Abstract
Experimental autoimmune encephalomyelitis (EAE) is a mouse model of multiple sclerosis (MS) in which Th17 cells have a crucial but unclear function. Here we show that choline acetyltransferase (ChAT), which synthesizes acetylcholine (ACh), is a critical driver of pathogenicity in EAE. Mice with ChAT-deficient Th17 cells resist disease progression and show reduced brain-infiltrating immune cells. ChAT expression in Th17 cells is linked to strong TCR signaling, expression of the transcription factor Bhlhe40, and increased Il2, Il17, Il22, and Il23r mRNA levels. ChAT expression in Th17 cells is independent of IL21r signaling but dampened by TGFβ, implicating ChAT in controlling the dichotomous nature of Th17 cells. Our study establishes a cholinergic program in which ACh signaling primes chronic activation of Th17 cells, and thereby constitutes a pathogenic determinant of EAE. Our work may point to novel targets for therapeutic immunomodulation in MS.
Subject terms: Immunology, Inflammation, Experimental models of disease, Cytokines
Introduction
Choline acetyltransferase (ChAT) catalyzes the reversible synthesis of acetylcholine (ACh) from acetyl-CoA and choline [1]. Although ACh is best known as the prototypical neurotransmitter, ChAT is now known to be expressed by a range of non-neuronal cells, including immune cells [2, 3]. Previous work showed that ChAT in lymphoid lineage cells was linked to the paracrine release of cytokines and control of blood pressure [3–5]. We then demonstrated that ChAT expressed in CD8+ T cells and driven by IL-21 signaling was required to induce infection-associated vasodilation [6]. These observations, particularly our finding that ChAT was important in T cell responses to infection, prompted us to investigate if ChAT was also involved in T cell responses mediating autoimmunity.
The interleukin (IL)-17-producing T helper (Th17) cell subset is a primary driver of multiple sclerosis (MS) in humans and experimental autoimmune encephalomyelitis (EAE) in mice [7–11]. In the gut, Th17 cells exert homeostatic functions in maintaining the integrity of epithelial cell barriers [12]. However, some Th17 cells can exhibit pathogenicity, emerging as harmful inflammatory cells driving autoimmunity [13–16]. Although mechanisms of Th17 cell-associated pathogenicity have been investigated for years, the underlying determinants remain obscure [17–23]. Here we use EAE as a model to demonstrate that a cholinergic system involving ChAT and ACh is vital for inducing autoimmunity through the perpetuation of Th17 cell encephalitogenicity.
Methods
Mice
C57BL/6 wild-type (B6), ChatGFP (B6.Cg-Tg(RP23-268L19-EGFP)2Mik/J), Chatflox (B6.129-ChATtm1Jrs/J), CD4Cre (Tg(Cd4-cre)1Cwi/BfluJ), Il21r−/− (B6N.129-IL21rtm1Kopf/J), MOG35-55 specific TCR transgenic 2D2 [C57BL/6-Tg(Tcra2D2,Tcrb2D2)1Kuch/J], Rag1−/− (B6.129S7-Rag1tm1Mom/J), and IL17acre (Il17atm1.1(icre)Stck/J) mice were all from the Jackson Laboratory. Bhlhe40flox (C57BL/6N-Bhlhe40tm1b(KOMP)Wtsi/Wtsi) mice were from the Infrafrontier/EMMA repository. Il17aKatushka mice were the kind gift of Dr. Richard A. Flavell (Yale University). All animals were maintained in fully accredited facilities at the Princess Margaret Cancer Centre.
Induction of EAE and disease analysis
For active induction of EAE, mice were immunized by subcutaneous injection of 100 μg MOG35–55 peptide (MEVGWYRSPFSRVVHLYRNGK) in Complete Freund’s Adjuvant plus 4 mg ml−1 of Mycobacterium tuberculosis H37RA (Difco), followed by intraperitoneal (i.p.) administration of 200 ng pertussis toxin (List Biological Laboratory) on days 0 and 2 post-immunization. Cohorts consisted of age-matched male and female animals. Minimum and maximum ages were 8 and 14 weeks, respectively. Littermate mice were used wherever possible. For passive EAE induction, cells were in vitro-differentiated (see below) and adoptively transferred as described [24]. Briefly, Rag1-/- mice were irradiated with 400 rad 12 h before adoptive transfer and injected i.p. with 1.5×106 sorted cells. Mice were assigned scores daily for EAE development according to the following criteria: 0, no disease; 1, decreased tail tone or mild balance defects; 2, hind limb weakness, partial paralysis or severe balance defects that caused spontaneous falling over; 3, complete hind limb paralysis or very severe balance defects that prevented walking; 4, front and hind limb paralysis or inability to move the body into a different position; 5, moribund state [25]. All mice were scored blind to genotype.
CNS-infiltrating cell isolation
CNS-infiltrating cells were isolated as described [25]. Briefly, at various days post-immunization, mice were perfused through the left cardiac ventricle with PBS, brains were removed, and single-cell suspensions were passed through 70μm strainers. Mononuclear cells were acquired by Percoll gradient (37%/70%) centrifugation. Cells were washed, labeled with antibodies conjugated to fluorochrome dyes, and analyzed by flow cytometry.
Histology and immunohistochemistry
Immunohistochemical and immunofluorescent assessments were performed essentially as described [26, 27]. The anti-neurofilament antibody was purchased from Abcam (ab207176).
In vitro Th17 cell differentiation, stimulation, and treatments
For in vitro Th17 cell differentiation, CD4+ T cells were purified from the spleen using the mouse Naive CD4+ T Cell Isolation Kit (Miltenyi Biotech, 130-104-453), or using the Dynabeads FlowComp, Mouse CD4 Kit (for passive EAE induction experiments; Invitrogen, 11461D). Purified cells were activated in 96-well plates (U-bottom, Greiner) with plate-bound anti-CD3 (2 μg/ml; 1452C11; Biolegend) and anti-CD28 (soluble or plate-bound; 2 μg/ml; 37.51; Biolegend) in IMDM (Gibco) containing 10% heat-inactivated FCS, sodium pyruvate, penicillin-streptomycin, HEPES, glutamate and β-mercaptoethanol. Polarization cytokines and blocking antibodies were: recombinant mouse IL-1β (20 ng/ml), IL-6 (25 ng/ml), IL-23 (20 ng/ml), recombinant human TGFβ1 (3 ng/ml) (all from Biolegend or Peprotech); anti-IFNγ (3 µg/ml, XMG1.2, Biolegend); and anti-IL-4 (3 µg/ml, 11B11, Biolegend). The culture medium was replaced (80%) with freshly conditioned medium 48 h after plating and daily thereafter. Cells were cultured for 5-6 days before harvesting.
For peptide stimulation, splenocytes (5 × 105 cells/well) were cultured for 72 h in 96-well plates (F-Bottom; Falcon) in IMDM containing cytokines and blocking antibodies as above in the presence of 0.4 mM of either MOG35–55 peptide (MEVGWYRSPFSRVVHLYRNGK) or NFM15–35 peptide (RRVTETRSSFSRVSGSPSSGF).
For treatments with FDA-approved drugs, mecamylamine (M9020; Sigma) and oxybutynin (O5015; Sigma) were administered starting on day 3 after EAE induction. Oxybutynin (stock at 3.75 mg/ml sterile water) was diluted, and 0.75 mg in 200 µl per mouse was administered daily via gastric gavage as previously described [28]. As reported previously, Mecamylamine (6.5 mg/kg in PBS) was administered daily by subcutaneous injection [29].
Cell preparations, cell sorting, and flow cytometry
Cell preparations and flow cytometric analyses were performed essentially as described [30]. All single-cell suspensions were treated with Fc-blocking antibody (anti-CD16/32; 2.4G2; Tonbo) before staining with titered antibody dilutions. The following antibodies were used for cell surface staining: anti-CD3 (145-2c11), anti-CD4 (GK1.5), anti-CD8a (53-6.7), anti-TCRγδ (GL3), anti-NK1.1 (PK136), anti-CD19 (1D3), anti-CD25 (PC61), anti-CD11b (M1/70), anti-CD11c (N418), anti-CD44 (IM7), and anti-CD62L (MEL-14) (from BD Biosciences, BioLegend, or Thermo Fisher Scientific). Dead cells were stained and excluded using 12.5 ng/ml DAPI (Sigma).
RNA extraction and real-time PCR
Purified or sorted cells were lysed in Trizol (Invitrogen). RNA was extracted according to the manufacturer’s instructions and reverse-transcribed using the iScript cDNA Synthesis kit (Bio-Rad). For quantitative PCR, 10 μl reactions contained 5 μl 2x Power SYBR Green PCR Sample Mix (Applied Biosystems), 3 μl H2O, 0.5 μl of each primer (at 10 μM concentration), and 1 μl cDNA. Samples were run on an ABI 7900HT Fast Real-Time PCR system (Applied Biosystems).
Chromatin immunoprecipitation (ChIP)
ChIP was performed essentially as described [30]. Briefly, in vitro-polarized Th17 cells originating from Bhlhe40fl/flChATgfp and Bhlhe40fl/flChATgfpCD4cre mice were sorted into ChAT-GFP+ and ChAT-GFP- subsets and 106 Th17 cells were used per sample as input. Two biological replicate samples had to be pooled to obtain sufficient cells for the third Bhlhe40-deficient replicate. The anti-Bhlhe40 ChIP-grade antibody was from Novus Biologicals (NB100-1800), and the IgG isotype control antibody was from Santa Cruz (sc-2027). Primer sequences used for the qPCR analysis of ChIP’ed chromatin were as follows:
ChAT_BS1_fwd: CAAGACAGGGAAAGTCCAATCTCAA
ChAT_BS1_rev: TCAGAAGAGTTTTGACCCTAGATGG
ChAT_BS2_fwd: TTAGAATACTTGTGGGCATTCAAGC
ChAT_BS2_rev: GCAAAACTATATCTGCTGGGTTCAG
Cox6a1_fwd: GGTGCTGTCAGCGTCTCGGG
Cox6a1_rev: CGGGTACCTGAACCCTCCTCG
ChrX_fwd: GGCTTACAAGTAGACACCATCACACT
ChrX_rev: TTGCAAGCAGGAGAACCTTAGTT
RNA sequencing
RNA of flow cytometrically-sorted lymphocytes was processed, and a library was prepared using the Illumina NeoPrep System and protocols. Sequencing was performed at the Princess Margaret Genomics Centre (Toronto) on a Nextseq 500 instrument (Illumina) using a 75-cycle paired-end protocol and multiplexing to obtain ~40 million reads/sample. The raw 76-base pair paired-end reads were quality-checked using FASTQC v0.11.2 and MultiQC v0.8 and aligned to the Mus musculus genome assembly version GRCm38 (mm10) using Tophat v2.1.0. Aligned transcripts were processed using Cufflinks [31] for abundance estimation and differential expression tests using default settings. The differentially expressed gene lists were analyzed to produce overlap Venn diagrams and arrive at a statistically significant subset. Normalized FPKM values for the top genes were used for expression heatmaps.
ATAC sequencing
Sorted cells (5 × 104) were suspended in a 50 μl reaction mixture containing 25 μl 2X TD Buffer (Illumina Nextera kit), 2.5 μl transposase enzyme (Illumina Nextera kit, 15028252), and 22.5 μl nuclease-free water. After 1 h incubation at 37 °C, DNA was purified using the Qiagen MinElute PCR purification kit (28004) (10 μl final volume). ATAC-seq libraries were prepared using the Buenrostro protocol [32] and sequenced as 50 base paired-end reads on the Illumina HiSeq 4000 at the Genome Technology Center at NYU. The three replicates of Th17-polarized CD4+ T cells used for ATAC-seq studies were prepared under slightly different conditions. Cells of replicates 1 and 3 were stimulated using plate-bound antibodies against CD3 and CD28, whereas replicate 2 cells were stimulated using plate-bound anti-CD3 plus soluble anti-CD28. Replicate 3 cells were harvested in the presence of sodium butyrate (0.25 mM). The raw 51-base pair paired-end reads were quality-checked and processed using the ATACseq data-processing pipeline standardized by the ENCODE project [33]. A peak list for each replicate was obtained using MACS [34]. The functions “dba.count” and “dba.peakset” from the DiffBind R package v2.12.0 were used to generate consensus peaks. Read counts within a summit-centered 201-basepair window in these consensus peaks were used to compute fold changes between each pair’s corresponding ChATgfp+ and ChATgfp- samples. A master list for each pair was assembled that contained fold changes, peak locations, and read count values. A peak overlap analysis was conducted in two steps using ChIPpeakAnno v3.22.4. First, overlapping peaks between the ChATgfp+ and ChATgfp- samples were determined for each pair and excluded from further analysis. Peaks unique to the ChATgfp+ and ChATgfp- samples for each pair were retained. Second, a 3-way overlap analysis of the retained peak sets across the three ChATgfp+ samples was conducted to distill a total of 167 peaks that most strongly represented the ChATgfp+ condition. ChIPpeakAnno was utilized to annotate these 167 peaks. The Integrative Genomics Viewer (IGV) [35] tool was used to illustrate selected peak regions.
Limitations of the study
The experimental setup of the adoptive transfer experiment (passive EAE induction) has been employed similarly in many publications addressing Th17 cell biology [15, 24]. Although the results obtained from these experiments substantiate our in vivo findings achieved using active EAE induction, we cannot exclude the possibility that these outcomes instead reflect proper cell activation (in vitro-polarized ChAT-GFP+ Th17 cells) versus a cell state resembling anergy (in vitro-polarized ChAT-GFP- Th17 cells). Alternative passive EAE induction experiments using ex vivo purified ChAT-proficient and ChAT-deficient Th17 cells would eliminate potential artifacts stemming from prolonged in vitro culture or polarization.
Results
ChAT deficiency in Th17 cells confers EAE resistance
To determine if ChAT is required in T lineage cells to induce autoimmunity, we crossed mice harboring loxP-flanked ChAT alleles with CD4cre animals to generate ChATfl/flCD4cre mice lacking ChAT in all T lymphocytes. We induced EAE in these animals by immunization with a peptide containing amino acids 35-55 of myelin oligodendrocyte glycoprotein (MOG35-55) [25, 36]. Mice with ChAT-deficient T cells resisted EAE progression and showed milder symptoms than control littermates (Fig. 1A). In addition, the total number of viable cells isolated from whole brain preparations of these mutants was decreased compared to controls by day 30 post-EAE induction (Fig. 1B). Significantly, flow cytometric analysis showed that the number of brain-infiltrating CD4+ T helper (Th) cells in these samples was also reduced (Fig. 1C). Similar results were obtained for various other immune cell subsets normally recruited to the CNS in EAE, including monocytes, macrophages/microglia, regulatory T cells, and B cells (Supplemental Figs. S1A–D), whereas numbers of brain-infiltrating neutrophils, NK cells, dendritic cells, and CD8+ T cells were not affected (Fig. S1E–H).
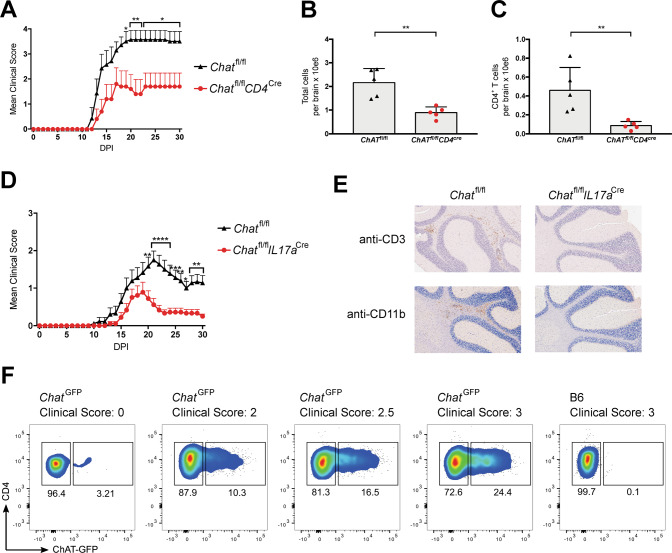
Fig. 1. T cell-specific ChAT deficiency reduces EAE severity.
A EAE clinical scores of Chatfl/fl (n = 7) and Chatfl/flCD4cre (n = 5) mice immunized with an emulsion containing MOG35–55 plus Complete Freund’s Adjuvant (CFA) and pertussis toxin (PTX). DPI, days post-immunization. Mean clinical score: 0, no disease; 1, decreased tail tone or mild balance defects; 2, hind limb weakness, partial paralysis or severe balance defects that caused spontaneous falling over; 3, complete hind limb paralysis or very severe balance defects that prevented walking; 4, front and hind limb paralysis or inability to move the body into a different position; 5, moribund state. B Quantitation of absolute numbers of viable brain-infiltrating immune cells per brain from the Chatfl/fl and Chatfl/flCD4cre mice in A at day 30 post-EAE induction as determined by flow cytometry. C Quantitation of absolute numbers of brain-infiltrating total Th cells (CD3+CD4+) in the brain suspensions in B. D EAE clinical scores of Chatfl/fl (n = 18) and Chatfl/flIl17acre (n = 18) mice immunized as in A. E Histological analyses of the brains of the mice in D on day 30 post-EAE induction. Cross-sections of the same brain areas were stained with anti-CD3 antibody (to detect T cells) or anti-Mac-1 (CD11b) antibody (macrophages, activated microglia). Scale bars, 200 μm. Results shown are for one mouse/genotype representative of three mice/group. F Representative flow cytometric analysis of ChatGFP mice or B6 wild type (WT) control animals (n = 4/group) that were immunized as in A and analyzed on day 21 post-immunization to detect ChAT-GFP+ Th cells. Corresponding clinical scores are indicated. For all applicable panels, data are the mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. For A, D, significance was determined by regression analysis with two-way analysis of variance (ANOVA) followed by Bonferroni post hoc multiple comparison test; (B, C) Student’s t-test (two-sided). Data are representative of three (A, F) or two (D) independent experiments.
Next, we generated ChATfl/flIL17acre mice lacking ChAT specifically in Th17 cells and subjected these animals to EAE induction. Once again, these mutants showed delayed EAE progression and significantly reduced clinical scores from day 20 post-immunization onwards (Fig. 1D). Immunohistochemical analysis (IHC) of brain sections at day 30 post-immunization revealed infiltration by CD3+ T cells and CD11b+ macrophages only in control EAE-induced mice bearing ChAT+ Th17 cells (Fig. 1E). Similarly, immunofluorescent histopathological assessment of spinal cord sections showed significantly more neurofilament (NF) staining in ChATfl/flIL17acre mice than in ChATfl/fl control animals (Fig. S1I, J). Loss of NF staining indicates advanced axon loss and neuronal degeneration [27], a feature we saw only in control EAE-induced mice bearing ChAT+ Th17 cells. We confirmed these results using ChAT-GFP reporter mice, where EAE disease scores correlated positively with the number of brain-infiltrating ChAT-GFP+ Th cells (Fig. 1F). Because the timing of differences in EAE-associated symptoms was virtually identical in ChATfl/flIL17acre and ChATfl/flCD4cre mice, we concluded that ChAT’s role in EAE induction was predominantly linked to Th17 cells.
Bhlhe40 controls ChAT expression in lymphocytes
Murine naïve T cells (CD62L+CD44-) at steady-state never express ChAT; however, ChAT is detected in <10% of follicular (FO) and marginal zone (MZ) B cells and antigen-experienced CD44+ Th cells [3]. We investigated whether these rare ChAT+ cells shared a gene expression program. We sorted ChAT-GFP+ and ChAT-GFP- fractions of FO B cells, MZ B cells, and splenic T cells from ChAT-GFP reporter mice and performed an unbiased comparative RNAseq analysis. Heat-mapping of the top 20 up- or down-regulated genes in ChAT-GFP+ T cells revealed that elevations of Apoe, Ascl2, Esm1, and Padi4 mRNAs were highly associated with ChAT expression, whereas Adh1, Cxcr6, Rras2, and Lad1 mRNAs were markedly increased in ChAT-GFP- T cells (Fig. 2A). We then identified four core genes linked to ChAT in ChAT-GFP+ T and B lymphocyte fractions: transcription factors Bhlhe40 and Fosb; phosphatase Dusp1; and ChAT itself (Fig. 2B). Bhlhe40, Fosb, and Dusp1 are known to contribute to EAE, Th17 cell pathogenicity, or proinflammatory processes [37–39]. Whereas Fosb regulates Th17 cell proliferation, Bhlhe40 is implicated in Th17 pathogenicity [40–42]. We therefore tested whether ChAT is a target gene of Bhlhe40 by generating Bhlhe40fl/flChATgfpCD4cre mice. Deletion of Bhlhe40 significantly reduced ChAT in naïve splenic CD4+ T cells at steady-state (Fig. S2A, B). The same was true for naïve splenic CD4+ T cells induced to differentiate into Th17 cells in vitro by culture with plate-bound anti-CD3 plus anti-CD28 antibodies in the presence of IL-1β + IL-6+IL-23, a cytokine cocktail (PM) that induces polarization towards the pathogenic Th17 cell fate [15] (Fig. 2C, D). Pertinently, an in silico motif analysis of the murine ChAT promoter identified a putative Bhlhe40 binding site (BS1) located ~100 bp upstream of the first non-coding exon (Fig. S2C). Chromatin immunoprecipitation (ChIP) analysis confirmed the direct binding of Bhlhe40 to BS1 in in vitro-polarized Th17 cells derived from Bhlhe40fl/flChATgfp and Bhlhe40fl/flChATgfpCD4cre mice (Fig. 1E). Another putative Bhlhe40 binding site (BS2) located about 500 bp upstream of the first coding exon showed an increased (but not significant) trend towards Bhlhe40 occupancy (Fig. 2F). No Bhlhe40 binding was detected in a negative control region located on the X chromosome (Fig. S2D). Bhlhe40 binding to the human ChAT promoter was verified using a publicly available ChiP-seq dataset (Fig. S2E). Thus, ChAT is a novel Bhlhe40 target in both humans and mice, and a key player in Th17 cell pathogenicity.
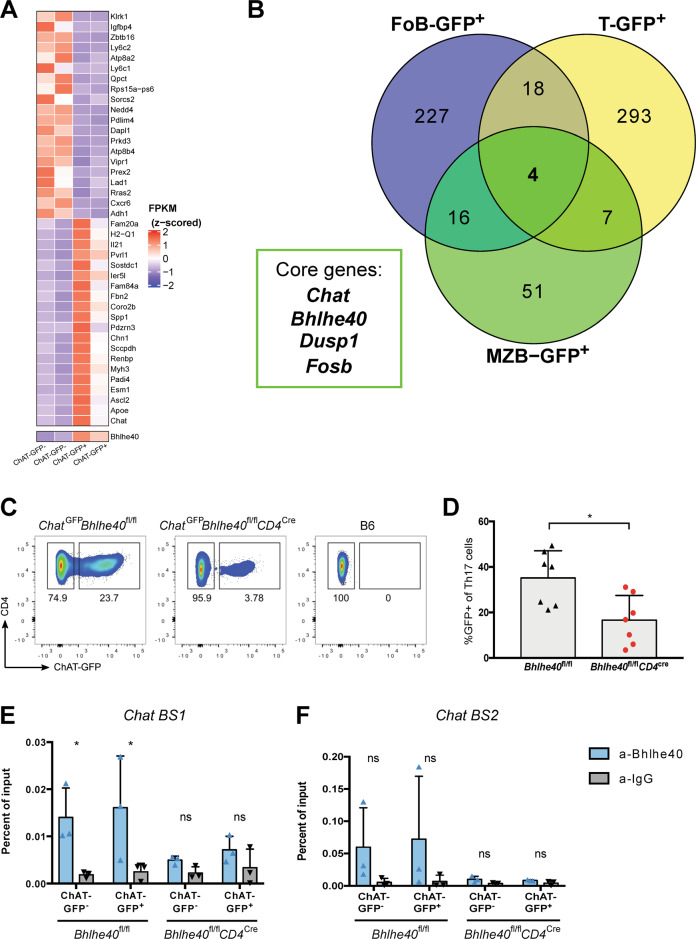
Fig. 2. Bhlhe40-associated Th17 gene expression pattern drives ChAT expression.
A Heat map showing the top 20 up- and down-regulated genes derived from comparative RNA-seq analysis of splenic ChAT-GFP+ and ChAT-GFP– CD4+ T cells. B Venn diagram showing numbers of up-regulated genes (compared to ChAT-GFP- counterparts) shared among ChAT-GFP+ splenic CD4+ T cells (T-GFP+), follicular B cells (FoB-GFP+), and marginal zone B cells (MZB-GFP+) at steady-state as identified by comparative RNA-seq analysis. The four core genes shared by all these ChAT-GFP+ subsets are indicated. C Representative flow cytometric analysis of ChAT-GFP expression in vitro-differentiated Th17 cells derived from ChatGFPBhlhe40fl/fl and ChatGFPBhlhe40fl/flCD4cre mice. Cells were stimulated for 6d with plate-bound anti-CD3 plus anti-CD28 in the presence of IL-1β + IL-6+IL-23 (PM). B6, WT control. D Quantitation of the ChAT-GFP+ Th17 cells in C. *P < 0.05, by Student’s t-test (two-sided). Relative chromatin enrichment as percentages of input in in vitro-differentiated Th17 cells from the mice in C that showed direct binding of Bhlhe40 to BS1 (E) or BS2 (F) in the murine Chat promoter region as revealed by ChIP. *P < 0.05, determined by regression analysis with two-way analysis of variance (ANOVA) followed by Sidak post hoc multiple comparison test. Data in B, and D are compiled from two independent experiments. Data in E, F are from one experiment.
TCR signaling governs ChAT expression in Th17 cells
We next determined if TCR signaling was associated with ChAT induction in Th17 cells. To monitor the kinetics of ChAT-GFP expression, we isolated naïve splenic CD4+ T cells from ChAT-GFP+ mice and subjected them to in vitro Th17 cell differentiation utilizing plate-bound anti-CD3 plus soluble anti-CD28 antibodies plus PM. About 20% of Th17 cells expressed ChAT-GFP after 5-6 days of culture under these conditions (Fig. 3A, Fig. S3A). Utilizing plates coated with both anti-CD3 and anti-CD28 antibodies yielded ~60% ChAT-GFP+ Th17 cells within 6 days (Fig. S3A), and the amount of anti-CD3 antibody used for plate-coating correlated positively with the percentage of ChAT-GFP+ Th17 cells present on day 6 (Fig. S3B, top). Flow cytometric examination of intracellular cytokines revealed that more IL-17a+, GM-CSF+, and IL-17a+GM-CSF+ cells were present in the ChAT-GFP+ fraction than among ChAT-GFP− cells across all anti-CD3 antibody levels (Fig. S3B, middle, bottom). Therefore, the affinity of antigen binding to the TCR, and thus the strength of TCR signaling, may influence ChAT expression.
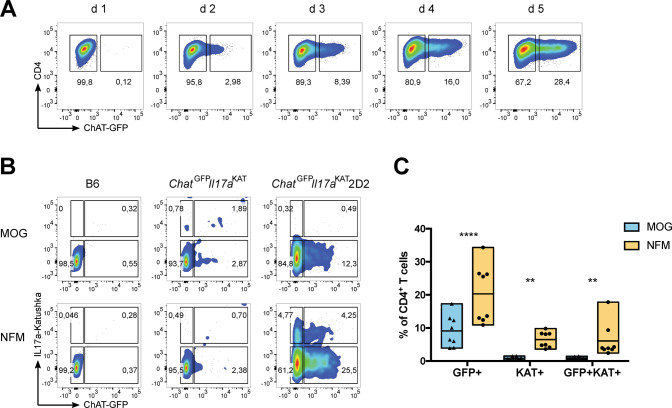
Fig. 3. Antigen affinity determines ChAT expression in Th17 cells.
A Representative flow cytometric analysis of the kinetics over 5 days of ChAT-GFP expression in naïve CD4+ T cells that were polarized towards the Th17 cell fate through stimulation with plate-bound anti-CD3 plus soluble anti-CD28 in the presence of PM. B Representative flow cytometric analysis of ChAT-GFP expression by CD4+ T cells among total splenocytes that were isolated from mice of the indicated genotypes and cultured for 3 days in the presence of PM plus either MOG or NFM peptide (0.4 mM). C Quantitation of the data shown in B. Each data point represents a technical replicate, with a total of 4 biological replicates, from a combination of two independent experiments. Data in A and B are representative of two independent experiments.
To test this hypothesis, we crossed 2D2 mice [which harbor transgenic (tg) expression of a TCR specific for MOG35-55] with reporter mice expressing both ChAT-GFP and IL17a-Katushka [10, 43] to generate ChATGFPIL17aKat2D2 animals. Naïve ChATGFPIL17aKat2D2 splenocytes were Th17-polarized in vitro using either MOG35-55 peptide or a peptide containing amino acids 15-35 of neurofilament medium chain (NFM15-35). MOG35-55 shows a low affinity for 2D2-tg T cells, whereas NFM15-35 binds with high affinity [44, 45]. We found that Th17 polarization in the presence of NFM15-35 yielded ~20% ChAT-GFP+ Th17 cells, whereas polarization in the presence of MOG35-55 resulted in only ~8% ChAT-GFP+ Th17 cells (Fig. 3B, C). Notably, IL17a-Kat+ cells were readily detected in NFM15-35-stimulated cultures but rare in MOG35-55-stimulated cultures. These data suggest that the degree of TCR stimulation determines a T cell’s level of ChAT, and that this ChAT is associated with both Th17 cell differentiation and IL-17 production.
Although both IL21−/− and IL21r−/− mice can develop EAE, IL-21 has been linked to some Th17 cell-associated functions [46, 47]. We recently showed that IL-21 induces ChAT-GFP expression in ~5% of in vitro-stimulated CD8+ T cells [6]. To determine if IL-21 induces ChAT in Th17 cells, we isolated naïve T cells from B6, ChATGFPIL21r+/+ and ChATGFPIL21r−/− mice and polarized them in vitro in the presence of either PM or an alternative cocktail (HM) of TGFβ + IL-6 that induces the differentiation of homeostatic Th17 cells [15, 16, 48–50]. We found no effect of IL-21R deficiency on ChAT-GFP expression in either case (Fig. S4A, B). To confirm these results in vivo, we examined CD4+ Th cells isolated from spleen and mesenteric lymph nodes (MLN) of B6, ChATGFPIL21r+/+ and ChATGFPIL21r−/− mice. Unlike the spleen, MLNs contain Th17 cells even in naïve mice [51]. We found that ChAT-GFP was comparable in splenic and MLN CD4+ Th cells of ChATGFPIL21r+/+ and ChATGFPIL21r−/− mice (Fig. S4 C, D), ruling out a major role for IL21r signaling in inducing ChAT in CD4+ Th cells in vivo.
ChAT determines Th17 cell pathogenicity through specific patterns of gene expression and chromatin state
While the addition of IL-23 to TGFβ + IL-6 (HM) did not affect ChAT-GFP+ Th17 cell differentiation in vitro, the addition of TGFβ to PM significantly reduced their generation (Fig. 4A, B). Th17 cells polarized in HM are much less pathogenic than Th17 cells generated in PM [15, 16], a difference that could be due to differing ChAT levels. To test this theory, we performed passive EAE induction experiments in which Rag1-deficient mice undergo adoptive transfer of pathogenic Th17 cells [16, 52]. Strikingly, fatal EAE disease occurred when Rag1-deficient mice received 2D2-tg ChAT-GFP+ Th17 cells generated in vitro in PM for 6 days (Fig. 4C, D). Rag1-deficient recipients transplanted with 2D2-tg ChAT-GFP- Th17 cells showed only mild EAE signs. Thus, ChAT is a critical determinant of Th17 cell encephalitogenicity.
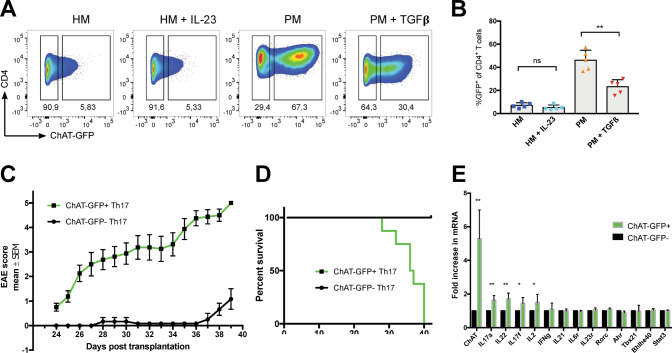
Fig. 4. ChAT is a key determinant of Th17 cell pathogenicity.
A Representative flow cytometric analysis of ChAT-GFP expression by CD4+ splenocytes polarized towards the Th17 cell fate through stimulation with plate-bound anti-CD3 plus anti-CD28 in the presence of either HM (TGFβ + IL-6), HM + IL-23, PM, or PM + TGFβ. Data are representative of at least two independent experiments. B. Quantitation of the data shown in A. Each data point represents one biological replicate (mean of three technical replicates). C EAE clinical scores of Rag1-deficient recipients transplanted with in vitro-generated and sorted 2D2 ChAT-GFP+ (green, n = 8) or 2D2 ChAT-GFP- (black, n = 7) Th17 cells. Disease severity was scored daily. Data are the mean score ± s.e.m. and are representative of three experiments. D Percent survival of the mice in C. E qPCR determination of mRNA levels (relative to Actin) of the indicated genes in ChAT-GFP+ and ChAT-GFP- splenic T cells polarized towards the Th17 fate by incubation in PM for 6 days. Data are the mean ± s.e.m. of 5 biological replicates and representative of 2 experiments.
To explore the molecular basis of ChAT-driven Th17 pathogenicity, we performed qPCR analysis of selected target genes previously implicated in Th17 biology. Naïve ChatGFP T cells were in vitro Th17-polarized in PM for 6 days, followed by isolation of ChAT-GFP- and ChAT-GFP+ Th17 cells. mRNA abundance analysis revealed significant increases in ChAT, Il17a, Il17f, Il22, and Il2 in ChAT-GFP+ Th17 cells compared with their ChAT-GFP- counterparts (Fig. 4E). When we repeated this polarization but followed it by resting the cells in IL-2lo medium for 6 days, ~20% remained ChAT-GFP+ (Fig. S5A). qPCR analysis revealed a 4-fold increase in IL23r in these ChAT-GFP+ Th17 cells, with less dramatic increases in Il21 and Il2 mRNAs (Fig. S5B).
We next performed a limited ATAC-seq analysis of in vitro-differentiated (PM) ChAT-GFP- and ChAT-GFP+ Th17 cells (see Methods). We identified 167 loci shared by ChAT-GFP+ Th17 fractions of three samples (Fig. S6A). Open chromatin stretches predominately associated with ChAT-GFP+ Th17 cells occurred within several genes of potential interest, including Bcl6, Tab2, and Hdac4 (Fig. S6B; Table S1).
Taken together, these data shed light on the specific patterns of gene expression and chromatin state underlying Th17 pathogenicity.
AChR antagonists diminish EAE severity and reduce ChAT-GFP+ T cells in mice
Our data suggested that interference with cholinergic signaling in Th17 cells might decrease their pathogenicity and block EAE progression, a concept that might ultimately be translated to MS therapy. Oxybutynin and mecamylamine are FDA-approved mAChR and nAChR antagonists, respectively, that are well-characterized and known to cross the blood-brain barrier (BBB). Importantly, both drugs ameliorate EAE symptoms in mice: oxybutynin enhances remyelination, whereas mecamylamine reduces inflammation and brain infiltration by T cells [29, 53].
We subjected ChAT-GFP reporter mice to EAE induction and administered oxybutynin or mecamylamine daily starting on day 3. Both treatments delayed disease onset and drastically impeded disease progression, as expected (Fig. 5A). On day 17 (the usual peak of EAE disease), we examined immune cell infiltration and T cell-associated ChAT-GFP expression by flow cytometry and found brain-infiltrating immune cells (Fig. S7A). When calculating cohort disease scores, we observed one outlier animal in each experimental cohort of 5 animals (Fig. S7B), likely mice that either developed only very mild EAE or did not respond to drug treatment.; these outliers were removed from subsequent analyses. While we did not detect differences in total numbers of brain-infiltrating cells in our drug-treated mice (Fig. 5B), administration of mecamylamine or oxybutynin significantly reduced the numbers of brain-infiltrating CD4+ T helper cells (Fig. 5C). Strikingly, only mecamylamine treatment reduced numbers of ChAT-GFP+ T helper cells at disease peak (Fig .5D). These results indicate that multifaceted, non-redundant mechanisms exist to interfere with the cholinergic signaling that controls autoimmune inflammation, Th17 pathogenicity, and licensing of T cells to cross the BBB. Our data further demonstrate that modulation of cholinergic T cells via AchR antagonist drugs is a viable means of decreasing EAE severity, a therapeutic strategy that may eventually be suitable for the MS clinic.
Fig. 5. AChR antagonists impair brain infiltration by Th cells in EAE-induced mice.
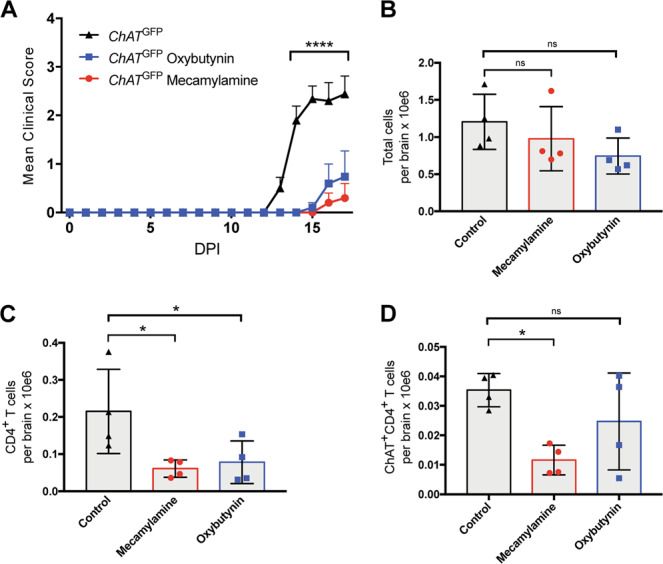
A Clinical scores of ChatGFP mice that were immunized to induce EAE and either left untreated or treated daily with mecamylamine or oxybutynin starting on day 3 (n = 5 mice/group). B Quantitation of absolute numbers of total viable brain-infiltrating immune cells per brain from the ChatGFP mice in A at day 17 post-EAE induction as determined by flow cytometry. C Quantitation of absolute numbers of brain-infiltrating Th cells (CD3+CD4+) in the brain suspensions in B. D Quantitation of absolute numbers of brain-infiltrating ChAT-expressing Th cells (ChAT-GFP+CD3+CD4+) in the brain suspensions in C. For B–D, n = 4 mice/group. For A significance was determined by regression analysis with two-way analysis of variance (ANOVA) followed by Bonferroni post hoc multiple comparison test; for B–D, significance was determined using ordinary one-way ANOVA followed by False Discovery Rate (FDR) post hoc multiple comparisons correction. The FDR was controlled by employing the two-stage step-up method of Benjamini, Krieger, and Yekutieli. Data are from one experiment.
Discussion
Our study has elucidated that ChAT expression is a key feature driving Th17 pathogenicity, both in vivo (EAE induction) and in vitro (under inflammatory conditions lacking TGFβ). Furthermore, we demonstrated that TGFβ antagonizes ChAT induction and that ChAT expression is associated with increased intrinsic Th17 cell pathogenicity. In vitro, we found up-regulation of important proinflammatory mediators such as IL-17f, IL-22, IL-23r, and Tab2. In vivo, our examination of ChAT-GFP reporter mice revealed that EAE severity correlates positively with the number of brain-infiltrating ChAT+ T cells and that ChAT-deficiency reduces this infiltration as well as disease symptoms. Our experiments using AChR antagonists confirmed that Th cells need ACh-mediated signaling to breach the BBB. Although these drugs did not alter the total number of all immune cell types infiltrating the brain, they did decrease the number of ChAT+ (and thus potentially pathogenic) Th cells in brain tissue.
ACh controls endothelial cell activation and BBB integrity [54–62], but it is unclear whether ACh acts alone during BBB permeabilization. The increased brain infiltration phenotype we observed in ChAT-GFP+ mice is reminiscent of the enhanced vasodilation capacity of ChAT+ CD8+ T cells responding to chronic LCMV infection [6]. While IL-21 signaling was critical for ChAT expression in CD8+ T cells, it does not induce ChAT in Th17 cells. Thus, there are multiple routes to achieving ChAT expression in immune cells, necessitating their delineation in specific disease scenarios. With respect to autoreactive Th17 cells, it appears that strong TCR activation induces ChAT expression, which might exert vasodilative effects on the BBB that facilitate infiltration of Th cells into the brain. The emergence of ChAT-GFP+ Th cells was blocked only by treatment with mecamylamine (a pan-nAChR antagonist), suggesting that mAChRs and nAChRs have non-redundant functions in this context. We propose that signaling through nAChRs drives the expression of a neuronal-associated pathogenic gene expression pattern (e.g., Hdac4, Gdf7, Tab2, Il1rl2, Bcl6) in an autocrine manner [63], whereas signaling through mAChRs might work in a paracrine fashion. However, this hypothesis and the delineation of potential target cells (endothelial cells, macrophages, neutrophils, or others) responding to the ACh production by ChAT+ Th17 cells remain to be determined in future studies.
Interestingly, in a recent report, increased EAE severity was associated with ACh released by the vagus nerve rather than by ChAT+ T cells [64]. Although both ChAT- and ChAT+ T cells were reduced upon vagotomy in mice, these subsets were not investigated for their functions. Vagotomy did, however, reduce Th17 cell frequency and proliferation. We speculate that vagus nerve-induced ACh may have propelled the differentiation of pathogenic Th17 cells that bolstered EAE progression.
Our work has pinpointed four key genes expressed in ChAT+ lymphocytes, namely ChAT, Bhlhe40, Fosb, and Dusp1. We identified ChAT as a novel Bhlhe40 target in Th17 cells, in line with Bhlhe40’s known role in shaping of Th17 cell pathogenicity [37, 40]. Because Bhlhe40 function is vital in human memory Th cells [65] as well as in tissue-resident memory and tumor-infiltrating T cells [66], it will be interesting to investigate ChAT in these cell populations. The MAPK phosphatase Dusp1 is implicated in human chronic inflammatory diseases and is a positive regulator of T cell activation [39, 67]. Notably, Dusp1-deficient mice show reduced sensitivity to EAE induction [39, 67]. Fosb is linked to inflammation, Th17 pathogenicity, and pro-inflammatory, excitotoxic microglia [38, 68–72]. Importantly, the products of all four core genes (including ChAT) can localize to the nucleus under certain conditions [42, 73–76]. Although ChAT-GFP- and ChAT-GFP+ Th17 cells showed comparable mRNA levels of these genes, a ChAT-associated cholinergic system might induce post-translational modifications of the corresponding proteins that would allow them to participate in a gene expression program linked to Th17 cell pathogenicity.
Our initial ATAC-seq results identified open chromatin stretches in the Gdf7 and Hdac genomic regions in pathogenic ChAT+ Th17 cells. Gdf7 induces a program driving sensory neuron differentiation [77], whereas Hdac4 is a histone deacetylase that controls the expression of cholinergic components in neuromuscular junctions [78]. Both Bhlhe40 and Hdac4 negatively regulate Pgc-1a, which increases mitochondrial efficiency [79–82]. Upregulation of Hdac4 and Bhlhe40 in pathogenic ChAT+ Th17 cells might thereby decrease ROS generated by the enhanced metabolism of these cells. In this way, ACh signaling may safeguard the pathogenic Th17 cell fate under hypermetabolic conditions, a hypothesis to be explored in future work.
In conclusion, our data indicate that strong TCR activation in the absence of TGFβ signaling initiates a cholinergic program that drives Th17 cell pathogenicity. In other words, ChAT expression in Th17 cells appears to be a consequence of chronic activation of these cells, a scenario strikingly similar to the inflammatory setting in MS patients; that is, incessant autoimmune T cell stimulation coupled with prolonged inflammation. Our study may therefore point to promising starting points for the design of novel MS treatments.
Supplementary information
Acknowledgements
We thank the Princess Margaret Genomics Centre and the UHN Bioinformatics and HPC Core for RNA sequencing; the Weill Cornell Medicine and Genome Technology Center (GTC) at NYU for ATAC sequencing; and the Pathology Core at The Centre for Phenogenomics in Toronto for all histology services, antibody optimization, and immunohistochemistry. We are grateful to M.E.S. for scientific editing and Dr. Thorsten Berger for careful reading of the manuscript. We also thank the Genotyping Facility and the Animal Resource Centre at the Princess Margaret Cancer Centre (Toronto).
Author contributions
R.N. and T.W.M conceived the project. R.N. and D.N. initiated the project, designed and performed experiments, and analyzed data. G.S.D., C.Z., C.G., V.B.C., and M.G. performed experiments and analyzed data. K.T.G., J.H., A.C.W., and B.S. assisted with experiments. P.R. and Z.L. performed bioinformatic analyses. R.N. wrote the original draft of the manuscript with input from D.N., P.R., M.E.S., and T.W.M. R.A.F. kindly provided Il17a-Katushka mice. T.W.M supervised the research. All authors have reviewed and approved the manuscript in its current form.
Funding
This work was supported by a grant to T.W.M. and R.N. from the Canadian Institutes of Health Research (CIHR) and a postdoctoral fellowship to R.N. from EMBO and Marie Curie (ALTF 725-2015).
Data availability
The bulk RNA-seq and the ATAC-seq raw and processed data files can be accessed using the GEO SuperSeries with accession GSE215673. The Bhlhe40 TFBS track shown in Fig. S2C is from the JASPAR CORE 2022 Predicted Transcription Factor Binding Sites repository incorporated as a standard track into the UCSC Genome Browser. The Bhlhe40 ChIPseq track shown in Fig. S2E corresponds to the IgG-rab ChIP-seq Signal from the Snyder Lab dataset (ENCODE/SYDH, GSM935440). In addition, all data needed to evaluate the conclusions in this publication are present in the main article or the Supplementary Information.
Code availability
Critical scripts used to analyze the RNAseq and ATACseq data presented in the paper are available at https://github.com/CodeInTheSkies/Th17-Cell-Pathogenicity-Data.
Competing interests
The authors do not have competing financial interests concerning the work described. T. Mak owns equity in Treadwell Therapeutics Inc. and Agios Pharmaceuticals and is a consultant for AstraZeneca and Tessa Therapeutics.
Ethics approval
The Ontario Cancer Institute Animal Care Committee approved all animal research and mouse experiments.
Footnotes
Edited by G Melino
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Robert Nechanitzky, Duygu Nechanitzky.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-01092-y.
References
- 1.Oda Y. Choline acetyltransferase: the structure, distribution and pathologic changes in the central nervous system. Pathol Int. 1999;49:921–37. doi: 10.1046/j.1440-1827.1999.00977.x. [DOI] [PubMed] [Google Scholar]
- 2.Cox MA, Bassi C, Saunders ME, Nechanitzky R, Morgado-Palacin I, Zheng C, et al. Beyond neurotransmission: acetylcholine in immunity and inflammation. J Intern Med. 2020;287:120–33. doi: 10.1111/joim.13006. [DOI] [PubMed] [Google Scholar]
- 3.Reardon C, Duncan GS, Brustle A, Brenner D, Tusche MW, Olofsson PS, et al. Lymphocyte-derived ACh regulates local innate but not adaptive immunity. Proc Natl Acad Sci USA. 2013;110:1410–5. doi: 10.1073/pnas.1221655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olofsson PS, Steinberg BE, Sobbi R, Cox MA, Ahmed MN, Oswald M, et al. Blood pressure regulation by CD4(+) lymphocytes expressing choline acetyltransferase. Nat Biotechnol. 2016;34:1066–71. doi: 10.1038/nbt.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosas-Ballina M, Olofsson PS, Ochani M, Valdes-Ferrer SI, Levine YA, Reardon C, et al. Acetylcholine-synthesizing T cells relay neural signals in a vagus nerve circuit. Science. 2011;334:98–101. doi: 10.1126/science.1209985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox MA, Duncan GS, Lin GHY, Steinberg BE, Yu LX, Brenner D, et al. Choline acetyltransferase-expressing T cells are required to control chronic viral infection. Science. 2019;363:639–44. doi: 10.1126/science.aau9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korn T, Kallies A. T cell responses in the central nervous system. Nat Rev Immunol. 2017;17:179–94. doi: 10.1038/nri.2016.144. [DOI] [PubMed] [Google Scholar]
- 8.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–8. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 9.Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140:845–58. doi: 10.1016/j.cell.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 10.Gagliani N, Amezcua Vesely MC, Iseppon A, Brockmann L, Xu H, Palm NW, et al. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature. 2015;523:221–5. doi: 10.1038/nature14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Jarjour AA, Boyd A, Williams A. Central nervous system remyelination in culture—a tool for multiple sclerosis research. Exp Neurol. 2011;230:138–48. doi: 10.1016/j.expneurol.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 13.Patel DD, Kuchroo VK. Th17 cell pathway in human immunity: lessons from genetics and therapeutic interventions. Immunity. 2015;43:1040–51. doi: 10.1016/j.immuni.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Stockinger B, Omenetti S. The dichotomous nature of T helper 17 cells. Nat Rev Immunol. 2017;17:535–44. doi: 10.1038/nri.2017.50. [DOI] [PubMed] [Google Scholar]
- 15.Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–71. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–9. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–40. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kullberg MC, Jankovic D, Feng CG, Hue S, Gorelick PL, McKenzie BS, et al. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J Exp Med. 2006;203:2485–94. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–49. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung Y, Chang SH, Martinez GJ, Yang XO, Nurieva R, Kang HS, et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009;30:576–87. doi: 10.1016/j.immuni.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol. 2011;12:255–63. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komuczki J, Tuzlak S, Friebel E, Hartwig T, Spath S, Rosenstiel P, et al. Fate-mapping of GM-CSF expression identifies a discrete subset of inflammation-driving T helper cells regulated by Cytokines IL-23 and IL-1beta. Immunity. 2019;50:1289–304 e1286. doi: 10.1016/j.immuni.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 23.McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, et al. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10:314–24. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thakker P, Leach MW, Kuang W, Benoit SE, Leonard JP, Marusic S. IL-23 is critical in the induction but not in the effector phase of experimental autoimmune encephalomyelitis. J Immunol. 2007;178:2589–98. doi: 10.4049/jimmunol.178.4.2589. [DOI] [PubMed] [Google Scholar]
- 25.Brustle A, Brenner D, Knobbe CB, Lang PA, Virtanen C, Hershenfield BM, et al. The NF-kappaB regulator MALT1 determines the encephalitogenic potential of Th17 cells. J Clin Invest. 2012;122:4698–709. doi: 10.1172/JCI63528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mak TW, Grusdat M, Duncan GS, Dostert C, Nonnenmacher Y, Cox M, et al. Glutathione Primes T cell metabolism for inflammation. Immunity. 2017;46:675–89. doi: 10.1016/j.immuni.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 27.Mei F, Lehmann-Horn K, Shen YA, Rankin KA, Stebbins KJ, Lorrain DS, et al. Accelerated remyelination during inflammatory demyelination prevents axonal loss and improves functional recovery. Elife. 2016;5:1–21. [DOI] [PMC free article] [PubMed]
- 28.Klausner AP, Sharma S, Fletcher S, Neff P, Yang SK, Son H, et al. Does oxybutynin alter plaques, amyloid beta peptides and behavior in a mouse model of Alzheimer’s disease? J Urol. 2008;179:1173–7. doi: 10.1016/j.juro.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 29.Godin JR, Roy P, Quadri M, Bagdas D, Toma W, Narendrula-Kotha R, et al. A silent agonist of alpha7 nicotinic acetylcholine receptors modulates inflammation ex vivo and attenuates EAE. Brain Behav Immun. 2020;87:286–300. doi: 10.1016/j.bbi.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nechanitzky R, Akbas D, Scherer S, Gyory I, Hoyler T, Ramamoorthy S, et al. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol. 2013;14:867–75. doi: 10.1038/ni.2641. [DOI] [PubMed] [Google Scholar]
- 31.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. [DOI] [PMC free article] [PubMed]
- 32.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–21.29.9. [DOI] [PMC free article] [PubMed]
- 33.https://github.com/ENCODE-DCC/atac-seq-pipeline.
- 34.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. [DOI] [PMC free article] [PubMed]
- 35.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bornancin F, Renner F, Touil R, Sic H, Kolb Y, Touil-Allaoui I, et al. Deficiency of MALT1 paracaspase activity results in unbalanced regulatory and effector T and B cell responses leading to multiorgan inflammation. J Immunol. 2015;194:3723–34. doi: 10.4049/jimmunol.1402254. [DOI] [PubMed] [Google Scholar]
- 37.Lin CC, Bradstreet TR, Schwarzkopf EA, Jarjour NN, Chou C, Archambault AS, et al. IL-1-induced Bhlhe40 identifies pathogenic T helper cells in a model of autoimmune neuroinflammation. J Exp Med. 2016;213:251–71. doi: 10.1084/jem.20150568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoppmann N, Graetz C, Paterka M, Poisa-Beiro L, Larochelle C, Hasan M, et al. New candidates for CD4 T cell pathogenicity in experimental neuroinflammation and multiple sclerosis. Brain. 2015;138:902–17. doi: 10.1093/brain/awu408. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Reynolds JM, Chang SH, Martin-Orozco N, Chung Y, Nurieva RI, et al. MKP-1 is necessary for T cell activation and function. J Biol Chem. 2009;284:30815–24. doi: 10.1074/jbc.M109.052472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin CC, Bradstreet TR, Schwarzkopf EA, Sim J, Carrero JA, Chou C, et al. Bhlhe40 controls cytokine production by T cells and is essential for pathogenicity in autoimmune neuroinflammation. Nat Commun. 2014;5:3551. doi: 10.1038/ncomms4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown JR, Nigh E, Lee RJ, Ye H, Thompson MA, Saudou F, et al. Fos family members induce cell cycle entry by activating cyclin D1. Mol Cell Biol. 1998;18:5609–19. doi: 10.1128/MCB.18.9.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milde-Langosch K, Kappes H, Riethdorf S, Loning T, Bamberger AM. FosB is highly expressed in normal mammary epithelia, but down-regulated in poorly differentiated breast carcinomas. Breast Cancer Res Treat. 2003;77:265–75. doi: 10.1023/A:1021887100216. [DOI] [PubMed] [Google Scholar]
- 43.Bettelli E, Pagany M, Weiner HL, Linington C, Sobel RA, Kuchroo VK. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J Exp Med. 2003;197:1073–81. doi: 10.1084/jem.20021603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenthal KM, Edwards LJ, Sabatino JJ, Jr, Hood JD, Wasserman HA, Zhu C, et al. Low 2-dimensional CD4 T cell receptor affinity for myelin sets in motion delayed response kinetics. PLoS One. 2012;7:e32562. doi: 10.1371/journal.pone.0032562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krishnamoorthy G, Saxena A, Mars LT, Domingues HS, Mentele R, Ben-Nun A, et al. Myelin-specific T cells also recognize neuronal autoantigen in a transgenic mouse model of multiple sclerosis. Nat Med. 2009;15:626–32. doi: 10.1038/nm.1975. [DOI] [PubMed] [Google Scholar]
- 46.Lee Y, Mitsdoerffer M, Xiao S, Gu G, Sobel RA, Kuchroo VK. IL-21R signaling is critical for induction of spontaneous experimental autoimmune encephalomyelitis. J Clin Invest. 2015;125:4011–20. doi: 10.1172/JCI75933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonderegger I, Kisielow J, Meier R, King C, Kopf M. IL-21 and IL-21R are not required for development of Th17 cells and autoimmunity in vivo. Eur J Immunol. 2008;38:1833–8. doi: 10.1002/eji.200838511. [DOI] [PubMed] [Google Scholar]
- 48.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 49.Lee JY, Hall JA, Kroehling L, Wu L, Najar T, Nguyen HH, et al. Serum amyloid a proteins induce pathogenic Th17 cells and promote inflammatory disease. Cell. 2020;180:79–91. doi: 10.1016/j.cell.2019.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–74. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 51.Kawabe T, Sun SL, Fujita T, Yamaki S, Asao A, Takahashi T, et al. Homeostatic proliferation of naive CD4+ T cells in mesenteric lymph nodes generates gut-tropic Th17 cells. J Immunol. 2013;190:5788–98. doi: 10.4049/jimmunol.1203111. [DOI] [PubMed] [Google Scholar]
- 52.Sheng W, Yang F, Zhou Y, Yang H, Low PY, Kemeny DM, et al. STAT5 programs a distinct subset of GM-CSF-producing T helper cells that is essential for autoimmune neuroinflammation. Cell Res. 2014;24:1387–402. doi: 10.1038/cr.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deshmukh VA, Tardif V, Lyssiotis CA, Green CC, Kerman B, Kim HJ, et al. A regenerative approach to the treatment of multiple sclerosis. Nature. 2013;502:327–32. doi: 10.1038/nature12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kirschner H. [Surgical endodontic tooth preservation with implant material] Zahnarzt Prax. 1991;42:246–50. [PubMed] [Google Scholar]
- 55.Ignarro LJ, Buga GM, Wood KS, Byrns RE, Chaudhuri G. Endothelium-derived relaxing factor produced and released from artery and vein is nitric oxide. Proc Natl Acad Sci USA. 1987;84:9265–9. doi: 10.1073/pnas.84.24.9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saeed RW, Varma S, Peng-Nemeroff T, Sherry B, Balakhaneh D, Huston J, et al. Cholinergic stimulation blocks endothelial cell activation and leukocyte recruitment during inflammation. J Exp Med. 2005;201:1113–23. doi: 10.1084/jem.20040463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beny JL, Nguyen MN, Marino M, Matsui M. Muscarinic receptor knockout mice confirm involvement of M3 receptor in endothelium-dependent vasodilatation in mouse arteries. J Cardiovasc Pharm. 2008;51:505–12. doi: 10.1097/FJC.0b013e31816d5f2f. [DOI] [PubMed] [Google Scholar]
- 58.Radu BM, Osculati AMM, Suku E, Banciu A, Tsenov G, Merigo F, et al. All muscarinic acetylcholine receptors (M1-M5) are expressed in murine brain microvascular endothelium. Sci Rep. 2017;7:5083. doi: 10.1038/s41598-017-05384-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu MH, Jin HK, Floten HS, Yang Q, Yim AP, Furnary A, et al. Vascular endothelial growth factor-mediated endothelium-dependent relaxation is blunted in spontaneously hypertensive rats. J Pharm Exp Ther. 2001;296:473–7. [PubMed] [Google Scholar]
- 60.Mayhan WG. VEGF increases permeability of the blood-brain barrier via a nitric oxide synthase/cGMP-dependent pathway. Am J Physiol. 1999;276:C1148–1153. doi: 10.1152/ajpcell.1999.276.5.C1148. [DOI] [PubMed] [Google Scholar]
- 61.Arnold WP, Mittal CK, Katsuki S, Murad F. Nitric oxide activates guanylate cyclase and increases guanosine 3’:5’-cyclic monophosphate levels in various tissue preparations. Proc Natl Acad Sci USA. 1977;74:3203–7. doi: 10.1073/pnas.74.8.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Furchgott RF, Zawadzki JV. The obligatory role of endothelial cells in the relaxation of arterial smooth muscle by acetylcholine. Nature. 1980;288:373–6. doi: 10.1038/288373a0. [DOI] [PubMed] [Google Scholar]
- 63.Simard AR, Gan Y, St-Pierre S, Kousari A, Patel V, Whiteaker P, et al. Differential modulation of EAE by alpha9*- and beta2*-nicotinic acetylcholine receptors. Immunol Cell Biol. 2013;91:195–200. doi: 10.1038/icb.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou L, Lin X, Ma X, Liu Y, Ma L, Chen Z, et al. Acetylcholine regulates the development of experimental autoimmune encephalomyelitis via the CD4+ cells proliferation and differentiation. Int J Neurosci. 2020;130:788–803. doi: 10.1080/00207454.2019.1706504. [DOI] [PubMed] [Google Scholar]
- 65.Emming S, Bianchi N, Polletti S, Balestrieri C, Leoni C, Montagner S, et al. A molecular network regulating the proinflammatory phenotype of human memory T lymphocytes. Nat Immunol. 2020;21:388–99. doi: 10.1038/s41590-020-0622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li C, Zhu B, Son YM, Wang Z, Jiang L, Xiang M, et al. The Transcription Factor Bhlhe40 programs mitochondrial regulation of resident CD8(+) T cell fitness and functionality. Immunity. 2019;51:491–507.e497. doi: 10.1016/j.immuni.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Korhonen R, Moilanen E. Mitogen-activated protein kinase phosphatase 1 as an inflammatory factor and drug target. Basic Clin Pharm Toxicol. 2014;114:24–36. doi: 10.1111/bcpt.12141. [DOI] [PubMed] [Google Scholar]
- 68.Moon YM, Lee SY, Kwok SK, Lee SH, Kim D, Kim WK, et al. The Fos-related antigen 1-JUNB/Activator Protein 1 Transcription Complex, a downstream target of signal transducer and activator of Transcription 3, induces T Helper 17 differentiation and promotes experimental autoimmune arthritis. Front Immunol. 2017;8:1793. doi: 10.3389/fimmu.2017.01793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nomaru H, Sakumi K, Katogi A, Ohnishi YN, Kajitani K, Tsuchimoto D, et al. Fosb gene products contribute to excitotoxic microglial activation by regulating the expression of complement C5a receptors in microglia. Glia. 2014;62:1284–98. doi: 10.1002/glia.22680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holtman IR, Skola D, Glass CK. Transcriptional control of microglia phenotypes in health and disease. J Clin Invest. 2017;127:3220–9. doi: 10.1172/JCI90604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cervantes-Madrid DL, Nagi S, Asting Gustafsson A. FosB transcription factor regulates COX-2 expression in colorectal cancer cells without affecting PGE2 expression. Oncol Lett. 2017;13:1411–6. doi: 10.3892/ol.2017.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H, Bradbury JA, Dackor RT, Edin ML, Graves JP, DeGraff LM, et al. Cyclooxygenase-2 regulates Th17 cell differentiation during allergic lung inflammation. Am J Respir Crit Care Med. 2011;184:37–49. doi: 10.1164/rccm.201010-1637OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huynh JP, Lin CC, Kimmey JM, Jarjour NN, Schwarzkopf EA, Bradstreet TR, et al. Bhlhe40 is an essential repressor of IL-10 during Mycobacterium tuberculosis infection. J Exp Med. 2018;215:1823–38. doi: 10.1084/jem.20171704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jeffrey KL, Camps M, Rommel C, Mackay CR. Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat Rev Drug Disco. 2007;6:391–403. doi: 10.1038/nrd2289. [DOI] [PubMed] [Google Scholar]
- 75.Tian J, Wu J, Chen X, Guo T, Chen ZJ, Goldstein JL, et al. BHLHE40, a third transcription factor required for insulin induction of SREBP-1c mRNA in rodent liver. Elife. 2018;7:1–18. [DOI] [PMC free article] [PubMed]
- 76.Gill SK, Bhattacharya M, Ferguson SS, Rylett RJ. Identification of a novel nuclear localization signal common to 69- and 82-kDa human choline acetyltransferase. J Biol Chem. 2003;278:20217–24. doi: 10.1074/jbc.M213153200. [DOI] [PubMed] [Google Scholar]
- 77.Lo L, Dormand EL, Anderson DJ. Late-emigrating neural crest cells in the roof plate are restricted to a sensory fate by GDF7. Proc Natl Acad Sci USA. 2005;102:7192–7. doi: 10.1073/pnas.0502581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cohen TJ, Waddell DS, Barrientos T, Lu Z, Feng G, Cox GA, et al. The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J Biol Chem. 2007;282:33752–9. doi: 10.1074/jbc.M706268200. [DOI] [PubMed] [Google Scholar]
- 79.Chang HC, Kao CH, Chung SY, Chen WC, Aninda LP, Chen YH, et al. Bhlhe40 differentially regulates the function and number of peroxisomes and mitochondria in myogenic cells. Redox Biol. 2019;20:321–33. doi: 10.1016/j.redox.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luo L, Martin SC, Parkington J, Cadena SM, Zhu J, Ibebunjo C, et al. HDAC4 controls muscle homeostasis through deacetylation of myosin heavy chain, PGC-1alpha, and Hsc70. Cell Rep. 2019;29:749–763. doi: 10.1016/j.celrep.2019.09.023. [DOI] [PubMed] [Google Scholar]
- 81.Qian X, Li X, Shi Z, Bai X, Xia Y, Zheng Y, et al. KDM3A senses oxygen availability to regulate PGC-1alpha-mediated mitochondrial biogenesis. Mol Cell. 2019;76:885–895. doi: 10.1016/j.molcel.2019.09.019. [DOI] [PubMed] [Google Scholar]
- 82.Fu G, Xu Q, Qiu Y, Jin X, Xu T, Dong S, et al. Suppression of Th17 cell differentiation by misshapen/NIK-related kinase MINK1. J Exp Med. 2017;214:1453–69. doi: 10.1084/jem.20161120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bulk RNA-seq and the ATAC-seq raw and processed data files can be accessed using the GEO SuperSeries with accession GSE215673. The Bhlhe40 TFBS track shown in Fig. S2C is from the JASPAR CORE 2022 Predicted Transcription Factor Binding Sites repository incorporated as a standard track into the UCSC Genome Browser. The Bhlhe40 ChIPseq track shown in Fig. S2E corresponds to the IgG-rab ChIP-seq Signal from the Snyder Lab dataset (ENCODE/SYDH, GSM935440). In addition, all data needed to evaluate the conclusions in this publication are present in the main article or the Supplementary Information.
Critical scripts used to analyze the RNAseq and ATACseq data presented in the paper are available at https://github.com/CodeInTheSkies/Th17-Cell-Pathogenicity-Data.