Atlastins regulate the structure and function of the endoplasmic reticulum (ER). In this study, using reconstituted proteoliposomes with a lipid composition mimicking that of the ER, Jang et al. show that all human atlastins are sufficient to drive membrane fusion.
Abstract
The dynamin-like GTPase atlastin is believed to be the minimal machinery required for homotypic endoplasmic reticulum (ER) membrane fusion, mainly because Drosophila atlastin is sufficient to drive liposome fusion. However, it remains unclear whether mammalian atlastins, including the three human atlastins, are sufficient to induce liposome fusion, raising doubts about their major roles in mammalian cells. Here, we show that all human atlastins are sufficient to induce fusion when reconstituted into liposomes with a lipid composition mimicking that of the ER. Although the fusogenic activity of ATL1, which is predominantly expressed in neuronal cells, was weaker than that of ATL2 or ATL3, the addition of M1-spastin, a neuron-specific factor, markedly increased ATL1-mediated liposome fusion. Although we observed efficient fusion between ER microsomes isolated from cultured, non-neuronal cells that predominantly express ATL2-1, an autoinhibited isoform of ATL2, ATL2-1 failed to support liposome fusion by itself as reported previously, indicating that cellular factors enable ATL2-1 to mediate ER fusion in vivo.
Introduction
The shape and structure of intracellular organelles are believed to closely correlate with their functions; however, the structure–function relationship of most organelles remains to be elucidated (Heald and Cohen-Fix, 2014). Such organelles include the ER, which is a single-unit organelle composed of three major subregions (ER sheets, ER tubules, and the nuclear envelope; Friedman and Voeltz, 2011; Westrate et al., 2015). ER sheets are located in the perinuclear region and provide a large surface area for ribosomal binding, and hence protein synthesis and translocation. ER tubules spread across nearly all of the cytoplasm, forming a mesh-like network and mediating interorganellar contacts (English and Voeltz, 2013; Wu et al., 2018). These ER-mediated organelle contact sites are believed to mediate lipid exchange and regulate organelle dynamics (Phillips and Voeltz, 2016; Wu et al., 2018). The perinuclear space is directly connected with the lumen of the ER and is thus classified as an ER subregion. The hallmark of the characteristic mesh-like ER network structure is the presence of three-way junctions, which are formed via membrane fusion between two ER tubules: fusion of the tip of an ER tubule to the side of another tubule generates a new polygon within the ER network (Moss et al., 2011). This homotypic fusion between ER tubules is believed to be mainly mediated by the dynamin-like GTPase family protein, termed atlastin, which is evolutionarily conserved from yeast to humans (Anwar et al., 2012; Hu et al., 2009; Orso et al., 2009). Humans have three atlastins (ATL1, ATL2, and ATL3; Moss et al., 2011; Zhu et al., 2003), whereas the fruit fly Drosophila melanogaster has a single atlastin (dATL) protein (Moss et al., 2011). In addition, Saccharomyces cerevisiae and Arabidopsis thaliana have one atlastin-like protein, named Sey1p and Root Hair Defective 3 (RHD3), respectively. dATL, Sey1p, and RHD3 are sufficient to drive membrane fusion when reconstituted into liposomes in the presence of GTP (Anwar et al., 2012; Lee et al., 2015; Orso et al., 2009; Zhang et al., 2013), suggesting that atlastin is the major fusogen for ER membrane fusion. Consistently, loss-of-function studies suggest that human atlastins are critical for the branched tubular structure of the ER in cultured human cells (Hu et al., 2009; Morin-Leisk et al., 2011; Wu et al., 2015). Human ATL1 is expressed primarily in neurons, whereas ATL2 and ATL3 are found more ubiquitously in human tissues (Hu et al., 2015; Rismanchi et al., 2008). There is considerable homology between human atlastins and dATL as well as its distant cousins (Sey1p and RHD3) in terms of their amino acid sequences and domain organization, and they have comparable GTPase activities. Interestingly, however, mammalian atlastins, including the three human atlastins, have not been shown to drive proteoliposome fusion until recently (Betancourt-Solis et al., 2018; Wu et al., 2015). While this article was under revision, Lee and colleagues reported that human ATL1 and an isoform of ATL2 purified from cultured human cells support liposome fusion (Crosby et al., 2022). Although Lee and colleagues did not explain why the use of cultured human cells for the preparation of human atlastins is critical to reconstitute atlastin-mediated membrane fusion, they showed that phosphorylation of ATL1 is not essential for its fusion activity. In the current study, we speculated that one of the main reasons why previous studies failed to detect human atlastin-mediated liposome fusion was because liposomes were prepared with phosphatidylcholine (PC) and phosphatidylserine (PS) instead of a lipid mix mimicking the lipid composition of the ER (Betancourt-Solis et al., 2018; Wu et al., 2015). In support of this idea, physiological lipids, including sterols, are critical for Sey1p-mediated liposome fusion (Lee et al., 2015; Lee et al., 2019; Sugiura and Mima, 2016), and all human atlastins contain potential cholesterol-binding motifs in their transmembrane domains (Lee et al., 2019). Furthermore, lipids not only play a structural role but also a functional role in various intracellular membrane fusion events (Moon and Jun, 2020; Starr and Fratti, 2019; Wickner, 2010). Using liposomes with a physiological lipid composition, we showed that all human atlastins were sufficient to drive liposome fusion in the presence of GTP, even though they were expressed and purified from Escherichia coli. Another important finding made by Crosby et al. was that the C-terminal extension of the canonical splicing variant of ATL1 (ATL1-1) and the most common splicing isoform of ATL2 (ATL2-1) has autoinhibitory activity: full-length ATL1-1 exhibits relatively low activity, but deletion of the C-terminal extension increases fusion by more than twofold. More strikingly, full-length ATL2-1 exhibits only marginal fusion, but ATL2-1 with deletion of the C-terminal extension exhibits full fusion activity. The ATL2 isoform used in our study is an ATL2-2 variant, which fully supports liposome fusion (Crosby et al., 2022). Consistently, we observed full fusion when it was reconstituted into liposomes with a lipid composition mimicking that of the ER. Lee and colleagues further explored the molecular mechanisms underlying the C-terminal extension-mediated fusion inhibition and found that the C-terminal extension inhibits G domain-mediated trans-dimerization, the GTPase activity of atlastins, and atlastin-mediated membrane tethering; however, it remains elusive how the C-terminal extension elicits these effects. Furthermore, it is puzzling how ATL2-1, which is predominantly expressed in non-neuronal cells, supports ER fusion in vivo. In the current study, we established an in vitro fusion assay using ER microsomes isolated from HEK293T cells and found that ATL2-1 plays a major role in ER microsome fusion in vitro. Based on the data presented here, we propose a working model of how ATL3 at a non-fusogenic concentration enables autoinhibited-ATL2-1 to support ER fusion in non-neuronal cells that predominantly express ATL2-1.
Results
Human atlastins are sufficient to drive liposome fusion
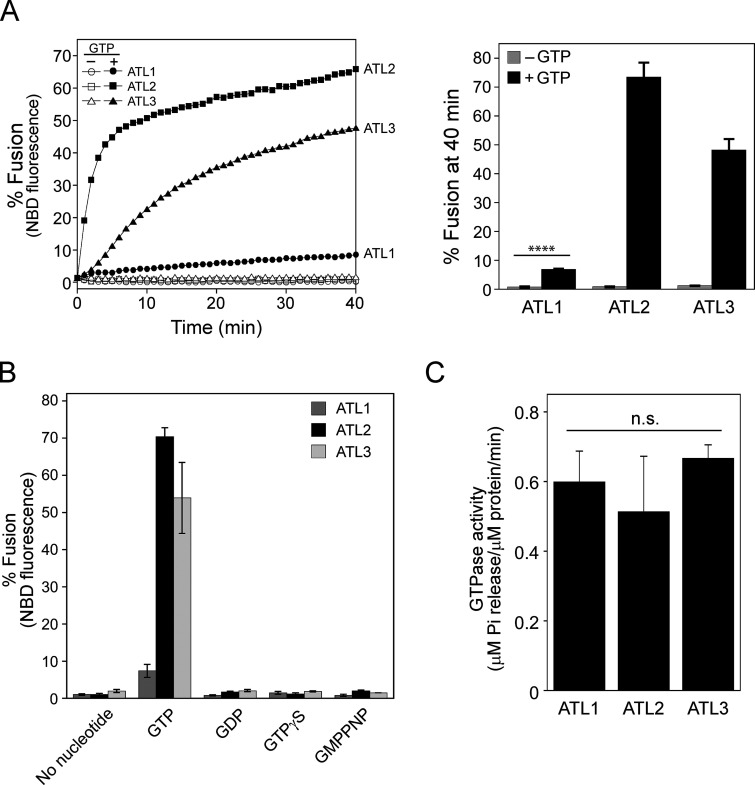
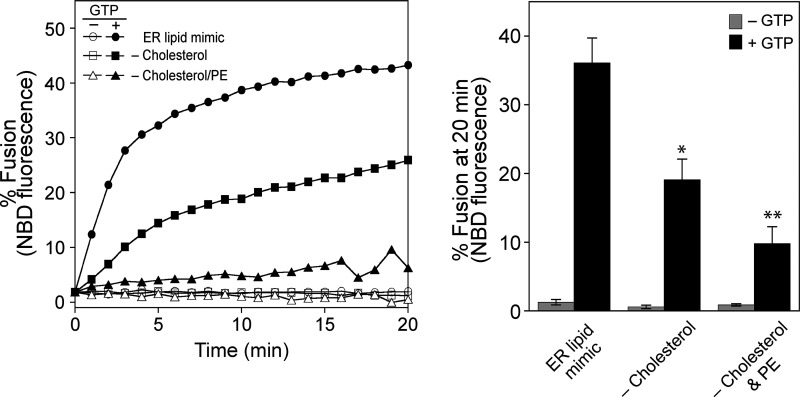
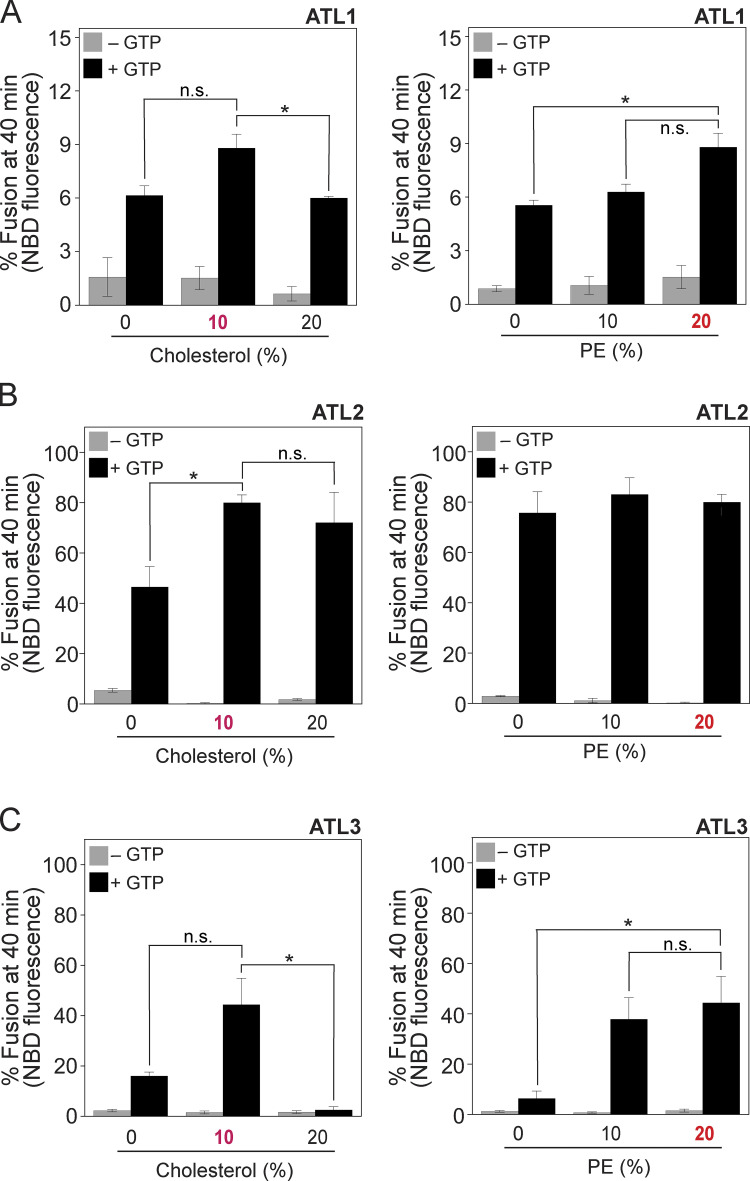
The dynamin-like GTPase atlastin is believed to mediate ER membrane fusion in eukaryotic cells. This idea is strongly supported by the observation that dATL and Sey1p are sufficient to drive liposome fusion without any additional protein factors. Intriguingly, however, it has never been shown that mammalian atlastins, including the three human atlastins, induce liposome fusion, although their depletion in cultured cells severely disrupts ER morphology, which requires homotypic ER membrane fusion, raising doubts about the contribution of atlastins to the ER structure in mammalian cells. In previous studies reporting that liposomes containing human atlastins are resistant to fusion, liposomes were prepared using only PC and PS instead of a lipid mix mimicking the lipid composition of the ER (Betancourt-Solis et al., 2018; Wu et al., 2015). The physiological lipid composition is critical for Sey1p to induce liposome fusion (Sugiura and Mima, 2016); therefore, we first tested whether human atlastins induce membrane fusion when reconstituted into liposomes with a physiological lipid composition. To this end, we purified the three human atlastins that were recombinantly expressed in E. coli and generated liposomes with a mixed lipid composition mimicking that of the ER in mammalian cells (Keenan and Morré, 1970; Sugiura and Mima, 2016) and bearing human ATL1, ATL2, or ATL3 at a molar ratio to lipids of 1:1,000 (Fig. 1 A). Strikingly, all human atlastins supported lipid mixing between donor and acceptor liposomes, although liposome fusion mediated by ATL1 was markedly lower than that mediated by ATL2 or ATL3 (Fig. 1 A), suggesting that a physiological lipid composition is critical for human atlastin-mediated membrane fusion. In support of this idea, the omission of cholesterol or cholesterol/phosphatidylethanolamine (PE) markedly reduced ATL2-mediated liposome fusion (Fig. S1). This atlastin-mediated liposome fusion required GTP and its hydrolysis because lipid mixing was not observed in the presence of GDP or non-hydrolysable GTP analogs (GTPγS and GMP-PNP; Fig. 1 B). The differential fusogenic activities of the three human atlastins did not correlate with their GTPase activities (Fig. 1 C), consistent with a previous study (Hu et al., 2015). While this study was under revision, Lee and colleagues reported that human ATL1 and ATL2-2, an alternatively spliced isoform, prepared from HEK293T cells are sufficient to induce the fusion of PC/PS liposomes, indicating that the physiological lipid composition per se is not essential to reconstitute human atlastin-mediated fusion (Crosby et al., 2022). Although neutral lipids with small head groups that tend to form non-bilayer structures, such as PE and cholesterol, generally enhanced human atlastin-mediated fusion, they differentially affected the fusion activity of different atlastin paralogs (Fig. S2).
Figure 1.
Human atlastins support proteoliposome fusion. (A) Human atlastins (ATL1, ATL2, and ATL3) are sufficient to drive liposome fusion. ATL1, ATL2, or ATL3 recombinantly expressed and purified from E. coli was reconstituted into liposomes with a protein-to-lipid ratio of 1:1,000 as described in the Materials and methods. Donor and acceptor proteoliposomes bearing ATL1, ATL2, or ATL3 were mixed in the presence of Mg2+ and incubated for 10 min at 30°C. After the addition of GTP, NBD fluorescence was measured every minute for 40 min. To determine total fluorescence, β-octylglucoside was added at the end of the reaction and NBD fluorescence was measured. Fusion is expressed as the percentage of total NBD fluorescence. The kinetics graph (left) is representative of three independent results, which are presented as a bar graph (right). Data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. ****P < 0.0001, two-tailed unpaired Student’s t test. (B) Human atlastin-mediated fusion requires GTP hydrolysis. Fusion assays were performed in the absence or presence of GTP, GDP, GTPγS, or GMPPNP for 40 min. Data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. (C) Human atlastins have comparable GTPase activities. The GTPase activities of human atlastins were determined by measuring the release of inorganic phosphate using the Malachite Green Phosphate Assay Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Briefly, proteoliposomes bearing ATL1, ATL2, or ATL3 were incubated with 0.25 mM GTP at 37°C. After incubation for 30 min with malachite green reagents, OD655 was measured and the readings were normalized using a phosphate standard curve. Data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. The difference between samples was not statistically significant (n.s.), two-way ANOVA with Tukey’s multiple comparisons test.
Figure S1.
Cholesterol and PE are critical for ATL2-mediated liposome fusion. Liposome fusion was assayed as in Fig. 1 A using ATL2-proteoliposomes bearing various sets of lipids (complete ER mix, complete ER mix without cholesterol, and complete ER mix without PE and cholesterol). The omission of cholesterol only or cholesterol/PE markedly reduces ATL2-driven liposome fusion. Donor and acceptor proteoliposomes were mixed in the presence of Mg2+ and incubated for 10 min at 30°C. After the addition of GTP, NBD fluorescence was measured every minute for 20 min. To determine total fluorescence, β-octylglucoside was added at the end of the reaction and NBD fluorescence was measured. Fusion is expressed as the percentage of total NBD fluorescence. The kinetics graph (left) is representative of three independent results, which are presented as a bar graph (right). Data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. *P < 0.05, **P < 0.01, one-way ANOVA with Tukey’s multiple comparisons test.
Figure S2.
Differential effects of cholesterol and PE on atlastin-mediated liposome fusion. (A–C) Liposome fusion was assayed as in Fig. 1 A using ATL1-liposomes (A), ATL2-liposomes (B), or ATL3-liposomes (C) bearing different concentrations of cholesterol and PE. Donor and acceptor proteoliposomes were mixed and incubated in the absence or presence of GTP at 30°C for 40 min. Fusion is expressed as the percentage of total NBD fluorescence. Data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. *P < 0.05, one-way ANOVA with Tukey’s multiple comparisons test.
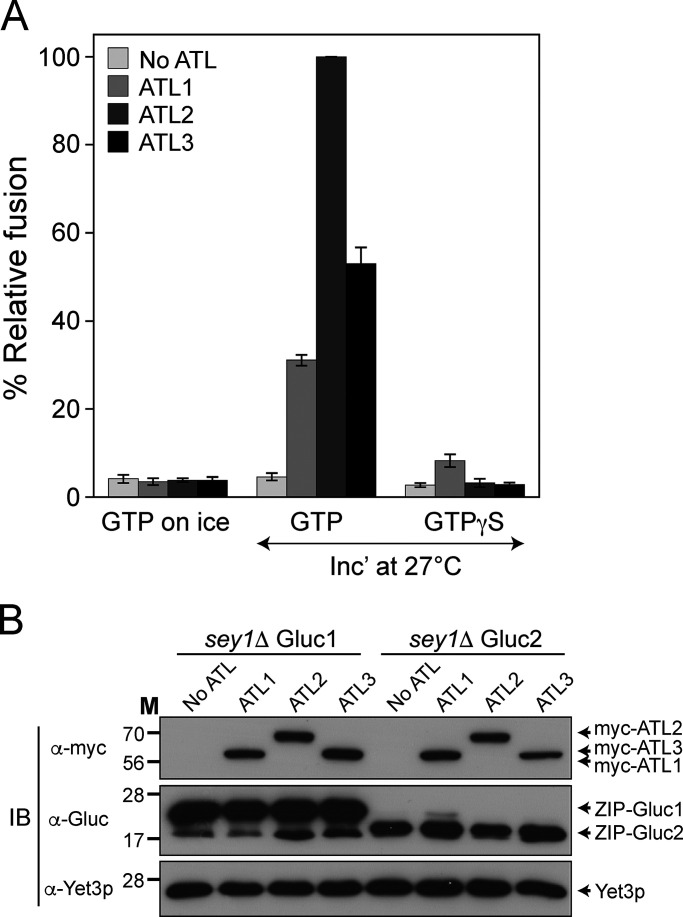
Human atlastins can reportedly replace Sey1p, the yeast ortholog of mammalian atlastins, to maintain the normal ER structure in budding yeast (Anwar et al., 2012; Hu et al., 2015). Although consistent with the idea that human atlastins are important for establishing or maintaining the normal ER structure, these results do not prove that human atlastins are directly responsible for ER membrane fusion. Using an in vitro ER membrane fusion assay incorporating isolated yeast ER microsomes (Lee et al., 2015), we examined whether human atlastins can replace Sey1p for ER microsome fusion in vitro. The gene encoding human ATL1, ATL2, or ATL3 was introduced into sey1Δ yeast cells, and ER microsomes were isolated for the in vitro ER membrane fusion assay (Fig. 2). Consistent with human atlastin-driven liposome fusion (Fig. 1), all three human atlastins restored the fusion of sey1Δ ER microsomes (Fig. 2).
Figure 2.
Human atlastins can replace Sey1p to support ER microsome fusion. (A) ER microsomes were isolated from the BJ-Gluc1 sey1Δ and BJ-Gluc2 sey1Δ yeast strains expressing ATL1, ATL2, or ATL3, and incubated on ice or at 27°C in the presence of GTP or GTPγS. After 90 min, luciferase activity was measured as previously described (Lee et al., 2015). Data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. (B) Protein profiles of ER microsomes used in the experiments are shown in A. Expression of Gluc protein fragment complementation assay fragments and myc-tagged human atlastins was analyzed by immunoblotting using the indicated antibodies. The ER-resident protein Yet3p was used as a loading control. Source data are available for this figure: SourceData F2.
ATL1 may require additional factors for its enrichment at three-way junctions and optimal fusogenic activity
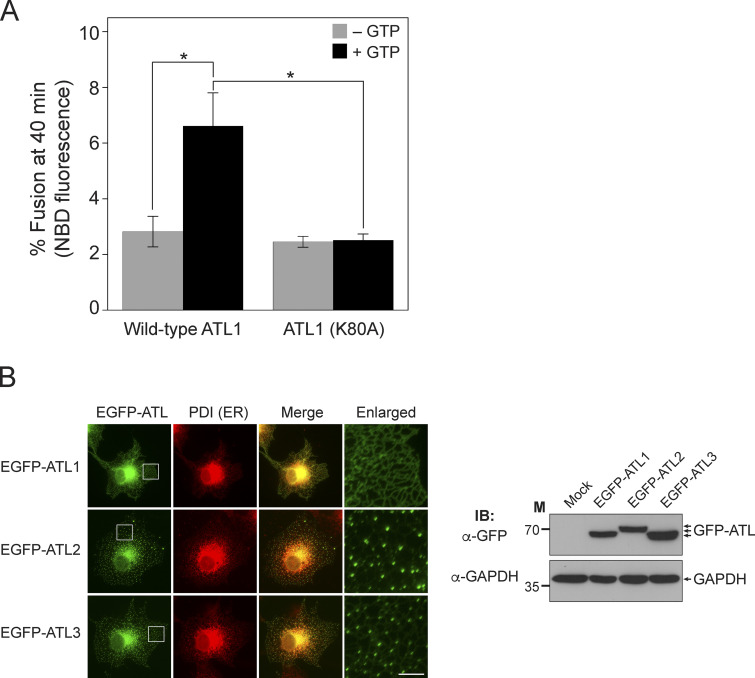
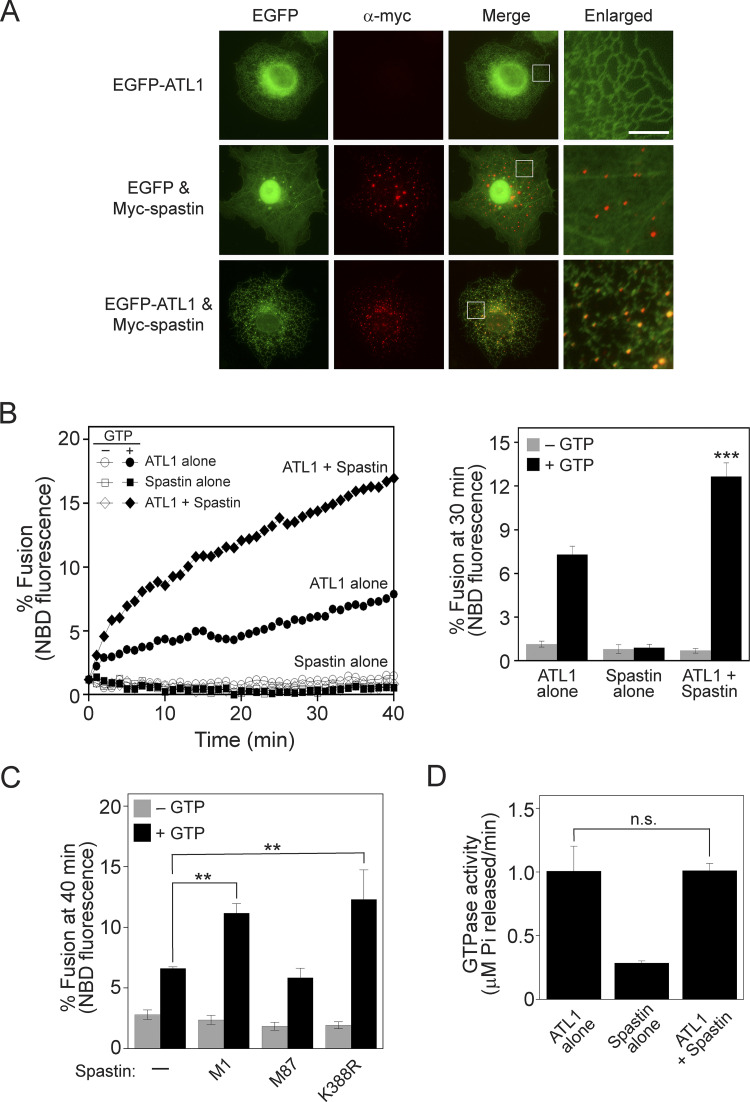
As revealed by in vitro assays using liposomes reconstituted with human atlastins (Fig. 1), ATL1 poorly supported fusion compared with ATL2 and ATL3, although their GTPase activities were comparable (Fig. 1 C). To confirm that the relatively weak signal of ATL1-mediated fusion was dependent on the weak fusogenic activity of ATL1, liposome fusion mediated by the ATL1 (K80A) mutant, which is defective for GTP binding, was compared with that mediated by wild-type ATL1 (Fig. 3 A). The signal with the ATL1 (K80A) mutant was not higher than the background signal (fusion without GTP), confirming that ATL1 alone is sufficient to induce liposome fusion. ATL1 is predominantly expressed in neuronal cells (Zhu et al., 2006); therefore, one plausible scenario is that ATL1 requires additional and/or neuron-specific factors to function as an effective fusogen for ER membrane fusion. Intriguingly, when GFP-fused human atlastins were expressed in non-neuronal cells, GFP-ATL2 and GFP-ATL3 were clearly enriched at three-way junctions, which are characteristic structures of the tubular ER network generated by membrane fusion between ER tubules, whereas GFP-ATL1 was largely dispersed along ER tubules instead of forming distinct puncta at three-way junctions (Fig. 3 B). These results led us to hypothesize that additional factors (that are preferentially expressed in neuronal cells) are needed for ATL1 to accumulate at sites of fusion (three-way junctions) and to effectively mediate fusion between ER tubules. The M1 isoform of spastin, which is preferentially expressed in neuronal tissues, was reported to interact with ATL1 and be involved in shaping the ER structure (Evans et al., 2006; Park et al., 2010; Rao et al., 2016; Sanderson et al., 2006), and thus is one candidate for a neuron-specific factor that is involved in ATL1-mediated ER fusion. In support of this idea, the coexpression of M1-spastin with GFP-ATL1 in non-neuronal cells drastically increased the formation of GFP-ATL1 puncta at three-way junctions (Fig. 4 A). Furthermore, M1-spastin markedly increased ATL1-driven liposome fusion when reconstituted together with ATL1 into liposomes (Fig. 4 B). This increase required the direct binding of spastin to ATL1, but not the ATPase activity of spastin, because M87-spastin, which is defective for ATL1 binding, failed to stimulate ATL1-mediated fusion, whereas the spastin (K388R) mutant, which is defective for ATPase activity, increased ATL1-mediated fusion comparably with wild-type M1-spastin (Fig. 4 C). M1-spastin did not stimulate the GTPase activity of ATL1 (Fig. 4 D).
Figure 3.
ATL2 and ATL3, but not ATL1, accumulate at three-way junctions in cultured non-neuronal cells. (A) Donor and acceptor liposomes bearing wild-type ATL1 or ATL1 (K80A) at a protein/lipid ratio of 1:1,000 were mixed and incubated in the absence or presence of GTP. Fusion is represented as the percentage of total NBD fluorescence. Data represent the mean ± SEM (error bars; n = 3). n refers to the number of independent experiments. *P < 0.05, two-way ANOVA with Tukey’s multiple comparisons test. (B) COS7 cells were transfected with genes encoding EGFP-conjugated human atlastins, and the intracellular localizations of these proteins were analyzed by fluorescence microscopy (left). PDI, an ER-resident protein, was visualized using an anti-PDI antibody conjugated with Alexa Fluor 594 and used as an ER marker. Scale bar: 5 μm. The expression of EGFP-tagged human atlastins was analyzed by immunoblotting (right). GAPDH was used as a loading control. Source data are available for this figure: SourceData F3.
Figure 4.
M1-spastin, a neuronal protein, recruits ATL1 to three-way junctions in cultured non-neuronal cells and stimulates ATL1-mediated liposome fusion. (A) COS7 cells were transfected with genes encoding the indicated proteins, and the intracellular localizations of EGFP-ATL1 and myc-tagged M1-spastin were analyzed by fluorescence microscopy. Myc-tagged M1-spastin was visualized using an anti-myc antibody conjugated with Alexa Fluor 594. Scale bar: 5 μm. (B) Donor and acceptor liposomes bearing ATL1 alone, M1-spastin alone, or both ATL1 and M1-spastin at a protein-to-lipid ratio of 1:1,000 were mixed and incubated in the absence or presence of GTP. Fusion is represented as the percentage of total NBD fluorescence. The kinetics graph (left) is representative of three independent results, which are presented as a bar graph (right). Data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. ***P < 0.001, two-way ANOVA with Tukey’s multiple comparisons test. (C) Donor and acceptor liposomes bearing ATL1 alone, ATL1 and M1-spastin, ATL1 and M87-spastin, or ATL1 and M1-spastin (K388R) at a ratio of each protein/lipid of 1:1,000 were mixed and incubated in the absence or presence of GTP. Fusion is represented as the percentage of total NBD fluorescence. Data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. **P < 0.01, two-tailed unpaired Student’s t test. (D) The GTPase activity of ATL1 alone, spastin alone, or ATL1 in the presence of spastin was determined by measuring the release of inorganic phosphate using a Malachite Green Phosphate Assay Kit (Sigma-Aldrich). Data represent the means ± SEM (error bars; n = 3). The difference between samples was not statistically significant (n.s.), two-tailed unpaired Student’s t test.
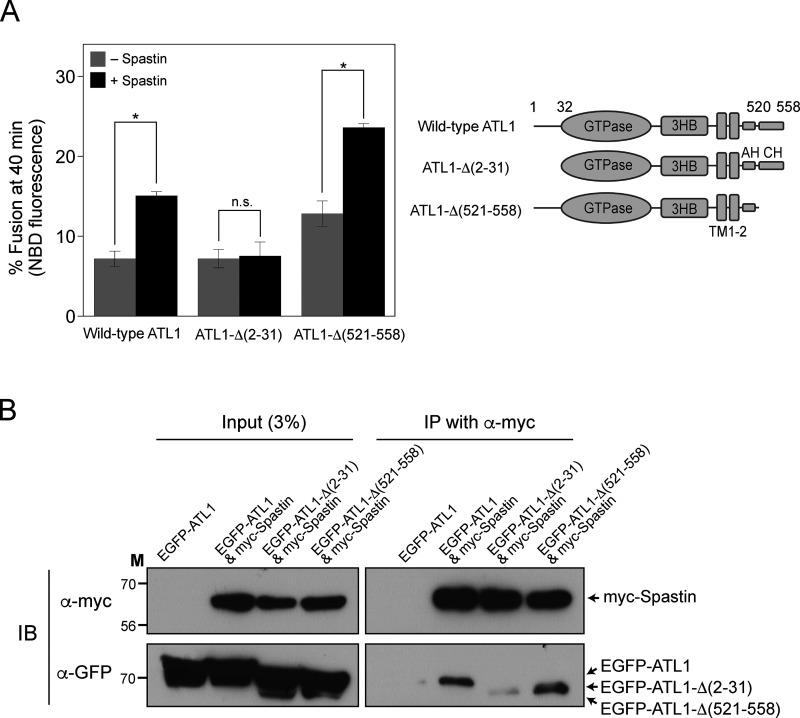
To further explore the molecular mechanism by which M1-spastin stimulates ATL1-mediated fusion, we first attempted to determine which region of ATL1 is required for M1-spastin-mediated stimulation. To this end, we coreconstituted M1-spastin into liposomes bearing wild-type ATL1, ATL1-Δ(2−31; which lacks amino acids 2–31), or ATL1-Δ(521–558; which lacks amino acids 521–558) and compared their fusion. Spastin-mediated fusion enhancement required the N-terminal region of ATL1 (Fig. 5 A). As reported by Lee and colleagues (Crosby et al., 2022), deletion of the C-terminal extension of ATL1 enhanced fusion, which was further stimulated by M1-spastin, suggesting that M1-spastin associates with the N-terminal region of ATL1 to upregulate ATL1-mediated fusion. Consistently, co-immunoprecipitation experiments revealed that the N-terminal region of ATL1 was needed for its optimal interaction with M1-spastin (Fig. 5 B).
Figure 5.
The N-terminal 31 amino acids of ATL1 are critical for interaction with spastin and spastin-mediated enhancement of ATL1-mediated liposome fusion. (A) Wild-type ATL1, ATL1-Δ(2–31), or ATL1-Δ(521–558) was reconstituted into liposomes with or without M1-spastin. Donor and acceptor liposomes were then mixed and incubated in the presence of GTP. Fusion is represented as the percentage of total NBD fluorescence. Data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. *P < 0.05, two-tailed unpaired Student’s t test. (B) HEK293T cells were transfected with genes encoding EGFP-conjugated ATL1 (EGFP-ATL1) alone, EGFP-ATL1 and myc-tagged Spastin (myc-Spastin), EGFP-ATL1-Δ(2–31) and myc-Spastin, or EGFP-ATL1-Δ(521–558) and myc-Spastin, and the physical interaction between M1-spastin and wild-type or mutant ATL1 was analyzed by co-immunoprecipitation. Source data are available for this figure: SourceData F5.
ATL2-1, the major splice isoform of ATL2 expressed in non-neuronal cells, does not support liposome fusion
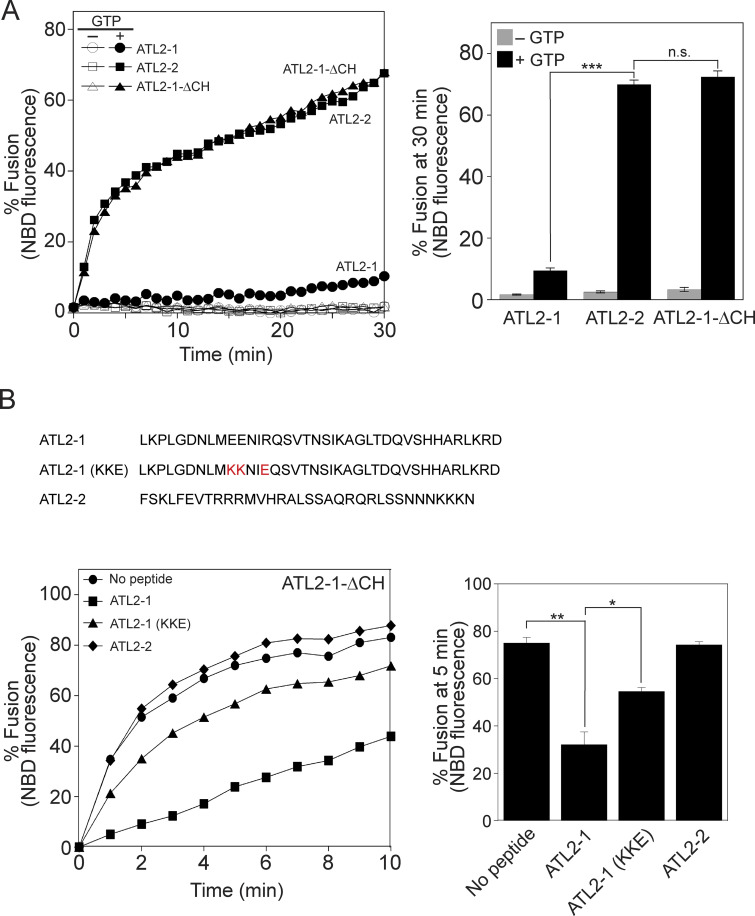
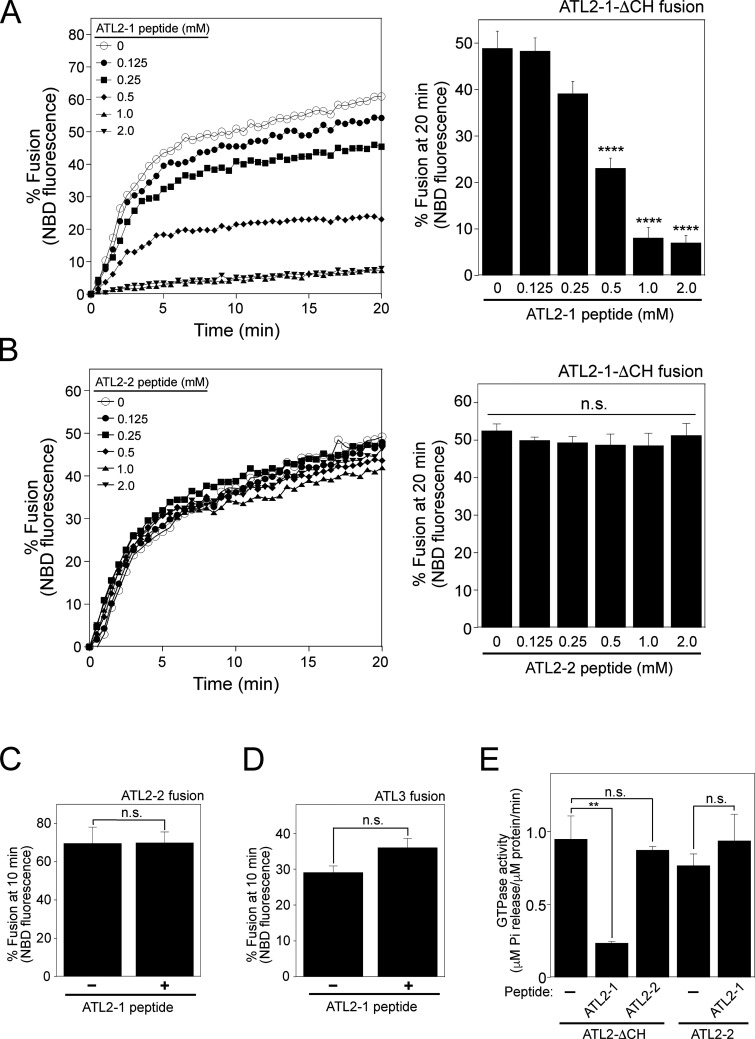
Lee and colleagues (Crosby et al., 2022) found that ATL2-1, the major splice isoform of ATL2 that is predominantly expressed in non-neuronal cells, does not support liposome fusion because its C-terminal extension has an autoinhibitory activity. By contrast, ATL2-2, another isoform of ATL2, efficiently induces liposome fusion. The only difference between ATL2-1 and ATL2-2 is in their C-terminal extensions (amino acids 545–583 of ATL2-1 vs. amino acids 545–579 of ATL2-2). Mutant ATL2-1 (ATL2-ΔCH) that lacks the C-terminal 39 amino acids (hereafter termed the C-terminal helix or CH) supported fusion comparably with ATL2-2, indicating that the CH of ATL2-1 has an autoinhibitory function. We were able to reproduce the results of Lee and colleagues (Crosby et al., 2022): ATL2-1 did not support liposome fusion, but deletion of its CH restored fusion comparably with ATL2-2 (Fig. 6 A). Although Lee and colleagues reported that the C-terminal extension of ATL2-1 markedly suppresses G domain-mediated trans-dimerization and tethering, and reduces GTPase activity, which likely underlies fusion inhibition (Crosby et al., 2022), it remains elusive how the CH inhibits GTPase activity and thereby ATL2-1-mediated fusion. One plausible mechanism is that the CH directly associates with the G-domain of ATL2-1, hindering its trans-dimerization and GTPase activity. To test this idea, we custom-synthesized three peptides: the ATL2-1 peptide derived from the CH of ATL2-1; the ATL2-1 (KKE) peptide, which bears mutations that largely disrupt the autoinhibitory activity (Crosby et al., 2022); and the ATL2-2 peptide derived from the CH of ATL2-2. The ATL2-1 mutant (ATL2-ΔCH) lacking the CH efficiently supported fusion (circle) in the absence of any peptide (Fig. 6 B). The addition of the ATL2-1 peptide markedly inhibited fusion (square, also see Fig. S3 A), whereas the ATL2-1 (KKE) peptide largely lacked inhibitory activity (triangle). By contrast, the addition of the ATL2-2 peptide did not affect fusion (diamond, also see Fig. S3 B), indicating that the inhibitory activity of the ATL2-1 peptide is specific and is preserved even when the CH acts in trans. The specificity of ATL2-1 peptide-mediated inhibition was further confirmed by the observation that the ATL2-1 peptide did not affect ATL2-2- or ATL3-mediated liposome fusion (Fig. S3, C and D). These data, together with the finding that the ATL2-1 peptide inhibited ATL2-ΔCH-mediated fusion, suggest that the CH of ATL2-2 or ATL3 prevents ATL2-1 peptide-mediated inhibition. Consistently, the ATL2-1 peptide inhibited the GTPase activity of ATL2-ΔCH but not that of the full-length ATL2-2 (Fig. S3 E).
Figure 6.
The CH of ATL2-1 has auto-inhibitory activity. (A) Donor and acceptor liposomes bearing the ATL2-1 isoform, the ATL2-2 isoform, or mutant ATL2-1 lacking its CH (ATL2-1-ΔCH) at a protein/lipid ratio of 1:1,000 were mixed and incubated for 30 min at 30°C in the absence or presence of GTP. Fusion is represented as the percentage of total NBD fluorescence. Data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. ***P < 0.001, two-tailed unpaired Student’s t test. (B) Donor and acceptor liposomes bearing ATL2-1-ΔCH at a protein:lipid ratio of 1:1,000 were mixed and incubated in the presence of GTP and synthetic peptides (500 µM) derived from the CH of ATL2-1, the CH of ATL2-2, or the mutant CH of ATL2-1 reported to lack autoinhibitory activity (Crosby et al., 2022). Fusion is represented as the percentage of total NBD fluorescence. The kinetics graph (left) represents three independent results, which are presented as a bar graph (right). Data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. **P < 0.01, *P < 0.05, two-tailed unpaired Student’s t test.
Figure S3.
The CH of ATL2-1 does not inhibit ATL2-2- or ATL3-mediated liposome fusion. (A) ATL2-1 peptide inhibits ATL2-1-ΔCH-mediated fusion in a dose-dependent manner. Donor and acceptor liposomes bearing ATL2-1-ΔCH at a protein/lipid ratio of 1:1,000 were mixed and incubated in the presence of GTP and synthetic peptides derived from the CH of ATL2-1 at 30°C for 20 min. The kinetics graph (left) represents five independent results, which are presented as a bar graph (right). Data represent the means ± SEM (error bars; n = 5). n refers to the number of independent experiments. ****P < 0.0001, one-way ANOVA with Tukey’s multiple comparisons test. (B) ATL2-2 peptide does not inhibit ATL2-1-ΔCH-mediated fusion. Donor and acceptor liposomes bearing ATL2-1-ΔCH at a protein/lipid ratio of 1:1,000 were mixed and incubated in the presence of GTP and synthetic peptides derived from the CH of ATL2-2 at 30°C for 20 min. The kinetics graph (left) is representative of three independent results, which are presented as a bar graph (right). Data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. p = n.s., one-way ANOVA with Tukey’s multiple comparisons test. (C and D) ATL2-1 peptide does not inhibit ATL2-2- or ATL3-mediated fusion. Donor and acceptor liposomes bearing ATL2-2 (C) or ATL3 (D) at a protein/lipid ratio of 1:1,000 were mixed and incubated in the presence of GTP and synthetic peptides (500 μM) derived from the CH of ATL2-1 at 30°C for 10 min. Data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. p = n.s., one-way ANOVA with Tukey’s multiple comparisons test. (D) The CH of ATL2-1, but not that of ATL2-2, inhibits the GTPase activity of ATL2-1-ΔCH. The GTPase activities of ATL2-1-ΔCH and the full-length ATL2-2 in the absence or presence of the ATL2-1 or ATL2-2 peptide were determined by measuring the release of inorganic phosphate using the Malachite Green Phosphate Assay Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. **P < 0.01, one-way ANOVA with Tukey’s multiple comparisons test.
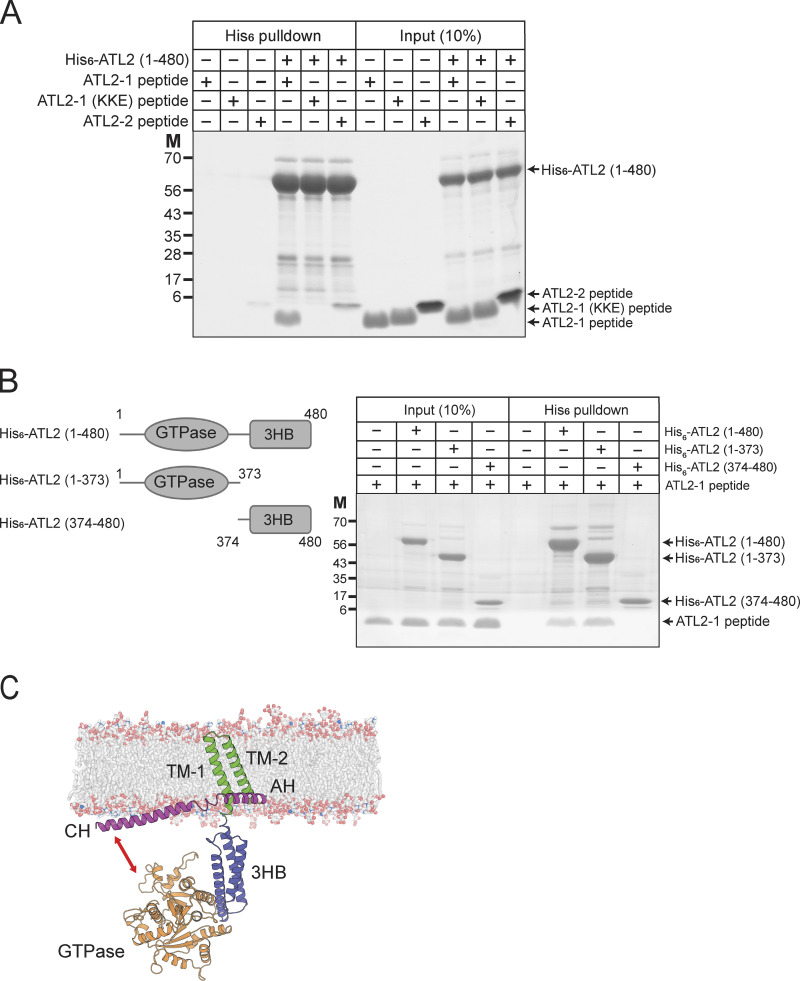
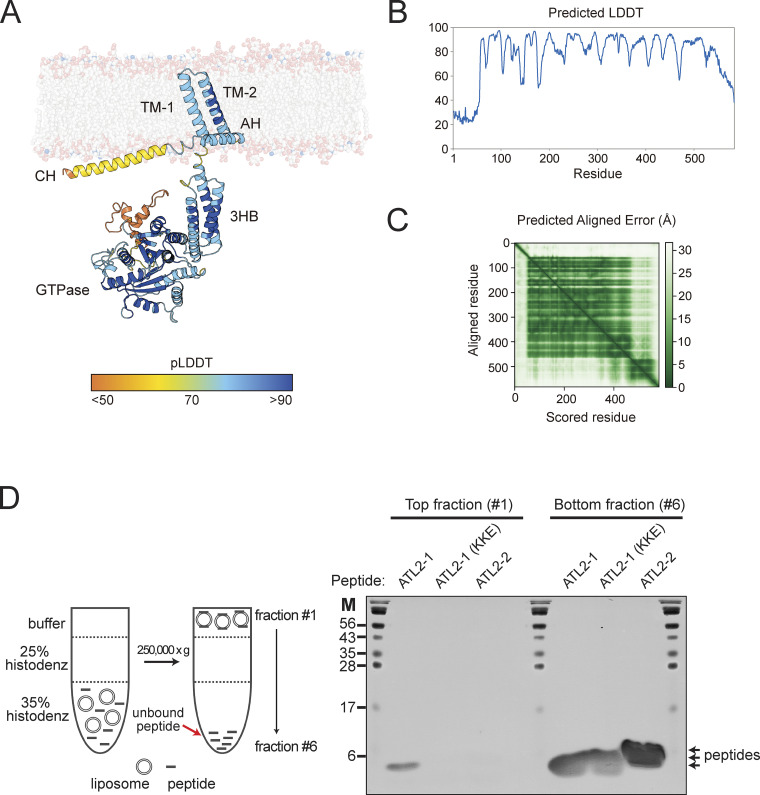
To examine whether the ATL2-1 peptide directly binds to the N-terminal cytosolic domain of ATL2-1, which is composed of the N-terminal extension, the GTPase domains, and the three-helical bundle (3HB) domain, we performed in vitro binding experiments (Fig. 7 A). The N-terminal cytoplasmic domain of ATL2 (1–480) was associated with the ATL2-1 peptide, but not with the ATL2-1 (KKE) peptide, which correlated well with their inhibitory effects on ATL2–ΔCH-mediated liposome fusion. Intriguingly, the ATL2-2 peptide weakly interacted with the N-terminal cytoplasmic domain of ATL2-1. This weak binding may be critical for the resistance of ATL2-2 to ATL2-1 peptide-mediated inhibition (Fig. S3 C) because the N-terminal cytosolic domain of ATL2-2 is identical to that of ATL2-1. To determine the domain that the ATL2-1 peptide interacts with, we prepared the GTPase domain (1–373) and 3HB (374–480) domain and compared their binding to the ATL2-1 peptide by performing in vitro binding experiments. The ATL2-1 peptide is specifically associated with the GTPase domain (1–373), but not with the 3HB domain (374–480), demonstrating that the CH of ATL2-1 directly interacts with the GTPase domain of ATL2-1 (Fig. 7 B). Consistently, the ATL2-1 peptide inhibits the GTPase activity of ATL2-ΔCH (Fig. S3 E). These results led us to propose the following working model (Fig. 7 C, also see Fig. S4, A–C): according to the potential three-dimensional structure of ATL2-1 predicted using the protein structure prediction tool AlphaFold (Jumper et al., 2021), for the CH to associate with the GTPase domain, either the linker between the CH and the amphiphilic helix (AH) or the linker between the 3HB and the TM1 should be flexible. Our liposome co-floatation assay indicated that the CH of ATL2-1 interacted with protein-free liposomes composed of ER-mimicking lipids (Fig. S4 D). Moreover, the linker between the 3HB and the TM1, but not the linker between the CH and the AH, is likely flexible according to a software that predicts the secondary structure of proteins. Taken together, the GTPase domain may bend toward the CH, which lies on the ER membrane, resulting in a fusion-incompetent state.
Figure 7.
The CH of ATL2-1 directly interacts with the GTPase domain of ATL2-1. (A) His6-tagged ATL2 (1–480) was mixed with the ATL2-1 peptide, the ATL2-1 (KKE) peptide, or the ATL2-2 peptide at 4°C for 1 h. The protein–peptide mixtures were then further incubated with Ni-NTA agarose (Qiagen) at 4°C for 30 min. Proteins and peptides bound to Ni-NTA agarose were analyzed by SDS-PAGE followed by Coomassie Brilliant Blue staining. (B) His6-tagged ATL2 (1–480), his6-tagged ATL2 (1–373), or his6-tagged ATL2 (373–480) was mixed with the ATL2-1 peptide at 4°C for 1 h. The protein–peptide mixtures were then further incubated with Ni-NTA agarose (Qiagen) at 4°C for 30 min. Proteins and peptides bound to Ni-NTA agarose were analyzed by SDS-PAGE followed by Coomassie Brilliant Blue staining. (C) AlphaFold structure prediction of ATL2-1 rendered in cartoon form in PyMOL. The per-residue confidence score (pLDDT) and predicted aligned error (PAE) of this predicted structure are shown in Fig. S4. Source data are available for this figure: SourceData F7.
Figure S4.
The CH of ATL2-1 interacts with protein-free liposomes. (A and B) The per-residue confidence score (pLDDT) and (C) predicted aligned error (PAE) of the AlphaFold structure prediction of ATL2-1 are shown in Fig. 7 C. (D) The CH of ATL2-1, but not that of ATL2-2, interacts with protein-free liposomes. Protein-free liposomes (0.5 mM) were incubated with synthetic peptides (300 μM) derived from the CH of ATL2-1, the CH of ATL2-2, or the mutant CH of ATL2-1 [ATL2-1 (KKE)], which is reported to lack autoinhibitory activity (Crosby et al., 2022), at 4°C for 2 h. The mixtures were then mixed at a 1:1 ratio with 70% (w/v) Histodenz, layered with 25% (w/v) Histodenz, and topped with RB150 buffer containing 1 mM EDTA. After centrifugation at 250,000 × g at 4°C for 1 h, the sample was divided into six fractions. The top and bottom fractions were analyzed for the presence of peptides by SDS-PAGE followed by Coomassie Brilliant Blue staining. Source data are available for this figure: SourceData FS4.
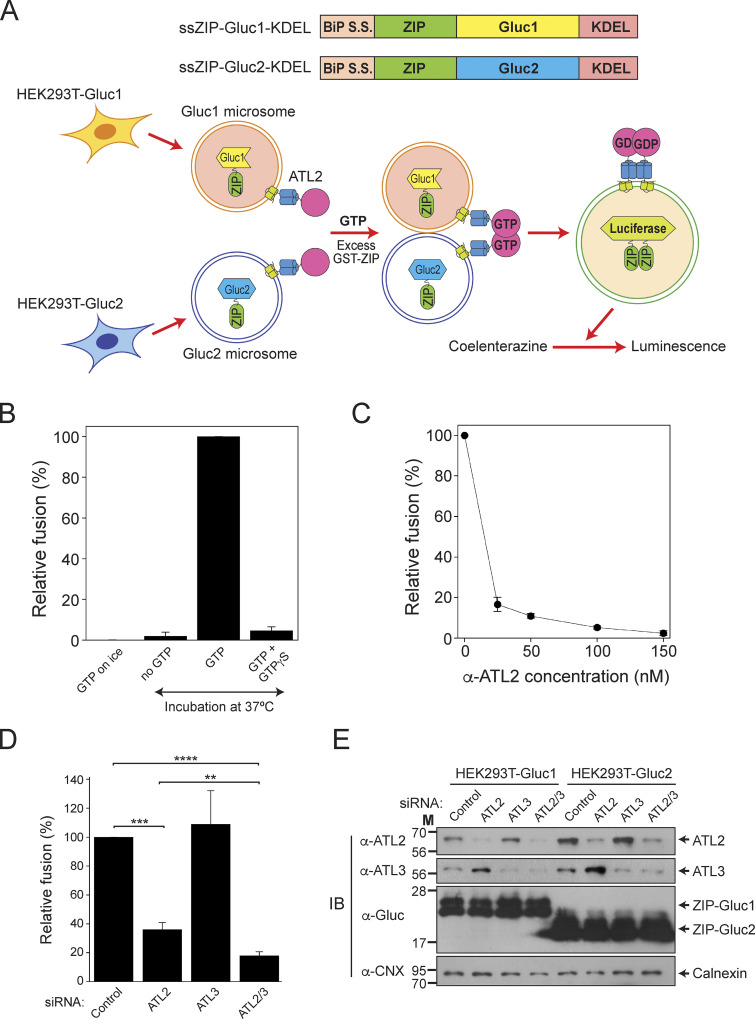
ATL2-1 is the major atlastin expressed in HEK293T cells and is required for efficient ER fusion
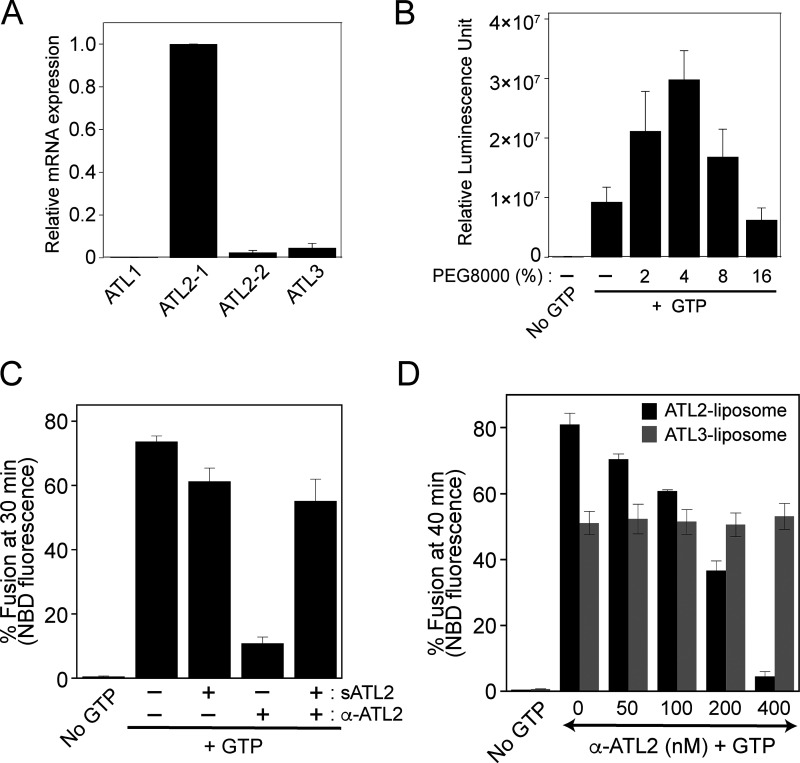
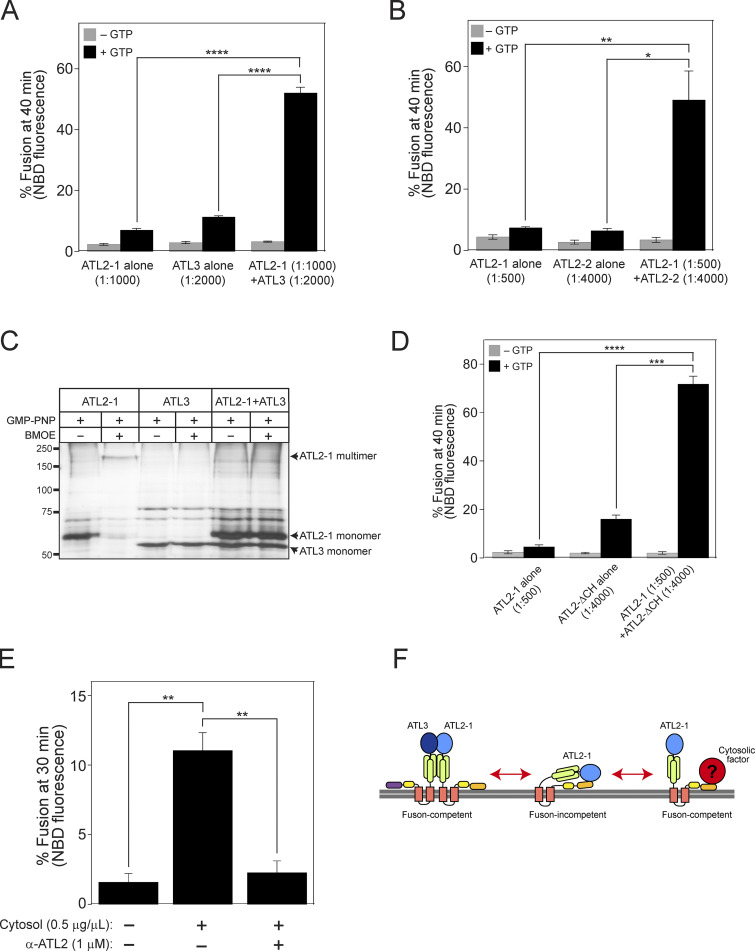
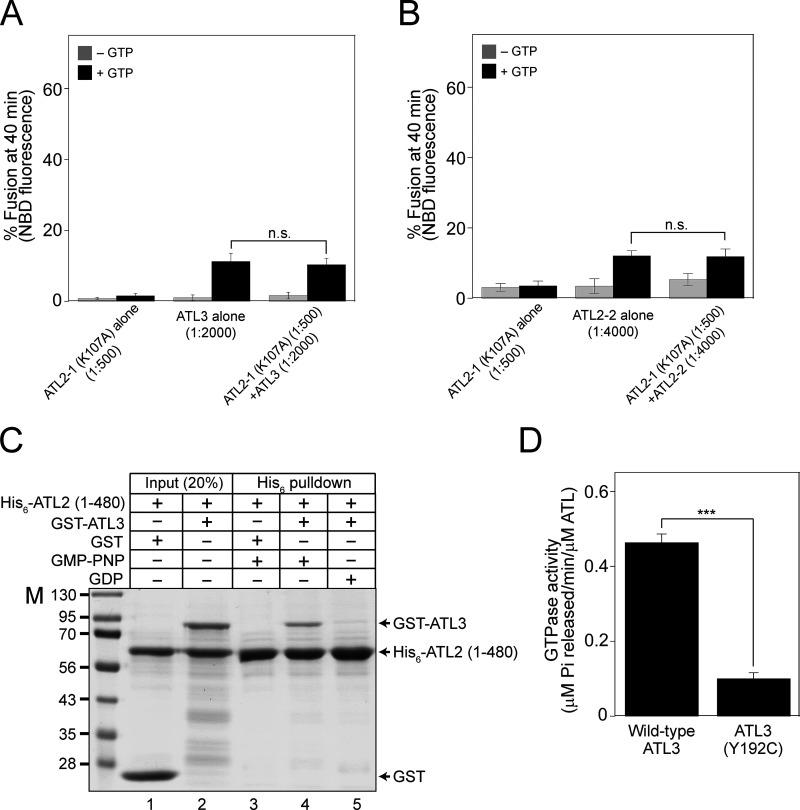
As Lee and colleagues reported, we showed that ATL2-1 fails to drive liposome fusion by itself; therefore, we attempted to examine whether this isoform is involved in ER fusion in a more physiological context. To select a cultured cell line in which ATL2, particularly ATL2-1, is predominantly expressed over the other atlastins, we referred to transcriptomic/proteomic analysis of all protein-coding genes in 32 human tissues and chose HEK293T cells for our current study because they predominantly express ATL2 over the other atlastin paralogs (Lin et al., 2014; Uhlén et al., 2015). Our quantitative RT-PCR analysis of human atlastins in HEK293T cells not only confirmed these results but also found that ATL2-1 is the major atlastin expressed in this cell line: expression of ATL2-1 was ∼50- and 20-fold higher than that of ATL2-2 and ATL3 in this cell line, respectively (Fig. S5 A). To examine whether ATL2-1, which failed to support liposome fusion, directly participates in ER fusion, we first developed an in vitro assay for membrane fusion using ER microsomes isolated from HEK293T cells (Fig. 8 A) similar to the in vitro ER fusion assay using microsomes isolated from the budding yeast S. cerevisiae (Fig. 2; Lee et al., 2015). To this end, we generated HEK293T cells stably expressing a chimeric protein (ssZIP-Gluc1-KDEL or ssZIP-Gluc2-KDEL) with the following four parts: (1) the signal sequence of the ER-resident chaperone BiP, which targets the chimeric protein to the lumen of the ER; (2) the leucine zipper domain of yeast Gcn4p (ZIP); (3) the amino-terminal half (Gluc1) or carboxylic-terminal half (Gluc2) of Gaussia princeps luciferase (Gluc; Remy and Michnick, 2006); and (4) the ER-retrieval signal KDEL. ER microsomes isolated from the two cell lines lacked luciferase activity, but membrane fusion between the two microsome populations allowed their lumenal contents to mix, resulting in ZIP homodimerization and subsequent reconstitution of functional luciferase (Fig. 8 A). As expected, luciferase activity was detected when the two microsome populations were mixed and incubated at the physiological temperature (37°C) in the presence of a GTP-regeneration system (Fig. 8 B). The efficiency of fusion between ER microsomes in this assay was about 25% of the maximal fusion, which was estimated as fusion induced by polyethylene glycol (PEG). PEG induces non-specific membrane fusion (Burgess et al., 1992; Lentz, 2007; Fig. S5 B). This microsome fusion required GTP and its hydrolysis because a luciferase signal was not detected in the absence of GTP or in the presence of GTPγS, a non-hydrolysable GTP analog (Fig. 8 B). To determine whether this luciferase signal originates mainly from ER microsome fusion mediated by ATL2-1, the major atlastin in HEK293T cells, we employed two approaches. First, we examined whether microsome fusion was blocked by the addition of an affinity-purified anti-ATL2 antibody (Fig. 8 C). This antibody was raised against the N-terminal cytosolic domain of ATL2 and was therefore expected to bind to all ATL2 isoforms, including ATL2-1 and ATL2-2, which are expressed in HEK293 cells. This antibody specifically recognized the N-terminal cytosolic domain of ATL2 because its inhibitory effect on the fusion between ATL2-containing liposomes was abrogated by coincubation with the cytosolic domain of ATL2 protein recombinantly expressed in E. coli (Fig. S5 C). Furthermore, this antibody inhibited ATL2-driven, but not ATL3-driven, liposome fusion (Fig. S5 D). ER microsome fusion was completely inhibited upon the addition of the anti-ATL2 antibody, indicating that ATL2, more specifically ATL2-1, plays a critical role in the fusion of ER microsomes purified from HEK293T cells (Fig. 8 C), which predominantly express ATL2-1. Second, ER microsomes were isolated from HEK293T cells pretreated with control, ATL2-targeting, ATL3-targeting, or ATL2/ATL3-targeting siRNAs (Fig. 8 D). Fusion between microsomes isolated from cells treated with ATL2-targeting siRNA (targeting both ATL2-1 and ATL2-2) was significantly lower than fusion between microsomes isolated from cells treated with control siRNA, whereas ATL3 knock-down did not markedly affect ER microsome fusion. Intriguingly, microsome fusion was reduced to a greater extent by the knock-down of both ATL2 and ATL3 than by the knock-down of ATL2 alone, suggesting that ATL3, although it barely contributes to ER fusion on its own in this cell line, promotes ATL2-driven ER fusion. Interestingly, ATL2 knock-down markedly enhanced the expression of ATL3 in HEK293T cells (Fig. 8 E), which partly explains why maximal inhibition of fusion required double knock-down of ATL2/ATL3 (Fig. 8 D). Collectively, these results suggest that ATL2, particularly ATL2-1, plays a major role in ER membrane fusion in HEK293T cells, although this ATL2 isoform by itself cannot efficiently support liposome fusion in vitro. The discrepancy between the inability of ATL2-1 to drive liposome fusion and the crucial role of ATL2-1 in ER microsome fusion indicates that autoinhibition of ATL2-1 by its CH is relieved during ER fusion in vivo. Additionally, siRNA-mediated knock-down experiments (Fig. 8, D and E) suggest that the main role of ATL3 is not to function directly in ER fusion in this cell line but to stimulate ATL2-mediated ER fusion. Thus, one potential scenario is that ATL3 relieves the autoinhibition of ATL2-1 or prevents ATL2-1 from being autoinhibited. To test this idea, we prepared liposomes containing ATL2-1 alone, ATL3 (at a sub-fusogenic concentration) alone, or both ATL2-1 and ATL3, and compared their fusion (Fig. 9 A). As expected, ATL2-1 or ATL3 alone did not support liposome fusion at a protein/lipid ratio of 1:1,000 or 1:2,000, respectively. Strikingly, however, efficient fusion was observed with liposomes containing both ATL2-1 (1:1,000) and ATL3 (1:2,000), suggesting that ATL3 prevents the autoinhibition of ATL2-1 and allows it to support ER fusion. Similarly, ATL2-2 at a subfusogenic concentration seemed to allow ATL2-1 to support efficient fusion because a significant fusion signal was observed with liposomes containing both ATL2-1 (1:500) and ATL2-2 (1:4,000), while liposomes bearing ATL2-1 (1:500) or ATL2-2 (1:4,000) alone supported little fusion (Fig. 9 B). By contrast, there was no synergistic fusion activity between ATL2-1 (K104A), a nucleotide binding-deficient mutant, and ATL3 or ATL2-2 (Fig. S6, A and B), further suggesting that the synergistic fusion activity shown in Fig. 9, A and B results from the alleviation of ATL2-1 autoinhibition by ATL3 or ATL2-2. Although the underlying mechanism remains elusive, the presence of ATL3 or ATL2-2 may prevent ATL2-1 from forming homotypic ATL2-1 dimers, which are fusion-incompetent, because both ATL2-1 molecules in the dimer are defective for GTPase activity and facilitate the formation of ATL2-1/ATL3 (Fig. S6 C) or ATL2-1/ATL2-2 heterotypic complexes, which may be fusion competent. Consistent with this idea, the presence of ATL3 markedly abolished the formation of homotypic multimers of ATL2-1 (Fig. 9 C). Another possibility is that the CH of ATL3 or ATL2-2 somehow prevents the inhibitory effect of the CH of ATL2-1 or is involved in relieving inhibition by the CH of ATL2-1. This possibility is ruled out by the observation that a subfusogenic concentration of ATL2-ΔCH enabled ATL2-1 to support liposome fusion, strongly suggesting that the CH of ATL3 or ATL2-2 plays no active roles in relieving inhibition by the CH of ATL2-1 (Fig. 9 D). Finally, another scenario is that a cytosolic factor, which is likely a peripheral ER membrane protein, relieves CH-mediated inhibition of ATL2-1 and enables it to support ER fusion in vivo. To test this possibility, we prepared cytosol from HEK293T cells and added it to fusion reactions containing liposomes bearing ATL2-1 alone. Strikingly, the addition of cytosol markedly stimulated the fusogenic activity of ATL2-1, which was otherwise largely autoinhibited by its CH (Fig. 9 E). The resulting fusion activity was dependent on the fusion activity of ATL2-1 because it was completely inhibited by the addition of an anti-ATL2 antibody. These results strongly indicate that a cytosolic protein relieves CH-mediated inhibition of ATL2-1 (Fig. 9 F).
Figure S5.
ATL2-1 is the major atlastin in HEK293T cells. (A) Total RNA was extracted from HEK293T cells, and cDNA was generated by RT-PCR. Real-time quantitative RT-PCR was performed to quantify the transcripts of human atlastins. All reactions were conducted in duplicate, and amplification signals from target genes were normalized to that from β-actin. The data represent the means ± SEM (error bars; n = 3). (B) The efficiency of in vitro fusion of ER microsomes isolated from HEK293T cells. Gluc1 and Gluc2 microsomes were mixed and incubated in the presence of GTP with increasing concentrations of PEG8000 at 37°C for 90 min. The data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. (C) An anti-ATL2 antibody inhibits fusion by specifically recognizing ATL2. Donor and acceptor ATL2-bearing liposomes were incubated with an affinity-purified anti-ATL2 antibody (0.25 μM) at 30°C for 10 min. Fusion reactions were initiated by adding GTP and further incubated in the absence or presence of the N-terminal cytosolic domain of ATL2 at a subinhibitory concentration (1 µM) for 30 min. Fusion is expressed as the percentage of total fluorescence. The data represent the means ± SEM (error bars; n = 3). (D) Donor and acceptor ATL2- or ATL3-bearing liposomes were incubated with the indicated concentrations of an affinity-purified anti-ATL2 antibody in the presence of GTP at 30°C for 40 min. The data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments.
Figure 8.
ATL2 is the major atlastin in HEK293T cells and is required for ER fusion. (A) Schematic representation of the in vitro membrane fusion assay using ER microsomes isolated from cultured HEK293 cells. ER microsomes containing ZIP-Gluc1-KDEL (Gluc1 microsome) or ZIP-Gluc2-KDEL (Gluc2 microsome) were isolated from HEK293 cells transfected with genes encoding ssZIP-Gluc1-KDEL or ssZIP-Gluc2-KDEL, mixed, and incubated in the presence of a GTP-regenerating system at 37°C for 90 min. Fusion was measured as reconstituted luciferase activity, which produces luminescence in the presence of coelenterazine. (B) ER microsome fusion in vitro requires GTP hydrolysis. Gluc1 and Gluc2 microsomes were mixed and incubated in the absence or presence of GTP or GTPγS on ice or at 37°C for 90 min. The data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. (C) ER microsome fusion is inhibited by an anti-ATL2 antibody. Gluc1 and Gluc2 microsomes were mixed and incubated in the presence of GTP with increasing concentrations of an anti-ATL2 antibody at 37°C for 90 min. The data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. (D) Gluc1 and Gluc2 microsomes were isolated from HEK293 cells treated with indicated siRNAs and incubated in the presence of GTP at 37°C for 90 min. Fusion values were normalized to fusion between microsomes isolated from HEK293 cells treated with control siRNA. The data represent the means ± SEM (error bars; n = 4). n refers to the number of independent experiments. ****P < 0.0001, ***P < 0.001, and **P < 0.01, one-way ANOVA with Tukey’s multiple comparisons test. (E) Protein profiles of microsomes used in D. The expression levels of ATL2, ATL3, ZIP-Gluc1-KDEL, and ZIP-Gluc2-KDEL were analyzed by immunoblotting using the indicated antibodies. Calnexin, an ER-resident protein, was used as a loading control. Source data are available for this figure: SourceData F8.
Figure 9.
ATL3 or a cytosolic factor relieves the autoinhibition by the CH and enables ATL2-1 to support ER fusion in vivo. (A) Donor and acceptor liposomes bearing ATL2-1 alone (1:1,000), ATL3 alone (1:2,000), or both ATL2-1 (1:1,000) and ATL3 (1:2,000) were mixed and incubated in the absence or presence of GTP. Fusion is presented as the percentage of total NBD fluorescence. The data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. ****P < 0.0001, two-way ANOVA with Tukey’s multiple comparisons test. (B) Donor and acceptor liposomes bearing ATL2-1 alone (1:500), ATL2-2 alone (1:4,000), or both ATL2-1 (1:500) and ATL2-2 (1:4,000) were mixed and incubated in the absence or presence of GTP. Fusion is presented as the percentage of total NBD fluorescence. The data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. *P < 0.05, **P < 0.01, two-way ANOVA with Tukey’s multiple comparisons test. (C) Liposomes containing ATL2-1 alone (1:1,000), ATL3 alone (1:2,000), or both ATL2-1 (1:1,000) and ATL3 (1:2,000) were incubated in the presence of GMP-PNP at 37°C for 30 min. Then, liposomes were treated or untreated with BMOE at 37°C for 1 h for crosslinking. Reactions were quenched by adding DTT (50 mM) at room temperature for 15 min. Samples were analyzed by SDS-PAGE followed by silver staining (Thermo Fisher Scientific). (D) Donor and acceptor liposomes bearing ATL2-1 alone (1:500), ATL2-ΔCH alone (1:4,000), or both ATL2-1 (1:500) and ATL2-ΔCH (1:4,000) were mixed and incubated in the absence or presence of GTP. Fusion is presented as the percentage of total NBD fluorescence. The data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. ***P < 0.001, ****P < 0.0001, two-way ANOVA with Tukey’s multiple comparisons test. (E) Donor and acceptor liposomes bearing ATL2-1 (1:500) were incubated with GTP in the absence or presence of cytosol (0.5 μg/μl) purified from HEK293T cells and affinity-purified anti-ATL2 antibodies (1 μM). Fusion is represented as the percentage of total NBD fluorescence. The data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. **P < 0.01, two-way ANOVA with Tukey’s multiple comparisons test. (F) Working model of how the CH-mediated autoinhibition is relieved during ER fusion in vivo. Source data are available for this figure: SourceData F9.
Figure S6.
ATL2 and ATL3 can form heterodimers. (A) Donor and acceptor liposomes bearing ATL2-1 (K107A) alone (1:500), ATL3 alone (1:2,000), or both ATL2-1 (K107A; 1:500) and ATL3 (1:2,000) were mixed and incubated in the absence or presence of GTP. Fusion is presented as the percentage of total NBD fluorescence. The data represent the means ± SEM (error bars; n = 3). (B) Donor and acceptor liposomes bearing ATL2-1 (K107A) alone (1:500), ATL2-2 alone (1:4,000), or both ATL2-1 (K107A; 1:500) and ATL2-2 (1:4,000) were mixed and incubated in the absence or presence of GTP. Fusion is presented as the percentage of total NBD fluorescence. The data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. The difference between samples was not statistically significant (n.s.), one-way ANOVA with Tukey’s multiple comparisons test. (C) The his6-tagged N-terminal cytosolic domain of ATL2 was incubated with free GST or GST-tagged ATL3 in the presence of GDP or GMP-PNP at 37°C for 1 h. The mixture was then further incubated with Ni-NTA agarose (Qiagen) at 4°C for 30 min, and proteins bound to Ni-NTA agarose were analyzed by SDS-PAGE followed by Coomassie Brilliant Blue staining. GST-ATL3, but not free GST, was co-precipitated with the his6-tagged N-terminal cytosolic domain of ATL2 in the presence of GMP-PNP, a non-hydrolyzable GTP analog. (D) GTPase activity is largely impaired in ATL3 (Y192C). The GTPase activity of wild-type ATL3 or ATL3 (Y192C) was determined by measuring the release of inorganic phosphate using a Malachite Green Phosphate Assay Kit (Sigma-Aldrich). Liposomes bearing wild-type ATL3 or ATL3 (Y192C) were incubated in the presence of GTP at 37°C for 30 min. After further incubation with malachite green reagents for 30 min, OD655 was measured, and the readings were normalized using a phosphate standard curve. The data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. ***P < 0.001, two-tailed unpaired Student’s t test. Source data are available for this figure: SourceData FS6.
ATL2-2 supports full fusion
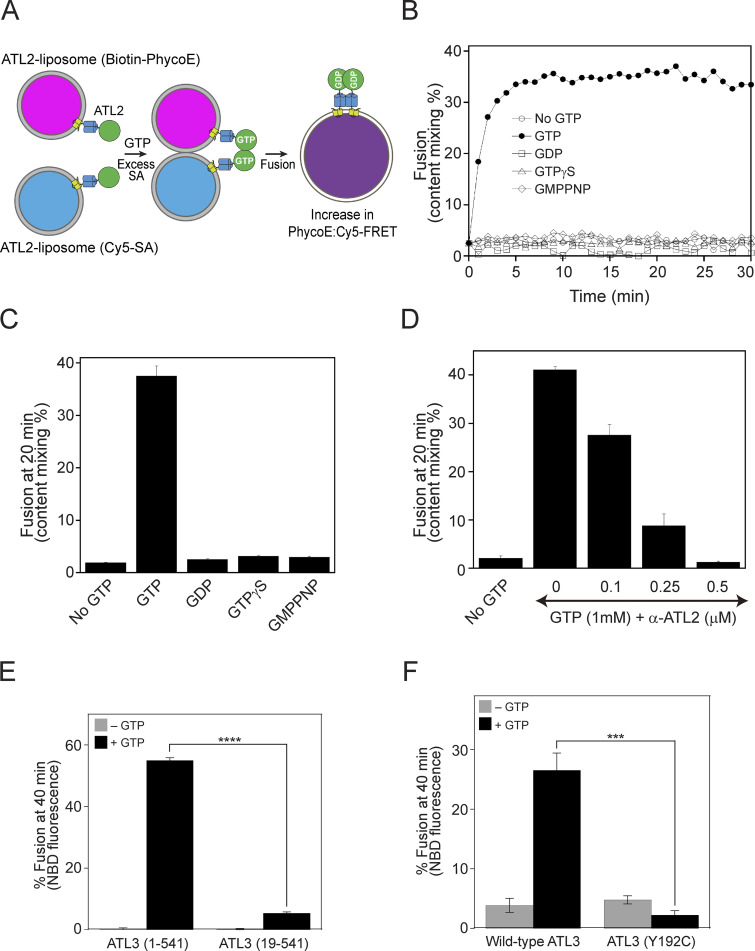
In this study, we examined the fusogenic activities of human atlastins by measuring fusion between proteoliposomes reconstituted with purified recombinant ATL1, ATL2, or ATL3. As previously described (Orso et al., 2009; Scott et al., 2003), one population of atlastin-bearing liposomes (donor) contains both 7-nitrobenzoxadiazole (NBD)-conjugated PE and rhodamine-labeled PE, while the other (acceptor) contains no fluorophore-tagged lipids. NBD and rhodamine constitute a fluorescence resonance energy transfer (FRET) pair, and fusion between these two liposome populations dilutes the two fluorescent probes and thereby decreases their FRET efficiency. Fusion between the two populations of proteoliposomes can be analyzed by measuring NBD fluorescence. Although this approach has been widely employed to analyze the fusogenic activities of various proteins, such as SNAREs and viral fusion proteins (Nunes-Correia et al., 2002; Weber et al., 1998), it cannot distinguish full fusion from hemifusion, an obligatory intermediate in the membrane fusion pathway where the outer leaflets of the two fusing membranes have mixed but the inner leaflets and internal contents remain separate, or membrane rupture and subsequent resealing (Zick and Wickner, 2014). Moreover, we reported that Sey1p-mediated fusion occurs through hemifusion (Kim et al., 2017), indicating that human atlastin-mediated fusion also involves hemifusion intermediates. To resolve this issue, we developed a content mixing assay using ATL2-2-reconstituted proteoliposomes bearing streptavidin derivatized with the fluorophore Cy5 (Cy5-SA) or biotinylated R-phycoerythrin (Biotin-PhycoE) in their lumens (Fig. 10 A), similar to that used for studying yeast vacuole fusion (Zick and Wickner, 2014). These proteoliposomes are mixed and incubated with Mg2+, GTP, and excess non-fluorescent streptavidin (to block any extra-lumenal interactions between Cy5-SA and Biotin-PhycoE that might have leaked out from the proteoliposomes). Similar to the observations in lipid mixing assays with human atlastin-bearing proteoliposomes (Fig. 1 C), robust content mixing was observed in the presence of GTP but not in the presence of GDP or non-hydrolyzable GTP analogs (Fig. 10, B and C). Furthermore, this content mixing was efficiently blocked by an affinity-purified anti-ATL2 antibody (Fig. 10 D). These results strongly suggest that human atlastins are sufficient to induce full fusion, consistent with the finding that dATL supports full fusion (Liu et al., 2012; Orso et al., 2009), as well as the recent report by Crosby et al. showing that a human ATL2-1 mutant (ATL2-1-ΔCH) lacking the inhibitory CH supports full fusion (Crosby et al., 2022).
Figure 10.
ATL2-2 supports content mixing. (A) Schematic representation of the content mixing assay using ATL2-containing liposomes. (B) ATL2-bearing liposomes containing Biotin-PhycoE and ATL2-bearing liposomes containing Cy5-SA were mixed and incubated at 30°C for 10 min. Reactions were further incubated in the absence or presence of GTP or a GTP analog at 30°C for 30 min, and FRET signals between phycoerythrin and Cy5 were measured every minute for 30 min. To determine total fluorescence, 1% Thesit was added to the mixture of ATL2-bearing liposomes containing Biotin-PhycoE and ATL2-bearing liposomes containing Cy5-SA in the absence of streptavidin. Experiments were performed multiple times, and representative data are shown. (C) Experiments were performed as described in B. Fusion after incubation for 20 min was determined. The data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. (D) Content mixing assays were performed in the presence of GTP and increasing concentrations of an anti-ATL2 antibody. The data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. (E) The short isoform [ATL3 (19–541)] of ATL3 failed to support liposome fusion. Donor and acceptor liposomes bearing the long isoform [ATL3 (1–541)] or the short isoform [ATL3 (19–541)] of ATL3 at a protein:lipid ratio of 1:1,000 were mixed and incubated in the absence or presence of GTP. Fusion is represented as the percentage of total NBD fluorescence. The data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. ****P < 0.0001, two-tailed unpaired Student’s t test. (F) Donor and acceptor liposomes bearing wild-type ATL3 (the long isoform) or mutant ATL3 (Y192C), which causes sensory neurodegeneration, at a protein/lipid ratio of 1:1,000 were mixed and incubated in the absence or presence of GTP. Fusion is represented as the percentage of total NBD fluorescence. The data represent the means ± SEM (error bars; n = 3). n refers to the number of independent experiments. ***P < 0.001, two-way ANOVA with Tukey’s multiple comparisons test.
The major isoform of ATL3, but not a short isoform, supports liposome fusion
Although we found that ATL3 was sufficient to induce liposome fusion (Fig. 1), a series of previous reports suggest that ATL3 does not function mainly as an ER fusogen. ATL3 functions as an ER-phagy receptor by interacting with GABARAP, and there are no obvious morphological changes in the ER network in ATL3-depleted cells (Chen et al., 2019a; Chen et al., 2019b; Hu et al., 2015; Pawar et al., 2017). These results are partly consistent with our observation that no fusion defect was seen with ER microsomes isolated from HEK293T cells treated with siATL3 (i.e., with ATL3 depletion by siRNA). Moreover, overexpression of ATL3 did not enhance ER microsome fusion, which heavily relies on ATL2-1. However, ATL3 seems to be essential for ATL2-1, an autoinhibited isoform of ATL2, to support membrane fusion according to our liposome reconstitution studies (Fig. 10 E). There are two known isoforms of ATL3: a long isoform (1–541 amino acids) and a short isoform (19–541 amino acids). To investigate whether these two ATL3 isoforms support membrane fusion comparably, we prepared liposomes containing the long or short isoform and compared their fusogenic activities (Fig. 10 E). Surprisingly, the short isoform did not support liposome fusion, although the only difference between the two isoforms is the presence of an additional 18 amino acids at the N-terminus in the long isoform. In addition, there are two autosomal-dominant missense mutations of ATL3, namely, Y192C and P338R, that cause sensory neurodegeneration. Interestingly, these mutants were reported to increase tethering when reconstituted into liposomes, but it has not been directly tested whether they affect ATL3-mediated membrane fusion. Taking advantage of our liposome fusion assay, we attempted to test whether one of the mutations that cause sensory neuropathy (Y192C) affects ATL3-mediated liposome fusion. The Y192C mutation of ATL3 markedly abolished liposome fusion (Fig. 10 F). Consistently, this mutation markedly abolished the GTPase activity of ATL3 (Fig. S6 D).
Discussion
In this study, we showed that human atlastins are sufficient to drive liposome fusion, demonstrating that atlastins are evolutionarily conserved fusogens for ER membrane fusion. Until recently (Crosby et al., 2022), mammalian atlastins, including the three human atlastins, have not been directly shown to have fusogenic activities, unlike their lower eukaryotic orthologs, such as dATL and yeast Sey1p. In previous studies that failed to observe human atlastin-driven liposome fusion (Betancourt-Solis et al., 2018; Wu et al., 2015), only human ATL1 was examined, and PC:PS liposomes, instead of liposomes with an ER-mimicking lipid composition, were used. The fusogenic activity of ATL1 was barely detectable in comparison with those of ATL2 and ATL3 (Fig. 1 A). Furthermore, the physiological lipid composition mimicking that of the ER seemed to be important, although not essential, for atlastin-mediated liposome fusion (Fig. S1). This idea is further supported by the requirement of physiological lipids for efficient Sey1p-driven liposome fusion and the presence of potential binding motifs of cholesterol, which stimulates Sey1p-mediated fusion and SNARE-mediated membrane fusion, in all human atlastins (Lee et al., 2019). Neutral lipids with small head groups that tend to form non-bilayer structures, such as PE and cholesterol, are important for SNARE-mediated membrane fusion (Zick et al., 2014). These findings may collectively explain the failure to reconstitute atlastin-mediated fusion in previous studies (Betancourt-Solis et al., 2018; Wu et al., 2015).
Lee and colleagues speculated that the previously observed lack of ATL1 fusion activity in vitro was because posttranslational modifications (PTMs), which are lacking in proteins purified from E. coli, are potentially required for the fusion activity of ATL1 (Crosby et al., 2022). Thus, they purified ATL1 from a HEK293-derived suspension cell line and successfully reconstituted ATL1-mediated fusion. Although they initially attributed the fusion activity to phosphorylation of the serine residues at positions 22 and 23 of ATL1, which are heavily phosphorylated, alanine substitution of both residues had no effect, demonstrating that phosphorylation is not required for the fusion activity of ATL1. This conclusion is further confirmed by our finding that ATL1 purified from E. coli can induce liposome fusion. Although our data suggest that PTMs are not essential for the fusion activity of all human atlastins (Fig. 1), it remains possible that both PTMs and the presence of curvature-inducing, non-bilayer-prone lipids cooperatively aid atlastin-mediated membrane fusion.
Why is the intrinsic fusogenic activity of ATL1 lower than those of the other human atlastins? One plausible idea is that fusion between ER tubules must be more tightly regulated in neuronal cells, which are subdivided into three distinct regions, namely, soma, axons, and dendrites. ER organization seems to differ between axons and dendrites (Wu et al., 2017), and thus additional factors that elaborately regulate the fusogenic activity of ATL1 in a spatiotemporal manner may be required. In contrast with ATL2 and ATL3, which accumulate at three-way junctions, ATL1 appears to be largely dispersed along ER tubules in non-neuronal cells. Co-expression of M1-spastin caused ATL1 to accumulate at three-way junctions, and co-reconstitution of M1-spastin increased the fusion efficiency of ATL1-bearing liposomes, indicating that there is a positive correlation between the fusogenic activities of atlastins and their accumulation at three-way junctions. In support of this idea, all atlastins and their orthologs that are sufficient to mediate liposome fusion accumulate at three-way junctions when expressed in vivo (Anwar et al., 2012; Hu et al., 2009; Lee et al., 2015; Orso et al., 2009; Zhang and Hu, 2013; Zhang et al., 2013; Zhou et al., 2019). Similarly, the enrichment of SNAREs and their accessory factors at fusion sites is a prerequisite for efficient fusion in various SNARE-mediated membrane fusion events (Fratti et al., 2004; Wang et al., 2003; Wickner and Rizo, 2017).
The major splice isoform of ATL2 predominantly expressed in non-neuronal cells, ATL2-1, is autoinhibited by its CH, resulting in little fusion when reconstituted into liposomes (Crosby et al., 2022; also see Fig. 6 A). Intriguingly, however, ATL2-1 is required for efficient fusion of ER microsomes isolated from HEK293T cells, in which ATL2-1 is predominantly expressed. How can autoinhibited ATL2-1 play a crucial role in ER fusion? Although liposomes bearing ATL2-1 or ATL3 alone at a sub-fusogenic concentration did not fuse, efficient fusion was observed in liposomes containing both ATL2-1 and ATL3 (Fig. 9 A), suggesting that ATL3, although its expression is marginal, enables ATL2-1 to support ER fusion. ATL2-2, which is barely expressed in HEK293T cells, may have a similar function (Fig. 9 B). Alternatively, a cytosolic factor may relieve the autoinhibition of ATL2-1 by the CH and allow ATL2-1 to support ER fusion (Fig. 9 E). This cytosolic factor is very likely a peripheral ER membrane protein because ER microsomes isolated from HEK293T cells fuse efficiently in the presence of GTP even without the addition of cytosol. Taken together, our data led us to propose the following working model of how ATL2-1 is autoinhibited by its CH and how the autoinhibition is relieved to enable ATL2-1 to support ER fusion in vivo (Fig. 9 F). The linker between the 3HB domain and the TM1 of ATL2-1 is flexible enough to allow the GTPase domain to bend toward the CH, which interacts with the ER membrane. This configuration may make ATL2-1 fusion incompetent. ATL3 can associate with ATL2-1 to form a heterodimer through the GTPase or 3HB domain (Fig. S6 C). This interaction may release the GTPase domain of ATL2-1 from the CH and the GTPase domain may become active. The resulting ATL2-1/ATL3 heterodimer is fusion-competent and can mediate ER fusion. Alternatively, a cytosolic factor may bind to the CH of ATL2-1, releasing the GTPase domain so that it becomes available for the formation of fusion-competent ATL2-1 homodimers. These two potential mechanisms are not necessarily mutually exclusive.
Materials and methods
DNA constructs and plasmids
The pHIS-ATL1 plasmid was generated by inserting a DNA fragment encoding human ATL1, which was PCR-amplified from pHAGE2-mCherry-ATL1 (#86678; Addgene), into the pHIS-parallel vector (Sheffield et al., 1999). The pHIS-ATL2 plasmid was generated by inserting a DNA fragment encoding human ATL2, which was cloned by RT-PCR of mRNA prepared from HeLa cells, into the pHIS-parallel vector. The open reading frame (ORF) of human ATL3 was PCR-amplified from pOTP7-ATL3 (hMU008830; Korea Human Gene Bank) and inserted into the pGST-parallel vector (Sheffield et al., 1999), generating pGST-ATL3. The pYJ408-myc-ATL1, pYJ408-myc-ATL2, and pYJ408-myc-ATL3 plasmids were generated by inserting DNA fragments, which were PCR-amplified from pHIS-ATL1, pHIS-ATL2, and pGST-ATL3, respectively, into pYJ408 (Starai et al., 2007). Similarly, the pEGFP-ATL1, pEGFP-ATL2, and pEGFP-ATL3 plasmids were generated using pEGFP-C1 (Invitrogen). The pMBP-spastin plasmid was generated by PCR amplification of the M1-spastin ORF from pCMV-Tag3A/myc-M1-spastin (#87719; Addgene), followed by insertion into the pMBP-parallel vector (Sheffield et al., 1999). The pcDNA3.1-puro-ssZIP-Gluc1-KDEL and pcDNA3.1-puro-ssZIP-Gluc2-KDEL plasmids were generated by inserting a DNA fragment encoding the signal sequence (the first 18 amino acids) of the ER-resident protein BiP and PCR products containing Gluc1-KDEL and Gluc2-KDEL, respectively, amplified from pYJ406-ssZIP-Gluc1-HDEL and pYJ406-ssZIP-Gluc2-HDEL (Lee et al., 2015) into pcDNA3.1-puro (Invitrogen). pAc-GFPC1-Sec61β was obtained from Addgene (#15108).
Antibodies
Anti-ATL2 rabbit polyclonal antibodies were custom-raised against the his6-tagged cytosolic domain (1−479 a.a.) of ATL2 (AbFrontier). These antibodies were affinity-purified using their antigens bound to SulfoLink resin (20401; Thermo Fisher Scientific). Anti-ATL3 rabbit polyclonal (ab117819; Abcam), anti-myc mouse monoclonal (2276S; Cell Signaling technology), anti-GFP mouse monoclonal (MA5-15256; Invitrogen), anti-Gluc rabbit polyclonal (E8023; New England BioLabs), anti-calnexin mouse monoclonal (sc-23954; Santa Cruz), anti-GAPDH mouse monoclonal (MA5-15738; Thermo Fisher Scientific), and anti-protein disulfide isomerase mouse monoclonal (MA3-019; Invitrogen) antibodies were purchased. Anti-Yet3p rabbit sera were a generous gift from C. Barlowe (Geisel School of Medicine at Dartmouth). Alexa Fluor 594-conjugated anti-mouse goat IgG (A-11005) was purchased from Invitrogen. Peroxidase-conjugated anti-mouse goat IgG (AB_10015289) and peroxidase-conjugated anti-rabbit goat IgG (AB_2313567) were purchased from Jackson ImmunoResearch.
Protein preparation
All recombinant proteins used in this study were produced in E. coli Rosetta 2 (λDE3; Novagen) grown in LB medium containing ampicillin by induction with 0.2 mM IPTG at 16°C for 16 h or at 37°C for 4 h. Cells were harvested, resuspended in ice-cold Buffer A (25 mM HEPES-NaOH, pH 7.4, 200 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, and a protease inhibitor cocktail), and lysed by sonication. After centrifugation at 23,000 g at 4°C for 30 min, the pellet was homogenized in Buffer B (25 mM HEPES-NaOH, pH 7.4, 200 mM NaCl, 10% glycerol, 1 mM EDTA, and 2% Triton X-100) containing 20 mM imidazole to prepare his6-tagged proteins or lacking imidazole to prepare GST- and MBP-tagged proteins. The homogenized pellet was incubated with gentle agitation at 4°C for 1 h and centrifuged at 23,000 g at 4°C for 30 min. The resulting supernatant was mixed with pre-washed Ni-NTA beads (QIAGEN) for his6-tagged proteins, with glutathione beads (GE Healthcare) for GST-tagged proteins, or with amylose beads (New England BioLabs) for MBP-tagged proteins at 4°C for 2 h. After washing with Buffer C (20 mM HEPES-NaOH, pH 7.4, 150 mM NaCl, 10% glycerol, 1 mM EDTA, and 0.4% Triton X-100) containing or lacking 20 mM imidazole, proteins were eluted with Buffer C containing 300 mM imidazole, 10 mM reduced glutathione, or 20 mM maltose. Eluates were then dialyzed against RB150 buffer (20 mM HEPES-NaOH, pH 7.4, 150 mM NaCl, and 10% glycerol) containing 1 mM EDTA and 0.4% Triton X-100.
Preparation of reconstituted atlastin-containing liposomes for the lipid mixing assay
Reconstitution of proteoliposomes bearing purified recombinant ATL1, ATL2, or ATL3 was performed as described previously with modifications (Kim et al., 2017; Orso et al., 2009). All non-fluorescent lipids were purchased from Avanti Polar Lipids, Inc., except for cholesterol (Sigma-Aldrich). The fluorescent lipids NBD-PE and N-(lissamine rhodamine B sulfonyl)-PE (Rh-PE) were purchased from Invitrogen. ER-mimicking lipid mixes for atlastin-bearing liposomes contained 1-palmitoyl-2-oleoyl-PC (50% [mol/mol]) for donor and acceptor atlastin-containing proteoliposomes), POPE (17 and 19% for donor and acceptor atlastin-containing liposomes, respectively), DOPS (19%), cholesterol (10%), diacylglycerol (1%), and fluorescent lipids (1.5% each of NBD-PE/Rh-PE or 1% dansyl-PE for donor and acceptor liposomes, respectively). These lipids in chloroform were mixed and dried under a stream of N2 gas. The resulting dried lipid films were dissolved in RB150 buffer containing 1 mM EDTA and incubated at 37°C for 30 min. Large unilamellar vesicles (LUVs) were formed by five freeze-thaw cycles in liquid N2 and water at 37°C. To form uniformly sized unilamellar liposomes, LUVs were extruded 18 times through a 100-nm polycarbonate filter (Avanti Polar Lipids, Inc.). Proteins were reconstituted into liposomes by detergent-assisted insertion as described previously (Rigaud and Lévy, 2003). In brief, purified proteins in Triton X-100 were mixed with preformed liposomes at the indicated protein-to-lipid ratio and an effective detergent-to-lipid ratio (Reff) of 0.384 and incubated at 4°C for 1 h. Reff is defined by the following equation: Reff = (Dtotal—Dwater)/[total lipid], where Dtotal is the total detergent concentration and Dwater is the aqueous monomeric detergent concentration (0.18 mM for Triton X-100). After mixing proteins and lipids at 4°C for 1 h, detergent was removed by incubation with Bio-Beads SM-2 adsorbent beads (Bio-Rad) at 4°C for 3 h with stirring. After further incubation with fresh beads at 4°C for 16 h with continuous stirring, proteoliposomes were adjusted to a final lipid concentration of 2 mM with RB150 buffer containing 1 mM EDTA. Small aliquots were frozen in liquid nitrogen and stored at −80°C.
Lipid mixing assay
Lipid mixing assays were performed as previously described with minor modifications (Lee et al., 2015; Lee et al., 2019). Briefly, labeled donor proteoliposomes and unlabeled acceptor proteoliposomes were mixed at a molar ratio of 1:5, transferred to a black polystyrene 384-well plate, and incubated at 30°C for 10 min. The reaction was initiated by adding 1 mM GTP and 1 mM Mg2+. NBD fluorescence was measured every minute using a SpectraMax Gemini XPS plate reader (Molecular Devices). After 40 min, β-octygludoside (final concentration 90 mM) was added to determine the total NBD fluorescence in the sample. Fusion was expressed as the percentage of total fluorescence.
GTPase activity assay
The GTPase activities of atlastins were determined using a Malachite Green Phosphate Assay Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Briefly, atlastin-bearing proteoliposomes were incubated with 0.25 mM GTP and 0.25 mM Mg2+ at 37°C for 30 min. Malachite Green reaction reagents were added to the reaction and further incubated for 30 min. The optical density at 655 nm (OD655) was measured using an iMark microplate reader (Bio-Rad). Absorbance was normalized to phosphate standards, and measurements of samples containing protein-free liposomes were used as background values.
Cell culture and transfection
COS7 and HEK293T cells were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco), 2 mM GlutaMAX-I (Invitrogen), 100 U/ml penicillin (Invitrogen), and 100 μg/ml streptomycin (Invitrogen) at 37°C with 5% CO2. Cells were transfected using Lipofectamine 3000 (Invitrogen). After 2 d, cells were imaged or harvested for immunoblotting. HEK293T cells stably expressing ssZIP-Gluc1-KDEL (HEK293T-Gluc1) or ssZIP-Gluc2-KDEL (HEK293T-Gluc2) were generated by transfection and long-term selection with puromycin (Sigma-Aldrich).
Preparation of ER microsomes
ER microsomes were prepared from cultured HEK293 cells as previously described (Farhan et al., 2010) with minor modifications. HEK293T-Gluc1 and HEK293T-Gluc2 cells were harvested, washed with PBS containing protease inhibitors, and centrifuged at 600 g for 10 min. The resulting pellets were resuspended in Buffer F (10 mM HEPES-KOH, pH 7.2, 250 mM sorbitol, 10 mM KOAc, and 1.5 mM MgOAc), lysed using a 26-gauge needle syringe, and centrifuged at 1,000 g for 10 min. The resulting supernatant was harvested and centrifuged at 6,000 g for 10 min. The pellet was resuspended in PS buffer (10 mM PIPES-KOH, pH 6.8, and 200 mM sorbitol) and centrifuged at 6,000 g for 10 min. The resulting pellet contained microsomes, which were obtained by gently resuspending the pellet in PS buffer. Yeast ER microsomes were prepared as previously described (Lee et al., 2015; Lee et al., 2019).
In vitro ER microsome fusion assay
ER microsome fusion assays using isolated mammalian microsomes were performed as described for the ER microsome fusion assay using isolated yeast microsomes (Lee et al., 2015; Lee et al., 2019) with modifications. The standard ER microsome fusion reactions contained 1 μg of Gluc1 microsomes, 1 μg of Gluc2 microsomes, reaction buffer (10 mM PIPES-KOH, pH 6.8, 5 mM MgCl2, and 200 mM sorbitol), an energy-regenerating system (0.5 mM MgCl2, 0.5 mg/ml creatine kinase, and 14.5 mM creatine phosphate), 10 μM coenzyme A, and 200 nM GST-ZIP. Fusion reactions were initiated by adding 1 mM GTP (Roche) and then incubated at 37°C. After 90 min, fusion reaction mixtures were mixed with coelenterazine (40 μM; GoldBio), and luminescence was measured using a luminometer (Centro XS3 LB 960; Berthold Technologies). ER microsome fusion assays using microsomes isolated from yeast cells expressing human atlastins were performed as previously described (Lee et al., 2015; Lee et al., 2019).
Real-time quantitative RT-PCR
Total RNA was extracted from HEK293T cells using RNAiso plus reagent (Takara), and cDNA was generated by RT-PCR using TOPscript RT DryMIX (Enzynomics). Real-time quantitative RT-PCR was performed using a CFX Connect Real-Time PCR Detection System (Bio-Rad) with SYBR Green TOPreal qPCR PreMIX (Enzynomics). All reactions were conducted in duplicate. Amplification signals from target genes were normalized to that from β-actin.
Immunofluorescence assay and fluorescence microscopy
COS7 cells grown on coverslips were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, treated with 5% normal goat serum to block non-specific antibody binding, and incubated with specific antibodies followed by Alexa Fluor 594-conjugated anti-mouse antibodies. Coverslips were mounted on glass slides using PermaFluor Aqueous Mounting Medium (Thermo Fisher Scientific). Cells were imaged at room temperature using a × 100/NA 1.45 oil immersion objective lens (Nikon Plan Apo λ) and a fluorescence microscope (Nikon Ti-U). Images were acquired using a Nikon camera (DS-Qi-2), analyzed using Nikon software (NIS-Element AR 4.40), and prepared using Photoshop (Adobe).
Construction of yeast strains
Budding yeast S. cerevisiae strains expressing human atlastins were generated by introducing pYJ408-myc-ATL1, pYJ408-myc-ATL2, or pYJ408-myc-ATL3 into BJ-Gluc1 sey1Δ or BJ-Gluc2 sey1Δ (Lee et al., 2015).
Proteoliposome reconstitution for the content mixing assay
ER-mimicking lipid mixes (47% PC, 20% PE, 10% soy phosphatidylinositol [PI], 8% PS, 3% phosphatidic acid [PA], 10% cholesterol, 1% diacylglycerol, and 1% PI(3)P) in chloroform were dried under a stream of N2 gas, resuspended in 2.5 × RB150 buffer by nutation at room temperature for 30 min, and mixed with β-octylglucoside (final concentration 0.2%) and biotinylated phycoerythrin (final concentration 4 μM) or Cy5-streptavidin (final concentration 8 μM). The mixture was dialyzed in RB150 buffer with Bio-Beads SM-2 (Bio-Rad) at 4°C for 5 h. Liposomes containing the fluorescent probe were then incubated with his6-tagged ATL2 in RB150 buffer containing 1 mM EDTA and 0.1% Triton X-100 at 4°C for 5 h. His6-tagged tobacco etch virus protease was added to cleave the his6-tag from ATL2. Triton X-100 was removed by incubation with Bio-Beads SM-2 at 4°C for 1 h. After further incubation with Bio-Beads SM-2 at 4°C for 16 h, ATL2-containing liposomes were obtained by flotation via density gradient centrifugation using HistoDenz (Sigma-Aldrich). Content mixing assays between atlastin-containing liposomes were performed as previously described (Zick and Wickner, 2014) with minor modifications. Briefly, Biotin-PhycoE-containing ATL2-bearing liposomes and Cy5-SA-containing ATL2-bearing liposomes were mixed at a molar ratio of 1:1 in the presence of 1 mM Mg2+ and 5 μM free streptavidin, transferred to a black 384-well plate (Corning), and incubated at 30°C for 10 min. Content mixing assays were initiated by adding 1 mM Mg2+ and 1 mM GTP. During incubation for 30 min at 30°C in a fluorescence plate reader (SpectraMax Gemini XPS, Molecular Devices), fluorophore FRET (phycoerythrin:Cy5 FRET; excitation, 565 nm; emission, 670 nm; cutoff, 630 nm) was measured each minute, and complete content mixing levels were assayed by adding 1% (wt/vol) Thesit to samples lacking streptavidin.
In vitro binding assay
Purified his6-sATL2 (N-terminal cytosolic domain of ATL2) and GST-ATL3 or GST proteins were mixed in RB150 buffer containing 0.5% Triton X-100 in the absence or presence of GMP-PNP or GDP (1 mM) and incubated at 37°C for 1 h with nutation. Thereafter, 20 μl of Ni-NTA beads (50% slurry, Qiagen), which had been pre-equilibrated with reaction buffer, was added to the protein mixture and further incubated at 4°C for 30 min in the presence of 20 mM imidazole. The beads were collected by centrifugation at 3,000 g for 1 min at 4°C and washed four times with 1 ml of RB150 buffer containing 0.5% Triton X-100 and 20 mM imidazole. Bound proteins were eluted with SDS sample buffer and analyzed by SDS-PAGE and Coomassie Brilliant Blue staining.
Crosslinking of ATL2 homodimers with BMOE
Proteoliposomes (1 mM) were incubated with 1.25 mM energy (GDP or GMP-PNP) at 37°C for 30 min to induce protein dimerization and tethering. Liposomes were then incubated with or without 5 mM BMOE for a further 1 h at 37°C to induce crosslinking. Reactions were quenched by the addition of DTT (final 50 mM) at room temperature for 15 min and mixed with SDS sample buffer lacking β-mercaptoethanol. Samples were analyzed by SDS-PAGE followed by silver staining (Thermo Fisher Scientific).
Protein–peptide pulldown
His6-tagged proteins (20 μM) were mixed with 200 μM C-terminal peptides at 4°C for 1 h. The protein–peptide mixtures were then further incubated with Ni-NTA agarose (Qiagen) in RB150 buffer containing 0.4% Triton X-100 and 20 mM imidazole for 30 min at 4°C. Ni-NTA agarose was washed with RB150 buffer containing 0.4% Triton X-100 and 20 mM imidazole. Proteins bound to Ni-NTA agarose were then analyzed by SDS-PAGE followed by Coomassie Brilliant Blue staining.
Liposome coflotation
Protein-free liposomes (0.5 mM) were incubated with 300 μM of the indicated peptides at 4°C for 2 h. The mixtures were then mixed at a 1:1 ratio with 70% (w/v) Histodenz, layered with 25% (w/v) Histodenz, and topped with RB150 buffer containing 1 mM EDTA. The gradient was then centrifuged at 250,000 g for 1 h at 4°C. The gradient was divided into six fractions, and the top fraction (fraction 1) and the bottom fraction (fraction 6) were analyzed by SDS-PAGE followed by Coomassie Brilliant Blue staining.
Statistical analysis
Statistical significance was determined from at least three independent experiments using GraphPad Prism 7 (GraphPad Software) for one-way or two-way ANOVA with Tukey’s test (for multiple comparisons) or Microsoft Excel 2016 for unpaired two-tailed Student’s t test. Data distribution was assumed to be normal, but this was not formally tested. All data represent the means ± SEM (error bars).
Online supplemental material
Fig. S1 shows that cholesterol and PE are critical for ATL2-mediated liposome fusion. Fig. S2 shows the differential effects of cholesterol and PE on ATL1-, ATL2-, or ATL3-mediated liposome fusion. Fig. S3 shows the effects of ATL2-1 or ATL2-2 peptide on ATL2-1-ΔCH-, ATL2-2-, or ATL3-mediated liposome fusion. Fig. S4 shows the per-residue confidence score (pLDDT) and predicted aligned error (PAE) of the AlphaFold structure prediction of ATL2-1 shown in Fig. 7 C. Fig. S5 shows that ATL2-1 is the major atlastin in HEK293T cells and is crucial for the in vitro fusion of microsomes isolated from HEK293T cells. Fig. S6 shows that ATL2 and ATL3 can form heterodimers.
Supplementary Material
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 5.
is the source file for Fig. 7.
is the source file for Fig. 8.
is the source file for Fig. 9.
is the source file for Fig. S4.
is the source file for Fig. S6.
Acknowledgments
We thank Dr. Charles Barlowe (Dartmouth College) for his generous gift of rabbit antisera against Yet3p.
This research was supported by the Cell Logistics Research Center (2016R1A5A1007318) funded by the National Research Foundation (NRF) of Korea and by grants NRF-2021M3A9G8022959 and NRF-2022R1A2C1007314 from the NRF of Korea.
Author contributions: E. Jang, Y. Moon, and Y. Jun conceived and designed the experiments. E. Jang, Y. Moon, S.Y. Yoon, J.A.R. Diaz, M. Lee, and N. Ko performed the experiments. E. Jang, Y. Moon, S.Y. Yoon, J.A.R. Diaz, M. Lee, N. Ko, J. Park, S.H. Eom, C. Lee, and Y. Jun analyzed and interpreted the data. E. Jang, Y. Moon, and Y. Jun wrote the manuscript.
References
- Anwar, K., Klemm R.W., Condon A., Severin K.N., Zhang M., Ghirlando R., Hu J., Rapoport T.A., and Prinz W.A.. 2012. The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J. Cell Biol. 197:209–217. 10.1083/jcb.201111115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt-Solis, M.A., Desai T., and McNew J.A.. 2018. The atlastin membrane anchor forms an intramembrane hairpin that does not span the phospholipid bilayer. J. Biol. Chem. 293:18514–18524. 10.1074/jbc.RA118.003812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, S.W., McIntosh T.J., and Lentz B.R.. 1992. Modulation of poly(ethylene glycol)-induced fusion by membrane hydration: Importance of interbilayer separation. Biochemistry. 31:2653–2661. 10.1021/bi00125a004 [DOI] [PubMed] [Google Scholar]
- Chen, Q., Teng J., and Chen J.. 2019a. ATL3, a cargo receptor for reticulophagy. Autophagy. 15:1465–1466. 10.1080/15548627.2019.1609862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q., Xiao Y., Chai P., Zheng P., Teng J., and Chen J.. 2019b. ATL3 is a tubular ER-phagy receptor for GABARAP-mediated selective Autophagy. Curr. Biol. 29:846–855.e6. 10.1016/j.cub.2019.01.041 [DOI] [PubMed] [Google Scholar]
- Crosby, D., Mikolaj M.R., Nyenhuis S.B., Bryce S., Hinshaw J.E., and Lee T.H.. 2022. Reconstitution of human atlastin fusion activity reveals autoinhibition by the C terminus. J. Cell Biol. 221:e202107070. 10.1083/jcb.202107070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, A.R., and Voeltz G.K.. 2013. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb. Perspect. Biol. 5:a013227. 10.1101/cshperspect.a013227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, K., Keller C., Pavur K., Glasgow K., Conn B., and Lauring B.. 2006. Interaction of two hereditary spastic paraplegia gene products, spastin and atlastin, suggests a common pathway for axonal maintenance. Proc. Natl. Acad. Sci. USA. 103:10666–10671. 10.1073/pnas.0510863103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan, H., Wendeler M.W., Mitrovic S., Fava E., Silberberg Y., Sharan R., Zerial M., and Hauri H.P.. 2010. MAPK signaling to the early secretory pathway revealed by kinase/phosphatase functional screening. J. Cell Biol. 189:997–1011. 10.1083/jcb.200912082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti, R.A., Jun Y., Merz A.J., Margolis N., and Wickner W.. 2004. Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J. Cell Biol. 167:1087–1098. 10.1083/jcb.200409068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, J.R., and Voeltz G.K.. 2011. The ER in 3D: A multifunctional dynamic membrane network. Trends Cell Biol. 21:709–717. 10.1016/j.tcb.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald, R., and Cohen-Fix O.. 2014. Morphology and function of membrane-bound organelles. Curr. Opin. Cell Biol. 26:79–86. 10.1016/j.ceb.2013.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J., Shibata Y., Zhu P.P., Voss C., Rismanchi N., Prinz W.A., Rapoport T.A., and Blackstone C.. 2009. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 138:549–561. 10.1016/j.cell.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, X., Wu F., Sun S., Yu W., and Hu J.. 2015. Human atlastin GTPases mediate differentiated fusion of endoplasmic reticulum membranes. Protein Cell. 6:307–311. 10.1007/s13238-015-0139-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper, J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature. 596:583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan, T.W., and Morré D.J.. 1970. Phospholipid class and fatty acid composition of golgi apparatus isolated from rat liver and comparison with other cell fractions. Biochemistry. 9:19–25. 10.1021/bi00803a003 [DOI] [PubMed] [Google Scholar]
- Kim, K.T., Moon Y., Jang Y., Lee K.T., Lee C., Jun Y., and Lee S.. 2017. Molecular mechanisms of atlastin-mediated ER membrane fusion revealed by a FRET-based single-vesicle fusion assay. Sci. Rep. 7:8700. 10.1038/s41598-017-09162-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M., Ko Y.J., Moon Y., Han M., Kim H.W., Lee S.H., Kang K., and Jun Y.. 2015. SNAREs support atlastin-mediated homotypic ER fusion in Saccharomyces cerevisiae. J. Cell Biol. 210:451–470. 10.1083/jcb.201501043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M., Moon Y., Lee S., Lee C., and Jun Y.. 2019. Ergosterol interacts with Sey1p to promote atlastin-mediated endoplasmic reticulum membrane fusion in Saccharomyces cerevisiae. FASEB J. 33:3590–3600. 10.1096/fj.201800779RR [DOI] [PubMed] [Google Scholar]
- Lentz, B.R. 2007. PEG as a tool to gain insight into membrane fusion. Eur. Biophys. J. 36:315–326. 10.1007/s00249-006-0097-z [DOI] [PubMed] [Google Scholar]
- Lin, Y.C., Boone M., Meuris L., Lemmens I., Van Roy N., Soete A., Reumers J., Moisse M., Plaisance S., Drmanac R., et al. 2014. Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat. Commun. 5:4767. 10.1038/ncomms5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T.Y., Bian X., Sun S., Hu X., Klemm R.W., Prinz W.A., Rapoport T.A., and Hu J.. 2012. Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc. Natl. Acad. Sci. USA. 109:E2146–E2154. 10.1073/pnas.1208385109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, Y., and Jun Y.. 2020. The effects of regulatory lipids on intracellular membrane fusion mediated by dynamin-like GTPases. Front. Cell Dev. Biol. 8:518. 10.3389/fcell.2020.00518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin-Leisk, J., Saini S.G., Meng X., Makhov A.M., Zhang P., and Lee T.H.. 2011. An intramolecular salt bridge drives the soluble domain of GTP-bound atlastin into the postfusion conformation. J. Cell Biol. 195:605–615. 10.1083/jcb.201105006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, T.J., Daga A., and McNew J.A.. 2011. Fusing a lasting relationship between ER tubules. Trends Cell Biol. 21:416–423. 10.1016/j.tcb.2011.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Correia, I., Eulálio A., Nir S., Düzgünes N., Ramalho-Santos J., and Pedroso de Lima M.C.. 2002. Fluorescent probes for monitoring virus fusion kinetics: Comparative evaluation of reliability. Biochim. Biophys. Acta. 1561:65–75. 10.1016/s0005-2736(01)00457-6 [DOI] [PubMed] [Google Scholar]
- Orso, G., Pendin D., Liu S., Tosetto J., Moss T.J., Faust J.E., Micaroni M., Egorova A., Martinuzzi A., McNew J.A., and Daga A.. 2009. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 460:978–983. 10.1038/nature08280 [DOI] [PubMed] [Google Scholar]
- Park, S.H., Zhu P.P., Parker R.L., and Blackstone C.. 2010. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J. Clin. Invest. 120:1097–1110. 10.1172/JCI40979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar, S., Ungricht R., Tiefenboeck P., Leroux J.C., and Kutay U.. 2017. Efficient protein targeting to the inner nuclear membrane requires Atlastin-dependent maintenance of ER topology. Elife. 6:e28202. 10.7554/eLife.28202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, M.J., and Voeltz G.K.. 2016. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 17:69–82. 10.1038/nrm.2015.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, K., Stone M.C., Weiner A.T., Gheres K.W., Zhou C., Deitcher D.L., Levitan E.S., and Rolls M.M.. 2016. Spastin, atlastin, and ER relocalization are involved in axon but not dendrite regeneration. Mol. Biol. Cell. 27:3245–3256. 10.1091/mbc.E16-05-0287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy, I., and Michnick S.W.. 2006. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods. 3:977–979. 10.1038/nmeth979 [DOI] [PubMed] [Google Scholar]
- Rigaud, J.L., and Lévy D.. 2003. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 372:65–86. 10.1016/S0076-6879(03)72004-7 [DOI] [PubMed] [Google Scholar]
- Rismanchi, N., Soderblom C., Stadler J., Zhu P.P., and Blackstone C.. 2008. Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum. Mol. Genet. 17:1591–1604. 10.1093/hmg/ddn046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson, C.M., Connell J.W., Edwards T.L., Bright N.A., Duley S., Thompson A., Luzio J.P., and Reid E.. 2006. Spastin and atlastin, two proteins mutated in autosomal-dominant hereditary spastic paraplegia, are binding partners. Hum. Mol. Genet. 15:307–318. 10.1093/hmg/ddi447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, B.L., Van Komen J.S., Liu S., Weber T., Melia T.J., and McNew J.A.. 2003. Liposome fusion assay to monitor intracellular membrane fusion machines. Methods Enzymol. 372:274–300. 10.1016/S0076-6879(03)72016-3 [DOI] [PubMed] [Google Scholar]
- Sheffield, P., Garrard S., and Derewenda Z.. 1999. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15:34–39. 10.1006/prep.1998.1003 [DOI] [PubMed] [Google Scholar]
- Starai, V.J., Jun Y., and Wickner W.. 2007. Excess vacuolar SNAREs drive lysis and Rab bypass fusion. Proc. Natl. Acad. Sci. USA. 104:13551–13558. 10.1073/pnas.0704741104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, M.L., and Fratti R.A.. 2019. The participation of regulatory lipids in vacuole homotypic fusion. Trends Biochem. Sci. 44:546–554. 10.1016/j.tibs.2018.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura, S., and Mima J.. 2016. Physiological lipid composition is vital for homotypic ER membrane fusion mediated by the dynamin-related GTPase Sey1p. Sci. Rep. 6:20407. 10.1038/srep20407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén, M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. 2015. Proteomics. Tissue-based map of the human proteome. Science. 347:1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Wang, L., Merz A.J., Collins K.M., and Wickner W.. 2003. Hierarchy of protein assembly at the vertex ring domain for yeast vacuole docking and fusion. J. Cell Biol. 160:365–374. 10.1083/jcb.200209095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, T., Zemelman B.V., McNew J.A., Westermann B., Gmachl M., Parlati F., Söllner T.H., and Rothman J.E.. 1998. SNAREpins: Minimal machinery for membrane fusion. Cell. 92:759–772. 10.1016/s0092-8674(00)81404-x [DOI] [PubMed] [Google Scholar]
- Westrate, L.M., Lee J.E., Prinz W.A., and Voeltz G.K.. 2015. Form follows function: The importance of endoplasmic reticulum shape. Annu. Rev. Biochem. 84:791–811. 10.1146/annurev-biochem-072711-163501 [DOI] [PubMed] [Google Scholar]
- Wickner, W. 2010. Membrane fusion: Five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu. Rev. Cell Dev. Biol. 26:115–136. 10.1146/annurev-cellbio-100109-104131 [DOI] [PubMed] [Google Scholar]
- Wickner, W., and Rizo J.. 2017. A cascade of multiple proteins and lipids catalyzes membrane fusion. Mol. Biol. Cell. 28:707–711. 10.1091/mbc.e16-07-0517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F., Hu X., Bian X., Liu X., and Hu J.. 2015. Comparison of human and Drosophila atlastin GTPases. Protein Cell. 6:139–146. 10.1007/s13238-014-0118-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Carvalho P., and Voeltz G.K.. 2018. Here, there, and everywhere: The importance of ER membrane contact sites. Science. 361:eaan5835. 10.1126/science.aan5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., Whiteus C., Xu C.S., Hayworth K.J., Weinberg R.J., Hess H.F., and De Camilli P.. 2017. Contacts between the endoplasmic reticulum and other membranes in neurons. Proc. Natl. Acad. Sci. USA. 114:E4859–E4867. 10.1073/pnas.1701078114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., and Hu J.. 2013. Homotypic fusion of endoplasmic reticulum membranes in plant cells. Front. Plant Sci. 4:514. 10.3389/fpls.2013.00514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., Wu F., Shi J., Zhu Y., Zhu Z., Gong Q., and Hu J.. 2013. ROOT HAIR DEFECTIVE3 family of dynamin-like GTPases mediates homotypic endoplasmic reticulum fusion and is essential for Arabidopsis development. Plant Physiol. 163:713–720. 10.1104/pp.113.224501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X., He Y., Huang X., Guo Y., Li D., and Hu J.. 2019. Reciprocal regulation between lunapark and atlastin facilitates ER three-way junction formation. Protein Cell. 10:510–525. 10.1007/s13238-018-0595-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, P.P., Patterson A., Lavoie B., Stadler J., Shoeb M., Patel R., and Blackstone C.. 2003. Cellular localization, oligomerization, and membrane association of the hereditary spastic paraplegia 3A (SPG3A) protein atlastin. J. Biol. Chem. 278:49063–49071. 10.1074/jbc.M306702200 [DOI] [PubMed] [Google Scholar]
- Zhu, P.P., Soderblom C., Tao-Cheng J.H., Stadler J., and Blackstone C.. 2006. SPG3A protein atlastin-1 is enriched in growth cones and promotes axon elongation during neuronal development. Hum. Mol. Genet. 15:1343–1353. 10.1093/hmg/ddl054 [DOI] [PubMed] [Google Scholar]
- Zick, M., Stroupe C., Orr A., Douville D., and Wickner W.T.. 2014. Membranes linked by trans-SNARE complexes require lipids prone to non-bilayer structure for progression to fusion. Elife. 3:e01879. 10.7554/eLife.01879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick, M., and Wickner W.T.. 2014. A distinct tethering step is vital for vacuole membrane fusion. Elife. 3:e03251. 10.7554/eLife.03251 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 5.
is the source file for Fig. 7.
is the source file for Fig. 8.
is the source file for Fig. 9.
is the source file for Fig. S4.
is the source file for Fig. S6.