Abstract
Genes encoding polypeptides of an ATP binding cassette (ABC)-type ferric iron transporter that plays a major role in iron acquisition in Synechocystis sp. strain PCC 6803 were identified. These genes are slr1295, slr0513, slr0327, and recently reported sll1878 (Katoh et al., J. Bacteriol. 182:6523–6524, 2000) and were designated futA1, futA2, futB, and futC, respectively, for their involvement in ferric iron uptake. Inactivation of these genes individually or futA1 and futA2 together greatly reduced the activity of ferric iron uptake in cells grown in complete medium or iron-deprived medium. All the fut genes are expressed in cells grown in complete medium, and expression was enhanced by iron starvation. The futA1 and futA2 genes appear to encode periplasmic proteins that play a redundant role in iron binding. The deduced products of futB and futC genes contain nucleotide-binding motifs and belong to the ABC transporter family of inner-membrane-bound and membrane-associated proteins, respectively. These results and sequence similarities among the four genes suggest that the Fut system is related to the Sfu/Fbp family of iron transporters. Inactivation of slr1392, a homologue of feoB in Escherichia coli, greatly reduced the activity of ferrous iron transport. This system is induced by intracellular low iron concentrations that are achieved in cells exposed to iron-free medium or in the fut-less mutants grown in complete medium.
Iron serves as an essential component of heme and iron sulfur centers integrated into a variety of proteins that function in basic physiological processes such as photosynthesis, respiration, and nitrogen metabolism (23). In the Earth's crust, iron is the fourth-most-abundant element. However, the biological availability of iron is severely reduced since in an aqueous oxygenic environment ferrous iron is quickly oxidized to ferric iron, which forms insoluble hydroxides at physiological pH (5, 6). Organisms have developed mainly two sophisticated systems for iron acquisition. One involves utilization of iron-chelating compounds including various siderophores and transport of chelated iron. The other system involves reduction of ferric iron to ferrous iron by a plasma membrane redox system, followed by uptake using specific transporters (11, 18).
Molecular analysis of iron transport systems has been carried out mostly on nonphotosynthetic bacteria (6). Escherichia coli has specific receptor proteins in the outer membrane that bind ferrichrome (FhuA), ferric aerobactin (IutA), ferric coprogen or ferric rhodotorulate (FhuE), and ferric dicitrate (FecA). FhuA, FhuE, and IutA are components of siderophore-mediated iron transport systems that involve typical ATP binding cassette (ABC)-type transporters consisting of a periplasmic iron-binding protein (FhuD) and cytoplasmic membrane proteins (FhuB and FhuC) (7). Ferric dicitrate is taken up via an ABC transporter system that consists of FecA to -E (22). E. coli also has a ferrous iron transport system consisting of polypeptides encoded by the feoA, -B, and -C genes. The product of the feoB gene has a typical ATP-binding motif at the N-terminal end. Mutants defective in feoA or feoB showed strongly reduced ferrous iron uptake activity (12). Transport systems for iron delivered as transferrin and lactoferrin, such as Sfu and Fbp systems in Serratia marcescens (3) and Neisseria gonorrhoeae (4), have been found in other bacteria. In these systems, ferric ion is transported across the inner membrane. The Sfu proteins constitute a typical ABC transporter in which SfuA is localized in the periplasm, SfuB is a cytoplasmic membrane protein, and SfuC is a membrane-bound protein carrying a nucleotide-binding motif.
In spite of these studies on nonphotosynthetic bacteria, little is known about the molecular mechanism of iron transport in photoautotrophic bacteria. We have recently demonstrated that one gene, registered as sll1878 in CyanoBase (http://www.kazusa.or.jp/cyano/), plays an important role in iron uptake in the cyanobacterium Synechocystis sp. strain PCC 6803 (14). The whole-genome sequence revealed that Synechocystis has 15 open reading frames (ORFs) whose putative products show high similarity with components of iron transporters identified in other bacteria (13). In order to understand the molecular mechanism of iron acquisition in Synechocystis, we have constructed mutants by disrupting these ORFs. Analysis of the mutants for growth and iron uptake both in nutrient-sufficient and iron-deprived conditions enabled us to identify the genes essential to ferric and ferrous iron transport.
MATERIALS AND METHODS
Growth conditions.
Wild-type and mutant cells of Synechocystis sp. strain PCC 6803 were grown at 30°C in BG-11 medium (21) buffered with 20 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES)–KOH at pH 8.0. Cultures were aerated with 3% (vol/vol) CO2 in air. Iron-free BG-11 medium was prepared as described previously (14). In order to simplify the assay system, sodium citrate, the ferric iron chelator, was omitted in iron-free BG-11 medium. To starve Synechocystis cells for iron, wild-type and mutant strains were grown in BG-11 medium, collected by centrifugation (1,600 × g, 8 min), and washed by resuspension in 20 mM TES-KOH buffer treated with Chelex 100 resin (Bio-Rad, Hercules, Calif.) at pH 8.0. After a second wash, cells were resuspended in fresh iron-free BG-11 medium. In order to deplete trace iron in the medium and in the cells (iron deprivation treatment), culture was continued for 18 h under continuous illumination of photosynthetically active radiation at 60 μmol of photons m−2 s−1 (400 to 700 nm).
Analysis of gene expression.
The amount of transcripts was evaluated by the reverse transcription-PCR (RT-PCR) method (8). RNAs were extracted from Synechocystis sp. strain PCC 6803 cells cultured in normal or iron-free BG-11 medium by the method of Aiba et al. (2), treated with RNase-free DNase I (Boehringer Mannheim), and then purified by phenol-chloroform extraction and ethanol precipitation. The RT reaction was performed using Superscript II (Gibco BRL) and reverse primers. The products were amplified by PCR and then analyzed by electrophoresis on a 0.8% agarose gel. Primers were designed so that the amplified products would be internal to the coding region of the gene. All the forward primers were designed for the sequences downstream of the translation initiation codon, and the reverse primers were designed to obtain PCR products of about 350 bp. The regions amplified by RT-PCR are summarized in Table 1 as base numbers counted from the initiation codons. The RNase P gene was used as a control template (1).
TABLE 1.
Regions of ORFs amplified by the RT-PCR method or replaced by drug resistance cassettes in the mutant strains
| ORF | Positionc of:
|
Drugd | Mutant strain | |
|---|---|---|---|---|
| RT-PCR producta | Deleted regionb | |||
| sll1206 | 28–368 | 444–458 | Kanamycin | M9 |
| sll1406 | 24–387 | 533–583 | Chloramphenicol | M9 |
| sll1409 | 73–439 | 581–719 | Hygromycin | M9 |
| slr1490 | 55–418 | 418–563 | Spectinomycin | M9 |
| sll1202 | 84–480 | 454–500 | Hygromycin | M8 |
| slr0513 | 60–483 | 448–530 | Kanamycin | M4 or M6 |
| slr1295 | 20–427 | 317–373 | Chloramphenicol | M3 or M6 |
| slr1319 | 66–441 | 473–477 | Kanamycin | M8 |
| slr1491 | 81–435 | 481–511 | Chloramphenicol | M8 |
| slr1492 | 15–395 | 166e | Spectinomycin | M8 |
| sll1878 | 32–400 | 134–479 | Kanamycin | M1 or M5 |
| slr0327 | 75–434 | 538–573 | Spectinomycin | M2 or M5 |
| slr1318 | 69–422 | 70–518 | Spectinomycin | M7 |
| slr1392 | 79–430 | 485–553 | Hygromycin | M10 |
Regions amplified by RT-PCR (from Fig. 2).
Regions replaced by drug resistance cassettes.
Positions of nucleotides are counted from the first nucleotide of the initiation codon.
Drug to which the cassette confers resistance.
Position at which cassette was inserted.
Construction of mutants.
Two DNA fragments from different regions in a given Synechocystis sp. strain PCC 6803 gene, each containing 500- to 700-bp nucleotides, were amplified by PCR. The primers used for the amplification contained different restriction endonuclease sites and an additional three nucleotides at their proximal ends for recognition of specific endonucleases. These sites were chosen based on sites present at both ends of the drug resistance marker gene used for constructing the gene disruption. Following digestion of the PCR products with appropriate endonucleases, the products were ligated to a cartridge carrying the drug resistance marker gene and to the pGEM-T vector (Promega, Madison, Wis.). The vector harboring the drug resistance cassette flanked by the PCR products on each side was amplified in E. coli and then transformed (26) into wild-type Synechocystis sp. strain PCC 6803 cells. The mutants constructed in this study have been deposited in website CyanoMutants (http://www.kazusa.or.jp/cyano/mutants/). The drug resistance cassette used for each inactivation and the sites of insertion into the target gene are shown in Table 1. The disrupted target gene in the transformants was segregated to homogeneity by successive streak purifications as determined by PCR amplification.
Determination of growth characteristics.
Iron-deprived wild-type and mutant cells were collected by centrifugation and resuspended in fresh iron-free BG-11 medium to optical densities at 730 nm (OD730) of 0.1, 0.01, and 0.001. The OD730 of the cell culture was determined using a recording spectrophotometer (model UV2200; Shimadzu Co., Kyoto, Japan). Two microliters of each of the cell suspensions was spotted onto normal or iron-free BG-11 agar plates. The plates were incubated in 3% (vol/vol) CO2 in air for 7 days with continuous illumination by fluorescent lamps providing photosynthetically active radiation at 60 μmol of photons m−2 s−1.
Measurements of ferric and ferrous iron uptake.
The amounts of iron taken up by wild-type and mutant cells were measured using radioactive tracer 59FeCl3 (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) as previously described (14). Ferric iron uptake was measured in the presence of 1 mM Ferrozine, which inhibits ferrous iron uptake (9). Uptake of ferrous iron was measured in the presence of 5 mM ascorbate, which reduces Fe3+ to Fe2+ (12). The uptake reaction was terminated by transferring the reaction mixture on ice, followed by centrifugation at 4°C. The pellet was washed twice with 20 mM TES-KOH (pH 8.0) containing 10 mM EDTA before being analyzed for the incorporation of 59Fe. The gamma emission from 59Fe in the cells was measured by the Auto Well gamma system (model ARC-380; Aloka, Tokyo, Japan). Cells at the late logarithmic stage of growth (OD730 = 1.0 to 1.3) were used for the iron uptake measurements. The concentration of FeCl3 in the uptake reaction solution was fixed at 10 μM, and the uptake reaction was performed with continuous light illumination. The light source was from a 600-W halogen lamp (Cabin Co., Tokyo, Japan), and the intensity of photosynthetically active radiation was 700 μmol of photons m−2 s−1 (400 to 700 nm)
Other methods.
Unless otherwise stated, standard techniques were used for DNA manipulation (19).
RESULTS
Location of gene products and genes inactivated in the mutants.
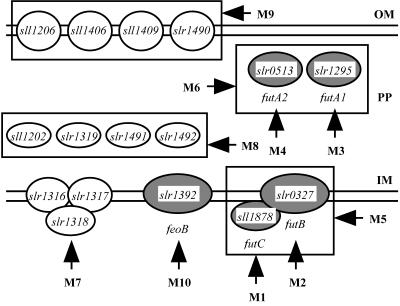
The whole-genome sequence of Synechocystis sp. strain PCC 6803 revealed the presence of 15 genes that are homologous to iron transporter genes identified in various nonphotosynthetic organisms (13). In addition, we have recently demonstrated that sll1878, whose putative product shows sequence similarity to HitC of an Sfu/Fbp-type ferric ion transporter in Haemophilus influenzae, plays an essential role in ferric iron transport (14, 20). Figure 1 depicts the possible locations of the products of these genes in cells, as predicted from the locations of their homologues. Sll1406, Sll1409, Slr1490, and Sll1206 are homologous to FhuA of E. coli (7) and IutA of Alcaligenes eutrophus (10), which are located in the outer membrane. Sll1202, Slr1491, Slr1492, and Slr1319 showed similarities with FhuD and FecB of E. coli (7, 22). Slr0513 (FutA2) and Slr1295 (FutA1) are homologous to SfuA of S. marcescens (3). These homologues are the substrate-binding proteins located in the periplasmic space. Slr1316 and Slr1317 showed similarities with FecC and FecD (22). Slr0327 (FutB) is homologous to HitB of H. influenzae (20). FecC and FecD are the inner membrane subunits of an ABC transporter that moves iron(III) dicitrate across the cytoplasmic membrane, and HitB is the inner membrane subunit of an Sfu/Fbp-type ferric ion transporter. Slr1392 showed similarity with FeoB of E. coli, which functions in ferrous iron transport (12). Figure 1 shows genes that were individually inactivated and groups of genes that were inactivated in a single strain. In all, 10 mutant strains (M1 to M10) were analyzed for their growth and iron uptake characteristics.
FIG. 1.
Mutants constructed by inactivating genes presumably involved in iron acquisition and the possible localization of the gene products. Open ovals, gene products that are positioned based on the localization of their homologues in nonphotosynthetic bacteria; shaded ovals, putative proteins that were experimentally shown to function in iron transport. For details see the text. Vertical arrows, genes that were individually inactivated; horizontal arrows, groups of genes (boxed) that were inactivated in a single strain. OM, outer membrane; PP, periplasmic space; IM, inner membrane.
In this paper, slr1295, sl0513, slr0327, and sll1878 are designated futA1, futA2, futB, and futC, respectively, for ferric iron uptake based on their functions as demonstrated below.
Expression of genes putatively involved in iron acquisition.
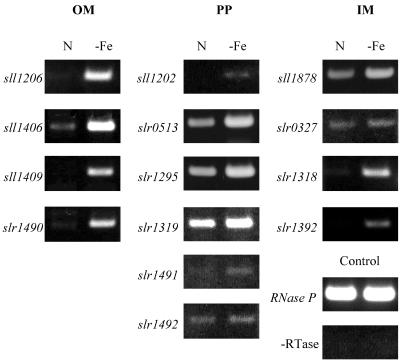
Figure 2 shows the levels of transcripts of 14 genes listed in Table 1 in wild-type cells grown in normal BG-11 medium and in iron deprived medium, as determined by the RT-PCR method. All four genes (sll1206, sll1406, sll1409, and slr1490) presumed to encode outer membrane receptor proteins were strongly expressed in iron-deprived cells. The slr1295 (futA1), slr0513 (futA2), and slr1319 genes encoding putative substrate-binding proteins were expressed at high levels in both iron-replete and iron-deprived cells, with the levels of transcripts higher in iron-deprived cells. The sll1878 (futC) and slr0327 (futB) genes were also expressed constitutively in iron-replete cells. The amounts of transcripts of sll1202, slr1491, and slr1492 were small, even in iron-deprived cells. The transcripts of slr1318 and slr1392 (feoB) were detected only in iron-deprived cells.
FIG. 2.
Expression profiles of putative iron transporter genes in Synechocystis sp. strain PCC 6803. The amounts of transcripts in cells grown in normal BG-11 medium (lanes N) or in iron-free BG-11 medium (lanes -Fe) were determined by the RT-PCR method. The regions of the genes amplified are summarized in Table 1. OM, PP, and IM are as indicated in Fig. 1. Absence of contamination of DNA was confirmed by performing the RT reaction without reverse transcriptase (-RTase) followed by PCR.
Growth characteristics.
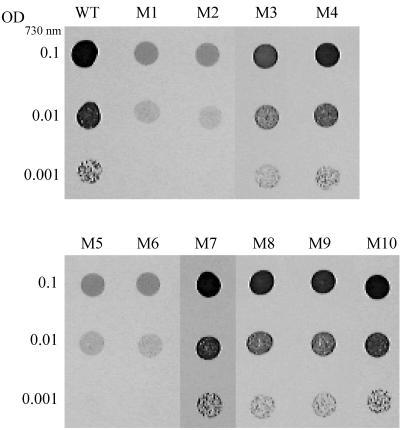
In a previous report, we showed that the futC-inactivated mutant (M1) grew much slower than the wild type on iron-free BG-11 agar plates (14). This was confirmed in this study (lanes M1 and WT in Fig. 3). The M2, M5, and M6 mutants showed growth characteristics similar to those of M1 and grew very poorly on iron-free plates, while all other mutants grew as well as the wild type under these conditions (Fig. 3). The growth of the M1, M2, M5, and M6 mutants appeared to be slower than that of the wild type on normal BG-11 plates, but the difference was not significant (not shown). The growth of the double mutant lacking futB and futC (M5) was similar to that of the single mutants (M1 and M2) in which these genes were individually inactivated. This suggested that futB and futC encode subunit polypeptides of a single transporter. The double mutant (M6), in which both futA1 and futA2 encoding putative substrate-binding proteins were inactivated, showed growth characteristics like those of the M1 strain, while single mutants M3 and M4, in which either of these genes was inactivated, grew as well as the wild type on the same iron-free medium. These results suggest that FutA1 and FutA2 are subunit proteins of an ABC transporter essential to iron acquisition in Synechocystis and that they have redundant or overlapping substrate-binding functions. These putative periplasmic iron-binding proteins may function in conjunction with FutB and FutC in the iron transporter complex. The M7, M8, M9, and M10 mutants showed wild-type growth characteristics under both normal and iron-free conditions. Thus, none of the genes inactivated in these mutants play a significant role in iron acquisition in wild-type cells under these conditions.
FIG. 3.
Growth of the wild type and mutants on solid iron-free BG-11 medium. Wild-type (WT) and mutant (M1 to M10 [Fig. 1]) cells of Synechocystis were pelleted by centrifugation and resuspended in iron-free BG-11 medium at pH 8.0. Two microliters each of cell suspensions, with OD730 values of 0.1, 0.01, and 0.001, were spotted on agar plates containing iron-free BG-11 buffered at pH 8.0, and the plates were incubated under 3% (vol/vol) CO2 in air for 7 days.
Uptake of ferric iron.
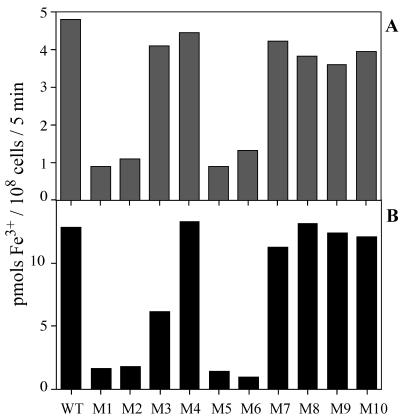
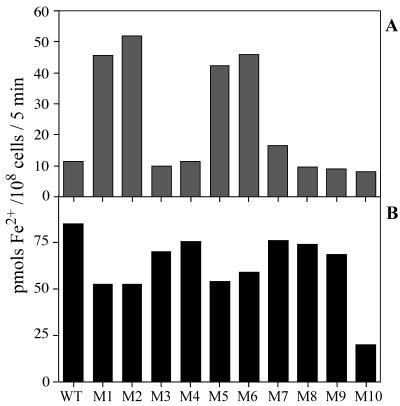
Figure 4 shows the amounts of Fe3+ taken up by the wild-type and mutant strains grown in nutrient-replete medium (A) or by iron-deprived cells (B) during 5 min of incubation in the light with 10 μM FeCl3 in the presence of 1 mM Ferrozine. The Fe3+ uptake activities of the M1, M2, M5, and M6 mutants, either maintained in nutrient-replete medium or starved for iron, were much lower than those of wild-type cells and of the other mutants. The activity of M5 was similar to that of M1 or M2. These results support the view that the fut genes disrupted in the mutants encode the subunits of a single Fe3+ transporter. These mutants still retained low levels of Fe3+ transport activity, suggesting the presence of an Fe3+ transporter(s) other than the fut-dependent system. The Fe3+ transport activity in the iron-deprived M3 cells was about half the activity of the M4 mutant (B), suggesting that FutA1 is a major iron-binding protein involved in fut-dependent Fe3+ transport.
FIG. 4.
Amounts of 59Fe3+ taken up by wild-type (WT) and mutant (M1 to M10 [Fig. 1]) cells. Cells grown in complete medium (A) and iron-deprived cells (B) were incubated with 10 μM 59FeCl3 for 5 min in the light in the presence of 1 mM Ferrozine.
Uptake of ferrous iron.
Figure 5 shows the amounts of Fe2+ taken up by the wild-type and mutant strains grown in nutrient-replete medium (A) and by iron-deprived cells (B) during 5 min of incubation in the light with 10 μM FeCl3 in the presence of 5 mM ascorbate (12). The activity of Fe2+ uptake was low in wild-type cells cultured in normal BG-11 medium and was increased about eightfold after iron deprivation. Thus, Synechocystis sp. strain PCC 6803 has an Fe2+ transporter that is induced by iron deprivation. The M3, M4, M7, M8, and M9 strains were similar to the wild type in that their activities were low in cells grown in normal medium and were increased six- to eightfold after iron deprivation. In contrast, the M1, M2, M5, and M6 mutants grown in nutrient-replete medium showed much higher activity of Fe2+ transport than wild-type cells, indicating that the Fe2+ transport system is induced in these mutants during growth in the complete medium. The activities in iron-deprived cells of these mutants were not significantly different from those in iron-replete cells and were two-thirds the activity in iron-deprived wild-type cells. The lower levels of Fe2+ uptake in the mutants may be due to changes in cell size or cellular composition. In fact, the chlorophyll contents of these four mutants were 2.7 to 3.1 μg/108 cells, while those in the wild-type cells and other mutant cells were 3.7 to 4.0 μg/108 cells. Therefore, the Fe2+ uptake activities in the iron-deprived cells of the mutants were similar to the wild-type activity when the values are expressed on a chlorophyll basis.
FIG. 5.
Amounts of 59Fe2+ taken up by wild-type (WT) and mutant (M1 to M10 [Fig. 1]) cells. Cells grown in complete medium (A) and iron-deprived cells (B) were incubated with 10 μM 59FeCl3 for 5 min in the light in the presence of 5 mM ascorbate.
The Fe2+ uptake activity of the M10 mutant was similar to the wild-type activity when cells were grown in normal medium but was much lower, about one-fourth of the wild-type activity, after iron deprivation. Thus, slr1392 (feoB) encodes a protein that is induced under iron deprivation and functions to transport ferrous iron.
DISCUSSION
In this study, we identified four genes that play a major role in iron acquisition in Synechocystis sp. strain PCC 6803, both in iron-replete and iron-deficient conditions. These four genes appear to encode subunits of a single Fe3+ transporter. The Synechocystis genome contains many genes that are homologous to the genes involved in iron acquisition in nonphotosynthetic bacteria. Ten mutants were constructed by inactivating single or multiple genes putatively involved in iron uptake (Fig. 1). Out of these mutants, only four mutants, with slr1295 (futA1), slr0513 (futA2), slr0327 (futB), and/or sll1878 (futC) in-activated, exhibited a mutant phenotype. Computer analysis indicated that all of these genes possess an ABC transporter gene family signature. The following results strongly suggest that these genes may encode subunit proteins of a single Fe3+ transporter. (i) The M1, M2, M5, and M6 mutants all exhibited the same growth characteristics (Fig. 3) and showed a marked reduction in their ability to take up Fe3+ (Fig. 4). All of these mutants showed high Fe2+ uptake activity when grown in normal BG-11 medium, indicating that the intracellular iron concentrations were low enough to induce the expression of the feoB gene even when the cells were grown in iron-replete medium (Fig. 5A). (ii) Inactivation of futB against the background of futC mutation did not further decrease the Fe3+ uptake activity (M1 versus M5 in Fig. 4), which strongly suggested that FutB and FutC are the subunit polypeptides of a single transporter. An ATP/GTP-binding motif (consensus: [AG]-x(4)-G-K-[ST] [25]) was identified in FutB (AARSLGKS: positions 458 to 465) as well as in FutC (GPSGCGKT; positions 53 to 60). It is well known that the archetypal ABC transporter consists of two ABC domains and two transmembrane domains. In many bacterial ABC transporters, each of these four domains is encoded as an independent polypeptide, although in other transporters the domains can be fused in any one of a number of ways into multidomain polypeptides (17). However, association of one subunit carrying the membrane domain and the ABC domain fused on one polypeptide, with a second peripheral subunit carrying only the ABC domain, has not been reported. The present results for M1, M2, and M5 strains imply the presence of such a unique combination in the ABC transporter family. (iii) Both futA1 and futA2 are homologous to sfuA in S. marcescens, which encodes a substrate-binding protein (Fig. 1). The M3 and M4 strains, in which these genes were inactivated individually, showed growth characteristics similar to those of the wild type (Fig. 3). Inactivation of other genes that encode putative substrate-binding proteins had no effect on growth and Fe3+ uptake activity (lanes M8 in Fig. 3 and 4). Only the M6 strain, lacking both futA1 and futA2, showed a mutant phenotype similar to those of the M1 and M2 strains. These results suggest that FutA1 and FutA2 have redundant or overlapping Fe3+ binding activities that function in the ABC transporter containing FutB and FutC as the inner membrane and membrane-associated subunits, respectively. (iv) All the fut genes were expressed constitutively in cells grown in normal medium (Fig. 2). This is consistent with the observation that inactivation of these genes significantly lowered the activity of Fe3+ uptake in cells grown in iron-replete medium (Fig. 4A). The form of iron transported across the inner membrane by the Fut system is most probably the ferric ion since this system is related to the Sfu/Fbp family, which transports ferric ion (3, 4). Our preliminary result for recombinant FutA1 expressed in E. coli indicated that the protein binds ferric ion directly.
The possibility of polar effects of the inactivation of fut genes is ruled out for the following reasons. Such polar effects may happen for the genes that constitute an operon structure. The insertion of a kanamycin resistance cassette does not suppress the expression of the gene(s) downstream of the cassette. As a result, the only possible gene whose inactivation causes a polar effect is slr0327 (futB). slr0328 and slr0329 are downstream of slr0327 on the genome. These genes are supposed to encode protein phosphatase and the xylose repressor, and it is unlikely that these proteins are involved in iron transport.
Inactivation of putative iron transporter genes other than fut genes did not have a significant effect on growth and Fe3+ uptake activity, suggesting that the contribution of these genes, if any, to iron acquisition is small. Synechococcus sp. strain PCC 7942 and Anabaena variabilis are known to produce unique hydroxamate-type siderophores (24). Morganella morganii takes up iron chelated to the fungal siderophore rhizoferrin (15). It is possible that the above putative iron transporter genes are involved in the transport of iron siderophores. The transport of iron in the form of complexes with siderophores may not be essential for iron acquisition under laboratory conditions, but it may give great advantage to Synechocystis cells in the natural environment. The low activities of Fe3+ uptake in M1, M2, M5, and M6 indicate the presence of another Fe3+ transporter(s), which could be encoded by the above putative iron transporter genes.
The mechanism by which ferric iron moves across the outer membrane to the periplasmic space is not known. Since the M9 strain did not show a mutant phenotype, a gene(s) other than those inactivated in this mutant may be involved in this process.
Synechocystis showed very high ability to take up ferrous iron when iron in the medium was reduced (Fig. 5). However, in the absence of ascorbate in the medium, the activity was much smaller. Since the activity in the absence of ascorbate fluctuates so much, depending on the growth conditions and stage of the growth, we did not include the results in this paper. The M10 strain grew as well as the wild type under iron-replete and iron-deficient conditions (Fig. 3), suggesting that the transport of Fe2+ is not essential for iron acquisition in the wild type. However, it is possible that FeoB-dependent Fe2+ uptake is essential for iron acquisition when Fut-dependent Fe3+ transport was impaired. In fact, we were unable to inactivate sll1392 (feoB) against the background of mutations of fut genes. The low Fe2+ transport activity in wild-type and M10 cells grown in normal medium and in iron-deprived M10 cells indicates that a second Fe2+ transporter is present constitutively in iron-replete cells of wild-type and mutant strains used in this study (16).
ACKNOWLEDGMENTS
This work was supported by grants for the Research for the Future program (JSPS-RFTF97R16001 and JPSP-RFTF96L00105), a grant-in aid for scientific research (B) (2)(12440228), and a grant from the Human Frontier Science Program (RG0051/1997M) to T.O. and National Science Foundation grant MCB 9727836 to A.R.G.
REFERENCES
- 1.Agustin V. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 1992;20:6331–6337. doi: 10.1093/nar/20.23.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 3.Angerer A, Gaisser S, Braun V. Nucleotide sequences of the sfuA, sfuB, and sfuC genes of Serratia marcescens suggest a periplasmic-binding-protein-dependent iron transport mechanism. J Bacteriol. 1990;172:572–578. doi: 10.1128/jb.172.2.572-578.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berish S A, Mietzner T A, Mayer L W, Genco C A, Holloway B P, Morse S A. Molecular cloning and characterization of the structual gene for the major iron-regulated protein expressed by Neisseria gonorrhoeae. J Exp Med. 1990;171:1535–1546. doi: 10.1084/jem.171.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer G L, Gillam A H, Trick C. Iron chelation and uptake. In: Fay P, van Baalen C, editors. The cyanobacteria. Amsterdam, The Netherlands: Elsevier; 1987. pp. 415–436. [Google Scholar]
- 6.Braun V, Hantke K, Koster W. Bacterial iron transport: mechanisms, genetics, and regulation. Met Ions Biol Syst. 1998;35:67–145. [PubMed] [Google Scholar]
- 7.Braun V, Gross R, Koester W, Zimmermann L. Plasmid and chromosomal mutants in the iron(III)-aerobactin transport system of Escherichia coli. Use of streptonigrin for selection. Mol Gen Genet. 1983;192:131–139. doi: 10.1007/BF00327658. [DOI] [PubMed] [Google Scholar]
- 8.Chelly J, Kahn A. RT-PCR and mRNA quantitation. In: Mullis K B, Ferré F, Gibbs R A, editors. The polymerase chain reaction. Boston, Mass: Birkhauser; 1994. pp. 97–109. [Google Scholar]
- 9.Ecker D J, Emery T. Iron uptake from ferrichrome A and iron citrate in Ustilago sphaerogena. J Bacteriol. 1983;155:616–622. doi: 10.1128/jb.155.2.616-622.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilis A, Khan M A, Cornelis P, Meyer J M, Mergeay M, van der Lelie D. Siderophore-mediated iron uptake in Alcaligenes eutrophus CH34 and identification of aleB encoding the ferric iron-alcaligin E receptor. J Bacteriol. 1996;178:5499–5507. doi: 10.1128/jb.178.18.5499-5507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerinot M L, Yi Y. Iron: nutritious, noxious, and not readily available. Plant Physiol. 1994;104:815–820. doi: 10.1104/pp.104.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kammler M, Schon C, Hantke K. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol. 1993;175:6212–6219. doi: 10.1128/jb.175.19.6212-6219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 14.Katoh H, Grossman A R, Hagino N, Ogawa T. A gene of Synechocystis sp. strain PCC 6803 encoding a novel iron transporter. J Bacteriol. 2000;182:6523–6524. doi: 10.1128/jb.182.22.6523-6524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn A, Braun V, Koester W. Ferric rhizoferrin uptake into Morganella morganii: characterization of genes involved in the uptake of a polyhydroxycarboxylate siderophore. J Bacteriol. 1996;178:496–504. doi: 10.1128/jb.178.2.496-504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lesuisse E, Raguzzi F, Crichton R R. Iron uptake by the yeast Saccharomyces cerevisiae: involvement of a reduction step. J Gen Microbiol. 1987;133:3229–3236. doi: 10.1099/00221287-133-11-3229. [DOI] [PubMed] [Google Scholar]
- 17.Linton K J, Higgins C F. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol Microbiol. 1998;28:5–13. doi: 10.1046/j.1365-2958.1998.00764.x. [DOI] [PubMed] [Google Scholar]
- 18.Marschner H, Römheld V. Strategies of plants for acquisition of iron. Plant Soil. 1994;165:261–274. [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Sanders J D, Cope L D, Hansen E J. Identification of a locus involved in the utilization of iron by Haemophilus influenzae. Infect Immun. 1994;62:4515–4525. doi: 10.1128/iai.62.10.4515-4525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanier R Y, Kunisawa R, Mandel M, Cohen-Bazire G. Purification and properties of unicellular blue-green algae (order Chroococcales) Bacteriol Rev. 1971;35:171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staudenmaier H, van Hove B, Yaraghi Z, Braun V. Nucleotide sequences of the fecBCDE genes and locations of the proteins suggest a periplasmic-binding-protein-dependent transport mechanism for iron(III) dicitrate in Escherichia coli. J Bacteriol. 1989;171:2626–2633. doi: 10.1128/jb.171.5.2626-2633.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Straus N A. Iron deprivation: physiology and gene regulation. In: Bryant D A, editor. The molecular biology of Cyanobacteria. Dordrecht, The Netherlands: Kluwer; 1994. pp. 731–750. [Google Scholar]
- 24.Trick C G, Kerry A. Isolation and purification of siderophores produced by cyanobacteria, Synechococcus sp. strain PCC7942 and Anabaena variabilis ATCC29413. Curr Microbiol. 1992;24:241–245. [Google Scholar]
- 25.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams J G K, Szalay A A. Stable integration of foreign DNA into the chromosome of the cyanobacterium Synechococcus R2. Gene. 1983;24:37–51. doi: 10.1016/0378-1119(83)90129-4. [DOI] [PubMed] [Google Scholar]