Abstract
Objectives
Juvenile-onset systemic lupus erythematosus (jSLE) affects 15–20% of lupus patients. Clinical heterogeneity between racial groups, age groups and individual patients suggests variable pathophysiology. This study aimed to identify highly penetrant damaging mutations in genes associated with SLE/SLE-like disease in a large national cohort (UK JSLE Cohort Study) and compare demographic, clinical and laboratory features in patient sub-cohorts with ‘genetic’ SLE vs remaining SLE patients.
Methods
Based on a sequencing panel designed in 2018, target enrichment and next-generation sequencing were performed in 348 patients to identify damaging gene variants. Findings were integrated with demographic, clinical and treatment related datasets.
Results
Damaging gene variants were identified in ∼3.5% of jSLE patients. When compared with the remaining cohort, ‘genetic’ SLE affected younger children and more Black African/Caribbean patients. ‘Genetic’ SLE patients exhibited less organ involvement and damage, and neuropsychiatric involvement developed over time. Less aggressive first line treatment was chosen in ‘genetic’ SLE patients, but more second and third line agents were used. ‘Genetic’ SLE associated with anti-dsDNA antibody positivity at diagnosis and reduced ANA, anti-LA and anti-Sm antibody positivity at last visit.
Conclusion
Approximately 3.5% of jSLE patients present damaging gene variants associated with younger age at onset, and distinct clinical features. As less commonly observed after treatment induction, in ‘genetic’ SLE, autoantibody positivity may be the result of tissue damage and explain reduced immune complex-mediated renal and haematological involvement. Routine sequencing could allow for patient stratification, risk assessment and target-directed treatment, thereby increasing efficacy and reducing toxicity.
Keywords: SLE, juvenile-onset, paediatric, childhood, jSLE, genetic, monogenic, stratification
Rheumatology key messages.
Using panel sequencing, rare variants predicted to be damaging were identified in 3.5% of jSLE patients.
The majority of SLE-associated gene mutations detected affect type I interferon expression.
‘Genetic’ SLE associates with reduced persistent antibody positivity, less organ involvement, fewer disease flares and reduced damage.
Introduction
SLE is a complex autoimmune/inflammatory disease that can affect any organ system [1]. Approximately 15–20% of patients develop disease during childhood and are diagnosed with juvenile-onset SLE (jSLE) [2]. When compared with adult-onset SLE (aSLE), paediatric patients exhibit higher disease activity, more organ involvement and damage, and increased need for immunosuppressive treatment [3].
Variability in clinical presentations, treatment response and outcomes suggests that ‘SLE’ comprises a range of conditions [4]. Across all ages, an estimated 1–3% of SLE patients experience disease that is caused by mutations in single genes [5], and this discovery has improved our understanding of the pathophysiology of more common ‘classical’ SLE and explained shared characteristics between SLE and conditions caused by uncontrolled expression of type 1 interferons (T1IFN; ‘type 1 interferonopathies’), including Aicardi–Goutières syndrome (AGS), Singleton–Merten syndrome, and STING-associated vasculopathy with onset in infancy [6].
While the exact pathophysiology of SLE remains unknown [1], genetic heterogeneity contributes to clinical variation and inconsistent outcomes [7, 8]. Thus, detailed molecular phenotyping may allow for patient stratification towards individualized and target-directed treatment [4].
This study linked genetic factors with clinical phenotypes and outcomes in jSLE. Target enrichment followed by next-generation sequencing was chosen to screen for variants in lupus-associated genes in a large national cohort (UK JSLE Cohort Study). This paper introduces the sequencing platform, and presents data from the sub-cohort experiencing ‘genetic’ SLE.
Methods
Study cohort
Three hundred and forty-eight patients with jSLE enrolled in the UK JSLE Cohort Study were included [9]. JSLE patients were classified using the 1997 ACR SLE classification criteria [10] and ≤18 years at diagnosis. The study received ethical approval from the National Research Ethics Service North West (REC 06/Q1502/77). Written informed patient assent/consent and parental consent was obtained to participate in the UK JSLE Cohort Study. Research was carried out in accordance with the Declaration of Helsinki.
Data collected
Demographic, clinical and serological information was analysed, including: (i) ACR-1997 scores; (ii) Systemic Lupus International Collaborating Clinics standardized damage index (SLICC-SDI) scores; (iii) The SLE Disease Activity Index 2000 (SLEDAI-2K); (iv) paediatric British Isles Lupus Assessment Grade 2004 numerical scores (pBILAG) with organ/system domains (alphabetical score A–E); (v) key laboratory findings (Supplementary methods, available at Rheumatology online), and (vi) treatments received (Supplementary methods, available at Rheumatology online).
Gene panel selection
Sequencing targets were selected based on a literature search targeting known Mendelian disease, and risk alleles previously identified through GWAS. For this, a literature search was performed (March 2018) using PubMed, OMIM and Ensembl.
Target sequencing and variant identification
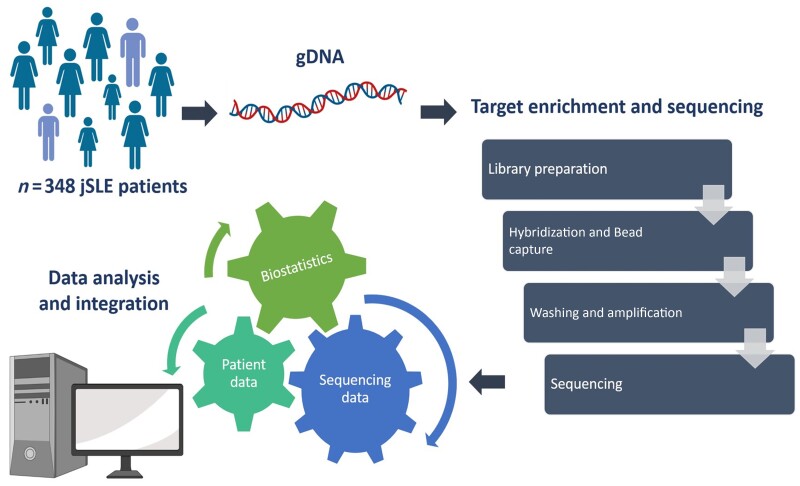
Sequence capture enrichment (NimbleGen/Roche, Switzerland) was followed by sequencing (MiSeq2500 technology, Illumina, San Diego, CA, USA). Demultiplexing, adaptor and quality trimming [Cutadapt v1.2.1 (https://doi.org/10.14806/ej.17.1.200), Sickle v1.2 (https://github.com/najoshi/sickle)] of reads was performed. Sequencing data were aligned using BWA (Burrows-Wheeler Alignment), and variants were subsequently detected, filtered and annotated with Genome Analysis Toolkit Software (GATK) (https://doi.org/10.1101/gr.107524.110) and SnpEFF (https://doi.org/10.4161/fly.19695) (Fig. 1). Variants were considered to be of ‘high impact’ when predicted to disrupt proteins encoded, cause truncation, loss-of-function (LOF) or to trigger decay (Supplementary methods, available at Rheumatology online).
Fig. 1.
Analytic workflow
gDNA: genomic DNA; jSLE: juvenile SLE. Created using BioRender.com.
Bioinformatic analysis
Publicly available databases were used in combination to predict pathogenicity of variants, including SIFT (Sorting Intolerant From Tolerant), polyphen-2, CADD (Combined Annotation Dependent Depletion), and REVEL (Rare Exome Variant Ensemble Learner) scores. Clinical associations were investigated using OMIM and ClinVar. SNPnexus was used to complete interpretation of sequence variants. Allele frequencies and counts were extracted from the 1000 Genomes Project Phase 3 and gnomAD genomes v3.1.1. Variant and reference allele counts within this cohort were subsequently compared with those of the control datasets using Fisher’s exact test with false discovery rate correction of P-values to identify variants with significantly different frequencies between cohort and control groups.
Statistical analysis
Demographics and baseline characteristics were analysed using descriptive and summary statistics. The χ2 test was used to compare frequencies between patient subgroups. Analyses were completed using GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA).
Results
Sequencing panel
Twenty-three genes associated with Mendelian forms of SLE/SLE-like disease, and 40 genomic regions/59 genes covering >130 SLE-associated risk alleles were identified and included (Supplementary Table S1, available at Rheumatology online).
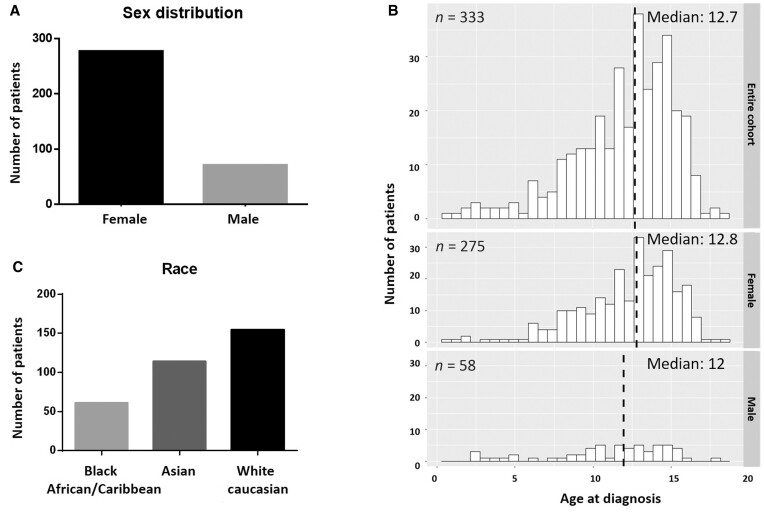
Demographic information
Three hundred and forty-eight jSLE patients were recruited to this study, of whom 82.6% (n = 275) were girls. The mean age at disease onset was 12 years (SD: 3; 0.7–17.7). Two sub-cohorts were defined: (i) ‘genetic’ SLE including 12 patients (3.5%, below), and (ii) ‘remaining’ SLE patients (336; 96.5%).
When compared with the remaining UK jSLE cohort, ‘genetic’ SLE patients were characterized by a higher proportion of patients with Black African/Caribbean background (36.3% vs 18.2%, P = 0.004) (Supplementary Table S2, available at Rheumatology online). Although the median age at disease onset was comparable between sub-cohorts, a higher proportion of patients with ‘genetic’ SLE developed disease around puberty (81.2% vs 56.5%, P = 0.0002; Supplementary Table S2, available at Rheumatology online, Fig. 2).
Fig. 2.
Demographic composition of the study cohort
Variant selection strategy
Using in silico (SnpEFF) screening for ‘high impact’ variants with low minor allele frequencies (MAF; <5%) delivered 78 candidate variants in 36 genes. This was followed by analysis of inheritance patterns using OMIM, ClinVar and SNPnexus. Patients with predicted ‘high impact’ variants and autosomal dominant (AD) inheritance were included in the study (n = 8). Patients with variants following autosomal recessive inheritance were included, if ≥1 additional ‘high impact’ mutations were present on the second allele (compound heterozygous) or ≥1 other gene associated with SLE within the same immunological pathway exhibited a ‘high impact’ mutation (n = 4). This delivered 18 variants in 14 genes of 12 (3.5%) jSLE patients (Table 1).
Table 1.
Patients with ‘genetic’ SLE and disease-causing variants
| Patient ID | Gene (variant) | Likely effects | Function affected/predicted effect (snpEFF, ClinVar, snpNexus and PubMed) | Zygosity | Inheritance pattern | Reference |
|---|---|---|---|---|---|---|
| 15009 | C1S (rs117907409) | Disease-causing | Deleterious | Heterozygous | AD | [14] |
| SAMHD1 (c.811C>A) | Disease-causing and/or -modifying | Impacts catalytic activity | Heterozygous | AD/AR | [12] | |
| 15015 | C3 (rs117793540) | Disease-causing and/or -modifying | Gain-of-function | Heterozygous | AD/AR | [33, 34] |
| TREX1 (rs1331920811) | Disease-causing and/or -modifying | Loss-of-function | Heterozygous | AD/AR | [13] | |
| 15033 | SAMHD1 (c.811C>A) | Disease-causing and/or -modifying | Impacts catalytic activity | Heterozygous | AD | [12] |
| IRF7 (c.−283-1G>T) | Disease-modifying | Loss-of-function | Heterozygous | AR | — | |
| 15049 | C3 (c.4341C>A) | Disease-causing | Loss-of-function | Heterozygous | AD/AR | — |
| RNASEH2B (c.184G>T) | Disease-modifying | Deleterious | Heterozygous | AR | — | |
| 15068 | BANK1 (rs928173624) | Disease-causing | Loss-of-function | Heterozygous | N/A | [16, 18] |
| 15070 | PTPN22 (c.369 + 1G>T) | Disease-causing | In silico predicted ‘high impact’ | Heterozygous | AD | — |
| 15087 | TNFAIP3 (rs776714084), RNASEH2A (rs549586181) | Disease-causing | Loss-of-function | Heterozygous | AD | — |
| Disease-modifying | Loss-of-function | Heterozygous | AR | [13] | ||
| 15111 | C3 (c.1303G>T) | Disease-causing | Loss-of-function | Heterozygous | AD/AR | — |
| TNFSF4 (c.368_369delAG) | Disease-modifying | Loss-of-function | Heterozygous | N/A | — | |
| RNASEH2C (rs759118175) | Disease-modifying | Loss-of-function | Heterozygous | AR | — | |
| 21058 | TREX1 (rs749323787) | Disease-causing | Loss-of-function | Heterozygous | AD/AR | [18] |
| 21067 | SAMHD1 (rs1400380009) | Disease-causing | Impacts catalytic activity | Heterozygous | AD/AR | — |
| 21123 | PEPD (rs529315200) | Disease-modifying | Loss-of-function | Heterozygous | AR | [24] |
| 33007 | DNASE1 (rs201571412) | Disease-causing | Loss-of-function | Heterozygous | AD | [11] |
AD: autosomal dominant inheritance; AR: autosomal recessive inheritance, BANK1: B cell scaffold protein with ankyrin repeats 1; C1S: complement C1S; C3: complement C3; DNASE1: deoxyribonuclease 1; IRF7: interferon regulatory factor 7; N/A: not available; PEPD: peptidase D; PTPN22: protein tyrosine phosphatase non-receptor type 22; RNASEH2A: ribonuclease H2 subunit A; RNASEH2B: ribonuclease H2 subunit B; RNASEH2C: ribonuclease H2 subunit C; SAMHD1: SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1; TNFAIP3: TNF-α induced protein 3; TNFSF4: TNF superfamily member 4; TREX1: three prime repair exonuclease 1.
Allele counts were calculated for all sub-selected variants and compared with allele frequencies in the general population (Supplementary Table S3, available at Rheumatology online). The frequency of rare, predicted damaging variants was significantly increased in the UK JSLE cohort for 8/18 variants, for three differences were not statistically significant [1] or comparable between the JSLE cohort and general populations. For 7/18 variants, no allele frequencies were available in the general population.
Notably, the most ‘common’ variant included had a reported MAF of 0.007 in the general population (gnomAD) (Supplementary Table S4, available at Rheumatology online).
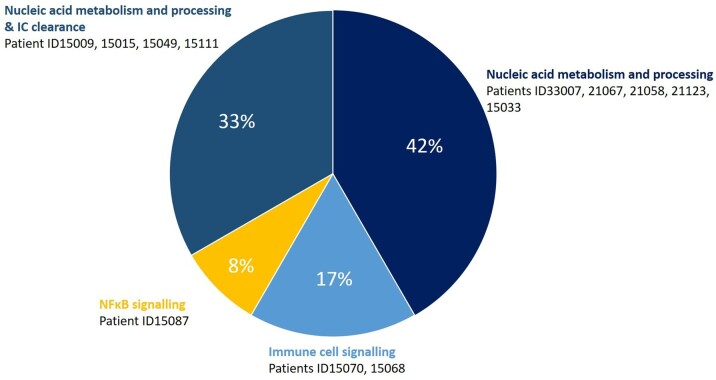
Pathways affected
Four main signalling pathways were defined: (i) nucleic acid sensing and processing (42% of ‘genetic’ SLE), (ii) a combination of (i) and immune complex (IC) clearance (33%), (iii) immune cell signalling (17%), and (iv) the nuclear factor κB (NF-κB) pathway (8%; Fig. 3, Table 2 and Supplementary Table S5, available at Rheumatology online).
Fig. 3.
Stratification of gene variants by immunological pathway
Pie chart summarizing immunological pathways identified in 12 patients with ‘genetic’ SLE carrying 18 variants in 14 genes. Variants following autosomal inheritance were considered for the association with immunological pathways, and patients may carry additional disease-modifying alleles (summarized in Table 1 and Supplementary Tables S3 and S4, available at Rheumatology online). IC: immune complex; NF-κB: nuclear factor κB.
Table 2.
Genetic variants and pathways affected
| Pathway | Gene | Disease associated | Reference |
|---|---|---|---|
| IC clearance | C1S | SLE-like disease, aHUS, primary angioedema | [30] |
| C3 | SLE-like disease | [30] | |
| PEPD | SLE-like disease | [23] | |
| Immune cell signalling | BANK1 | AITD, CLL, SSc, RA, SLE | [38] |
| PTPN22 | RA, T1D, CD, JIA, HT, SLE | [15] | |
| TNFSF4 | SSc, SLE | [37] | |
| NF-κB signalling | TNFAIP3 | SS, RA, SSc, SLE, A20 haploinsufficiency | [36] |
| Nucleic acid sensing and processing | DNASE1 | AGS, ChLE | [6] |
| RNASEH2A | AGS, ChLE | [13] | |
| RNASEH2B | AGS, ChLE | [13] | |
| RNASEH2C | AGS, ChLE | [13] | |
| SAMHD1 | AGS, ChLE | [6] | |
| TREX1 | AGS, ChLE | [6] | |
| TLR/IFN signalling | IRF7 | SLE | [28] |
aHUS: atypical haemolytic uremic syndrome; BANK1: B cell scaffold protein with ankyrin repeats 1; C1S: complement C1S; C3: complement C3; ChLE: chilblain lupus erythematosus; DNASE1: deoxyribonuclease 1; HT: Hashimoto thyroiditis; IFN: (type 1) interferon; IC: immune complex; IRF7: interferon regulatory factor 7; NF-κB: nuclear factor κB; PEPD: peptidase D; PTPN22: protein tyrosine phosphatase non-receptor Type 22; RNASEH2A: ribonuclease H2 subunit A; RNASEH2B: ribonuclease H2 subunit B; RNASEH2C: ribonuclease H2 subunit C; SAMHD1: SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1; T1D: type 1 diabetes; TLR: Toll-like receptor; TNFAIP3: TNF-α induced protein 3; TNFSF4: TNF superfamily member 4; TREX1: three prime repair exonuclease 1.
Nucleic acid sensing and processing
One patient carried a previously reported mono-allelic endonuclease LOF variant in DNASE1 (rs201571412) [11].
Two patients had a structural interaction variant in SAMHD1 (c.811C>A) affecting the HD [histidine (H) and Aspartate (D)] domain [12]. One additional variant was reported in these patients: one mono-allelic variant in IRF7 (c.−283-1G>T), and C1S (rs117907409). One patient had a monoallelic mutation in SAMHD1 (rs1400380009) affecting the aforementioned HD domain. A non-synonymous variant in TREX1 (rs749323787), encoding the 3′ repair exonuclease three prime repair exonuclease 1, introduces a splice site. Another previously unreported frameshift variant affects the DNase domain (rs1331920811).
Mono-allelic mutations affecting RNASEH2 subunits were present in patients with additional variants in other genes. In a patient with an additional mutation in TNFAIP3, RNASEH2A (rs549586181), previously reported in AGS, resulted in frameshift and LOF [13]. One individual had a ‘stop’ mutation in RNASEH2B (c.184G>T) in addition to a mutation in C3. Another patient carried a missense mutation in RNASEH2C (rs759118175), previously associated with AGS (ClinVar), and mutations in C3 (c.1303G>T) and TNFSF4 (c.368_369delAG).
Immune complex clearance
One patient with a homozygous variant in SAMDH1 also exhibited a mono-allelic C1S variant (rs117907409) recently associated with ‘genetic’ SLE [14].
Three mono-allelic variants were identified in C3; one patient carried a variant predicted to result in a gain-of-function (GOF) (rs117793540). One variant introducing a stop codon (c.4341C>A) affecting the A2M (Alpha-2-Macroglobulin) receptor binding domain and one not previously described variant results in LOF (c.1303G>T) were found in this sub-cohort.
Immune cell signalling
The cytoplasmic lymphoid-specific phosphatase (Lyp, PTPN22), inhibits T-cell activation and is involved in the termination of antigen responses [15]. A previously undescribed non-synonymous mutation in PTPN22 was predicted to mediate LOF.
One previously unreported LOF mutation in B-cell scaffold protein with ankyrin repeats 1 (BANK1; rs928173624) was identified in another patient [16].
A not previously reported LOF variant was found in the tumour necrosis factor superfamily 4 (TNFSF4) gene of one patient (c.368_369delAG) who carried additional mutations in C3 and RNASEH2C.
NF-κB signalling pathway
An autoinflammatory disease resembling Behçet’s disease was linked with mutations in TNFAIP3 [17]. A mono-allelic missense TNFAIP3 variant (rs776714084), predicted to cause LOF, was identified in one individual also carrying a RNASEH2A mutation.
Clinical and laboratory characteristics
Considering pBILAG organ domains, at diagnosis, smaller proportions of ‘genetic’ SLE patients exhibited ‘severe’ and ‘moderate’ (pBILAG A, B) or ‘mild’ (C) activity in the neuropsychiatric (P = 0.002), cardiorespiratory (P = 0.0006), gastrointestinal (P = 0,01), musculoskeletal (P = 0.0005), renal (P = 0.004) and haematological (P = 0.004) domains when compared with the remaining cohort. A smaller proportion of ‘genetic’ SLE patients exhibited ‘mild’ ophthalmic involvement (P = 0.009; Table 3). Moreover, a lower proportion of patients had a high SLEDAI score (27.3% vs 41.5%; P = 0.03).
Table 3.
Clinical picture at diagnosis and last visit
| At diagnosis |
At last visit |
Test used | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | SLE patient with rare variants predicted to be damaging (‘genetic’ SLE) | Remaining SLE patients | P-value | Total | SLE patient with rare variants predicted to be damaging (‘genetic’ SLE) | Remaining SLE patients | P-value | ||
| SLICC, median (IQR) | 0.00 (0.00–3) | 0.00 (0.00–1) | 0.00 (0.00–3) | 0.62 | 0.00 (0.00–12) | 0.00 (0.00–1) | 0.00 (0.00–12) | 0.07 | Mann–Whitney |
| SLEDAI, median (IQR) | 8.00 (0–34) | 6 (2–23) | 8.00 (0–34) | 0.22 | 2 (0–24) | 2 (0–14) | 2 (0–24) | 0.77 | Mann–Whitney |
| SLEDAI high (≥10), n (%) | 140/341 (41) | 3/11 (27.3) | 137/330 (41.5) | 0.026 | 40/338 (11.8) | 3/11 (27.3) | 37/327 (11.3) | 0.004 | χ2 |
| SLEDAI intermediate (5–9), n (%) | 87/341 (25.5) | 3/11 (27.3) | 84/330 (25.4) | 0.75 | 56/338 (16.6) | 1/11 (9.1) | 55/327 (16.8) | 0.09 | χ2 |
| SLEDAI mild (0–4), n (%) | 114/341 (33.4) | 5/11 (45.5) | 109/330 (33) | 0.06 | 242/338 (71.6) | 7/11 (63.6) | 235/327 (71.9) | 0.22 | χ2 |
| Missing information, n (%) | 7 (2) | 1 (8.3) | 6 (1.8) | — | 10 (2.9) | 1 (8.3) | 9 (2.7) | — | — |
| BILAG2004score, median (IQR) | 7.00 (0–51) | 5 (1–16) | 7.00 (0–51) | 0.52 | 1 (0–30) | 2 (0–4) | 1 (0–30) | 0.70 | Mann–Whitney |
| Active organ/system involvement, n (%) | |||||||||
| pBILAG constitutional domain | |||||||||
| A/B (‘severe’ and ‘moderate’ activity) | 94/339 (27.7) | 2/11 (18.2) | 91/328 (27.7) | 0.09 | 6/338 (1.8) | 0/11 (0) | 6/327 (1.8) | 0.15 | χ2 |
| C (‘mild’ activity) | 40/339 (11.8) | 2/11 (18.2) | 38/328 (11.6) | 0.24 | 18/338 (5.3) | 0/11 (0) | 18/327 (5.5) | 0.01 | χ2 |
| D and E (no activity) | 205/339 (60.5) | 7/11 (63.6) | 198/328 (60.4) | 0.56 | 314/338 (92.9) | 11/11 (100) | 296/327 (90.5) | 0.002 | χ2 |
| pBILAG mucocutaneous domain | |||||||||
| A/B (‘severe’ and ‘moderate’ activity) | 122/339 (36) | 4/11 (36.4) | 118/328 (36) | 1 | 38/338 (11.2) | 1/11 (9.1) | 37/327 (11.3) | 0.64 | χ2 |
| C (‘mild’ activity) | 75/339 (22.1) | 3/11 (27.3) | 72/328 (21.9) | 0.41 | 50/338 (14.8) | 2/11 (18.2) | 48/327 (14.7) | 0.57 | χ2 |
| D and E (no activity) | 142/339 (41.9) | 4/11 (36.4) | 138/328 (42.1) | 0.38 | 250/338 (74) | 8/11 (72.7) | 242/327 (74) | 0.87 | χ2 |
| pBILAG neuropsychiatric domain | |||||||||
| A/B (‘severe’ and ‘moderate’ activity) | 30/339 (8.8) | 0/11 (0) | 30/328 (9.1) | 0.002 | 14/338 (4.1) | 2/11 (18.2) | 12/327 (3.7) | 0.0016 | χ2 |
| C (‘mild’ activity) | 2/329 (0.6) | 0/11 (0) | 2/329 (0.6) | 0.32 | 3/338 (0.9) | 0/11 (0) | 3/327 (0.9) | 0.32 | χ2 |
| D and E (no activity) | 307/329 (93.3) | 11/11 (100) | 289 (87.8) | 0.0004 | 321/338 (95) | 9/11 (81.8) | 305/327 (93.3) | 0.02 | χ2 |
| pBILAG musculoskeletal domain | |||||||||
| A/B (‘severe’ and ‘moderate’ activity) | 93/339 (27.4) | 4/11 (36.4) | 89/328 (27.1) | 0.17 | 17/338 (5) | 0/11 (0) | 17/327 (5.2) | 0.02 | χ2 |
| C (‘mild’ activity) | 92/339 (27.1) | 1/11 (9.1) | 91/328 (27.7) | 0.0005 | 51/338 (15.1) | 1/11 (9.1) | 49/327 (15) | 0.19 | χ2 |
| D and E (no activity) | 154/339 (45.4) | 6/11 (54.5) | 148/328 (45.1) | 0.16 | 270/338 (79.9) | 10 (90.9) | 255/327 (78) | 0.01 | χ2 |
| pBILAG cardiorespiratory domain | |||||||||
| A/B (‘severe’ and ‘moderate’ activity) | 38/339 (11.2) | 0/11 (0) | 38/328 (11.6) | 0.0004 | 6/338 (1.8) | 0/11 (0) | 6/327 (1.8) | 0.15 | χ2 |
| C (‘mild’ activity) | 2/339 (0.6) | 0/11 (0) | 2/328 (0.6) | 0.32 | 0/338 (0.0) | 0/11 (0) | 0/327 (0) | — | χ2 |
| D and E (no activity) | 299/339 (88.2) | 11/11 (100) | 288/328 (87.8) | 0.0004 | 332/338 (98.2) | 11 (100) | 321/327 (98.1) | 0.15 | χ2 |
| pBILAG gastrointestinal domain | |||||||||
| A/B (‘severe’ and ‘moderate’ activity) | 19/339 (5.6) | 0/11 (0) | 19/328 (5.8) | 0.01 | 5/338 (1.5) | 0/11 (0) | 5/327 (1.5) | 0.15 | χ2 |
| C (‘mild’ activity) | 3/339 (0.9) | 0/11 (0) | 3/328 (0.9) | 0.32 | 1/338 (0.3) | 0/11 (0) | 1/327 (0.3) | 1 | χ2 |
| D and E (no activity) | 317/339 (93.5) | 11/11 (100) | 306/328 (93.3) | 0.007 | 332/338 (95.4) | 11/11 (100) | 321/327 (98.1) | 0.15 | χ2 |
| pBILAG ophthalmic domain | |||||||||
| A/B (‘severe’ and ‘moderate’ activity) | 2/339 (0.6) | 0/11 (0) | 2/328 (0.6) | 0.32 | 1/338 (0.3) | 0/11 (0) | 1/327 (0.3) | 1 | χ2 |
| C (‘mild’ activity) | 4/339 (1.2) | 1/11 (9.1) | 3/328 (0.9) | 0.009 | 1/338 (0.3) | 0/11 (0) | 1/327 (0.3) | 1 | χ2 |
| D and E (no activity) | 333/339 (98.2) | 10 (90.9) | 323/328 (98.5) | 0.03 | 336/338 (99.4) | 11/11 (100) | 325/327 (99.4) | 0.32 | χ2 |
| pBILAG renal domain | |||||||||
| A/B (‘severe’ and ‘moderate’ activity) | 102/339 (30) | 3/11 (27.3) | 99/328 (30.2) | 0.64 | 52/338 (15.4) | 1/11 (9.1) | 51/327 (15.6) | 0.13 | χ2 |
| C (‘mild’ activity) | 26/339 (7.7) | 0/11 (0) | 26/328 (7.9) | 0.004 | 26/338 (7.7) | 1/11 (9.1) | 25/327 (7.6) | 0.80 | χ2 |
| D and E (no activity) | 211/339 (62.2) | 8/11 (72.7) | 203/328 (61.9) | 0.17 | 260/338 (76.9) | 9/11 (81.8) | 251/327 (76.8) | 0.38 | χ2 |
| pBILAG haematological domain | |||||||||
| A/B (‘severe’ and ‘moderate’ activity) | 90/339 (26.5) | 4/11 (36.4) | 86/328 (26.2) | 0.13 | 24/338 (7.1) | 0/11 (0) | 24/327 (7.3) | 0.007 | χ2 |
| C (‘mild’ activity) | 120/339 (35.4) | 2/11 (18.2) | 118/328 (36) | 0.004 | 127/338 (37.6) | 3/11 (27.3) | 123/327 (37.6) | 0.097 | χ2 |
| D and E (no activity) | 129/339 (38) | 5/11 (45.5) | 124/328 (37.8) | 0.25 | 187/338 (55.3) | 8/11 (72.7) | 179/327 (54.7) | 0.008 | χ2 |
P-values indicating statistical significance are displayed in bold italic. IQR: interquartile range.
At last visit, a higher proportion of ‘genetic’ SLE exhibited a ‘high’ SLEDAI score (SLEDAI ≥ 10; 27.3% vs 11.3%; P = 0.004); 2/3 of these patients (15049 and 15111) sustained a ‘high’ SLEDAI score from the first to last visit. Moreover, ‘genetic’ SLE patients exhibited less ‘severe’ or ‘moderate’ activity in the pBILAG haematological (P = 0.008) and musculoskeletal (P = 0.01) domains, and a smaller proportion of patient had ‘no activity’ in the neuropsychiatric domain (P = 0.02, Table 3). Lastly a higher proportion demonstrated a severe to moderate activity for the neuropsychiatric domain (18.2% vs 3.7%; P = 0.0016) and an absence of activity for the haematological domain (72.7% vs 54.7%, P = 0.008) in the ‘genetic’ SLE group (Table 3).
To assess disease progression, patients were followed over time. The median disease duration across the entire cohort was 9.1 (range: 2.8–22.6) years. Notably, median follow-up duration was comparable between sub-cohorts [‘genetic SLE’: 10.97 (range: 7.2–14.8) years; remaining cohort: 9.12 (range: 2.8–22.7) years].
Considering disease progression from first to last visits, fewer patients with ‘genetic’ SLE experienced ‘deterioration’ in the mucocutaneous and renal domains (P = 0.013 and P = 0.007, respectively) when compared with the remaining cohort. Persistent activity in the haematological domain was less common among ‘genetic’ SLE patients (P = 0.04). Likely because of lower disease activity at diagnosis, ‘improvement’ in the neuropsychiatric (P = 0.004) and gastrointestinal (P = 0.02) domains was less common among patients with ‘genetic’ SLE when compared with the remaining cohort. More ‘genetic SLE’ patients experienced ‘deterioration’ in the neuropsychiatric and musculoskeletal domains (P = 0.0005 and P = 0.03, respectively; Supplementary Table S6, available at Rheumatology online).
At first visit, while ANA positivity did not vary between sub-cohorts, a higher proportion of ‘genetic’ SLE patients tested positive for anti-dsDNA antibodies (P = 0.02), and fewer exhibited anti-Sm and anti-Ro antibodies (P = 0.03 and P = 0.0004, respectively; Table 4). Furthermore, ‘genetic’ SLE patients less frequently experienced anaemia [P = 0.0002 (overall 18.2% vs 41.6%)] when compared with the remaining cohort (Table 4). No patient in the ‘genetic’ SLE group presented severe anaemia (P = 0.001), which associated with the absence of mild and moderate leukopenia (0% vs 23.2%; P < 0.0001). Notably, a higher proportion of ‘genetic’ SLE patients experienced severe thrombocytopenia (P = 0.003), which was, however, caused by one individual patient.
Table 4.
Laboratory findings at diagnosis and last visit
| At diagnosis |
At last visit |
Test used | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | SLE patient with rare variants predicted to be damaging (‘genetic’ SLE) | Remaining SLE patients | P-value | Total | SLE patient with rare variants predicted to be damaging (‘genetic’ SLE) | Remaining SLE patients | P-value | ||
| Immunology | |||||||||
| ANA (when tested), n (%) | 219/275 (79.6) | 8/9 (88.9) | 211/266 (79.3) | 0.054 | 143/228 (62.7) | 1/5 (20) | 141/223 (63.2) | <0.0001 | χ2 |
| Anti-dsDNA negative (0–15), n (%) | 111/314 (35.4) | 2/10 (20) | 109/304 (35.8) | 0.04 | 172/316 (54.4) | 2/10 (20) | 170/306 (55.5) | <0.0001 | χ2 |
| Anti-dsDNA positive (15–>400), n (%) | 203/314 (64.6) | 8/10 (80) | 195/304 (64.1) | 0.02 | 144/316 (43.6) | 8/10 (80) | 136/306 (40.5) | <0.0001 | χ2 |
| Anti-Sm, n (of 238 tested at first visit, of 200 tested at last visit) (%) | 56/238 (23.5) | 1/8 (12.5) | 55/230 (23.9) | 0.03 | 31/200 (15.5) | 0/4 (0.0) | 31/196 (15.8) | <0.0001 | χ2 |
| Anti-RNP, n (of 238 tested at first visit, of 200 tested at last visit) (%) | 74/238 (31.1) | 2/8 (25) | 72/230 (31.3) | 0.34 | 44/200 (22) | 2/4 (50) | 42/196 (21.4) | <0.0001 | χ2 |
| Anti-Ro, n (of 238 tested at first visit, of 200 tested at last visit) (%) | 76/238 (31.9) | 1/8 (12.5) | 75/230 (32.6) | 0.0004 | 47/200 (23.5) | 1/4 (25) | 46/196 (23.5) | 0.87 | χ2 |
| Anti-La, n (of 238 tested at first visit, of 200 tested at last visit) (%) | 33/238 (13.9) | 1/8 (12.5) | 32/230 (13.9) | 0.63 | 15/200 (7.5) | 0/4 (0.0) | 15/196 (7.6) | 0.004 | χ2 |
| C3, median (IQR) | 0.94 (0.12–3.47) | 0.82 (0.21–1.44) | 0.95 (0.12–3.47) | 0.38 | 1.06 (0.12–2.08) | 0.9 (0.39–1.36) | 1.07 (0.12–2.08) | 0.08 | Mann–Whitney |
| C4, median (IQR) | 0.12 (0.01–1.95) | 0.1 (0.01–0.28) | 0.12 (0.01–1.95) | 0.31 | 0.17 (0.01–0.66) | 0.18 (0.01–0.22) | 0.17 (0.03–0.66) | 0.47 | Mann–Whitney |
| Haemoglobin level, n (%) | |||||||||
| Severe anaemia (<8 g/dl) | 31/309 (10) | 0/11 (0) | 31/298 (10.4) | 0.001 | 15/300 (5) | 0/8 (0.0) | 15/292 (5.1) | 0.02 | χ2 |
| Moderate anaemia (8–8.9 g/dl) | 24/309 (6.9) | 1/11 (9.1) | 23/298 (7.7) | 0.80 | 4/300 (1.3) | 0/8 (0.0) | 4/292 (1.4) | 0.32 | χ2 |
| Mild anaemia (9–10.9 g/dl) | 72/309 (23.3) | 1/11 (9.1) | 70/298 (23.5) | 0.004 | 34/300 (11.3) | 2/8 (25) | 32/292 (11) | 0.01 | χ2 |
| Normal (>10.9 g/dl) | 182/309 (58.9) | 8/11 (72.7) | 167/298 (56) | 0.02 | 247/300 (82.3) | 6/8 (75) | 241/292 (82.5) | 0.16 | χ2 |
| White cell count, n (%) | |||||||||
| Severe leukopenia (<1.0 × 109/l) | 0/307 (0.0) | 0/10 (0.0) | 0/297 (0.0) | 1 | 0/302 (0.0) | 0/9 (0.0) | 0/293 (0.0) | 1 | χ2 |
| Moderate leukopenia (1–1.9 × 109/l) | 6/307 (1.9) | 0/10 (0.0) | 6/297 (2) | 0.15 | 2/302 (0.7) | 0/9 (0.0) | 2/293 (0.7) | 0.32 | χ2 |
| Mild leukopenia (2–3.9 × 109/l) | 63/307 (20.5) | 0/10 (0.0) | 63/297 (21.2) | <0.0001 | 62/302 (20.5) | 2/9 (22.2) | 60/293 (20.5) | 0.86 | χ2 |
| Normal (>3.9 × 109/l) | 238/307 (77.5) | 9/10 (90) | 229/297 (77.1) | 0.01 | 238/302 (78.8) | 6/9 (66.7) | 232/293 (79.2) | 0.056 | χ2 |
| Platelets, n (%) | |||||||||
| Severe thrombocytopenia (<25 × 109/l) | 4/305 (1.3) | 1/9 (11.1) | 3/295 (1) | 0.003 | 0/301 (0.0) | 0/8 (0.0) | 0/292 (0.0) | 1 | χ2 |
| Moderate thrombocytopenia (25–49 × 109/l) | 8/305 (2.6) | 0/9 (0.0) | 8/295 (2.7) | 0.08 | 3/301 (1) | 0/8 (0.0) | 3/292 (1) | 0.32 | χ2 |
| Mild thrombocytopenia (50–149 × 109/l) | 30/305 (9.8) | 1/9 (11.1) | 29/295 (9.8) | 0.82 | 14/301 (4.6) | 1/8 (12.5) | 13/292 (4.4) | 0.02 | χ2 |
| Normal (>149 × 109/l) | 263/305 (86.2) | 7/9 (77.8) | 256/295 (86.8) | 0.09 | 284/301 (94.3) | 7/8 (87.5) | 277/292 (94.9) | 0.08 | χ2 |
| ESR, n (%) | |||||||||
| Severe elevation (>50 mm/h) | 98/263 (37.3) | 4/11 (36.4) | 94/252 (37.3) | 0.97 | 14/277 (5.0) | 0/6 (0) | 14/271 (5.2) | 0.02 | χ2 |
| Mild–moderate elevation (≥10, ≤50 mm/h) | 112/263 (42.6) | 5/11 (45.5) | 107/252 (42.5) | 0.67 | 130/277 (46.9) | 3/6 (50) | 127/271 (46.9) | 0.67 | χ2 |
| Normal (<10 mm/h) | 53/263 (20.1) | 1/11 (9.1) | 52/252 (20.6) | 0.02 | 133/277 (48) | 3/6 (50) | 130/271 (48) | 0.78 | χ2 |
P-values indicating statistical significance are displayed in bold italic. IQR: interquartile range.
At last visit, when compared with the remaining cohort, fewer ‘genetic’ SLE patients remained positive for ANA (P < 0.0001), anti-Sm (P < 0.0001), and anti-La (P = 0.004) antibodies (Table 4). And a higher proportion of patients remained positive for dsDNA (P < 0.0001) and anti-RNP antibodies (P < 0.0001).
Next, we asked whether reduced organ involvement in ‘genetic’ SLE associated with more aggressive induction treatment. Notably, patients with ‘genetic’ SLE did not receive cytotoxic cyclophosphamide (CPM) as first-line treatment, while 7.7% of the remaining cohort did (Supplementary Table S7, available at Rheumatology online). Later in the disease course, however, fever ‘genetic’ SLE patients were off steroid-sparing agents, while more third-line DMARDs were used as compared with the remaining cohort.
Discussion
Using a panel sequencing approach limited to genes chosen during the design stage in 2018, we identified rare damaging gene mutations in 3.5% of jSLE patients. This is above the 1–3% of SLE patients with ‘genetic disease’ previously reported across all ages [4]. The majority of rare, likely damaging variants identified in the UK JSLE cohort, were significantly more common than in the general population (8/18), not previously reported and therefore no data available, 1/18 variants was more common while failing to reach significance level, and two showed comparable frequencies. While results should be interpreted with caution because of the relatively small size of the cohort, and different sequencing approaches were used across studies, this suggests increased incidence and prevalence of ‘genetic’ SLE in the paediatric age group [18]. In agreement with previous reports, ‘genetic’ SLE patients exhibited mutations nucleic acid sensing and processing (42%), combination of nucleic acid sensing and processing with IC clearance (33%), immune cell signalling (17%) and NF-κB (8%) signalling pathways. This translates to a majority (80%) of mutations affecting T1IFN production [19]. In agreement with previous reports [7], ‘genetic’ SLE was associated with an earlier disease onset, peaking in peri-pubertal patients [8], and less widespread organ involvement. As previously suggested by case reports and small case series, ‘genetic’ SLE more frequently affects young children (namely peripubertal onset) when compared with other forms of jSLE [7]. The relative over-representation of Black African/Caribbean patients in the ‘genetic’ (36.3%) when compared with the remaining SLE cohort (18.2%) further supports reports on genetic factors contributing to increased incidence of SLE in this racial group [4, 8, 20, 21].
While genetic factors in SLE are not limited to rare damaging variants, but include disease predisposing factors such as copy number variants, common nucleotide polymorphisms, and others [4, 20, 22], this study focussed on the group of jSLE patients experiencing disease caused by rare damaging variants, here referred to as ‘genetic’ SLE. The study identified a number of variants previously linked with AGS (9/348 patients, 2.6%), a type I interferonopathy that shares clinical and laboratory characteristics with SLE [6]. Why some patients develop AGS while others experience SLE remains unknown, but several ‘genetic’ SLE patients carried additional variants in SLE-associated genes (6/9, 66.7%) that may explain a shift from AGS to SLE phenotypes [6]. One patient exhibited a heterozygous ‘stop’ mutation in PEPD. Peptidase D is a prolidase involved in cleavage of dipeptides with a proline or hydroxyproline at their C-terminus. It contributes to collagen metabolism and matrix remodelling. Associations between prolidase deficiency and SLE-like disease and hypocomplementaemia have been reported [23, 24]. Thus, the identified heterozygous PEPD variant was classified as likely ‘disease-modifying’. The cytoplasmic enzyme SAMHD1 depletes deoxynucleoside triphosphates (dNTPs) necessary for viral cDNA synthesis and has nuclease activity against single-stranded DNA/RNA associated with its HD domain [25]. Damaging mutations in SAMHD1 have been linked with AGS, familial chilblain lupus and ‘genetic’ SLE [26]. Three patients in this cohort exhibit mutations that likely impact on phosphohydrolase activity, and two carried additional variants in genes previously associated with ‘genetic’ SLE (C1S, IRF7). Notably, no patient with SAMHD1 variants in this cohort had neurological, but two had severe mucocutaneous involvement. The absence of neurological disease supports reports of mild delay or normal development, and increased skin disease and glaucoma in patients with SAMHD1 mutations [27]. An additional variant in IRF7 was present in one patient with a SAMHD1 mutation. The IRF7 transcription factor regulates T1IFN-dependent immune responses against DNA and RNA viruses. While the here-identified IRF7 variant was not reported before, other variants in IRF7 were linked with increased IFN-α serum levels in SLE [28]. Two mutations affecting the endonuclease subunit of TREX1 were identified here. LOF mutations reducing DNase activity are associated with cytoplasmic accumulation of single-stranded DNA and ‘genetic’ SLE [14].
Gene variants affecting IC clearance can result in Toll-like receptor activation and T1IFN expression [29]. Defective IC clearance compromises immune tolerance and contributes to the production of autoantibodies, mainly directed against nuclear components [30]. Here, mutations affecting IC clearance were found in C1S, C3 and PEPD. The C1S AD variant rs117907409 has been associated with ‘genetic’ SLE [14]. Deficiencies of C1r and C1S impact upon the classic complement pathway. C1S contributes to C4 and C2 activation, and formation of the C3-convertase enzyme complex [30]. Two patients had not previously reported predicted LOF variants in C3 (c.4341C>A; c.1303G>T). Both experienced severe mucocutaneous involvement and high disease activity at diagnosis (SLEDAI: 23,12), mirroring reports of jSLE patients with low complement and high frequency of mucocutaneous manifestations [31]. One patient exhibited a GOF mutation in C3 (c.2203C>T, rs117793540) predicted to cause hyper-activation of C3 through the alternative C3-convertase pathway, interfering with the anaphylatoxin-mediated response to infection. Previously, this mutation was reported in atypical haemolytic uremic syndrome (aHUS), C3 glomerulopathy [32], and increased risk of age-related macular degeneration [33, 34]. A significant proportion of aHUS patients exhibit C3 GOF mutations that associate with poor prognosis. The patient reported here did not experience renal involvement or haemolytic anaemia (yet). It remains unknown how many patients with C3 GOF mutations exhibit SLE-like disease in the absence of aHUS. Furthermore, patients reported in the literature developed aHUS in adulthood (>30 years of age), while the patient reported here is still now an adolescent/young adult [35]. The patient here is a carrier of an additional variant in TREX1, which may shift the phenotype to jSLE. One patient carried a predicted ‘high impact’ TNFAIP3 variant. The TNFAIP3 gene encodes the A20 protein, a negative regulator of the NF-κB pathway. Loss-of-function mutations in TNFAIP3 were reported in haploinsufficiency A20 (HA20), phenotypically resembling Behçet’s disease. Recent reports suggest the HA20 clinical picture may be more variable, including SLE-like phenotypes [36]. Interestingly, the patient here carried an additional variant in RNASEH2A previously associated with AGS that may contribute to the SLE phenotype [13].
Monoallelic mutations in genes involved in T- and B-cell activation were observed in three patients. One patient carried a PTPN22 mutation predicted to introduce a splice site abrogating protein function. Lymphoid-specific phosphatase (Lyp) is a key regulator of T cell activation, effector responses during infection, anti-tumour responses and autoimmunity. Risk alleles in PTPN22 have been associated with SLE risk [15]. Another patient had a predicted LOF mutation in TNFSF4, accompanied by mutations in C3 and RNASEH2C. The TNFSF4 gene is implicated in T cell–antigen-presenting cell interactions, T/B cell proliferation, and adhesion of activated T lymphocytes to endothelial cells. Polymorphisms are associated with pathological T cell responses [37]. One patient carried a mutation in BANK1. BANK1 is a positive regulator of B cell signalling, and variants in the gene are associated with SLE and other autoimmune/inflammatory disorders [38]. The here-identified variant was localized at the exon 2–intron 2 junction encoding a TIR domain involved in aggregation with MyD88, K63-linked polyubiquitination and IL-8 induction [16].
Antibody patterns across jSLE cohorts are of particular interest, as revised ACR/EULAR classification criteria include ‘ANA positivity’ as an entry criterion [39]. Compared with the remaining cohort, a higher proportion of ‘genetic’ SLE patients tested positive at diagnosis for anti-dsDNA antibodies that contribute to the pathogenesis through IC formation and T1IFN expression [40]. Antibodies against nuclear components can result from tissue damage and release of (auto-)antigens [41]. Type I interferonopathies and complement deficiencies involve pathological cytokine expression, sometimes in the absence of autoantibodies, resulting in tissue damage that, in turn, may trigger autoantibody production [4]. Thus, anti-dsDNA antibody production may be an early event in more common, ‘classical’ forms of SLE, contributing to disease expression, whereas in patients with ‘genetic’ SLE they may be secondary to tissue damage and reflect disease activity as previously reported in ‘classical’ SLE [42]. This is supported by the observation that, while comparable at diagnosis, ‘genetic’ SLE patients more frequently tested negative for ANA, anti-Sm and anti-La antibodies at last visit, which associated with reduced damage and fewer flares over time when compared with the remaining UK jSLE cohort.
In line with differential autoantibody patterns between sub-cohorts, and in agreement with previous reports, at diagnosis, ‘genetic’ SLE patients less frequently exhibited renal, neuropsychiatric and gastrointestinal involvement when compared with the remaining UK jSLE cohort, and deterioration in the renal domain and persistent significant activity in the haematological domain were less common [7]. In fact, persistent autoantibody positivity in the ‘remaining’ SLE cohort may contribute to, when compared with ‘genetic’ SLE, higher incidences and severity of renal and haematological involvement as autoantibodies have been linked to the pathophysiology and damage accrual in lupus nephritis, cytopenia and haemolysis [43].
Deterioration in the neuropsychiatric domain was more common in ‘genetic’ SLE when compared with the remaining patient cohort. Between diagnosis and last visit, two ‘genetic’ SLE patients developed neuropsychiatric involvement; both carried gene variants affecting genes involved in immune cell signalling and nucleic acid sensing and processing (BANK1 and DNASE1). Though no in vitro data are available for the two mutations reported here, mutations in these genes were reported in SLE and AGS [6]. Development of neuropsychiatric symptoms may result from uncontrolled T1IFN expression as seen in AGS [44]. Generally, patients with AGS develop neurological symptoms early in life that are the result of pathological T1IFN expression, among others, caused by mutations in TREX1 (60%), SAMHD1 (15%), IFIH1 and DNASE1 [6]. In this cohort, six ‘genetic’ SLE patients exhibited six individual or combined variants affecting these genes; 33.3% (2/6) exhibited moderate or severe neuropsychiatric involvement by their last visit, in contrast to none of the remaining ‘genetic’ SLE and 3.6% of the remaining cohort. Recently, Blokland et al. reported a mechanism involving the AKT–dopamine signalling pathway, suggesting a mechanism for the involvement of BANK1 in the BOLD (blood oxygen level-dependent) response to working memory and thus indirect impact on common disorders affecting brain function [45]. Moreover, BANK1 was identified as a modulator of gut microbiota composition, which affects susceptibility to schizophrenia [46].
Considering that, across the available literature, the cumulative jSLE patient population experiences more renal and haematological involvement when compared with adult-onset SLE patients [1], jSLE patients without rare damaging gene variants (‘classical SLE’) appear to be primarily responsible for this. Notably, this is not caused by more aggressive first-line treatment in ‘genetic SLE’. Initially, DMARD use (including rituximab) was comparable between groups, and ‘genetic’ SLE patients did not receive CPM as first-line agent, while 7.7% of the remaining SLE patients did. However, later during the disease course, ‘genetic’ SLE patients less frequently received ‘no treatment’, while more frequently requiring third-line DMARDs. This suggests that persisting inflammation and accumulation of tissue damage may trigger increased use of second- or third-line treatments, and that early effective treatment may prevent disease progression. This study investigates the largest number of jSLE patients to date targeting a comprehensive panel of genetic variants, but is limited by patient numbers and the insilico methodology. While for some variants in vitro data were available and considered for data interpretation, previously not reported variants were assessed using combined computational tools. Study design and ethics precluded investigation of family members, which may have assisted in identifying additional rare mutations and to assess the de novo character of variants [47]. The size of the ‘genetic’ and ‘remaining’ SLE cohorts varied and may limit statistical comparisons. Variants included were selected in 2018, based on literature available at the time. As expected, in the meantime, additional rare variants were identified suggesting likely underestimation of the number of ‘genetic SLE’ patients [14, 18]. Furthermore, protein kinase Cδ (PKCδ), a regulator of B cell survival and proliferation that has been associated with SLE-like disease, was not included in the panel [48].
Conclusions
In the UK, a minimum of 3.5% of jSLE patients experience genetic disease, exceeding the previously reported 1–3% across all age groups. Variants identified affect nucleic acid sensing and processing, IC clearance, and immune cell and NF-κB signalling pathways. Notably, mutations previously associated with other autoimmune/inflammatory conditions were identified, suggesting that additional factors may shift phenotypes to SLE. Patients with ‘genetic’ SLE more frequently develop disease around puberty, exhibit distinct antibody patterns and dynamics, develop fewer flares and damage, exhibit less haematological and renal involvement, while developing neuropsychiatric involvement over time. Thus, achieving a molecular diagnosis aiding patient stratification will enable personalized treatment, prevention of damage, individualized disease-monitoring, patient education and (genetic) counselling.
Supplementary Material
Acknowledgements
The authors would like to acknowledge all patients and their families for participating in this study. Specifically, the authors are grateful to all the support given by the entire multi-disciplinary team within each of the paediatric centres who are part of the UK JSLE Study Group (https://www.liverpool.ac.uk/translational-medicine/research/ukjsle/jsle/). Special recognition also goes to Dr Duncan Appleby for database and information technology support. We thank Myriam Lorenz and Marita Führer for excellent technical support. C.M.H., M.W.B. and A.C. conceived the study; A.C., S.H., E.M.D.S., N.E., L.O., J.G.K., K.S., C.R. and C.H. interpreted data; S.H., L.O., J.G.K. and A.C. were involved in sequencing and variant analysis, filtering and analyses on genomic datasets. E.A., K.A., K.B., C.C., J.G.-M., K.H., D.D.P.H., A.L., V.L., F.M., G.M., C.P., A.V.R., S.R., P.R., A.S., M.W.B. and C.M.H. provided clinical samples and critically reviewed patient data; A.C. and C.M.H. wrote the initial draft, all authors reviewed the manuscript and agreed to submission. The UK JSLE Cohort Study and Repository is supported by LUPUS UK and Versus Arthritis UK, and the University of Liverpool, and the EATC4Children by Versus Arthritis UK, University of Liverpool and Alder Hey Children’s Foundation Trust. The sequencing project was supported by LUPUS UK and the University of Liverpool. This work was partially carried out at the NIHR Alder Hey Clinical Research Facility. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health and Social Care.
Funding: This research was funded by LUPUS UK (to C.M.H.). This work was further supported by the UK’s Experimental Arthritis Treatment Centre for Children (supported by Versus Arthritis, Alder Hey Children’s NHS Foundation Trust, the Alder Hey Charity, and the University of Liverpool) and partially carried out at the NIHR Alder Hey Clinical Research Facility.
Disclosure statement: The authors report no financial conflicts of interest exist in relation to the work presented.
Contributor Information
Amandine Charras, Department of Women's & Children's Health, Institute of Life Course and Medical Sciences.
Sam Haldenby, Centre for Genomic Research, Institute of Infection, Veterinary & Ecological Sciences, University of Liverpool.
Eve M D Smith, Department of Women's & Children's Health, Institute of Life Course and Medical Sciences; Department of Paediatric Rheumatology, Alder Hey Children's NHS Foundation Trust Hospital, Liverpool, UK.
Naomi Egbivwie, Department of Women's & Children's Health, Institute of Life Course and Medical Sciences; Department of Paediatric Rheumatology, Alder Hey Children's NHS Foundation Trust Hospital, Liverpool, UK.
Lisa Olohan, Centre for Genomic Research, Institute of Infection, Veterinary & Ecological Sciences, University of Liverpool.
John G Kenny, Centre for Genomic Research, Institute of Infection, Veterinary & Ecological Sciences, University of Liverpool; Teagasc Food Research Centre, Moorepark, Cork, Ireland.
Klaus Schwarz, Institut for Transfusion Medicine, University Ulm, Ulm; Institute for Clinical Transfusion Medicine and Immunogenetics Ulm, German Red Cross Blood Service Baden-Württemberg—Hessen, Ulm, Germany.
Carla Roberts, Department of Women's & Children's Health, Institute of Life Course and Medical Sciences.
Eslam Al-Abadi, Department of Rheumatology, Birmingham Children’s Hospital, Birmingham.
Kate Armon, Department of Paediatric Rheumatology, Cambridge University Hospitals, Cambridge.
Kathryn Bailey, Department of Paediatric Rheumatology, Oxford University Hospitals NHS Foundation Trust, Oxford.
Coziana Ciurtin, Centre for Adolescent Rheumatology, University College London, London.
Janet Gardner-Medwin, Department of Child Health, University of Glasgow, Glasgow.
Kirsty Haslam, Department of Paediatrics, Bradford Royal Infirmary, Bradford.
Daniel P Hawley, Department of Paediatric Rheumatology, Sheffield Children’s Hospital, Sheffield.
Alice Leahy, Department of Paediatric Rheumatology, Southampton General Hospital, Southampton.
Valentina Leone, Department of Paediatric Rheumatology, Leeds Children Hospital, Leeds.
Flora McErlane, Paediatric Rheumatology, Great North Children’s Hospital, Royal Victoria Infirmary, Institute of Cellular Medicine, Newcastle University, Newcastle upon Tyne.
Gita Modgil, Department of Paediatrics, Musgrove Park Hospital, Taunton.
Clarissa Pilkington, Department of Paediatric Rheumatology, Great Ormond Street Hospital, London.
Athimalaipet V Ramanan, University Hospitals Bristol NHS Foundation Trust & Bristol Medical School, University of Bristol, Bristol.
Satyapal Rangaraj, Department of Paediatric Rheumatology, Nottingham University Hospitals, Nottingham.
Phil Riley, Department of Paediatric Rheumatology, Royal Manchester Children’s Hospital, Manchester.
Arani Sridhar, Department of Paediatrics, Leicester Royal Infirmary, Leicester, UK.
Michael W Beresford, Department of Women's & Children's Health, Institute of Life Course and Medical Sciences; Centre for Genomic Research, Institute of Infection, Veterinary & Ecological Sciences, University of Liverpool.
Christian M Hedrich, Department of Women's & Children's Health, Institute of Life Course and Medical Sciences; Centre for Genomic Research, Institute of Infection, Veterinary & Ecological Sciences, University of Liverpool.
Data availability statement
Access to raw data will be considered by the UK JSLE Cohort Study board upon reasonable request. At the end of the wider study, sequencing datasets will be deposited in publicly available databases.
Supplementary data
Supplementary data are available at Rheumatology online.
References
- 1. Tarvin SE, O'Neil KM.. Systemic lupus erythematosus, Sjögren syndrome, and mixed connective tissue disease in children and adolescents. Pediatr Clin North Am 2018;65:711–37. [DOI] [PubMed] [Google Scholar]
- 2. Brunner HI, Huggins J, Klein-Gitelman MS.. Pediatric SLE—towards a comprehensive management plan. Nat Rev Rheumatol 2011;7:225–33. [DOI] [PubMed] [Google Scholar]
- 3. Tucker LB, Uribe AG, Fernández M. et al. Adolescent onset of lupus results in more aggressive disease and worse outcomes: results of a nested matched case-control study within LUMINA, a multiethnic US cohort (LUMINA LVII). Lupus 2008;17:314–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Charras A, Smith E, Hedrich CM.. Systemic lupus erythematosus in children and young people. Curr Rheumatol Rep 2021;23:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Costa-Reis P, Sullivan KE.. Monogenic lupus: it’s all new! Curr Opin Immunol 2017;49:87–95. [DOI] [PubMed] [Google Scholar]
- 6. Kim H, Montealegre Sanchez GA, Goldbach-Mansky R.. Insights from Mendelian interferonopathies: comparison of CANDLE, SAVI with AGS, monogenic lupus. J Mol Med (Berl) 2016;94:1111–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Massias JS, Smith EMD, Al-Abadi E. et al. Clinical and laboratory characteristics in juvenile-onset systemic lupus erythematosus across age groups. Lupus 2020;29:474–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Massias JS, Smith EM, Al-Abadi E. et al. Clinical and laboratory phenotypes in juvenile-onset Systemic Lupus Erythematosus across ethnicities in the UK. Lupus 2021;30:597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Watson L, Leone V, Pilkington C. et al. ; UK Juvenile-Onset Systemic Lupus Erythematosus Study Group. Disease activity, severity, and damage in the UK juvenile-onset systemic lupus erythematosus cohort. Arthritis Rheum 2012;64:2356–65. [DOI] [PubMed] [Google Scholar]
- 10. Hartman EAR, van Royen-Kerkhof A, Jacobs JWG, Welsing PMJ, Fritsch-Stork RDE.. Fritsch-Stork RDE. Performance of the 2012 Systemic Lupus International Collaborating Clinics classification criteria versus the 1997 American College of Rheumatology classification criteria in adult and juvenile systemic lupus erythematosus. A systematic review and meta-analysis. Autoimmun Rev 2018;17:316–22. [DOI] [PubMed] [Google Scholar]
- 11. Kimura-Kataoka K, Ueki M, Takeshita H. et al. Identification of the functional alleles of the nonsynonymous single-nucleotide polymorphisms potentially implicated in systemic lupus erythematosus in the human deoxyribonuclease I gene. DNA Cell Biol 2014;33:492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goldstone DC, Ennis-Adeniran V, Hedden JJ. et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 2011;480:379–82. [DOI] [PubMed] [Google Scholar]
- 13. Rice GI, Forte GMA, Szynkiewicz M. et al. Assessment of interferon-related biomarkers in Aicardi-Goutières syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. Lancet Neurol 2013;12:1159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sandling JK, Pucholt P, Hultin Rosenberg L. et al. ; ImmunoArray Development Consortium and DISSECT consortium. Molecular pathways in patients with systemic lupus erythematosus revealed by gene-centred DNA sequencing. Ann Rheum Dis 2021;80:109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu L-Y, Cheng Z, Zhang B. et al. Associations between PTPN22 and TLR9 polymorphisms and systemic lupus erythematosus: a comprehensive meta-analysis. Arch Dermatol Res 2017;309:461–77. [DOI] [PubMed] [Google Scholar]
- 16. Georg I, Díaz-Barreiro A, Morell M, Pey AL, Alarcón-Riquelme ME.. BANK1 interacts with TRAF6 and MyD88 in innate immune signaling in B cells. Cell Mol Immunol 2020;17:954–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aeschlimann FA, Batu ED, Canna SW. et al. A20 haploinsufficiency (HA20): clinical phenotypes and disease course of patients with a newly recognised NF-kB-mediated autoinflammatory disease. Ann Rheum Dis 2018;77:728–35. [DOI] [PubMed] [Google Scholar]
- 18. Belot A, Rice GI, Omarjee SO. et al. Contribution of rare and predicted pathogenic gene variants to childhood-onset lupus: a large, genetic panel analysis of British and French cohorts. Lancet Rheumatol 2020;2:e99–109. [DOI] [PubMed] [Google Scholar]
- 19. Uggenti C, Lepelley A, Crow YJ.. Self-awareness: nucleic acid-driven inflammation and the type I interferonopathies. Annu Rev Immunol 2019;37:247–67. [DOI] [PubMed] [Google Scholar]
- 20. Smith EMD, Lythgoe H, Midgley A, Beresford MW, Hedrich CM.. Juvenile-onset systemic lupus erythematosus: update on clinical presentation, pathophysiology and treatment options. Clin Immunol (Orlando) 2019;209:108274. [DOI] [PubMed] [Google Scholar]
- 21. Webb R, Kelly JA, Somers EC. et al. Early disease onset is predicted by a higher genetic risk for lupus and is associated with a more severe phenotype in lupus patients. Ann Rheum Dis 2011;70:151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsokos GC. Systemic Lupus Erythematosus. N Engl J Med 2011;365:2110–21. [DOI] [PubMed] [Google Scholar]
- 23. Butbul Aviel Y, Mandel H, Avitan Hersh E. et al. Prolidase deficiency associated with systemic lupus erythematosus (SLE): single site experience and literature review. Pediatr Rheumatol Online J 2012;10:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Besio R, Maruelli S, Gioia R. et al. Lack of prolidase causes a bone phenotype both in human and in mouse. Bone 2015;72:53–64. [DOI] [PubMed] [Google Scholar]
- 25. Coquel F, Neumayer C, Lin Y-L, Pasero P.. SAMHD1 and the innate immune response to cytosolic DNA during DNA replication. Curr Opin Immunol 2019;56:24–30. [DOI] [PubMed] [Google Scholar]
- 26. Alperin JM, Ortiz-Fernández L, Sawalha AH.. Monogenic lupus: a developing paradigm of disease. Front Immunol 2018;9:2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Livingston JH, Crow YJ.. Neurologic phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR1, and IFIH1: Aicardi–Goutières syndrome and beyond. Neuropediatrics 2016;47:355–60. [DOI] [PubMed] [Google Scholar]
- 28. Salloum R, Franek BS, Kariuki SN. et al. Genetic variation at the IRF7/PHRF1 locus is associated with autoantibody profile and serum interferon-α activity in lupus patients. Arthritis Rheum 2010;62:553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ivashkiv LB, Donlin LT.. Regulation of type I interferon responses. Nat Rev Immunol 2014;14:36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lintner KE, Wu YL, Yang Y. et al. Early components of the complement classical activation pathway in human systemic autoimmune diseases. Front Immunol 2016;7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Afzali P, Isaeian A, Sadeghi P. et al. Complement deficiency in pediatric-onset systemic lupus erythematosus. J Lab Physicians 2018;10:232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tortajada A, Marín AV, Arjona E. et al. The C3-R735W variant results in a hyperactive C3 that associates with atypical haemolytic uremic syndrome and C3-glomerulopathy. Mol Immunol 2018;102:221. [Google Scholar]
- 33. Shoshany N, Weiner C, Safir M. et al. Rare genetic variants in Jewish patients suffering from age-related macular degeneration. Genes 2019;10:825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Duvvari MR, Paun CC, Buitendijk GHS. et al. Analysis of rare variants in the C3 gene in patients with age-related macular degeneration. PLoS One 2014;9:e94165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ellithi M, Shahid M, Abdullah HM, Bleeker J.. Complement C3 mutation causing atypical hemolytic uremic syndrome successfully treated with eculizumab. Hematol Transfus Cell Ther 2021;43:364–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu M-P, Xu X-S, Zhou Q, Deuitch N, Lu M-P.. Haploinsufficiency of A20 (HA20): updates on the genetics, phenotype, pathogenesis and treatment. World J Pediatr WJP 2020;16:575–84. [DOI] [PubMed] [Google Scholar]
- 37. Croft M. The role of TNF superfamily members in T-cell function and diseases. Nat Rev Immunol 2009;9:271–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dieudé P, Wipff J, Guedj M. et al. BANK1 is a genetic risk factor for diffuse cutaneous systemic sclerosis and has additive effects with IRF5 and STAT4. Arthritis Rheum 2009;60:3447–54. [DOI] [PubMed] [Google Scholar]
- 39. Aringer M, Costenbader K, Daikh D. et al. 2019 European League Against Rheumatism/American College of Rheumatology classification criteria for systemic lupus erythematosus. Ann Rheum Dis 2019;78:1151–9. [DOI] [PubMed] [Google Scholar]
- 40. Bai Y, Tong Y, Liu Y, Hu H.. Self-dsDNA in the pathogenesis of systemic lupus erythematosus. Clin Exp Immunol 2018;191:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang X, Xia Y.. Anti-double stranded DNA antibodies: origin, pathogenicity, and targeted therapies. Front Immunol 2019;10:1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Isenberg D, I Giles, JE Hansen, Rahman A.. 27 – Antinuclear antibodies, antibodies to DNA, histones, and nucleosomes. In: Wallace DJ, Hahn BH, eds. Dubois’ Lupus Erythematosus and Related Syndromes. 9th edn. London: Elsevier, 2019, 355–65. [Google Scholar]
- 43. Petri M, Orbai A-M, Alarcón GS. et al. Derivation and validation of systemic lupus international collaborating clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum 2012;64:2677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saeed M. Lupus pathobiology based on genomics. Immunogenetics 2017;69:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blokland GAM, Wallace AK, Hansell NK. et al. Genome-wide association study of working memory brain activation. Int J Psychophysiol 2017;115:98–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martins-Silva T, Salatino-Oliveira A, Genro JP. et al. Host genetics influences the relationship between the gut microbiome and psychiatric disorders. Prog Neuropsychopharmacol Biol Psychiatry 2021;106:110153. [DOI] [PubMed] [Google Scholar]
- 47. Demirkaya E, Sahin S, Romano M, Zhou Q, Aksentijevich I.. New horizons in the genetic etiology of systemic lupus erythematosus and lupus-like disease: monogenic lupus and beyond. J Clin Med 2020;9:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Belot A, Kasher PR, Trotter EW. et al. Protein kinase Cδ deficiency causes Mendelian systemic lupus erythematosus with B cell-defective apoptosis and hyperproliferation. Arthritis Rheum 2013;65:2161–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Access to raw data will be considered by the UK JSLE Cohort Study board upon reasonable request. At the end of the wider study, sequencing datasets will be deposited in publicly available databases.