Abstract
Retron reverse transcriptases are unusual procaryotic enzymes capable of synthesis of low-molecular-weight DNA by reverse transcription. All of the so-far-described DNA species synthesized by retron reverse transcriptases have been identified as multicopy single-stranded DNA. We have shown that Salmonella enterica serovar Enteritidis is also capable of synthesis of the low-molecular-weight DNA by retron reverse transcriptase. Surprisingly, Salmonella serovar Enteritidis-produced low-molecular-weight DNA was shown to be a double-stranded DNA with single-stranded overhangs (sdsDNA). The sdsDNA was 72 nucleotides (nt) long, of which a 38-nt sequence was formed by double-stranded DNA with 19- and 15-nt single-stranded overhangs, respectively. Three open reading frames (ORFs), encoded by the 4,053-bp plasmid, were essential for the production of sdsDNA. These included an ORF with an unknown function, the retron reverse transcriptase, and an ORF encoding the cold shock protein homologue. This plasmid was also able to confer phage resistance onto the host cell by a mechanism which was independent of sdsDNA synthesis.
Salmonella enterica serovar Enteritidis frequently contains plasmids. The best understood is the serovar-specific plasmid which encodes for virulence genes, e.g., spv, pef, or rck (6, 12, 29, 30, 39). Besides this plasmid, wild-type Salmonella serovar Enteritidis strains occasionally contain additional plasmids, frequently of low molecular weight. They have been extensively used in molecular typing (5, 10, 26, 33), but only a few reports describing their biological functions have appeared. They have been suspected to influence resistance to antibiotics (34, 36) and phages (11), and when transformed into Escherichia coli, these plasmids affect its growth (16). During our recent studies in molecular typing (31, 32), we have observed that in a particular serovar Enteritidis strain, a single low-molecular-weight plasmid was responsible for the resistance to phage infection. Loss of this plasmid resulted in conversion of phage type PT21 to phage type PT1 (32). The difference between PT1 and PT21 strains is mainly in resistances to phages P3, P5, and P7 of the standard phage typing set, to which strains of PT21 are resistant while strains belonging to PT1 are sensitive (38). Therefore we sequenced the whole plasmid, and sequence analysis revealed that it coded for an open reading frame (ORF) similar to those of the retron reverse transcriptases (RRT) (35).
RRTs are unusual enzymes that were first described for Myxococcus xanthus (18) and later for E. coli (21, 23). They catalyze the synthesis of multicopy single-stranded DNA (msDNA) (8, 40), which is usually 50 to 150 nucleotides (nt) in length, present freely in the cytoplasm of bacterial cells (7, 28). Biosynthesis of msDNA by the RRT has been described in detail using in vitro systems (15), but the biological role of msDNA is still unknown. RRTs are present in most of the M. xanthus strains (8, 19) but in only 10 to 15% of E. coli isolates (14). Genes for RRT were localized on the chromosomal DNA. In some cases, mainly in E. coli, the chromosomal loci resembled structures of bacteriophages (9), and some bacteriophages were even found to encode RRT as well (17). So far, RRT has never been described to be plasmid encoded. There was only a single report on its presence in Salmonella (27).
MATERIALS AND METHODS
Bacterial strains.
Two field strains of Salmonella serovar Enteritidis, previously characterized by plasmid profiling (31, 32) and by phage typing (38), were used as the donors and final recipients of plasmids. Salmonella serovar Enteritidis strain 2159 was of phage type PT21 and plasmid type SE55IJ, i.e., it contained the 55-kb virulence plasmid, plasmid I, and plasmid J (31). After prolonged storage at 4°C (for about a year), Salmonella serovar Enteritidis strain 2160, originally of the same plasmid profile, SE55IJ, spontaneously lost the plasmid designated I. This resulted in a strain of plasmid type SE55J and conversion of phage type PT21 to phage type PT1 (see Fig. 1). Strains were grown in Luria broth (Difco) at 37°C in a shaking incubator at 200 rpm.
FIG. 1.
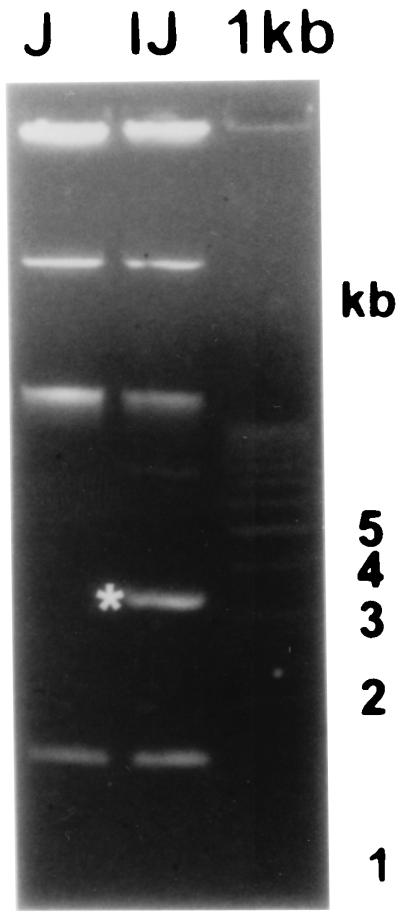
Plasmid profiles of Salmonella serovar Enteritidis 2159 (lane IJ) and serovar Enteritidis 2160 (lane J) after electrophoresis on an 0.8% agarose gel. Plasmid I is marked with an asterisk. A 1-kb ladder was used as the molecular size standard.
Sequencing of plasmid I.
Plasmid DNA was purified with a QIAprep Spin Miniprep kit (Qiagen, Hilden, Germany) from the strain serovar Enteritidis 2159. Total plasmid DNA was digested with the restriction endonuclease TaqI and ligated into the ClaI-digested plasmid pBluescript SK(−). Clones containing fragments from plasmid I were selected by colony hybridization using plasmid I, labeled by the ECL Direct Nucleic Acid Labelling and Detection System (Amersham, Little Chalfont, United Kingdom) in low-melting-point agarose, as a probe. Six clones were selected for the initial sequencing. Obtained sequences were used for the design of walking primers, which were used in sequencing reactions with serovar Enteritidis 2159 total plasmid DNA as a template. Sequencing was carried out with an ABI Prism 310 Genetic Analyzer. Sequence alignments and basic analysis were carried out using Gene Compar software (Applied Maths, Kortrijk, Belgium). MFold calculation was done with the GCG software package. Sequence comparison with GenBank entries was done by basic BLAST (http://www.ncbi.nlm.nih.gov).
Detection and initial characterization of low-molecular-weight DNA.
The DNA produced by RRT was isolated as described previously (3, 19). During the purification procedure, RNase A (100 μg/ml) was present in the cell resuspension buffer, unless otherwise stated. After electrophoresis on a 10% polyacrylamide gel, the DNA was visualized by staining with Sybr Gold fluorescent dye (Molecular Probes). Prior to being electroblotted onto a nylon membrane (Hybond N; Amersham), the samples were denatured in 0.25 M NaOH for 15 min. Using a PCR Master Mix kit (Qiagen), a PCR product (658 bp in size) spanning the expected low-molecular-weight DNA locus was amplified, labeled by the ECL Direct Nucleic Acid Labelling kit, and used as a probe in hybridization. Sequences of the primers used for the probe amplification were as follows: P2, 5′ AAT TAT CCT GAG TGC CGA TG 3′; P3, 5′ TAA ACC TGG GTT TAT TCA TG 3′.
To determine the nature of the low-molecular-weight DNA, it was treated with 10 μg of RNase A/ml for 30 min at 37°C, 10 μg of DNase I/ml for 30 min at 37°C, 20 U of S1 nuclease for 30 min at 23°C, or 10 U of RNase H for 30 min at 23°C. The low-molecular-weight DNA was also tested for heat resistance for 10 min at 99°C in a thermocycler.
Sequence characterization of low-molecular-weight DNA.
Two independent protocols were used for sequence determination. First, the low-molecular-weight DNA was used as an in vivo-produced primer in a PCR to which only one standard PCR primer was added. Primers of both possible orientations were tested in these PCRs. Two selected PCR products were cloned into pcDNA3.1/V5/His-TOPO (Invitrogen), recombinant plasmids were purified, and the sequence of the cloned PCR product was determined. The same protocol was used for sequence determination of the S1 nuclease-treated low-molecular-weight DNA.
In a second protocol, the low-molecular-weight DNA was excised and extracted from a polyacrylamide gel using the QIAEX II Gel Extraction kit (Qiagen), incubated with Taq polymerase for 20 min at 72°C to add 3′-end A overhangs, and cloned into the pCR4 TOPO plasmid (Invitrogen). The sequence of the cloned DNA was determined by sequencing with standard M13 forward and reverse primers.
Identification of genes essential for the synthesis of low-molecular-weight DNA.
Two independent protocols were used. In the first protocol, two fragments of plasmid I were PCR amplified. The forward primer in both reactions was identical and was located 795 bp upstream from the start codon of retron reverse transcriptase. The reverse primer in the first kind of PCR allowed amplification of the low-molecular-weight DNA locus together with retron reverse transcriptase. The second reverse primer allowed amplification of the above-mentioned region, including an ORF5 downstream from the retron reverse transcriptase. Amplification products were cloned into pcDNA3.1/V5/His-TOPO and transformed into host E. coli TOP10 (provided with the cloning kit by Invitrogen). After multiplication in E. coli, the recombinant plasmids (pRT7 where ORF5 was missing and pRT21 including the ORF5) were purified and electrotransformed (E. coli Pulser; Bio-Rad) into Salmonella serovar Enteritidis 2160 to check for restoration of low-molecular-weight DNA synthesis and also for the restoration of the original phage type.
In a second experiment, insertional mutagenesis was applied. An ampicillin resistance gene cassette was prepared by amplification of the bla gene from the pBluescript plasmid with the following primers: AmpF, 5′ GTT AAG GGA TTT TGG TCA TG 3′; AmpR, 5′ GCA CTT TTC GGG GAA ATG TG 3′.
Four pairs of these primers differed in the modifications of their 5′ ends by the presence of either the NsiI, EcoRI, SacI, or EcoRV restriction sites, respectively. The 1,092-bp PCR products were purified with the QIAquick Gel Extraction kit (Qiagen), digested with the appropriate restriction endonuclease, purified with the Gel Extraction kit again, and cloned into the plasmid I, digested and purified in the same way with only one exception–the EcoRV blunt-ended PCR product was cloned into the SfcI site of the plasmid I, which was filled by Klenow fragment to produce a blunt-ended linear plasmid molecule. Ligation mixtures were used for electrotransformation of the Salmonella serovar Enteritidis 2160 strain, selecting for ampicillin-resistant colonies. Clones containing plasmid I with the inserted ampicillin resistance gene cassette were selected by brief phenol plasmid extraction (1) followed by PCR verification.
Nucleotide sequence accession number.
The complete sequence of the plasmid I encoding RRT is available in GenBank under accession number AF218051.
RESULTS
Sequencing of plasmid I.
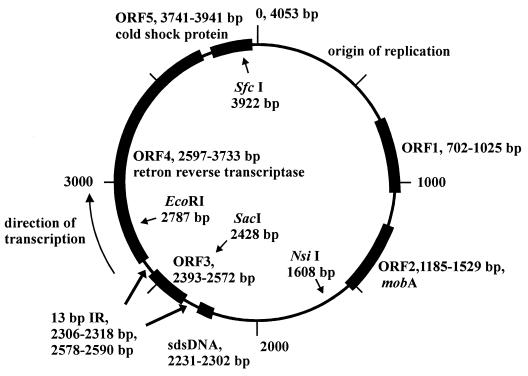
We have determined the complete sequence of the low-molecular-weight plasmid of Salmonella serovar Enteritidis which was capable of protecting the bacterium against phage infection. The original field strain containing the plasmid was of phage type PT21; after the spontaneous loss of this plasmid, the resulting strain was of phage type PT1 (Fig. 1). The size of the plasmid is 4,053 bp, and it codes for five possible ORFs (Fig. 2).
FIG. 2.
Map of plasmid I derived from the sequence analysis and BLAST comparison.
ORF1 encoded a protein (12.2 kDa) showing homology to ORFs already identified in similar-sized plasmids of E. coli and Salmonella enterica serovar Typhimurium; however, its function was not determined. ORF3 (6.8 kDa) was of a sequence which did not match any sequence available in GenBank. For the remaining three ORFs, the function could be predicted based on the similarities to proteins with already-identified functions. ORF2 was homologous to mobA, a protein involved in the initiation and termination of conjugal DNA transfer (2); ORF4 shared homology with RRTs of E. coli (for retron Ec107, 30% identity and 52% similarity; for Ec86, 27% identity and 48% similarity; and for Ec67, 27% identity and 46% similarity) and also with the M. xanthus retron Mx65 (23% identity; 42% similarity). ORF5 was similar to cold-shock proteins from multiple bacterial species, such as Pseudomonas aeruginosa, Lactobacillus plantarum, Staphylococcus aureus, Listeria monocytogenes, or Bacillus subtilis, displaying identities and similarities around 35 and 55%, respectively
ORF1 and ORF2 were located in the part of the plasmid which showed extensive homology to sequences of other low-molecular-weight plasmids and were therefore quite probably linked with the replication and maintenance of the plasmid I in the bacterial cell (position, bp 1 to 1600). ORFs 3, 4, and 5 were located in the remaining part of the plasmid. Since we did not expect that phage resistance could be influenced by ORF1 and ORF2, the remaining three ORFs were analyzed further in detail.
Detection and initial characterization of low-molecular-weight DNA.
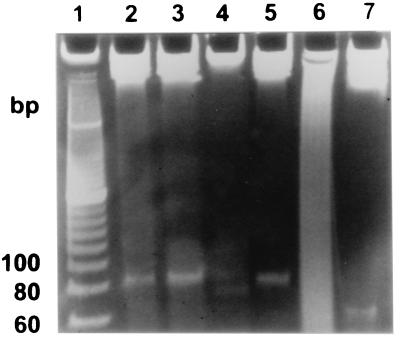
The products of almost all RRTs described so far were multicopy single-stranded DNAs (14, 19, 27, 28). As expected, similar low-molecular-weight DNA was present in the plasmid-containing Salmonella serovar Enteritidis 2159 strain but was missing in the Salmonella serovar Enteritidis 2160 strain (Fig. 3). The DNA was estimated to be approximately 82 nt in length. The DNA was resistant to RNase A and RNase H treatment. On the other hand, it was sensitive to DNase I and partially sensitive to S1 nuclease treatment (Fig. 4). Treatment with S1 nuclease resulted in an increase in polyacrylamide gel electrophoresis mobility, corresponding to an approximately 20-bp decrease in molecular size. The results therefore indicated that the product of RRT was either a single-stranded DNA with a complex secondary structure or a double-stranded DNA species with a single-stranded overhang(s) at the end(s) of the molecule. Such results were obtained for the low-molecular-weight DNA purified with the protocol in which the RNase A was present in first cell resuspension buffer. In a second experiment, we purified the low-molecular-weight DNA by the same protocol, however, without RNase A. As can be seen in Fig. 5, the DNA was present, although in lower quantities. The quantity of the low-molecular-weight DNA was increased considerably after RNase A treatment. However, the other enzymes had standard effects on this sample—S1 nuclease treatment resulted in increased mobility, and DNase I treatment considerably decreased the amount of the low-molecular-weight DNA (Fig. 5).
FIG. 3.
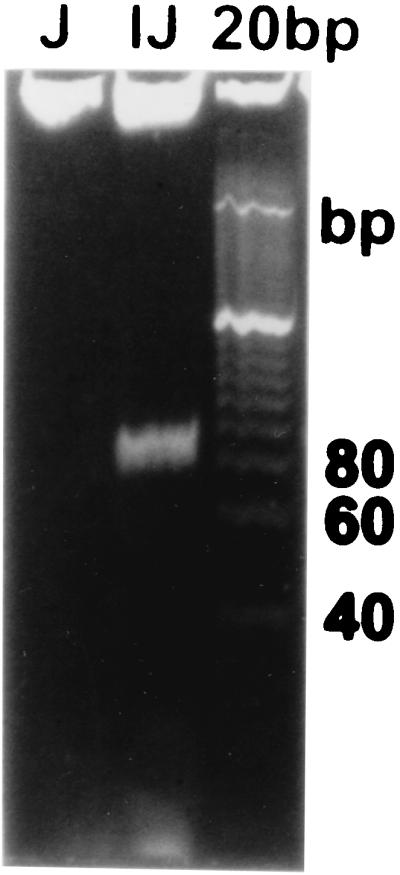
Low-molecular-weight DNA produced by Salmonella serovar Enteritidis 2159 (lane IJ) and Salmonella serovar Enteritidis 2160 (lane J) after electrophoresis on a 10% polyacrylamide gel. A 20-bp ladder was used as a molecular size standard.
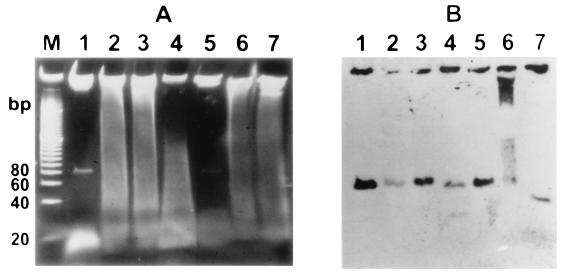
FIG. 4.
Low-molecular-weight DNA electrophoresed on a 10% polyacrylamide gel after treatment with different enzymes. Lane 1, a 20-bp ladder used as a molecular size standard; lane 2, untreated control DNA; lane 3, DNA treated with RNase H; lane 4, heat-treated DNA; lane 5, DNA treated with RNase A; lane 6, DNA treated with DNase I; lane 7, DNA treated with S1 nuclease.
FIG. 5.
(A) Low-molecular-weight DNA isolated in the absence of RNase A in the purification protocol, treated with different enzymes, and electrophoresed on a 10% polyacrylamide gel. Lane M, a 20-bp ladder used as a molecular size standard; lane 1, control DNA purified in the presence of RNase A; lane 2, untreated DNA; lane 3, DNA treated with RNase H; lane 4, heat-treated DNA; lane 5, DNA treated with RNase A; lane 6, DNA treated with DNase I; lane 7, DNA treated with S1 nuclease. (B) Since the presence of low-molecular-weight DNA is hidden by the presence of RNA, hybridization was carried out to detect the low-molecular-weight DNA.
Sequence characterization of low-molecular-weight DNA.
To identify the sequence of the low-molecular-weight DNA and to localize it on the plasmid map, we used the approximately 82-nt DNA as one of the primers in PCR together with only a single in vitro-synthesized primer. Surprisingly, the PCRs repeatedly resulted in a positive amplification regardless of the orientation of the in vitro-synthesized primer added to the reaction. The PCRs were positive even in the presence of RNase A in the PCR tube (not shown). Two PCR products, each originating from oppositely oriented PCRs, were cloned into pcDNA3.1/V5/His-TOPO and sequenced. Comparison of both sequences revealed an overlap of 72 bp which must have served as an in vivo primer in PCRs and therefore represented the sequence of low-molecular-weight DNA. Exactly the same protocol was applied to samples which were first treated with S1 nuclease. After cloning and sequencing of two PCR products, the overlap was only 38 bp in size and this sequence was internal to the 72-bp sequence of nontreated sample.
The sequence of the full-size low-molecular-weight DNA was independently confirmed by its direct cloning after Taq polymerase extension. In this experiment, the identified 72-bp sequence was identical to the sequence determined by the PCR-based protocol. All of this led us to the conclusion that the low-molecular-weight DNA synthesized by serovar Enteritidis is a small double-stranded DNA (sdsDNA) with single-stranded overhangs (Fig. 6).
FIG. 6.
Expected structure of sdsDNA. The total length was determined to be 72 nt, with a central double-stranded core of 38 bp and 19- or 15-nt-long single-stranded overhangs.
Sequence analysis of the locus encoding sdsDNA.
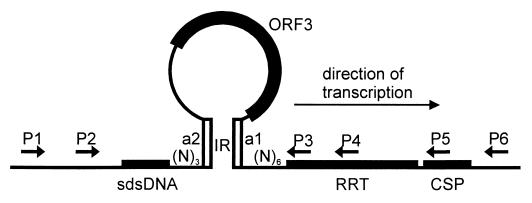
Sequence analysis of the part of the plasmid encoding the RRT and sdsDNA revealed a 13-bp-long inverted repeat which started 3 nt downstream from the sdsDNA (positions 2306 to 2318 included), and the complementary repeat (positions 2578 to 2590 included) ended 6 bp upstream of the start codon of the RRT. The loop formed by the inverted repeat contained the whole ORF3, the termination codon of which was located 2 bp upstream of the start of the inverted repeat (Fig. 7). This loop was also predicted by the MFold calculation (not shown).
FIG. 7.
Detailed description of RRT locus. Primers P1 and P4 were used together with low-molecular-weight DNA in PCRs. Finally, amplification products originating from P1-sdsDNA and P4-sdsDNA PCRs were cloned and sequenced. The P1-P5 and P1-P6 primer pairs were used in the cloning of the DNA fragments necessary for sdsDNA production and phage resistance. Cloning of the P1-P5 PCR product resulted in the pRT7 plasmid, and cloning of the P1-P6 PCR product resulted in the pRT21 plasmid. MFold calculation confirmed the predicted ORF3 loop. Calculation was performed on the sequence starting from the position 2000 of the plasmid and ending at bp 3000. The figure illustrates the ORF3 predicted loop.
Identification of genes essential for the sdsDNA production and phage resistance.
Finally we verified the function of ORF3, ORF4 (rrt), and ORF5 (csp) in the production of the sdsDNA and in the resistance to phage infection. Using PCR, two different fragments of plasmid I were amplified and cloned to form plasmids pRT7 and pRT21. Plasmid pRT7 contained the sdsDNA region, ORF3, and ORF4, and plasmid pRT21 contained the same genes and also ORF5. Only plasmid pRT21, which contained the sdsDNA region together with RRT and the ORF5 located downstream of the RRT, allowed the synthesis of sdsDNA in Salmonella serovar Enteritidis 2160. However, these plasmids (pRT7 and pRT21) were quite unstable at 37°C in Salmonella serovar Enteritidis 2160 in the absence of ampicillin and made the phage typing difficult to assay.
Therefore we performed insertional mutagenesis on plasmid I, which enabled a proper investigation of the role of individual ORFs in the biosynthesis of the sdsDNA and also in phage resistance. An ampicillin resistance gene cassette was inserted into the NsiI site where no ORF was predicted, into the SacI site to interrupt the ORF3 sequence, into the EcoRI site to interrupt the RRT, and into the SfcI site of the plasmid to inactivate the cold-shock protein homologue (Fig. 2). When these plasmids were transformed into Salmonella serovar Enteritidis 2160, the sdsDNA synthesis was restored only when the ampicillin resistance gene cassette was inserted in the NsiI site, outside all the predicted ORFs. The original phage type, PT21, was not restored when the ampicillin resistance gene cassette was inserted in the SacI site in the ORF3 (Table 1). The remaining three recombinant plasmids restored phage resistance to the host strain, Salmonella serovar Enteritidis 2160, although plasmid I with the ampicillin resistance cassette inserted in the NsiI site gave sometimes ambiguous results in phage typing.
TABLE 1.
List of Salmonella strains used in the study and their abilities to synthesize sdsDNA and to resist phage infection
| Strain | Plasmid type (characteristic[s]) | Interrupted ORF | Presence of sdsDNA synthesis | Phage type |
|---|---|---|---|---|
| Serovar Enteritidis 2159 | SE55IJ | None | + | PT21 |
| Serovar Enteritidis 2160 | SE55J | − | PT1 | |
| Serovar Enteritidis 2160 | SE55J (pRT21) | + | ?a | |
| Serovar Enteritidis 2160 | SE55J (pRT7) | − | ? | |
| Serovar Enteritidis 2160 | SE55J (I-Amp-NsiI) | None | + | PT1/PT21b |
| Serovar Enteritidis 2160 | SE55J (I-Amp-SacI) | ORF3 | − | PT1 |
| Serovar Enteritidis 2160 | SE55J (I-Amp-EcoRI) | ORF4/rrt | − | PT21c |
| Serovar Enteritidis 2160 | SE55J (I-Amp-SfcI) | ORF5/csp | − | PT21c |
?, virtually impossible to phage type due to the plasmid instability.
Phage typing of this strain gave ambiguous results, sometimes resulting in the phage type PT21 and in another experiment retaining the phage type PT1.
Phage typing of these strains occasionally resulted in the phage type PT6a, which is, however, similar to PT21, since strains of both the phage types are resistant to the phages P3, P5, and P7, to which strains of the phage type PT1 are sensitive.
DISCUSSION
In the present study we report on the low-molecular-weight plasmid which encodes the RRT and influences resistance to phage infection in Salmonella serovar Enteritidis. RRTs are quite ubiquitous in Myxococcus spp. (8, 19) and rather rare in field strains of E. coli and Salmonella (14, 27). They have been reported to be chromosomally encoded or phage encoded. Here the first evidence for plasmid-encoded reverse transcriptase is presented. Furthermore, we have shown that the enzyme is encoded by a very small plasmid, the size of which was only 4,053 bp.
Most of the so-far-described RRTs catalyze synthesis of msDNA. Our results surprisingly show that the DNA produced by the RRT of serovar Enteritidis is double-stranded DNA with single-stranded overhangs. Because RNase A treatment of natural sdsDNA repeatedly resulted in an increase in sdsDNA quantity, it seems probable that during its biosynthesis it is bound to an RNA molecule. However, this RNA molecule must be of a greater molecular size, definitively more than several hundred nucleotides, since we should have detected molecules below this limit after blotting and hybridization (Fig. 5). The sdsDNA was also free of any DNA-RNA hybrid molecule, since the sdsDNA was totally resistant to RNase H treatment. S1 nuclease treatment, on the other hand, always increased the mobility of sdsDNA in polyacrylamide gel electrophoresis, indicating that single-stranded structures are present in the molecule. We found that the sdsDNA was efficiently used by Taq polymerase as a primer in PCR and, consistent with the suggested model of double-stranded DNA, the sdsDNA could prime the PCR in both possible directions. Successful direct cloning of the sdsDNA after incubation with Taq polymerase indicates that the single-stranded overhangs are 5′ ends of both strands of the molecule—only in this case, the 3′ recessing ends could be efficiently used by Taq polymerase as a template. This could be also a reason that we did not succeed in direct cloning of the sdsDNA treated with the S1 nuclease—already blunt-ended sdsDNA was probably not as effective a template for the Taq polymerase as the molecule with 3′ recessing ends. We succeeded, however, in amplifying, cloning, and sequencing the PCR products derived from the PCR in which sdsDNA treated with S1 nuclease and external primers of both possible orientations were used. Comparison of the sequences of the two PCR products allowed us to identify an overlap of 38 bp, which represented the S1 nuclease treatment-resistant part of the sdsDNA. Double-stranded DNA with 5′-end single-stranded overhangs has not been reported so far to be a product of RRT. Although most of the reports describe msDNA as a single-stranded DNA bound by an unusual 2′-5′ phosphodiester bond to RNA (15, 27), there are studies reporting that the final product could be RNA-free single-stranded DNA (24), and at least one study on in vitro synthesis showed that RRT is capable of synthesis of double-stranded DNA molecules (22).
The RRT of Salmonella serovar Enteritidis possessed additional unique properties. The RRT itself was not enough to initiate the production of sdsDNA. A functional, downstream-located sequence coding for a peptide of 86 amino acids homologous to cold-shock proteins was necessary for the production of the sdsDNA. In M. xanthus, the msDNA was shown to form a complex with multiple proteins in the cell (37), and so it is tempting to speculate on the similar role of csp in the biosynthesis of the sdsDNA, although it might be also possible that the mere insertion of an approximately 1-kb sequence of the Ampr gene cassette influences the secondary structure of the RNA transcript to an extent incompatible with the reverse transcription. Next, the structure of the whole locus was different from that described in previous reports on RRTs. The inverted repeats a1 and a2 (15, 18, 23), 13 bp long in this case, could be identified, but the sdsDNA coding region was located not inside but outside the loop formed by the inverted repeats. Instead, inside the loop formed by the a1 and a2 inverted repeats, an additional ORF3, of no homology to GenBank entries, was located.
Plasmid I was the first identified as being linked with resistance to phage infection. Strains bearing this plasmid were of phage type PT21, while a strain which spontaneously lost this plasmid was of phage type PT1. The difference between PT1 and PT21 strains is mainly in resistances to the phages P3, P5, and P7, to which strains of the PT21 group are resistant and strains belonging to the PT1 group are sensitive (38). Surprisingly, the resistance to phage infection was independent of the sdsDNA production, and it was also independent of functional ORF4 (rrt) and ORF5 (csp). It was, on the other hand, absolutely dependent on functional ORF3. Insertion upstream of the ORF3 into the NsiI site, although it did not influence the sdsDNA synthesis, gave ambiguous results in phage resistance (Table 1). We therefore conclude that the phage resistance and sdsDNA production do not correlate. How the phage resistance is expressed and the sdsDNA is synthesized are currently unknown; however, we speculate that the 13-bp inverted repeats and the loop structure containing the ORF3 are of particular importance. Formation of such a loop on the mRNA level could lead to a single gene translation of ORF3 and could also put the sdsDNA locus in a close proximity to the rrt gene.
The biological role of msDNA in Myxococcus and E. coli remains unclear (28). It was considered to be mutagenic (4, 20, 25), and it was also shown to be induced in stationary phage (13). In our study, we have shown that the plasmid I encodes the RRT and the presence of the plasmid correlates with the synthesis of sdsDNA. The plasmid is also involved in resistance to phage infection. Plasmid I therefore represents a very interesting, self-replicating model of a DNA molecule encoding the RRT with multiple unique features.
ACKNOWLEDGMENTS
This work has been supported by grants EP6076 and QC0195 from the Ministry of Agriculture of the Czech Republic.
REFERENCES
- 1.Akada R. Quick-check method to test the size of Escherichia coli plasmids. BioTechniques. 1994;17:58. [PubMed] [Google Scholar]
- 2.Bhattacharjee M K, Meyer R J. Specific binding of MobA, a plasmid-encoded protein involved in the initiation and termination of conjugal DNA transfer, to single-stranded oriT DNA. Nucleic Acids Res. 1993;21:4563–4568. doi: 10.1093/nar/21.19.4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridges B A. Starvation-associated mutation in E. coli strains with and without reverse transcriptase. Mutat Res. 1995;347:13–15. doi: 10.1016/0165-7992(95)90026-8. [DOI] [PubMed] [Google Scholar]
- 5.Brown D J, Threlfall E J, Hampton M D, Rowe B. Molecular characterization of plasmids in Salmonella enteritidis phage types. Epidemiol Infect. 1993;110:209–216. doi: 10.1017/s0950268800068126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buisan M, Rodriguez-Pena J M, Rotger R. Restriction map of the Salmonella enteritidis virulence plasmid and its homology with the plasmid of Salmonella typhimurium. Microb Pathog. 1994;16:165–169. doi: 10.1006/mpat.1994.1017. [DOI] [PubMed] [Google Scholar]
- 7.Dhundale A, Lampson B, Furuichi T, Inouye M, Inouye S. Structure of msDNA from Myxococcus xanthus: evidence for a long, self-annealing RNA precursor for the covalently linked, branched RNA. Cell. 1987;51:1105–1112. doi: 10.1016/0092-8674(87)90596-4. [DOI] [PubMed] [Google Scholar]
- 8.Dhundale A R, Furuichi T, Inouye S, Inouye M. Distribution of multicopy single-stranded DNA among myxobacteria and related species. J Bacteriol. 1985;164:914–917. doi: 10.1128/jb.164.2.914-917.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodd I B, Egan J B. The Escherichia coli retrons Ec67 and Ec86 replace DNA between the cos site and a transcription terminator of a 186-related prophage. Virology. 1996;219:115–124. doi: 10.1006/viro.1996.0228. [DOI] [PubMed] [Google Scholar]
- 10.Dorn C R, Silapanuntakul R, Angrick E J, Shipman L D. Plasmid analysis of Salmonella enteritidis isolated from human gastroenteritis cases and from epidemiologically associated poultry flocks. Epidemiol Infect. 1993;111:239–243. doi: 10.1017/s0950268800056946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gado I, Laszlo V G, Nagy B, Milch H, Drin I, Awad-Masalmeh M, Horvath J. Phage restriction and the presence of small plasmids in Salmonella enteritidis. Int J Med Microbiol. 1998;287:509–519. doi: 10.1016/s0934-8840(98)80192-8. [DOI] [PubMed] [Google Scholar]
- 12.Halavatkar H, Barrow P A. The role of a 54-kb plasmid in the virulence of strains of Salmonella enteritidis of phage type 4 for chickens and mice. J Med Microbiol. 1993;38:171–176. doi: 10.1099/00222615-38-3-171. [DOI] [PubMed] [Google Scholar]
- 13.Herzer P J. Starvation-induced expression of retron-Ec107 and the role of ppGpp in multicopy single-stranded DNA production. J Bacteriol. 1996;178:4438–4444. doi: 10.1128/jb.178.15.4438-4444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herzer P J, Inouye S, Inouye M, Whittam T S. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu M Y, Eagle S G, Inouye M, Inouye S. Cell-free synthesis of the branched RNA-linked msDNA from retron-Ec67 of Escherichia coli. J Biol Chem. 1992;267:13823–13829. [PubMed] [Google Scholar]
- 16.Ibanez M, Rotger R. Characterization of a small cryptic plasmid from Salmonella enteritidis that affects the growth of Escherichia coli. FEMS Microbiol Lett. 1993;109:225–230. doi: 10.1111/j.1574-6968.1993.tb06172.x. [DOI] [PubMed] [Google Scholar]
- 17.Inouye S, Sunshine M G, Six E W, Inouye M. Retronphage phi R73: an E. coli phage that contains a retroelement and integrates into a tRNA gene. Science. 1991;252:969–971. doi: 10.1126/science.1709758. [DOI] [PubMed] [Google Scholar]
- 18.Lampson B C, Inouye M, Inouye S. Reverse transcriptase with concomitant ribonuclease H activity in the cell-free synthesis of branched RNA-linked msDNA of Myxococcus xanthus. Cell. 1989;56:701–707. doi: 10.1016/0092-8674(89)90592-8. [DOI] [PubMed] [Google Scholar]
- 19.Lampson B C, Inouye M, Inouye S. Survey of multicopy single-stranded DNAs and reverse transcriptase genes among natural isolates of Myxococcus xanthus. J Bacteriol. 1991;173:5363–5370. doi: 10.1128/jb.173.17.5363-5370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lampson B C, Rice S A. Repetitive sequences found in the chromosome of the myxobacterium Nannocystis exedens are similar to msDNA: a possible retrotransposition event in bacteria. Mol Microbiol. 1997;23:813–823. doi: 10.1046/j.1365-2958.1997.2671627.x. [DOI] [PubMed] [Google Scholar]
- 21.Lampson B C, Sun J, Hsu M Y, Vallejo-Ramirez J, Inouye S, Inouye M. Reverse transcriptase in a clinical strain of Escherichia coli: production of branched RNA-linked msDNA. Science. 1989;243:1033–1038. doi: 10.1126/science.2466332. [DOI] [PubMed] [Google Scholar]
- 22.Lampson B C, Viswanathan M, Inouye M, Inouye S. Reverse transcriptase from Escherichia coli exists as a complex with msDNA and is able to synthesize double-stranded DNA. J Biol Chem. 1990;265:8490–8496. [PubMed] [Google Scholar]
- 23.Lim D, Maas W K. Reverse transcriptase-dependent synthesis of a covalently linked, branched DNA-RNA compound in E. coli B. Cell. 1989;56:891–904. doi: 10.1016/0092-8674(89)90693-4. [DOI] [PubMed] [Google Scholar]
- 24.Lima T M, Lim D. A novel retron that produces RNA-less msDNA in Escherichia coli using reverse transcriptase. Plasmid. 1997;38:25–33. doi: 10.1006/plas.1997.1298. [DOI] [PubMed] [Google Scholar]
- 25.Maas W K, Wang C, Lima T, Zubay G, Lim D. Multicopy single-stranded DNAs with mismatched base pairs are mutagenic in Escherichia coli. Mol Microbiol. 1994;14:437–441. doi: 10.1111/j.1365-2958.1994.tb02178.x. [DOI] [PubMed] [Google Scholar]
- 26.Millemann Y, Lesage M C, Chaslus-Dancla E, Lafont J P. Value of plasmid profiling, ribotyping, and detection of IS200 for tracing avian isolates of Salmonella typhimurium and S. enteritidis. J Clin Microbiol. 1995;33:173–179. doi: 10.1128/jcm.33.1.173-179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice S A, Bieber J, Chun J Y, Stacey G, Lampson B C. Diversity of retron elements in a population of rhizobia and other gram-negative bacteria. J Bacteriol. 1993;175:4250–4254. doi: 10.1128/jb.175.13.4250-4254.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rice S A, Lampson B C. Bacterial reverse transcriptase and msDNA. Virus Genes. 1996;11:95–104. doi: 10.1007/BF01728651. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Pena J M, Alvarez I, Ibanez M, Rotger R. Homologous regions of the Salmonella enteritidis virulence plasmid and the chromosome of Salmonella typhi encode thiol: disulphide oxidoreductases belonging to the DsbA thioredoxin family. Microbiology. 1997;143:1405–1413. doi: 10.1099/00221287-143-4-1405. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Pena J M, Buisan M, Ibanez M, Rotger R. Genetic map of the virulence plasmid of Salmonella enteritidis and nucleotide sequence of its replicons. Gene. 1997;188:53–61. doi: 10.1016/s0378-1119(96)00776-7. [DOI] [PubMed] [Google Scholar]
- 31.Rychlik I, Karpiskova R, Faldynova M, Sisak F. Computer-assisted restriction endonuclease analysis of plasmid DNA in field strains of Salmonella enteritidis. Can J Microbiol. 1998;44:1183–1185. [PubMed] [Google Scholar]
- 32.Rychlik I, Svestkova A, Karpiskova R. Subdivision of Salmonella enterica serovar Enteritidis phage-types PT21 and PT14b by plasmid profiling. Vet Microbiol. 2000;74:217–225. doi: 10.1016/s0378-1135(00)00185-1. [DOI] [PubMed] [Google Scholar]
- 33.Stubbs A D, Hickman-Brenner F W, Cameron D N, Farmer J J. Differentiation of Salmonella enteritidis phage type 8 strains—evaluation of three additional phage typing systems, plasmid profiles, antibiotic susceptibility patterns, and biotyping. J Clin Microbiol. 1994;32:199–201. doi: 10.1128/jcm.32.1.199-201.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tassios P T, Markogiannakis A, Vatopoulos A C, Katsanikou E, Velonakis E N, Kourea-Kremastinou J, Legakis N J. Molecular epidemiology of antibiotic resistance of Salmonella enteritidis during a 7-year period in Greece. J Clin Microbiol. 1997;35:1316–1321. doi: 10.1128/jcm.35.6.1316-1321.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Temin H M. Reverse transcriptases. Retrons in bacteria. Nature. 1989;339:254–255. doi: 10.1038/339254a0. [DOI] [PubMed] [Google Scholar]
- 36.Vatopoulos A C, Mainas E, Balis E, Threlfall E J, Kanelopoulou M, Kalapothaki V, Malamoulada H, Legakis N J. Molecular epidemiology of ampicillin-resistant clinical isolates of Salmonella enteritidis. J Clin Microbiol. 1994;32:1322–1325. doi: 10.1128/jcm.32.5.1322-1325.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viswanathan M, Inouye M, Inouye S. Myxococcus xanthus msDNA. Mx162 exists as a complex with proteins. J Biol Chem. 1989;264:13665–13671. [PubMed] [Google Scholar]
- 38.Ward L R, de Sa J D H, Rowe B. A phage-typing scheme for Salmonella enteritidis. Epidemiol Infect. 1987;99:291–294. doi: 10.1017/s0950268800067765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woodward M J, Allen-Vercoe E, Redstone J S. Distribution, gene sequence and expression in vivo of the plasmid encoded fimbrial antigen of Salmonella serotype Enteritidis. Epidemiol Infect. 1996;117:17–28. doi: 10.1017/s0950268800001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yee T, Furuichi T, Inouye S, Inouye M. Multicopy single-stranded DNA isolated from a gram-negative bacterium, Myxococcus xanthus. Cell. 1984;38:203–209. doi: 10.1016/0092-8674(84)90541-5. [DOI] [PubMed] [Google Scholar]