Abstract
Background
Due to the national dynamic zero-COVID strategy in China, there were no persistent local transmissions of SARS-CoV-2 in Beijing before December, 2022. However, imported cases have been frequently detected over the past 3 years. With soaring growth in the number of COVID-19 cases in China recently, there are concerns that there might be an emergence of novel SARS-CoV-2 variants. Routine surveillance of viral genomes has been carried out in Beijing over the last 3 years. Spatiotemporal analyses of recent viral genome sequences compared with that of global pooled and local data are crucial for the global response to the ongoing COVID-19 pandemic.
Methods
We routinely collected respiratory samples covering both imported and local cases in Beijing for the last 3 years (of which the present study pertains to samples collected between January and December, 2022), and then randomly selected samples for analysis. Next-generation sequencing was used to generate the SARS-CoV-2 genomes. Phylogenetic and population dynamic analyses were performed using high-quality complete sequences in this study.
Findings
We obtained a total of 2994 complete SARS-CoV-2 genome sequences in this study, among which 2881 were high quality and were used for further analysis. From Nov 14 to Dec 20, we sequenced 413 new samples, including 350 local cases and 63 imported cases. All of these genomes belong to the existing 123 Pango lineages, showing there are no persistently dominant variants or novel lineages. Nevertheless, BA.5.2 and BF.7 are currently dominant in Beijing, accounting for 90% of local cases since Nov 14 (315 of 350 local cases sequenced in this study). The effective population size for both BA.5.2 and BF.7 in Beijing increased after Nov 14, 2022.
Interpretation
The co-circulation of BF.7 and BA.5.2 has been present in the current outbreak since Nov 14, 2022 in Beijing, and there is no evidence that novel variants emerged. Although our data were only from Beijing, the results could be considered a snapshot of China, due to the frequent population exchange and the presence of circulating strains with high transmissibility.
Funding
National Key Research and Development Program of China and Strategic Priority Research Program of the Chinese Academy of Sciences.
Translation
For the Chinese translation of the abstract see Supplementary Materials section.
Introduction
The COVID-19 pandemic has been ongoing for nearly 3 years, and remains a global concern.1, 2, 3 Given the wide spread and continuous evolution of SARS-CoV-2, numerous variants have emerged globally. The emergence of variants of concern (VOCs), such as alpha (B.1.1.7), beta (B.1.351), gamma (P.1), delta (B.1.617.2), and omicron (B.1.1.529), has caused multiple waves.4 Omicron was first identified in South Africa and designated as a VOC by WHO; it had an unprecedented number of mutations and increased transmissibility.5 It quickly took over other co-circulating variants across the globe and increased the diversification of viral lineages.6
China took a different strategy in outbreak response to other countries before December, 2022, from the lockdown in Wuhan in 2020 to the dynamic zero-COVID policy, and employed precise prevention and control tactics to stop the transmission of SARS-CoV-2.7, 8, 9, 10 Consequently, China quickly controlled regional outbreak clusters of COVID-19 from early 2020 to 2022.9 With the attenuated pathogenicity of omicron subvariants, the popularisation of vaccination, and the accumulation of prevention and control experience,10, 11 China adjusted and optimised the prevention and control strategies for COVID-19 in mid-November, 2022.
Following the adjustment of prevention and control policies in China before December, 2022, the rapid development of the epidemic in China has attracted worldwide attention, and raised concerns about whether this outbreak is being driven by the emergence of novel SARS-CoV-2 variants. This highlights the importance of long-term and continuous monitoring of SARS-CoV-2 at the genomic level.12, 13 Consequently, a comprehensive spatiotemporal study of circulating SARS-CoV-2 variants is crucial for the global response to the ongoing COVID-19 pandemic. Beijing, with a permanent population of 21 million, became one of the Chinese cities with the highest case numbers. The prevalence of SARS-CoV-2 variants in Beijing could therefore be considered a snapshot of China. Here, we describe the epidemiology and phylogeny of high-quality complete genome sequences of SARS-CoV-2 collected in 2022. In particular, we report the genomic characteristics of SARS-CoV-2 after the adjusted policy, providing important information on the current epidemic situation in Beijing.
Research in context.
Evidence before this study
COVID-19 has been a global pandemic for nearly 3 years. The emergence of variants of concern (VOCs), such as alpha, beta, gamma, delta, and omicron, has caused multiple waves of cases. The omicron VOC quickly took over other co-circulating variants across the globe. With soaring growth of COVID-19 cases in China recently after the adjustment of prevention and control policies, whether cases were caused by novel, emerging SARS-CoV-2 variants is an important area of study. We searched PubMed for studies in English and published as of Dec 28, 2022, using the search term “Chinese SARS-CoV-2 epidemic in late 2022”. We found 22 articles in total. However, none described the SARS-CoV-2 epidemic situation in China at the end of 2022. On Jan 3, 2023, the Chinese Center for Disease Control and Prevention reported the epidemic situation in late 2022 to WHO, indicating that the omicron subvariants BA.5.2 and BF.7 are responsible for the epidemic since late 2022, accounting for 97·5% of all local infections as per genomic sequencing.
Added value of this study
This study describes the epidemiological characteristics and phylogenetic analyses of SARS-CoV-2 in Beijing during 2022. A total of 2881 SARS-CoV-2 genome sequences were obtained from routine surveillance and analysed. The composition of SARS-CoV-2 variants changed over time during 2022. BA.5.2 and BF.7 became dominant and increased in genetic diversity in Beijing since the adjustment of prevention and control policies against COVID-19, accounting for 90% of local cases. However, there is no evidence that novel variants emerged in Beijing during 2022. Since Beijing is one of the hardest-hit cities after the adjustment of policies, the temporal compositional dynamic of SARS-CoV-2 variants in Beijing can be considered a snapshot of the situation in China.
Implications of all the available evidence
This study suggests that the current surge in Beijing was caused by co-circulation of two pre-existing omicron subvariants, BA.5.2 and BF.7, rather than novel SARS-CoV-2 variants. However, the persistent and large-scale circulation of SARS-CoV-2 variants in China should be monitored continuously to detect novel VOCs at the earliest opportunity.
Methods
Sample and data sources
The number of laboratory-confirmed SARS-CoV-2 cases was ascertained from the daily report of Beijing Municipal Health Commission from Jan 1 to Dec 28, 2022. Laboratory-confirmed tests were completed by the district Center for Disease Prevention and Control (CDC), clinical laboratories in hospitals, and third-party testing laboratories outside the hospital, following national guidelines.14 Cases testing positive for both target genes (open reading frame 1ab and nucleocapsid protein) were classified as laboratory-confirmed cases; otherwise, they were treated as negative results or inconclusive, for which further tests were required for validation.14 The demographic data of individuals with laboratory-confirmed SARS-CoV-2 infection were obtained using a standardised questionnaire by interviewing infected individuals or their family members or relatives, attending doctors, and other health-care providers, supplemented by case medical records. All data were scrutinised by two professionals. For local infections, 1686 cases were selected and sequenced out of a total of 4845 local infections before Nov 14. The remaining 3159 confirmed cases were not sequenced, mainly due to low viral loads in samples, insufficient sample volume, or missed sampling during the outbreak investigation. A total of 350 local infections were then randomly selected and successfully sequenced from Nov 14 to Dec 20. Additionally, 824 imported cases were randomly selected for sequencing. Finally, a total of 3745 samples were enrolled from all laboratory-confirmed SARS-CoV-2 cases for genomic sequencing between Jan 1 and Dec 20 in Beijing. The upper and lower respiratory tract specimens, including nasopharyngeal swabs, oropharyngeal swabs, sputum, etc, were obtained from infected individuals.
All samples used in this study were based on residual oropharyngeal and nasal swab collections from district CDCs in Beijing, sentinel hospitals, and airport quarantine in Beijing between Jan 1 and Dec 20, 2022. All samples were de-identified before receipt by the researchers. Ethical approval for this study was provided by the ethical review board of Beijing CDC.
Next-generation sequencing
Viral RNA was extracted from 200 μL of sample and eluted in 90 μL elution buffer by KingFisher Flex Purification System (Thermo Fisher, Waltham, MA, USA). cDNA synthesis was performed from the extracted RNA using random hexamers, and the LunaScript RT SuperMix Kit (New England Biolabs, Hertfordshire, UK). Subsequently, the revised ARTIC Network SARS-CoV-2 version 4.1 primer scheme and Q5 High-Fidelity DNA polymerase (New England Biolabs, UK) were used for SARS-CoV-2 whole-genome multiplex PCR amplification.15 Commercial SARS-CoV-2 whole-genome multiplex PCR kits (MicroFuture, Beijing, China; JuJi, Hangzhou, China; and Laboratory Biology Technology, Beijing, China), based on a similar amplicon-enrichment strategy to that used in the ARTIC Network pipeline, were also used to amplify the SARS-CoV-2 whole genome. AmpureXP beads (Beckman Coulter, Brea, CA, USA) were used for PCR product purification, and fluorimetry-based quantification was carried out using the Qubit dsDNA High Sensitivity assay on the Qubit 3.0 Fluorometer (Life Technologies, Austin, TX, USA). The PCR products were used to prepare a library for next-generation sequencing using a Nextera XT DNA Sample Preparation and Index kit and DNA Prep Sample Preparation and Index kit (Illumina, San Diego, CA, USA) following manufacturer instructions, and the sequencing was carried out on an Illumina MiSeq or MiniSeq platform using the 2 × 150 cycles paired-end sequencing protocol. SARS-CoV-2 genome assembly was performed using CLC Genomics Workbench, version 21.0. Briefly, reads with length less than 60 nucleotides were trimmed, then trimmed reads were mapped to reference sequences (accession number MN908947.3). The minimum percentage of the total alignment length and similarity was set as 80%. Local realignment and primers and dimers trimming were then performed to improve mapping results. Low frequency variant detection was performed to call variants. Consensus sequences were created from variants, and the regions with read depth less than 20× were not used for consensus generation.
Phylogenetic and phylodynamic analysis
The evaluation of the quality of genomes, genomic alignment, clade, and Pango lineage assignment, and the genetic variation annotations of SARS-CoV-2 genomes were performed by Nextclade version 2.9.1.16 After quality control, we found 113 out of 2994 SARS-CoV-2 genomes were of low quality. Consequently, 2881 SARS-CoV-2 genomes were used for further analysis. A phylogenetic tree containing the remaining SARS-CoV-2 genomes was also reconstructed by Nextclade version 2.9.1.16
Before phylodynamic analysis of BA.5.2 and BF.7 collected in Beijing after Nov 14, 2022, RDP417 was first used to detect recombination events. No evidence for recombination was found in either dataset. We used modeltest-ng version 0.1.7 to find the best substitution model for each dataset according to the Bayesian information criterion.18, 19 The best substitution model for both datasets was TrN (variable base frequencies, equal transversion rates, variable transition rates) with a discrete (four categories) gamma-distributed rate heterogeneity among sites. In addition, we also tested whether there was enough temporal molecular signal in both datasets before phylodynamic analysis. IQ-TREE version 2.0.320 was used to reconstruct phylogenetic trees for both datasets with 1000 ultrafast bootstrap replicates.21 The linear regression of root-to-tip divergence (from the phylogeny above) against sampling dates for genomic data was performed using TempEst version 1.5.322 (appendix 2 p 1). The results indicated that there was sufficient temporal signal in both datasets after discarding several outliers to infer the population dynamics over time. We then used the Bayesian Markov Chain Monte Carlo (MCMC) approach implemented in BEAST version 1.10.423 to infer the effective population size for both BA.5.2 (n=44) and BF.7 (n=262). Since the time interval is about 1 month for both datasets and genome sequences from each dataset belong to the same Pango lineage, the evolutionary rates of branches within each dataset were thus expected to be constant. Consequently, we used a strict molecular clock model as prior to perform the phylodynamic analysis. We then tested which coalescent tree priors were more suitable for these two datasets by using path sampling and stepping-stone sampling to estimate marginal likelihood.24 Three coalescent tree priors—a constant-size population, an exponential growth population, and a Bayesian skyline tree prior (ten groups, piecewise-constant model)—were tested in this study. The Bayesian skyline tree prior was the best fit to both datasets (appendix 2 p 5). The Bayesian skyline plot (piecewise-constant model with ten groups), a non-parametric method which is independent on particular demographic history, was then used as the tree prior to estimate the median effective population size through time with a 95% highest posterior density.25 At least five replicate runs for each 10 million and 50 million MCMC steps, sampling parameters, and trees every 1000 and 5000 steps were performed for BA.5.2 and BF.7, respectively. Tracer version 1.7.126 was then used to check the convergence of MCMC chains (effective sample size >200) and compute marginal posterior distributions of parameters, after discarding the first 10% of the MCMC chain as burn-in. TreeAnnotator was then used to summarise the maximum clade credibility tree based on the posterior distribution of trees.
Statistical analysis
Data were analysed using SPSS 20.0. We determined that the data satisfied the conditions for using Pearson's χ2 test. Therefore, differences between groups were evaluated using Pearson's χ2 and odds ratio (OR) estimates with 95% CIs. Statistical significance was defined as p<0·05.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
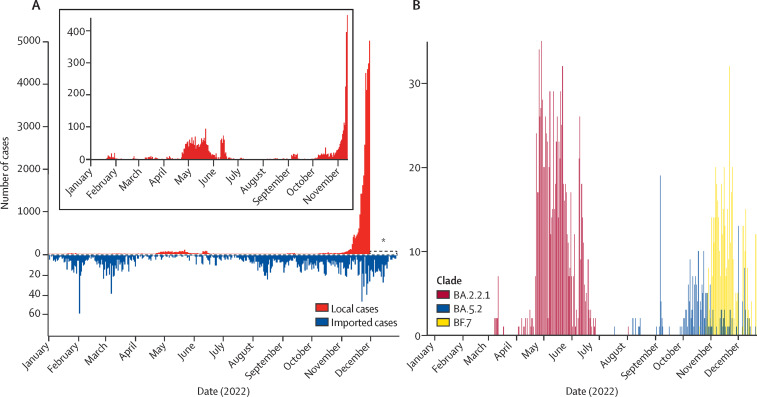
A total of 39 007 local cases were observed in Beijing, from Jan 1 to Nov 30, 2022 (figure 1A ). The average age of all cases was 40·16 years. The male to female sex ratio was 1·29. Individuals younger than 5 years and older than 60 years accounted for 663 (1·70%) and 4380 (11·23%) of the 39 007 local cases, respectively. Due to the adjusted strategies, the number of infections increased markedly from Nov 14; thus, the accurate number of local infections for Nov 30 to Dec 28 was unknown, and could only be obtained by statistical inference. Only one local outbreak was observed in Beijing before Nov 14, involving a total of 2230 local infections in all 17 districts of Beijing, starting from April 22 and lasting for 73 days. However, imported cases were frequently identified by airport quarantine surveillance of COVID-19. A total of 2600 overseas imported cases were observed in Beijing from Jan 1 to Dec 28. The number of imported cases was relatively low from April to August, showing limited spatiotemporal consistency with the local infections.
Figure 1.
Temporal dynamics of local and imported SARS-CoV-2 cases in Beijing in 2022
(A) Number of reported SARS-CoV-2 cases in Beijing, identified by local and imported cases. An asterisk is used to indicate the uncertainty on the number of infected cases since Nov 30, the start of the current COVID-19 surge in Beijing. (B) Number of SARS-CoV-2 cases caused by major variants in Beijing.
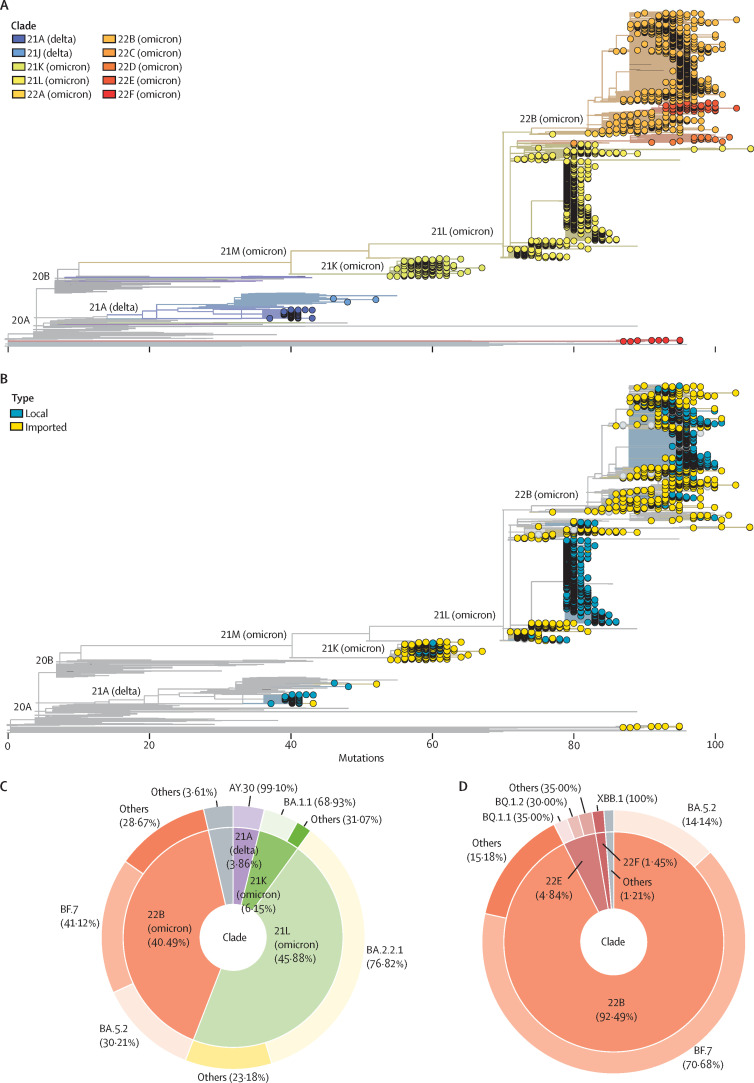
Among all local and imported cases detected in Beijing in 2022, a total of 3745 laboratory-confirmed COVID-19 cases were randomly selected for genomic sequencing. In total, we obtained 2994 complete SARS-CoV-2 genomes in this study, of which 2881 were high quality and used for further analysis. The sampling time for these sequences spanned the whole year, and sampling locations covered all 17 districts of Beijing (figure 1B, appendix 2 pp 2–3, 6). All of the sequences belonged to VOCs: delta (n=114) and omicron (n=2767). Detailed analysis indicates that they came from ten clades (figure 2A , appendix 2 p 4) and 123 Pango lineages. Imported cases had a wider clade range compared with the local cases (figure 2B). No novel Pango lineages were found in our dataset. All imported cases came from 63 countries and regions (appendix 2 pp 7–8). In addition, we also found a small number of previously reported recombinant SARS-CoV-2 subvariants XBB (n=1), XBB.1 (n=9), and XBB.2 (n=1) in Beijing. From a year-round perspective, variants from clade 21L (BA.2.2.1) and 22B (BF.7 and BA.5.2) dominated in Beijing during 2022 (figure 2C). However, 22B became absolutely dominant in Beijing after mid-November, 2022 (figure 2D).
Figure 2.
Phylogeny and statistics of SARS-CoV-2 variants circulating in Beijing during the whole year 2022
(A) The phylogenetic tree of complete high-quality SARS-CoV-2 genomes generated in this study. The phylogenetic tree was visualised by the Auspice online tool. Tips with a circle indicate the genomic sequences generated in this study. Others are reference sequences. The colour of the circle corresponds to different clades. (B) The phylogenetic tree of complete high-quality SARS-CoV-2 genomes generated in this study. The phylogenetic tree was visualised by the Auspice online tool. Tips with a circle indicate the genomic sequences generated in this study. Others are reference sequences. The colour of the circle corresponds to local or imported cases. (C) The composition of SARS-CoV-2 variants circulating in Beijing during the whole year 2022. The inner doughnut represents the composition of clades. The outer doughnut represents the detailed Pango lineage within each clade. (D) The composition of SARS-CoV-2 variants circulating in Beijing after Nov 14, 2022. The inner doughnut represents the composition of clades. The outer doughnut represents the detailed Pango lineage within each clade.
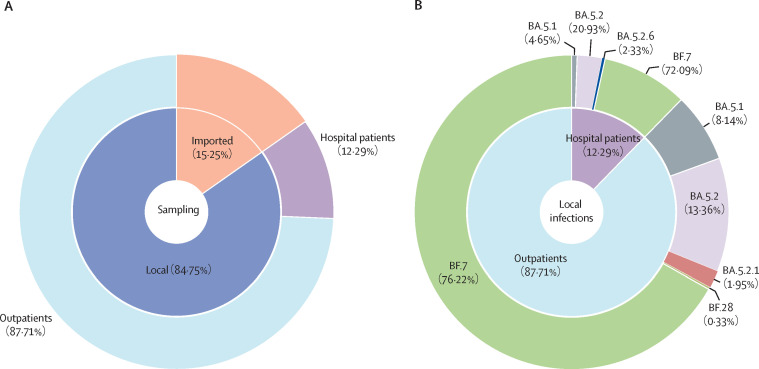
From Nov 14, Beijing faced a significant surge of new infections and we sequenced 413 new infections, including 350 local cases and 63 imported cases (figure 3A ). Among these local infections, BF.7 accounted for 265 (75·71%) and became the dominant strain in Beijing after Nov 14, while BA.5.2-like (57 [16·29%]) and BA.5.1 (27 [7·71%]) took the second and third spots (figure 3B). We further classified the local infections into outpatients and hospitalised patients; the dominant strain was BF.7 among both groups, which was consistent with the local infections overall (figure 3B). There was no significant difference in the proportions of virus strains between the outpatients and hospitalised patients (OR 1·24, 95% CI 0·61–2·54; p=0·55).
Figure 3.
Sampling and distribution of omicron subvariants in local cases and imported cases since Nov 14, 2022
(A) The sampling distribution of local cases (outpatient and inpatient cases) and imported cases from Nov 14 to Dec 20. (B) The distribution of omicron subvariants in 350 local cases.
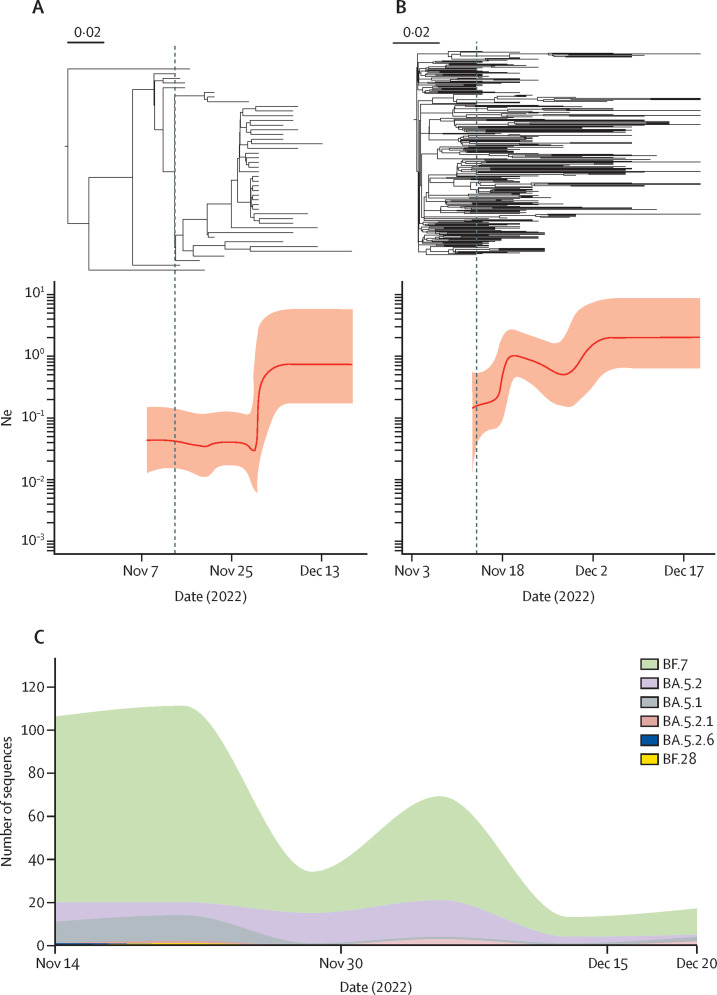
Further phylodynamic analysis showed that the effective population size for both BA.5.2 and BF.7 in Beijing experienced expansions after Nov 14, 2022, indicating an increase in genetic diversity within these two lineages (figure 4A, B ). However, different expansion patterns were found for BA.5.2 and BF.7. The effective population size of BA.5.2 did not change substantially between Nov 14 and Nov 25, but increased sharply around Nov 30, while that of BF.7 increased gradually from Nov 14. The sudden expansion of effective population size of BA.5.2 in Beijing is consistent with the increased number of BA.5.2 infections around Nov 30 (figure 4C). In addition, we did not find any novel SARS-CoV-2 variants circulating in Beijing in the recent outbreak. On the other hand, there were up to 16 types of subvariants identified in the imported cases (n=63) in the same period (appendix 2 p 9). Among them, 20 (31·75%) strains belonged to BQ.1, followed by BA.5.2 with 14 (22·22%) and XBB.1 with six (9·52%).
Figure 4.
Temporal population dynamics of BA.5.2 and BF.7 in Beijing after Nov 14, 2022
(A) The dated phylogeny (top) and corresponding estimation of effective population size, Ne (bottom) during the same period for BA.5.2 in Beijing. These two plots share the same timeline axis. (B) The dated phylogeny (top) and corresponding estimation of effective population size, Ne (bottom) during the same period for BF.7 in Beijing. These two plots share the same timeline axis. (C) The distribution of local subvariants identified in Beijing over time from Nov 14 to Dec 20.
Overall, local and imported infections exhibited substantial differences in the lineage distribution from Nov 14 to Dec 20. BF.7 and BA.5.2 were found in the majority of local infections and became the dominant variants, while the co-circulation of BQ.1 and other variants was observed in imported infections. SARS-CoV-2 variants found to be dominant internationally during the same period, including XBB and BQ.1, were not detected in local infections in Beijing.
Discussion
In this study, we report the trend of COVID-19 cases and the spread of SARS-CoV-2 variants in Beijing in 2022. The curve showed only one major cluster outbreak before mid-November, with a maximum number of 96 cases per day on May 22. Epidemic investigation and phylogenetic analysis indicated domestic transmission as a common cause for most of these cluster outbreaks during the study period. Several peaks of imported cases were also observed, which is consistent with the global COVID-19 wave caused by omicron subvariants in 2022,27 and is also linked to the number of flights that arrived in Beijing. However, no surge of secondary local infection caused by imported cases was found during the whole year. In general, our data showed a blocking of local transmission with continuing imported infection before December, which highlights the effectiveness of the dynamic zero-COVID policy implemented in China, considering the high transmissibility of omicron subvariants.
Before December, 2022, BA.2.2.1 was the most common subvariant in Beijing during April and July. That was because the only local outbreak was caused by imported cases from Shanghai Municipality, and is in line with the fact that omicron subvariant BA.2.2.1 was the dominant variant responsible for the outbreak in Shanghai Municipality during spring, 2022.28 A rapidly increasing number of cases has been observed in Beijing since December, 2022. In particular, BF.7 and BA.5.2 accounted for the current surge of cases beginning in mid-November; BF.7 and BA.5.2 have been demonstrated to have increased fitness of the prototype by approximately 24 and 20 times, respectively.29 This result is also in line with the current situation in the Chinese mainland.30, 31 A recent report also indicated that BA.5.2 was the dominant subvariant in Hebei Province, Guangzhou Municipality, and Shanghai Municipality. 32 In addition, the composition of omicron subvariants varied between Hebei Province and Shanghai Municipality. Taken together, even though the dominant omicron subvariant is the same, the composition of the rest of the subvariants is still different among provinces and cities, which deserves further attention and in-depth study. However, these two variants have been found in Beijing before November, 2022, and the potential secondary transmission had not been observed under the dynamic zero-COVID strategy. Consequently, we speculated that the outbreak after November was the combined effect of policy adjustment and the high fitness of variants circulating in Beijing. Despite the short time-span, sufficient temporal signal has been detected within both BF.7 and BA.5.2 datasets collected after mid-November, making it possible to reliably infer the population dynamics of these two lineages after the adjustment of prevention and control policies. We also found that the effective population size of both BA.5.2 and BF.7 increased in Beijing, indicating higher within-lineage genetic diversity. We found the effective population size of BA.5.2 exponentially expanded around Nov 30 (figure 4A). The exponential expansion of effective population size usually occurred during the early phase of the outbreak. Since a local outbreak of BA.5.2 in Beijing had been found around this time (figure 4C), we speculated that the outbreak might lead to an exponential increase in the effective population size of BA.5.2 in Beijing. However, BF.7 has been persistently circulating in Beijing since October, 2022. With the change of prevention and control policies and its high fitness, the effective population size increased gradually. In addition, the difference in the dynamic patterns of the effective population size of these two omicron subvariants might also be affected by other factors, such as the different fitness, as well as the cases imported from outside of Beijing (both in and outside of China). The increased genetic diversity within each currently circulating lineage could also lead to the emergence of novel subvariants in the future, posing a potential unknown threat to human health.
Currently, the BA.5-derived subvariant BQ.1, its subvariant BQ.1.1, and XBB (a recombinant of two BA.2 subvariants) are spreading globally.33 However, XBB, BQ.1, and BQ.1.1 were not detected in local cases and no novel recombinant strains were detected in circulating subvariants in Beijing, which might be due to the quarantine measures adopted. We noticed that a proportion of delta VOC was identified from imported cases and local cases in early 2022, but was quickly cleared and has not been detected since March, 2022.
For imported infections, the number of cases has increased and multiple subvariants were detected before December after the control policy adjustment. There were 16 omicron subvariants identified from overseas, including XBB, BQ.1, and BQ.1.1. However, these imported subvariants have not yet become the dominant strains, which might be due to the current effective quarantine measures for the imported cases, and the potential protective effect that is offered by the outbreak in progress. Thus, the immune evasion ability and growth advantages of the imported strains need to be continuously monitored. With the relaxation of the isolation policy for foreign passengers and the upcoming Spring Festival travel rush (large-scale population mobility during a short period), SARS-CoV-2 variants with high transmissibility or high immune escape will pose a threat to Chinese public health, which can be expanded globally. Therefore, close monitoring is crucial during this time.
Since its emergence, omicron rapidly became dominant worldwide, generating hundreds of subvariants with more mutations, such as BF.7 and BA.5.2 (approximately 100 mutations compared with the prototype). Among them, novel subvariants with advanced fitness continue to replace older ones and then cause new rounds of infections, which was also the case with the previous VOCs.34, 35 However, the accumulation speed of SARS-CoV-2 genomes is far less than its evolutionary rate, preventing us from truly understanding the dynamics. Consequently, it is vital to conduct timely and continuous large-scale monitoring of mutations during epidemics by sequencing as many SARS-CoV-2 genomes as possible. This is why we are addressing this particular scientific question in this study, and we will continue with such an approach in the future.
Our study has some limitations. Only the epidemic and viral genetic data in Beijing in 2022, rather than the rest of the Chinese mainland, were analysed. The number of laboratory-confirmed COVID-19 cases in December is unavailable at present since large-scale nucleic acid testing has been adjourned, and the actual number of infections would likely be underestimated, which will also lead to a certain degree of sampling bias in our dataset. However, our dataset represented the real-world data well. More sampling is required for investigation of the competitive transmission power and pathogenicity of omicron subvariants. In addition, a strict clock model was used as prior in the phylodynamic analysis. Although the assumption that the evolutionary rate of a virus is constant during the initial stage of an outbreak is usually reasonable, it might ignore the potential heterogeneity of evolutionary rate among branches.
In conclusion, we report the co-circulation of BF.7 and BA.5.2 in the current outbreak in Beijing and did not observe the existence of any novel variants. This study could be considered a snapshot of China, due to both the frequent population exchange and the circulating strains with high transmissibility.
Data sharing
Data have been made publicly available via the Global Initiative on Sharing Avian Influenza Data (GISAID) database.
Declaration of interests
We declare no competing interests.
Acknowledgments
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2021ZD0114103) and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29010202). We are grateful for the valuable contribution of the Nucleic Acid Testing Group of Beijing CDC, 17 district CDCs in Beijing, sentinel hospitals in Beijing, and the airport quarantine department in Beijing. We appreciate Sun Qiang, from Capital Medical University Electric Power Teaching Hospital, for his work on part of the figures in this work.
Contributors
GFG and QW designed and coordinated the study, with input from YP, LW, ZF, HX, FL, YS, DZ, and WJL, reviewed published literature, and wrote the first draft. YP, LW, ZF, HX, FL, and YS accessed and verified the data and made the tables and figures. GFG, QW, YP, LW, ZF, HX, FL, YS, DZ, and WJL reviewed and revised the report. All authors approved the final version. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Supplementary Materials
References
- 1.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan W, Zhao X, Ma X, et al. A novel coronavirus genome identified in a cluster of pneumonia cases—Wuhan, China 2019–2020. China CDC Wkly. 2020;2:61–62. [PMC free article] [PubMed] [Google Scholar]
- 4.WHO Tracking SARS-CoV-2 variants. https://www.who.int/activities/tracking-SARS-CoV-2-variants
- 5.Viana R, Moyo S, Amoako DG, et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Lai S, Gao GF, Shi W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature. 2021;600:408–418. doi: 10.1038/s41586-021-04188-6. [DOI] [PubMed] [Google Scholar]
- 7.Zhou L, Wu Z, Li Z, et al. One hundred days of coronavirus disease 2019 prevention and control in China. Clin Infect Dis. 2021;72:332–339. doi: 10.1093/cid/ciaa725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moeti M, Gao GF, Herrman H. Global pandemic perspectives: public health, mental health, and lessons for the future. Lancet. 2022;400:e3–e7. doi: 10.1016/S0140-6736(22)01328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Liu M, Liang W. The dynamic COVID-zero strategy in China. China CDC Wkly. 2022;4:74–75. doi: 10.46234/ccdcw2022.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shuai H, Chan JFW, Hu B, et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022;603:693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 11.Halfmann PJ, Iida S, Iwatsuki-Horimoto K, et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu WJ, Wu G. Convincing the confidence to conquer COVID-19: from epidemiological intervention to laboratory investigation. Biosafety Health. 2020;2:185–186. doi: 10.1016/j.bsheal.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu WJ, Liu S. A tale of two cities: from influenza HxNy to SARS-CoV-z. China CDC Wkly. 2021;3:1052–1056. doi: 10.46234/ccdcw2021.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Health Commission of the People's Republic of China and National Administration of Traditional Chinese Medicine of the People's Republic of China. Guidance for corona virus disease 2019: prevention, control, diagnosis and management, 9th edn. Beijing: State Council Joint COVID-19 Prevention And Control Mechanism Team, 2022.
- 15.Tyson JR, James P, Stoddart D, et al. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv. 2020 doi: 10.1101/2020.09.04.283077. published online Sept 4. (preprint). [DOI] [Google Scholar]
- 16.Aksamentov I, Roemer C, Hodcroft EB, Neher RA. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw. 2021;6:3773. [Google Scholar]
- 17.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 2020;37:291–294. doi: 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flouri T, Izquierdo-Carrasco F, Darriba D, et al. The phylogenetic likelihood library. Syst Biol. 2015;64:356–362. doi: 10.1093/sysbio/syu084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minh BQ, Schmidt HA, Chernomor O, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen) Virus Evol. 2016;2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baele G, Li WL, Drummond AJ, Suchard MA, Lemey P. Accurate model selection of relaxed molecular clocks in bayesian phylogenetics. Mol Biol Evol. 2013;30:239–243. doi: 10.1093/molbev/mss243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- 26.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.WHO Weekly epidemiological update on COVID-19—21 December 2022. https://www.who.int/publications/m/item/covid-19-weekly-epidemiological-update-21-december-2022
- 28.Ling Y, Lu G, Liu F, et al. The Omicron BA.2.2.1 subvariant drove the wave of SARS-CoV-2 outbreak in Shanghai during spring 2022. Cell Discov. 2022;8:97. doi: 10.1038/s41421-022-00468-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obermeyer F, Jankowiak M, Barkas N, et al. Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness. Science. 2022;376:1327–1332. doi: 10.1126/science.abm1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Technical Advisory Group on Virus Evolution TAG-VE statement on the meeting of 3 January on the COVID-19 situation in China. https://www.who.int/news/item/04-01-2023-tag-ve-statement-on-the-3rd-january-meeting-on-the-covid-19-situation-in-china/
- 31.Chinese Center for Disease Control and Prevention National novel coronavirus infection epidemic situation. Jan 25, 2023. https://www.chinacdc.cn/jkzt/crb/zl/szkb_11803/jszl_13141/202301/t20230125_263519.html
- 32.Gang L, Yun L, Minghao J, et al. Primary assessment of the diversity of Omicron sublineages and the epidemiologic features of autumn/winter 2022 COVID-19 wave in Chinese mainland. Front Med (in press). [DOI] [PMC free article] [PubMed]
- 33.Singapore Ministry of Health COVID-19 situation at a glance. https://www.moh.gov.sg/
- 34.Colson P, Fournier PE, Chaudet H, et al. Analysis of SARS-CoV-2 variants from 24,181 patients exemplifies the role of globalization and zoonosis in pandemics. Front Microbiol. 2022;12:786233. doi: 10.3389/fmicb.2021.786233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colson P, Gautret P, Delerce J, et al. The emergence, spread and vanishing of a French SARS-CoV-2 variant exemplifies the fate of RNA virus epidemics and obeys the Mistigri rule. J Med Virol. 2023;95:e28102. doi: 10.1002/jmv.28102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data have been made publicly available via the Global Initiative on Sharing Avian Influenza Data (GISAID) database.