Abstract
Background
Identifying sarcopenia’s causally associated plasma proteins would provide potential therapeutic targets.
Methods
We screened out sarcopenia-related proteins with genome-wide association studies (GWAS) summary data and cis-protein loci genetic instruments. Summary data of sarcopenia were obtained from a GWAS of 256,523 Europeans aged 60 years and over. The causal effects of the proteins were investigated by cis-Mendelian Randomisation (MR) and multiverse sensitivity analysis. We also explored the robust proteins’ causal associations with appendicular lean mass (ALM) and surveyed their druggability and clinical development activities.
Results
In sum, 60 proteins from plasma proteome analysis studies and 12 from other studies were enrolled for MR analysis. In the whole population, four proteins (HPT, AT1B2, ISLR2 and TNF12) showed causal associations with the risk of sarcopenia according to the European Working Group on Sarcopenia in Older People (EWGSOP) criterion. In the female population, AT1B2 and TNFSF12 revealed causal associations with sarcopenia risk according to the EWGSOP criterion; HGF revealed a negative association according to the National Institutes of Health criterion. All of them were druggable, and the inhibitors of TNF12 and HGF were evaluated in clinical trials for other diseases. TNF12 also revealed a negative causal association with ALM, whereas HGF was positively causally associated with ALM.
Conclusions
Five druggable plasma proteins revealed causal associations with sarcopenia in the whole or female populations. TNF12 and HGF were the targets of therapeutic agents evaluated in clinical trials, and they were also causally associated with ALM. Our study suggested the potential mechanisms and therapeutic targets for sarcopenia.
Keywords: sarcopenia, therapeutic targets prediction, plasma proteomics, Mendelian randomisation analysis, older people
Key Points
Five druggable plasma proteins revealed causal associations with sarcopenia by Mendelian randomisation analysis.
Tumour Necrosis Factor Ligand superfamily member 12 (TNF12) and Hepatocyte growth factor (HGF) were also causally associated with appendicular lean mass.
Our study suggested the potential mechanisms and therapeutic targets for sarcopenia.
Introduction
Sarcopenia has been recognised as a muscle disease in the International Classification of Disease (ICD-10: M62[84]) and is defined as muscle mass and strength loss [1]. It is an age-related process in older people and is associated with increased adverse outcomes, including functional decline, frailty and mortality. Besides the risk factors at the time, it is also influenced by genetic and lifestyle factors throughout the life course [2]. Large sample genome-wide association studies (GWAS) [3–6] have determined some novel loci of muscle mass and revealed their genetic effects on muscle functions. Genomics and metabolomics studies [5, 6] also suggested accessible biomarkers genetically associated with muscle mass. Currently, clinical practice guidelines recommend physical activity as the primary treatment of sarcopenia [7]. Despite the rapid increase in understanding of its pathophysiology in recent years, no specific pharmacological approach for sarcopenia has been established [2, 8]. Developing drugs for this complex age-related disease is very challenging. However, some recent studies [9, 10] provided an approach to seek clues in this regard.
Proteins are the main regulators of molecular pathways and the targets of most drugs. Plasma contains proteins derived from almost all cells and tissues that are secreted into the circulation or released during cellular injury or turnover [9, 11]. Thus, disentangling the causal inference of plasma proteins and sarcopenia would aid in its mechanism and drug target identification. Several studies [12–14] that included participants with sarcopenia or weakness performed plasma proteomic analyses and identified numerous differential proteins. Furthermore, other studies [15–17] provided some biomarkers to improve the accuracy of differential diagnosis and prognostic assessment. However, the causal effects of these proteins on sarcopenia remained undetermined.
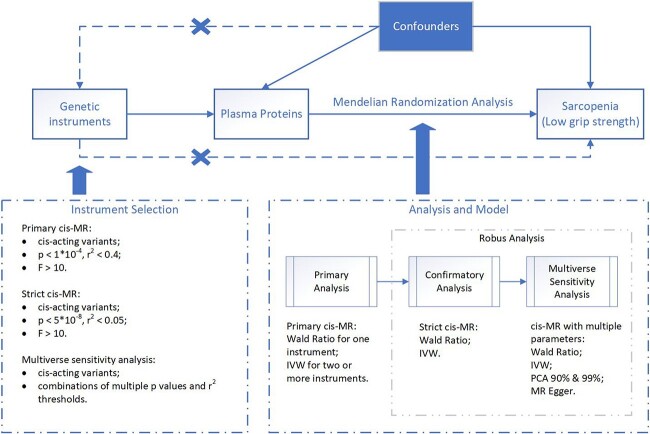
Mendelian randomisation (MR) is a genetic epidemiological study design with a great superior ability to investigate causality between traits and diseases [18] on the condition that three core assumptions are met: (i) genetic instruments are robustly associated with the exposure; (ii) genetic instruments are not associated with any confounders; (iii) Genetic instruments influence the risk of the outcome through exposure rather than other pathways [19]. Using the protein trait locus variants, derived from large-scale GWAS, as instruments in MR to estimate the causal effect of plasma protein levels on sarcopenia has the desirable properties to satisfy these assumptions [10]. To further reduce the horizontal pleiotropy, cis-MR [9, 10, 20] was proposed. Its feature was the selection of genetic instruments mapped to the vicinity of the transcriptional gene unit (defined as cis-acting variants). It has been used to identify novel therapeutic targets and predict the efficacy of drug targets for cardiovascular diseases and some other circumstances [9, 10, 21].
Herein, by summarising statistics from proteinic studies and leveraging data from multiple GWAS on plasma proteins [22–30] and sarcopenia [31], we implemented two-sample cis-MR studies to investigate the causal associations between plasma proteins and sarcopenia. We identified several potential proteins and surveyed their druggable characteristics.
Methods
Proteins screening
The sarcopenia-related proteome articles were mainly obtained from the PubMed database. Other databases include Web of Science and Google Scholar. The controlled vocabulary and the following keywords were used: ((‘sarcopenia’) OR (‘frailty’) OR (‘muscle weakness’)) AND ((‘proteomics’) OR (‘proteome’)). Secondary sources included papers cited by articles retrieved from the above mentioned sources. No limitations on the publication year and language were imposed. Studies that measured only in vitro parameters or used animal models were excluded.
Given that only a small number of publications with various study populations were included, we relaxed the criterion for protein enrollment. For the studies [12, 32–35] performed the plasma proteome analysis in which participants with the condition of sarcopenia or frailty or muscle weakness were recruited, the reported differential expression proteins (N = 178) were included. Additionally, from the muscular proteome analysis studies [13, 36] and review articles [37–41], we also collected the proteins associated with sarcopenia (N = 216) not reported in previous plasma studies. We also searched the PubMed database with the keywords as ((‘sarcopenia’) OR (‘frailty’) OR (‘muscle weakness’)) AND (‘biomarker’). The plasma protein biomarkers (N = 5) related to sarcopenia significantly, reported within 3 years and not collected previously, were recorded.
Then we searched the Uniprot database (https://www.uniprot.org/) to get the Entry, Entry Names, Recommended Name, Alternative Names and Gene Name of the proteins. Referred to the Genome Reference Consortium Human Build 37 (GRCh37) [42], we got the location of the gene in the chromosome. Then the sex-chromosome proteins or those without Gene names and available location of the genes (N = 10) were excluded. We queried the GWAS summary data of the proteins from the IEU OpenGWAS (https://gwas.mrcieu.ac.uk/) and GWAS Catalogue databases (https://www.ebi.ac.uk/gwas). The population of GWAS data was limited to ‘European’. For those proteins with more than one summary data, the one with the largest sample size was chosen. The GWAS summary data were available for 140 of 178 plasma differential proteins (Group1) and 80 of 221 other associated proteins (Group2). All of their GWAS data traits were plasma protein level measurements.
Then we extracted the cis-acting genetic variants as instrumental variables for each protein trait at the genome-wide significance level (P < 5 × 10−8). Those proteins without instrumental variables were excluded. Eventually, 60 and 12 proteins survived for further analyses in two groups, respectively. The flow chart of protein screening and the list of proteins enrolled were shown in Figure 1 and supplemental files (Tables S1 and S2).
Figure 1.
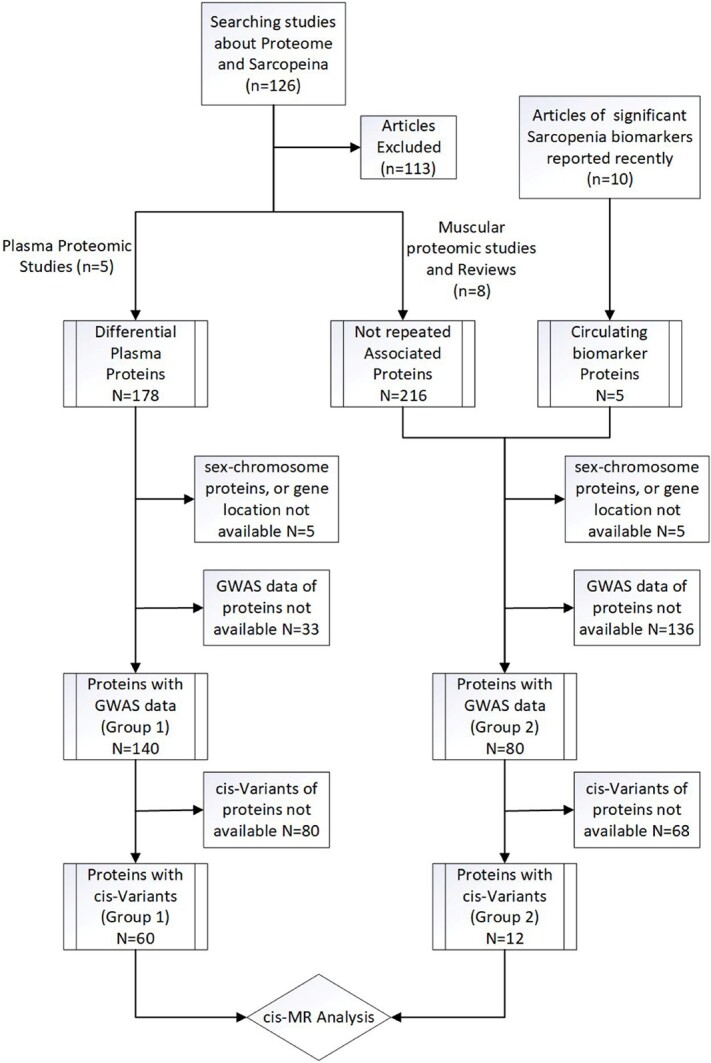
The flow chart of protein screening.
Outcome data
We obtained the GWAS summary data of sarcopenia (adjusted by age, sex and population substructure, accounting for relatedness and technical covariates) from the GWAS Catalogue database. It was obtained from the GWAS meta-analysis of 256,523 Europeans aged 60 years and over by Garan Jones et al. [31]. The data were summarised according to the low grip strength criteria of the European Working Group on Sarcopenia in Older People (EWGSOP) [43] (grip strength <30 kg Male; <20 kg Female) or Foundations of the National Institutes of Health (FNIH) [44] (<26 kg for males and < 16 kg for females), respectively. However, there were no available GWAS data for muscle mass with cut-off values in the European population. We chose appendicular lean mass (ALM) as another outcome to explore the effects of the proteins that survived in previous analysis on the muscle mass. ALM was proved to account for ≥75% of skeletal muscle in the body and was also a widely studied indicator among sarcopenia patients [43, 45]. It was obtained from the GWAS meta-analysis of 450,243 UK Biobank participants [46]. Moreover, these GWAS data also were stratified by sex. The study accessions and reported traits are listed in Table S3.
Statistical analysis
Primary cis-MR analysis
We assessed the causality of associations between the plasma levels of these proteins and sarcopenia by performing a two-sample cis-MR refer to the study by Henry et al. [9]. The selected genetic instruments were located within 200 kbp upstream or downstream of the cognate protein-encoding transcription start and stop sites. A less strict genome-wide significance P-value threshold (P < 1 × 10−4) and linkage disequilibrium (LD) r2 threshold (r2 < 0.4) was primarily applied [47, 48]. The referred LD model was derived from individual-level genotype data imputed against the Haplotype Reference Consortium [49] reference panel from a random sample of 10,000 UK Biobank [50] participants. Additionally, we calculated F-statistics for each SNP, and the weak instruments were excluded (F-statistics < 10) [51]. Then, MR models accounting for residual correlation were implemented. Considering the multiple testing, we also performed the Bonferroni correction (0.05/number of associated proteins with one or more instruments).
For proteins with only one single instrument, the Wald ratio Method was used. The inverse-variance weighted (IVW) method was used for those with more than one instrument. It approximated a risk ratio (RR) of sarcopenia per 1 standard deviation (SD) increase in normalised expression unit of proteins.
Robustness analysis of primary results
The proteins surviving multiple testing corrections from the primary cis-MR analysis underwent a conventional MR analysis with strict threshold parameters (P < 5 × 10−8; r2 < 0.05). For those proteins that showed consistently causal effects from strict MR analysis, multiverse sensitivity (MVS) analysis [9, 52] with multiple instrument selection parameters and MR models was performed. Besides Wald ratio and IVW models, the Egger regression and principal components with 99% variance and 90% variance explained were also used to calculate the estimates. Egger regression model was used for those with three or more instruments [48]. For each MR model, causal estimates were analysed with all combinations of five P-value thresholds (5 × 10−8, 1 × 10−5, 1 × 10−4, 1 × 10−3 and 1 × 10−2) and five LD r2 thresholds (0.05, 0.1, 0.2, 0.4 and 0.6). The association was considered robust if all point estimates from the above analyses accorded directionally with the primary and strict cis-MR analysis estimates. Outlier estimates outside 1.5 times the interquartile range above the upper quartile or below the lower quartile were dropped.
The overview of the study design is displayed in Figure 2. All MR analyses were implemented using the TwoSampleMR [25] and MendelianRandomization [53] package in R Software 4.1.3 (https://www.R-project.org). As TwoSampleMR package could not estimate the MR effects accounting for the correlation between variants yet, we mainly used it to extract, format and harmonise data. And then, the data were converted into the format required by the MendelianRandomization package for further calculations.
Figure 2.
Overview of MR framework. MR, Mendelian randomisation; IVW, inverse-variance weighted; PCA, principal components analysis; MR Egger, MR with Egger regression.
Druggability and clinical development activities survey
Additionally, the proteins’ druggability was queried from a list of druggable genes [11]. And the ChEMBL database [54] (release 30, https://www.ebi.ac.uk/chembl/) and ClinicalTrials (https://www.ClinicalTrials.gov) were searched to get the drug molecule types, indications and development activities of protein targets.
MR analysis on muscle mass
To preliminarily explore the causal effects of the robustly associated proteins on muscle mass, we performed an MR using ALM as the outcome. MR and MVS analyses were performed as described above.
Data availability and ethical statement
All data used in this genetic association study are obtained from published studies and open-access databases. The analysis codes used in the main analysis have been made available at https://github.com/tjh-chiangwei/cis-MR-codes. The analysis codes are modified from those shared online by Henry et al. [9] and cited R packages. The supporting data sets are available in the article, supplemental files and noted public datasets. This study was approved by the Ethics Committee of Tongji Hospital.
Results
Associated proteins screened by primary cis-MR analysis
Of the 60 differential proteins identified by plasma proteomics (Group 1), the primary MR analysis suggested the causal effect of three proteins on the risk of sarcopenia according to the EWGSOP criterion (P < 0.05/60) in the whole population and one protein in the female population. According to the FNIH criterion, one protein showed a causal association with sarcopenia risk in the female population. In the male population, no protein survived the multiple testing correction.
Of the 12 proteins collected from the other studies (Group 2), the primary MR analysis suggested the causal effect of one same protein on the risk of sarcopenia according to the EWGSOP criterion (P < 0.05/12) in the whole and the female populations. No protein showed a causal association with sarcopenia risk according to the FNIH criterion. Full MR results are provided inTable S4 and S5.
Robustly associated proteins identified by strict cis-MR and MVS analysis
When conventional MR was performed with strict parameters, the effect directions for the proteins that survived from primary MR analysis were consistent with that calculated in primary MR with P < 0.05/number of proteins. All of them underwent further MVS analysis. And robust causal associations were indicated as all MR point estimates from MVS analysis were directionally concordant (Table S6). Additionally, the R2 values of the robust proteins in primary cis-MR were calculated (Table S7). We also conducted the colocalization analysis of these proteins and the outcome traits (Table S8).
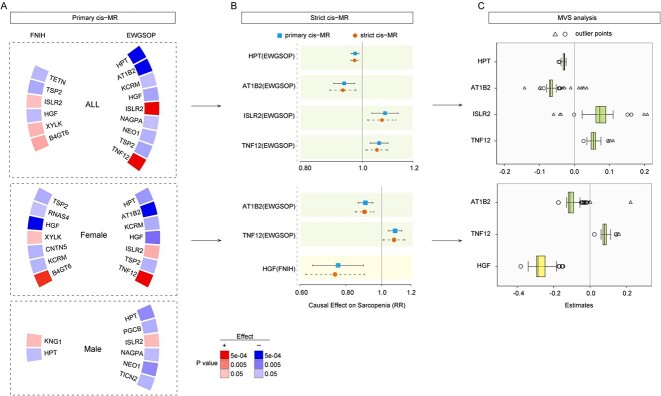
Thus, in the whole population, haptoglobin (HPT) (primary cis-MR: exp(beta) =0.97, P = 2.11E-04) and sodium/potassium-transporting ATPase subunit beta-2 (AT1B2) (0.94, P = 5.43E-04) showed negative causal associations with the risk of sarcopenia according to the EWGSOP criterion, whereas immunoglobulin superfamily containing leucine-rich repeat protein 2 (ISLR2) (1.08, P = 7.74E-04) and tumour necrosis factor ligand superfamily member 12 (TNF12) (1.06, P = 2.35E-04) showed positive causal associations.
In the female population, AT1B2 (0.90, P = 1.28E-05) and TNFSF12 (1.08, P = 8.53E-05) revealed causal associations with sarcopenia risk according to the EWGSOP criterion; hepatocyte growth factor (HGF) (0.76, P = 7.83E-04) revealed a negative association according to the FNIH criterion (Figure 3).
Figure 3.
The MR estimates of the association between plasma proteins and the risk of sarcopenia. (A) Circular heatmaps of statistical significance for estimates in primary cis-MR analysis. The proteins with P < 0.05 was presented. Colour represents the direction of effect and strength of the associations. (B) Forest plots of risk ratio from the proteins associated with sarcopenia in primary MR analysis. The odds ratios from primary and strict MR analysis were displayed. Coloured dots and error bars represent the point estimates and 95% CIs. (C) The box plots of MR estimates (beta value) from multiverse sensitivity analysis. The square represents the outlier estimates outside 1.5 times the interquartile range above the upper quartile or below the lower quartile. The triangle represents the outlier estimates outside three times the interquartile range above the upper quartile or below the lower quartile. MR, Mendelian randomisation; ALM, appendicular lean mass; CIs, confidence intervals; MVS, multiverse sensitivity; HPT, haptoglobin; AT1B2, sodium/potassium-transporting ATPase subunit beta-2; KCRM, creatine kinase M-type; HGF, hepatocyte growth factor; ISLR2, immunoglobulin superfamily containing leucine-rich repeat protein 2; NAGPA, N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase; NEO1, neogenin; TSP2, thrombospondin-2; TNF12, tumour necrosis factor ligand superfamily member 12; PGCB, brevican core protein; TICN2, testican-2; B4GT6, beta-1,4-galactosyltransferase 6; XYLK, glycosaminoglycan xylosylkinase; TETN, tetranectin; CNTN5, contactin-5; RNAS4, ribonuclease 4; KNG1, kininogen-1.
Survey of druggabilities and clinical development activities
The druggability and clinical development activities were assessed for those robustly associated proteins. Although no compounds were currently available in the ChEMBL database, HP, ATP1B2 and ISLR2 were listed as druggable targets in the druggable gene list [11]. TNF12 inhibitors, BIIB-023 and RO-5458640, were evaluated in clinical trials. The effect of BIIB-023 on muscle atrophy was explored in a phase one study, but no result was presented. HGF was suggested to have a protective effect on sarcopenia; however, currently, two available compounds that targeted HGF were its inhibitors (Table 1).
Table 1.
Druggablilities and clinical development activities of causally associated proteins [54]
| Proteins | Gene | Status | Molecule type | Compounds | Action type | Clinical development activities |
|---|---|---|---|---|---|---|
| HPT | HP | Druggable | Antibody | - | - | - |
| AT1B2 | ATP1B2 | Druggable | Small molecule | - | - | - |
| ISLR2 | ISLR2 | Druggable | Small molecule | - | - | - |
| TNF12 | TNFSF12 | In development | Antibody | BIIB-023; RO-5458640 |
Inhibitor | BIIB-023: phase 2 for lupus nephritis, phase 1 for rheumatoid arthritis and muscle atrophy. RO-5458640: phase 1 for neoplasm. |
| HGF | HGF | In development | Small molecule, Antibody | Rilotumumab; Ficlatuzumab |
Inhibitor | Rilotumumab: phase 1/2/3 for neoplasms; Ficlatuzumab: phase 1/2 for neoplasms. |
HPT, haptoglobin; AT1B2, sodium/potassium-transporting ATPase subunit beta-2; HGF, hepatocyte growth factor; ISLR2, immunoglobulin superfamily containing leucine-rich repeat protein 2; TNF12, tumour necrosis factor ligand superfamily member 12.
Investigation of proteins’ effects on muscle mass
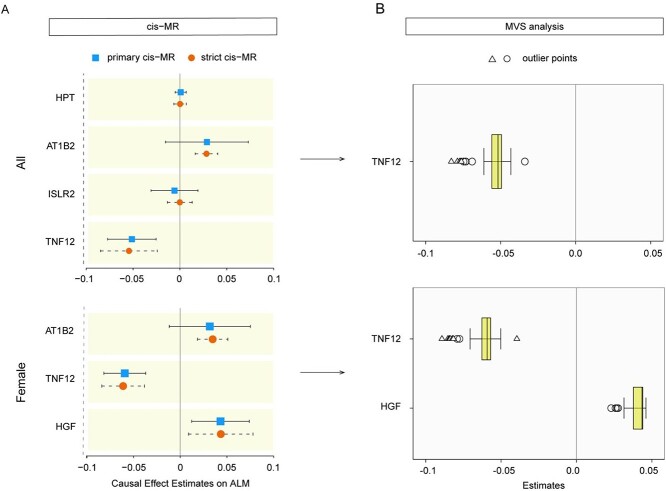
The causal effects of the robustly associated proteins on ALM were explored using the primary and strict MR analysis. Only TNF12 (primary cis-MR: beta = −0.05, P = 1.01E-04) showed a negative association with ALM (P < 0.05/4) in the whole population, whereas the other three proteins were not causally associated with ALM. In the female population, TNF12 (−0.06, P = 2.23E-07) also revealed a negative association with ALM (P < 0.05/2), whereas HGF (0.04, P = 6.14E-03) was positively causally associated with ALM. Moreover, the MVS analysis also showed that the associations were robust (Figure 4).
Figure 4.
The MR estimates of the association between robustly associated proteins and ALM. (A) Forest plots of risk ratio from the proteins associated with ALM. The beta value from primary and strict MR analysis were displayed. Coloured dots and error bars represent the point estimates and 95% CIs. (B) The box plots of MR estimates (beta value) from multiverse sensitivity analysis. The square represents the outlier estimates outside 1.5 times the interquartile range above the upper quartile or below the lower quartile. The triangle represents the outlier estimates outside three times the interquartile range above the upper quartile or below the lower quartile. MR, Mendelian randomisation; ALM, appendicular lean mass; CIs, confidence intervals; MVS, multiverse sensitivity; HPT, haptoglobin; AT1B2, sodium/potassium-transporting ATPase subunit beta-2; HGF, hepatocyte growth factor; ISLR2, immunoglobulin superfamily containing leucine-rich repeat protein 2; TNF12, tumour necrosis factor ligand superfamily member 12.
Discussion
To our knowledge, our study is the first large-scale analysis of the causal effects of plasma proteins on the risk of sarcopenia. A total of 220 proteins with GWAS data and associated with sarcopenia reported in previous studies were collected for MR analysis in this study. Five proteins revealed genetic support of causal effects on the risk of sarcopenia in the whole or female population. All of them were druggable, and two proteins were the targets of therapeutic agents evaluated in clinical trials. In some sense, our study provided clues to the prevention, diagnosis and treatment of sarcopenia.
Sarcopenia is a complex and progressive disease associated with a higher burden of morbidity and mortality [1]. Despite the more in-depth understanding of this disease in recent years, the effective biomarkers, preventive interventions and therapeutic drugs have not yet been established in clinical practice [2]. And it was considered occurrence with complex mechanisms, and the pathogenesis was not wholly understood [55]. Birth cohort studies [56] suggested its origin in early life and the influence of genetics. Our study used genetic instruments to calculate the causal estimates of plasma proteins on the risk of sarcopenia in MR, which validated this influence. Moreover, our study showed that multiple proteins with different functions presented potential causal effects, confirming its comprehensive development mechanisms.
In our study, we used the GWAS data summarised according to two criteria of sarcopenia. EWGSOP criterion was more commonly used, according to which more proteins showed causal associations with the risk of sarcopenia. According to the FNIH criterion, which was stricter, only HGF in the female population survived the multiple test correction. Additionally, in 2019, a revised European consensus (EWGSOP2) on definition and diagnosis was published. Its low grip strength cut-off values were lower than the EWGSOP criterion, and the female cut-off value was the same as the FNIH criterion. The comparison studies between EWGSOP and EWGSOP2 criteria [57, 58] showed that it was controversial which standard was more effective in clinical practice. EWGSOP2 criterion might produce fewer associations with adverse health outcomes in UK Biobank cohort [57]. Our study presented that different plasma proteins revealed causal effects on the risk of sarcopenia according to different criteria. Therefore, the study about the diagnostic criteria of sarcopenia might need to be more in-depth and refined.
As the gender disparity in skeletal muscle morphology and function, sarcopenia was defined by the gender-specific cut points of various parameters. Our study presented different results between the male and female populations. Associated estimates were calculated for several plasma proteins with P < 0.05 from primary MR in the male population, but no one survived multiple test correction. It might be because proteins encoded by sex-chromosome genes or hormones played an essential role in maintaining skeletal muscle homeostasis in the male population. There were different responses to catabolic conditions between males and females [59]. Further studies should be conducted for different genders, and the interventions need to be gender-specific.
Besides the proteomic analysis study, only a small number of clinical observational studies [14, 60, 61] presented the associations between sarcopenia and HP, TNF12 and HGF. However, they failed to demonstrate the cause-and-effect relationships and the mechanisms. Only one animal study [62] suggested that the TNFSF12-Fn14 signalling axis potentially contributed to age-associated muscle atrophy and fibrosis. In previous studies, the other two proteins (AT1B2 and ISLR2) were not reported to be related to sarcopenia. Our study provided new evidence of the causal associations between these plasma proteins and the risk of sarcopenia in the corresponding population. More attention needs to be paid to exploring the potential mechanism of these proteins in the pathogenesis of sarcopenia.
It suggested that TNF12 and HGF were causally associated with the relevant muscle mass trait. Other proteins did not show the causal effects on ALM. No available GWAS data for muscle mass with cut-off thresholds might be the reason, or they were causally associated with low muscle strength through other related traits. Furthermore, for some proteins (HP, ISLR2 and HGF), the associations with sarcopenia risk presented opposite directions in proteomic and MR analysis. It might be because the alterations of these proteins’ levels occurred during the preclinical phase or as an adaptive protective response to mitigate the disease process.
In our study, all robustly associated proteins were evaluated as druggable. The inhibitors of TNF12 and HGF were the targets of therapeutic agents evaluated in clinical trials. In addition, TNF12 showed positive associations with sarcopenia risk in both observational studies and MR analysis. And TNF12 was causally associated with both muscle strength and mass traits. Thus, the TNF12 inhibitor might be a promising therapeutic agent for sarcopenia. More research was needed before other proteins were used as therapeutic targets.
There are some limitations in our study. First, the participants were of European origin. Our results’ generalisation needs to be confirmed. Second, although our MR framework was based on previous high-impact studies [9, 21], the lack of consensus about the optimal approach to cis-MR [9] is an unavoidable limitation. Third, we could not directly assess associations of individual genetic variants with potential confounders due to the lack of knowledge and the unavailability of individual-level data. A type 2 error caused by negative confounding or the imprecision of MR estimates could not be excluded. Furthermore, as our study showed, the diagnostic criteria significantly impacted the results. However, the current public database does not contain the summary-level statistics for sarcopenia diagnosed by the newest algorithm. Finally, although this study suggests that some plasma proteins are causally associated with sarcopenia risk, it merely provides a prediction without verification. The causal associations need to be further explored and verified in future studies.
Conclusions
This cis-MR analysis study evaluated the causal effects of the plasma proteins with cis-variants on the risk of sarcopenia. Five of them revealed causal associations in the whole or female populations. These proteins were shown to be druggable, and two (TNF12 and HGF) were the targets of therapeutic agents evaluated in clinical trials. Our results suggested the potential mechanisms and therapeutic targets for sarcopenia.
Supplementary Material
Acknowledgements
Thanks to the investigators who performed GWAS analyses and summarised the data available in this study. We thank Dr. Henry Albert and others for sharing the analysis code.
Contributor Information
Wei Jiang, Department of Gastrointestinal Surgery, Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology, Wuhan 4300030, PR China.
Wenli Zhan, Department of Gastrointestinal Surgery, Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology, Wuhan 4300030, PR China.
Luoqi Zhou, Department of Neurology, Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology, Wuhan 4300030, PR China.
Minghao Dong, Department of Neurology, Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology, Wuhan 4300030, PR China.
Liang Liu, Department of Gastrointestinal Surgery, Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology, Wuhan 4300030, PR China.
Xiangshang Xu, Department of Gastrointestinal Surgery, Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology, Wuhan 4300030, PR China.
Zhixin Cao, Department of Gastrointestinal Surgery, Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology, Wuhan 4300030, PR China.
Declaration of Conflicts of Interest
None.
Declaration of Sources of Funding
This study was supported by the Chen Xiao-Ping Foundation for the Development of Science and Technology of Hubei Province (CXPJJH121003-2104) for Z.-X.C.
References
- 1. Petermann-Rocha F, Balntzi V, Gray SR et al. Global prevalence of sarcopenia and severe sarcopenia: a systematic review and meta-analysis. J Cachexia Sarcopenia Muscle 2022; 13: 86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cruz-Jentoft AJ, Sayer AA. Sarcopenia. The Lancet 2019; 393: 2636–46. [DOI] [PubMed] [Google Scholar]
- 3. Zillikens MC, Demissie S, Hsu YH et al. Large meta-analysis of genome-wide association studies identifies five loci for lean body mass. Nat Commun 2017; 8: 80. 10.1038/s41467-017-00031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Karasik D, Zillikens MC, Hsu YH et al. Disentangling the genetics of lean mass. Am J Clin Nutr 2019; 109: 276–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Livshits G, Gao F, Malkin I et al. Contribution of heritability and epigenetic factors to skeletal muscle mass variation in United Kingdom twins. J Clin Endocrinol Metab 2016; 101: 2450–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Korostishevsky M, Steves CJ, Malkin I, Spector T, Williams FM, Livshits G. Genomics and metabolomics of muscular mass in a community-based sample of UK females. Eur J Hum Genet 2016; 24: 277–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cruz-Jentoft AJ, Bahat G, Bauer J et al. Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing 2019; 48: 16–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Spiegeleer A, Beckwee D, Bautmans I, Petrovic M, Sarcopenia Guidelines Development group of the Belgian Society of Gerontology and Geriatrics (BSGG) . Pharmacological interventions to improve muscle mass, muscle strength and physical performance in older people: an umbrella review of systematic reviews and meta-analyses. Drugs Aging 2018; 35: 719–34. [DOI] [PubMed] [Google Scholar]
- 9. Henry A, Gordillo-Maranon M, Finan C et al. Therapeutic targets for heart failure identified using proteomics and Mendelian randomization. Circulation 2022; 145: 1205–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmidt AF, Finan C, Gordillo-Maranon M et al. Genetic drug target validation using Mendelian randomisation. Nat Commun 2020; 11: 3255. 10.1038/s41467-020-16969-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Finan C, Gaulton A, Kruger FA et al. The druggable genome and support for target identification and validation in drug development. Sci Transl Med 2017; 9: eaag1166. 10.1126/scitranslmed.aag1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sathyan S, Ayers E, Gao T, Milman S, Barzilai N, Verghese J. Plasma proteomic profile of frailty. Aging Cell 2020; 19: e13193. 10.1111/acel.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou D, Zhang Y, Mamtawla G et al. Iron overload is related to muscle wasting in patients with cachexia of gastric cancer: using quantitative proteome analysis. Med Oncol 2020; 37: 113. 10.1007/s12032-020-01439-w. [DOI] [PubMed] [Google Scholar]
- 14. Li CW, Yu K, Shyh-Chang N et al. Circulating factors associated with sarcopenia during ageing and after intensive lifestyle intervention. J Cachexia Sarcopenia Muscle 2019; 10: 586–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim M, Walston JD, Won CW. Associations between elevated growth differentiation Factor-15 and sarcopenia among community-dwelling older adults. J Gerontol A Biol Sci Med Sci 2022; 77: 770–80. [DOI] [PubMed] [Google Scholar]
- 16. Ingenbleek Y. Plasma transthyretin as a biomarker of sarcopenia in elderly subjects. Nutrients 2019; 11: 895. 10.3390/nu11040895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jung HW, Park JH, Kim DA et al. Association between serum FGF21 level and sarcopenia in older adults. Bone 2021r; 145: 115877. 10.1016/j.bone.2021.115877. [DOI] [PubMed] [Google Scholar]
- 18. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014; 23: R89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Davies NM, Holmes MV, Davey SG. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 2018; 362: k601. 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chong M, Sjaarda J, Pigeyre M et al. Novel drug targets for ischemic stroke identified through Mendelian randomization analysis of the blood proteome. Circulation 2019; 140: 819–30. [DOI] [PubMed] [Google Scholar]
- 21. Ference BA, Ray KK, Catapano AL et al. Mendelian randomization study of ACLY and cardiovascular disease. N Engl J Med 2019; 380: 1033–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Folkersen L, Fauman E, Sabater-Lleal M et al. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet 2017; 13: e1006706. 10.1371/journal.pgen.1006706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sun BB, Maranville JC, Peters JE et al. Genomic atlas of the human plasma proteome. Nature 2018; 558: 73–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suhre K, Arnold M, Bhagwat AM et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun 2017; 8: 14357. 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hemani G, Zheng J, Elsworth B et al. The MR-base platform supports systematic causal inference across the human phenome. Elife 2018; 7: e34408. 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gudjonsson A, Gudmundsdottir V, Axelsson GT et al. A genome-wide association study of serum proteins reveals shared loci with common diseases. Nat Commun 2022; 13: 480. 10.1038/s41467-021-27850-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pietzner M, Wheeler E, Carrasco-Zanini J et al. Genetic architecture of host proteins involved in SARS-CoV-2 infection. Nat Commun 2020; 11: 6397. 10.1038/s41467-020-19996-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gilly A, Park YC, Png G et al. Whole-genome sequencing analysis of the cardiometabolic proteome. Nat Commun 2020; 11: 6336. 10.1038/s41467-020-20079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Savvoulidis P, Kalogeropoulos AP, Raptis V et al. Calcification of coronary arteries and aortic valve and circulating a-klotho levels in patients with chronic kidney disease. J Thorac Dis 2020; 12: 431–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Caron B, Patin E, Rotival M et al. Integrative genetic and immune cell analysis of plasma proteins in healthy donors identifies novel associations involving primary immune deficiency genes. Genome Med 2022; 14: 28. 10.1186/s13073-022-01032-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jones G, Trajanoska K, Santanasto AJ et al. Genome-wide meta-analysis of muscle weakness identifies 15 susceptibility loci in older men and women. Nat Commun 2021; 12: 654. 10.1038/s41467-021-20918-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Verghese J, Ayers E, Sathyan S et al. Trajectories of frailty in aging: prospective cohort study. PLoS One 2021; 16: e0253976. 10.1371/journal.pone.0253976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Landino K, Tanaka T, Fantoni G, Candia J, Bandinelli S, Ferrucci L. Characterization of the plasma proteomic profile of frailty phenotype. Geroscience 2021; 43: 1029–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin CH, Liao CC, Huang CH et al. Proteomics analysis to identify and characterize the biomarkers and physical activities of non-frail and frail older adults. Int J Med Sci 2017; 14: 231–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shamsi KS, Pierce A, Ashton AS, Halade DG, Richardson A, Espinoza SE. Proteomic screening of glycoproteins in human plasma for frailty biomarkers. J Gerontol A Biol Sci Med Sci 2012; 67: 853–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ebhardt HA, Degen S, Tadini V et al. Comprehensive proteome analysis of human skeletal muscle in cachexia and sarcopenia: a pilot study. J Cachexia Sarcopenia Muscle 2017; 8: 567–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu JC, Dong SS, Shen H et al. Multi-omics research in sarcopenia: current progress and future prospects. Ageing Res Rev 2022; 76: 101576. 10.1016/j.arr.2022.101576. [DOI] [PubMed] [Google Scholar]
- 38. Murphy S, Zweyer M, Mundegar RR, Swandulla D, Ohlendieck K. Proteomic serum biomarkers for neuromuscular diseases. Expert Rev Proteomics 2018; 15: 277–91. [DOI] [PubMed] [Google Scholar]
- 39. Pan Y, Ji T, Li Y, Ma L. Omics biomarkers for frailty in older adults. Clin Chim Acta 2020; 510: 363–72. [DOI] [PubMed] [Google Scholar]
- 40. Danese E, Montagnana M, Lippi G. Proteomics and frailty: a clinical overview. Expert Rev Proteomics 2018; 15: 657–64. [DOI] [PubMed] [Google Scholar]
- 41. Ohlendieck K. Proteomic identification of biomarkers of skeletal muscle disorders. Biomark Med 2013; 7: 169–86. [DOI] [PubMed] [Google Scholar]
- 42. Yates AD, Achuthan P, Akanni W et al. Ensembl 2020. Nucleic Acids Res 2020; 48: D682–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cruz-Jentoft AJ, Baeyens JP, Bauer JM et al. Sarcopenia: European consensus on definition and diagnosis: report of the European working group on sarcopenia in older people. Age Ageing 2010; 39: 412–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Studenski SA, Peters KW, Alley DE et al. The FNIH sarcopenia project: rationale, study description, conference recommendations, and final estimates. J Gerontol A Biol Sci Med Sci 2014; 69: 547–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brown JC, Harhay MO, Harhay MN. Appendicular lean mass and mortality among prefrail and frail older adults. J Nutr Health Aging 2017; 21: 342–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pei YF, Liu YZ, Yang XL et al. The genetic architecture of appendicular lean mass characterized by association analysis in the UK Biobank study. Commun Biol 2020; 3: 608. 10.1038/s42003-020-01334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med 2016; 35: 1880–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Burgess S, Zuber V, Valdes-Marquez E, Sun BB, Hopewell JC. Mendelian randomization with fine-mapped genetic data: choosing from large numbers of correlated instrumental variables. Genet Epidemiol 2017; 41: 714–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McCarthy S, Das S, Kretzschmar W et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016; 48: 1279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bycroft C, Freeman C, Petkova D et al. The UK biobank resource with deep phenotyping and genomic data. Nature 2018; 562: 203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol 2016; 40: 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Steegen S, Tuerlinckx F, Gelman A, Vanpaemel W. Increasing transparency through a multiverse analysis. Perspect Psychol Sci 2016; 11: 702–12. [DOI] [PubMed] [Google Scholar]
- 53. Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol 2017; 46: 1734–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mendez D, Gaulton A, Bento AP et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res 2019; 47: D930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Riuzzi F, Sorci G, Arcuri C et al. Cellular and molecular mechanisms of sarcopenia: the S100B perspective. J Cachexia Sarcopenia Muscle 2018; 9: 1255–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sayer AA, Syddall HE, Gilbody HJ, Dennison EM, Cooper C. Does sarcopenia originate in early life? Findings from the Hertfordshire cohort study. J Gerontol A Biol Sci Med Sci 2004; 59: M930–4. [DOI] [PubMed] [Google Scholar]
- 57. Petermann-Rocha F, Chen M, Gray SR, Ho FK, Pell JP, Celis-Morales C. New versus old guidelines for sarcopenia classification: what is the impact on prevalence and health outcomes? Age Ageing 2020; 49: 300–4. [DOI] [PubMed] [Google Scholar]
- 58. Costanzo L, De Vincentis A, Di Iorio A et al. Impact of low muscle mass and low muscle strength according to EWGSOP2 and EWGSOP1 in community-dwelling older people. J Gerontol A Biol Sci Med Sci 2020; 75: 1324–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Anderson LJ, Liu H, Garcia JM. Sex differences in muscle wasting. Adv Exp Med Biol 2017; 1043: 153–97. [DOI] [PubMed] [Google Scholar]
- 60. Yuan J, Watanabe M, Suliman M et al. Serum hepatocyte growth factor is associated with truncal fat mass and increased mortality in chronic kidney disease stage 5 patients with protein-energy wasting. Nephrol Dial Transplant 2015; 30: 274–82. [DOI] [PubMed] [Google Scholar]
- 61. Karim A, Muhammad T, Ustrana S, Qaisar R. Intestinal permeability marker zonulin as a predictor of sarcopenia in chronic obstructive pulmonary disease. Respir Med 2021; 189: 106662. 10.1016/j.rmed.2021.106662. [DOI] [PubMed] [Google Scholar]
- 62. Tajrishi MM, Sato S, Shin J, Zheng TS, Burkly LC, Kumar A. The TWEAK-Fn14 dyad is involved in age-associated pathological changes in skeletal muscle. Biochem Biophys Res Commun 2014; 446: 1219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this genetic association study are obtained from published studies and open-access databases. The analysis codes used in the main analysis have been made available at https://github.com/tjh-chiangwei/cis-MR-codes. The analysis codes are modified from those shared online by Henry et al. [9] and cited R packages. The supporting data sets are available in the article, supplemental files and noted public datasets. This study was approved by the Ethics Committee of Tongji Hospital.