Abstract
There are two distinct nickel resistance loci on plasmid pTOM9 from Achromobacter xylosoxidans 31A, ncc and nre. Expression of the nreB gene was specifically induced by nickel and conferred nickel resistance on both A. xylosoxidans 31A and Escherichia coli. E. coli cells expressing nreB showed reduced accumulation of Ni2+, suggesting that NreB mediated nickel efflux. The histidine-rich C-terminal region of NreB was not essential but contributed to maximal Ni2+ resistance.
Nickel is the 24th most abundant element in the earth's crust and has been detected in different media in all parts of the biosphere. Nickel is classified as a borderline metal ion because it has both soft and hard metal properties and can bind to sulfur, nitrogen, and oxygen groups (3). In many bacteria, nickel is required for enzymes such as urease, CO dehydrogenase, and hydrogenase (5, 10). However, excess nickel is toxic. Nickel binds to proteins and nucleic acids and frequently inhibits enzymatic activity, DNA replication, transcription, and translation (1). Several nickel-resistant bacteria have been isolated from heavy-metal-contaminated sites. Well-studied examples include Ralstonia metallidurans CH34 and Achromobacter xylosoxidans 31A (8, 24). The determinant responsible for nickel resistance in R. metallidurans CH34, cnr (cobalt-nickel resistance), encodes three regulatory genes (cnrY, cnrX, and cnrH) and three structural genes encoding the subunits of the Co-Ni efflux pump (cnrC, cnrB, and cnrA) (8, 26). The cnr determinant is similar to the ncc determinant (nickel-cobalt-cadmium resistance) of A. xylosoxidans 31A. The proposed gene products for the efflux system CnrCBA and NccCBA are largely homologous to the gene products for the three subunits of the better-characterized CzcCBA cation-proton antiporter and probably have a similar function (16, 17, 27). In addition to the ncc locus, A. xylosoxidans 31A contains another distinct nickel resistance locus, nre, located on plasmid pTOM9. The nre locus confers low-level nickel resistance on both Ralstonia and Escherichia coli strains (24). The closest homologue of the deduced nreB gene product is NrsD from Synechocystis sp. strain PCC 6803 (6). Both NreB and NrsD belong to the major facilitator superfamily (MFS), and computer analysis indicates 12 putative transmembrane helices in each (11, 20). Additionally, both proteins possess histidine-rich C termini possibly implicated in metal binding (6).
In this study, we characterized the nre locus of A. xylosoxidans 31A and showed that only nreB is required for nickel resistance. In A. xylosoxidans, nreB was specifically induced by nickel but not by cobalt or zinc. The histidine-rich C terminus was not essential for NreB function but was necessary for maximum nickel resistance. E. coli cells harboring nreB showed reduced uptake of nickel compared to that of wild-type cells. The data support our hypothesis that NreB is a Ni2+ transporter responsible for Ni2+ efflux and resistance in A. xylosoxidans 31A and E. coli.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used are listed in Table 1. E. coli strains were grown in Luria-Bertani broth or agar. Antibiotics were added, as appropriate, to the following final concentrations: ampicillin, 100 μg/ml; tetracycline, 12.5 μg/ml; kanamycin, 25 μg/ml for E. coli and 1 mg/ml for A. xylosoxidans. To induce gene expression under the ptetA promoter anhydrotetracycline (AHT; 0.2 μg/ml; Sigma-Genosys) was added. A. xylosoxidans 31A and R. metallidurans AE104 were grown in N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES)- or Tris-buffered mineral salt medium containing a final concentration of 2 g of sodium gluconate per liter (13). NiCl2 was added as indicated. For conjugative gene transfer, overnight cultures of donor strain E. coli S17-1 (25) and recipient strain R. metallidurans AE104 or A. xylosoxidans 31A were grown at 30°C in complex medium, mixed (1:1), and plated onto Luria-Bertani plates. After overnight growth, the bacteria were suspended in saline (8.5 g of NaCl/liter), diluted, and plated onto selective medium as previously described (15).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype | Reference or source |
|---|---|---|
| Strains | ||
| A. xylosoxidans 31A | Nickel resistant (nre+ncc+) pTOM8 pTOM9 | 24 |
| A. xylosoxidans AX1 | Φ(nreB-lacZ); transcriptional fusion on pTOM8 | This work |
| R. metallidurans AE104 | Nis | 15 |
| E. coli DH5α | F−end-1 hsdR17 supE44 thi-1 recA1 deoR gyrA96 relA1 Δ(argF-lacZYA)U169 | New England Biolabs |
| E. coli S17-1 | thi pro hsdR hsdM+recA tra+ | 25 |
| E. coli W3110 | Wild type | Laboratory stock |
| Plasmids | ||
| pASK-IBA3 | Overexpression vector (STREP TagII); Ampr | IBA GmbH, Göttingen, Germany |
| pNREB | pASK-IBA3 with nreB under ptet control | This work |
| pNREB2 | pASK-IBA3 with truncated nreB (Δ aaa 404–446) under ptet control | This work |
| pGEM-T-Easy | Cloning vector; Ampr | Promega, Madison, Wis. |
| pLO18 | Suicide vector; sacB | G. Grass and D. H. Nies, unpublished data |
| pNREB4 | Φ(nreB-lacZ) transcriptional fusion in pLO18 | This work |
| pVDZ′2 | incP mob+ Tcr lacZα | 4 |
| pVDZ′2::TEC9 | nre+; 4.2-kb EcoRI fragment, ntb 1–4189 (accession no. L31491) | 23 |
| pVDZ′2::TBK9 | ncc+; 11.9-kb BamHI/KpnI fragment | 23 |
| pVDZ′2::TBA9 | ncc+nre+; 14.5-kb BamHI fragment | 23 |
| pVDZ′2::TBA9-EcoRI | ncc+nre+; 14.5-kb BamHI fragment with deleted EcoRI site in ncc; parallel orientation of ncc and nre | This work |
| pVDZ′2::TBA9-EcoRI-anti | ncc+nre+; 13.5-kb BamHI/EcoRI fragment with inverted internal EcoRI fragment of pVDZ′2::TBA9-EcoRI resulting in opposite orientation of ncc and nre | This work |
aa, amino acids.
nt, nucleotides.
63Ni uptake.
Uptake experiments were performed by filtration. Following growth in Luria-Bertani medium, cells were harvested and washed once with a buffer consisting of 20 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (pH 7.0), 100 mM sucrose, and 100 mM potassium phosphate. Transport assays were conducted by addition of 63Ni2+ to a final concentration of 5 μM, with filtration at the indicated times. The nitrocellulose filters (0.45-μm pore size; Whatman) were washed with 10 ml of the same buffer. A blank value, obtained by filtering 0.1 ml of the assay mixture without cells and washing it with 10 ml of buffer, was subtracted from all measured values. 63NiCl2 (1.25 μCi/ml) was from Amersham/Pharmacia.
Genetic techniques.
Standard molecular genetic techniques were used (21). Transposon mutagenesis was conducted as described previously (24). Total RNA of A. xylosoxidans was isolated as described previously (8, 9). Northern blot analysis was performed as described by Große et al. (9). PCR was performed using Pwo (Roche) or Taq (Qiagen) DNA polymerase. DNA sequences were obtained by the dideoxy-mediated chain termination method of Sanger et al. using 35S (22) or using an ABISEQ automatic sequencer.
Construction of the truncated nreB gene lacking the coding region for the histidine rich C terminus.
To construct the truncated nreB gene, PCR amplification was performed using the primer pair 5′-GAGGAATTC2460ATGCTTGATGTATTGAAGAACCGGA-3′ and 5′-CTCCTGCAG3649GATGACATCTTCGTCGCGTGACG (the underlined sequences are restriction sites), resulting in a fragment lacking 3′ positions 3650 to 3786. The PCR fragment was then cloned into overexpression vector pASK-IBA3 using the EcoRI and PstI restriction sites introduced by PCR. The complete nreB gene was amplified using the primer pair 5′ GAGGAATTC2460ATGCTTGATGTATTGAAGAACCGGA-3′ and 5′-ATTCTGCAG3780ATGCGCGTCGGGCCATCG-3′ and also cloned into vector pASK-IBA3.
Reporter gene fusion.
To construct a Φ(nreB-lacZ) transcriptional fusion in strain A. xylosoxidans 31A, a 1,273-bp fragment, nreB' (amplified with the primer pair 5′-CCGGTCGAC2498GTTCACGGCACAGGTGATCCC-3′ and 5′-3783ATTCTAATGCGCGTCGGGCCA-3′ ), containing the 3′ end of nreB was amplified as a SalI/PstI fragment from A. xylosoxidans 31A and cloned into pGEM T-Easy (Promega, Madison, Wis.). A promoterless lacZ gene was cloned into the single PstI site directly downstream of nreB'. The lacZ gene was amplified from chromosomal DNA of E. coli W3110, introducing PstI sites at each end. The correct orientation of lacZ was confirmed by restriction analysis, and the Φ(nreB-lacZ) fragment was subcloned as a SalI-fragment into suicide vector pLO18 taking advantage of an additional SalI site in pGEM-T-Easy. The resulting plasmid was used to insert the lacZ gene by recombination immediately downstream of nreB in A. xylosoxidans as described by Groβe et al. (9), creating strain AX1 Φ(nreB-lacZ). The β-galactosidase activity in permeabilized cells was determined as described previously (14).
RESULTS
The nre locus is a second distinct nickel resistance determinant of A. xylosoxidans 31A.
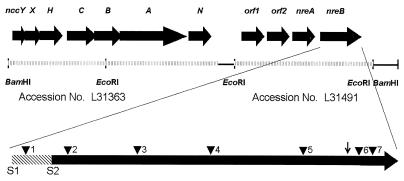
Previously, the ncc (nickel, cobalt, cadmium) determinant on plasmid pTOM9 of A. xylosoxidans 31A was identified as a nickel resistance determinant (23, 24). However, a 14.5-kb BamHI fragment containing not only the ncc determinant but also a 4.2-kb EcoRI fragment downstream of ncc rendered cells of AE104 more nickel resistant than a fragment containing only ncc (Table 2). This suggested the presence of an additional nickel resistance determinant downstream of ncc. Expression of this determinant is independent of ncc, because inversion of the 4.2-kb EcoRI fragment did not alter the resistance pattern (Table 2). The 4.2-kb EcoRI fragment alone was able to confer nickel resistance on both R. metallidurans AE104 and E. coli (Table 2). Sequence analysis of the 4.2-kb EcoRI fragment suggested the presence of four open reading frames (accession no. L31491). Subsequent subcloning (12) of this 4.2-kb EcoRI fragment showed that only the 3′ 2-kb fragment is necessary for nickel resistance in A. xylosoxidans 31A. This distinct nickel resistance determinant possessing two putative genes, nreA and nreB, was termed nre (Fig. 1).
TABLE 2.
MICs for different constructs in R. metallidurans AE104 and E. coli S17-1
| Plasmida | MICb of Ni2+ (mM) for:
|
|
|---|---|---|
| R. metallidurans AE104 | E. coli S17-1 | |
| pVDZ′2 | 0.5 | 0.3 |
| pVDZ′2::TEC9a | 3 | 5 |
| pVDZ′2::TEC9b | 3 | 5 |
| pVDZ′2::TBK9a | 50 | 0.3 |
| pVDZ′2::TBK9b | 45 | 0.3 |
| pVDZ′2::TBA9a | 70 | 5 |
| pVDZ′2::TBA9b | 70 | 5 |
| pVDZ′2::TBA9a-EcoRI | 70 | 5 |
| pVDZ′2::TBA9b-EcoRI | 70 | 5 |
| pVDZ′2::TBA9b-antiTEC9a | 70 | 5 |
Fragments were cloned in both orientations as follows: a; under the control of plac; b, opposite orientation with respect to plac.
The MIC was defined as the concentration at which no growth was observed. Cells were streaked on TES agar with NiCl2 at 0, 0.3, 0.5, 1.5, 3, 5, 7, 9, 11, 15, 20, 30, 40, 45, 50 60, or 70 mM, and growth was monitored after 4 days at 30°C (R. metallidurans) or 37°C (E. coli).
FIG. 1.
Genetic organization of the nickel resistance determinants ncc and nre from A. xylosoxidans 31A (accession no. L31491 and L31363) on a 14.5-kb BamHI fragment. Arrows indicate the transcriptional orientation of the respective genes. Triangles represent loci of Tn5 insertions (nucleotide numbering according to the sequence with accession number L31491) as follows: 1, 2397; 2, 2535; 3, 2876; 4, 3239; 5, 3573; 6, 3703; 7, 3718. The two possible start codons are indicated by S1 (nucleotide 2376) and S2 (nucleotide 2460). The vertical arrow marks the 3′ end of the truncated nreB gene (nucleotides 2460 to 3649). Accession number L31363 is a 4.2-kb EcoRI fragment (shaded), and accession number L31491 is an 8.1-kb fragment (shaded). Distances are not drawn to scale.
The nreB gene is induced by nickel but not by zinc and cobalt in A. xylosoxidans 31A.
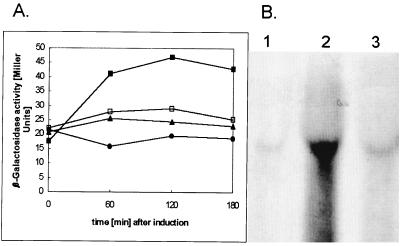
The nre determinant has metal ion specificity different from that of the ncc determinant. Whereas the ncc determinant confers Ni2+, Co2+, and Cd2+ resistance, the nre determinant confers only Ni2+ resistance. Regulation of nre transcription was examined by Northern blot analysis and by a Φ(nreB-lacZ) operon fusion. For Northern blot analysis, cells of A. xylosoxidans 31A were grown in Tris minimal medium (13) to mid-log phase, at which point either NiCl2 or CoCl2 was added. As a control, a culture was grown with no added metal. An increase in an nreB-specific transcript was detected only after addition of Ni2+ (Fig. 2B). Addition of Co2+ did not result in an increase in transcription compared to that of the control with no metal added (Fig. 2B). The actual size of the transcript could not be determined because of compressions due to rRNA and unstable transcripts. Transcription of a Φ(nreB-lacZ) operon fusion was specifically induced by nickel. Neither zinc nor cobalt induced transcription (Fig. 2A). These results show specific induction of nreB expression by nickel and not by cobalt or zinc, consistent with the specificity of metal resistance. Induction of nreB expression was also dependent on the nickel concentration in the medium. After 1 h, the β-galactosidase activity (in Miller units) of a Φ(nreB-lacZ) operon fusion was 28 without added metal, 55.85 with 0.5 mM Ni2+ added, and 78.2 with 1 mM Ni2+ added. Maximal induction was obtained after 2 h with 3 and 0.5 mM Ni2+ and after 1 h with 1 mM Ni2+. After 3 h, a slight decrease in nreB transcription could be observed for all of the concentrations tested (data not shown).
FIG. 2.
Induction of nreB. Induction of β-galactosidase activity in an nreB-lacZ mutant strain. (A) Cells of A. xylosoxidans AX1 containing a φ(nreB-lacZ) operon fusion on plasmid pTOM9 were diluted 15-fold to an optical density at 600 nm of 0.15 into fresh medium containing no added metal (□) or were induced after 5 h of growth with 0.3 mM Ni2+ (■), Co2+ (▴), or Zn2+ (●). Incubation was continued with shaking at 30°C, and the β-galactosidase activity was determined as Miller units (14). (B) Northern blot analysis of nreB transcription. Total RNA was separated by electrophoresis in 1.5% agarose, transferred to a nylon membrane, and hybridized with an nreB-specific probe (all lanes). Total RNA was isolated from nreB-containing strain A. xylosoxidans 31A that was cultivated without toxic concentrations of heavy-metal cations (lane 1) or induced for 30 min with 300 μM Ni2+ (lane 2) or Co2+ (lane 3). Blebs and compressions are due to the 16S and 23S rRNAs. The original photograph was scanned with Ofoto 2.0 (Light Source Computer Images, Inc.) and processed with Adobe Photoshop 3.0 (Adobe Systems, Inc.).
NreB confers nickel resistance on E. coli.
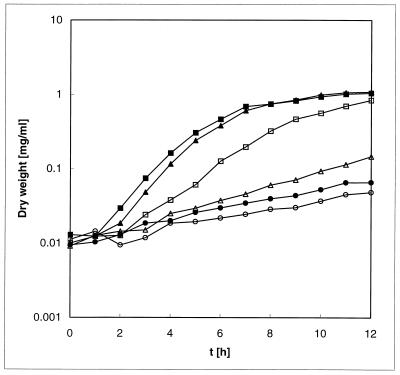
There are two possible ATG start codons for NreB. However, transposon mutagenesis (Fig. 1; Table 3) and codon usage (2) suggest that the second start codon is used. Therefore, nreB starting with the second start codon was cloned into the expression vector pASK-IBA3 (Sigma-Genosys, The Woodlands, Tex.) to create pNREB. Expression of nreB in E. coli W3110(pNREB) rendered cells more nickel resistant than E. coli W3110(pASK-IBA3) lacking the nreB gene (Fig. 3). Although maximal nickel resistance was obtained after addition of the inducer AHT, a substantial increase in nickel resistance was also observed in the absence of induction, indicating that the ptet promoter was leaky and that only minute amounts of NreB were necessary for an increase in nickel resistance (Fig. 3). Expression of the NreB protein could not be visualized by silver-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown).
TABLE 3.
Effects of different transposon insertions in nreB on nickel resistance of E. coli strain S17-1
| Plasmid, Tn5 insertiona (nucleotide no.) |
MICb of Ni2+ (mM) |
|---|---|
| pVDZ'2 | 0.3 |
| pVDZ'2::TEC9 | 5 |
| pVDZ'2::TEC9, Tn5-1 (2397) | 3 |
| pVDZ'2::TEC9, Tn5-2 (2535) | 0.3 |
| pVDZ'2::TEC9, Tn5-3 (2876) | 0.3 |
| pVDZ'2::TEC9, Tn5-4 (3239) | 0.3 |
| pVDZ'2::TEC9, Tn5-5 (3573) | 0.3 |
| pVDZ'2::TEC9, Tn5-6 (3703) | 3 |
| pVDZ'2::TEC9, Tn5-7 (3718) | 3 |
Tn5 insertion loci were determined by DNA sequencing. The nucleotide numbers shown correspond to the sequence with accession no. L31491.
The MIC was defined as the concentration at which no growth was observed. Cells were grown overnight, resuspended in saline (0.85%, wt/vol), and streaked on TES agar with NiCl2 at 0, 0.3, 0.5, 1.5, 3, 5, or 7 mM, and growth was monitored at 37°C.
FIG. 3.
Effect of 3 mM Ni2+ on the growth of E. coli W3110 containing specific constructs. Cells were grown overnight in Luria-Bertani medium, diluted 200-fold into fresh Luria-Bertani medium containing 3 mM NiCl2, and grown at 37°C with shaking. The optical density at 600 nm was monitored hourly for 12 h and converted to dry weight (milligrams per milliliter) and is presented as a semilogarithmic plot. Symbols: ● and ○, pASK-IBA3; ■ and □, pNREB; ▴ and ▵, pNREB2. The inducer AHT was either not added (open symbols) or was added to a final concentration of 0.2 μg/ml (filled symbols).
The C-terminal histidine-rich region of NreB is not essential but contributes to maximal nickel resistance.
The C-terminal histidine-rich region in the homologous nrsD protein from Synechocystis sp. strain PCC 6803 had previously been shown to bind metals (6). To assess if the C-terminal region is essential for NreB, the C-terminal region was deleted (amino acids 404 to 446) and the truncated NreB protein was expressed in E. coli W3110(pNREB2). The truncated NreB protein was still able to confer a significant increase in nickel resistance compared to E. coli W3110(pASK-IBA3) (Fig. 3). Indeed, the truncated NreB protein was able to confer nearly the same resistance as the full-length protein when induced with AHT (Fig. 3). However, a significant loss of nickel resistance was observed when no inducer (AHT) was added (Fig. 3). These results are also in agreement with insertional mutants generated by transposon mutagenesis. Transposon insertions into the nreB gene at sites encoding the histidine-rich C-terminal domain resulted in only a slight decrease in nickel resistance, whereas insertions into other parts of the gene completely abolished nickel resistance (Table 3).
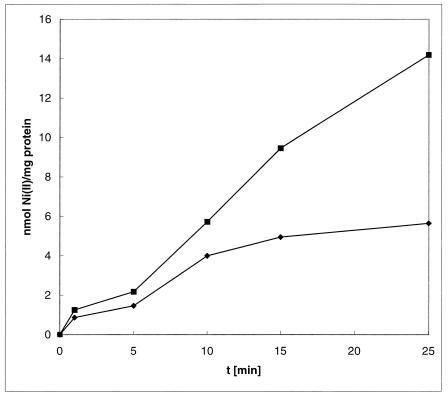
Expression of NreB is responsible for reduced uptake of 63Ni2+ in E. coli W3110. Since NreB is predicted to be a cytoplasmic membrane protein and responsible for Ni2+ resistance, we examined whether cells expressing nreB exhibit decreased Ni2+ uptake in E. coli W3110, reflecting increased efflux. Alternately, resistance could be due to binding of the metal by the C-terminal histidine-rich domain of NreB. Binding should increase the amount of metal ions in resistant cells compared to that in sensitive ones, while efflux should decrease the concentration of cytosolic metal ions. When levels of cell-bound metal ions in cells with and without expression of nreB were compared, resistant cells contained only 40% of the 63Ni2+ that the respective control cells contained after 25 min (Fig. 4). Thus, expression of nreB resulted in reduced accumulation of Ni2+.
FIG. 4.
63Ni2+ uptake by E. coli W3110 expressing nreB. Cells were grown overnight in Luria-Bertani medium and diluted 100-fold into fresh Luria-Bertani medium. The cells were grown to an optical density at 600 nm of 0.8 and induced with AHT at 0.2 μg/ml. After growth for 2.5 h, the cells were washed with buffer and concentrated fourfold in buffer. 63Ni2+ was added to a final concentration of 5 μM. The cells were incubated at 37°C, and 0.1-ml aliquots were filtered through a nitrocellulose membrane (0.45-μm pore size) at various times and immediately washed with 10 ml of buffer. The membranes were dried, and radioactivity was determined using a liquid scintillation counter. The protein concentration was determined using the bicinchoninic acid kit (Sigma), and the Ni2+ concentration per milligram of protein was calculated. E. coli W3110(pASK-IBA3), ■; E. coli W3110(pNREB), ⧫.
DISCUSSION
The results of this study suggest that NreB from A. xylosoxidans is responsible for nickel resistance by efflux. NreB is most closely related to proteins of the DHA3 family of the MFS (TC no. 2.A.1.21.5). MFS transporters have been shown to transport small solutes in response to a chemiosmotic gradient (20). Prior to this study, proteins of the MFS had not been shown to transport metals. The closest homolog of NreB is the NrsD protein from Synechocystis sp. strain PCC 6803 (6). Computer analysis of both NreB and NrsD suggests the presence of 12 transmembrane helices and histidine-rich C-termini in both proteins. Metal binding of the histidine-rich domain from NrsD was evaluated using metal affinity chromatography and showed that this domain has a low specificity for metal binding (6). In this report, we show that the histidine-rich domain is not essential for NreB function. Transposon insertions into the nreB gene at sequences encoding the histidine-rich domain slightly reduced nickel resistance but did not abolish it. However, insertions into other regions of the nreB gene resulted in complete loss of nickel resistance. Furthermore, truncated NreB still conferred nickel resistance on E. coli when the gene encoding it was induced with AHT. However, in cells where nreB was not induced, there was a significant loss of nickel resistance compared to cells producing the complete protein. These results suggest that this domain binds nickel, thereby making nickel transport more efficient.
The mechanism of NreB resistance is most likely the result of nickel efflux coupled to a chemiosmotic gradient. Consistent with this hypothesis, cells of E. coli W3110 expressing nreB exhibited reduced uptake of nickel compared to a control without nreB. This is the first example of an MFS protein catalyzing metal ion transport. However, at this point, the actual substrate of NreB is not known, as it might be the free cation or a metal conjugate. Other nickel resistance determinants, such as those encoded by cnr and ncc, are close homologues of the better-studied czc determinant from R. metallidurans CH34. The czcCBA gene products form a membrane-bound protein complex catalyzing Co2+, Zn2+, and Cd2+ efflux by a proton-cation antiporter in R. metallidurans CH34 (7, 16, 19). The actual substrate transported is also not known for CzcCBA, NccCBA, or CnrCBA. NreB is more specific for nickel, while NccCBA and CnrCBA have a broad range of metal ion substrates. In addition to nickel, they also transport cobalt and cadmium (ncc) and cobalt and zinc (cnr) (28). It is not known how nreB is regulated; however, nreB is induced by nickel and not by cobalt or zinc, consistent with the specificity of metal resistance. Transcriptional analysis was performed with A. xylosoxidans 31A harboring both the ncc and nre determinants. Therefore, over time, the decrease in transcription of a φ(nreB-lacZ) operon fusion can be attributed to nickel efflux by NreB and NccCBA. This is similar to the self-repression of the ars operon that occurs when the ArsAB pump extrudes the inducer arsenite and transcript levels return to uninduced levels (18).
ACKNOWLEDGMENTS
This work was supported by hatch project 136713 to C.R. and U.S. Public Health Service grant GM 55425 to B.P.R.
We thank T. Schmidt for providing nreB Tn5 mutants.
REFERENCES
- 1.Babich H, Stotzky G. Toxicity of nickel to microbes: environmental aspects. Adv Appl Microbiol. 1983;29:195–265. doi: 10.1016/s0065-2164(08)70358-7. [DOI] [PubMed] [Google Scholar]
- 2.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 3.Costa M, Klein C B. Nickel carcinogenesis, mutation, epigenetics, or selection. Environ Health Perspect. 1999;107:A438–439. doi: 10.1289/ehp.99107a438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deretic V, Chandrasekharappa S, Gill J F, Chatterjee D K, Chakrabarty A. A set of cassettes and improved vectors for genetic and biochemical characterization of Pseudomonas genes. Gene. 1987;57:61–72. doi: 10.1016/0378-1119(87)90177-6. [DOI] [PubMed] [Google Scholar]
- 5.Friedrich C G, Schneider K, Friedrich B. Nickel is in the catalytically active hydrogenase of Alcaligenes eutrophus. J Bacteriol. 1982;152:42–48. doi: 10.1128/jb.152.1.42-48.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Dominguez M, Lopez-Maury L, Florencio F J, Reyes J C. A gene cluster involved in metal homeostasis in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2000;182:1507–1514. doi: 10.1128/jb.182.6.1507-1514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldberg M, Pribyl T, Juhnke S, Nies D H. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J Biol Chem. 1999;274:26065–26070. doi: 10.1074/jbc.274.37.26065. [DOI] [PubMed] [Google Scholar]
- 8.Grass G, Große C, Nies D H. Regulation of the cnr cobalt and nickel resistance determinant from Ralstonia sp. strain CH34. J Bacteriol. 2000;182:1390–1398. doi: 10.1128/jb.182.5.1390-1398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Große C, Grass G, Anton A, Franke S, Navarrete Santos A, Lawley B, Brown N L, Nies D H. Transcriptional organization of the czc heavy metal homesotasis determinant from Alcaligenes eutrophus. J Bacteriol. 1999;181:2385–2393. doi: 10.1128/jb.181.8.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hausinger R P. Nickel utilization by microorganisms. Microbiol Rev. 1987;51:22–42. doi: 10.1128/mr.51.1.22-42.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 12.Lemke K. Schwermetallresistenz in den zwei Alcaligenes-Stämmen A. eutrophus CH34 und A. xylosoxydans 31A: Transposonmutagenese, Klonierung und Sequenzierung. Ph. D. dissertation. Germany: Universität Göttingen, Göttingen; 1993. [Google Scholar]
- 13.Mergeay M, Nies D, Schlegel H G, Gerits J, Charles J, van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985;162:328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 15.Nies D H, Mergeay M, Friedrich B, Schlegel H G. Cloning of plasmid genes encoding resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus CH34. J Bacteriol. 1987;169:4865–4868. doi: 10.1128/jb.169.10.4865-4868.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nies D H, Nies A, Chu L, Silver S. Expression and nucleotide sequence of a plasmid-determined divalent efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci USA. 1989;86:7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nies D H. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J Bacteriol. 1995;177:2707–2712. doi: 10.1128/jb.177.10.2707-2712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Owolabi J B, Rosen B P. Differential mRNA stability controls relative gene expression within the plasmid-encoded arsenical resistance operon. J Bacteriol. 1990;172:2367–2371. doi: 10.1128/jb.172.5.2367-2371.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rensing C, Pribyl T, Nies D H. New functions for the three subunits of the CzcCBA cation-proton antiporter. J Bacteriol. 1997;179:6871–6879. doi: 10.1128/jb.179.22.6871-6879.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saier M H, Jr, Beatty J T, Goffeau A, Harley K T, Heijne W H, Huang S C, Jack D L, Jahn P S, Lew K, Liu J, Pao S S, Paulsen I T, Tseng T T, Virk P S. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt T. Die plasmidkodierte Nickelresistenz von Alcaligenes xylosoxydans 31A: Klonierung und Expression beteiligter Gene. Ph. D. dissertation. Germany: Universität Göttingen, Göttingen; 1991. [Google Scholar]
- 24.Schmidt T, Schlegel H G. Combined nickel-cobalt-cadmium resistance encoded by the ncc locus of Alcaligenes xylosoxidans 31A. J Bacteriol. 1994;176:7045–7054. doi: 10.1128/jb.176.22.7045-7054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 26.Tibazarwa C, Wuertz S, Mergeay M, Wyns L, van der Lelie D. Regulation of the cnr cobalt and nickel resistance determinant of Ralstonia eutropha (Alcaligenes eutrophus) CH34. J Bacteriol. 2000;182:1399–1409. doi: 10.1128/jb.182.5.1399-1409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng T T, Gratwick K S, Kollman J, Park D, Nies D H, Goffeau A, Saier M H. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 28.Varma A K, Sensfuss C, Schlegel H G. Inhibitor effects on the accumulation and efflux of nickel ions in plasmid pMOL28-harboring strains of Alcaligenes eutrophus. Arch Microbiol. 1990;154:42–49. [Google Scholar]