Summary
Ectodermal dysplasias including skin abnormalities and cleft lip/palate result from improper surface ectoderm (SE) patterning. However, the connection between SE gene regulatory networks and disease remains poorly understood. Here, we dissect human SE differentiation with multiomics and establish GRHL2 as a key mediator of early SE commitment, which acts by skewing cell fate away from the neural lineage. GRHL2 and master SE regulator AP2a balance early cell fate output, with GRHL2 facilitating AP2a binding to SE loci. In turn, AP2a restricts GRHL2 DNA binding away from de novo chromatin contacts. Integration of these regulatory sites with ectodermal dysplasia-associated genomic variants annotated within the Biomedical Data Commons identifies 55 loci previously implicated in craniofacial disorders. These include ABCA4/ARHGAP29 and NOG regulatory regions where disease-linked variants directly affect GRHL2/AP2a binding and gene transcription. These studies elucidate the logic underlying SE commitment and deepen our understanding of human oligogenic disease pathogenesis.
Subject areas: Biological sciences, Cell biology, Developmental biology
Graphical abstract

Highlights
-
•
GRHL2 is required for surface ectoderm formation and neural cell identity repression
-
•
GRHL2 promotes AP2a binding and AP2a regulates the level and sites of GRHL2 binding
-
•
Craniofacial-associated SNPs disproportionately fall in ectopic GRHL2 binding sites
-
•
rs1211213 regulates GRHL2 and AP2a binding in its enhancer and gene target expression
Biological sciences; Cell biology; Developmental biology
Introduction
Ectodermal dysplasias, including skin, craniofacial, visual, and hearing abnormalities are among the most common birth defects.1 Proper formation of craniofacial features, in particular, relies on the precise balance of neural, mesodermal, and surface ectoderm (SE) lineage interactions that lead to proper embryonic patterning and folding.2,3,4 Previous investigations have established lineage-specific master transcription factors which open chromatin and regulate gene expression required for proper lineage contributions to neural tube and SE maturation.4,5,6,7 However, experimental systems to link high resolution genomics and functional studies during lineage commitment in ectodermal dysplasia have been lacking.
To understand the SE contribution to disease, we interrogated human embryonic stem cell (hESC) in vitro differentiation with retinoic acid (RA) and bone morphogenetic protein 4 (BMP4).8 Previous inference network modeling identified key transcription factors such as AP2a/c, GRHL2, and GATA3 that initiated SE chromatin and TP63 expression to drive the epidermal lineage.8,9 The central upstream role for AP2c, which is associated with orofacial clefts and ectodermal dysplasia, came from the demonstration that it was sufficient to induce SE in the absence of RA/BMP4. Subsequent analysis demonstrated the importance of cell-cell communication between the germ layer cell types in SE differentiation.10
Recently, we created the Biomedical Data Commons (BMDC), a community-based knowledge graph that enables complex queries across multiple types of publicly available biomedical data.11 BMDC addresses GWAS limitations such as linkage disequilibrium (LD), activity across diverse cell types, and loci with multiple genes.12 These issues lead to overemphasis of variants with relatively rare minor allele frequencies, overlooking those in non-coding regions, and failing to identify many variants associated with common oligogenic diseases. In particular, the lack of functionalization of non-coding regions, which compose 98.5% of the genome, forces many studies to instead rely on evolutionary conservation and linear relationships between coding and regulatory regions.13,14 Our workflow prioritizes disease-related genetic variants within BMDC and has been applied to Type 1 diabetes to identify non-coding, poorly conserved, common, cell type–specific variants.11
In this study, we find that GRHL2 skews ectodermal fate away from the neural lineage toward SE by promoting AP2a DNA binding, which then feeds back to restrict GRHL2 activity. BMDC prioritization of human craniofacial genetic variants revealed a surprisingly large overlap of single nucleotide polymorphisms (SNPs) within GRHL2 or AP2a enhancer binding sites, altering their binding and downstream target gene expression. Together these results illustrate a mechanism for how reciprocal transcription factor interactions precisely pattern gene expression during ectodermal lineage specification and provide a reference framework for genetic variant prioritization for ectodermal dysplasia.
Results
GRHL2 promotes surface ectoderm and inhibits neural cell differentiation
We previously demonstrated that RA/BMP4 treated hESCs recapitulate regional development of the SE and reciprocal signaling with the underlying mesoderm8,9,10 (Figure 1A). To uncover regulators of these lineages in our system, we performed single cell multiome sequencing (scRNA + scATAC-seq) after addition of RA/BMP4. Weighted nearest neighbor analysis revealed that EPCAM+ SE and PDGFRA+ mesoderm were the two major cell types (Figures 1B and S1A–S1D), with a small population of neural cells. The top enriched motif in the early SE was GRHL2 (Figures 1C and 1D, Table S1). Although GRHL2 loss-of-function studies have been reported in human cancer cells, epiblasts, mouse embryos, zebrafish, and fly eye disks,2,15,16,17,18 its role in the differentiating human SE has not been studied. GRHL1 and GRHL3 mutations cause epidermal defects in mice,17,18,19,20 but GRHL3 is not expressed in our system. Importantly, GRHL2 craniofacial expression precedes that of the other isoforms during mouse development,21 and it has been implicated in ectoderm development in both mice and Drosophila.2,18
Figure 1.
GRHL2 promotes surface ectoderm and represses neural identities
(A) Differentiation strategy to produce early SE. RA and BMP4 are added to hESC colonies for 7 days, resulting in a mixture of SE and supporting mesenchymal cell types.
(B) Joint neighbor UMAP representing both single cell gene expression and DNA accessibility measurements from hESCs after 7 days of RA/BMP4.
(C) Gene expression of GRHL2 on joint UMAP.
(D) GRHL2 motif enrichment on joint UMAP.
(E) CRISPR strategy and confirmation immunoblot for GRHL2 deletion in hESCs.
(F) Representative immunofluorescent images of WT and GRHL2 KO hESCs after 7 days of RA/BMP4 using antibodies against GRHL2, Keratin-18 (KRT18) or DAPI for nuclei. Scale bars in each micrograph represent 25 μm.
(G) Expression of GRHL2-dependent or repressed genes across clusters identified using integrated scRNA from WT and GRHL2 KO cells.
(H) UMAP of integrated WT and GRHL2 KO cells colored by cell type.
(I) Integrated UMAP colored by genotype.
(J) Proportion of each identity in WT or GRHL2 KO cells.
(K) UMAPs showing gene expression of individual transcripts colored by log2-transformed rendering of UMI counts.
(L) UMAPs of motif enrichment colored by motif Z score.
(M) Integrated UMAP colors by k-means clusters based on gene expression or (N) ATAC accessibility.
(O) Pseudotime plots of WT and GRHL2 KO cells colored by integrated identities.
We therefore used CRISPR to deplete GRHL1 or GRHL2 protein in hESCs. Gene expression analyses suggested that loss of GRHL2, but not GRHL1, had a significant effect on lineage-specific transcripts in our system (Figures S1E and S1F). This is consistent with our previous network inference modeling which implicated GRHL2 (as opposed to GRHL1),9 and the observation that GRHL1 does not appear to play a role in ectoderm or craniofacial development in mice and zebrafish.19,22 Upon addition of RA/BMP4, GRHL2 KO hESCs (Figures 1E and S1G) failed to flatten and produce characteristic KRT18 networks, exhibiting striking abnormalities (Figure 1F). By RNA-seq, the expression of thousands of transcripts was altered (Figure S1H, Table S2), including 70% of those reported previously in a GRHL2 null mouse model.2 Transcripts encoding epithelial adhesion and extracellular matrix proteins such as CDH1 and CXCL14 were severely reduced in the KO, and neural development markers such as ZIC2 and PAX3 were increased (Figures S1I–S1K). GRHL1 expression was reduced in the KO, suggesting it likely acts downstream of GRHL2. GRHL2-dependent genes were enriched in the SE population identified by scRNA-seq (Figure 1G). Of interest, GRHL2-repressed genes were expressed primarily in the small neural population.
Loss of GRHL2 resulted in a striking reduction in the SE at the expense of 4-fold expansion of the neuroectoderm (Figures 1H–1J). SE markers such as TP63, CXCL14, and GRHL1 were virtually absent in the KO (Figures 1K and S2A) alongside expansion of the neural transcripts PAX6 and PAX3. There was a small de novo neural population that appeared only in the GRHL2 mutant, which was enriched for ASCL motifs and neural progenitor markers such as STMN2 and TUBB3 (Figures 1L and S2B). PCA analysis suggested this cluster of cells is a maturing neural progenitor population (Figure S2C). An advantage of multiomic scRNA/scATAC is the ability to generate 3D chromatin information, linking a gene and enhancer using the statistically significant correlation between gene expression and chromatin accessibility across all cells in the dataset (Table S3). Consistent with neuroectoderm expansion was the appearance of GRHL2-repressed links between neural genes and distal enhancers (Figure S2D, Table S3) likely originating from new open chromatin sites at ZIC, SOX, and RFX motifs (Figure 1L).
Through k-means clustering of the integrated WT/GRHL2 KO projection, we discovered that GRHL2 KO gene expression and chromatin accessibility identities do not overlap with WT cell types (Figures 1M and 1N). Gene expression that was once distinct to the SE (EPCAM, PDGFRA, PERP, etc.) became reduced and diffused across all cells in the GRHL2 KO, indicating the inability of GRHL2 to effectively resolve canonical SE (Figure S2E). GRHL2-deficient cells also fail to produce distinct branches of pseudotime-based differentiation trajectories (Figures 1O and S2F). Together these results suggest that GRHL2 skews stem cell differentiation away from neural/neuroectoderm cell fates and toward SE commitment.
GRHL2 facilitates AP2a DNA binding
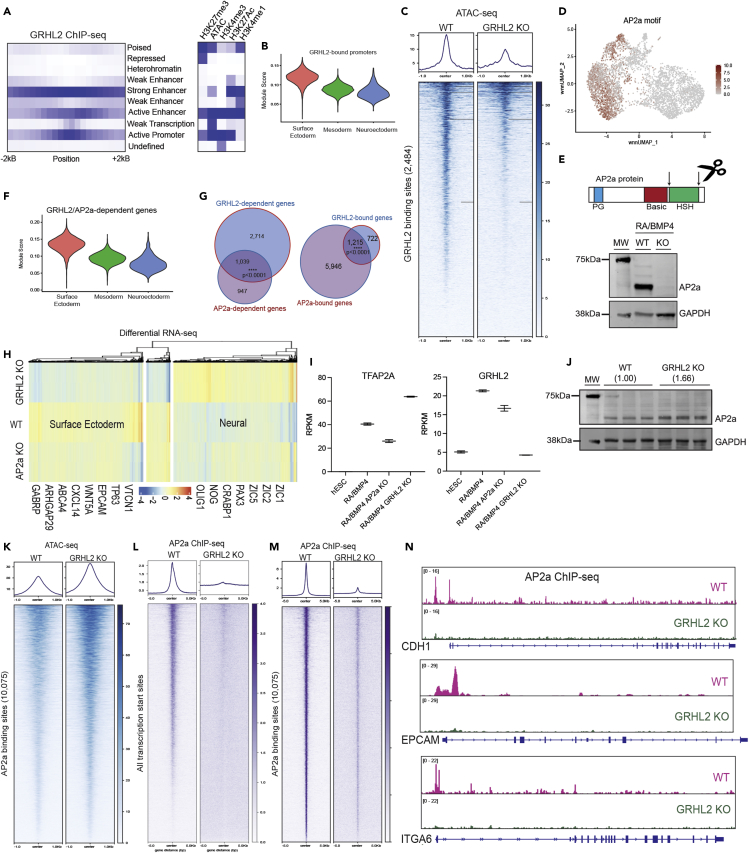
We elucidated the mechanism by which GRHL2 enforces SE differentiation by integrating GRHL2 ChIP-seq with chromatin states defined by histone marks and chromatin accessibility (Figure 2A, Table S2). Consistent with binding site profiles in human epiblast and Drosophila studies,15,18 we found that GRHL2 binds to enhancers and promoters at SE loci as well as opens chromatin at its binding sites (Figures 2B and 2C). Importantly, we observed that GRHL2 directly binds to only 16% of its regulated gene promoters (Table S2), suggesting it acts at a distance or in coordination with one or more upstream transcription factors.
Figure 2.
GRHL2 is required for AP2a activity at SE loci
(A) Enrichment of various chromatin states in relation to GRHL2 ChIP-seq coordinates. Chromatin states were defined by ChromHMM using previously published histone mark ChIP-seq and ATAC-seq datasets in hESCs treated with RA/BMP4 for 7 days.
(B) Expression of GRHL2-bound genes across integrated scRNA-seq identities.
(C) Distribution of ATAC-seq signal (score based on number of reads per bin) relative to GRHL2 binding sites in WT and GRHL2 KO cells.
(D) AP2a motif enrichment joint UMAP.
(E) CRISPR strategy and immunoblot confirming AP2a KO hESCs.
(F) Expression of GRHL2 and AP2a-dependent genes across integrated scRNA-seq identities.
(G) Overlap of GRHL2 and AP2a-dependent and bound genes. Significance was calculated by Fisher exact test, ∗∗∗ indicates p value <0.0001.
(H) Heatmap of read counts from WT, GRHL2 KO, and AP2a KO RNA-seq.
(I) Expression of TFAP2A or GRHL2 in hESCs or WT, GRHL2 KO or AP2a KO hESCs treated with RA/BMP4. Error bars represent mean +/−SD, n= 2 biological replicates.
(J) Immunoblot showing levels of AP2a protein in WT and GRHL2 KO cells treated with RA/BMP4.
(K) Distribution of ATAC-seq signal (score based on number of reads per bin) relative to AP2a binding sites in WT and GRHL2 KO cells. Distribution of AP2a ChIP-seq signal (score based on number of reads per bin) relative to (L) all transcription star sites or (M) AP2a binding sites in WT and GRHL2 KO cells.
(N) Representative bedgraphs of AP2a ChIP-seq in WT or GRHL2 KO cells.
Because previous work in our lab showed that AP2 transcription factors are sufficient to drive SE and keratinocyte formation in the absence of RA/BMP4,9 we investigated the GRHL2/AP2 genetic network interaction. Our multiome data showed that all three AP2 family members (AP2a/TFAP2A, AP2b/TFAP2B, and AP2c/TFAP2C) were enriched in the SE, with TFAP2B expression at much lower levels (Figures 2D and S3A). Further, AP2a and AP2b have been implicated in ectodermal processes including human epidermal signaling, mouse and zebrafish neural crest and craniofacial development, as well as human branchiooculofacial syndrome.5,23,24,25,26,27,28 To investigate the relationship between GRHL2 and AP2 family members, we used CRISPR to deplete AP2a or AP2b protein (AP2c depletion was lethal). Consistent with mouse mutant data,28 AP2a-depleted cells display a much more prominent mutant phenotype than AP2b (Figures S3B and S3C), which provided rationale to specifically interrogate the interaction between GRHL2 and AP2a (Figure 2E).
By RNA-seq, we found that GRHL2 and AP2a regulate a statistically significant set of overlapping SE genes (Figures 2F–2H, Table S1) without affecting each other’s expression (Figures 2I, 2J, and S3D). Loss of GRHL2 caused a significant reduction in both TFAP2B and TFAP2C expression, suggesting it acts upstream of these transcripts (Figure S3E) Strikingly, despite unchanged AP2a protein levels or chromatin accessibility at AP2a binding sites (Table S1, Figure 2K), we observed dramatically reduced AP2a DNA binding in the GRHL2 KO (Figures 2L and 2M). Differential AP2a binding strength was observed at 9,560 sites, with only 10% remaining in the GRHL2 KO (p value<0.01, Table S2, Figures 2L and 2M). This observation was consistent across SE markers, including CDH1, EPCAM, and ITGA6 (Figure 2N). These data demonstrate that GRHL2 promotes AP2a binding at open chromatin sites to enforce SE gene expression.
AP2a restricts GRHL2 DNA binding
Although GRHL2 promotes AP2a DNA binding, we were surprised to find that loss of AP2a in turn caused a marked increase, rather than decrease, in GRHL2 DNA binding without affecting its expression levels (Figure 3A, Table S2). Although GRHL2 can be detected binding to a modest 2,500 loci genome-wide, AP2a loss caused new GRHL2 binding sites at over 15,000 loci in the absence of any changes in chromatin accessibility (Figure 3B). Half of these new sites directly overlap with AP2a (Table S2), and the rest are enriched at promoters and enhancers connected to AP2a in 3D space (Figures 3C and 3D, p value <0.0001). Furthermore, the increase in GRHL2 DNA binding occurred alongside the appearance of de novo 3D chromatin contacts, a near 3-fold increase compared to WT (Figures 3E and 3F). This suggests chromatin contact insulation as a possible mechanism by which AP2a restricts GRHL2 binding. Promoters connected to these ectopic binding sites were mis-regulated in AP2a KO cells and enriched for cell migration, wnt signaling, and chromatin remodeling genes (Figures 3G and 3H). Furthermore, the loss of AP2a caused a marked increase in the neural lineage at the expense of the SE by scRNA-seq, similar to the GRHL2 KO (Figures 3H and 3I). Together these results suggest that AP2a prevents GRHL2 from binding to and forcing the expression of inappropriate neuroectodermal lineage factors during SE development (Figure 3J).
Figure 3.
AP2a restricts GRHL2 binding to appropriate target genes
(A) Distribution of GRHL2 ChIP-seq signal (score based on number of reads per bin) relative to GRHL2 or AP2a binding sites in WT and AP2a KO cells.
(B) Distribution of ATAC-seq signal (score based on number of reads per bin) relative to the new GRHL2 binding sites in WT or AP2a KO cells.
(C) Enrichment of various chromatin states in relation to AP2a KO GRHL2 ChIP-seq coordinates. Chromatin states were defined by ChromHMM using histone mark ChIP-seq and ATAC-seq datasets in hESCs treated with RA/BMP4 for 7 days.
(D) Percentage of GRHL2 peaks found at or connected to promoters or AP2a binding sites in both WT and AP2a KO cells. p values were calculated with two-tailed Fisher’s exact tests. ∗∗∗∗ indicates a p value <0.0001.
(E) Number of chromatin contacts in WT and AP2a KO cells as measured by cohesin HiChIP.
(F) Chromatin contact strength at ectopic GRHL2 binding sites in WT and AP2a KO cells. Boxes represent the median with interquartile range and error bars represent the minimum and maximum.
(G) Empirical cumulative distribution function of the log2FoldChange in gene expression of promoters bound or looped to GRHL2 or AP2a binding sites (n = 6,564, purple) compared to all protein coding genes (n = 19,923, black) in WT vs. AP2a KO cells. p value <0.01 calculated by Student’s two-tailed test.
(H) UMAP of integrated WT and AP2a KO cells colored by cell type.
(I) Fold change in AP2a KO cell type proportions compared to WT, as measured by scRNA-seq.
(J) Model of GRHL2 and AP2a regulation of gene expression. GRHL2 acts by blocking neural gene expression and promoting AP2a binding at surface ectoderm genes. In the absence of AP2a, GRHL2 binds unrestricted to inappropriate loci.
(K) MA plot of differential expression analysis between WT and GRHL overexpression bulk RNA-seq. Transcript expression is either unchanged (gray), increased (blue, >2-fold), or decreased (red, <2-fold). p values for differentially expressed genes were calculated using DESEQ2 with a cutoff of 0.05. All differential genes and statistics can be found in Table S5.
(L) Overlap of GRHL2-regulated genes and genes altered by GRHL2 overexpression. p value was calculated with a Fisher exact test.
(M) UMAP of integrated WT and GRHL2 over expression cells colored by cell type.
(N) Fold change in GRHL2 overexpression cell type proportions compared to WT, as measured by scRNA-seq.
To determine whether increased GRHL2 levels are sufficient to alter gene expression, we introduced a doxycycline-inducible GRHL2 into the CLYBL safe harbor locus.29 Overexpression of GRHL2 indeed caused abnormal expression of many key developmental regulatory genes including DLX6, TBX1, NOG, and PAX6 all of which are associated with known human craniofacial phenotypes30,31,32,33 (Figure 3K, Table S2). Gene expression changes in response to ectopic GRHL2 expression are consistent with previous reports on chromatin accessibility in Drosophila brain and mouse embryos.2,18 However, GRHL2 fails to induce a statistically significant (p = 0.06) number of its target genes likely because of the presence of AP2a (Figure 3L). Furthermore, by scRNA-seq, GRHL2 overexpression alone induces a modest increase in the neuroectoderm lineage (Figures 3M and 3N). We conclude that proper SE lineage specification depends on dosage-dependent GRHL2/AP2a regulatory interactions.
GRHL2 and AP2a binding sites overlap with craniofacial SNPs
Both GRHL2 and TFAP2A loss-of-function mutations cause craniofacial and neurulation defects including cleft lip/palate in numerous species and recent murine data implicate AP2a specifically in SE.28 Further, vertebrate embryologic studies demonstrate that facial morphogenesis requires not only neural crest, but also interactions with the overlying SE.28,34,35 We therefore sought to validate the relevance of our GRHL2/AP2a multimodal network by integrating these data with previously published cleft lip/palate GWAS variants.2,7,21,36,37 We compiled a list of 500 variants with 13,378 in LD,38 followed by integration of the GRHL2/AP2a ChIP-seq, ATAC-seq, and chromatin conformation (cohesin Hi-ChIP) datasets using a previously developed informatic pipeline11 (Table S4). We identified 141 genetic variants located in 175 unique enhancers, which are distally looped to 165 unique genes in 55 genomic regions (Figures 4A and 4B, Tables S5 and S6). Of interest, the majority of these variants have no previously reported clinical significance (Figure 4C) and had a minor allele frequency (MAF) above 2% (Figures 4D and S4A–S4E) indicating they are relatively common. In addition, Combined Annotation Dependent Depletion (CADD) scores were low, suggesting the variants are not evolutionarily conserved (Figure 4E) and would have been missed by previous conservation-based approaches.28
Figure 4.
Cleft lip/palate genetic variants prioritized by integrating multiomic GRHL2/AP2a functional datasets
(A) Pipeline overview, which connects single nucleotide polymorphisms (SNPs) in enhancers to distal genes via chromatin looping.
(B) Flow chart illustrating how genes of interest were filtered using functional data.
(C) Clinical significance of pipeline-identified output genetic variants.
(D) Histogram of the minor allele frequency or (E) CADD scores of the genetic variants. Red dashed lines indicate standard cutoffs for minor allele frequency (MAF <0.02) or CADD score (CADD ≥15) for identifying disease-associated genetic variants.
(F) Proportion of pipeline input or output genetic variants which were found directly from GWAS studies or are in linkage disequilibrium (LD). Significance was calculated with a Fisher’s exact test, ∗ indicates a p value <0.05.
(G) The chromosomal location and (H) functional categories of pipeline-identified genetic variants.
(I) Number of pipeline-identified genetic variants falling within GRHL2 or AP2a binding sites, or AP2a-dependent ATAC sites in WT (black) or AP2a KO cells (white). p values were calculated with two-tailed Fisher’s exact tests, ∗∗∗∗ indicates p value <0.0001.
(J) Overlap of genetic variants identified in our day 7 RA/BMP4 cells compared to published neural crest studies.39,40
Over 90% of the pipeline-identified genetic variants were in LD with GWAS SNPs and significantly increased compared with that expected from the input (Figure 4F). The 141 pipeline-identified variants are in 55 loci across 16 different chromosomes primarily in introns or near promoters of genes (Figures 4G and 4H), and thus were not in gene duplicated clusters. These loci include known and previously recognized regulators of SE, neural, or craniofacial development including the genes TACC1, IRF6, and MTHFR (Figures S5A–S5D).41,42,43,44 Of interest, the number of variants within GRHL2 binding sites was increased over 10-fold in the AP2a KO (Figure 4I). Genes associated with these variants had minimal overlap with SNPs identified using enhancer ChIP-seq from neural crest cells (Figure 4J), emphasizing the cell-type specificity of this pipeline and the unique SE regulatory elements.39,40 Only variant rs227727 in the NOG locus was identified by both our pipeline and by the FaceBase Consortium, which enriched for craniofacial variants in cranial neural crest-specific enhancers.39 We conclude that the GRHL2/AP2a binding sites mark an SE-focused set of cleft lip/palate enhancer variants.
Of interest, an A>G SNP (rs1211213) within a GRHL2 binding site was found at the ABCA4-ARHGAP29 locus, which is one of the most commonly associated cleft lip-palate disease loci. This specific variant has not previously been identified by GWAS, but was found in our analysis through its LD with cleft lip/palate SNPs rs3789432 and rs481931.45,46 Importantly, none of the nearby GWAS-identified SNPs45,47,48,49,50,51 overlapped with GRHL2 or AP2a binding sites (Figure 5A), suggesting rs1211213 may have been overlooked because of the lack of SE-specific transcription factor network analyses. Chromatin accessibility at this variant is specific to the SE and is significantly correlated with ABCA4 and ARHGAP29 expression, suggesting that it is a major driver of ABCA4 and ARHGAP29 in the SE (Figures 5B and 5C).
Figure 5.
Cleft lip/palate genetic variants alter GRHL2 binding and downstream gene expression
(A) Table of SNPs identified from previous GWAS studies showing whether they fall within GRHL2 or AP2a binding sites.
(B) Feature linkage between the ABCA4 promoter and accessible chromatin from WT and GRHL2 KO single cell multiome datasets. Arc height corresponds to the absolute value of each linkage. Red = negative correlation and Blue = positive correlation.
(C) Chromatin accessibility at the ABCA4 locus in each cell type identified by scATAC.
(D) AP2a and GRHL2 ChIP-seq signal in WT and AP2a KO cells at the ABCA4 locus.
(E) Raw cohesion HiChIP signal at the ABCA4/ARHGAP29 locus in WT and AP2a KO cells.
(F) Genetic variant preference at the SNP rs1211213 for AP2a or GRHL2 DNA binding as identified by ChIP-seq. Boxes represent median with interquartile ranges.
(G) Expression of ABCA4 or (H) ARHGAP29 from bulk RNA-seq data. Error bars represent mean +/−SD, n= 2 biological replicates.
(I) UMAP of ABCA4 or (J) ARHGAP29 expression from integrated scRNA-seq.
(K) Raw sequencing data illustrating the single nucleotide change at rs1211213 made with CRISPR genome editing.
(L) qPCR comparing homozygous vs heterozygous rs1211213 alleles. Bars indicated mean with SD. p values were calculated with two-tailed Student’s ttest, n= 5 biological replicates run in triplicate. ∗∗∗∗ indicates p value <0.0001.
Consistent with our model, GRHL2 is required for AP2a binding at this locus (Figure 5D). Loss of AP2a in turn removes a GRHL2-bound 3D chromatin contact at ARHGAP29 (Figure 5E). ChIP-seq in H9 hESCs, which are heterozygous for rs1211213, exhibits altered GRHL2 and AP2a DNA binding as both proteins preferentially binding to the major allele (Figures 5F–5H). Furthermore, GRHL2 loss results in lowered gene expression of both ABCA4 and ARHGAP29 (Figures 5I and 5J). To confirm the role of this variant, we used CRISPR genome editing to alter a single base pair in H9 hESCs, converting the remaining major allele (A) at rs1211213 to the minor allele (G) (Figure 5K). Indeed, RA/BMP4-treated homozygous minor (G/G) cells exhibited decreased expression of both ABCA4 and ARHGAP29, independent of changes in flanking genes ABCD3 and DNTTIP2 (Figure 5L). This example illustrates how a single base pair variant in the SE-associated GRHL2/AP2a regulatory network alters the cooperative balance of transcription factor binding, resulting in altered expression of disease-associated genes.
As a second example, we interrogated the NOG locus, which makes well-established contributions to craniofacial development and disease52,53 and is repressed by both AP2a and GRHL2 during SE differentiation (Figure S6D). We found a cleft lip/palate SNP (rs227727) located within an AP2a binding site at a distal NOG enhancer54 that is accessible and connected to the promoter by a 3D chromatin contact (Figure S6A). AP2a loss results in ectopic GRHL2 DNA binding (Figure S6B) and increased NOG expression (Figure S6D). As with the ARHGAP29 locus, AP2a preferentially binds to the major allele (Figure S6C), and we infer that the resulting GRHL2 binding is responsible for the observed increased expression of NOG. Intriguingly, loss of GRHL2 results in decreased AP2a binding at rs227727 and a further increase in NOG expression (Figure S6D). Analysis of the NOG locus further illustrates the delicate balance of GRHL2 and AP2a feedback required for lineage specification in the developing embryo.
Discussion
In this study we investigate early SE commitment with multimodal single cell analysis and use BMDC to correlate our results with disease-associated variants. We identify GRHL2 and AP2a as key players responsible for SE specification, clarify their mechanistic relationship, and use their regulatory network to identify 55 genomic regions associated with human craniofacial disorders. Our approach demonstrates how single base pair genetic variation contributes to altered lineage commitment in human disease, and how multidimensional genomic data can be used to nominate known and previously unrecognized loci for downstream studies in model systems and in the clinic.
GRHL2 loss-of-function experiments showed that this protein is not only vital to SE formation, but it is required to repress aberrant neural gene expression and chromatin accessibility (Figure 1). Our previous studies have demonstrated that cell-cell communication between the early germ layers is crucial to development of more mature tissues like skin and facial structure.10 These new results suggest that GRHL2 controls this balance of cell types branching out from differentiation, likely affecting communication with the supporting mesenchyme. In elucidating GRHL2 gene regulation, we uncover a surprising regulatory interaction where GRHL2 promotes binding of AP2a, which in turn restricts GRHL2 DNA binding. This newly described regulatory mechanism controls the balance of neural and SE formation, and directly implicates GRHL2 and AP2a in human ectodermal dysplasia. We hypothesize that protein-protein interaction domains in GRHL2 are key to its interaction with AP2a, as genetic variants in GRHL2 non DNA-binding domains can result in human phenotypes.55,56,57 Furthermore, previous studies have shown that AP2a can negatively control the transcriptional activity of other factors including c-Myc and Max by interacting with BR/HLH/LZ domains.58,59
We extend the utility of BMDC as we demonstrate the value and ease of connecting a functional chromatin dynamic map with disease-causing genetic variants. Although whole genome sequencing has been instrumental in identifying SNPs, we found that integration of functional data is crucial in pinpointing relevant variants to investigate complex epistasis in oligogenic syndromes. Many cleft lip/palate genetic variants have been reported in GWAS studies,45,48 but we demonstrate that functional ABCA4/ARHGAP29 SNPs were actually in LD and had not been previously identified owing to the lack of relevant cell type-specific experimental data (Figure 5). Combining our functional data with GWAS studies has allowed us to better understand allele frequencies of common and rare variants in oligogenic diseases. The GRHL2/AP2a map created in this study will serve as a reference frame in future studies of ectodermal dysplasia and developmental disorders to prioritize genetic variants and predict disease in individuals.
Limitations of the study
Although using an isogenic human pluripotent cell-derived skin differentiation system enables us to study the function of non-coding regions in humans, this model does have some limitations. Our cell differentiation system mimics general ectodermal lineage commitment, but limits interpretation of our results in the context of multi-lineage craniofacial development. Although we were gratified that a base pair mutation of one SNP in the ABC4A locus sufficiently impacts GRHL2 and AP2a binding and expression of target genes ABC4A and ARHGAP29, the rs1211213 minor allele frequency in the general population is ∼30%. This indicates that it is unlikely to be sufficient on its own for a penetrant craniofacial phenotype in the absence of other variants. This highlights the need for oligogenic analysis of craniofacial dysfunction.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-rabbit GRHL2 | Sigma | RRID: AB_1857928 |
| Anti-sheep KRT18 | R&D Systems | RRID: AB_1935896 |
| Anti-mouse AP2A | Santa Cruz | RRID: AB_667767 |
| Anti-mouse GAPDH | Santa Cruz | RRID: AB_627679 |
| Anti-rabbit SMC1A | Bethyl Biosciences | RRID: AB_2192467 |
| Bacterial and virus strains | ||
| Stellar Competent Cells | Takara Biosciences | CAT# 636766 |
| Chemicals, peptides, and recombinant proteins | ||
| BMP4 | R&D Systems | Cat #314-BP-050 |
| RA | Sigma | Cat #R2625 |
| Essential 8 Medium | Life Technologies | Cat #A1517001 |
| Essential 6 Medium | Life Technologies | Cat #A1516401 |
| BD hESC Qualified Matrigel | Fisher | Cat #354277 |
| Accutase | Innovative Cell Technologies | Cat #AT104 |
| DTT | Sigma | Cat #646563 |
| Protector RNase Inhibitor | Sigma | Cat #3335399001 |
| Digitonin | Thermo Fisher | Cat #BN2006 |
| MACS BSA Stock Solution | Miltenyi Biotech | Cat #130-091-376 |
| BamBanker | Wako Chemicals | Cat #30214681 |
| Normal Horse Serum Blocking Solution | Vector Laboratories | Cat #S-2000 |
| ProLong Gold Antifade Mountant | Life Technologies | Cat #P36930 |
| TruCut Cas9 Protein | Thermo Fisher | Cat #A36498 |
| Y-27632 (ROCKi) | Stem Cell Technologies | Ca t#72304 |
| Protein-G Dynal magnetic beads | Life Technologies | Cat #10004D |
| Ampure XP beads | Beckman Coulter | Cat #A63881 |
| Biotin D-ATP | Thermo Fisher | Cat #19524016 |
| DNA Polymerase I, Large (Klenow) Fragment | NEB | Cat #M0210 |
| T4 DNA Ligase | NEB | Cat #M0202 |
| GreenTaq | GenScript | Cat #E00043 |
| Doxyclycine | Sigma | Cat #D9891 |
| Critical commercial assays | ||
| Chromium Next GEM Single Cell Multiome ATAC + Gene Expression Reagent Kits | 10X Genomics | Cat #PN-1000283 |
| Chromium Next GEM Chip J Single Cell Kit | 10X Genomics | Cat #PN-1000234 |
| Human Stem Cell Nucleofector Kit | Lonza | Cat #VPH-5012 |
| Rneasy Kit | Qiagen | Cat #74106 |
| RNAse-free DNAse set | Qiagen | Cat #79254 |
| QIAquick PCR purification Kit | Qiagen | Cat #28106 |
| DNeasy Blood and Tissue Kit | Qiagen | Cat #69506 |
| Tagment DNA Enzyme and Buffer Kit | Illumina | Cat #20034197 |
| NEBNext ChIP-Seq Library Prep kit | NEB | Cat #E6240S/L |
| MinElute PCR Purification Kit | Qiagen | Cat # 28006 |
| In-Fusion HD Cloning Kit | Takara Biosciences | Cat #638910 |
| Dead Cell Removal Kit | Miltenyi | Cat #130-090-101 |
| KAPA kit for PolyA enriched mRNA-seq Library prep | Roche | Cat #KK8420 |
| Qbit dsDNA assay kit | Thermo Fisher | Cat #Q32851 |
| TaqMan™ RNA-to-CT™ 1-Step Kit | Thermo Fisher | Cat #392938 |
| Deposited data | ||
| Deep sequencing data | This paper | GEO:GSE165714 |
| Previously published sequencing data (chromHMM) | Pattison et al. 201960 | GEO:GSE114846 |
| Experimental models: Cell lines | ||
| H9 (WA09) human embryonic stem cell line | Stanford Stem Cell Bank | NIHhESC-10-0062 |
| Oligonucleotides (all oligo sequences provided inTable S7) | ||
| GRHL2 KO genotyping F GACTAG TGGCCTTAGTGCCC |
This Paper | N/A |
| GRHL2 KO genotyping R ACTTCCTC TGGATGGGTGAT |
This Paper | N/A |
| AP2A KO genotyping F TGGGGTAG GTAAGTAGGGGG |
This Paper | N/A |
| AP2A KO genotyping R GGCACTG TAGGTCAATCTCCC |
This Paper | N/A |
| GRHL2 KO sgRNA 1 UAGGCU CUUCGGGUAUUGAA |
This Paper | N/A |
| GRHL2 KO sgRNA 2 AGUAGUCAU AGAGCAGGCCG |
This Paper | N/A |
| GRH1 KO sgRNA 1 CUGAAGCAA ACGGCCAGUGU |
This Paper | N/A |
| GRH1 KO sgRNA 2 GCGGCGGU CCUACACUAGUG |
This Paper | N/A |
| GRH1 KO sgRNA 3 CUUUGGUC GCUGCAGUGAGA |
This Paper | N/A |
| GRHL1 KO genotyping F TGGTCTGCA CTTACGTGGTT |
This Paper | N/A |
| Recombinant DNA | ||
| GRHL2 over-expression plasmid | This study | N/A |
| pUCM-CLYBL-hNIL | Addgene | Cat #105841 |
| Software and algorithms | ||
| CellRanger Arc 2.0.0 | 10X Genomics | https://support.10xgenomics.com/single-cell-multiome-atac-gex/software/downloads/latest |
| Seurat 4.0.5 | Hao et al. 202161 | https://satijalab.org/seurat/ |
| Loupe Browser 6.0.0 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest#loupe |
| Monocle 2.22.0 | Trapnell et al. 201462 | http://cole-trapnell-lab.github.io/monocle-release/docs/ |
| Kallisto v0.46.1 | Bray et al. 201663 | https://pachterlab.github.io/kallisto/download.html |
| DESEQ2 1.34.0 | Love et al. 201464 | http://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| bedtools 2.27.1 | Quinlan et al. 201065 | Stanford Sherlock |
| IDR 2.0.3 | Li et al. 201166 | https://github.com/nboley/idr |
| Bowtie 2.3.4.1 | Langmead et al. 201367 | Stanford Sherlock |
| samtools 1.8 | Li et al. 200968 | Stanford Sherlock |
| Macs2 v2.2.7.1 | Zhang et al. 200869 | https://github.com/taoliu/MACS |
| Deeptools 3.5.1 | Ramirez et al. 201470 | Ramirez et al. 2014 |
| HiCPro 2.11.4 | Servant et al. 201571 | https://github.com/nservant/HiC-Pro |
| FitHiChIP 9.0 | Bhattacharyya et al. 201972 | https://github.com/ay-lab/FitHiChIP |
| Diffloop 1.23.1 | Lareu et al. 2021 | 2- |
| GenomicRanges 1.46.1 | Lawrence et al. 201373 | https://bioconductor.org/packages/release/bioc/html/GenomicRanges.html |
| Genova 1.0.0 | Haarhuis et al. 201774 | https://github.com/robinweide/GENOVA |
| chromHMM v1.23 | Ernst et al. 201775 | http://compbio.mit.edu/ChromHMM/ |
| Limma 3.50.0 | Ritchie et al. 2015 | https://github.com/gangwug/limma |
| ImageStudioLite 5.2.5 | Li-Cor | https://www.licor.com/bio/image-studio-lite/ |
| BioRender | Biorender | https://www.Biorender.com |
| Prism 9 | GraphPad | https://www.graphpad.com |
| Leica Application Suite for Advanced Fluorescence v2.7.9 | Leica | https://www.leica-microsystems.com/products/microscope-software/p/leica-application-suite/ |
| EnrichR | Chen et al., 201376 | https://maayanlab.cloud/Enrichr/ |
| Pheatmap 1.0.12 | Kolde, 201977 | https://rdrr.io/cran/pheatmap/ |
| Matplotlib 3.5.0 | Hunter et al. 200778 | https://matplotlib.org/ |
| ggplot2 3.3.5 | Wickham et al. 2016 | https://ggplot2.tidyverse.org/ |
| Networkx 2.5.1 | Hagberg et al. 200879 | https://networkx.org/ |
| Custom SNP pipeline | Piekos et al. 202111 | https://github.com/OroLabStanford/SNP_Prioritization_Pipeline |
| Scipy 1.6.2 | Virtanen et al. 202080 | https://scipy.org/ |
| Numpy 1.20.2 | Harris et al. 202081 | https://numpy.org/ |
| Pandas 1.2.4 | Reback et al. 202082 | https://pandas.pydata.org/ |
| Seaborn 0.11.1 | Waskom et al. 202183 | https://seaborn.pydata.org/index.html |
| Rpy2 3.4.5 | https://rpy2.github.io/ | |
| Re 2.2.1 | Python library 3.7.9 | https://pypi.org/project/regex/ |
| Jupyter 1.0.0 | Kluyver et al. 201684 | https://jupyter.org/ |
| Datacommons 1.4.3 | https://datacommons.org/ | |
| Pywaffle 0.6.4 | https://pypi.org/project/pywaffle/ | |
| Mygene 3.2.2 | Xin et al. 201685 | https://pypi.org/project/mygene/ |
| Stats 3.4.3 | R version 3.4.2 | R Core Team |
| Awk 4.0.2 | ||
| GNU coreutils 8.22 | https://www.gnu.org/software/coreutils/manual/coreutils.html | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Anthony Oro (oro@stanford.edu).
Materials availability
All materials generated in this study are available from the lead contact without restriction.
Experimental model and subject details
ES cell maintenance and differentiation
Human embryonic stem cells (H9, female) were maintained in E8 medium (Life Technologies) on Matrigel (Stem Cell Technologies) coated plates, dissociated using 0.5 mM EDTA, and passaged at 1:10 ratio. For differentiations, the media was changed to Essential 6 (Life Technologies) supplemented with 1 μM retinoic acid (Sigma) and 5 ng/mL Recombinant Human BMP4 (R&D Systems) and replaced every two days for seven days. On day 7, cells were collected for downstream analysis.
Cell line generation
ES cells were singularized by Accutase treatment, washed with PBS, counted (1 million cells) and nucleofected using Lonza Human Stem Cell Nucleofector Kit 1 (Lonza VPH-5012). CRISPR sgRNAs (Synthego) were precomplexed to Cas9 protein (Thermo Fisher) at room temperature for 10 minutes. For GRHL2-overexpression, an sgRNA targeting the CLYBL locus was nucleofected alongside a doxycycline-inducible expression plasmid with CLYBL homology arms.29 Immediately post-nucleofection, cells were grown in E8 supplemented with Rock inhibitor (10 μm Y-27632) for up to 4 days. Post-nucleofection pool and subcloned colonies were genotyped. GRHL2 overexpressing cells were also selected using neomycin. sgRNAs, plasmids, and primer sequences are listed in the key resources table and in Table S7.
Method details
Single cell multiome
Single cell suspensions were prepared with Accutase, and cells were frozen in BamBanker (Wako Chemicals). Cells were thawed in cryovials in a 37°C water bath for 1-2 min, spun out of freezing media, and any large clumps were removed with 40 μM Flowmi Cell Strainers. Dead cells were eliminated with a dead cell removal kit (Miltenyi) per manufacturer instructions. 1 million cells per sample were lysed for 5 min in 1X chilled lysis buffer (10 mN Tris-HCl ph 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% Tween-20, 0.1% NP40 substitute, 0.01% digitonin, 1% BSA, 1 mM DTT, and 1U/μL RNase inhibitor). Cells were washed 3 times with chilled wash buffer (10 mN Tris-HCl ph 7.4, 10 mM NaCl, 3 mM MgCl2, 1% BSA, 0.1% Tween-20, 1 mM DTT, and 1U/μL RNase inhibitor) and resuspended at a concentration of 2,700 nuclei/μL in Diluted Nuclei Buffer (1X Nuclei Buffer (10x Genomics), 1 mM DTT, 1U/μL RNase Inhibitor). 6,000 nuclei were targeted for multiome library prep per the manufacturer’s instructions (10x Genomics Multiome ATAC + Gene Expression User Guide CG000338). scATAC libraries were sequenced on a NovaSeq SP100 lane at a depth of 25,000 read pairs per nucleus. scRNA libraries were sequenced on an S4 NovaSeq at a depth of 20,000 read pairs per nucleus.
Immunoblotting
Whole cell lysates were isolated in RIPA buffer supplemented with protease inhibitors (Roche) and separated on gradient SDS-PAGE gels (Life Technologies). Proteins were wet transferred onto nitrocellulose membranes (0.45microns, BioRad) at 100V for 1h. Membranes were blocked in 5% BSA + TBST for 1h. Primary antibodies were diluted in 5% BSA + TBST and incubated, shaking, with the membranes overnight at 4°C. Primary antibodies used in this study are listed in the key resources table. Fluorescent secondary antibodies compatible with Odyssey CLx (Li-Cor) were used for 2-color imaging of membranes. Image analysis was performed with ImageStudioLite (Li-Cor).
Immunofluorescence
Cells were cultured/differentiated on glass cover slips in 12 well plates, fixed for 10minat room temperature in 4% paraformaldehyde in PBS. Cells were permeabilized for 10 min with permeabilization buffer (0.1% Triton-X + 0.05% Tween-20 in PBS) and blocked for 30 min with 10% Normal Horse Serum (Vector Laboratories) in permeabilization buffer. Antibodies were incubated overnight at 4°C and are listed in the key resources table. Secondary antibodies (Thermo Fisher) were added at 1:500 dilution and incubated at room temperature for 1h. Cells were washed twice in PBS and once in Hoechst (Thermo Fisher) 1:10,000 in PBS. Glass cover slips were mounted onto glass slides with ProLong Gold mounting medium (Life Technologies) before imaging on Leica SP8 confocal microscope. Image analysis was performed on Leica LAS X software.
RNA isolation
Cells were lysed in Trizol (Invitrogen), and the aqueous layer was isolated as indicated by the manufacturer. RNA was purified with a RNeasy kit (Qiagen). DNase (Qiagen) treatment was added to the column prior to elution as per manufacturer instructions. Real time PCR was performed using the ThermoFisher TaqMan RNA-CT 1-Step kit with using a Stratagene real time PCR machine. Probes are listed in Table S7. RNA-seq libraries were prepared using the KAPA kit for PolyA enriched mRNA-seq (Roche) according to the manufacturer’s protocol. Libraries were pooled and sequenced on a NovaSeq. Two (RNA-seq) or 3-5 (qPCR) independent, biological replicates were sequenced per cell type.
ChIP-seq
Cells were grown on 15-cm dishes (25 million cells per replicate), singularized by Accutase (Innovative Cell Technologies), crosslinked by 1% formaldehyde for 10 minutes, lysed (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 0.5% SDS, 1X protease inhibitors), and sonicated to 200-300 bp size using a Bioeruptor (Diagenode). Samples were centrifuged to remove insoluble debris and diluted in dilution buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1X protease inhibitors) to final concentration of 0.1% SDS. Sheared chromatin was incubated overnight at 4°C with appropriate antibodies, followed by incubation on rotator with 30 μL of pre-washed agarose G beads (Invitrogen) for 4h at 4°C. AP2A ChIP antibody (Santa Cruz: sc-12726) used at 20 μg per 25 million cells and GRHL2 ChIP antibody (Sigma Prestige: HPA004820) used at 6 μg per 25 million cells. Beads were washed twice each with low salt buffer (50 mM Tris-HCl pH 8.0, 0.15M NaCl, 1 mM EDTA pH 8.0, 0.1% SDS, 1% triton X-100, 0.1% sodium deoxycholate), high salt buffer (50 mM Tris-HCl pH 8.0, 0.5M NaCl, 1 mM EDTA pH 8.0, 0.1% SDS, 1% triton X-100, 0.1% sodium deoxycholate), and LiCl buffer (50 mM Tris-HCl pH 8.0, 0.15M LiCl, 1 mM EDTA pH 8.0, 1% NP-40, 0.1% sodium deoxycholate). DNA was eluted in 100 μL of elution buffer (50 mM NaHCO3, 1% SDS) and crosslinks were reversed with 4 μL of 5M NaCl incubated overnight at 67°C. RNA was removed by adding 1 μL of 10 mg/mL RNase A and incubating for 30minat 37°C. DNA was cleaned using the Qiagen Qiaquick PCR purification kit and quantified using Qubit (Invitrogen). Between 5 ng and 1 μg of pooled DNA were used for library preparation using NEBNext Ultra™ II DNA Library Prep Kit for Illumina kit (New England Biolabs) and AMPure XP beads (Beckman) according to the manufacturer’s protocol. Single-read libraries were sequenced on Illumina NextSeq or NovaSeq sequencer. Two independent, biological replicates were sequenced per cell type.
ATAC-seq
ATAC-seq was performed as described previously86 and as follows. 7x104 cells were washed with cold PBS and lysed in 0.1% NP40 RSB buffer. Nuclei were transposed with Nextera Transposase at 37°C for 30 minutes, then purified with the Qiagen MinElute PCR Purification Kit. Libraries were amplified for 9 total cycles using the Nextera Ad1 and Ad 2.1–2.16 barcodes. Libraries were purified and eluted using the MinElute columns (Qiagen). Library concentrations were determined with Bioanalyzer High-Sensitivity DNA analysis (Agilent). Paired-end libraries for all samples analyzed were pooled and sequenced on an Illumina NextSeq 500 and two independent, biological replicates were sequenced per sample.
Cohesin HiChIP
Cohesin HiChIP library prep was performed with standard methods.87 25x106 cells were crosslinked and digested with MboI (NEB). After digest, biotin was incorporated into the sticky ends of fragments before ligation. Cohesin ChIP was performed to enrich for proximity ligations bound to cohesin, using an SMC1 antibody (Bethyl, A300-055A). The paired-end library quality was assessed on a MiSeq sequencer before sequencing on an Illumina HiSeq 4000. Three replicates were pooled and sequenced across two HiSeq lanes for a total of 1200 million reads per sample.
GRHL2 expression vector
A doxycycline-inducible GRHL2 expression construct was introduced into the CLYBL29 safe harbor locus to ensure controlled and consistent expression levels of GRHL2. GRHL2 cDNA was first cloned into a CLYBL targeting vector (Addgene #105841) using In-Fusion HD cloning (Takara Biosciences). The construct was nucleofected along with a CLYBL targeting sgRNA (Synthego) and Cas9 (Thermo Fisher) as described above. GRHL2 over-expression was induced with 2 μg/μL doxycycline (Sigma) for 7 days. All sgRNA and plasmids are listed in the key resources table.
Quantification and statistical analysis
Gene expression by qPCR
Significance between groups was calculated using a student’s two-tailed t-test with at least three technical and three biological replicates per experiment.
Gene overlaps
Significance between overlapping gene lists was calculated using a Fisher’s Exact Test.
Single cell multiome processing
FASTQ files were processed using 10x Genomics Cell Ranger Arc 2.0.0 and the human reference genome hg38. Cells with UMI counts between 1000 and 8500 were used for further analysis and cells with mitochondrial percentages above 20% were excluded. For each cell type, Seurat88 was used to create a multimodal object with paired RNA and ATAC profiles, followed by weighted neighbor clustering. To directly compare WT and KO cell types, Seurat objects from each genotype were merged together and then integrated based on transcriptome data. The merged object was split by sample and anchors were identified between samples using FindIntegrationAnchors based on transcripts. 2,000 highly variable RNA features were identified, objects were scaled to regress out cell cycle stages, and PCA was performed using variable features. Cells were clustered using 10 dimensions and a resolution of 0.05. The resulting projection coordinates and RNA-based clusters were imported on top of multiome Loupe files from each cell type in order to simultaneously analyze scATAC and scRNA in the same cells. Differential gene expression, motif analysis, and feature linkage of integrated datasets was performed using the 10X genomics Loupe browser. For pseudotime analysis, data was extracted from the integrated Seurat object to create a Monocle cds.62 2,000 variable features were used to order the pseudotime process, and the dimension was reduced using DDRTree and Monocle scaling.
RNA-seq processing
Pseudoalignment index via kallisto index was generated using Gencode v35 transcript and annotation ref.63 Pseudoalignment count tables in TPM (transcripts per million) were generated using kallisto quant. Resulting files were piped into R package DESEQ264 using the same annotation reference files as alignment. R package limma89 was used to remove batch effects, and adjusted counts were used for differential RNA analysis by DESEQ2 to determine p values and fold changes (p<0.05 was used as a cuttoff). GO term analysis was performed using EnrichR.76
ChIP-seq processing
Fastq files were quality controlled and trimmed using trim_galore (parameters: trim_galore-q 10). Alignment to hg38 was performed using bowtie290 (parameters: bowtie2-p 4--very-sensitive). Reads were subsequently sorted and deduplicated using samtools.68 Macs269 with default settings was used to call peaks. Irreproducible peaks were filtered using IDR (Irreproducible Discovery Rate).66 Total comparable IDR peaks were pooled into a bed file and merged to eliminate overlaps. Bedtools65 multicov was used to populate a count table based on read coverage for use in downstream differential analysis by Deseq2. ChromHMM software91 was used to learn and identify chromatin states. Histone mark sequencing data was published previously.8 Enrichment of each state at ChIP-seq summits was calculated using the NeighborhoodEnrichment command. Enrichments were plotted using Python matplotlib. ChIP-seq heatmaps were plotted using Deeptools.92 To measure the binding strength of GRHL2 or AP2A at genetic variants, individual reads for each allele were counted manually from raw bam files.
ATAC-seq processing
Fastq files were quality controlled and trimmed using trim_galore (parameters: trim_galore-q 10--dont_gzip –fastqc). Alignment to hg38 was performed using bowtie2 (parameters: bowtie2-p 4--very-sensitive). Reads were subsequently sorted and deduplicated using samtools. Following removal of mitochondrial reads, bam files were converted to bed using bedtools bamtobed. Macs2 was used to call peaks (parameters: macs2 callpeak-f BED--nomodel--extsize 73--shift -37-g hs-p 0.05). Resulting peak files from all samples were pooled into a bed file and merged to eliminate overlaps. Bedtools multicov was used to populate a count table based on read coverage for use in downstream differential analysis by Deseq2. ATAC-seq heatmaps were plotted using Deeptools.
Cohesin HiChIP processing
Paired end reads were aligned to hg38 using HiC-Pro.71 10 kb binned matrices from HiC-Pro were used to call high confidence contacts using FitHiChIP.72 Total loops were subset to remove loops smaller than 5000 bases. Loops were further subset by loops that had fewer than 5 contacts in 2 out of the 3 replicates in each genotype. Diffloop93 was used to call differential loops between WT and KO. GenomicRanges73 was used to identify overlaps between GRHL2 ChIP peaks and HiChIP loops and anchors. Differential loops filtered for FDR <0.1 were used for correlational association with differential GRHL2 ChIP binding sites. Genova74 was used to quantify chromatin contact strength at GRHL2 binding sites using 10 kb interaction matrices. 5 kb interaction matrices were used to visualize contacts by Virtual 4C.
Chromatin contact analyses
Two transcription factors and/or genes (TSS +/− 1 kb) were considered connected via chromatin looping if an element was present in one 10 kb bin and the other element was in the corresponding bin of a cohesin contact. Two elements were considered overlapping if they were both present at the same genomic location. Difference in rates of transcription factor participation, LD prevalence, and SNP overlap with ChIP/ATAC sites were evaluated using a Fisher’s Exact Tests.
Empirical cumulative distribution function analysis (ECDF)
ECDF was performed to determine whether the cumulative log2FoldChange in a subset of genes was significantly different compared to all protein-coding genes using a student’s two tailed t-test (scipy v1.6.2). Plots were generated using matplotlib v3.4.1. Only protein-coding genes for which there was a fold change value calculated using DESEQ2 were used in the analysis.
SNP prioritization pipeline
We prioritized genetic variants associated with craniofacial phenotypes.11 Normalized significant genetic variants associated with cleft lip/palate were obtained for craniofacial traits from NHGRI-GBI GWAS Catalog.38 Variants in linkage disequilibrium for each ethnicity were obtained using SNiPA94 using genome assembly GRCh37, variant set 1000 Genomes Project Phase 3 version 5, genome annotation Ensembl 87, and linkage disequilibrium threshold 0.8. A combined list of GWAS significant genetic variants and variants in linkage disequilibrium were converted into GRCh38 using the UCSC Genome Browser liftover tool. Cohesin HiChIP datasets (FDR <0.001) from WT or AP2a KO cells were used as chromatin contact input, GRHL ChIP-seq and AP2a ChIP-seq (FDR <0.01), WT or AP2a KO cells as well as AP2a-dependent ATAC-seq peaks were used as regulatory element input. A bed 5+ file of protein coding genes +/− 1 kB was used as promoter input. The pipeline first subset regulatory elements participating in 3D chromatin connections, followed by filtering for regulatory elements containing one or more genetic variants. The algorithm then associated the paired regulatory element and genetic variant to distal genes. The output is a list of regulatory element-SNP-gene trios which can be found in Tables S2 and S3. Bipartite graphs were generated using the python package network v2.5.1 and visualized using maplotlib v3.4.1.
The clinical significance (ClinVar) and functional category (dbSNP) for each genetic variant were obtained by querying Google Data Commons using the python API.11 CADD scores were obtained from https://cadd.gs.washington.edu.14 Global MAF frequencies as reported by dbGAP and/or the 1000 Genomes Project were retrieved from dbSNP whereas population-specific MAFs were obtained from SNiPA. Code for running the pipeline was published previously11 and is available at https://github.com/OroLabStanford/SNP_Prioritization_Pipeline.
Graphics and gene ontology
Graphics were created with BioRender.com. Gene ontology analysis was performed using EnrichR.com. Volcano and MA plots were made using ggplot2.95 RNA-seq heatmaps were made using pheatmap.77 Bar graphs were made using Prism 9 GraphPad software. ChIP-seq and ATAC-seq Heatmaps were made using deeptools.92 ChromHMM enrichment plots were made using matplotlib.96
Acknowledgments
We thank the Oro Laboratory members for helpful comments. This work was funded by NIH grant RO1AR073170 (AEO), F32ARO70565 (JMP, AEC), Maternal and Child Health Research Institute (AEC).
Author contributions
Conceptualization and methodology, A.C., S.P., A.L., J.P., and T.O. Investigation and validation, A.C., A.L., J.P., F.F., A.B., E.S., and H.Z. Formal analysis, A.C., S.P., A.L., J.P., F.F., A.B., E.S., and S.G. Visualization, A.C., S.P., A.B., E.S., and F.F. Data curation and software, A.C., S.P., and S.G. Writing – original draft, A.C. Writing – review and editing, A.C., S.P., A.L., J.P., F.F., and T.O. Funding acquisition, A.C., S.P., J.P., and T.O. Resources, project administration, and supervision, T.O.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: February 3, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106125.
Supplemental information
Data and code availability
Sequencing data have been deposited at GEO:GSE165714 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper also analyzes existing, publicly available data.8 Any custom code was published previously8,11 and is available at https://github.com/OroLabStanford. All code is publicly available as of the date of publication. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Ganske I.M., Irwin T., Langa O., Upton J., Tan W.H., Mulliken J.B. Cleft lip and palate in ectodermal dysplasia. Cleft Palate. Craniofac. J. 2021;58:237–243. doi: 10.1177/1055665620949124. [DOI] [PubMed] [Google Scholar]
- 2.Nikolopoulou E., Hirst C.S., Galea G., Venturini C., Moulding D., Marshall A.R., Rolo A., De Castro S.C.P., Copp A.J., Greene N.D.E. Spinal neural tube closure depends on regulation of surface ectoderm identity and biomechanics by Grhl2. Nat. Commun. 2019;10:2487. doi: 10.1038/s41467-019-10164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larsen W.J., Lawrence S., Sherman W.J.L. 3rd Ed. Hum. Embryol.; 2002. Ectoderm: Neurulation, Neural Tube, Neural Crest. [Google Scholar]
- 4.Liem K.F., Tremml G., Roelink H., Jessell T.M. Dorsal differentiation of neural plate cells induced by BMP-mediated signals from epidermal ectoderm. Cell. 1995;82:969–979. doi: 10.1016/0092-8674(95)90276-7. [DOI] [PubMed] [Google Scholar]
- 5.Wang W.D., Melville D.B., Montero-Balaguer M., Hatzopoulos A.K., Knapik E.W. Tfap2a and Foxd3 regulate early steps in the development of the neural crest progenitor population. Dev. Biol. 2011;360:173–185. doi: 10.1016/j.ydbio.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tchieu J., Zimmer B., Fattahi F., Amin S., Zeltner N., Chen S., Studer L. A modular platform for differentiation of human PSCs into all major ectodermal lineages. Cell Stem Cell. 2017;21:399–410.e7. doi: 10.1016/j.stem.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrof G., Nanda A., Howden J., Takeichi T., McMillan J.R., Aristodemou S., Ozoemena L., Liu L., South A.P., Pourreyron C., et al. Mutations in GRHL2 result in an autosomal-recessive ectodermal dysplasia syndrome. Am. J. Hum. Genet. 2014;95:308–314. doi: 10.1016/j.ajhg.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pattison J.M., Melo S.P., Piekos S.N., Torkelson J.L., Bashkirova E., Mumbach M.R., Rajasingh C., Zhen H.H., Li L., Liaw E., et al. Retinoic acid and BMP4 cooperate with p63 to alter chromatin dynamics during surface epithelial commitment. Nat. Genet. 2018;50:1658–1665. doi: 10.1038/s41588-018-0263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L., Wang Y., Torkelson J.L., Shankar G., Pattison J.M., Zhen H.H., Fang F., Duren Z., Xin J., Gaddam S., et al. TFAP2C- and p63-dependent networks sequentially rearrange chromatin landscapes to drive human epidermal lineage commitment. Cell Stem Cell. 2019;24:271–284.e8. doi: 10.1016/j.stem.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collier A., Liu A., Torkelson J., Pattison J., Gaddam S., Zhen H., Patel T., McCarthy K., Ghanim H., Oro A.E. Gibbin mesodermal regulation patterns epithelial development. Nature. 2022;606:188–196. doi: 10.1038/s41586-022-04727-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piekos S.N., Gaddam S., Bhardwaj P., Radhakrishnan P., Guha R.V., Oro A.E. Biomedical data commons (BMDC) prioritizes B-lymphocyte non-coding genetic variants in type 1 Diabetes. PLoS Comput. Biol. 2021;17:e1009382. doi: 10.1371/journal.pcbi.1009382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cano-Gamez E., Trynka G. From GWAS to function: using functional genomics to identify the mechanisms underlying complex diseases. Front. Genet. 2020;11:424. doi: 10.3389/fgene.2020.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mather C.A., Mooney S.D., Salipante S.J., Scroggins S., Wu D., Pritchard C.C., Shirts B.H. CADD score has limited clinical validity for the identification of pathogenic variants in noncoding regions in a hereditary cancer panel. Genet. Med. 2016;18:1269–1275. doi: 10.1038/gim.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rentzsch P., Schubach M., Shendure J., Kircher M. CADD-Splice—improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13:31. doi: 10.1186/s13073-021-00835-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen A.F., Liu A.J., Krishnakumar R., Freimer J.W., DeVeale B., Blelloch R. GRHL2-Dependent enhancer switching maintains a pluripotent stem cell transcriptional subnetwork after exit from naive pluripotency. Cell Stem Cell. 2018;23:226–238.e4. doi: 10.1016/j.stem.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung V.Y., Tan T.Z., Ye J., Huang R.L., Lai H.C., Kappei D., Wollmann H., Guccione E., Huang R.Y.J. The role of GRHL2 and epigenetic remodeling in epithelial–mesenchymal plasticity in ovarian cancer cells. Commun. Biol. 2019;2:272. doi: 10.1038/s42003-019-0506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carpinelli M.R., De Vries M.E., Auden A., Butt T., Deng Z., Partridge D.D., Miles L.B., Georgy S.R., Haigh J.J., Darido C., et al. Inactivation of Zeb1 in GRHL2-deficient mouse embryos rescues mid-gestation viability and secondary palate closure. Dis. Model. Mech. 2020;13:dmm042218. doi: 10.1242/dmm.042218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs J., Atkins M., Davie K., Imrichova H., Romanelli L., Christiaens V., Hulselmans G., Potier D., Wouters J., Taskiran I.I., et al. The transcription factor Grainy head primes epithelial enhancers for spatiotemporal activation by displacing nucleosomes. Nat. Genet. 2018;50:1011–1020. doi: 10.1038/s41588-018-0140-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilanowski T., Caddy J., Ting S.B., Hislop N.R., Cerruti L., Auden A., Zhao L.L., Asquith S., Ellis S., Sinclair R., et al. Perturbed desmosomal cadherin expression in grainy head-like 1-null mice. EMBO J. 2008;27:886–897. doi: 10.1038/emboj.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Z., Lin K.K., Bhandari A., Spencer J.A., Xu X., Wang N., Lu Z., Gill G.N., Roop D.R., Wertz P., Andersen B. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev. Biol. 2006;299:122–136. doi: 10.1016/j.ydbio.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 21.Carpinelli M.R., De Vries M.E., Jane S.M., Dworkin S. Grainyhead-like transcription factors in craniofacial development. J. Dent. Res. 2017;96:1200–1209. doi: 10.1177/0022034517719264. [DOI] [PubMed] [Google Scholar]
- 22.Jänicke M., Renisch B., Hammerschmidt M. Zebrafish grainyhead-like1 is a common marker of different non-keratinocyte epidermal cell lineages, which segregate from each other in a Foxi3-dependent manner. Int. J. Dev. Biol. 2010;54:837–850. doi: 10.1387/ijdb.092877mj. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Bolotin D., Chu D.H., Polak L., Williams T., Fuchs E. AP-2á: a regulator of EGF receptor signaling and proliferation in skin epidermis. J. Cell Biol. 2006;172:409–421. doi: 10.1083/jcb.200510002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W., Cornell R.A. Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Dev. Biol. 2007;304:338–354. doi: 10.1016/j.ydbio.2006.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J., Hagopian-Donaldson S., Serbedzija G., Elsemore J., Plehn-Dujowich D., McMahon A.P., Flavell R.A., Williams T. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238–241. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]
- 26.Schorle H., Meier P., Buchert M., Jaenisch R., Mitchell P.J. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- 27.Milunsky J.M., Maher T.A., Zhao G., Roberts A.E., Stalker H.J., Zori R.T., Burch M.N., Clemens M., Mulliken J.B., Smith R., Lin A.E. TFAP2A mutations result in Branchio-Oculo-facial syndrome. Am. J. Hum. Genet. 2008;82:1171–1177. doi: 10.1016/j.ajhg.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Otterloo E., Milanda I., Pike H., Thompson J.A., Li H., Jones K.L., Williams T. AP-2α and AP-2β cooperatively function in the craniofacial surface ectoderm to regulate chromatin and gene expression dynamics during facial development. Elife. 2022;11:e70511. doi: 10.7554/eLife.70511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandopulle M.S., Prestil R., Grunseich C., Wang C., Gan L., Ward M.E. Transcription factor–mediated differentiation of human iPSCs into neurons. Curr. Protoc. Cell Biol. 2018;79:e51. doi: 10.1002/cpcb.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Compagnucci C., Fish J.L., Schwark M., Tarabykin V., Depew M.J. Pax6 regulates craniofacial form through its control of an essential cephalic ectodermal patterning center. Genesis. 2011;49:307–325. doi: 10.1002/dvg.20724. [DOI] [PubMed] [Google Scholar]
- 31.Wang J., Bai Y., Li H., Greene S.B., Klysik E., Yu W., Schwartz R.J., Williams T.J., Martin J.F. MicroRNA-17-92, a direct Ap-2α transcriptional target, modulates T-box factor activity in orofacial clefting. PLoS Genet. 2013;9:e1003785. doi: 10.1371/journal.pgen.1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakatomi M., Ludwig K.U., Knapp M., Kist R., Lisgo S., Ohshima H., Mangold E., Peters H. Msx1 deficiency interacts with hypoxia and induces a morphogenetic regulation during mouse lip development. Development. 2020;147:dev189175. doi: 10.1242/dev.189175. [DOI] [PubMed] [Google Scholar]
- 33.Lo Iacono N., Mantero S., Chiarelli A., Garcia E., Mills A.A., Morasso M.I., Costanzo A., Levi G., Guerrini L., Merlo G.R. Regulation of Dlx5 and Dlx6 gene expression by p63 is involved in EEC and SHFM congenital limb defects. Development. 2008;135:1377–1388. doi: 10.1242/dev.011759. [DOI] [PubMed] [Google Scholar]
- 34.Chai Y., Maxson R.E. Recent advances in craniofacial morphogenesis. Dev. Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- 35.Ferretti E., Li B., Zewdu R., Wells V., Hebert J.M., Karner C., Anderson M.J., Williams T., Dixon J., Dixon M.J., et al. A conserved Pbx-wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev. Cell. 2011;21:627–641. doi: 10.1016/j.devcel.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan Q., Blanton S.H., Hecht J.T. Genetic causes of nonsyndromic cleft lip with or without cleft palate. Adv. Oto-Rhino-Laryngol. 2011;70:107–113. doi: 10.1159/000322486. [DOI] [PubMed] [Google Scholar]
- 37.Knight R.D., Javidan Y., Zhang T., Nelson S., Schilling T.F. AP2-dependent signals from the ectoderm regulate craniofacial development in the zebrafish embryo. Development. 2005;132:3127–3138. doi: 10.1242/dev.01879. [DOI] [PubMed] [Google Scholar]
- 38.Buniello A., Macarthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White J.D., Indencleef K., Naqvi S., Eller R.J., Hoskens H., Roosenboom J., Lee M.K., Li J., Mohammed J., Richmond S., et al. Insights into the genetic architecture of the human face. Nat. Genet. 2021;53:45–53. doi: 10.1038/s41588-020-00741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naqvi S., Sleyp Y., Hoskens H., Indencleef K., Spence J.P., Bruffaerts R., Radwan A., Eller R.J., Richmond S., Shriver M.D., et al. Shared heritability of human face and brain shape. Nat. Genet. 2021;53:830–839. doi: 10.1038/s41588-021-00827-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kousa Y.A., Schutte B.C. Toward an orofacial gene regulatory network. Dev. Dyn. 2016;245:220–232. doi: 10.1002/dvdy.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kousa Y.A., Fuller E., Schutte B.C. IRF6 and AP2A interaction regulates epidermal development. J. Invest. Dermatol. 2018;138:2578–2588. doi: 10.1016/j.jid.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 43.Amooee A., Dastgheib S.A., Niktabar S.M., Noorishadkam M., Lookzadeh M.H., Mirjalili S.R., Heiranizadeh N., Neamatzadeh H. Association of fetal MTHFR 677C > T polymorphism with non-syndromic cleft lip with or without palate risk: asystematic Review and meta-analysis. Fetal Pediatr. Pathol. 2021;40:337–353. doi: 10.1080/15513815.2019.1707918. [DOI] [PubMed] [Google Scholar]
- 44.Sasaki Y., Taya Y., Saito K., Fujita K., Aoba T., Fujiwara T. Molecular contribution to cleft palate production in cleft lip mice. Congenit. Anom. 2014;54:94–99. doi: 10.1111/cga.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H., Leslie E.J., Carlson J.C., Beaty T.H., Marazita M.L., Lidral A.C., Cornell R.A. Identification of common non-coding variants at 1p22 that are functional for non-syndromic orofacial clefting. Nat. Commun. 2017;8:14759. doi: 10.1038/ncomms14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Q., Deng Q., Fu F., Li R., Zhang W., Wan J., Yang X., Wang D., Li F., Wu S., et al. A novel splicing mutation of ARHGAP29 is associated with nonsyndromic cleft lip with or without cleft palate. J. Matern. Fetal Neonatal Med. 2022;35:2499–2506. doi: 10.1080/14767058.2020.1786523. [DOI] [PubMed] [Google Scholar]
- 47.Leslie E.J., Mansilla M.A., Biggs L.C., Schuette K., Bullard S., Cooper M., Dunnwald M., Lidral A.C., Marazita M.L., Beaty T.H., Murray J.C. Expression and mutation analyses implicate ARHGAP29 as the etiologic gene for the cleft lip with or without cleft palate locus identified by genome-wide association on chromosome 1p22. Birth Defects Res. A Clin. Mol. Teratol. 2012;94:934–942. doi: 10.1002/bdra.23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beaty T.H., Murray J.C., Marazita M.L., Munger R.G., Ruczinski I., Hetmanski J.B., Liang K.Y., Wu T., Murray T., Fallin M.D., et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat. Genet. 2010;42:525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vsevolozhskaya O.A., Shi M., Hu F., Zaykin D.V. DOT: gene-set analysis by combining decorrelated association statistics. PLoS Comput. Biol. 2020;16:e1007819. doi: 10.1371/journal.pcbi.1007819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu-Chou Y.H., Chen K.T.P., Lu Y.C., Lin Y.T., Chang H.F., Lo L.J. The SNP rs560426 within ABCA4-ARHGAP29 locus and the risk of nonsyndromic oral clefts. Cleft Palate. Craniofac. J. 2020;57:671–677. doi: 10.1177/1055665619899764. [DOI] [PubMed] [Google Scholar]
- 51.Leslie E.J., Carlson J.C., Shaffer J.R., Butali A., Buxó C.J., Castilla E.E., Christensen K., Deleyiannis F.W.B., Leigh Field L., Hecht J.T., et al. Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Hum. Genet. 2017;136:275–286. doi: 10.1007/s00439-016-1754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsui M., Klingensmith J. Multiple tissue-specific requirements for the BMP antagonist Noggin in development of the mammalian craniofacial skeleton. Dev. Biol. 2014;392:168–181. doi: 10.1016/j.ydbio.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He F., Xiong W., Wang Y., Matsui M., Yu X., Chai Y., Klingensmith J., Chen Y. Modulation of BMP signaling by Noggin is required for the maintenance of palatal epithelial integrity during palatogenesis. Dev. Biol. 2010;347:109–121. doi: 10.1016/j.ydbio.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mangold E., Ludwig K.U., Birnbaum S., Baluardo C., Ferrian M., Herms S., Reutter H., De Assis N.A., Chawa T.A., Mattheisen M., et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet. 2010;42:24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- 55.Peters L.M., Anderson D.W., Griffith A.J., Grundfast K.M., San Agustin T.B., Madeo A.C., Friedman T.B., Morell R.J. Mutation of a transcription factor, TFCP2L3, causes progressive autosomal dominant hearing loss, DFNA28. Hum. Mol. Genet. 2002;11:2877–2885. doi: 10.1093/hmg/11.23.2877. [DOI] [PubMed] [Google Scholar]
- 56.Van Laer L., Van Eyken E., Fransen E., Huyghe J.R., Topsakal V., Hendrickx J.J., Hannula S., Mäki-Torkko E., Jensen M., Demeester K., et al. The grainyhead like 2 gene (GRHL2), alias TFCP2L3, is associated with age-related hearing impairment. Hum. Mol. Genet. 2008;17:159–169. doi: 10.1093/hmg/ddm292. [DOI] [PubMed] [Google Scholar]
- 57.Lin X., Teng Y., Lan J., He B., Sun H., Xu F. GRHL2 genetic polymorphisms may confer a protective effect against sudden sensorineural hearing loss. Mol. Med. Rep. 2016;13:2857–2863. doi: 10.3892/mmr.2016.4871. [DOI] [PubMed] [Google Scholar]
- 58.Gaubatz S., Imhof A., Dosch R., Werner O., Mitchell P., Buettner R., Eilers M. Transcriptional activation by Myc is under negative control by the transcription factor AP-2. EMBO J. 1995;14:1508–1519. doi: 10.1002/j.1460-2075.1995.tb07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu L., Hitchler M.J., Sun W., Sarsour E.H., Goswami P.C., Klingelhutz A.J., Domann F.E. AP-2α inhibits c-MYC induced oxidative stress and apoptosis in HaCaT human keratinocytes. J. Oncol. 2009;2009:780874. doi: 10.1155/2009/780874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pattison J.M., Melo S.P., Piekos S.N., Torkelson J.L., Bashkirova E., Mumbach M.R., Rajasingh C., Zhen H.H., Li L., Liaw E., et al. Retinoic acid and BMP4 cooperate with p63 to alter chromatin dynamics during surface epithelial commitment. Nat. Genet. 2018;50:1658–1665. doi: 10.1038/s41588-018-0263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 64.Love M.I., Anders S., Huber W. 2014. Differential Analysis of Count Data - the DESeq2 Package. [Google Scholar]
- 65.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Q., Brown J.B., Huang H., Bickel P.J. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 2011;5:1752–1779. doi: 10.1214/11-AOAS466. [DOI] [Google Scholar]
- 67.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramirez F., Dundar F., Diehl S., Gruning B.A., Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.J., Vert J.P., Heard E., Dekker J., Barillot E. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhattacharyya S., Chandra V., Vijayanand P., Ay F. Identification of significant chromatin contacts from HiChIP data by FitHiChIP. Nat. Commun. 2019;10:4221. doi: 10.1038/s41467-019-11950-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haarhuis J.H.I., van der Weide R.H., Blomen V.A., Yáñez-Cuna J.O., Amendola M., van Ruiten M.S., Krijger P.H.L., Teunissen H., Medema R.H., van Steensel B., et al. The cohesin release factor WAPL restricts chromatin loop extension. Cell. 2017;169:693–707.e14. doi: 10.1016/j.cell.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ernst J., Kellis M. Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc. 2017;12:2478–2492. doi: 10.1038/nprot.2017.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kolde R. 2019. Pretty Heatmaps R Package. Version 1.0.12. [Google Scholar]
- 78.Hunter J.D. Comput. Sci. Eng. Vol. 9. 2007. Matplotlib: A 2D Graphics Environment; pp. 90–95. [Google Scholar]
- 79.Hagberg A.A., Schult D.A., Swart P.J. In: Exploring network structure, dynamics, and function using networkx. Varoquaux G., Vaught T., Millman J., editors. United States; 2008. pp. 11–15.https://networkx.org/ [Google Scholar]
- 80.Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harris C.R., Millman K.J., van der Walt S.J., Gommers R., Virtanen P., Cournapeau D., Wieser E., Taylor J., Berg S., Smith N.J., et al. Array programming with NumPy. Nature. 2020;585:357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reback, J., McKinney, W., brockmendel, J., et al. (2020). pandas-dev/pandas: Pandas 1.1.3 (zenodo). 10.5281/ZENODO.4067057. [DOI]
- 83.Waskom M.L. seaborn: statistical data visualization. J. open source softw. 2021;6:3021. doi: 10.21105/joss.03021. [DOI] [Google Scholar]
- 84.Kluyver T., Ragan-Kelley B., Pérez F., Granger B., Bussonnier M., et al. In: Positioning and Power in Academic Publishing: Players, Agents and Agendas. Fernando L., Birgit S., editors. IOS Press; 2016. Jupyter Notebooks - a publishing format for reproducible computational workflows; pp. 87–90. [DOI] [Google Scholar]
- 85.Xin J., Mark A., Afrasiabi C., Tsueng G., Juchler M., Gopal N., Stupp G.S., Putman T.E., Ainscough B.J., Griffith O.L., et al. High-performance web services for querying gene and variant annotation. Genome Biol. 2016;17:91. doi: 10.1186/s13059-016-0953-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buenrostro J.D., Wu B., Chang H.Y., Greenleaf W.J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mumbach M.R., Rubin A.J., Flynn R.A., Dai C., Khavari P.A., Greenleaf W.J., Chang H.Y. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods. 2016;13:919–922. doi: 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smyth G.K. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. 2005. Limma: linear models for microarray data; pp. 397–420. [DOI] [Google Scholar]
- 90.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ernst J., Kellis M. Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc. 2017;12:2478–2492. doi: 10.1038/nprot.2017.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T. DeepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lareau C.A., Aryee M.J. Diffloop: a computational framework for identifying and analyzing differential DNA loops from sequencing data. Bioinformatics. 2018;34:672–674. doi: 10.1093/bioinformatics/btx623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arnold M., Raffler J., Pfeufer A., Suhre K., Kastenmüller G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics. 2015;31:1334–1336. doi: 10.1093/bioinformatics/btu779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vinet L., Zhedanov A. A “missing” family of classical orthogonal polynomials. J. Phys. A: Math. Theor. 2011;44:085201. doi: 10.1088/1751-8113/44/8/085201. [DOI] [Google Scholar]
- 96.Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited at GEO:GSE165714 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper also analyzes existing, publicly available data.8 Any custom code was published previously8,11 and is available at https://github.com/OroLabStanford. All code is publicly available as of the date of publication. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.