostra B., Meco G., and Heutink P.. 2003. DJ-1( PARK7), a novel gene for autosomal recessive, early onset parkinsonism. Neurol. Sci. 24:159–160. 10.1007/s10072-003-0108-0 [DOI] [PubMed] [Google Scholar]
- Buckner, C.M., Moir S., Kardava L., Ho J., Santich B.H., Kim L.J.Y., Funk E.K., Nelson A.K., Winckler B., Chairez C.L., et al. 2014. CXCR4/IgG-expressing plasma cells are associated with human gastrointestinal tissue inflammation. J. Allergy Clin. Immunol. 133:1676–1685.e5. 10.1016/j.jaci.2013.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker, J.J., Erickson S.A., Flynn T.M., Henry C., Koval J.C., Meisel M., Jabri B., Antonopoulos D.A., Wilson P.C., and Bendelac A.. 2017. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science. 358:eaan6619. 10.1126/science.aan6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker, J.J., Drees C., Watson A.R., Plunkett C.H., Nagler C.R., Schneewind O., Eren A.M., and Bendelac A.. 2019. B cell superantigens in the human intestinal microbiota. Sci. Transl. Med. 11:eaau9356. 10.1126/scitranslmed.aau9356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Dopico, T., Dennison T.W., Ferdinand J.R., Mathews R.J., Fleming A., Clift D., Stewart B.J., Jing C., Strongili K., Labzin L.I., et al. 2019. Anti-commensal IgG drives intestinal inflammation and type 17 immunity in ulcerative colitis. Immunity. 50:1099–1114.e10. 10.1016/j.immuni.2019.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, A., Gotelli N.J., Hsieh T.C., Sander E.L., Ma K.H., Colwell R.K., and Ellison A.M.. 2014. Rarefaction and extrapolation with hill numbers: A framework for sampling and estimation in species diversity studies. Ecol. Monogr. 84:45–67. 10.1890/13-0133.1 [DOI] [Google Scholar]
- Chao, A., Hsieh T.C., Chazdon R.L., Colwell R.K., and Gotelli N.J.. 2015. Unveiling the species-rank abundance distribution by generalizing the Good-Turing sample coverage theory. Ecology. 96:1189–1201. 10.1890/14-0550.1 [DOI] [PubMed] [Google Scholar]
- Christmann, B.S., Abrahamsson T.R., Bernstein C.N., Duck L.W., Mannon P.J., Berg G., Björkstén B., Jenmalm M.C., and Elson C.O.. 2015. Human seroreactivity to gut microbiota antigens. J. Allergy Clin. Immunol. 136:1378–1386.e1–e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyster, J.G. 2003. Homing of antibody secreting cells. Immunol. Rev. 194:48–60. 10.1034/j.1600-065x.2003.00041.x [DOI] [PubMed] [Google Scholar]
- Cyster, J.G., and Allen C.D.C.. 2019. B cell responses: Cell interaction dynamics and decisions. Cell. 177:524–540. 10.1016/j.cell.2019.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuisson, M., Vander Stricht D., Clippe A., Etienne F., Nauser T., Kissner R., Koppenol W.H., Rees J.-F., and Knoops B.. 2004. Human peroxiredoxin 5 is a peroxynitrite reductase. FEBS Lett. 571:161–165. 10.1016/j.febslet.2004.06.080 [DOI] [PubMed] [Google Scholar]
- Franzosa, E.A., Sirota-Madi A., Avila-Pacheco J., Fornelos N., Haiser H.J., Reinker S., Vatanen T., Hall A.B., Mallick H., McIver L.J., et al. 2019. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 4:293–305. 10.1038/s41564-018-0306-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido, C., Brunet M., Didelot C., Zermati Y., Schmitt E., and Kroemer G.. 2006. Heat shock proteins 27 and 70: Anti-apoptotic proteins with tumorigenic properties. Cell Cycle. 5:2592–2601. 10.4161/cc.5.22.3448 [DOI] [PubMed] [Google Scholar]
- Gaudette, B.T., Jones D.D., Bortnick A., Argon Y., and Allman D.. 2020. mTORC1 coordinates an immediate unfolded protein response-related transcriptome in activated B cells preceding antibody secretion. Nat. Commun. 11:723. 10.1038/s41467-019-14032-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, N.T., Vander Heiden J.A., Uduman M., Gadala-Maria D., Yaari G., and Kleinstein S.H.. 2015. Change-O: A toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 31:3356–3358. 10.1093/bioinformatics/btv359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, C.W.Y., Martin A., Sepich-Poore G.D., Shi B., Wang Y., Gouin K., Humphrey G., Sanders K., Ratnayake Y., Chan K.S.L., et al. 2020. Translocation of viable gut microbiota to mesenteric adipose drives formation of creeping fat in humans. Cell. 183:666–683.e17. 10.1016/j.cell.2020.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A.B., Yassour M., Sauk J., Garner A., Jiang X., Arthur T., Lagoudas G.K., Vatanen T., Fornelos N., Wilson R., et al. 2017. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 9:103. 10.1186/s13073-017-0490-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, M.O. 1973. Diversity and evenness: A unifying notation and its consequences. Ecology. 54:427–432. 10.2307/1934352 [DOI] [Google Scholar]
- Hiller, M.M., Finger A., Schweiger M., and Wolf D.H.. 1996. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 273:1725–1728. 10.1126/science.273.5282.1725 [DOI] [PubMed] [Google Scholar]
- Høydahl, L.S., Richter L., Frick R., Snir O., Gunnarsen K.S., Landsverk O.J.B., Iversen R., Jeliazkov J.R., Gray J.J., Bergseng E., et al. 2019. Plasma cells are the most abundant gluten peptide MHC-expressing cells in inflamed intestinal tissues from patients with celiac disease. Gastroenterology. 156:1428–1439.e10. 10.1053/j.gastro.2018.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., Fang M., Jostins L., Umićević Mirkov M., Boucher G., Anderson C.A., Andersen V., Cleynen I., Cortes A., Crins F., et al. 2017. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 547:173–178. 10.1038/nature22969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, W.E., Li C., and Rabinovic A.. 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 8:118–127. 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- Kabbert, J., Benckert J., Rollenske T., Hitch T.C.A., Clavel T., Cerovic V., Wardemann H., and Pabst O.. 2020. High microbiota reactivity of adult human intestinal IgA requires somatic mutations. J. Exp. Med. 217:e20200275. 10.1084/jem.20200275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotliar, D., Veres A., Nagy M.A., Tabrizi S., Hodis E., Melton D.A., and Sabeti P.C.. 2019. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. Elife. 8:e43803. 10.7554/eLife.43803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwada, T., Shiokawa M., Kodama Y., Ota S., Kakiuchi N., Nannya Y., Yamazaki H., Yoshida H., Nakamura T., Matsumoto S., et al. 2021. Identification of an anti-integrin αvβ6 autoantibody in patients with ulcerative colitis. Gastroenterology. 160:2383–2394.e21. 10.1053/j.gastro.2021.02.019 [DOI] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle R.F.. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132. 10.1016/0022-2836(82)90515-0 [DOI] [PubMed] [Google Scholar]
- de Lange, K.M., Moutsianas L., Lee J.C., Lamb C.A., Luo Y., Kennedy N.A., Jostins L., Rice D.L., Gutierrez-Achury J., Ji S.-G., et al. 2017. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 49:256–261. 10.1038/ng.3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc, M.-P., Giudicelli V., Duroux P., Jabado-Michaloud J., Folch G., Aouinti S., Carillon E., Duvergey H., Houles A., Paysan-Lafosse T., et al. 2015. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 43:D413–D422. 10.1093/nar/gku1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, J.D., Chen E.Z., Baldassano R.N., Otley A.R., Griffiths A.M., Lee D., Bittinger K., Bailey A., Friedman E.S., Hoffmann C., et al. 2015. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe. 18:489–500. 10.1016/j.chom.2015.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., Gould J., Yang Y., Sarkizova S., Tabaka M., Ashenberg O., Rosen Y., Slyper M., Kowalczyk M.S., Villani A.-C., et al. 2020. Cumulus provides cloud-based data analysis for large-scale single-cell and single-nucleus RNA-seq. Nat. Methods. 17:793–798. 10.1038/s41592-020-0905-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisak, D.A., Schacht T., Enders V., Habicht J., Kiviluoto S., Schneider J., Henke N., Bultynck G., and Methner A.. 2015. The transmembrane Bax inhibitor motif (TMBIM) containing protein family: Tissue expression, intracellular localization and effects on the ER CA²⁺-filling state. Biochim. Biophys. Acta. 1853:2104–2114. 10.1016/j.bbamcr.2015.03.002 [DOI] [PubMed] [Google Scholar]
- Liu, C., Li X., Li C., Zhang Z., Gao X., Jia Z., Chen H., Jia Q., Zhao X., Liu J., et al. 2018. SLC3A2 is a novel endoplasmic reticulum stress-related signaling protein that regulates the unfolded protein response and apoptosis. PLoS One. 13:e0208993. 10.1371/journal.pone.0208993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T., et al. 2015. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47:979–986. 10.1038/ng.3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price, J., Arze C., Ananthakrishnan A.N., Schirmer M., Avila-Pacheco J., Poon T.W., Andrews E., Ajami N.J., Bonham K.S., Brislawn C.J., et al. 2019. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 569:655–662. 10.1038/s41586-019-1237-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubos, E., Loscalzo J., and Handy D.E.. 2011. Glutathione peroxidase-1 in health and disease: From molecular mechanisms to therapeutic opportunities. Antioxid. Redox Signal. 15:1957–1997. 10.1089/ars.2010.3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, M.J. 2014. DirichletReg: Dirichlet regression for compositional data in R. Research Report Series, Department of Statistics and Mathematics. 125. [Google Scholar]
- McInnes, L., Healy J., Saul N., and Großberger L.. 2018. Umap: Uniform manifold approximation and projection. J. Open Source Softw. 3:861. 10.21105/joss.00861 [DOI] [Google Scholar]
- Minnich, M., Tagoh H., Bönelt P., Axelsson E., Fischer M., Cebolla B., Tarakhovsky A., Nutt S.L., Jaritz M., and Busslinger M.. 2016. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat. Immunol. 17:331–343. 10.1038/ni.3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misiewicz, M., Déry M.-A., Foveau B., Jodoin J., Ruths D., and LeBlanc A.C.. 2013. Identification of a novel endoplasmic reticulum stress response element regulated by XBP1. J. Biol. Chem. 288:20378–20391. 10.1074/jbc.M113.457242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, D.S. 1985. Amino acid and peptide net charges: A simple calculational procedure. Biochem. Educ. 13:10–11. 10.1016/0307-4412(85)90114-1 [DOI] [Google Scholar]
- Mouquet, H., Scheid J.F., Zoller M.J., Krogsgaard M., Ott R.G., Shukair S., Artyomov M.N., Pietzsch J., Connors M., Pereyra F., et al. 2010. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 467:591–595. 10.1038/nature09385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurath, M.F. 2019. Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat. Immunol. 20:970–979. 10.1038/s41590-019-0415-0 [DOI] [PubMed] [Google Scholar]
- Nouri, N., and Kleinstein S.H.. 2018. A spectral clustering-based method for identifying clones from high-throughput B cell repertoire sequencing data. Bioinformatics. 34:i341–i349. 10.1093/bioinformatics/bty235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst, O., and Slack E.. 2020. IgA and the intestinal microbiota: The importance of being specific. Mucosal Immunol. 13:12–21. 10.1038/s41385-019-0227-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm, N.W., de Zoete M.R., Cullen T.W., Barry N.A., Stefanowski J., Hao L., Degnan P.H., Hu J., Peter I., Zhang W., et al. 2014. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 158:1000–1010. 10.1016/j.cell.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson, E.K., Uronen-Hansson H., Semmrich M., Rivollier A., Hägerbrand K., Marsal J., Gudjonsson S., Håkansson U., Reizis B., Kotarsky K., and Agace W.W.. 2013. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 38:958–969. 10.1016/j.immuni.2013.03.009 [DOI] [PubMed] [Google Scholar]
- Price, M.J., Patterson D.G., Scharer C.D., and Boss J.M.. 2018. Progressive upregulation of oxidative metabolism facilitates plasmablast differentiation to a T-independent antigen. Cell Rep. 23:3152–3159. 10.1016/j.celrep.2018.05.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roco, J.A., Mesin L., Binder S.C., Nefzger C., Gonzalez-Figueroa P., Canete P.F., Ellyard J., Shen Q., Robert P.A., Cappello J., et al. 2019. Class-switch recombination occurs infrequently in germinal centers. Immunity. 51:337–350.e7. 10.1016/j.immuni.2019.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubelt, F., Sievert V., Knaust F., Diener C., Lim T.S., Skriner K., Klipp E., Reinhardt R., Lehrach H., and Konthur Z.. 2012. Onset of immune senescence defined by unbiased pyrosequencing of human immunoglobulin mRNA repertoires. PLoS One. 7:e49774. 10.1371/journal.pone.0049774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasako, T., Ohsugi M., Kubota N., Itoh S., Okazaki Y., Terai A., Kubota T., Yamashita S., Nakatsukasa K., Kamura T., et al. 2019. Hepatic Sdf2l1 controls feeding-induced ER stress and regulates metabolism. Nat. Commun. 10:947. 10.1038/s41467-019-08591-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazonovs, A., Stevens C.R., Venkataraman G.R., Yuan K., Avila B., Abreu M.T., Ahmad T., Allez M., Ananthakrishnan A.N., Atzmon G., et al. 2021. Sequencing of over 100,000 individuals identifies multiple genes and rare variants associated with Crohns disease susceptibility. medRxiv. 10.1101/2021.06.15.21258641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid, J.F., Barnes C.O., Eraslan B., Hudak A., Keeffe J.R., Cosimi L.A., Brown E.M., Muecksch F., Weisblum Y., Zhang S., et al. 2021. B cell genomics behind cross-neutralization of SARS-CoV-2 variants and SARS-CoV. Cell. 184:3205–3221.e24. 10.1016/j.cell.2021.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, K.A., Bertha M., Hofmekler T., Chopra P., Vatanen T., Srivatsa A., Prince J., Kumar A., Sauer C., Zwick M.E., et al. 2016. Dysbiosis, inflammation, and response to treatment: A longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med. 8:75. 10.1186/s13073-016-0331-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie, C.S., Biton M., Ordovas-Montanes J., Sullivan K.M., Burgin G., Graham D.B., Herbst R.H., Rogel N., Slyper M., Waldman J., et al. 2019. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell. 178:714–730.e22. 10.1016/j.cell.2019.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, J., and Sollid L.M.. 2016. The human intestinal B-cell response. Mucosal Immunol. 9:1113–1124. 10.1038/mi.2016.59 [DOI] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. III, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive integration of single-cell data. Cell. 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, C., Hiwa R., Mueller J.L., Vykunta V., Hibiya K., Noviski M., Huizar J., Brooks J.F., Garcia J., Heyn C., et al. 2020. NR4A nuclear receptors restrain B cell responses to antigen when second signals are absent or limiting. Nat. Immunol. 21:1267–1279. 10.1038/s41590-020-0765-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller, T., Tsuiji M., Yurasov S., Velinzon K., Nussenzweig M.C., and Wardemann H.. 2007. Autoreactivity in human IgG+ memory B cells. Immunity. 26:205–213. 10.1016/j.immuni.2007.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller, T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., and Wardemann H.. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 329:112–124. 10.1016/j.jim.2007.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag, V.A., Waltman L., and van Eck N.J.. 2019. From Louvain to Leiden: Guaranteeing well-connected communities. Sci. Rep. 9:5233. 10.1038/s41598-019-41695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treanor, B., Depoil D., Bruckbauer A., and Batista F.D.. 2011. Dynamic cortical actin remodeling by ERM proteins controls BCR microcluster organization and integrity. J. Exp. Med. 208:1055–1068. 10.1084/jem.20101125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trychta, K.A., Bäck S., Henderson M.J., and Harvey B.K.. 2018. KDEL receptors are differentially regulated to maintain the ER proteome under calcium deficiency. Cell Rep. 25:1829–1840.e6. 10.1016/j.celrep.2018.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, L., Ma C., Dulai P.S., Prokop L.J., Eisenstein S., Ramamoorthy S.L., Feagan B.G., Jairath V., Sandborn W.J., and Singh S.. 2020. Contemporary risk of surgery in patients with ulcerative colitis and Crohn’s disease: A meta-analysis of population-based cohorts. Clin. Gastroenterol. Hepatol. 19. 2031–2045.e11. 10.1016/j.cgh.2020.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzzan, M., Martin J.C., Mesin L., Livanos A.E., Castro-Dopico T., Huang R., Petralia F., Magri G., Kumar S., Zhao Q., et al. 2022. Ulcerative colitis is characterized by a plasmablast-skewed humoral response associated with disease activity. Nat. Med. 28:766–779. 10.1038/s41591-022-01680-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann, H., Yurasov S., Schaefer A., Young J.W., Meffre E., and Nussenzweig M.C.. 2003. Predominant autoantibody production by early human B cell precursors. Science. 301:1374–1377. 10.1126/science.1086907 [DOI] [PubMed] [Google Scholar]
- Yaari, G., Uduman M., and Kleinstein S.H.. 2012. Quantifying selection in high-throughput immunoglobulin sequencing data sets. Nucleic Acids Res. 40:e134. 10.1093/nar/gks457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, K., Wang L., Chou C.-H., Liu K., Nakata T., Jaiswal A., Yao J., Lefkovith A., Omar A., Perrigoue J.G., et al. 2021. QRICH1 dictates the outcome of ER stress through transcriptional control of proteostasis. Science. 371:eabb6896. 10.1126/science.abb6896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G., Wang L.-G., Han Y., and He Q.-Y.. 2012. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows patient characteristics.
shows differentially expressed genes in UC-inflamed compared to healthy samples.
shows the scores of each cell for the 10 latent factors identified by the non-negative factorization of the scRNA-seq count matrix.
shows the gene loadings per each of the 10 latent factors identified by the non-negative factorization of the scRNA-seq count matrix.
shows all mAbs cloned and produced from human colon tissue in this study.
shows mAb reactivities in ELISA.
shows the composition of all 13 mAb mixes used in Fig. S5 and the characteristics of the mAbs in each mix as indicated.
shows all antigens used in the protein and bacterial extract microarray.
shows heatmap and phylogenetic trees of expanded clones with IgG and IgA isotypes. Heatmap displaying the percentage of IGHA, IGHG, and IGHM isotype usage (columns) within expanded clones (n >9). Lineage trees of the expanded clones (n >9) which have both IgG and IgA cells. Nodes are colored based on the isotype of the majority of cells within each given node. Node size corresponds to the number of identical cells written in the center of the node.
Data Availability Statement
Antibody sequences and scRNA-seq count matrices will be deposited in the Single Cell Portal of the Broad Institute. Raw scRNA-seq files will be deposited on the platform DUOS. Source codes of the scRNA-seq and Ab repertoire analyses are deposited on GitHub (https://github.com/EraslanBas/UCPlasmaCellRepertoire). Antibody sequences and scRNA-seq count matrices are available under Single Cell Portal accession no. SCP1690 (https://singlecell.broadinstitute.org/single_cell/study/SCP1690/remodeling-of-human-colon-plasma-cell-repertoire-in-ulcerative-colitis). Raw FASTQ files are available under controlled access at DUOS accession no. DUOS-000148 (https://duos.broadinstitute.org/dataset_catalog).