Abstract
To establish an intimate interaction with the host epithelial cell surface, enteropathogenic Escherichia coli (EPEC) produces Tir, a bacterial protein that upon translocation and insertion into the epithelial cell membrane constitutes the receptor for intimin. The tir gene is encoded by the locus for enterocyte effacement (LEE), where it is flanked upstream by orf19 and downstream by the cesT and eae genes. With the use of a series of cat transcriptional fusions and primer extension analysis, we confirmed that tir, cesT, and eae form the LEE5 operon, which is under the control of a promoter located upstream from tir, and found that the orf19 gene is transcribed as a monocistronic unit. We also demonstrated that the LEE-encoded regulator Ler was required for efficient activation of both the tir and the orf19 promoters and that a sequence motif located between positions −204 and −157 was needed for the Ler-dependent activation of the tir operon. Sequence elements located between positions −204 and −97 were determined to be required for the differential negative modulatory effects exerted by unknown regulatory factors under specific growth conditions. Upon deletion of the upstream sequences, the tir promoter was fully active even in the absence of Ler, indicating that tir expression is subject to a repression mechanism that is counteracted by this regulatory protein. However, its full activation was still repressed by growth in rich medium or at 25°C, suggesting that negative regulation also occurs at or downstream of the promoter. Expression of orf19, but not of the tir operon, became Ler independent in an hns mutant strain, suggesting that Ler overcomes the repression exerted by H-NS (histone-like nucleoid structuring protein) on this gene.
Enteropathogenic Escherichia coli (EPEC) is a major cause of acute and persistent infantile diarrhea and a leading cause of infant death in developing countries (35, 43, 45). The interaction of EPEC with the host cell, which has been the subject of several recent reports, has been divided into three different stages characterized by two distinctive phenotypes, localized adherence and the attaching-and-effacing (A/E) lesion (reviewed in references 9, 11, 18, and 43). The virulence determinants required for the induction of the A/E lesion in EPEC are encoded in a 35.6-kb pathogenicity island, denoted LEE (for locus of enterocyte effacement), which contains 41 predicted open reading frames (16, 38, 39). Based on recent studies and sequence analyses, most of the LEE-encoded genes have been divided into three functional regions: the esc and sep genes, which code for a type III secretion-translocation apparatus (25); the tir, cesT, and eae genes, coding for the proteins involved in intimate attachment (1, 14, 26, 28); and the esp genes, which encode effector proteins that are involved in the formation of a translocon for delivering effector molecules to the host cell (17, 29, 30, 32, 33, 51).
Recent studies have indicated that the LEE-encoded genes are organized into five major operons (14, 41): the LEE1, LEE2, and LEE3 operons, which contain the esc and sep genes; the LEE4 operon, which encodes secreted Esp proteins; and the tir, cesT, and eae cluster, herein denoted LEE5. Ler acts as a positive transcriptional regulator of the LEE2, LEE3, LEE4, and LEE5 operons (5, 19, 41). Furthermore, integration host factor (IHF) (19) and a quorum-sensing autoinducer (LuxS) (47) are also required for efficient activation of the LEE-encoded genes. In addition, it has been proposed that Ler overcomes the negative regulation exerted by H-NS (histone-like nucleoid structuring protein) on the expression of at least the LEE2, LEE3, and LEE4 operons (5).
We studied the transcriptional organization and regulation of the orf19, tir, cesT, and eae genes of EPEC. The tir gene codes for Tir (translocated intimin receptor), which is transferred by the type III secretion system into host cells, where it is phosphorylated and inserted into the host cell membrane (28). Tir is the receptor for intimin, an eae-encoded outer membrane protein necessary for intimate attachment to epithelial cells (26). cesT, previously known as orfU, codes for a chaperone that is required for the stable secretion of Tir (1, 14). The orf19 gene encodes a protein that exhibits similarity to IpgB of Shigella flexneri (16) and to TrcA and TrcP of EPEC (48, 50). In the present study we have demonstrated that tir, cesT, and eae constitute an operon and that orf19 is a monocistronic unit. We show that transcription of the tir operon and the orf19 gene requires Ler, which seems to overcome the negative regulation exerted by a repressor protein that is also present in E. coli K-12. In addition, we show that distinct cis-acting elements are involved in negative and positive regulation of tir expression by different regulatory elements and environmental cues.
(A preliminary account of this work was presented at the 99th General Meeting of the American Society for Microbiology, Chicago, Ill., 30 May to 3 June 1999 [C. Sanchez-SanMartín, M. G. Sosa, V. H. Bustamante, E. Calva, and J. L. Puente, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. B/D-227, p. 74, 1999].)
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and plasmids used in this study are listed in Table 1. Overnight cultures were grown at 37°C in Luria-Bertani (LB) broth medium (46). Dulbecco's modified Eagle's medium (DMEM) containing 0.45% (wt/vol) glucose and l-glutamine (584 mg/l) without sodium pyruvate (Gibco Life Technologies) and supplemented with pyridoxal (4 μg/ml) was used for growth at 37°C. Where indicated, 20 mM ammonium sulfate was added. An overnight LB culture was pelleted, and the bacteria were resuspended in phosphate-buffered saline, pH 7.4, to an optical density at 600 nm (OD600) of 1. Fifty milliliters of DMEM or LB was inoculated with 1 ml of the bacterial phosphate-buffered saline suspension and incubated in an orbital shaker water bath (Amerex Instruments) at 200 rpm and various temperatures. When necessary, antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; nalidixic acid, 15 μg/ml, kanamycin, 40 μg/ml, chloramphenicol, 50 μg/ml, gentamicin, 15 μg/ml; and tetracycline, 25 μg/ml. Samples were collected every hour, or when the cultures reached OD600s of 0.8, 1.0, 1.2, and 1.4, to determine chloramphenicol acetyltransferase (CAT) activity or for RNA extraction.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E2348/69 | Wild-type EPEC O127:H6 | 36 |
| JPN15 | E2348/69 lacking the EAF plasmid | 26 |
| E2348/69Δler | E2348/69; Δler mutant | 5 |
| B171-8 | Wild-type EPEC O111:NM | 44 |
| B171-10 | B171-8 lacking the EAF plasmid | 4 |
| MC4100 | F−araD139 Δ(argF-lac)U169 prsL150 relA1 deoC1 rbsR fthD5301 fruA25λ− | 7 |
| RO64 | MC4100 lrp-201::Tn10 | 34 |
| MCG007 | MC4100 rpoS::Tn10 | C. Gómez |
| MCG009 | MC4100 fis::kan | C. Gómez |
| GS480Δ900 | MC4100 malP::neo Δ(envZ-malP)900 | 20 |
| CSH56 | Δlac-pro supD nalA thi | 52 |
| CSH56 Δhns | CSH56 carrying a deletion of hns | 52 |
| N99 | F−galK2 rpsL λ− | 22 |
| K5185 | N99 ΔhimA82 | 42 |
| ET8000 | rbs gyrA hutCKlacZ::IS1 Mu cts62 | 23 |
| ET8045 | ET8000 rpoN208::Tn10 | 8 |
| SM796 | F−araD139 Δ(araABC-leu)7697 galE galK Δ(lac) X74 rpsL thi phoA ΔPvull phpR | 6 |
| SBC796 | SM796 fur::Tn5 | 6 |
| Plasmids | ||
| pKK232-8 | pBR322 derivative containing a promoterless cat gene | 3 |
| pCAT232 | pKK232-8 derivative containing a bfpA-cat transcriptional fusion from nt −232 to +36 | 44 |
| pKORF1 | pMPM-K3 derivative carrying the ler gene expressed under the control of the tac promoter | 5 |
| pCS-TVW | pMPM-K3 derivative carrying the per (bfpTVW) locus expressed under the control of the tac promoter | Sánchez-SanMartín et al. (unpublished) |
| pORF19 | pKK232-8 derivative containing the orf19-cat transcriptional fusion from nt −175 to +166 | This study |
| pTIR394 | tir-cat transcriptional fusion from nt −394 to +138 | This study |
| pTIR243 | tir-cat transcriptional fusion from nt −243 to +138 | This study |
| pTIR204 | tir-cat transcriptional fusion from nt −204 to +138 | This study |
| pTIR157 | tir-cat transcriptional fusion from nt −157 to +138 | This study |
| pTIR122 | tir-cat transcriptional fusion from nt −122 to +138 | This study |
| pTIR97 | tir-cat transcriptional fusion from nt −97 to +138 | This study |
| pTIR80 | tir-cat transcriptional fusion from nt −80 to +138 | This study |
| pTIR45 | tir-cat transcriptional fusion from nt −45 to +138 | This study |
| pTIR22 | tir-cat transcriptional fusion from nt −22 to +138 | This study |
| pTIREAE | tir-cat transcriptional fusion from nt −294 of tir to the fourth codon of eae | This study |
| pTIR-EAE-DEL1 | tir-cat transcriptional fusion from nt −294 of tir to the 15th codon of cesT | This study |
| pTIRCAM-10 | pTIR-EAE derivative in which the −10 promoter hexamer sequence of tir was replaced by a SacI recognition sequence | This study |
| pTIRCAM-10-DEL1 | pTIRCAM-10 derivative from which the cesT-eae intergenic region was deleted | This study |
| pCEST | cesT-cat transcriptional fusion containing the intergenic region between tir and cesT | This study |
| pEAE1800 | eae-cat transcriptional fusion containing 1,800 bp upstream from the eae start codon | This study |
| pEAE1629 | eae-cat transcriptional fusion containing 1,629 bp upstream from the eae start codon | This study |
| pCEST-EXT-EAE | pCEST derivative, equivalent to pEAE1629 | This study |
| pEAE1422 | eae-cat transcriptional fusion containing 1,422 bp upstream from the eae start codon | This study |
| pEAE532 | eae-cat transcriptional fusion containing 532 bp upstream from the eae start codon | This study |
Molecular biology techniques.
DNA manipulations were performed according to standard protocols (46). Restriction and DNA-modifying enzymes were obtained from Boehringer Mannheim, New England Biolabs, or Gibco BRL and used according to the manufacturer's instructions. [α-32P]dCTP (3,000 Ci mmol−1) was purchased from Amersham Corp. Oligonucleotides were purchased from BioSynthesis or provided by the Oligonucleotide Synthesis Facility at our institute. PCRs were performed in 100- or 50-μl volumes, with AmpliTaq (Perkin-Elmer) being used according to the manufacturer's instructions. Double-stranded DNA sequencing of the plasmids generated in this work was carried out by the dideoxy-chain termination procedure with a Thermo Sequenase cycle sequencing kit according to the manufacturer's (Amersham) instructions.
Construction of cat transcriptional fusions.
PCR fragments of different lengths that spanned the region between orf19 and eae were amplified using as a template chromosomal DNA from wild-type EPEC strain E2348/69. The forward and reverse oligonucleotides were designed to introduce BamHI or HindIII restriction sites, respectively (Table 2). The PCR-amplified fragments were digested with BamHI and HindIII and cloned into pKK232-8, a vector, digested with the same enzymes, containing a promoterless CAT gene (cat) (Pharmacia LKB Biotechnology). Each ligation reaction product was electroporated into E. coli MC4100, after which ampicillin-resistant colonies were selected. The resulting plasmids (Table 1) were sequenced to confirm the fidelity of the PCR amplification and introduced into different strains by electroporation, using a Gene Pulser apparatus (Bio-Rad) at settings of 2.5 kV, 25 μF, and 200 Ω, or by CaCl2 transformation according to standard protocols (46).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| CAM10TIR2R | ACCAGAGCTCTCACAGTATAATTTTTG |
| CAM10TIR1F | GTGAGAGCTCTGGTTTATGCTCAGTTTG |
| EAE1669REG | GGGATAAAAAGCTTGAGTA |
| EAEREG06 | GTTCGCTCAGATCCTAAATTCTG |
| EAEREG1052L | CAACGTTGCAGCATGGGTAAC |
| ORFU-H3R | CCAACACCAATTTTTTCCGC |
| ORF19-H3R | CCGCCTAAGCTTACGCTCTAC |
| RORF10-H3R | TGTTATCCCAAGCTTATGT |
| TIR-H3R | CTGTTTGTGAAGCTTGTGGC |
| TIR22F-E23 | CATCAAAAAGGATCCTGTGATTTATTTG |
| TIR45F-E23 | CATTTCTGGATCCTTATTTTGC |
| TIR80F-E23 | GATTTTGATTATGGGATCCTTTA |
| TIR97F-E23 | GATAATTATAGGATCCTTTTGATTATG |
| TIR122F-E23 | CATATAAAAATTAGGATCCTTTTTTTTC |
| TIR157F-E23 | ATATTATTTTTGGATCCACAATTAAATTTC |
| TIR204F-E23 | TTTTTTCTGGATCCAAAAAGGTCTC |
| TIR243F-E23 | GATACTCGGATCCAGGGGGAAAC |
| 16S | CACAGATTGTCTGATAAATTG |
Mutagenesis of the −10 promoter sequence.
Oligonucleotide CAM10tirR plus pKKampi and oligonucleotide CAM10tirF plus pKKcat (Table 2) were used to amplify two fragments encompassing the tir regulatory region contained in pTIREAE. Oligonucleotides CAM10tirR and -F introduce a SacI restriction site that replaces the −10 promoter hexamer. Both fragments were digested with SacI and with either BamHI (left fragment) or HindIII (right fragment) and cloned into pKK232-8 digested with BamHI and HindIII, creating pTIRCAM-10. The insert in this plasmid was sequenced to verify that only the −10 hexamer sequence was modified.
RNA isolation and primer extension analysis.
Total bacterial RNA from samples obtained from DMEM or LB cultures was isolated using a commercial kit (RNeasy [Qiagen] or Boehringer Mannheim High Pure RNA isolation kit). End-labeled oligonucleotides complementary to the 5′ end of the cat structural gene or the tir, cesT, eae, or orf19 structural gene were used for primer extension reactions as previously described (5, 37). The extended products were purified with a Microcon-10 microconcentrator (Amicon) and analyzed by electrophoresis in 8% polyacrylamide–urea gels. Primer extension using a 16S rRNA-specific oligonucleotide was included as a control to monitor RNA quality and loading concentration. Sequence ladders were generated with the same primers and DNA of the different pKK232-8 derivatives or plasmids carrying the genes being studied in this work.
CAT assay.
The CAT assay was performed as described previously (5, 37, 44).
RESULTS
Transcriptional organization of the orf19, tir, cesT, and eae genes.
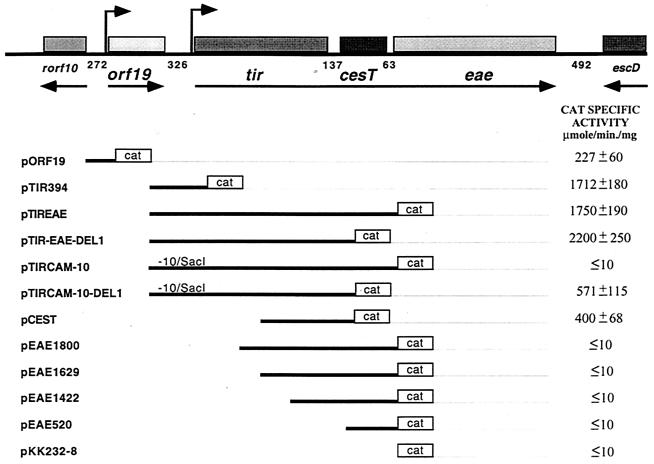
The start codon of tir is located 326 bp downstream from the stop codon of orf19, while the tir-cesT and cesT-eae intergenic regions consist of 137 and 63 bp, respectively (Fig. 1). To confirm and further analyze the transcriptional organization of the orf19, tir, cesT, and eae genes and to identify intergenic regions with promoter activity, a series of transcriptional fusions to the cat reporter gene was constructed (Fig. 1). These plasmids were transformed into EPEC wild-type strains E2348/69 and B171-8, and CAT activity directed by each fusion was determined from samples collected from cultures grown in DMEM at 37°C, conditions that induce expression of different virulence factors in EPEC (5, 27, 37, 44). The fusions carried by plasmids pTIR394, pTIREAE, and pTIREAE-DEL1, which all contain the 5′ upstream region of tir, expressed significant levels of CAT (Fig. 1). In contrast, fusions pEAE1800, pEAE1629, pEAE1422, and pEAE520, which contain different lengths of the region upstream from the eae start codon but lack the tir promoter region, expressed only background levels of CAT activity (Fig. 1 and data not shown). These results confirmed that tir, cesT, and eae constitute an operon that is expressed under the control of a promoter located upstream of tir and indicated that eae does not have an independent promoter.
FIG. 1.
tir, cesT, and eae constitute an operon. (A) Schematic representation of the organization of the orf19, tir, cesT, and eae genes and of the transcriptional fusions constructed to study their regulation. The horizontal arrows indicate the direction of transcription. Bent-tailed arrows denote the transcriptional start sites identified in this work (see the text). The sizes (in base pairs) of the intergenic regions are shown below the thick horizontal line. Plasmid denominations are indicated in the left-hand column below. The fragments cloned into the promoterless cat gene vector pKK232-8 are denoted by solid lines, and the cat gene is indicated by an open box at the end of each fragment. The −10/SacI label indicates the presence of a mutation that replaced the putative tir −10 promoter hexamer by a SacI restriction site. The right-hand column shows the CAT activity expressed by each fusion in EPEC E2348/69 grown in DMEM at 37°C. The data obtained for EPEC B171-8 are not shown, for simplicity, but rendered the same conclusions. The CAT specific activity was determined from cells harvested at an OD600 of 1.4. The results reported are the averages ± standard deviations of data from at least four different experiments.
To confirm that the eae gene is transcribed from the tir promoter, the putative −10 hexamer (see below) was replaced by the CTCGAG sequence in pTIREAE, as described in Materials and Methods, generating pTIRCAM-10. As expected, this fusion did not express CAT (Fig. 1). Fusion plasmid pORF19, which contains 308 bp upstream from the translational start codon of orf19, was active (Fig. 1), indicating that this gene is transcribed from its own promoter.
A fusion containing just the tir-cesT intergenic region (pCEST) rendered activity levels that suggested the existence of an additional active promoter for the cesT gene (Fig. 1). However, fusions pEAE1800, pEAE1629, and pEAE1422, containing the tir-cesT and cesT-eae intergenic regions, were inactive (Fig. 1), suggesting a role for the cesT-eae intergenic region in terminating transcription originating at the cesT putative promoter. To further analyze this possibility, the cesT-eae intergenic region was cloned into pCEST to generate plasmid pCEST-EXT-EAE (see Materials and Methods). In this case, no activity was detected (data not shown). When this region was deleted from the inactive fusions carried by plasmids pEAE1629 and pTIRCAM-10, thus recreating the pCEST fusion and generating pTIRCAM-10-DEL1 (which lacks a functional tir promoter), respectively, the cesT transcriptional activity was recovered (Fig. 1).
Two transcriptional start sites were identified for the putative cesT promoter when primer extension experiments were performed with total RNA of EPEC E2348/69 carrying either pCEST, pEAE1629, or pTIRCAM-10 fusions (Fig. 2C). These results confirmed the existence of active cesT promoters in these fusions and suggested that the eae upstream region contains elements involved in terminating transcription originating from the cesT promoter.
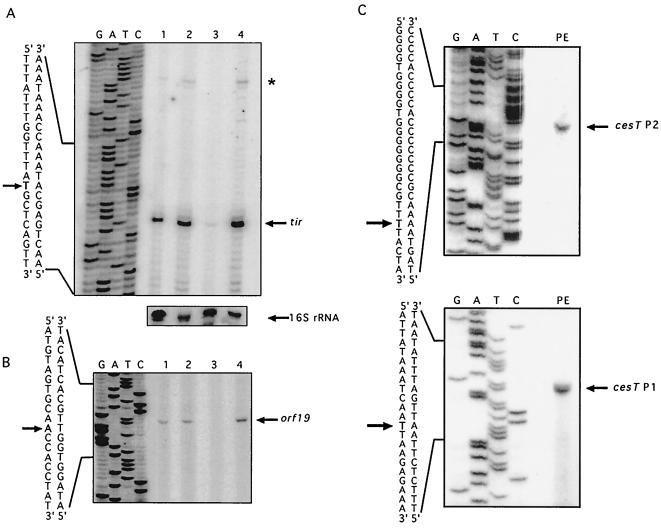
FIG. 2.
Primer extension analysis of the tir, orf19, and cesT promoter regions. (A) Total RNA was obtained from culture samples of strains EPEC E2348/69 wild type (lane 1), JPN15 (pEAF cured) (lane 2), EPEC E2348/69Δler (lane 3), and EPEC E2348/69Δler carrying pKORF1 (lane 4) growing in DMEM at 37°C (OD600 = 0.8). A primer specific for the tir structural gene was used, and primer extension was performed as described in Materials and Methods. A primer extension assay using a primer specific for the 16S rRNA gene was performed as a control. (B) Primer extension analysis was performed as described for panel A but with a primer specific for the orf19 structural gene. (C) Total RNA from EPEC E2348/69 carrying pCEST (cesT-cat fusion) and a cat-specific primer were used for primer extension reactions. The upper and bottom panels show the two extended products. Lanes G, A, T, and C correspond to the DNA sequence ladder obtained with the corresponding primer. The sequences spanning the transcription start site are shown, and the transcription start sites (in bold) are marked with arrows.
Determination of the transcriptional start sites of the orf19 and tir genes.
The potential transcriptional activity from the 5′ upstream regions of the four genes considered for this study was tested by primer extension analysis (Fig. 2 and data not shown). A transcriptional start site corresponding to a T residue was located 85 bp upstream from the translational start codon of tir (Fig. 2A), one base further downstream from where it was previously mapped (14).
Examination of the 5′ upstream region revealed the presence of putative −35 (TTGCAT) and −10 (TTTATT) promoter sequences (Fig. 3A). When the putative −10 promoter sequence was replaced by the CTCGAG sequence, fusions carrying this mutation became inactive (Fig. 1), indicating that this sequence motif was essential for transcriptional activation of the tir operon. An additional transcriptional start site for tir, corresponding to a T residue, was identified 24 bp upstream from the first reported site (Fig. 2A); however, the relevance of this weaker potential promoter is unknown. A transcriptional start site was also identified for orf19, on an A residue located 133 bp upstream from its translational start codon (Fig. 2B). Analysis of its 5′ upstream region revealed the presence of putative −35 (TTGCAT) and −10 (ATAAAT) promoter sequences (Fig. 3B). In contrast, no transcriptional start site was observed for the chromosomal cesT and eae genes under the conditions tested.
FIG. 3.
Nucleotide sequences of the orf19-tir (A), rorf10-orf19 (r, reverse) (B), and tir-cesT (C) intergenic regions. The transcriptional start sites (+1) are indicated by the bent-tailed open-headed arrows. The bent-tailed filled-head arrows indicate the 5′ ends of the different tir-cat fusions. The predicted −10 and −35 promoter sequences are underlined. The sequences containing motifs potentially involved in Ler activation and ammonium-dependent repression, in negative regulation by an as-yet-unidentified repressor, and in repression at high temperature are indicated by solid, broken, and dotted underlines, respectively.
Ler is required for transcriptional activation of tir and orf19.
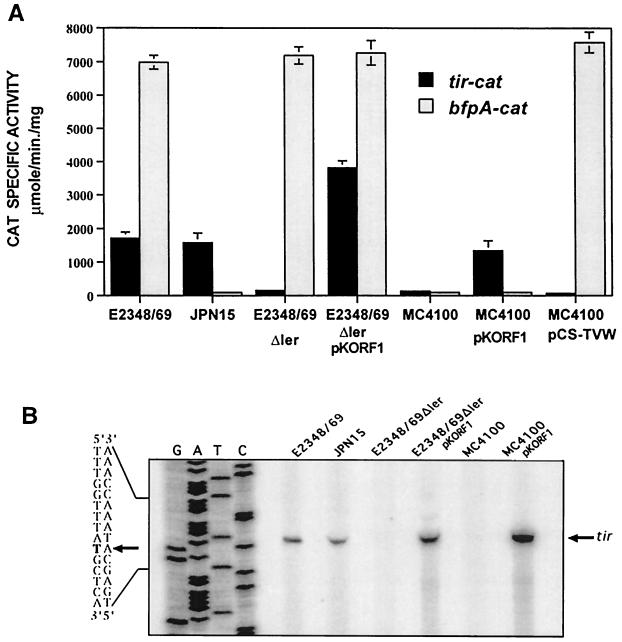
As described above, tir-cat fusions were active in two different EPEC wild-type strains (Fig. 1). However, when plasmid pTIR394, carrying the tir-cat fusion, was transformed into E. coli MC4100 (a K-12 derivative laboratory strain), its transcriptional activity was reduced (Fig. 4A), indicating that full expression of the tir operon required a regulatory factor that was present only in wild-type EPEC. To identify the activator(s) involved in tir expression, we first examined the potential role of the EAF plasmid by transforming pTIR394 into EPEC strains JPN15 and B171-10 (pEAF-minus derivatives of EPEC wild-type strains E2348/69 and B171-8, respectively) (Table 1). Expression of the tir-cat fusion in the plasmid-cured strains rendered levels of CAT activity similar to those obtained in the wild-type strains (Fig. 4A and data not shown), indicating that the transcriptional activation of tir was independent of the EAF plasmid. This observation was confirmed by determining that the transcriptional activities of the chromosomal tir promoters of wild-type EPEC and its pEAF-minus derivative were similar when compared by primer extension (Fig. 2A, lanes 1 and 2).
FIG. 4.
Expression of tir requires the Ler protein. (A) The transcriptional activity directed by the tir-cat fusion in pTIR394 and the bfpA-cat fusion contained in pCAT232 was tested in EPEC strains E2348/69 (wild type), JPN15 (an EAF-minus derivative), and Δler (a ler in-frame deletion mutant of E2348/69) carrying or not carrying pKORF1 (ler+), as well as in E. coli K-12 strain MC4100 (wild type) carrying or not carrying either pKORF1 (ler+) or pCS-TVW (per/bfpTVW+). The CAT specific activity was determined from cells grown in DMEM at 37°C and harvested at an OD600 of 1.4. The data are the averages of results from at least three different experiments. Error bars indicate standard deviations. (B) Primer extension analysis of the tir-cat fusion in pTIR394 was carried out with total RNA extracted from samples obtained at an OD600 of 0.8 from the cultures described above. The primer extension reactions were performed as described in the legend to Fig. 2, using a primer specific for cat. The arrow on the left indicates the transcriptional start site of tir-cat.
It has been shown that the product of the ler gene (previously known as orf1) is a positive regulator of the expression of sepZ and orf12, two genes also located in the LEE (5, 41). This observation prompted us to investigate the role of Ler in tir expression by performing primer extension experiments with total RNA obtained from DMEM cultures of an EPEC Δler strain which carries an in-frame deletion of ler (5). Expression of tir was considerably reduced in this mutant, while complementation with ler on a plasmid restored its expression (Fig. 2A, lanes 3 and 4). In agreement with these results, the activity directed by the tir-cat fusion (pTIR394) was reduced about 10-fold in the Δler mutant strain, while its activation was restored and enhanced by supplementing ler in trans with plasmid pKORF1 (Fig. 4A). Furthermore, activation was restored in E. coli MC4100 when supplemented with pKORF1, but not with a plasmid carrying the entire per locus (pCS-TVW), which, in contrast, complemented the expression of the bfpA-cat fusion that was used as a control (Fig. 4A).
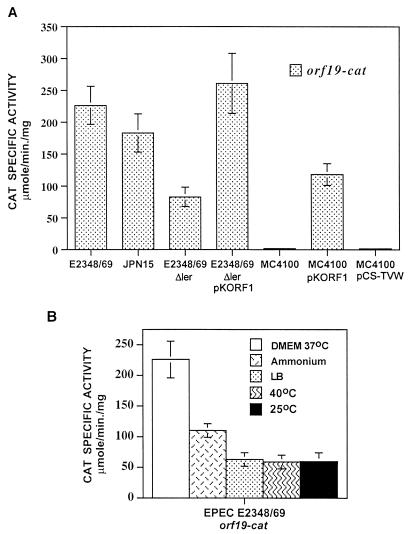
Consistent with these results, primer extension analysis revealed that Ler-dependent activation of the tir-cat fusion in pTIR394 was directed by the same putative promoter predicted for the wild-type gene (Fig. 4B). Similar experiments, using the same set of strains carrying the pORF19 fusion, revealed that orf19 expression also required a functional ler gene (Fig. 2B; see also Fig. 7A). Taken together, these results indicated that Ler was required for the positive regulation of tir and orf19 expression.
FIG. 7.
Regulation of orf19. (A) Expression of orf19 requires the Ler protein. The transcriptional activity directed by the orf19-cat fusion in pORF19 was tested in different strains as described in the legend to Fig. 4A. (B) Effect of the growth medium, the presence of ammonium, and temperature on orf19 expression. EPEC E2348/69 carrying the orf19-cat fusion (pORF19) was grown under different conditions to determine CAT activity, as described in the legend to Fig. 6.
Analysis of the tir promoter region.
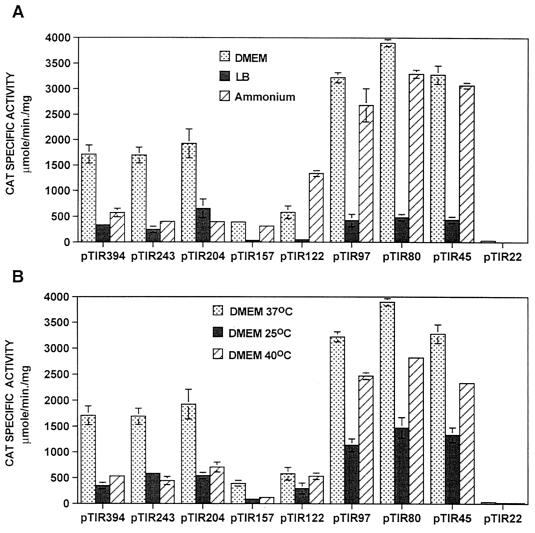
To define the minimal regulatory sequence required for tir expression, a series of fusions containing segments of the tir 5′ promoter region ranging from 394 to 22 bp in length, with respect to the transcriptional start site (Fig. 3 and 5), was constructed. Fusions containing sequences up to position −204 (pTIR204) or longer were found to express similar amounts of CAT in EPEC E2348/69 (Fig. 5A). However, fusions to position −157 (pTIR157) or −122 (pTIR122) showed a significant reduction of CAT activity compared with that obtained with pTIR394 (Fig. 5A). Fusions containing up to position −97, −80, or −45 (pTIR97, pTIR80, and pTIR45, respectively) expressed a twofold increase in CAT activity in comparison to that obtained with pTIR394 (Fig. 5A). As expected, a fusion containing up to position −22 which lacks the putative −35 promoter hexamer did not express CAT.
FIG. 5.
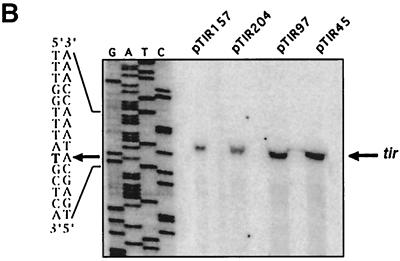
cis-acting elements involved in tir expression. (A) Schematic representation of the tir regulatory region and tir-cat fusions. Numbering is relative to the transcription start site, which is indicated by a bent-tailed arrow. Regions containing sequences potentially involved in Ler-dependent activation (subdivided boxes) or in negative regulation (shaded boxes) are indicated. EPEC strains E2348/69 and E2348/69Δler, as well as E. coli MC4100, were transformed with different tir-cat fusions contained in plasmids pTIR394, pTIR243, pTIR204, pTIR157, pTIR122, pTIR97, pTIR80, pTIR45, and pTIR22. The resulting strains were grown in DMEM at 37°C, and the CAT specific activity was determined from samples obtained at an OD600 of 1.4. The plots of the activities determined from samples obtained along the growth curve at other OD600 values showed the same pattern. Values are the averages of data from at least three different experiments; error bars indicate standard deviations. (B) Total RNA was obtained from EPEC E2348/69 carrying the tir-cat fusions in pTIR204, pTIR157, pTIR80, and pTIR45, and primer extension assays were performed as described in the legend to Fig. 2. The arrow on the left indicates the transcriptional start site of the tir-cat fusions, which corresponds to that identified for the wild-type gene.
In the absence of Ler (in EPEC Δler or E. coli MC4100), tir-cat fusions in pTIR394, pTIR243, and pTIR204 showed only background levels of tir promoter activity (Fig. 5A). In the absence of sequence elements located between positions −157 and −97, activation of the tir promoter became Ler independent (Fig. 5A). These results suggested that Ler could interact with a DNA sequence motif located between positions −204 and −157 and that this interaction overcame the repressing effect exerted by a negative regulatory factor, also present in E. coli K-12, that interacts with sequence elements located between positions −157 and −97. To rule out the possibility that an additional promoter was responsible for the Ler-independent activity shown by the shorter fusions, primer extension experiments were performed with total RNA obtained from culture samples of strains EPEC E2348/69, EPEC Δler, and E. coli MC4100 carrying fusions pTIR204, pTIR157, pTIR97, or pTIR45. The resulting primer extension products demonstrated that the transcriptional activity observed for these fusions was directed by the same promoter predicted for the wild-type gene (Fig. 5B and data not shown).
H-NS represses the expression of orf19 but not tir.
The experiments described above suggested that the putative repressor for tir and orf19 was conserved between EPEC and E. coli K-12. To identify putative repressors, different E. coli K-12-derived strains carrying mutations in well-characterized genes coding for global regulators were transformed with fusion plasmids pORF19 and pTIR394. Only background levels of CAT activity were obtained for both fusions in strains carrying mutations in the genes coding for LRP (leucine-responsive regulatory protein), IHF (integration host factor), Fis (factor for inversion stimulation), StpA (suppressor of td− phenotype A), factor, FIS, StpA, RpoN, RpoS, and OmpR as well as in their corresponding parental strains (Table 1 and data not shown). In contrast, expression of the orf19-cat fusion resulted in a 25-fold increase in CAT activity in E. coli CSH56 Δhns (470 ± 36 CAT units [mean ± standard deviation]) in comparison with its parental strain, CSH56 (18 ± 5 CAT units). Repression was restored when a plasmid carrying the hns gene was introduced into CSH56 Δhns/pORF19 (11 ± 7 CAT units). The absence of H-NS did not have any effect on expression of the tir-cat fusion.
Regulation of tir and orf19 expression in response to environmental conditions.
Expression and secretion of different virulence factors in EPEC are optimal in tissue culture medium (DMEM) at 37°C and are negatively regulated in response to growth in a rich medium such as LB at temperatures above or below 37°C or in the presence of ammonium salts (4, 5, 27, 37, 44). To determine whether tir expression was modulated by changing the growth conditions, EPEC E2348/69 carrying plasmid pTIR394 was grown in DMEM at 37°C, in LB at 37°C, in DMEM containing 20 mM ammonium sulfate at 37°C, and in DMEM at 25°C and 40°C, as described previously (44). tir expression was significantly reduced by growth in LB or in DMEM containing ammonium, as well as by growth at temperatures above or below 37°C (Fig. 6).
FIG. 6.
Effect of the growth medium, the presence of ammonium, and temperature on tir expression. EPEC E2348/69 derivatives carrying the tir-cat fusions described in the legend to Fig. 5 were grown at 37°C in DMEM, DMEM plus 20 mM ammonium sulfate, or LB (A) or in DMEM at 37, 25, or 40°C (B). CAT specific activities from samples obtained at an OD600 of 1.4 were determined and plotted. The plots of the activities determined from samples obtained along the growth curve at other OD600 values showed the same pattern. Values are the averages of data from at least three different experiments; error bars indicate standard deviations.
When the same experiment was carried out with EPEC E2348/69 carrying the shorter tir-cat transcriptional fusions, it was observed that growth in LB (Fig. 6A) and in DMEM at 25°C (Fig. 6B) resulted in negative modulation of the expression of all the active fusions (pTIR243 to pTIR45). This indicated that repression under these conditions acts on or downstream of the promoter. In contrast, the repression mediated by the presence of ammonium or by growth at 40°C was observed only for fusions containing the sequence between positions −204 and −157 (Fig. 6A) or between positions −157 and −122 (Fig. 6B), respectively. These observations suggested that tir expression is subjected to different levels of negative regulation, probably involving one or more factors that act on different segments of the tir regulatory region. In addition, CAT activity assays of samples obtained from cultures of EPEC E2348/69/pORF19 grown under the conditions described above revealed that expression of orf19 is regulated coordinately with the expression of the tir operon (Fig. 7B) and suggested that Orf19 plays an important role in EPEC pathogenesis.
DISCUSSION
In this work, we studied the regulation of the tir-cesT-eae operon (henceforth referred as the LEE5 operon), whose expression is directed by a promoter located upstream of the tir gene (Fig. 1 to 3). Recently, it was reported that these genes are expressed in the same transcript, which initiates 86 nucleotides (nt) upstream of the tir start codon (14), one base upstream from where the transcriptional start site was determined to be located (Fig. 3A). The existence of an eae transcriptional start site located inside the structural cesT gene was previously proposed (21). However, neither fusions carrying different fragments of the eae upstream region (pEAE1800, pEAE1629, pEAE1422, and pEAE520) nor a tir-eae-cat fusion (pTIRCAM-10) carrying a mutation in the −10 tir promoter sequence demonstrated promoter activity (Fig. 1). Different attempts to define a transcriptional start site for eae by primer extension rendered only a variable ladder of undefined reverse transcription products (data not shown). We cannot rule out the possibility that additional promoters allow the differential expression of the components of the LEE5 operon under different conditions. However, our initial data suggest that the observed eae primer extension products could be the result of posttranscriptional mRNA processing events.
A transcriptional start site for cesT has been reported (14). Consistent with this observation, cesT-cat fusions carrying sequences of the upstream region of cesT without the tir promoter (pCEST and pTIRCAM-10-DEL1) directed the expression of significant levels of CAT activity (Fig. 1). Primer extension experiments with the cat-specific primer revealed two different transcriptional start sites (Fig. 2C). The first transcriptional start site was located 15 nt upstream of the cesT start codon and allowed the prediction of a good putative −10 (TATTAT) promoter sequence and a poor −35 (TGGGTA) hexamer. The sequence preceding the second start site did not show any homology with known promoter sequences (Fig. 3C). Because of its weak activity, we were unable to detect primer extension products derived from the transcript of the chromosomal cesT gene. This Ler-independent promoter activity does not seem to read through the eae gene, since the presence of the cesT-eae intergenic region rendered fusions inactive (compare pTIRCAM-10 with pTIRCAM-10-DEL1 and pEAE1422 with pCEST [Fig. 1]). These results suggest that Ptir-initiated transcription may be antiterminated while PcesT-initiated transcription is not. Further detailed analysis is required to establish the significance of these observations; however, it is tempting to speculate that the independent expression of cesT may ensure the presence of the chaperone when Tir is translated or that CesT has additional, as-yet-undefined functions.
Two major mechanisms regulate virulence gene expression in EPEC (5, 15, 19, 21, 41, 49). One involves a classical activator (PerA/BfpT) that is fully required for the activation of the bfpA and perA (bfpT) promoters (37, 49). per is also proposed to be involved in the modulation of eae expression (21), protein secretion (27), the down-regulation of intimin during A/E adhesion (31), and the direct activation of the LEE1 operon, which encodes the ler gene (41). The other involves an antagonist protein (Ler) (19, 41, 47) that is required to overcome the repression exerted by negative regulators on the expression of several LEE-encoded genes, which is directed by promoters that in the absence of upstream regulatory elements are constitutively expressed (5). Ler seems to act as a master key which modulates the expression of different virulence factors that allow the intimate colonization of the proximal small intestine and the generation of A/E lesions by EPEC. Based on these observations, we analyzed the role of the ler and per (bfpTVW)-encoded products in LEE5 and orf19 expression. Ler was needed for the efficient expression of both the LEE5 operon and the orf19 gene (Fig. 2 and 4 and data not shown). In contrast, wild-type EPEC, its EAF-cured derivative, and a per mutant strain did not show significant differences in their expression (Fig. 2 and 4 and data not shown). Hence, the LEE5 operon and the orf19 gene are part of the Ler regulon.
Deletion analysis of the LEE5 upstream regulatory region indicated that nucleotides up to position −45, which include only the tir promoter, were sufficient for maximal activation even in the absence of Ler (Fig. 5). Full activation was maintained with sequences up to position −97, but not with sequences up to position −122 or −157, which sustained low levels of activation. This suggested that a sequence motif involved in negative regulation is located upstream of position −97 and that the negative regulator was conserved in nonpathogenic E. coli strains. Fusions up to position −204 or longer were activated in a Ler-dependent manner but only reached intermediate levels of activity with respect to those directed by the shorter fusions (Fig. 5). This established that sequences between positions −204 and −157 were necessary for Ler-dependent activation of the LEE5 promoter, which was still modulated by a putative negative regulator that probably interacts with the region between positions −157 and −97.
Ler shows amino acid similarity to the DNA-binding global transcriptional regulator H-NS and its paralogue, StpA (12, 16, 41). The H-NS protein has been implicated in the negative regulation of several virulence factors and housekeeping genes (2). Considering its similarity to H-NS, it is likely that Ler binds DNA sequences between positions −204 and −157. The H-NS protein is involved in the negative regulation of the LEE2 and LEE3 operons (5) and the orf19 gene but not LEE5 expression (see Results). Ler might overcome the repression exerted by H-NS or an unknown factor by directly or indirectly (e.g., changing the DNA topology) interfering with its binding. However, further work is required to distinguish between these and other possibilities.
Environmental conditions regulate the expression of several virulence factors in different bacteria, such as Vibrio cholerae, Shigella spp., Yersinia spp., and pathogenic E. coli strains (10, 24, 40). In EPEC, the expression of virulence factors is also regulated in response to culture conditions that mimic in vivo regulatory signals (5, 27, 37, 44). We found that distinct regulatory sequences in the LEE5 promoter region are involved in the regulatory response to these conditions (Fig. 6). Negative regulation by growth in rich medium or at temperatures below 37°C involves factors that act directly on the promoter (Fig. 6). The repression mechanism mediated by temperatures above 37°C and by the presence of ammonium in the culture medium is dependent on sequences located upstream from the putative promoter, between positions −157 and −122 and positions −204 and −157, respectively (Fig. 6). These motifs overlap with the sequence presumably required for negative regulation by an unknown factor and the putative Ler-binding region, respectively (Fig. 3A), suggesting the existence of different regulatory mechanisms that probably involve additional trans-acting factors. Neither ammonium nor high temperature is a common regulatory signal for the regulation of virulence factors. However, both can be found during transit along the intestinal lumen, and they may represent signals indicating harmful or inappropriate niches for colonization (13, 44).
ACKNOWLEDGMENTS
We particularly thank Susana López, Mario Rocha, Joaquín Sánchez, and Y. Martínez-Laguna for helpful discussions. We also thank Martha G. Sosa, Francisco Santana, and Alejandra Vázquez for technical assistance.
C.S.-S. was supported by a Ph.D. fellowship from the Consejo Nacional de Ciencia y Tecnología (CONACYT), México (no. 91904), and from the Universidad Nacional Autónoma de México. This research was supported by grants from the Consejo Nacional de Ciencia y Tecnología, México (CONACyT 27831-N), and from the Universidad Nacional Autónoma de México (DGAPA IN206594).
REFERENCES
- 1.Abe A, de Grado M, Pfuetzner R A, Sánchez-SanMartín C, DeVinney R, Puente J L, Strynadka N C J, Finlay B B. Enteropathogenic Escherichia coli translocated intimin receptor, Tir, requires a specific chaperone for stable secretion. Mol Microbiol. 1999;33:1162–1174. doi: 10.1046/j.1365-2958.1999.01558.x. [DOI] [PubMed] [Google Scholar]
- 2.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 3.Brosius J. Plasmid vectors for the selection of promoters. Gene. 1984;27:151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- 4.Bustamante V H, Calva E, Puente J L. Analysis of cis-acting elements required for bfpA expression in enteropathogenic Escherichia coli. J Bacteriol. 1998;180:3013–3016. doi: 10.1128/jb.180.11.3013-3016.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bustamante V H, Santana F J, Calva E, Puente J L. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli (EPEC): Ler antagonizes H-NS-dependent repression. Mol Microbiol. 2001;39:664–677. doi: 10.1046/j.1365-2958.2001.02209.x. [DOI] [PubMed] [Google Scholar]
- 6.Calderwood S B, Mekalanos J J. Confirmation of the Fur operator site by insertion of a synthetic oligonucleotide into an operon fusion plasmid. J Bacteriol. 1988;170:1015–1017. doi: 10.1128/jb.170.2.1015-1017.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 8.Cava J R, Elias P M, Turowski D A, Noel K D. Rhizobium leguminosarum CFN42 genetic regions encoding lipopolysaccharide structures essential for complete nodule development on bean plants. J Bacteriol. 1989;171:8–15. doi: 10.1128/jb.171.1.8-15.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeVinney R, Gauthier A, Abe A, Finlay B B. Enteropathogenic Escherichia coli: a pathogen that inserts its own receptor into host cells. Cell Mol Life Sci. 1999;55:961–976. doi: 10.1007/PL00013202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiRita V J, Engleberg N C, Heath A, Miller A, Crawford J A, Yu R. Virulence gene regulation inside and outside. Philos Trans R Soc Lond B Biol Sci. 2000;355:657–665. doi: 10.1098/rstb.2000.0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg M S, Kaper J B, Finlay B B. Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 1997;5:109–114. doi: 10.1016/S0966-842X(97)01000-7. [DOI] [PubMed] [Google Scholar]
- 12.Dorman C J, Hinton J C, Free A. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 1999;7:124–128. doi: 10.1016/s0966-842x(99)01455-9. [DOI] [PubMed] [Google Scholar]
- 13.Edwards R A, Puente J L. Fimbrial expression in enteric bacteria: a critical step in intestinal pathogenesis. Trends Microbiol. 1998;6:282–287. doi: 10.1016/s0966-842x(98)01288-8. [DOI] [PubMed] [Google Scholar]
- 14.Elliott S J, Hutcheson S W, Dubois M S, Mellies J L, Wainwright L A, Batchelor M, Frankel G, Knutton S, Kaper J B. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol Microbiol. 1999;33:1176–1188. doi: 10.1046/j.1365-2958.1999.01559.x. [DOI] [PubMed] [Google Scholar]
- 15.Elliott S J, Sperandio V, Girón J A, Shin S, Mellies J L, Wainwright L, Hutcheson S W, McDaniel T K, Kaper J B. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 2000;68:6115–6126. doi: 10.1128/iai.68.11.6115-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott S J, Wainwright L A, McDaniel T K, Jarvis K G, Deng Y K, Lai L C, McNamara B P, Donnenberg M S, Kaper J B. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 17.Foubister V, Rosenshine I, Donnenberg M S, Finlay B B. The eaeB gene of enteropathogenic Escherichia coli is necessary for signal transduction in epithelial cells. Infect Immun. 1994;62:3038–3040. doi: 10.1128/iai.62.7.3038-3040.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frankel G, Phillips A D, Rosenshine I, Dougan G, Kaper J B, Knutton S. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 19.Friedberg D, Umanski T, Fang Y, Rosenshine I. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol Microbiol. 1999;34:941–952. doi: 10.1046/j.1365-2958.1999.01655.x. [DOI] [PubMed] [Google Scholar]
- 20.Garrett S, Taylor R K, Silhavy T J, Berman M L. Isolation and characterization of ΔompB strains of Escherichia coli by a general method based on gene fusions. J Bacteriol. 1985;162:840–844. doi: 10.1128/jb.162.2.840-844.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gómez-Duarte O G, Kaper J B. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect Immun. 1995;63:1767–1776. doi: 10.1128/iai.63.5.1767-1776.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottesman M E, Yarmolinsky M B. Integration-negative mutants of bacteriophage lambda. J Mol Biol. 1968;31:487–505. doi: 10.1016/0022-2836(68)90423-3. [DOI] [PubMed] [Google Scholar]
- 23.Guterman S K, Howitt C L. Rho and ribosome mutation interaction: lethality of rho-15 in rpsL or rpsE strains, and rho-15 methionine auxotrophy in rps+ strains of Escherichia coli. Genetics. 1979;93:353–360. doi: 10.1093/genetics/93.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harel J, Martin C. Virulence gene regulation in pathogenic Escherichia coli. Vet Res. 1999;30:131–155. [PubMed] [Google Scholar]
- 25.Jarvis K G, Giron J A, Jerse A E, McDaniel T K, Donnenberg M S, Kaper J B. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci USA. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jerse A E, Yu J, Tall B D, Kaper J B. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenny B, Abe A, Stein M, Finlay B B. Enteropathogenic Escherichia coli protein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect Immun. 1997;65:2606–2612. doi: 10.1128/iai.65.7.2606-2612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenny B, DeVinney R, Stein M, Reinscheid D J, Frey E A, Finlay B B. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 29.Kenny B, Finlay B B. Protein secretion by enteropathogenic Escherichia coli is essential for transducing signals to epithelial cells. Proc Natl Acad Sci USA. 1995;92:7991–7995. doi: 10.1073/pnas.92.17.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenny B, Lai L C, Finlay B B, Donnenberg M S. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol Microbiol. 1996;20:313–323. doi: 10.1111/j.1365-2958.1996.tb02619.x. [DOI] [PubMed] [Google Scholar]
- 31.Knutton S, Adu-Bobie J, Bain C, Phillips A D, Dougan G, Frankel G. Down regulation of intimin expression during attaching and effacing enteropathogenic Escherichia coli adhesion. Infect Immun. 1997;65:1644–1652. doi: 10.1128/iai.65.5.1644-1652.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knutton S, Rosenshine I, Pallen M J, Nisan I, Neves B C, Bain C, Wolff C, Dougan G, Frankel G. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai L-C, Wainwright L A, Stone K D, Donnenberg M S. A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect Immun. 1997;65:2211–2217. doi: 10.1128/iai.65.6.2211-2217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lange R, Barth M, Hengge-Aronis R. Complex transcriptional control of the ςS-dependent stationary-phase-induced and osmotically regulated osmY (csi-5) gene suggests novel roles for Lrp, cyclic AMP (cAMP) receptor protein-cAMP complex, and integration host factor in the stationary-phase response of Escherichia coli. J Bacteriol. 1993;175:7910–7917. doi: 10.1128/jb.175.24.7910-7917.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levine M M. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J Infect Dis. 1987;155:377–389. doi: 10.1093/infdis/155.3.377. [DOI] [PubMed] [Google Scholar]
- 36.Levine M M, Bergquist E J, Nalin D R, Waterman D H, Hornick R B, Young C R, Sotman S. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;i:1119–1122. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- 37.Martínez-Laguna Y, Calva E, Puente J L. Autoactivation and environmental regulation of bfpT expression, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coli. Mol Microbiol. 1999;33:153–165. doi: 10.1046/j.1365-2958.1999.01460.x. [DOI] [PubMed] [Google Scholar]
- 38.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDaniel T K, Kaper J B. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol Microbiol. 1997;23:399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- 40.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mellies J L, Elliott S J, Sperandio V, Donnenberg M S, Kaper J B. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 42.Nash H A. Integration and excision of bacteriophage lambda: the mechanism of conservation site specific recombination. Annu Rev Genet. 1981;15:143–167. doi: 10.1146/annurev.ge.15.120181.001043. [DOI] [PubMed] [Google Scholar]
- 43.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puente J L, Bieber D, Ramer S W, Murray W, Schoolnik G K. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol Microbiol. 1996;20:87–100. doi: 10.1111/j.1365-2958.1996.tb02491.x. [DOI] [PubMed] [Google Scholar]
- 45.Robins-Browne R M. Traditional enteropathogenic Escherichia coli of infantile diarrhea. Rev Infect Dis. 1987;9:28–53. doi: 10.1093/clinids/9.1.28. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Sperandio V, Mellies J L, Nguyen W, Shin S, Kaper J B. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl Acad Sci USA. 1999;96:15196–15201. doi: 10.1073/pnas.96.26.15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tobe T, Hayashi T, Han C G, Schoolnik G K, Ohtsubo E, Sasakawa C. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect Immun. 1999;67:5455–5462. doi: 10.1128/iai.67.10.5455-5462.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tobe T, Schoolnik G K, Sohel I, Bustamante V H, Puente J L. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:963–975. doi: 10.1046/j.1365-2958.1996.531415.x. [DOI] [PubMed] [Google Scholar]
- 50.Tobe T, Tatsuno I, Katayama E, Wu C Y, Schoolnik G K, Sasakawa C. A novel chromosomal locus of enteropathogenic Escherichia coli (EPEC), which encodes a bfpT-regulated chaperone-like protein, TrcA, involved in microcolony formation by EPEC. Mol Microbiol. 1999;33:741–752. doi: 10.1046/j.1365-2958.1999.01522.x. [DOI] [PubMed] [Google Scholar]
- 51.Wolff C, Nisan I, Hanski E, Frankel G, Rosenshine I. Protein translocation into host epithelial cells by infecting enteropathogenic Escherichia coli. Mol Microbiol. 1998;28:143–155. doi: 10.1046/j.1365-2958.1998.00782.x. [DOI] [PubMed] [Google Scholar]
- 52.Yamada H, Yoshida T, Tanaka K, Sasakawa C, Mizuno T. Molecular analysis of the Escherichia coli hns gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol Gen Genet. 1991;230:332–336. doi: 10.1007/BF00290685. [DOI] [PubMed] [Google Scholar]