Abstract
Candida dubliniensis is a recently described opportunistic fungal pathogen that is closely related to Candida albicans but differs from it with respect to epidemiology, certain virulence characteristics, and the ability to develop fluconazole resistance in vitro. A comparison of C. albicans and C. dubliniensis at the molecular level should therefore provide clues about the mechanisms used by these two species to adapt to their human host. In contrast to C. albicans, no auxotrophic C. dubliniensis strains are available for genetic manipulations. Therefore, we constructed homozygous ura3 mutants from a C. dubliniensis wild-type isolate by targeted gene deletion. The two URA3 alleles were sequentially inactivated using the MPAR-flipping strategy, which is based on the selection of integrative transformants carrying a mycophenolic acid resistance marker that is subsequently deleted again by site-specific, FLP-mediated recombination. The URA3 gene from C. albicans (CaURA3) was then used as a selection marker for targeted integration of a fusion between the C. dubliniensis MDR1 (CdMDR1) promoter and a C. albicans-adapted GFP reporter gene. Uridine-prototrophic transformants were obtained with high frequency, and all transformants of two independent ura3-negative parent strains had correctly integrated the reporter gene fusion into the CdMDR1 locus, demonstrating that the CaURA3 gene can be used for efficient and specific targeting of recombinant DNA into the C. dubliniensis genome. Transformants carrying the reporter gene fusion did not exhibit detectable fluorescence during growth in yeast extract-peptone-dextrose medium in vitro, suggesting that CdMDR1 is not significantly expressed under these conditions. Fluconazole had no effect on MDR1 expression, but the addition of the drug benomyl strongly activated the reporter gene fusion in a dose-dependent fashion, demonstrating that the CdMDR1 gene, which encodes an efflux pump mediating resistance to toxic compounds, is induced by the presence of certain drugs.
Several yeast species within the genus Candida are opportunistic pathogens of humans, Candida albicans being by far the most frequently isolated and also the most pathogenic species (20). Recently, a previously unrecognized Candida species, Candida dubliniensis, has been described (28). C. dubliniensis is closely related to C. albicans, and before the phylogenetic separation of the two species was recognized and reliable phenotypic and molecular markers to distinguish them became established, C. dubliniensis was often misidentified as C. albicans (27). Whereas C. albicans is a member of the normal microflora in most healthy persons, C. dubliniensis has been isolated primarily from the oral cavities of human immunodeficiency virus (HIV)-infected individuals and AIDS patients. However, recent epidemiological studies have also identified C. dubliniensis in cases of oral infection in non-HIV-infected individuals and in cases of septicemia, although at a lower incidence than C. albicans (3, 13, 21, 23, 29). These observations still have to be confirmed on a broad scale using recently identified, easily screenable, species-specific phenotypic markers (25). However, the current evidence suggests that C. albicans and C. dubliniensis differ with respect to their epidemiology and, consequently, in their capacity to colonize and infect specific host niches. Of particular interest in this respect are the reported differences in certain phenotypic characteristics that are linked to virulence, such as adherence to epithelial cells, formation of hyphae, and proteinase production (9, 12), the relative importance of which probably depends on the colonization site and the infection stage. Of potential clinical significance also is the ability of C. dubliniensis to develop fluconazole resistance very rapidly in vitro, which is difficult to achieve with C. albicans, although fluconazole-resistant C. albicans is frequently isolated from patients (16). Similar mechanisms seem to be responsible for fluconazole resistance in the two species (15, 24), but the molecular basis for the higher propensity of C. dubliniensis compared with C. albicans to develop drug resistance in vitro is unknown.
Comparative analysis of two such related, but nevertheless distinct, species should facilitate a better understanding of their biology and mechanisms of pathogenicity. By revealing differences, for example, in the relative importance of virulence traits or in the regulation of the corresponding genes, insights into the specific adaptation mechanisms of each of the two species to their host should be gained. This goal can best be achieved if the full arsenal of molecular methods becomes available for both species.
The molecular genetic analysis of C. albicans is based mainly on the use of the URA3 gene as a marker for the selection of prototrophic transformants of ura3-auxotrophic host strains. Since the URA3 marker can be both positively and negatively selected for, it can be used as a single, recyclable marker, for example, in the construction of specific mutants by sequential, targeted gene disruptions, but also for the introduction of reporter gene fusions to analyze gene expression. The construction of a ura3 mutant by targeted gene deletion from a C. albicans wild-type strain by Fonzi and Irwin was a significant milestone in C. albicans genetics (6). Insertion of recombinant DNA into a specific locus in the C. albicans genome is straightforward, since homologous recombination is very efficient in this organism. Nevertheless, at that time the construction of such a strain required considerable effort because C. albicans is a diploid organism without a known sexual cycle, and two rounds of mutagenesis are necessary to obtain homozygous mutants. Deletion of the first URA3 allele would not result in a detectable phenotype and, in the absence of available dominant selection markers, the desired heterozygous mutants had to be identified and isolated by a tedious PCR-based approach combined with sibling selection (6). Subsequent inactivation of the second URA3 allele was then more readily achieved because ura3 mutants could be identified by selection with 5-fluoro-orotic acid (FOA) (2). The resulting ura3 strain, CAI4, is otherwise isogenic with the clinical C. albicans isolate SC5314 from which it was derived, and it has since been used as a model strain for the majority of molecular work in C. albicans. The development of CAI4 resulted in an explosion in the number of studies addressing many aspects of C. albicans biology and pathogenicity and, therefore, contributed much to our present knowledge about this fungus (4, 22).
For a comparison of C. albicans and C. dubliniensis at the molecular level, a similar ura3-negative C. dubliniensis host strain for use in genetic experiments would be extremely valuable. Recently, a dominant selection marker for the transformation of C. albicans wild-type strains became available (30), and it has been shown that this marker, which confers resistance to mycophenolic acid (MPA), can also be used for the selection of integrative C. dubliniensis transformants (26), which should allow the direct selection of heterozygous mutants. Although there is no negative selection scheme for deletion of the MPAR marker from heterozygous mutants to allow its use for inactivation of the second allele of the target gene, this problem was circumvented in C. albicans by the development of the MPAR-flipping strategy (18, 31). This mutagenesis scheme is based on the use of a cassette that contains, in addition to the selection marker, a C. albicans-adapted FLP gene (caFLP) encoding the site-specific recombinase FLP from Saccharomyces cerevisiae under the control of the inducible C. albicans SAP2 promoter. The cassette is flanked by direct repeats of the FLP recognition site such that it can be excised from the genome by induced, FLP-mediated recombination after specific chromosomal insertion, leaving behind a disrupted copy of the target gene. A second round of mutagenesis then generates the desired homozygous mutants, which are otherwise isogenic with the wild-type parent strain. The findings that the MPAR marker functions in C. dubliniensis and that the SAP2 promoter from C. albicans is regulated in the same way also in the heterologous species C. dubliniensis (26) suggested that the MPAR-flipping strategy could be used to generate specific mutants from C. dubliniensis wild-type strains by sequential, targeted gene disruption. Therefore, we set out to construct a C. dubliniensis ura3 mutant that could serve as a host strain for efficient genetic manipulations aimed at comparing various aspects in which C. dubliniensis may differ from C. albicans, such as morphogenesis, virulence, and drug resistance.
MATERIALS AND METHODS
Strains and growth media.
The C. dubliniensis strains used in this study are listed in Table 1. For routine growth of the strains, YPD liquid medium (containing, per liter, 10 g of yeast extract, 20 g of peptone, and 20 g of glucose) was used. Cells were grown overnight in YCB-BSA (23.4 g of yeast carbon base and 4 g of bovine serum albumin per liter [pH 4.0]) to induce the SAP2 promoter for excision of the MPAR flipper from MPA-resistant transformants. To screen for MPA-sensitive derivatives, 100 to 200 CFU was plated on minimal agar (containing, per liter, 6.7 g of yeast nitrogen base without amino acids [YNB; Bio 101, Vista, Calif.], 20 g of glucose, 0.77 g of complete supplement medium [CSM; Bio 101], and 15 g of agar) containing 1 μg of MPA ml−1, which resulted in the generation of large MPAr and small MPAs colonies, respectively (31). Uridine (100 μg ml−1) was added to the media to support the growth of ura3 mutant strains.
TABLE 1.
C. dubliniensis strains used in this study
| Strain | Parent | Relevant genotypea | Source or reference |
|---|---|---|---|
| Wü284 | Clinical isolate | 19 | |
| CdUM1A | Wü284 | ura3Δ1::MPAR-FLIP/URA3 | This study |
| CdUM1B | Wü284 | ura3Δ1::MPAR-FLIP/URA3 | This study |
| CdUM2A | CdUM1A | ura3Δ1::FRT/URA3 | This study |
| CdUM2B | CdUM1B | ura3Δ1::FRT/URA3 | This study |
| CdUM3A | CdUM2A | ura3Δ1::FRT/ ura3Δ2::MPAR-FLIP | This study |
| CdUM3B | CdUM2B | ura3Δ1::FRT/ ura3Δ2::MPAR-FLIP | This study |
| CdUM4A | CdUM3A | ura3Δ1::FRT/ ura3Δ2::FRT | This study |
| CdUM4B | CdUM3B | ura3Δ1::FRT/ ura3Δ2::FRT | This study |
| CdMGFP1A | CdUM4A | MDR1/mdr1::PCdMDR1-GFP | This study |
| CdMGFP1B | CdUM4B | MDR1/mdr1::PCdMDR1-GFP | This study |
MPAR-FLIP denotes the MPAR flipper cassette.
Cloning of the CdURA3 gene.
A genomic library from C. dubliniensis strain CD36 (5, 15) was screened with a C. dubliniensis URA3 (CdURA3) gene probe, generated using primers derived from the C. albicans URA3 (CaURA3) gene. A 2-kb SpeI-HindIII fragment from a positive clone was subcloned into pBluescript and completely sequenced. In a parallel approach, the CdURA3 downstream region was PCR amplified with primers URA17 (5′-CTATTTACAATCTCGAGGTGGTCCTTC-3′) and URA18 (5′-CCATTAATTGCGAGCTCTGCTACTGGAG-3′), derived from the CaURA3 sequence (http://www-sequence.stanford.edu/group/candida), using genomic DNA from C. dubliniensis strain Wü284 as a template. The PCR product was digested at the introduced XhoI and SacI sites (underlined), cloned into pBluescript, and sequenced. The overlapping sequences of the genomic clone and the PCR product were combined and used to select primers for the amplification of CdURA3-flanking sequences in gene disruption experiments.
Construction of CdURA3 gene deletion cassettes.
Two different deletion constructs were made to inactivate both URA3 alleles in C. dubliniensis. For pSFIcdU2, CdURA3 sequences from position −799 to +53 and from position +797 to +1712 (with respect to the start codon) were PCR amplified with the primer pair CdURA1 (5′-AGAACGCATGCCAAGTTTGATAGTACTG-3′) and CdURA2 (5′-TGTGCCCGCGGTGAAGCATGAGTCTCTGC-3′) and the primer pair URA21 (5′-ATGCTTATTTGCTCGAGACTGGCCAA-3′) and CdURA4 (5′-GATACTTGGAGAGCTCGATTTGTTGCTGGTGC-3′), respectively, thereby introducing SphI, SacII, XhoI, and SacI restriction sites (underlined). The SphI-SacII CdURA3 upstream fragment and the XhoI-SacI CdURA3 downstream fragment were then cloned on both sides of a SacII-XhoI fragment containing the MPAR flipper from pSFI1 (31) in the SphI/SacI-digested vector pBluescript to generate pSFIcdU2 (Fig. 1A, top). Similarly, for pSFIcdU4, CdURA3 sequences from position −799 to +225 and from position +620 to +1712 were amplified with the primer pair CdURA1 and CdURA5 (5′-CTAATAATGGTTCCGCGGTAGATTCATA-3′) and the primer pair CdURA6 (5′-TATTATGACTCCTCGAGTTGGATTAGA-3′) and CdURA4, respectively (the introduced SacII and XhoI restriction sites are underlined). The SphI-SacII CdURA3 upstream fragment and the XhoI-SacI CdURA3 downstream fragment were substituted for the corresponding CdURA3-flanking regions in pSFIcdU2, resulting in pSFIcdU4 (Fig. 1A, bottom). The SphI-SacI fragments from pSFIcdU2 and pSFIcdU4 were used in transformation experiments.
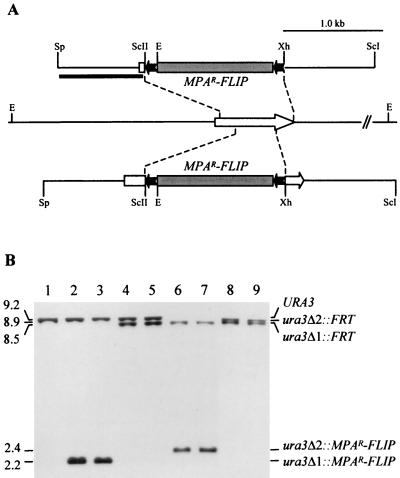
FIG. 1.
Inactivation of the CdURA3 gene by MPAR flipping. (A) Structure of the CdURA3 locus in strain Wü284 and allelic replacements using the inserts from pSFIcdU2 (upper part) or pSFIcdU4 (lower part). Open arrow, CdURA3 coding region; solid lines, CdURA3 upstream and downstream sequences. Only relevant restriction sites are shown: E, EcoRI; ScI, SacI; ScII, SacII; Sp, SphI; Xh, XhoI. The 5.6-kb MPAR flipper, details of which have been presented elsewhere (31), is not drawn to scale. Solid bar, DNA fragment used as a probe to verify the correct allelic replacements by Southern hybridization. (B) Southern hybridization of EcoRI-digested genomic DNA of the ura3 mutants using the 5′ CdURA3 fragment from pSFIcdU2 as a probe. The identities of the fragments are shown to the right of the blot, and molecular sizes are given on the left. Lanes: 1, Wü284 (URA3/URA3); 2, CdUM1A (ura3Δ1::MPAR-FLIP/URA3); 3, CdUM1B (ura3Δ1::MPAR-FLIP/URA3); 4, CdUM2A (ura3Δ1::FRT/URA3); 5, CdUM2B (ura3Δ1::FRT/URA3); 6, CdUM3A (ura3Δ1::FRT/ura3Δ2::MPAR-FLIP); 7, CdUM3B (ura3Δ1::FRT/ura3Δ2::MPAR-FLIP); 8, CdUM4A (ura3Δ1::FRT/ura3Δ2::FRT); 9, CdUM4B (ura3Δ1::FRT/ ura3Δ2::FRT). ura3Δ1 and ura3Δ2 indicate the deletions generated using the inserts from pSFIcdU2 and pSFIcdU4, respectively.
Construction of a PCdMDR1-GFP reporter gene fusion.
A CdMDR1 promoter fragment (positions −943 to −8 with respect to the CdMDR1 start codon) was obtained by PCR amplification from genomic DNA of strain Wü284 with primers CdMDR5 (5′-ATATATCTAGACTTACAAGATAGGTAAAG-3′) and CdMDR6 (5′-GCATTGTCGACAATTGTGTTTTCTTTGGTTGAG-3′), thereby introducing XbaI and SalI restriction sites (underlined). A CdMDR1 downstream fragment (positions +1564 to +2382) was amplified with primers CdMDR7 (5′-CCCGAATATCCTGCAGCCTGGGGTAGTTC-3′) and CdMDR8 (5′-ACTCACCCCACTAGAGCTCCAGTTTACAC-3′), thereby introducing PstI and SacI sites (underlined). The CdMDR1 fragments were used to replace the flanking SAP2 sequences in the previously described plasmid pGFP41 (17), yielding pCdMGFP2 (Fig. 2A). The XbaI-SacI fragment from this plasmid was used to transform the C. dubliniensis ura3 mutants.
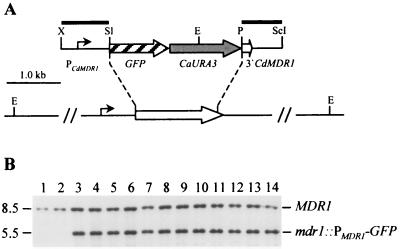
FIG. 2.
Integration of the PCdMDR1-GFP reporter gene fusion into the MDR1 locus of strains CdUM4A and CdUM4B. (A) Genomic structure of the MDR1 locus in strain Wü284 and its ura3 derivatives and structure of the inserted reporter gene fusion from plasmid pCdMGFP2. Open arrow, MDR1 coding region; solid lines, flanking upstream and downstream sequences. The position of the CdMDR1 promoter (PCdMDR1) and the direction of transcription are indicated by the solid arrow. Only relevant restriction sites are shown: E, EcoRI; P, PstI; ScI, SacI; Sl, SalI; X, XbaI. Solid bars indicate the DNA fragments used as probes for verification of the correct allelic replacement by Southern hybridization. (B) Southern hybridization of EcoRI-digested genomic DNA of the parent strains CdUM4A (lane 1) and CdUM4B (lane 2) and transformants carrying the PCdMDR1-GFP reporter gene fusion inserted into one of the MDR1 alleles using the PCdMDR1 fragment as a probe. Lanes 3 to 8 transformants of CdUM4A; lanes 9 to 14, transformants of CdUM4B. The identities of the fragments are shown to the right of the blot, and molecular sizes are given on the left.
C. dubliniensis transformation.
C. dubliniensis strains were transformed by electroporation (11). Cells from a YPD preculture were diluted 10−4 in 50 ml of fresh YPD medium and grown overnight at 30°C to an optical density at 600 nm (OD600) of 1.6 to 2.2, which yielded the best transformation efficiency in our hands. The cells were collected by centrifugation and resuspended in 8 ml of water. After addition of 1 ml of 10× TE (100 mM Tris-HCl, 10 mM EDTA [pH 7.5]) and 1 ml of 1 M lithium acetate (pH 7.5), the suspension was incubated in a rotary shaker at 150 rpm for 60 min at 30°C. A 250-μl volume of 1 M dithiothreitol was then added, and the cells were incubated for a further 30 min at 30°C with shaking. After addition of 40 ml of water, the cells were centrifuged, washed sequentially in 50 ml of ice-cold water and 10 ml of ice-cold 1 M sorbitol, resuspended in 50 μl of 1 M sorbitol, and kept on ice. The inserts from the plasmids described above were excised with appropriate restriction enzymes and purified by agarose gel electrophoresis and elution with the GeneClean kit from Bio 101. Five microliters (approximately 1 μg) of the linear DNA fragments was mixed with 40 μl of electrocompetent cells, and electroporation was carried out in a Bio-Rad Gene Pulser (0.2 cm cuvette, 1.6 kV, 200 Ω, 25 μF) with a Bio-Rad Pulse Controller included in the circuit. MPA-resistant transformants were selected on minimal agar plates containing 10 μg of MPA ml−1. Single colonies were picked after 5 to 7 days of growth at 30°C, restreaked on the same medium, and, after verification of the correct allelic replacement, maintained on YPD agar plates. Uridine-prototrophic transformants of ura3 mutants were selected on minimal agar plates without uridine and picked after 2 days of growth at 30°C. To determine the proportion of transformed cells, the number of viable cells after electroporation also was determined by plating an appropriate dilution of the cell suspension onto medium supplemented with uridine.
Isolation of chromosomal DNA and Southern hybridization.
Genomic DNA from C. dubliniensis strains was isolated as described by Millon et al. (14). DNA (10 μg) was digested with EcoRI, separated on a 1% (wt/vol) agarose gel, and, after ethidium bromide staining, transferred by vacuum blotting onto a nylon membrane and fixed by UV cross-linking. Southern hybridization with enhanced chemiluminescence (ECL)-labeled probes was performed with the ECL labeling and detection kit from Amersham (Braunschweig, Germany) according to the instructions of the manufacturer.
Drug stock solutions.
Fluconazole (a kind gift from Pfizer UK) was dissolved in water (5 mg ml−1), and benomyl (Sigma-Aldrich Chemie, Taufkirchen, Germany) was dissolved in dimethyl sulfoxide (DMSO; 10 mg ml−1).
Fluorescence microscopy.
Cells from an overnight culture were inoculated into fresh YPD medium and grown at 30°C to mid-log phase, when the indicated amount of drug was added. Aliquots of the cultures were taken at various times of growth in the absence or presence of drugs and spotted on microscope slides. Fluorescence was detected with a Zeiss Axiolab microscope equipped for epifluorescence microscopy with a 50-W mercury high-pressure bulb and the Zeiss fluorescein-specific filter set 09.
Nucleotide sequence accession numbers.
The sequence of CdURA3 obtained from the genomic library of strain CD36 and the CdURA3 downstream sequence obtained from strain Wü284 have been deposited in the EMBL and GenBank nucleotide sequence databases under accession no. AJ302032 and AF328138, respectively.
RESULTS
Cloning of the C. dubliniensis URA3 gene.
Initial attempts to disrupt the CdURA3 gene using a mutagenesis cassette in which the MPAR flipper was flanked by upstream and downstream sequences of the CaURA3 gene were unsuccessful, presumably because of sequence divergence between the two species (26; also unpublished data). Therefore, we cloned and sequenced the CdURA3 gene and flanking sequences in order to identify suitable sequences for the targeted integration of the MPAR flipper cassette into the CdURA3 locus by homologous recombination. PCR primers based on the CaURA3 gene sequence successfully amplified a fragment comprising 1 kb of sequence downstream of the CdURA3 gene of Wü284; however, we were unable to amplify upstream sequences using C. albicans-derived primers. Therefore, a PCR product containing 390 bp of the CdURA3 gene was used as a probe to identify a clone carrying the complete CdURA3 gene from a genomic library of C. dubliniensis strain CD36 (5), and a SpeI-HindIII fragment comprising the CdURA3 coding region plus an 807-bp upstream sequence and a 410-bp downstream sequence was completely sequenced. The CdURA3 sequences obtained from the library clone and the PCR fragment overlapped by 292 bp in the downstream region. These sequences were completely identical, despite the fact that they were derived from two unrelated C. dubliniensis strains. The CdURA3 coding region exhibited 93% similarity with the corresponding C. albicans sequence obtained from the genome sequencing project (http://www-sequence.stanford.edu/group/candida), with 98% identity at the amino acid level. However, the flanking regions showed a much higher sequence divergence, with 88% similarity within the sequenced 1,062-bp downstream region and only 70% similarity within the cloned 807-bp upstream region. This sequence divergence explains our failure to specifically integrate a C. albicans-based disruption construct into the C. dubliniensis genome and to amplify the CdURA3 upstream region with primers derived from the C. albicans sequence.
Construction of C. dubliniensis ura3 mutants by targeted gene disruption.
To facilitate the genetic manipulation of C. dubliniensis we constructed a ura3 mutant of the previously characterized strain Wü284 (19, 25, 26) by targeted gene disruption using the MPAR-flipping strategy (31). Two different deletion constructs were made for inactivation of the two URA3 alleles. In plasmids pSFIcdU2 and pSFIcdU4, the CdURA3 coding sequence from position +54 (with respect to the start codon) to +796 (15 bp in front of the stop codon), or from position +226 to +619, respectively, was replaced by the MPAR flipper, such that the flanking CdURA3 upstream and downstream sequences were of different lengths in the two plasmids (Fig. 1A). Strain Wü284 was first transformed with the insert from pSFIcdU2, and MPA-resistant transformants were analyzed by Southern hybridization. In the parent strain, Wü284, an EcoRI fragment of about 9.2 kb hybridized with the CdURA3 probe (Fig. 1B, lane 1). In 11 out of 12 transformants tested, a single new EcoRI fragment of 2.2 kb appeared, suggesting correct replacement of one of the CdURA3 alleles by the deletion cassette. In the remaining transformant an ectopic integration had occurred in addition to the desired allelic replacement. Two independent transformants that exhibited the new EcoRI fragment of 2.2 kb, termed CdUM1A and CdUM1B (Fig. 1B, lanes 2 and 3), were used to excise the MPAR flipper by induced, FLP-mediated recombination as described previously (31), resulting in strains CdUM2A and CdUM2B, in which the 2.2-kb EcoRI fragment was replaced by an expected 8.5-kb fragment, 673 bp smaller than the original wild-type fragment (Fig. 1B, lanes 4 and 5). The insert from pSFIcdU4 was then used to delete the remaining CdURA3 wild-type allele in strains CdUM2A and CdUM2B. MPA-resistant transformants were selected and screened for uridine auxotrophy. About half of the MPA-resistant transformants were auxotrophic, demonstrating that in both heterozygous parent strains, integration of the deletion cassette occurred with equal efficiency into the already disrupted or the remaining wild-type URA3 allele. Southern hybridization analysis demonstrated the correct replacement of the second CdURA3 allele in strains CdUM3A and CdUM3B (Fig. 1B, lanes 6 and 7), which is evident from the appearance of a 2.4-kb EcoRI fragment instead of the 9.2-kb fragment. Excision of the MPAR flipper from these strains resulted in strains CdUM4A and CdUM4B, in which the 2.4-kb EcoRI fragment was replaced by an expected 8.9-kb fragment, 335 bp smaller than the original wild-type fragment (Fig. 1B, lanes 8 and 9). All 10 tested MPA-sensitive derivatives of the two parent strains exhibited this hybridization pattern, demonstrating that loss of the MPAR marker was caused exclusively by intrachromosomal recombination of the FRT sites flanking the mutagenesis cassette and not by mitotic interchromosomal recombination of FRT sites on homologous chromosomes, which would have produced a different fragment (about 170 bp smaller).
The independently constructed ura3 mutants CdUM4A and CdUM4B had identical growth characteristics in YPD medium supplemented with uridine. In addition, reintroduction of a URA3 gene (see below) restored growth in medium without uridine to the same level as that of the original parent strain, Wü284.
Integration specificity in C. dubliniensis ura3 mutants using CaURA3 as a selection marker.
We next evaluated whether the URA3 gene could be used for selection of prototrophic transformants of the C. dubliniensis ura3 mutants and for targeted integration of recombinant DNA into a specific genomic locus by homologous recombination. The heterologous CaURA3 gene was used for this purpose, since it is widely used for C. albicans molecular genetics and many already existing genetic constructs could serve as the basis for future genetic manipulations of C. dubliniensis without the necessity to exchange the marker. Strains CdUM4A and CdUM4B were transformed by electroporation with a linear DNA fragment containing a C. albicans-adapted green fluorescent protein (GFP) gene under the control of the CdMDR1 promoter (see below), the CaURA3 selection marker, and CdMDR1 downstream sequences. The flanking CdMDR1 sequences served for integration of the reporter gene fusion into one of the CdMDR1 alleles by allelic exchange (Fig. 2A). Uridine-prototrophic transformants of the two parent strains used were obtained with similar frequencies: using approximately 1 μg of the linear DNA fragment, we obtained 300 transformed cells out of 5.4 × 107 viable cells for CdUM4A (5.5 × 10−6) and 950 transformed cells out of 1.36 × 108 viable cells for CdUM4B (7.0 × 10−6). For each parent strain, six independent transformants were analyzed by Southern hybridization. All 12 transformants had correctly integrated the reporter fusion into the CdMDR1 locus, as shown by the appearance of a new 5.5-kb EcoRI fragment in addition to the 8.5-kb wild-type fragment after hybridization with a probe from the CdMDR1 upstream region (Fig. 2B). The correct allelic replacement was also confirmed by hybridization with a probe from the CdMDR1 downstream region, which produced a 4.0-kb fragment in addition to the wild-type fragment in all 12 transformants (data not shown). These results demonstrate that the CaURA3 gene can be used for efficient and targeted integrative transformation of the C. dubliniensis ura3 mutants.
Expression of the CdMDR1 gene is induced by benomyl but not by fluconazole.
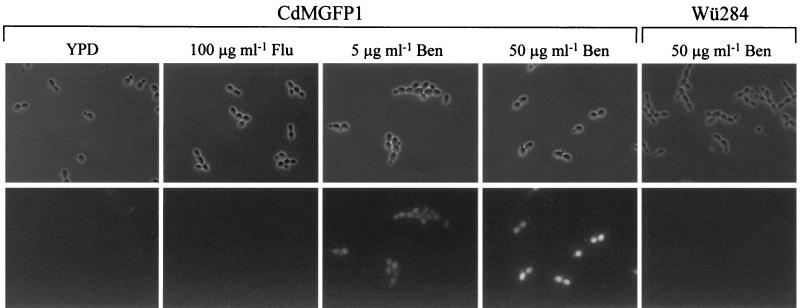
The MDR1 gene encodes a membrane transport protein of the major facilitator superfamily and is involved in the resistance of C. albicans and C. dubliniensis to fluconazole and several other, structurally unrelated drugs (1, 15, 24). Fluconazole-susceptible C. albicans strains do not significantly express the MDR1 gene in standard media in vitro, but MDR1 is constitutively activated in many fluconazole-resistant isolates (7, 8). Similar data have also been obtained for C. dubliniensis (15). Although fluconazole itself does not influence CaMDR1 expression, other drugs, such as benomyl, induce CaMDR1 transcription in fluconazole-susceptible C. albicans (10). Therefore, we investigated a possible induction of the CdMDR1 gene by the presence of these drugs using the C. dubliniensis strains with the chromosomally integrated PCdMDR1-GFP reporter gene fusion. After growth in YPD medium, no fluorescence of the reporter strains was observed, demonstrating that, as in C. albicans, the MDR1 gene was not significantly expressed in C. dubliniensis in the absence of drugs (Fig. 3). The addition of 100 μg of fluconazole ml−1 to the culture medium did not result in detectable fluorescence (Fig. 3), and the same result was obtained with a higher fluconazole concentration (200 μg ml−1) and during extended times of growth in the presence of the drug (data not shown). In contrast, benomyl induced the CdMDR1 promoter, resulting in maximal fluorescence of the reporter strains within 1 h following addition of the drug. CdMDR1 induction by benomyl was dose dependent, with a higher benomyl concentration resulting in stronger fluorescence of the reporter strains. This fluorescence was not caused by autofluorescence due to possible benomyl-induced cell damage, since the parent strain Wü284, which did not carry the GFP gene, produced no detectable fluorescent signal under the same conditions (Fig. 3).
FIG. 3.
Expression of the PCdMDR1-GFP reporter gene fusion in C. dubliniensis. Shown are phase-contrast and corresponding fluorescence micrographs of the parent strain, Wü284, or the reporter strains grown for 1 h in the absence or presence of the indicated drugs. Identical results were obtained with strains CdMGFP1A and CdMGFP1B. Flu, fluconazole; Ben, benomyl.
DISCUSSION
Until recently, the generation of specific mutants of C. albicans wild-type strains by targeted gene deletion was a laborious task and, to our knowledge, has been performed only once, to construct the widely used ura3 mutant strain CAI4 and similar derivatives from the clinical isolate SC5314 (6). The use of a dominant marker for the positive selection of integrative transformants, combined with its subsequent excision by FLP-mediated, site-specific recombination has made such an approach much more straightforward, and genes can now specifically be inactivated in any C. albicans strain, relieving the dependency on auxotrophic host strains for genetic manipulations (31). In the present work, using the same strategy, we successfully constructed a C. dubliniensis ura3 mutant by targeted gene replacement, thereby providing the first available auxotrophic C. dubliniensis strain. Our results suggest that in C. dubliniensis the MPAR marker is an even better tool than in C. albicans to direct the integration of recombinant DNA into a specific genomic locus. Since the MPAR marker is derived from the C. albicans IMH3 gene, MPA-resistant C. albicans transformants are frequently obtained that have not undergone the desired targeted recombination event but instead have presumably only integrated the marker by homologous recombination into the IMH3 locus (reference 30 and unpublished data). In contrast, almost all MPA-resistant C. dubliniensis transformants analyzed in this study had specifically integrated the MPAR flipper into one of the CdURA3 alleles. The more-specific integration of the mutagenesis cassette into the target locus was probably due to the divergence between C. albicans and C. dubliniensis sequences, which very likely prevented integration of the MPAR marker into the IMH3 locus in the heterologous host. Sequence divergence between the two species also caused our failure to specifically direct the integration of a reporter gene fusion or of the MPAR flipper into the C. dubliniensis genome when flanking sequences from C. albicans were used (26). These results are in line with a recent report demonstrating that even minor allelic differences within a given strain strongly bias integration specificity (32). Therefore, the C. albicans-derived MPAR marker appears to be a powerful tool for the genetic engineering of C. dubliniensis. This result also suggests that a similar marker derived from C. dubliniensis (or another species) should facilitate targeted integrations in C. albicans wild-type strains in the same way.
The MPAR-flipping strategy has now also been successfully used for inactivation of the MDR1 gene in a fluconazole-resistant, MDR1-overexpressing C. dubliniensis isolate to assess the contribution of this efflux pump to drug resistance (S. Wirsching et al., unpublished data), suggesting that the method is generally applicable for C. dubliniensis. Nevertheless, the availability of a ura3 mutant that is otherwise isogenic to a clinical isolate has several advantages for future genetic manipulations. First, using URA3 as a selection marker, prototrophic transformants can be recovered after only 2 days of selection on uridine-deficient medium, whereas the primary isolation of MPA-resistant transformants usually requires about 1 week, with additional time needed for clone purification. Therefore, the generation of mutants by sequential gene disruptions is significantly accelerated when the URA3 gene can be used as the selection marker. Second, many already available URA3-based genetic constructions that have been used for the molecular analysis of C. albicans can serve as the basis for analogous experiments in C. dubliniensis without the necessity for marker exchange; only homologous sequences from C. dubliniensis have to be substituted for the corresponding C. albicans fragments when genomic integration is required. Finally, the MPAR marker has also been used as a reporter gene in C. albicans (A. Strauß et al., submitted for publication). Introduction of such a reporter construct into a host strain requires a second marker like URA3 for the selection of transformants, and this approach is now feasible also for C. dubliniensis.
The two CdURA3 alleles were inactivated with different deletion constructs, such that any interchromosomal recombination between the two alleles would be recognized by the appearance of new bands. This experimental design ensured that the deletion of the two URA3 copies had occurred by independent, specific allelic replacements in our ura3 mutants. However, the ability to differentiate between the two URA3 alleles is also important with respect to future genetic manipulations, for example, when one is using the URA3-flipping strategy (18) to knock out additional genes. Since an FRT site is present in each of the disrupted URA3 alleles, FLP-mediated recombination could also involve these target sequences. Due to the different extent of the deletions on both sides of the FRT sites present in the inactivated URA3 alleles, any undesired recombination involving the URA3 locus would be easily detected.
It has been suggested that C. dubliniensis can develop fluconazole resistance in vitro more readily than C. albicans (16). As with many fluconazole-resistant clinical C. albicans isolates, resistance in in vitro-generated fluconazole-resistant C. dubliniensis derivatives seems to be caused most frequently by constitutive activation of the MDR1 gene, which also mediates resistance to several other compounds. The results of the present study do not provide an explanation for the higher propensity of C. dubliniensis to develop fluconazole resistance in vitro. As previously shown for C. albicans (7, 10), fluconazole itself did not induce the MDR1 promoter in C. dubliniensis. However, as described for C. albicans (10), the addition of benomyl to the culture medium resulted in strong activation of the CdMDR1 promoter. Therefore, certain drugs induce expression of the MDR1 gene in both species. One could envisage that there might be more regulatory pathways controlling MDR1 expression in C. dubliniensis than in C. albicans, which in turn would provide more targets for mutations that result in constitutive activation of the gene. The availability of reporter strains allowing easy detection of MDR1 expression will help to unravel these differences, for example, by screening for substances that induce MDR1 expression in C. dubliniensis but not in C. albicans.
In spite of the differences in the URA3 regulatory region, the CaURA3 gene was adequately expressed in C. dubliniensis, since we did not detect any difference in the growth of uridine-prototrophic transformants compared with that for the original C. dubliniensis wild-type isolate, Wü284. The transformation efficiency in the experiments described in this study, which required a double crossover event resulting in allelic replacement, was the same as we usually obtain in similar transformations of C. albicans strain CAI4 and derivatives. We also developed a URA3-based integrative vector for C. dubliniensis that requires only a single crossover for targeted insertion into the genome. Using this linearized plasmid, the transformation rate is enhanced about 10-fold (unpublished data). This transformation efficiency will allow the directed integration of genomic libraries, for example, to clone C. albicans genes that influence C. dubliniensis-specific characteristics or to complement mutant phenotypes generated by nonspecific mutagenization of a ura3 strain.
In conclusion, the full arsenal of molecular tools used in C. albicans genetics is now also available for its close relative C. dubliniensis. By taking advantage of species-specific differences, important questions regarding the biology and host adaptation mechanisms of these organisms can now be addressed at the molecular level.
ACKNOWLEDGMENTS
This study was supported by the Bundesministerium für Bildung und Forschung (BMBF grant O1 K1 8906-0). P. Staib is the recipient of a grant from the Studienstiftung des deutschen Volkes. Work performed in Dublin was supported by the Irish Health Research Board (grant 04/99).
REFERENCES
- 1.Ben-Yaacov R, Knoller S, Caldwell G A, Becker J M, Koltin Y. Candida albicans gene encoding resistance to benomyl and methotrexate is a multidrug resistance gene. Antimicrob Agents Chemother. 1994;38:648–652. doi: 10.1128/aac.38.4.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeke J D, Lacroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 3.Brandt M E, Harrison L H, Pass M, Sofair A N, Huie S, Li R-K, Morrison C J, Warnock D W, Hajjeh R H. Candida dubliniensis fungemia: the first four cases in North America. Emerg Infect Dis. 2000;6:46–49. doi: 10.3201/eid0601.000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Backer M D, Magee P T, Pla J. Recent developments in molecular genetics of Candida albicans. Annu Rev Microbiol. 2000;54:463–498. doi: 10.1146/annurev.micro.54.1.463. [DOI] [PubMed] [Google Scholar]
- 5.Donnelly S A, Sullivan D J, Shanley D B, Coleman D C. Phylogenetic analysis and rapid identification of Candida dubliniensis based on analysis of ACT1 intron and exon sequences. Microbiology. 1999;145:1871–1882. doi: 10.1099/13500872-145-8-1871. [DOI] [PubMed] [Google Scholar]
- 6.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franz R, Kelly S L, Lamb D C, Kelly D E, Ruhnke M, Morschhäuser J. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob Agents Chemother. 1998;42:3065–3072. doi: 10.1128/aac.42.12.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franz R, Ruhnke M, Morschhäuser J. Molecular aspects of fluconazole resistance development in Candida albicans. Mycoses. 1999;42:453–458. doi: 10.1046/j.1439-0507.1999.00498.x. [DOI] [PubMed] [Google Scholar]
- 9.Gilfillan G D, Sullivan D J, Haynes K, Parkinson T, Coleman D C, Gow N A R. Candida dubliniensis: phylogeny and putative virulence factors. Microbiology. 1998;144:829–838. doi: 10.1099/00221287-144-4-829. [DOI] [PubMed] [Google Scholar]
- 10.Gupta V, Kohli A, Krishnamurthy S, Puri N, Aalamgeer S A, Panwar S, Prasad R. Identification of polymorphic mutant alleles of CaMDR1, a major facilitator of Candida albicans which confers multidrug resistance, and its in vitro transcriptional activation. Curr Genet. 1998;34:192–199. doi: 10.1007/s002940050385. [DOI] [PubMed] [Google Scholar]
- 11.Köhler G A, White T C, Agabian N. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J Bacteriol. 1997;179:2331–2338. doi: 10.1128/jb.179.7.2331-2338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCullough M, Ross B, Reade P. Characterization of a genetically distinct subgroup of Candida albicans strains isolated from oral cavities of patients infected with human immunodeficiency virus. J Clin Microbiol. 1995;33:696–700. doi: 10.1128/jcm.33.3.696-700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meis J F, Ruhnke M, De Pauw B E, Odds F C, Siegert W, Verweij P E. Candida dubliniensis candidemia in patients with chemotherapy-induced neutropenia and bone marrow transplantation. Emerg Infect Dis. 1999;5:150–153. doi: 10.3201/eid0501.990119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millon L, Manteaux A, Reboux G, Drobacheff C, Monod M, Barale T, Michel-Briand Y. Fluconazole-resistant recurrent oral candidiasis in human immunodeficiency virus-positive patients: persistence of Candida albicans strains with the same genotype. J Clin Microbiol. 1994;32:1115–1118. doi: 10.1128/jcm.32.4.1115-1118.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moran G P, Sanglard D, Donnelly S M, Shanley D B, Sullivan D J, Coleman D C. Identification and expression of multiple drug transporters responsible for fluconazole resistance in Candida dubliniensis. Antimicrob Agents Chemother. 1998;42:1819–1830. doi: 10.1128/aac.42.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moran G P, Sullivan D J, Henman M C, McCreary C E, Harrington B J, Shanley D B, Coleman D C. Antifungal drug susceptibilities of oral Candida dubliniensis isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. Antimicrob Agents Chemother. 1997;41:617–623. doi: 10.1128/aac.41.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morschhäuser J, Michel S, Hacker J. Expression of a chromosomally integrated, single-copy GFP gene in Candida albicans, and its use as a reporter of gene regulation. Mol Gen Genet. 1998;257:412–420. doi: 10.1007/s004380050665. [DOI] [PubMed] [Google Scholar]
- 18.Morschhäuser J, Michel S, Staib P. Sequential gene disruption in Candida albicans by FLP-mediated site-specific recombination. Mol Microbiol. 1999;32:547–556. doi: 10.1046/j.1365-2958.1999.01393.x. [DOI] [PubMed] [Google Scholar]
- 19.Morschhäuser J, Ruhnke M, Michel S, Hacker J. Identification of CARE-2-negative Candida albicans isolates as Candida dubliniensis. Mycoses. 1999;42:29–32. doi: 10.1046/j.1439-0507.1999.00259.x. [DOI] [PubMed] [Google Scholar]
- 20.Odds F C. Candida and candidosis: a review and bibliography. London, United Kingdom: Bailliere Tindall; 1988. [Google Scholar]
- 21.Pinjon E, Sullivan D, Salkin I, Shanley D, Coleman D. Simple, inexpensive, reliable method for differentiation of Candida dubliniensis from Candida albicans. J Clin Microbiol. 1998;36:2093–2095. doi: 10.1128/jcm.36.7.2093-2095.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pla J, Gil C, Monteoliva L, Navarro-Garcia F, Sanchez M, Nombela C. Understanding Candida albicans at the molecular level. Yeast. 1996;12:1677–1702. doi: 10.1002/(SICI)1097-0061(199612)12:16%3C1677::AID-YEA79%3E3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 23.Polacheck I, Strahilevitz J, Sullivan D, Donnelly S, Salkin I F, Coleman D C. Recovery of Candida dubliniensis from non-human immunodeficiency virus-infected patients in Israel. J Clin Microbiol. 2000;38:170–174. doi: 10.1128/jcm.38.1.170-174.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanglard D, Kuchler K, Ischer F, Pagani J-L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staib P, Morschhäuser J. Chlamydospore formation on Staib agar as a species-specific characteristic of Candida dubliniensis. Mycoses. 1999;42:521–524. doi: 10.1046/j.1439-0507.1999.00516.x. [DOI] [PubMed] [Google Scholar]
- 26.Staib P, Michel S, Köhler G, Morschhäuser J. A molecular genetic system for the pathogenic yeast Candida dubliniensis. Gene. 2000;242:393–398. doi: 10.1016/s0378-1119(99)00512-0. [DOI] [PubMed] [Google Scholar]
- 27.Sullivan D, Coleman D. Candida dubliniensis: characteristics and identification. J Clin Microbiol. 1998;36:329–334. doi: 10.1128/jcm.36.2.329-334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sullivan D J, Westerneng T J, Haynes K A, Bennett D E, Coleman D C. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141:1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 29.Willis A M, Coulter W A, Sullivan D J, Coleman D C, Hayes J R, Bell P M, Lamey P-J. Isolation of C. dubliniensis from insulin-using diabetes mellitus patients. J Oral Pathol Med. 2000;29:86–90. doi: 10.1034/j.1600-0714.2000.290206.x. [DOI] [PubMed] [Google Scholar]
- 30.Wirsching S, Michel S, Köhler G, Morschhäuser J. Activation of the multiple drug resistance gene MDR1 in fluconazole-resistant, clinical Candida albicans strains is caused by mutations in a trans-regulatory factor. J Bacteriol. 2000;182:400–404. doi: 10.1128/jb.182.2.400-404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wirsching S, Michel S, Morschhäuser J. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol Microbiol. 2000;36:856–865. doi: 10.1046/j.1365-2958.2000.01899.x. [DOI] [PubMed] [Google Scholar]
- 32.Yesland K, Fonzi W A. Allele-specific gene targeting in Candida albicans results from heterology between alleles. Microbiology. 2000;146:2097–2104. doi: 10.1099/00221287-146-9-2097. [DOI] [PubMed] [Google Scholar]