Abstract
The planorbid gastropod genus Bulinus consists of 38 species that vary in their ability to vector Schistosoma haematobium (the causative agent of human urogenital schistosomiasis), other Schistosoma species, and non-schistosome trematodes. Relying on sequence-based identifications of bulinids (partial cox1 and 16S) and Schistosoma (cox1 and ITS), we examined Bulinus species in the Lake Victoria Basin in Kenya for naturally acquired infections with Schistosoma species. We collected 6,133 bulinids from 11 sites between 2014–2021, 226 (3.7%) of which harbored Schistosoma infections. We found 4 Bulinus taxa from Lake Victoria (B. truncatus, B. tropicus, B. ugandae, and B. cf. transversalis), and an additional 4 from other habitats (B. globosus, B. productus, B. forskalii, and B. scalaris). S. haematobium infections were found in B. globosus and B. productus (with infections in the former predominating) whereas S. bovis infections were identified in B. globosus, B. productus, B. forskalii, and B. ugandae. No nuclear/mitochondrial discordance potentially indicative of S. haematobium/S. bovis hybridization was detected. We highlight the presence of Bulinus ugandae as a distinct lake-dwelling taxon closely related to B. globosus yet, unlike all other members of the B. africanus species group, is likely not a vector for S. haematobium, though it does exhibit susceptibility to S. bovis. Other lake-dwelling bulinids also lacked S. haematobium infections, supporting the possibility that they all lack compatibility with local S. haematobium, thereby preventing widespread transmission of urogenital schistosomiasis in the lake’s waters. We support B. productus as a distinct species from B. nasutus, B. scalaris as distinct from B. forskalii, and add further evidence for a B. globosus species complex with three lineages represented in Kenya alone. This study serves as an essential prelude for investigating why these patterns in compatibility exist and whether the underlying biological mechanisms may be exploited for the purpose of limiting schistosome transmission.
Author summary
Human schistosomiasis is a neglected tropical disease caused by members of the trematode genus Schistosoma. Every schistosome species is dependent on a particular species, or array of species, of intermediate gastropod host(s) for their transmission. In the Lake Victoria Basin in Kenya, two related schistosome species (Schistosoma haematobium and Schistosoma bovis) utilize multiple species within the genus Bulinus as intermediate hosts. Discerning which bulinid species vector S. haematobium or S. bovis, or both, and identifying the habitats for each, is critical to understanding local transmission patterns. Closely related bulinids cannot be confidently distinguished using morphological criteria so this study used DNA sequence-based methods to identify local bulinid species and to identify schistosomes shed from infected snails. We implicate two bulinid species in the transmission of S. haematobium and four species in the transmission of S. bovis. Both S. haematobium associated species were found exclusively in streams and dams in the Lake Victoria Basin thereby seemingly keeping the shores of Lake Victoria largely free of S. haematobium transmission. Further study as to why some species like B. globosus are susceptible to S. haematobium whereas other close relatives like B. ugandae are apparently refractory may reveal underlying resistance factors potentially useful for control programs.
Introduction
One of the fascinating aspects of the biology of infectious diseases is that, in some cases, the parasites responsible and their hosts (including vectors) do not comprise a single parasite or host species, but complex arrays of related species [1–4]. Such arrays might reveal a checkerboard of host-parasite interactions, ranging from pairs of host and parasite species being fully compatible and supporting transmission to marginally compatible and fully incompatible pairs. The Schistosoma haematobium species group and species within the genus Bulinus comprise such an array [5]. We have directed our attention to representatives of these two groups of organisms that occupy the Kenyan waters of the Lake Victoria Basin (LVB), in hopes of eventually revealing the factors that dictate the various outcomes of such associations.
Like most digenetic trematodes, schistosomes depend on a gastropod intermediate host to complete their life cycles, within which vertebrate-infective cercariae are asexually produced in prolific numbers. Such gastropods are often termed intermediate hosts because of their obligatory role in schistosome larval development, and although they do not directly deliver parasites to their vertebrate hosts as, for example, a mosquito transmits malaria parasites, they nonetheless play an indispensable vector role. Successful transmission of vector-borne parasites like schistosomes is dependent on a variety of factors, including ecological circumstances which impact the encounter rates between parasite and vector [6–8], associations with particular symbionts including those that prey on the free-living forms of certain parasites [9], facilitated susceptibility where prior infection with a specific parasite allows a second parasite to develop in a non-typical host [10,11], and nuanced physiological and immunological interactions which dictate the outcome of an infection [2,12–20].
To begin to fully appreciate the intimate relationships between vector and parasite that influence compatibility and ultimately transmission, a sound understanding of the underlying systematics of both parasite and vector host is critical. This task is complicated when the species involved cannot consistently be accurately differentiated using morphology alone, or when a lack of clear morphological differences belie the presence of large genetic differences, that is, when cryptic species are involved [21].
With rare exceptions, members of the Schistosoma haematobium group depend on freshwater snails of the planorbid genus Bulinus for their transmission. This species group includes 9 species: S. margrebowiei, S. leiperi, S. mattheei, S. intercalatum, S. guineensis, S. curassoni, S. bovis, S. kisumuensis, and S. haematobium [22–24]. Collectively, they pose a persistent threat to human and domestic animal health throughout Africa, in parts of the Mediterranean region, and the Middle East. The agent of urogenital schistosomiasis, S. haematobium, is the most common human schistosome [25]. In general, anatomical similarities and a history of apparent hybridization among members of the S. haematobium group, [12,26–30] coupled with the changes posed by present-day events such as climate change [31] further highlight the need to clarify both the systematic status of schistosomes and the snails that transmit them.
There are 38 currently recognized species of Bulinus [32–35]. Bulinids have proven particularly challenging to identify because variable morphological and conchological traits make species identification and differentiation difficult [32,36–39] and the group is inherently complex with various mating systems represented [40] and some polyploid species [41].
Means to differentiate and reliably identify bulinid snails have improved considerably with the use of sequence-based genetic markers [34,38,42–44], mitochondrial genomes [35,45,46], and a recently published nuclear genome [47]. Modern phylogenetic analyses have provided a more firm systematic foundation for Bulinus [34,38,44–46,48,49], improved discrimination among morphologically similar species [39,50], better defined the four species groups within Bulinus [34,38], and improved understanding of how the genus diversified and evolved [34,35,44,45,51,52]. Such contributions have provided tools to determine what particular Bulinus species are involved in the transmission of the various species of Schistosoma [49,53,54] and other trematodes including several species of livestock-infective amphistomes, [55,56], and echinostomes [57]. Such investigations have greatly expanded our knowledge regarding the diversity of parasites that a snail species can transmit and have additionally revealed novel and presently unstudied parasite species [57,58].
As shown in numerous studies to date, the relationships between the S. haematobium group species and Bulinus species are complex [5,32]. A particular species of Bulinus may act as vector for multiple schistosome species, a single schistosome species, or not be involved in schistosome transmission at all [59–61]. In some cases, local adaptation of bulinids and schistosomes has resulted in a given schistosome species utilizing different intermediate host species in different regions. Previously, this was thought to be due to the existence of at least two S. haematobium strains which differ in their compatibility with Bulinus species [62–64]. More recently, it has been shown that S. haematobium isolates across Africa have low genetic diversity as compared to S. bovis, and that all tested isolates of S. haematobium (with the exception of the Madagascar isolate) have been observed to contain various levels of S. bovis introgression in their genomes [28–30]. Variable intermediate host-use patterns among modern S. haematobium isolates may be influenced by the particular segments of the S. bovis genome they retain [30].
The focus of this study is primarily on the relationships between bulinids and schistosomes and includes some insight into the relationships between bulinids and non-schistosome trematodes, in western Kenya, in the LVB. Lake Victoria is the world’s largest tropical lake and connects Kenya, Tanzania, and Uganda, with the surrounding LVB including a variety of smaller water bodies such as streams, dams, papyrus swamplands, rain-fed pools, ponds, and springs [65]. Based on considerations of conchology, anatomy, ploidy, and enzyme electrophoresis, Brown [32] recognized 38 Bulinus species divided into 4 species groups. 12 of which he reported from the LVB: from the B. africanus group, B. africanus, B. nasutus productus, B. globosus and B. ugandae; from the B. forskalii group, B. forskalii, B. browni and B. scalaris; from the B. tropicus/truncatus group, B. transversalis, B. tropicus, B. truncatus and B. trigonus; and from the B. reticulatus group, B. reticulatus. Several of the species he reported are hard to differentiate from one another, and some are rarely encountered or studied and in general are poorly known, including browni, scalaris and reticulatum. More recently, based largely on the useful discrimination provided by the cytochrome c oxidase subunit 1 (cox1) gene, Chibwana et al. [50] found 7 species in the LVB, including B. globosus (described as a complex), B. truncatus, B. tropicus, B. nasutus productus and B. forskalii as well as two taxa, Bulinus sp. 1 and 2, provisionally identified as B. trigonus and B. ugandae, respectively. Currently accepted species are associated (with some variation) with ephemeral pools or ponds (e.g. B. forskalii, B. scalaris, B. reticulatus), seasonal ponds or springs (Bulinus productus), more permanent habits such as streams or dams (Bulinus globosus), the lakeshore and associated papyrus swamps (Bulinus ugandae), or the deeper waters of the lake (B. tropicus, B. truncatus and B. trigonus). Of particular note is a growing body of evidence that the pan-African species B. globosus is not a single species, but a complex of multiple species [35,38,46,50].
Based on an examination of the relevant literature, coupled with sequence data of marker genes to aid in the identification of both snails and the schistosomes they host, we provide an overview of the bulinid species we have recovered from various habitats in the LVB. We highlight some difficulties regarding Bulinus systematics and identify some peculiarities regarding the role of bulinids in the transmission of S. haematobium and S. bovis in western Kenya. This study serves as a prelude to investigations aimed at understanding the underlying causes dictating the patterns of compatibility posed by the complex interacting arrays of Schistosoma and Bulinus species in western Kenya.
Materials and methods
Ethics statement
Informed written consent was obtained from all individual participants included in the study. The Kenya Medical Research Institute Scientific and Ethics Review Unit (KEMRI/SERU/CBRD/173/3540) and the University of New Mexico Institution Review Board (IRB 821021–12, IRB 821021–9) approved all aspects of this project involving human subjects. Ethical approval for the collection and analyses of snail and schistosome samples were obtained from the National Commission for Science, Technology and Innovation (permits number NACOSTI/P/21/9648 and NACOSTI/P/22/17142), and National Environmental Management Authority (permit number NEMA/AGR/149/2021).
Sampling
We collected Bulinus snails from 11 different localities (S1 Table and S1 Fig); some localities include endemic transmission sites where we have collected from Jan 2014 –Mar 2021. Two methods were used to collect snails: scooping from the shore and dredging from a boat [66]. From the shore, two experienced lab members scooped snails for 30 minutes per sampling site using long-handled scoops (steel sieve with a mesh size of 2 × 2 mm, supported on an iron frame). Offshore from a boat, snails were collected for 30 min by passing a dredge (0.75 m long and 0.4 m wide with an attached sieve, 2 × 2 mm mesh size) along the bottom. Dredge hauls were made, beginning at 1 m depth and extending perpendicular to the shore to a maximum of 10 m depth, typically covering a distance of about 150 m. Live snails were transported to the Kenya Medical Research Institute (KEMRI), Center for Global Health Research, Kisian, Kisumu.
Snails were provisionally identified using keys [32,67]. Snails were rinsed and placed one snail per well in 12-well cell culture plates in 3 ml of aged tap water. The plates were placed in ambient outdoor lighting for 2 hr to induce cercarial shedding. Cercariae were identified morphologically [68]. Each shedding snail was preserved in one sample tube, and the cercariae they released in a corresponding tube, all in 95% ethanol. Non-shedding snails were maintained in the lab to allow cercariae-shedding infections to develop and re-shed 1–5 weeks later. Snails were maintained in 20 L tanks with oyster shells, aeration, and fed boiled lettuce and shrimp pellets.
S. haematobium miracidia were sourced from the urines of local schoolchildren enrolled in this study (see ethics statement below) or from discarded clinical samples and were used for phylogenetic analyses and comparisons with schistosomes shed from infected snails.
Additional sampling records
Additional specimens were obtained by a loan from collections held at the Division of Parasites, Museum of Southwestern Biology, University of New Mexico.
Molecular characterization
Snail sequences
Prior to extraction, snails to be processed for sequencing were photographed to provide a record of shell size and shape. Snail genomic DNA was extracted from a small portion of the head foot using the E.Z.N.A. Mollusc DNA Kit (Omega Bio-Tek, Norcross, GA) according to manufacturer’s instructions. Partial sequences of the cytochrome c oxidase subunit I (cox1) and 16S rRNA genes were obtained for molecular identification and differentiation among Bulinus species.
Cox1 partial sequences (706 bp) were amplified using universal primers [69] and occasionally using reverse primer COR722b [70]. 16S partial sequences (481 bp) were amplified using forward primer 16Sar and reverse primer 16Sbr [71]. Thermocycling conditions for both cox1 and 16S were as follows: preheat at 94°C for 5 min followed by 45 cycles of denaturation at 94°C for 15 sec, annealing at 45°C for 30 sec and extension at 72°C for 1 min; final extension step at 72°C for 10 min. All snail and parasite PCR reactions had a volume of 25 μL with 1 μL of 40 ng of DNA, 0.8 mM/l dNTPs, 2.5 mM/l MgCl2, 0.25 units of Ex Taq DNA (Clontech, Mountain View, CA), and 0.4 μM/L of each primer.
Schistosome sequences
Partial cox1 mtDNA and partial internal transcribed spacer 1 (ITS1) + 5.8S + partial internal transcribed spacer 2 (ITS2) rRNA sequences were used to identify and differentiate among Schistosoma species. A single cercaria was removed from the ethanol preserved cercariae obtained from a single snail and used for DNA extraction. Genomic DNA was extracted from parasite specimens using QIAamp DNA Micro kit (Qiagen, Valencia, CA) according to manufacturer’s instructions with a 40 μL final elution volume.
Cox1 partial mtDNA (423 bp) sequences were generated using a modified forward primer designed from the S. bovis/S. haematobium universal primer [72] (ModShAsmit1: 5’ TTTTTTGGKCATCCTGAGGTGTAT3’), and the reverse primer Cox1_schist_3’ [73]. Thermocycling conditions were as follows: preheat at 94°C for 5 min followed by 30 cycles of denaturation at 94°C for 30 sec, annealing at 40°C for 30 sec and extension at 72°C for 2 min followed by a final extension period of 72°C for 5 min.
ITS1 + 5.8S + ITS2 partial rRNA (981 bp) sequences were amplified using forward primer ITS5 and reverse primer ITS4 [74]. Thermocycling conditions were as follows: preheat at 94°C for 5 min followed by 30 cycles of denaturation at 94°C for 30 sec, annealing at 54°C for 45 sec and extension at 72°C for 1 min; followed by a final extension period of 72°C for 5 min.
For both snails and schistosomes
PCR fragments were separated by 1% agarose gel electrophoresis and visualized with 0.5% GelRed Nucleic acid gel stain (Biotium, Hayward, CA). PCR products were purified using ExoSap-IT (Affymetrix, Santa Clara, CA). Both strands were sequenced using an Applied Biosystems 3130 automated sequencer and BigDye terminator cycle sequencing kit Version 3.1 (Applied Biosystems, Foster City, CA). DNA sequences were verified by aligning reads from the 5′ and 3′ directions using Sequencher 5.1 and manually corrected for ambiguous base calls (Gene Codes, Ann Arbor, MI).
Additional Bulinus (MT707420.1, AM286295.2, AM286296.2, LT671915.1, LT671916.1, MK414452.1, MK414453.1, MK414454.1, AM286286.2, AM286299.2, AM286300.2, AM286303.2, AM921814.1, AM286308.2, AM286309.2, MN551559.1, AM286318.2, MT707391.1, MT707392.1, MT707382.1, AM286311.2, AM921838.1, MT707425.1, GU451744.1, MH037061.1) sequences were retrieved from NCBI [38,46,48,50,75–77]. Additional Schistosoma sequences were used to represent S. mattheei (MW046871.1, AJ519518.1), S. guineensis (Z21717.1, AJ519523.1), and S. curassoni (MT580946.1, MT579422.1) for the cox1 + ITS concatenated phylogenetic analysis [78–81]. Indoplanorbis exustus and Schistosoma mattheei were selected as outgroups for the Bulinus and Schistosoma analyses, respectively. Genbank sequences MH037061 and MH037083, and GU451744 and GU451726 were concatenated to produce outgroup sequences for the bulinid cox1 + 16S concatenated alignment [76,77]. GenBank accession numbers for bulinid sequences provided in this study can be found in Table 1.
Table 1. Bulinus specimens.
| Species | MSB:Host: | Collection location | Habitat type | Latitude | Longitude | Date (YYYY/MM/DD) | Infection | Cercariae type | GenBank accession COI | GenBank accession 16S |
|---|---|---|---|---|---|---|---|---|---|---|
| B. globosus | 24516 | Asao Stream | R | −0.31810 | 35.0069 | 2016-08-02 | y | Schistosoma | OP233119 | |
| B. globosus | 24525 | Asao Stream | R | −0.31810 | 35.0069 | 2016-08-02 | y | Schistosoma | OP233113 | OP244943 |
| B. globosus | 24526 | Asao Stream | R | −0.31810 | 35.0069 | 2016-08-02 | y | Schistosoma | OP233114 | OP244944 |
| B. globosus | 24776 | Asao Stream | R | −0.31810 | 35.0069 | 2015-11-13 | n | OP233135 | OP244911 | |
| B. globosus | 24777 | Asao Stream | R | −0.31810 | 35.0069 | 2017-05-20 | y | Schistosoma | OP233098 | |
| B. globosus | 24778 | Asao Stream | R | −0.31810 | 35.0069 | 2017-05-20 | y | Schistosoma | OP233099 | |
| B. ugandae | 24542 | Power House | LS | −0.09410 | 34.7076 | 2017-05-23 | y | xiphidiocercariae | OP233117 | OP244923 |
| B. ugandae | 24543 | Usenge Beach | LS | -0.072636 | 34.059956 | 2016-04-16 | n | OP233118 | OP244920 | |
| B. ugandae | 24544 | Kagwa Beach | LS | −0.356594 | 34.68358 | 2016-04-04 | n | OP233120 | OP244921 | |
| B. ugandae* | 24558 | Mnazi Mmoja Beach, Ukerewe Island, TZ | LS | -2.1075 | 33.08361 | 2001-04-21 | n | OP233103 | OP244922 | |
| B. ugandae* | 24559 | Ukerewe Island, Kagera Stream, TZ | R | 2001-04-21 | y | strigeid | OP233107 | OP244915 | ||
| B. ugandae | 24549 | Usenge Beach | LS | -0.072636 | 34.059956 | 2017-08-03 | y | echinostome | OP233123 | OP244908 |
| B. ugandae* | 24573 | ADC Farm, Kisumu | S | -0.3333333 | 34.65 | 1987-01-21 | n | OP233143 | OP244933 | |
| B. ugandae | 24550 | Usenge Beach | LS | -0.072636 | 34.059956 | 2016-04-16 | n | OP233124 | OP244907 | |
| B. ugandae | 24510 | Koriang Beach | LS | -0.3548806 | 34.65903 | 2017-02-24 | y | xiphidiocercariae | OP233125 | OP244906 |
| B. ugandae | 24527 | Dunga Beach | LS | -0.14532 | 34.73633 | 2017-02-07 | y | strigeid | OP242173 | OP244902 |
| B. ugandae | 24529 | Kagwa Beach | LS | −0.356594 | 34.68358 | 2016-04-13 | n | OP233115 | OP244925 | |
| B. ugandae | 24770 | Gudwa Beach | LS | -0.3573667 | 34.3301 | 2018-11-20 | n | OP233142 | OP244912 | |
| B. ugandae* | 24564 | Kisumu | LS | −0.1091 | 34.775 | 1987-01-20 | n | OP242176 | OP244924 | |
| B. ugandae* | 24565 | Kagwell | LS | –0.191111 | 34.503333 | 2005-09-12 | n | OP233091 | OP244918 | |
| B. ugandae* | 24566 | Nawa | LS | –0.094051 | 34.707601 | 2005-09-28 | n | OP242177 | OP244919 | |
| B. ugandae* | 24567 | Asembo Bay | LS | −0.1885080 | 34.387534 | 2005-01-20 | n | OP233092 | OP244932 | |
| B. nasutus* | 24579 | Komarock | -1.26754 | 36.9094 | 1997-11-12 | n | OP233139 | OP244916 | ||
| B. nasutus* | 24580 | Kyenze | 1997-11-14 | n | OP233138 | OP244937 | ||||
| B. nasutus* | 24581 | Ng’alalia | −1.5357 | 37.2361 | 1997-11-12 | n | OP233137 | OP244938 | ||
| B. productus | 24561 | Tiengre | EP | -0.0898333 | 34.70313 | 2018-05-25 | n | OP233131 | OP244950 | |
| B. productus | 24562 | Tiengre | EP | -0.0898333 | 34.70313 | 2018-05-25 | n | OP233132 | OP244903 | |
| B. productus | 24563 | Tiengre | EP | -0.0898333 | 34.70313 | 2018-05-25 | n | OP233133 | OP244939 | |
| B. productus | 24522 | Tiengre | EP | -0.0898333 | 34.70313 | 2018-05-22 | y | Schistosoma | OP233106 | OP244910 |
| B. productus | 24524 | Tiengre, Kenya | EP | -0.0898333 | 34.70313 | 2018-05-21 | y | Schistosoma | OP233109 | OP244909 |
| B. productus | 24774 | Tiengre | EP | -0.0898333 | 34.70313 | 2018-05-21 | y | bent-bodied strigeid | OP233111 | OP244953 |
| B. tropicus | 24551 | Mwea | R | -0.6333333 | 37.46667 | 2015-06-20 | y | echinostome | OP242174 | OP244954 |
| B. tropicus* | 24766 | Usare | LS | –0.105712 | 34.67429 | 2005-09-08 | n | OP233094 | OP244929 | |
| B. tropicus* | 24767 | Iringa, Kilima Pond, TZ | P | -7.956333 | 35.86383 | 2001-04-30 | n | OP233110 | OP244905 | |
| B. tropicus* | 24568 | Sand Harvest, Adupe | L | −0.1013889 | 34.714722 | 2005-09-22 | n | OP233093 | OP244934 | |
| B. tropicus | 24545 | Minya Kochillo | L | -0.2363778 | 34.24605 | 2016-09-20 | n | OP242175 | OP244945 | |
| B. tropicus | 24546 | Gudwa Beach Dredge | L | -0.3573667 | 34.3301 | 2016-09-20 | n | OP233121 | OP244946 | |
| B. tropicus | 24547 | Kadidi Beach | LS | -0.2033583 | 34.15326 | 2016-04-07 | n | OP233122 | OP244947 | |
| B. tropicus | 24551 | Mwea | R | -0.6333333 | 37.46667 | 2015-06-20 | y | echinostome | OP233128 | |
| B. tropicus* | 24552 | Ukerewe Island, Kaseni-Shuleni, TZ | LS | -1.933333 | 32.85 | 2001-04-20 | n | OP233100 | OP244936 | |
| B. tropicus* | 24578 | Nyamlebi-Ngoma, Ukerewe Island, TZ | LS | -2.130333 | 3.1685 | 2001-04-22 | n | OP233105 | OP244904 | |
| B. tropicus* | 24530 | Tala | -1.270768 | 37.319472 | 1997-11-13 | n | OP233136 | OP244917 | ||
| B. tropicus | 24574 | Kanyibok | LS | -0.0895806 | 34.08593 | 2017-11-01 | y | echinostome | OP233126 | OP244948 |
| B. tropicus | 24775 | Eldoret | D | 0.4671 | 35.3517 | 2014-01-07 | n | OP233134 | OP244913 | |
| B. tropicus* | 24571 | Mbita Beach | LS | −0.4213889 | 34.2075 | 2005-10-04 | n | OP233088 | OP244959 | |
| B. truncatus* | 24553 | Ukerewe Island, Kaseni-Shuleni, TZ | LS | -1.933333 | 32.85 | 2001-04-20 | n | OP233101 | OP244940 | |
| B. truncatus* | 24554 | Kom Ombo, Southern Egypt | LS | 30.54558 | 32.21017 | 2003-03-01 | n | OP233112 | OP244935 | |
| B. truncatus* | 24569 | Mbita Beach | LS | −0.4213889 | 34.2075 | 2005-10-04 | n | OP233086 | OP244952 | |
| B. truncatus* | 24570 | Mbita Beach | LS | −0.4213889 | 34.2075 | 2005-10-04 | n | OP233087 | OP244960 | |
| B. truncatus* | 24572 | Mbita Beach | LS | −0.4213889 | 34.2075 | 2005-10-04 | n | OP233089 | OP244958 | |
| B. truncatus* | 24517 | Mbita Beach | LS | −0.4213889 | 34.2075 | 2005-10-04 | n | OP233090 | OP244957 | |
| Bul. cf. transversalis* | 24768 | Usare | LS | –0.105712 | 34.67429 | 2005-09-08 | n | OP233097 | OP244926 | |
| Bul. cf. transversalis* | 24769 | Usare | LS | –0.105712 | 34.67429 | 2005-09-08 | n | OP233096 | OP244927 | |
| Bul. cf. transversalis* | 24765 | Usare | LS | –0.105712 | 34.67429 | 2005-09-08 | n | OP233095 | OP244928 | |
| B. scalaris* | 24555 | Ukerewe Island, Kaseni-Shuleni, TZ | LS | -1.933333 | 32.85 | 2001-04-20 | n | OP233102 | OP244942 | |
| B. scalaris | 24514 | Tiengre | EP | -0.0898333 | 34.70313 | 2018-05-25 | y | amphistome | OP233127 | OP244955 |
| B. forskalii* | 24556 | Ukerewe Island, Mnazi Mmoja, TZ | LS | -2.1075 | 33.08361 | 2001-04-21 | n | OP233104 | OP244931 | |
| B. forskalii* | 24557 | Safisha Stream, Tunduma, TZ | R | -9.316667 | 32.76667 | 2001-04-29 | y | amphistome | OP233108 | OP244930 |
| B. forskalii | 24560 | Tiengre | EP | -0.0898333 | 34.70313 | 2018-05-25 | y | amphistome | OP233130 | OP244949 |
| B. forskalii | 24575 | Tiengre | EP | -0.0898333 | 34.70313 | 2018-05-29 | y | pigmented amphistome | OP233140 | OP244951 |
| B. forskalii | 24576 | Tiengre | EP | -0.0898333 | 34.70313 | 2018-05-29 | y | pigmented amphistome | OP233141 | OP244941 |
| B. forskalii | 24577 | Nyabera | S | −0.1091 | 34.775 | 2018-05-21 | Y | xiphidiocercariae | OP233116 | OP244956 |
| B. forskalii | 24771 | Nawa | LS | –0.094051 | 34.707601 | 2016-06-12 | Y | amphistome | OP233129 | OP244914 |
Table 1. Sequenced specimens with associated MSB:Host: numbers, collection locations, habitat type, GPS coordinates (when available), infection status, and associated GenBank accession numbers. Specimen names denoted with * indicate samples from archived specimens. Bulinus productus specimens are often designated as Bulinus nasutus productus in literature regarding this region. Habitat type abbreviations: LS = lakeshore, L = lake, R = river, EP = ephemeral pond, P = pond, D = Dam, S = Swamp. Samples were collected in Kenya unless otherwise indicated.
Multiple sequence alignments were performed using the program MUSCLE [82] in MEGA X [83]. The best fit maximum likelihood (ML) nucleotide substitution model was chosen for all genes in MEGA X using BIC criterion. Phylogenetic relationships were inferred using ML in MEGA X using 1000 bootstrap replicates. Uncorrected pairwise distance values (p-distances) were calculated in MEGA X [83]. Data were summarized within and between groups (Tables 2 and S2).
Table 2. Intra- and Interspecies p-distance values of partial cox1 of 68 bulinid sequences.
| B. globosus | B. ugandae | B. productus | B. nasutus | B. truncatus | B. tropicus | B. cf. transversalis | B. forskalii | B. scalaris | B. cf. trigonus | |
|---|---|---|---|---|---|---|---|---|---|---|
| B. globosus | 0.0227 | |||||||||
| B. ugandae | 0.0578 | 0.0077 | ||||||||
| B. productus | 0.1428 | 0.1178 | 0.0021 | |||||||
| B. nasutus | 0.1304 | 0.1083 | 0.0735 | 0.0053 | ||||||
| B. truncatus | 0.1650 | 0.1397 | 0.1780 | 0.1673 | 0.0095 | |||||
| B. tropicus | 0.1597 | 0.1375 | 0.1750 | 0.1657 | 0.0342 | 0.0097 | ||||
| B. cf. transversalis | 0.1833 | 0.1576 | 0.1910 | 0.1812 | 0.0485 | 0.0521 | 0.0011 | |||
| B. forskalii | 0.1383 | 0.1294 | 0.1674 | 0.1548 | 0.1778 | 0.1663 | 0.1850 | 0.0039 | ||
| B. scalaris | 0.1608 | 0.1408 | 0.1948 | 0.1626 | 0.1762 | 0.1803 | 0.1781 | 0.1126 | 0.0326 | |
| B. cf. trigonus | 0.1654 | 0.1424 | 0.1910 | 0.1750 | 0.0486 | 0.0446 | 0.0531 | 0.1718 | 0.1802 | 0.0000 |
Bolded values are intraspecific p-distance values. B. cf. trigonus consists of GenBank sequences MT707391.1 and MT707392.1 [50].
Specimens sequenced as part of this study were deposited as vouchers in the Division of Parasites, Museum of Southwestern Biology at the University of New Mexico. Snail and parasites specimens were designated a MSB:Host: or a MSB:Para: number, respectively (Tables 1 and 3).
Table 3. Schistosome samples.
| Species | MSB:Para: | Collection location | Stage (miracidium, cercaria, adult) | Latitude | Longitude | Date | Host species | GenBank ID COI | Genbank ID ITS |
|---|---|---|---|---|---|---|---|---|---|
| S. bovis | 32675 | Tiengre | cercaria | -0.089833 | 34.70313 | 5/21/2018 | B. productus | OP235447 | OP234419 |
| S. haematobium | 32678 | Asao | cercaria | −0.31810 | 35.0069 | 5/20/2017 | B. globosus | OP235442 | OP234418 |
| S. bovis | 32679 | Asao | cercaria | −0.31810 | 35.0069 | 5/20/2017 | B. globosus | OP235445 | OP234417 |
| S. haematobium | 32680 | Asao | cercaria | −0.31810 | 35.0069 | 8/2/2016 | B. globosus | OP235444 | OP234408 |
| S. haematobium | 32681 | Asao | cercaria | −0.31810 | 35.0069 | 5/20/2017 | B. globosus | OP235446 | OP234407 |
| S. haematobium | 32682 | Asao | cercaria | −0.31810 | 35.0069 | 8/2/2016 | B. globosus | OP235448 | OP234416 |
| S. haematobium | 32683 | Asao | cercaria | −0.31810 | 35.0069 | 3/1/2017 | B. globosus | OP235441 | OP234415 |
| S. haematobium | 32685 | Nyakango School | miracidium | -0.440004 | 34.640005 | 2/14/2019 | Homo sapiens | OP235443 | OP234405 |
| S. haematobium | 32686 | Asao | miracidium | -0.3169444 | 35.00611 | 11/2/2019 | Homo sapiens | OP235440 | OP234401 |
| S. bovis | 32687 | Asao | cercaria | −0.31810 | 35.0069 | 11/19/2018 | B. globosus | OP235431 | OP234400 |
| S. bovis | 32689 | Asao | cercaria | −0.31810 | 35.0069 | 3/19/2019 | B. globosus | OP235435 | OP234410 |
| S. bovis | 32690 | Asao | cercaria | −0.31810 | 35.0069 | 4/29/2019 | B. globosus | OP235430 | OP234413 |
| S. bovis | 32692 | Tiengre | cercaria | -0.089833 | 34.70313 | 5/21/2018 | B. forskalii | OP235449 | OP234420 |
| S. bovis | 32705 | Gudwa Beach | cercaria | -0.3573667 | 34.3301 | 5/15/2019 | B. ugandae | OP235434 | OP234409 |
| S. bovis | 32693 | Asao | cercaria | −0.31810 | 35.0069 | 3/20/2019 | B. globosus | OP235433 | OP234411 |
| S. bovis | 32694 | Tiengre | cercaria | -0.08983333 | 34.70313 | 5/18/2019 | B. forskalii | OP235436 | OP234412 |
| S. haematobium | 32695 | Asao | cercaria | −0.31810 | 35.0069 | 8/2/2016 | B. globosus | OP235439 | OP234414 |
| S. haematobium | 32696 | Asao | cercaria | −0.31810 | 35.0069 | 3/23/2017 | B. globosus | OP235432 | OP234406 |
| S. haematobium | 32697 | Nyakango School | miracidium | -0.440004 | 34.640005 | 2/14/2019 | Homo sapiens | OP235438 | OP234404 |
| S. haematobium | 32698 | Nyakango School | miracidium | -0.440004 | 34.640005 | 2/14/2019 | Homo sapiens | OP235437 | OP234403 |
| S. haematobium | 32699 | Asao | cercaria | −0.31810 | 35.0069 | 5/20/2017 | B. globosus | OP235429 | OP234402 |
| S. bovis | 32704 | Asao | miracidium | −0.31810 | 35.0069 | 8/2/2016 | Bos indicus | OP235425 | OP234397 |
| S. bovis | 32702 | Ayuka Dam | cercaria | -0.449 | 34.65532 | 5/24/2018 | B. productus | OP235427 | OP234396 |
| S. bovis | 32703 | Tiengre | cercaria | -0.08983333 | 34.70313 | 11/1/2019 | B. productus | OP235426 | OP234399 |
| S. bovis | 32701 | Tiengre | cercaria | -0.08983333 | 34.70313 | 11/1/2019 | B. productus | OP235428 | OP234398 |
| S. haematobium | 32706 | Ayuka Dam | cercaria | -0.449 | 34.65532 | 5/24/2018 | B. productus | OP235450 | OP234421 |
Sequenced schistosomes with associated MSB:Para: numbers, collection locations, life cycle stage, GPS coordinates (when available), collection date, host species, and GenBank accession numbers.
Results
Overview of Bulinus collections
A total of 6,133, Bulinus snails were collected from 11 locations (S1 Fig) in in the LVB between January 2014 and March 2021 and initially provisionally identified, in some cases just to species group (S1 Table). Recovered snails included B. globosus (n = 2994), B. ugandae (n = 889), B. productus (n = 1302), B. tropicus/truncatus group species (n = 245), and B. forskalii group species (n = 685). Bulinid species presence and trematode composition and prevalence varied by site (S1 Table). The highest schistosome prevalence was recovered from B. globosus at Asao stream (6.5% prevalence) and few to no schistosome infections were recovered from the various lake shore habitats. Further sequence-based specifications of species identities for both bulinids and schistosomes are found below.
Molecular identification of bulinids
Partial portions of the cox1 gene were sequenced from 62 bulinids. Because some clades were initially overrepresented, 58 sequences were used in the final phylogenetic analysis (Fig 1). 16S sequences were produced for 70 bulinid specimens. Some specimens did not produce amplicons for both genes and therefore concatenated (cox1 +16S) sequences were produced for 57 bulinid specimens (Fig 2). Specimens were chosen for sequencing to include representative species from the widest variety of habitats possible. Specimen information can be found in Table 1.
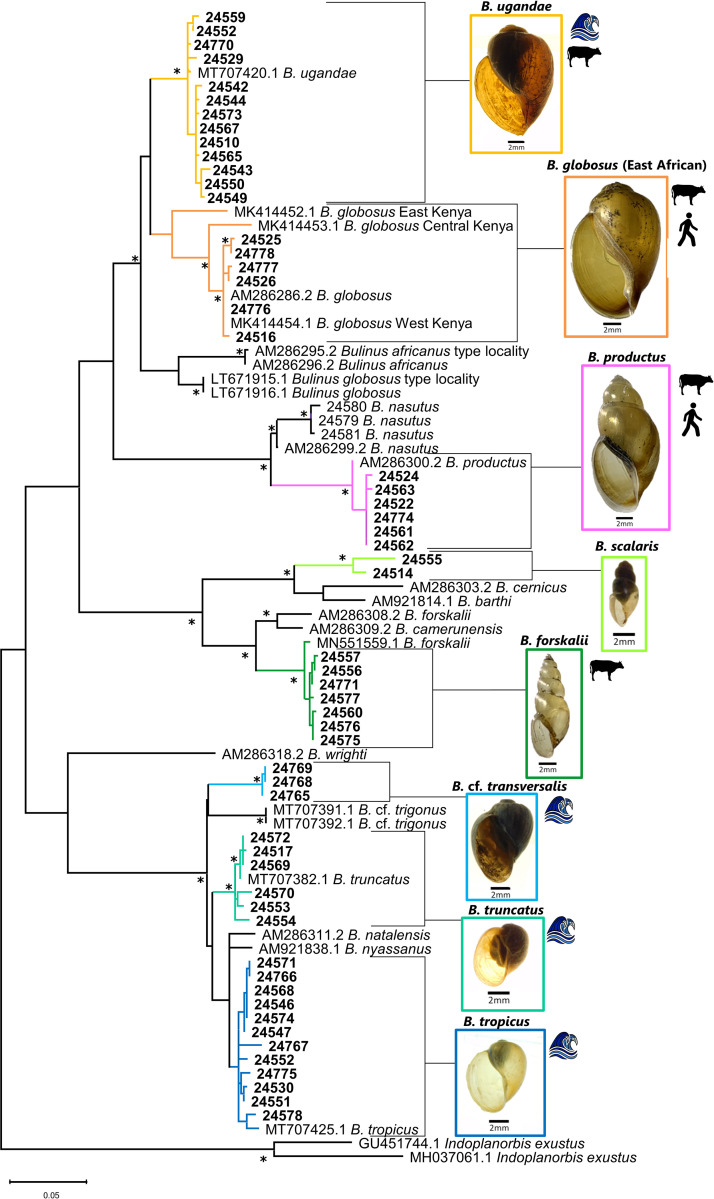
Fig 1. Phylogenetic relationships among Kenyan bulinids based on partial cox1 sequences.
Phylogenetic relationships of bulinids from this study and from GenBank (with accession numbers) based on 621 bp of the cytochrome oxidase subunit 1 gene inferred from ML analysis under the GTR+G+I model. Bootstrap values over 95% are indicated by an asterisk. Bolded sequences were generated during this study and listed by MSB:Host: number. Additional information for specimens can be found in Table 1. Specimens recovered within the LVB by this study are color coded by species. B. africanus group species are in warm colors, B. forskalii group species in greens, and B. truncatus/tropicus group species in blues. Wave icons indicates species found within Lake Victoria. Cow icons indicate species with naturally occurring S. bovis infections. Human icons indicate species with naturally occurring S. haematobium infections.
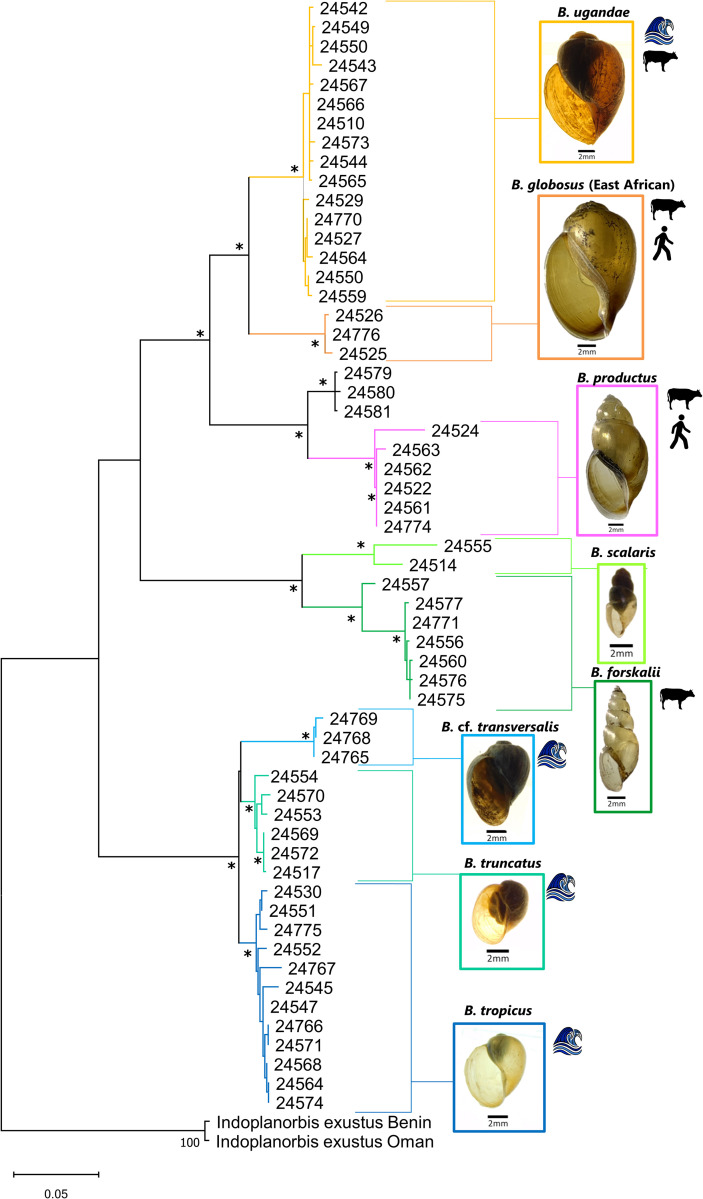
Fig 2. Phylogenetic relationships of bulinids based on concatenated cox1 + 16S sequences.
Phylogenetic relationships of bulinids from this study and from GenBank based on 1163 bp of combined cox1 and 16S sequences inferred from ML analysis under the GTR+G+I model. Bootstrap values above 95 are indicated by an asterisk. Sequences generated during this study are listed by MSB:Host number. Additional information for specimens can be found in Table 1. Specimens recovered within the LVB by this study are color coded by species. B. africanus group species are in warm colors, B. forskalii group species in greens, and B. truncatus/tropicus group species in blues. Wave icons indicates species found within Lake Victoria. Cow icons indicate species with naturally occurring S. bovis infections. Human icons indicate species with naturally occurring S. haematobium infections.
Phylogenetic analysis of bulinids using maximum likelihood methods
From our 11 study sites, we recovered seven named species of Bulinus based on cox1 and 16S sequences (B. globosus, B. ugandae, B. productus, B. forskalii, B. scalaris, B. truncatus, B. tropicus) and one distinct taxon we refer to as Bulinus cf. transversalis because it conforms in habitat and conchologically to B. transversalis [32] but for which no sequence references currently exist. The sequences we obtained for this taxon did not align with any known bulinid species in GenBank.
cox1 phylogenetic analysis
A total of 62 cox1 sequences were generated as a part of this study. Because some clades were overrepresented, 58 cox1 sequences from this study and 25 sequences from GenBank were used to hypothesize phylogenetic relationships among bulinid specimens we collected. The cox1 sequence analysis discriminated each of the four Bulinus species groups as well as each of the 17 species included in the phylogenetic analysis (Fig 1).
Intraspecific p-distances were less than 1% with the exceptions of the East African B. globosus complex (2.27%) and B. scalaris (3.26%). Interspecific p-distances ranged from 5.78% for within species group (ex. B. globosus and B. ugandae) to 19.48% between species groups (B. forskalii and B. productus). Exceptionally, members of the B. truncatus/tropicus group exhibited low interspecific p-distances as compared to other closely related bulinids (Table 2).
Combined (cox1 + 16S) dataset analysis
Concatenated cox1 and 16S sequences from GenBank representing outgroup sequences, and 58 sequences from this study (9 species) were used to infer phylogenetic relationships among bulinids. The concatenated sequence analysis discriminated among the three Bulinus species groups included in this analysis and additionally allowed interspecific discrimination (Fig 2) with greater resolution than the single cox1 dataset.
Intraspecific species p-distances were less than 1%, with the exception of B. scalaris. Interspecific p-distances were greater than 5%, with the exception of members of the B. truncatus/tropicus group which exhibited low intraspecific p-distances (S2 Table).
Trematode infections in field-collected snails
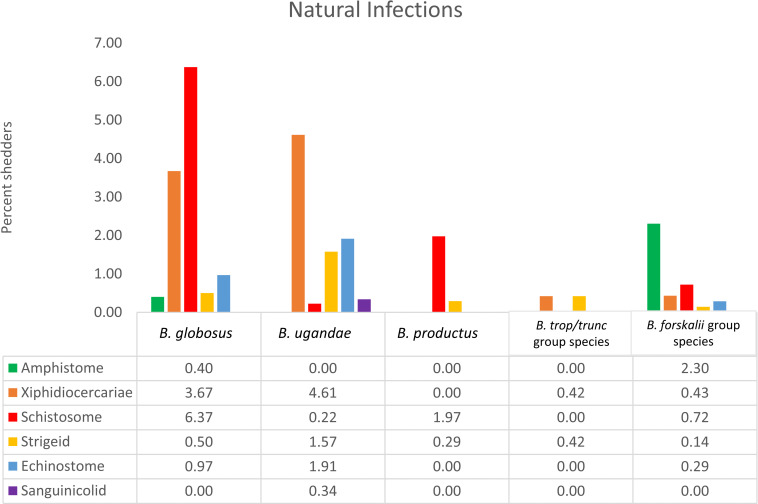
Natural infection prevalence varied by collection site and by host species (S1 Table). The highest prevalence of patent mammalian schistosome infections was found in B. globosus (6.37%) followed by B. productus (1.97%) and B. forskalii/scalaris (0.71%) with few infections found in B. ugandae (0.22%) and no schistosome infections observed among B. truncatus/tropicus specimens (S1 Table and Fig 3).
Fig 3. Prevalence of natural infections among field collected bulinids.
Natural infection prevalence of 6 cercarial types shed from Bulinus snails collected between January 2014 and March 2021 from various localities in Kenya (S1 Table).
Higher overall trematode diversity was observed among B. globosus, B. ugandae, and B. forskalii (minimum 5 trematode taxa per species) than was observed for B. truncatus/tropicus specimens (2 taxa) (Fig 3). This study did not seek to identify non-schistosome trematode cercariae to the species level and has therefore likely underestimated the diversity of trematode taxa coming from certain bulinid species including, for example, B. globosus and B. ugandae which are each known to host at least 2 species of echinostomes [57].
Phylogenetic analysis of schistosomes
Partial cox1 sequences provided by this study were primarily used to identify cercariae samples to the species level. Specimens were identified as either S. haematobium or S. bovis. Cox1 and ITS sequences were used to examine cercariae samples for nuclear/mitochondrial discordance, which was not observed. Concatenated (cox1 + ITS) alignments were used to infer relationships among specimens (Fig 4), and information relating to specimens provided by this study can be found in Table 3. S. haematobium cercariae were recovered from B. globosus and B. productus; S. bovis cercariae were recovered from B. globosus, B. productus, B. ugandae, and B. forskalii. The phylogenetic analysis did not indicate affiliations between intermediate host species and schistosome genotypes for S. bovis specimens (Fig 4).
Fig 4. Phylogenetic relationships among Kenyan schistosomes based on concatenated cox1 + ITS sequences.
Phylogenetic relationships of schistosomes from this study and from GenBank based on 1166 bp of concatenated cox1 + partial ITS1 + 5.8S + partial ITS2 sequences inferred from ML analysis. Bootstrap values over 95% are indicated by an asterisk. Specimens are listed by MSB:Para: number followed by the host species. Bolded sequences were generated during this study and additional information for specimens can be found in Table 3.
Discussion
Our long-term goal is to understand the underlying biological processes that influence the complex interrelationships between bulinid snails and trematodes, especially schistosomes, in the LVB. Towards that end, we identified 8 distinct Bulinus taxa, 2 of which were naturally infected with S. haematobium (B. globosus and B. productus) and 4 of which were naturally infected with S. bovis (B. globosus, B. productus, B. ugandae, and B. forskalii). Additionally, 5 broad categories of non-schistosome cercariae were found among the bulinids collected.
With respect to the B. africanus group, the species reported thus far in the LVB are B. africanus, B. globosus, B. nasutus, B. productus (often referred to as B. nasutus productus), and B. ugandae [32,33]. Our results are similar but noteworthy in that we did not find B. africanus or B. nasutus (Figs 1 and 2). The presence of these taxa in areas we did not sample surely cannot be ruled out. We note that many of the previous identifications of B. africanus in the LVB were based largely on morphological criteria [32,60,84–86], which for reasons noted [36,87], may be particularly unreliable in East Africa.
Based on several lines of evidence, we suggest that the widespread occurrence of B. africanus in the LVB needs to be reconsidered. Chibwana et al. [50] did not report B. africanus from the LVB, and Pennance [35] noted the taxon he represented as B. africanus sp. 1 (representing locations in the LVB) is likely to be B. ugandae, with which we agree (see more below).
The phylogenetic inferences we generated for west Kenyan specimens of the related species B. globosus grouped with East African specimens designated as B. globosus on GenBank. Similar to observations of Kane et al. [38], we found (Fig 1) that type locality specimens for B. africanus [88] from Port Durban, South Africa, and for B. globosus [89] from Angola, belong to lineages separate from the East African B. globosus lineage. This suggests that the East African B. globosus is not conspecific with those from the type localities. In our phylogenetic analysis, representatives from the LVB did not group with the type-locality specimens for either B. globosus or B. africanus. Thus, future work should be done to characterize the B. globosus from LVB as it may be a different species. For convenience, we refer to it in this paper as Bulinus globosus. This line of thinking is consistent with other recent papers [46,50,75] that refer to a “B. globosus complex”, with multiple lineages represented in Kenya alone [46], and rendered even more complex than what we found when specimens from other parts of Africa are included [35,38].
B. nasutus has traditionally been divided into two subspecies: B. nasutus nasutus distributed mainly along the coastal provinces of Kenya and Tanzania, and B. nasutus productus distributed further inland from Uganda to Tanzania [36]. Morphometric analysis [90], enzyme analysis [33], and sequence analysis [38] suggested that the B. nasutus complex consists of two separate species, B. nasutus and B. productus. The cox1 p-distances we calculated (Table 2) for B. nasutus and B. productus were comparable to the p-distances found between other Bulinus species and justifies the separation of the B. nasutus complex into two species, which is also supported by a mitogenome analysis [35]. Enzyme analysis indicated both taxa were present in the LVB [33], but we found B. nasutus specimens only from central Kenya and B. productus only from the LVB. Additionally, B. productus was found exclusively in ephemeral pools and dams within the LVB while the related B. globosus was found primarily in streams (Table 1).
A more detailed understanding of the underlying systematics for B. globosus and B. productus is important because both are vectors of S. haematobium and have been implicated in natural infections by this study and by others [59,60,91–93] in the LVB. These taxonomic difficulties are especially unfortunate for the B. globosus species complex, members of which are important hosts for S. haematobium across tropical Africa; it remains awkward as to how to accurately name these important vector snails.
As noted above, another enigmatic member of the B. africanus species group is B. ugandae, widely reported throughout the LVB [84,85,94,95] but more rarely identified using molecular criteria. However, Chibwana et al. [50] identified a Bulinus sp. 2 which they suggested was B. ugandae based on analysis of their sequence data. Likewise, Pennance [35] identified B. africanus sp. 1, also suggesting it might be B. ugandae. More recently Zhang et al. [46] assembled the mitogenome of a B. ugandae sample from Lake Victoria. Based on examination of the shell photographs, similarities in habitat types, and phylogenetic analysis of sequences, we agree that these sequences represent B. ugandae specimens.
The phylogenetic relationships inferred in our study indicate that B. ugandae is sister to the East African B. globosus lineage as we have described it above. Figs 1 and 2 differ slightly in their topology, which may be resolved in the future with increased taxon sampling. However, both phylogenetic analyses support B. ugandae and East African B. globosus as separate lineages. In agreement with earlier studies [32,36,84,85], we did not find B. globosus in lacustrine habitats, while B. ugandae was found commonly from the shore of Lake Victoria or in marshes and swamps along the lakes edge.
It is of more than passing interest to correctly discriminate B. ugandae from B. globosus [96], and the application of molecular criteria is recommended. Bulinus ugandae is the only member of the B. africanus group not implicated in the transmission of S. haematobium [59,60]. The relationship between B. ugandae and S. bovis is more nuanced with field studies suggesting that Kenyan B. ugandae is refractory to S. bovis [84] whereas other studies suggested that B. ugandae from Sudan or Uganda are vectors of S. bovis [94,97]. Bulinus ugandae from Western Kenya was found to be compatible with S. bovis in experimental infections [98], and we found B. ugandae to be naturally infected with S. bovis at two of our lakeshore study sites (S1 Table). The low prevalence of S. bovis in B. ugandae we observed may explain why some studies did not report natural infections. Alternatively, perhaps S. bovis relies on facilitation by other trematodes to successfully infect B. ugandae as has been reported in other bulinid species [10]. Pennance [35] also noted a natural infection of B. ugandae with an oft-overlooked member of the S. haematobium group, S. kisumuensis, previously known only from West Kenya based on anatomical characteristics and sequence data for adult worms recovered from rodents [24].
B. ugandae hosts a variety of other trematode species in the LVB (S1 Table and Fig 3). Amphistomes were not recovered during this study nor from a Tanzanian survey [85]. However, in Sudan, B. ugandae was found shedding amphistome cercariae [99], raising the possibility that significant intraspecific differences within B. ugandae may occur with resultant differences in compatibility with trematodes, further contributing to the complex patchwork of Bulinus-trematode compatibility so often noted.
B. forskalii species have received less attention than other bulinids in East Africa, likely because they are not associated with S. haematobium transmission in that area, unlike in West Africa [32,100,101]. Three B. forskalii group species: B. forskalii, B. scalaris, and B. browni have been reported from the LVB, and all have been observed to occur in sympatry [32]. In addition to finding B. forskalii commonly among our Kenyan samples, we found a juvenile of a second genetically distinct taxon that differed substantially from B. forskalii. It differed to a lesser extent from B. scalaris obtained from Ukerewe Island, Tanzania, the latter snail conforming conchologically to B. scalaris based on having rounded shoulders on the shell whorls [32]. The unknown juvenile tended to group with B. scalaris phylogenetically, yet intraspecific p-distances of these two sequences were higher than what has been reported within most Bulinus species (Tables 2 and S2). One possibility is that this snail is of the poorly known species B. browni, reported as being morphologically indistinguishable from B. forskalii but with unique enzyme banding patterns [102]. Its status remains uncertain as it has not been identified in any previous sequence-based analyses.
Neither B. forskalii nor B. scalaris are experimentally compatible with S. haematobium nor have been found to host natural infections in Western Kenya [59–61]. It is believed that B. browni similarly is not involved in transmission of S. haematobium [103], but both B. forskalii and B. browni have been implicated in the transmission of S. bovis [84,103,104]. These observations were supported by our findings which genetically identified S. bovis from natural infections in B. forskalii, yet we found no S. haematobium infections from any B. forskalii group snails. B. forskalii is known to vector a wide variety of other trematodes including amphistomes [55,105], echinostomes [106], and others [85]. Interestingly, the long periods of estivation that this species undergoes, which are associated with the ephemeral nature of their habitats, do not preclude it from frequently being parasitized by larval trematodes.
We found members of the B. truncatus/tropicus group only in Lake Victoria, an environment for which our accumulated taxonomic understanding for this species group is complicated. Based on morphological, enzymatic, and ploidy criteria, Brown [32] listed four members of the B. truncatus/tropicus group in Lake Victoria: B. truncatus, B. tropicus, B. transversalis, and B. trigonus. Chibwana et al. [50] recovered three taxa: B. truncatus, B. tropicus and Bulinus sp. 1 (considered to possibly be B. trigonus by the authors). Our efforts recovered three taxa: B. truncatus, B. tropicus and a third distinct taxon based on sequence criteria from Bulinus sp. 1 of Chibwana et al. [50]. Our third taxon most closely resembled B. transversalis conchologically [32,36], another bulinid species that remains poorly known.
The cox1 p-distances between B. truncatus and B. tropicus was the lowest among any two bulinid species we examined (Tables 2 and S2). Our presumptive B. transversalis and the presumptive B. trigonus of Chibwana et al. [50] differ to a greater extent from either B. tropicus or B. truncatus, and from each other, suggesting they are distinct species. The low p-distances between B. truncatus and B. tropicus has also been noted by others [39,43,107] and is somewhat paradoxical when considering their differences in ploidy, morphology and role as vectors of schistosomes.
Among the 245 individuals of the B. truncatus/tropicus group we examined, only 2 were positive for natural trematode infections (S1 Table and Fig 3). Neither Kenyan B. truncatus nor B. tropicus are known to vector local S. haematobium isolates [59,60,108]. However, Kenyan B. truncatus has been found compatible with allopatric S. haematobium isolates [60,109]. Experimental infections with what was likely a laboratory population of B. transversalis also proved refractory to East African S. haematobium infection [59]. B. truncatus has been found compatible with local isolates of S. bovis [84,110]. B. tropicus was found compatible with S. bovis only if it is previously infected with Calicophoron microbothrium [10,111]. No natural schistosome infections were documented for any member of the B. truncatus/tropicus group as part of our study.
As recently noted by Chibwana et al. [50], a range of Bulinus species are present in Lake Victoria and surrounding waters and they also noted that bulinid presence in the lake potentially implies the presence of S. haematobium and health risks from urogenital schistosomiasis for people living along the shore, or on the lake’s islands. A considerable body of work has been undertaken over the years to examine the role of lake-associated bulinids in schistosome transmission (see the several papers cited above). Evidence from surveys and experimental infections, in agreement with data provided by this study, indicate that common lake species like B. ugandae, B. tropicus and B. truncatus are not found to be infected with local S. haematobium isolates, nor are members of the B. forskalii species group. Common africanus group species members like B. productus and B. globosus found in habitats other than lake shore are found to naturally host S. haematobium. The lake-dwelling B. ugandae, along with B. forskalii, B. globosus and B. productus have been found to naturally host S. bovis infections. At this time, unlike the situation for S. mansoni, the shorelines of Lake Victoria do not seem to pose a strong risk of S. haematobium infection.
As has been noted [60,61], East African B. truncatus are susceptible to what was historically described as B. truncatus-adapted isolates of S. haematobium common to Western Africa and Egypt, and introduction of isolates from these regions into the lake region might pose a new lake-borne S. haematobium problem. Likewise, introductions of exotic species into the lake, altered thermal or water quality regimes or changing populations of snail predators might change the current picture of Bulinus species representation in the lake, as they have in other African lakes [112].
Of further interest to us is to understand the puzzling underlying factors that dictate compatibility with S. haematobium of one Bulinus species, like B. globosus, whereas its close relative, B. ugandae, is seemingly refractory? This characteristic has a great deal to do with keeping S. haematobium transmission from occurring in the lake, thereby averting what could be a massive public health problem. Can this natural resistance to S. haematobium infection, if explained, in some way be used to lessen the vector potential of other bulinid species as a novel means of schistosomiasis control?
Similarly, we are interested in the characteristics of the west Kenyan S. haematobium isolates which favor or disfavor compatibility with certain bulinid species. S. haematobium isolates from across Africa have recently been shown to be genetically homogenous as compared to S. bovis [30], a characteristic that belies the evident heterogeneity in compatibility shown by S. haematobium across Africa with respect to Bulinus species use. One possible explanation is that all S. haematobium isolates tested, with the exception of the Madagascar isolate, have been found to contain varying levels of S. bovis introgression in their genomes [28–30]. It will be of interest to determine if the content of such introgressed regions influence the compatibility of S. haematobium to different Bulinus species.
Other avenues of interest for disentangling the Bulinus-schistosome compatibility include the role of symbionts, such as annelids (Chaetogaster), which may prey upon the miracidia or cercariae of trematodes, thereby reducing transmission [9]. Chaetogasters are particularly conspicuous on field-derived specimens of Bulinus [113] and deserve further scrutiny with respect to their impact on influencing infection success of schistosome miracidia.
We are similarly interested in applying the notion of coevolutionary hot and cold spots [114,115] to Lake Victoria shorelines, owing to their intense use by many host species potentially carrying many trematode species [11]. Shoreline locations have been considered coevolutionary hot spots and may dictate certain type of immune or other avoidance strategies by snails to avoid high infection rates. In contrast, deep water locations are considered coevolutionary cold spots because fewer host species (and attendant trematodes) frequent them, which might select for different response strategies among snails living there. We are similarly interested to learn if species like B. forskalii that so often are found in ephemeral habitats and known to be preferential self-crossers [116] have fundamentally different strategies for dealing with pathogens like trematodes than snails that occupy far more stable conditions, like the shoreline habitats of Lake Victoria.
Conclusions
Based on cox1 sequence data, we found 8 distinct taxa of Bulinus in our west Kenyan sampling locations: B. globosus, B. productus, B. ugandae, B. forskalii, presumptive B. scalaris; B. tropicus, B. truncatus and presumptive B. transversalis. We found natural infections of S. haematobium in B. globosus and B. productus, and the ruminant schistosome S. bovis in these two species as well as in B. ugandae and B. forskalii, confirming the vector role for these species outlined in previous studies. We highlight the importance of providing molecularly-based identification, particularly in regards to discriminating S. haematobium vector species like B. globosus from related non-vector species like B. ugandae. Several outstanding issues with respect to Bulinus systematics were noted: the lack of bona fide B. africanus in our samples and the presence of a “B. globosus complex” requiring further resolution; the status of B. productus as a distinct species from B. nasutus; and the need for further collection and resolution among species in both the B. forskalii and B. tropics/truncatus groups, the latter especially as it pertains to the LVB. The complex patterns of Bulinus-Schistosoma compatibilities noted argue for more in-depth study to understand factors dictating the underlying patterns that, at least thus far, have fortuitously kept the immediate shoreline and waters of Lake Victoria largely free of S. haematobium transmission.
Supporting information
ExpertGPS Basemap of collection locations within the Lake Victoria Basin in Western Kenya for bulinid snails. Information regarding samples from these locations can be found in Tables 1 and S1. Base map and data from OpenStreetMap and OpenStreetMap Foundation. Base-layer retrieved from https://www.openstreetmap.org/relation/192798.
(TIF)
The number of snail specimens examined and number of observed trematode infections is listed per species and per survey sites. Total number of snail specimens and percent of individuals infected (in parentheses) is listed per cercarial type. Map of collection locations can be found in S1 Fig. Habitat types are: LS = lakeshore, L = lake, R = river, EP = ephemeral pond, D = Dam, S = Swamp.
(XLSX)
Bolded values are intraspecies p-distance values.
(XLSX)
Acknowledgments
We thank Ibrahim Mwangi, Joseph Kinuthia, Geoffrey Maina, and Boaz Oduor for their assistance with the collection of field samples and snail maintenance. We also thank Dr Stephen Munga, the Deputy Director of the Center for Global Health Research, Kenya Medical Research Institute (KEMRI), for providing the laboratory space to conduct these experiments. Technical assistance at the University of New Mexico Molecular Biology Facility was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number P30GM110907. This work was published with the approval of the Director-General, KEMRI.
The content for this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Data Availability
All sequence files are available from the Genbank database (accession numbers OP235425-OP235450; OP234396-OP234421; OP233086-OP233143; OP244902-OP244960; OP242173-OP242177).
Funding Statement
This study was funded by the National Institute of Health (https://www.nih.gov/) grant R37AI101438. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Perlman SJ, Jaenike J. Infection success in novel hosts: an experimental and phylogenetic study of Drosophila-parasitic nematodes. Evolution. 2003;57: 544–557. [DOI] [PubMed] [Google Scholar]
- 2.Medeiros MCI, Hamer GL, Ricklefs RE. Host compatibility rather than vector–host-encounter rate determines the host range of avian Plasmodium parasites. Proc R Soc B Biol Sci. 2013;280: 20122947. doi: 10.1098/rspb.2012.2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vanhove MPM, Pariselle A, Van Steenberge M, Raeymaekers JAM, Hablützel PI, Gillardin C, et al. Hidden biodiversity in an ancient lake: phylogenetic congruence between Lake Tanganyika tropheine cichlids and their monogenean flatworm parasites. Sci Rep. 2015;5: 13669. doi: 10.1038/srep13669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrow LN, McNew SM, Mitchell N, Galen SC, Lutz HL, Skeen H, et al. Deeply conserved susceptibility in a multi-host, multi-parasite system. Ecol Lett. 2019;22: 987–998. doi: 10.1111/ele.13263 [DOI] [PubMed] [Google Scholar]
- 5.Rollinson D, Stothard JR, Southgate VR. Interactions between intermediate snail hosts of the genus Bulinus and schistosomes of the Schistosoma haematobium group. Parasitology. 2001;123: 245–260. doi: 10.1017/S0031182001008046 [DOI] [PubMed] [Google Scholar]
- 6.Combes C. Parasitism: The ecology and evolution of intimate interactions. University of Chicago Press; 2001. [Google Scholar]
- 7.Medeiros MCI, Ricklefs RE, Brawn JD, Hamer GL. Plasmodium prevalence across avian host species is positively associated with exposure to mosquito vectors. Parasitology. 2015;142: 1612–1620. doi: 10.1017/S0031182015001183 [DOI] [PubMed] [Google Scholar]
- 8.Manzoli DE, Saravia-Pietropaolo MJ, Arce SI, Percara A, Antoniazzi LR, Beldomenico PM. Specialist by preference, generalist by need: availability of quality hosts drives parasite choice in a natural multihost–parasite system. Int J Parasitol. 2021;51: 527–534. doi: 10.1016/j.ijpara.2020.12.003 [DOI] [PubMed] [Google Scholar]
- 9.Hobart BK, Moss WE, McDevitt-Galles T, Stewart Merrill TE, Johnson PTJ. It’s a worm-eat-worm world: Consumption of parasite free-living stages protects hosts and benefits predators. J Anim Ecol. 2022;91: 35–45. doi: 10.1111/1365-2656.13591 [DOI] [PubMed] [Google Scholar]
- 10.Southgate VR, Brown DS, Warlow A, Knowles RJ, Jones A. The influence of Calicophoron microbothrium on the susceptibility of Bulinus tropicus to Schistosoma bovis. Parasitol Res. 1989;75: 381–391. doi: 10.1007/BF00931134 [DOI] [PubMed] [Google Scholar]
- 11.Laidemitt MR, Anderson LC, Wearing HJ, Mutuku MW, Mkoji GM, Loker ES. Antagonism between parasites within snail hosts impacts the transmission of human schistosomiasis. eLife. 2019;8: e50095. doi: 10.7554/eLife.50095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rollinson D, Southgate VR. Schistosome and snail populations: genetic variability and parasite transmission. Ecol Genet Host Parasite Interact Int Symp Keele Univ 12–13 July. 1985;1984: 91–109. [Google Scholar]
- 13.Panstruga R. Establishing compatibility between plants and obligate biotrophic pathogens. Curr Opin Plant Biol. 2003;6: 320–326. doi: 10.1016/s1369-5266(03)00043-8 [DOI] [PubMed] [Google Scholar]
- 14.Magez S, Caljon G. Mouse models for pathogenic African trypanosomes: unravelling the immunology of host–parasite–vector interactions. Parasite Immunol. 2011;33: 423–429. doi: 10.1111/j.1365-3024.2011.01293.x [DOI] [PubMed] [Google Scholar]
- 15.Mitta G, Adema CM, Gourbal B, Loker ES, Theron A. Compatibility polymorphism in snail/schistosome interactions: From field to theory to molecular mechanisms. Dev Comp Immunol. 2012;37: 1–8. doi: 10.1016/j.dci.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barribeau SM, Sadd BM, du Plessis L, Schmid-Hempel P. Gene expression differences underlying genotype-by-genotype specificity in a host–parasite system. Proc Natl Acad Sci. 2014;111: 3496–3501. doi: 10.1073/pnas.1318628111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodenburg J, Cissoko M, Kayongo N, Dieng I, Bisikwa J, Irakiza R, et al. Genetic variation and host–parasite specificity of Striga resistance and tolerance in rice: the need for predictive breeding. New Phytol. 2017;214: 1267–1280. doi: 10.1111/nph.14451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pila EA, Li H, Hambrook JR, Wu X, Hanington PC. Schistosomiasis from a snail’s perspective: advances in snail immunity. Trends Parasitol. 2017;33: 845–857. doi: 10.1016/j.pt.2017.07.006 [DOI] [PubMed] [Google Scholar]
- 19.Augusto R de C, Duval D, Grunau C. Effects of the environment on developmental plasticity and infection success of Schistosoma parasites–an epigenetic perspective. Front Microbiol. 2019;10: 1475. doi: 10.3389/fmicb.2019.01475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu L, Bu L, Zhang S-M, Buddenborg SK, Loker ES. An Overview of transcriptional responses of schistosome-susceptible (M line) or -resistant (BS-90) Biomphalaria glabrata exposed or not to Schistosoma mansoni infection. Front Immunol. 2022;12. Available: https://www.frontiersin.org/article/10.3389/fimmu.2021.805882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Struck TH, Feder JL, Bendiksby M, Birkeland S, Cerca J, Gusarov VI, et al. Finding evolutionary processes hidden in cryptic species. Trends Ecol Evol. 2018;33: 153–163. doi: 10.1016/j.tree.2017.11.007 [DOI] [PubMed] [Google Scholar]
- 22.Kaukas A, Neto ED, Simpson AJG, Southgate VR, Rollinson D. A phylogenetic analysis of Schistosoma haematobium group species based on randomly amplified polymorphic DNA. Int J Parasitol. 1994;24: 285–290. doi: 10.1016/0020-7519(94)90040-X [DOI] [PubMed] [Google Scholar]
- 23.Webster BL, Southgate VR, Littlewood DTJ. A revision of the interrelationships of Schistosoma including the recently described Schistosoma guineensis. Int J Parasitol. 2006;36: 947–955. doi: 10.1016/j.ijpara.2006.03.005 [DOI] [PubMed] [Google Scholar]
- 24.Hanelt B, Brant SV, Steinauer ML, Maina GM, Kinuthia JM, Agola LE, et al. Schistosoma kisumuensis n. sp. (Digenea: Schistosomatidae) from murid rodents in the Lake Victoria Basin, Kenya and its phylogenetic position within the S. haematobium species group. Parasitology. 2009;136: 987–1001. doi: 10.1017/S003118200900643X [DOI] [PubMed] [Google Scholar]
- 25.McManus DP, Dunne DW, Sacko M, Utzinger J, Vennervald BJ, Zhou X-N. Schistosomiasis. Nat Rev Dis Primer. 2018;4: 1–19. doi: 10.1038/s41572-018-0013-8 [DOI] [PubMed] [Google Scholar]
- 26.Brémond P, Sellin B, Sellin E, Naméoua B, Labbo R, Théron A, et al. Arguments for the modification of the genome (introgression) of the human parasite Schistosoma haematobium by genes from S. bovis, in Niger. C R Acad Sci III. 1993;316: 667–670. [PubMed] [Google Scholar]
- 27.Huyse T, Webster BL, Geldof S, Stothard RJ, Diaw OT, Polman K, et al. Bidirectional introgressive hybridization between a cattle and human schistosome species. PLoS Pathog. 2009;5. doi: 10.1371/journal.ppat.1000571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oey H, Zakrzewski M, Gravermann K, Young ND, Korhonen PK, Gobert GN, et al. Whole-genome sequence of the bovine blood fluke Schistosoma bovis supports interspecific hybridization with S. haematobium. PLOS Pathog. 2019;15: e1007513. doi: 10.1371/journal.ppat.1007513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Platt RN II, McDew-White M, Le Clec’h W, Chevalier FD, Allan F, Emery AM, et al. Ancient hybridization and adaptive introgression of an invadolysin gene in schistosome parasites. Mol Biol Evol. 2019;36: 2127–2142. doi: 10.1093/molbev/msz154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rey O, Toulza E, Chaparro C, Allienne J-F, Kincaid-Smith J, Mathieu-Begné E, et al. Diverging patterns of introgression from Schistosoma bovis across S. haematobium African lineages. PLOS Pathog. 2021;17: e1009313. doi: 10.1371/journal.ppat.1009313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stensgaard A-S, Vounatsou P, Sengupta ME, Utzinger J. Schistosomes, snails and climate change: Current trends and future expectations. Acta Trop. 2019;190: 257–268. doi: 10.1016/j.actatropica.2018.09.013 [DOI] [PubMed] [Google Scholar]
- 32.Brown DS. Freshwater snails of Africa and their medical importance. CRC Press; 1994. [Google Scholar]
- 33.Jelnes JE, Thiongo FW, Lwambo NSJ, Ouma JH. The Bulinus nasutus complex (Bulinus nasutus (Martens, 1879) and Bulinus productus (Mandahl-Barth, 1960)(Gastropoda: Planorbidae) in the Lake Victoria area elucidated by enzyme-profile electrophoresis and natural infections with Schistosoma spp. (Trematoda: Schistosomatidae). Steenstrupia. 2003;27: 257–262. [Google Scholar]
- 34.Jørgensen A, Madsen H, Nalugwa A, Nyakaana S, Rollinson D, Stothard JR, et al. A molecular phylogenetic analysis of Bulinus (Gastropoda: Planorbidae) with conserved nuclear genes. Zool Scr. 2011;40: 126–136. doi: 10.1111/j.1463-6409.2010.00458.x [DOI] [Google Scholar]
- 35.Pennance T. Genetic diversity and evolution within the genus Bulinus and species-level interactions with the transmission of Schistosoma haematobium group parasites. phd, Cardiff University. 2020. Available: https://orca.cardiff.ac.uk/135638/ [Google Scholar]
- 36.Mandahl-Barth G. The species of the genus Bulinus, intermediate hosts of Schistosoma. Bull World Health Organ. 1965;33: 33–44. [PMC free article] [PubMed] [Google Scholar]
- 37.Stothard JR, Mgeni AF, Alawi KS, Savioli I, Rollinson D. Observations on shell morphology, enzymes and random amplified polymorphic DNA (RAPD) in Bulinus africanus group snails (Gastropoda: Planorbidae) in Zanzibar. J Molluscan Stud. 1997;63: 489–503. doi: 10.1093/mollus/63.4.489 [DOI] [Google Scholar]
- 38.Kane RA, Stothard JR, Emery AM, Rollinson D. Molecular characterization of freshwater snails in the genus Bulinus: a role for barcodes? Parasit Vectors. 2008;1: 15. doi: 10.1186/1756-3305-1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nalugwa A, Jørgensen A, Nyakaana S, Kristensen T. Molecular phylogeny of Bulinus (Gastropoda: Planorbidae) reveals the presence of three species complexes in the Albertine Rift freshwater bodies. Int J Genet Mol Biol. 2010;2: 130–139. [Google Scholar]
- 40.Jarne P, Städler T. Population genetic structure and mating system evolution in freshwater pulmonates. Experientia. 1995;51: 482–497. doi: 10.1007/BF02143200 [DOI] [Google Scholar]
- 41.Goldman MA, LoVerde PT, Chrisman CL. Hybrid origin of polyploidy in freshwater snails of the genus Bulinus (Mollusca: Planorbidae). Evolution. 1983;37: 592–600. doi: 10.2307/2408272 [DOI] [PubMed] [Google Scholar]
- 42.Langand J, Barral V, Delay B, Jourdane J. Detection of genetic diversity within snail intermediate hosts of the genus Bulinus by using random amplified polymorphic DNA markers (RAPDs). Acta Trop. 1993;55: 205–215. doi: 10.1016/0001-706X(93)90078-P [DOI] [PubMed] [Google Scholar]
- 43.Stothard JR, Hughes S, Rollinson D. Variation within the Internal Transcribed Spacer (ITS) of ribosomal DNA genes of intermediate snail hosts within the genus Bulinus (Gastropoda: Planorbidae). Acta Trop. 1996;61: 19–29. doi: 10.1016/0001-706X(95)00137-4 [DOI] [PubMed] [Google Scholar]
- 44.Morgan JAT, DeJong RJ, Jung Y, Khallaayoune K, Kock S, Mkoji GM, et al. A phylogeny of planorbid snails, with implications for the evolution of Schistosoma parasites. Mol Phylogenet Evol. 2002;25: 477–488. doi: 10.1016/S1055-7903(02)00280-4 [DOI] [PubMed] [Google Scholar]
- 45.Young ND, Kinkar L, Stroehlein AJ, Korhonen PK, Stothard JR, Rollinson D, et al. Mitochondrial genome of Bulinus truncatus (Gastropoda: Lymnaeoidea): Implications for snail systematics and schistosome epidemiology. Curr Res Parasitol Vector-Borne Dis. 2021;1: 100017. doi: 10.1016/j.crpvbd.2021.100017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S-M, Bu L, Lu L, Babbitt C, Adema CM, Loker ES. Comparative mitogenomics of freshwater snails of the genus Bulinus, obligatory vectors of Schistosoma haematobium, causative agent of human urogenital schistosomiasis. Sci Rep. 2022;12: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young ND, Stroehlein AJ, Wang T, Korhonen PK, Mentink-Kane M, Stothard JR, et al. Nuclear genome of Bulinus truncatus, an intermediate host of the carcinogenic human blood fluke Schistosoma haematobium. Nat Commun. 2022;13: 977. doi: 10.1038/s41467-022-28634-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tumwebaze I, Clewing C, Dusabe MC, Tumusiime J, Kagoro-Rugunda G, Hammoud C, et al. Molecular identification of Bulinus spp. intermediate host snails of Schistosoma spp. in crater lakes of western Uganda with implications for the transmission of the Schistosoma haematobium group parasites. Parasit Vectors. 2019;12: 565. doi: 10.1186/s13071-019-3811-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pennance T, Allan F, Emery A, Rabone M, Cable J, Garba AD, et al. Interactions between Schistosoma haematobium group species and their Bulinus spp. intermediate hosts along the Niger River Valley. Parasit Vectors. 2020;13: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chibwana FD, Tumwebaze I, Mahulu A, Sands AF, Albrecht C. Assessing the diversity and distribution of potential intermediate hosts snails for urogenital schistosomiasis: Bulinus spp. (Gastropoda: Planorbidae) of Lake Victoria. Parasit Vectors. 2020;13: 418. doi: 10.1186/s13071-020-04281-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones CS, Rollinson D, Mimpfoundi R, Ouma J, Kariuki HC, Noble LR. Molecular evolution of freshwater snail intermediate hosts within the Bulinus forskalii group. Parasitology. 2001;123: 277–292. doi: 10.1017/S0031182001008381 [DOI] [PubMed] [Google Scholar]
- 52.Stothard JR, Brémond P, Andriamaro L, Sellin B, Sellin E, Rollinson D. Bulinus species on Madagascar: molecular evolution, genetic markers and compatibility with Schistosoma haematobium. Parasitology. 2001;123: 261–275. doi: 10.1017/S003118200100806X [DOI] [PubMed] [Google Scholar]
- 53.Jones CS, Noble LR, Ouma J, Kariuki HC, Mimpfoundi R, Brown DS, et al. Molecular identification of schistosome intermediate hosts: case studies of Bulinus forskalii group species (Gastropoda: Planorbidae) from Central and East Africa. Biol J Linn Soc. 1999;68: 215–240. doi: 10.1111/j.1095-8312.1999.tb01167.x [DOI] [Google Scholar]
- 54.Akinwale O, Oso O, Salawu O, Odaibo A, Tang P, Chen T, et al. Molecular characterization of Bulinus snails–intermediate hosts of schistosomes in Ogun State, South-western Nigeria. Folia Malacol. 2015;23: 137–147. doi: 10.12657/folmal.023.009 [DOI] [Google Scholar]
- 55.Laidemitt MR, Zawadzki ET, Brant SV, Mutuku MW, Mkoji GM, Loker ES. Loads of trematodes: discovering hidden diversity of paramphistomoids in Kenyan ruminants. Parasitology. 2016/10/20 ed. 2017;144: 131–147. doi: 10.1017/S0031182016001827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pfukenyi DM, Mukaratirwa S. Amphistome infections in domestic and wild ruminants in East and Southern Africa: A review. Onderstepoort J Vet Res. 2018;85: 1–13. doi: 10.4102/ojvr.v85i1.1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laidemitt MR, Brant SV, Mutuku MW, Mkoji GM, Loker ES. The diverse echinostomes from East Africa: With a focus on species that use Biomphalaria and Bulinus as intermediate hosts. Acta Trop. 2019;193: 38–49. doi: 10.1016/j.actatropica.2019.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chontananarth T, Tejangkura T, Wetchasart N, Chimburut C. Morphological characteristics and phylogenetic trends of trematode cercariae in freshwater snails from Nakhon Nayok province, Thailand. Korean J Parasitol. 2017;55: 47–54. doi: 10.3347/kjp.2017.55.1.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cridland CC. The experimental infection of several species of African freshwater snails with Schistosoma mansoni and S. hasmatobium. J Trop Med Hyg. 1955;58: 1–11. [PubMed] [Google Scholar]
- 60.Southgate VR, Knowles RJ. On the intermediate hosts of Schistosoma haematobium from Western Kenya. Trans R Soc Trop Med Hyg. 1977;71: 82–83. doi: 10.1016/0035-9203(77)90214-0 [DOI] [PubMed] [Google Scholar]
- 61.Frandsen F. Studies of the relationships between Schistosoma and their intermediate hosts. II. The genus Bulinus and Schistosoma haematobium from Sudan, Zaire and Zambia. J Helminthol. 1979;53: 205–212. doi: 10.1017/s0022149x00005988 [DOI] [PubMed] [Google Scholar]
- 62.McCullough FS. The susceptibility and resistance of Bulinus (Physopsis) globosus and Bulinus (Bulinus) truncatus rohlfsi to two strains of Schistosoma haematobium in Ghana. Bull World Health Organ. 1959;20: 75–85. [PMC free article] [PubMed] [Google Scholar]
- 63.Wright CA, Knowles RJ. Studies on Schistosoma haematobium in the laboratory III. Strains from Iran, Mauritius and Ghana. Trans R Soc Trop Med Hyg. 1972;66: 108–118. doi: 10.1016/0035-9203(72)90057-0 [DOI] [PubMed] [Google Scholar]
- 64.Véra C, Jourdane J, Sellin B, Combes C. Genetic variability in the compatibility between Schistosoma haematobium and its potential vectors in Niger: epidemiological implications. Trop Med Parasitol Off Organ Dtsch Tropenmedizinische Ges Dtsch Ges Tech Zusammenarbeit GTZ. 1990;41: 143–148. [PubMed] [Google Scholar]
- 65.Kendall RL. An ecological history of the Lake Victoria Basin. Ecol Monogr. 1969;39: 121–176. doi: 10.2307/1950740 [DOI] [Google Scholar]
- 66.Mutuku MW, Laidemitt MR, Beechler BR, Mwangi IN, Otiato FO, Agola EL, et al. A search for snail-related answers to explain differences in response of Schistosoma mansoni to praziquantel treatment among responding and persistent hotspot villages along the Kenyan shore of Lake Victoria. Am J Trop Med Hyg. 2019;101: 65–77. doi: 10.4269/ajtmh.19-0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kristensen TK. A field guide to African freshwater snails 2. East African species. Danish Bilharziasis Laboratory; 1987. [Google Scholar]
- 68.Frandsen F, Christensen NO. An introductory guide to the identification of cercariae from African freshwater snails with special reference to cercariae of trematode species of medical and veterinary importance. Acta Trop. 1984;41: 181–202. [PubMed] [Google Scholar]
- 69.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3: 294–299. [PubMed] [Google Scholar]
- 70.Wilke T, Davis GM. Infraspecific mitochondrial sequence diversity in Hydrobia ulvae and Hydrobia ventrosa (Hydrobiidae: Rissooidea: Gastropoda): Do their different life histories affect biogeographic patterns and gene flow? Biol J Linn Soc. 2000;70: 89–105. doi: 10.1111/j.1095-8312.2000.tb00202.x [DOI] [Google Scholar]
- 71.Palumbi SR, Andrew Martin, Sandra Romano, WOwen McMillan, Ligaya Stice, Gail Grabowski, et al. The simple fool’s guide to PCR. Version 2.0. Honolulu, HI: Dept. of Zoology and Kewalo Marine Laboratory, University of Hawaii; 2002. Available: http://palumbi.stanford.edu/SimpleFoolsMaster.pdf [Google Scholar]
- 72.Webster BL, Rollinson D, Stothard JR, Huyse T. Rapid diagnostic multiplex PCR (RD-PCR) to discriminate Schistosoma haematobium and S. bovis. J Helminthol. 2010;84: 107–114. doi: 10.1017/S0022149X09990447 [DOI] [PubMed] [Google Scholar]
- 73.Lockyer A, Olson P, Ostergaard P, Rollinson D, Johnston D, Attwood S, et al. The phylogeny of the Schistosomatidae based on three genes with emphasis on the interrelationships of Schistosoma Weinland, 1858. Parasitology. 2003;126: 203–24. doi: 10.1017/S0031182002002792 [DOI] [PubMed] [Google Scholar]
- 74.Barber KE, Mkoji GM, Loker ES. PCR-RFLP analysis of the ITS2 region to identify Schistosoma haematobium and S. bovis from Kenya. Am J Trop Med Hyg. 2000;62: 434–440. doi: 10.4269/ajtmh.2000.62.434 [DOI] [PubMed] [Google Scholar]
- 75.Allan F, Sousa-Figueiredo JC, Emery AM, Paulo R, Mirante C, Sebastião A, et al. Mapping freshwater snails in north-western Angola: distribution, identity and molecular diversity of medically important taxa. Parasit Vectors. 2017;10: 460. doi: 10.1186/s13071-017-2395-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu L, Mondal MM, Idris MA, Lokman HS, Rajapakse PJ, Satrija F, et al. The phylogeography of Indoplanorbis exustus (Gastropoda: Planorbidae) in Asia. Parasit Vectors. 2010;3: 57. doi: 10.1186/1756-3305-3-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mouahid G, Clerissi C, Allienne J-F, Chaparro C, Yafae SA, Nguema RM, et al. The phylogeny of the genus Indoplanorbis (Gastropoda, Planorbidae) from Africa and the French West Indies. Zool Scr. 2018;47: 558–564. doi: 10.1111/zsc.12297 [DOI] [Google Scholar]
- 78.Kane RA, Rollinson D. Repetitive sequences in the ribosomal DNA internal transcribed spacer of Schistosoma haematobium, Schistosoma intercalatum and Schistosoma mattheei. Mol Biochem Parasitol. 1994; 5. [DOI] [PubMed] [Google Scholar]
- 79.Léger E, Borlase A, Fall CB, Diouf ND, Diop SD, Yasenev L, et al. Prevalence and distribution of schistosomiasis in human, livestock, and snail populations in northern Senegal: a One Health epidemiological study of a multi-host system. Lancet Planet Health. 2020;4: e330–e342. doi: 10.1016/S2542-5196(20)30129-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kane RA, Southgate VR, Rollinson D, Littlewood DTJ, Lockyer AE, Pagès JR, et al. A phylogeny based on three mitochondrial genes supports the division of Schistosoma intercalatum into two separate species. Parasitology. 2003;127: 131–137. doi: 10.1017/s0031182003003421 [DOI] [PubMed] [Google Scholar]
- 81.Schols R, Mudavanhu A, Carolus H, Hammoud C, Muzarabani KC, Barson M, et al. Exposing the barcoding void: an integrative approach to study snail-borne parasites in a One Health context. Front Vet Sci. 2020;7. Available: https://www.frontiersin.org/articles/10.3389/fvets.2020.605280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35: 1547–1549. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Southgate VR, Knowles RJ. The intermediate hosts of Schistosoma bovis in Western Kenya. Trans R Soc Trop Med Hyg. 1975;69: 356–357. [DOI] [PubMed] [Google Scholar]
- 85.Loker ES, Moyo HG, Gardner SL. Trematode–gastropod associations in nine non-lacustrine habitats in the Mwanza region of Tanzania. Parasitology. 1981;83: 381–399. doi: 10.1017/S0031182000085383 [DOI] [Google Scholar]
- 86.Ofulla AV, Adoka SO, Anyona DN, Abuom PO, Karanja D, Vulule JM, et al. Spatial distribution and habitat characterization of schistosomiasis host snails in lake and land habitats of western Kenya. Lakes Reserv Sci Policy Manag Sustain Use. 2013;18: 197–215. doi: 10.1111/lre.12032 [DOI] [Google Scholar]
- 87.Raahauge P, Kristensen TK. A comparison of Bulinus africanus group species (Planorbidae; Gastropoda) by use of the internal transcribed spacer 1 region combined by morphological and anatomical characters. Acta Trop. 2000;75: 85–94. doi: 10.1016/S0001-706X(99)00086-8 [DOI] [PubMed] [Google Scholar]
- 88.Krauss F. Die südafrikanischen Mollusken; ein Beitrag zur Kenntniss der Mollusken des Kap- und Natallandes und zur geographischen Verbreitung derselben, mit Beschreibung und Abbildung der neuen Arten. Stuttgart: Ebner & Seubert; 1848. pp. 1–172. doi: 10.5962/bhl.title.13936 [DOI] [Google Scholar]
- 89.Morelet A. Coquilles nouvelles recueillies par le Dr. Fr. Welwitsch dans l’Afrique équatoriale, et particulièrement dans les provinces portugaises d’Angola et de Benguella. J Conchyliol. 1866;14: 153–163. [Google Scholar]
- 90.Kristensen TK, Frandsen F, Christensen AG. Bulinus africanus-group snails in East and South East Africa, differentiated by use of biometric multivariate analysis on morphological characters (Pulmonata: Planorbidae). Rev Zool Afr 1974. 1987;101: 55–67. [Google Scholar]
- 91.Webbe G. The transmission of Schistosoma haematobium in an area of Lake Province, Tanganyika. Bull World Health Organ. 1962;27: 59–85. [PMC free article] [PubMed] [Google Scholar]
- 92.Kinoti GK. The epidemiology of Schistosoma haematobium infection on the Kano Plain of Kenya. Trans R Soc Trop Med Hyg. 1971;65: 637–645. doi: 10.1016/0035-9203(71)90048-4 [DOI] [PubMed] [Google Scholar]
- 93.Pennance T, Ame SM, Amour AK, Suleiman KR, Muhsin MA, Kabole F, et al. Transmission and diversity of Schistosoma haematobium and S. bovis and their freshwater intermediate snail hosts Bulinus globosus and B. nasutus in the Zanzibar Archipelago, United Republic of Tanzania. PLoS Negl Trop Dis. 2022;16: e0010585. doi: 10.1371/journal.pntd.0010585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Berrie AD. Observations on the life-cycle of Bulinus (Physopsis) ugandae Mandahl-Barth, its ecological relation to Biomphalaria sudanica tanganyicensis (Smith), and its role as an intermediate host of Schistosoma. Ann Trop Med Parasitol. 1964;58: 457–466. doi: 10.1080/00034983.1964.11686268 [DOI] [PubMed] [Google Scholar]
- 95.Outa JO, Sattmann H, Köhsler M, Walochnik J, Jirsa F. Diversity of digenean trematode larvae in snails from Lake Victoria, Kenya: First reports and bioindicative aspects. Acta Trop. 2020;206: 105437. doi: 10.1016/j.actatropica.2020.105437 [DOI] [PubMed] [Google Scholar]
- 96.Opisa S, Odiere MR, Jura WG, Karanja DM, Mwinzi PN. Malacological survey and geographical distribution of vector snails for schistosomiasis within informal settlements of Kisumu City, western Kenya. Parasit Vectors. 2011;4: 226. doi: 10.1186/1756-3305-4-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Malek EA. Studies on bovine schistosomiasis in the Sudan. Ann Trop Med Parasitol. 1969;63: 501–513. doi: 10.1080/00034983.1969.11686654 [DOI] [PubMed] [Google Scholar]
- 98.Southgate VR, Knowles RJ. Observations on Schistosoma bovis Sonsino, 1876. J Nat Hist. 1975;9: 273–314. [Google Scholar]
- 99.Malek EA. The life cycle of Gastrodiscus aegyptiacus (Cobbold, 1876) Looss, 1896 (Trematoda: Paramphistomatidae: Gastrodiscinae). J Parasitol. 1971;57: 975–979. doi: 10.2307/3277847 [DOI] [PubMed] [Google Scholar]
- 100.Webster BL, Southgate VR. Compatibility of Schistosoma haematobium, S. intercalatum and their hybrids with Bulinus truncatus and B. forskalii. Parasitology. 2003;127: 231–242. doi: 10.1017/S0031182003003597 [DOI] [PubMed] [Google Scholar]
- 101.Labbo R, Djibrilla A, Zamanka H, Garba A, Chippaux J-P. Bulinus forskalii: a new potential intermediate host for Schistosoma haematobium in Niger. Trans R Soc Trop Med Hyg. 2007;101: 847–848. doi: 10.1016/j.trstmh.2007.03.016 [DOI] [PubMed] [Google Scholar]
- 102.Jelnes JE. Experimental taxonomy of Bulinus (Gastropoda: Planorbidae): II. Recipes for horizontal starch gel electrophoresis of ten enzymes in Bulinus and description of internal standard systems and of two new species of the Bulinus forskalii complex. J Chromatogr A. 1979;170: 405–411. doi: 10.1016/S0021-9673(00)95467-0 [DOI] [Google Scholar]
- 103.Jelnes JE. Bulinus browni Jelnes, 1979 (Gastropoda: Planorbidae), a member of the forskalii group, as intermediate host for Schistosoma bovis in western Kenya. Trans R Soc Trop Med Hyg. 1983;77: 566. doi: 10.1016/0035-9203(83)90143-8 [DOI] [PubMed] [Google Scholar]
- 104.Kinoti G. Observations on the transmission of Schistosoma haematobium and Schistosoma bovis in the Lake Region of Tanganyika. Bull World Health Organ. 1964;31: 815–823. [PMC free article] [PubMed] [Google Scholar]
- 105.Dinnik JA. Paramphistomum phillerouxi sp. nov. (Trematoda: Paramphistomatidae) and its development in Bulinus forskalii. J Helminthol. 1961;35: 69–90. doi: 10.1017/S0022149X00024792 [DOI] [PubMed] [Google Scholar]
- 106.Christensen NØ, Frandsen F, Roushdy MZ. The influence of environmental conditions and parasite-intermediate host-related factors on the transmission of Echinostoma liei. Z Für Parasitenkd. 1980;64: 47–63. doi: 10.1007/BF00927056 [DOI] [PubMed] [Google Scholar]
- 107.Nalugwa A, Kristensen TK, Nyakaana S, Jørgensen A. Mitochondrial DNA variations in sibling species of the Bulinus truncatus/tropicus complex in Lake Albert, western Uganda. Zool Stud. 2010;49: 515–522. [Google Scholar]
- 108.Cridland CC. Further experimental infection of several species of East African freshwater snails with Schistosoma mansoni and S. haematobium. J Trop Med Hyg. 1957;60: 18–23. [PubMed] [Google Scholar]
- 109.Brown DS, Wright CA. Letter: Bulinus truncatus as a potential intermediate host for Schistosoma haematobium on the Kano Plain, Kenya. Trans R Soc Trop Med Hyg. 1974;68: 341–342. doi: 10.1016/0035-9203(74)90049-2 [DOI] [PubMed] [Google Scholar]
- 110.Christensen NO, Mutani A, Frandsen F. A review of the biology and transmission ecology of African bovine species of the genus Schistosoma. Z Parasitenkd. 1983;69. doi: 10.1007/BF00926667 [DOI] [PubMed] [Google Scholar]
- 111.Southgate VR, Brown DS, Rollinson D, Ross GC, Knowles RJ. Bulinus tropicus from Central Kenya acting as a host for Schistosoma bovis. Z Für Parasitenkd. 1985;71: 61–69. doi: 10.1007/BF00932919 [DOI] [PubMed] [Google Scholar]
- 112.Evers BN, Madsen H, McKaye KM, Stauffer JR. The schistosome intermediate host, Bulinus nyassanus, is a “preferred” food for the cichlid fish, Trematocranus placodon, at Cape Maclear, Lake Malawi. Ann Trop Med Parasitol. 2006;100: 75–85. doi: 10.1179/136485906X78553 [DOI] [PubMed] [Google Scholar]
- 113.Ibrahim MM. Population dynamics of Chaetogaster limnaei (Oligochaeta: Naididae) in the field populations of freshwater snails and its implications as a potential regulator of trematode larvae community. Parasitol Res. 2007;101: 25–33. doi: 10.1007/s00436-006-0436-0 [DOI] [PubMed] [Google Scholar]
- 114.Gandon S, Buckling A, Decaestecker E, Day T. Host–parasite coevolution and patterns of adaptation across time and space. J Evol Biol. 2008;21: 1861–1866. doi: 10.1111/j.1420-9101.2008.01598.x [DOI] [PubMed] [Google Scholar]
- 115.King KC, Delph LF, Jokela J, Lively CM. Coevolutionary hotspots and coldspots for host sex and parasite local adaptation in a snail–trematode interaction. Oikos. 2011;120: 1335–1340. doi: 10.1111/j.1600-0706.2011.19241.x [DOI] [Google Scholar]
- 116.Gow JL, Noble LR, Rollinson D, Tchuem Tchuente L-A, Jones CS. High levels of selfing are revealed by a parent-offspring analysis of the medically important freshwater snail, Bulinus forskalii (gastropoda: pulmonata). J Molluscan Stud. 2005;71: 175–180. doi: 10.1093/mollus/eyi020 [DOI] [Google Scholar]