Abstract
Non-alcoholic fatty liver disease (NAFLD) has become a growing public health problem. However, the complicated pathogenesis of NAFLD contributes to the deficiency of effective clinical treatment. Here, we demonstrated that liver-specific loss of Arid2 induced hepatic steatosis and this progression could be exacerbated by HFD. Mechanistic study revealed that ARID2 repressed JAK2-STAT5-PPARγ signaling pathway by promoting the ubiquitination of JAK2, which was mediated by NEDD4L, a novel E3 ligase for JAK2. ChIP assay revealed that ARID2 recruited CARM1 to increase H3R17me2a level at the NEDD4L promoter and activated the transcription of NEDD4L. Moreover, inhibition of Jak2 by Fedratinib in liver-specific Arid2 knockout mice alleviated HFD-induced hepatic steatosis. Downregulation of ARID2 and the reverse correlation between ARID2 and JAK2 were also observed in clinical samples. Therefore, our study has revealed an important role of ARID2 in the development of NAFLD and provided a potential therapeutic strategy for NAFLD.
Subject terms: Metabolic disorders, Gene regulation, Ubiquitylation, Histone post-translational modifications, Genetics research
Introduction
Non-alcoholic fatty liver disease (NAFLD) and its more severe form non-alcoholic steatohepatitis (NASH) have become serious threats to world health. Hepatic steatosis, defined as the accumulation of triglycerides (TG) within hepatocytes, is the primary hallmark of NAFLD [1]. Obesity and diabetes contribute to the prevalence of NAFLD worldwide, which may cause severe hepatic outcomes including NASH, hepatic cirrhosis, liver failure and hepatocellular carcinoma, thus increase liver-related mortality [2–4]. However, no effective therapy has been approved for the treatment of NAFLD because of its complicated pathogenesis. Previous studies have underscored the importance of genetic factors in the development of NAFLD [5]. Hence, identification of the major genetic factors contributing to the development of NAFLD and clarification of their function will provide therapeutic benefit for this disease.
ARID2 is a subunit of chromatin remodeling complexes SWI/SNF, which is involved in numerous biological processes [6–8]. The SWI/SNF complex remodels nucleosomes and binds to specific DNA regions to regulate gene expression by influencing epigenetic modifications [9, 10]. Recently, increasing evidence has suggested that subunits of SWI/SNF show significant importance in metabolic homeostasis. For example, BAF60a was identified as a regulator of hepatic lipid metabolism [11]. Loss of Arid1a drove NAFLD in mice via epigenetic dysregulation of hepatic genes involved in lipogenesis and fatty acid oxidation [12]. However, the role of ARID2 in the development of NAFLD remains elusive.
Emerging evidence has shown that Janus kinase 2 (JAK2) and the downstream signal transducer and activator of transcription 5 (STAT5) can regulate the expression of PPARγ to modulate adipogenesis. It was reported that Ginsenoside Rg3 ameliorated HFD-induced hepatic steatosis through downregulation of STAT5-PPARγ signaling [13], and Cucurbitacin E reduced obesity and related metabolic dysfunction by targeting JAK2-STAT5 signaling [14]. These studies indicated the important role of JAK2-STAT5-PPARγ axis in lipid metabolism and NAFLD.
Coactivator-associated arginine methyltransferase 1 (CARM1), also known as PRMT4, is a type 1 protein arginine methyltransferase (PRMT) [15] and is reported to function as a transcriptional coactivator by asymmetrically dimethylating the arginine 17 of histone H3 (H3R17me2a) [16, 17]. The roles of CARM1 in cancer have been reported [18, 19], however, whether CARM1 participates in the development of NAFLD is still unknown.
In this study, we found that liver-specific Arid2 knockout mice developed spontaneous steatosis on chow diet and were more susceptible to HFD-induced hepatic steatosis. We explored the underlying mechanism and disclosed ARID2-mediated inhibition of JAK2-STAT5-PPARγ signaling. ARID2 promoted ubiquitination of JAK2 via upregulation of its E3 ligase NEDD4L. CARM1 was recruited by ARID2 to increase H3R17me2a level at the NEDD4L promoter. Accordingly, JAK2 inhibitor Fedratinib could restrain hepatic steatosis induced by HFD in Arid2 LKO mice. Decreased ARID2 level and the negative correlation between ARID2 and JAK2 were further confirmed in clinical samples. Therefore, our study elucidated the significant role of ARID2 in maintaining hepatic lipid homeostasis and provided potential therapeutic targets for NAFLD.
Results
Liver-specific loss of Arid2 leads to development of hepatic steatosis
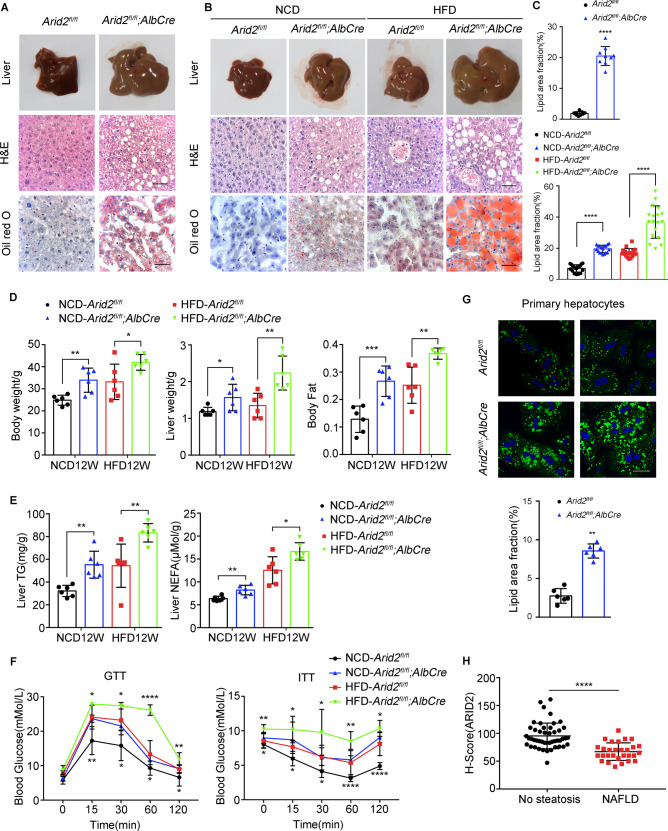
To determine the function of ARID2 in liver metabolism, we generated liver-specific Arid2 knockout mice by crossing Arid2 flox/flox mice with Albumin-Cre mice (Arid2 LKO). Arid2 LKO mice developed spontaneous hepatic steatosis at 7 months as indicated by liver pictures, H&E and Oil Red O staining (Fig. 1A, C). To further confirm this observation, both WT and Arid2 LKO mice were fed with either normal chow diet (NCD) or high-fat diet (HFD) for 12 weeks (Supplementary Fig. S1A). The livers of Arid2 LKO mice were larger than those of WT mice on either NCD or HFD. Histologically, H&E and Oil Red O staining demonstrated more lipid droplets with increased size in the livers of Arid2 LKO mice compared with WT mice, and HFD amplified this difference (Fig. 1B, C). Arid2 LKO mice showed higher body weight, liver weight and body fat content compared with WT mice (Fig. 1D). Consistently, hepatic triglyceride (TG) contents and non-esterified fatty acid (NEFA) levels were dramatically elevated in Arid2 LKO mice under both NCD and HFD conditions (Fig. 1E). Similar alterations were observed in serum TG contents and NEFA levels of the mice fed with NCD (Supplementary Fig. S1B). Glucose tolerance testing (GTT) and Insulin tolerance testing (ITT) were performed in 19-week-old mice. As shown in Fig. 1F, Arid2 LKO mice exhibited more severe glucose intolerance and insulin resistance compared with their WT counterparts.
Fig. 1. Arid2 LKO mice are more prone to the development of hepatic steatosis.
A Liver images, H&E and Oil Red O staining of the livers from WT (n = 3) and Arid2 LKO (n = 3) mice at 7 months. Scale bars, 45 μm. B Liver images, H&E and Oil Red O staining of the livers from WT (n = 6) and Arid2 LKO (n = 6) mice on either NCD or HFD at 20 weeks of age. Scale bars, 45 μm. C Statistical quantification of the Oil Red O staining in panel A and B, 3 images per mouse were randomly chosen for analysis. D Body weight, liver weight and body fat of WT (n = 6) and Arid2 LKO (n = 6) mice fed with NCD or HFD for 12 weeks. E Triglyceride (TG) and non-esterified fatty acid (NEFA) level in the livers of WT (n = 6) and Arid2 LKO (n = 6) mice at 20 weeks. F Glucose tolerance tests (GTTs) and Insulin tolerance tests (ITTs) of WT (n = 6) and Arid2 LKO (n = 6) mice at 19 weeks. The comparison was performed within HFD group and NCD group, respectively. G BODIPY staining and statistical quantification (n = 2 per group, 3 images per mouse) of the lipid droplets in primary hepatocytes isolated from 2-month-old WT and Arid2 LKO mice. Scale bars, 90 μm. Blue signal, nuclei stained with Hoechst; Green signal, lipid droplets stained with BODIPY. H H-scores of ARID2 expression in the livers from people without NAFLD (n = 47) and NAFLD patients (n = 29). All experiments were repeated 3 times independently. Quantification of the lipid droplets in the livers of 7-month old mice (C, upper panel) and primary hepatocytes (G, lower panel) was analyzed with Mann-Whitney test. Other data were analyzed with two-tailed unpaired Student’s t test. Results were represented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 ****p < 0.0001.
In addition, the inhibitory effect of ARID2 on hepatic lipid accumulation was validated in vitro. We isolated primary hepatocytes from 2-month-old WT and Arid2 LKO mice. BODIPY staining revealed increased accumulation of lipid droplets in the hepatocytes from Arid2 LKO mice compared with those from WT mice (Fig. 1G). To confirm this observation in human hepatic cells, we used shRNA to knockdown ARID2 in HepG2 cells and obtained similar results (Supplementary Fig. S1C). In contrast, Nile Red staining showed that lipid accumulation under normal condition or upon palmitic acid (PA) stimulation was reduced in ARID2-overexpressing Hep3B cells (Supplementary Fig. S1D). Inhibition of lipid droplets accumulation by ARID2 was further confirmed in ARID2 knockdown SNU387 and YY-8103 (Supplementary Fig. S1E) and ARID2-overexpressing Huh7 and SNU398 HCC cells (Supplementary Fig. S1F). Notably, decreased expression of ARID2 was detected by immunochemistry in the livers from NAFLD patients compared with those from people without NAFLD (Fig. 1H and Supplementary Fig. S1G). Therefore, both in vivo and in vitro studies suggested that ARID2 was required for the maintenance of hepatic lipid homeostasis and ARID2 deficiency led to hepatic steatosis, which was also indicated by the data from clinical samples.
ARID2 regulates hepatic JAK2-STAT5-PPARγ signaling
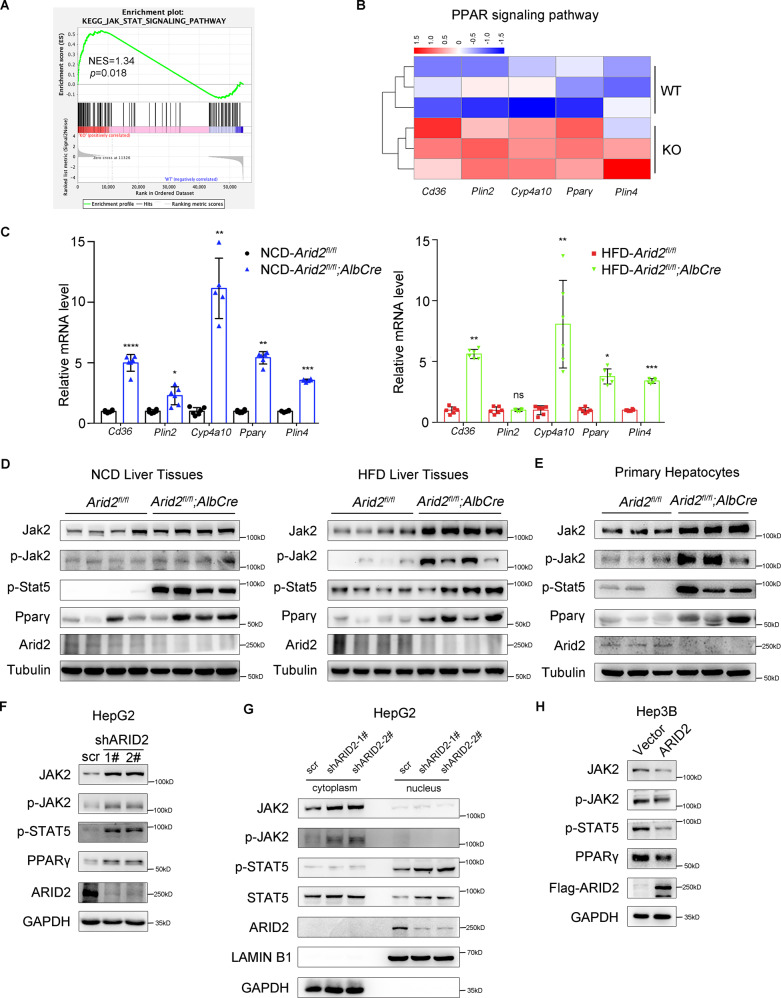
To explore the mechanism underlying the dysregulated lipid metabolism induced by Arid2 deficiency, we analyzed the transcriptome of the livers from WT and Arid2 LKO mice using RNA-seq. Reactome analysis indicated that genes differentially expressed in Arid2 LKO group were strongly associated with fatty acid metabolism (Supplementary Fig. S2A), which was consistent with hepatic steatosis phenotype. Gene set enrichment analysis (GSEA) revealed that JAK-STAT signaling pathway was significantly elevated in Arid2 LKO group (Fig. 2A). Considering that STAT5 regulates adipogenesis by activating PPARγ signaling [20], we further analyzed the expression of genes in PPAR signaling pathway and found that 5 genes were significantly upregulated in Arid2 LKO group, including Cd36, Pparγ, Plin2, Cyp4a10 and Plin4 (Fig. 2B). Altered gene expression was verified in the livers of mice fed with NCD and HFD (Fig. 2C). In addition, we also assessed the expression of genes related to hepatic inflammation and fibrosis in mouse livers. There were few changes in the mRNA levels of most genes under NCD feeding. However, under HFD, the expression of inflammatory genes showed an increasing trend and the expression of fibrotic genes including Colla1, Colla2 and Ctgf were apparently elevated in the livers of Arid2 LKO mice (Supplementary Fig. S2B, C), indicating that Arid2 LKO mice fed with HFD may be predisposed to develop hepatic steatohepatitis.
Fig. 2. ARID2 inhibits JAK2-STAT5-PPARγ signaling pathway.
A The gene set enrichment analysis (GSEA) plot of JAK-STAT signaling pathway based on the RNA seq analysis of the livers from WT and Arid2 LKO mice (n = 3 per group). NES, normalized enrichment score. B Heatmap of the differentially expressed genes in PPAR signaling pathway. C The relative mRNA levels of lipid metabolism-associated genes in the livers of NCD- or HFD-fed WT (n = 6) and Arid2 LKO (n = 6) mice at the age of 20 weeks. The mRNA expression levels of the indicated genes were normalized to that of Gapdh. D Expression of the indicated genes in the liver tissues from WT and Arid2 LKO mice fed with NCD and HFD were examined by western blot (n = 4 per group). E Expression of the indicated genes in the primary hepatocytes isolated from 2-month-old WT and Arid2 LKO mice were examined by western blot (n = 3 per group). F Protein levels of the indicated genes in control and ARID2 knockdown HepG2 cells. G The expression levels of JAK2, phosphorylated JAK2, STAT5, phosphorylated STAT5 and ARID2 in cytoplasm and nuclei were examined by nuclear and cytoplasmic separation assay in control and ARID2 knockdown HepG2 cells. H Protein levels of the indicated genes in control and ARID2-overexpressing Hep3B cells. All experiments were repeated 3 times independently. Data were analyzed by two-tailed unpaired Student’s t test and represented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 ****p < 0.0001, ns not significant.
Based on these results, we hypothesized that STAT5 and its upstream JAK2 may be involved in ARID2-regulated hepatic steatosis. The protein level of Jak2-Stat5-Pparγ axis was examined in the liver tissues of both NCD- and HFD-fed mice (Fig. 2D). The levels of phosphorylated Stat5 and Pparγ were remarkably elevated in the liver tissues of Arid2 LKO mice compared with their WT counterparts. Moreover, both total and phosphorylated Jak2 were increased as well, suggesting that the Jak2-Stat5-Pparγ signaling pathway was activated in the livers of Arid2 LKO mice. This activation was further validated in the primary hepatocytes from 2-month-old Arid2 LKO mice and their WT counterparts (Fig. 2E). Furthermore, we confirmed this observation in ARID2 knockdown HepG2 cells in vitro (Fig. 2F). Once activated, phosphorylated JAK2 phosphorylates downstream STAT5, which subsequently dimerizes, translocates to the nucleus and binds to target gene promoters, such as PPARγ [21, 22]. To verify this procedure, we performed nuclear and cytoplasmic separation assay in control and ARID2 knockdown HepG2 cells. As shown in Fig. 2G, increased total and phosphorylated JAK2 were mainly detected in the cytoplasm of ARID2 knockdown cells. Meanwhile, elevated level of p-STAT5 was detected in the nuclei of ARID2 knockdown cells. Consistently, we found that overexpression of ARID2 in Hep3B cells repressed JAK2-STAT5-PPARγ signaling (Fig. 2H). Taken together, these data indicated that ARID2 inhibited JAK2-STAT5-PPARγ signaling, which may account for its role in the regulation of lipid metabolism.
ARID2 regulates lipid accumulation via JAK2-STAT5 signaling pathway
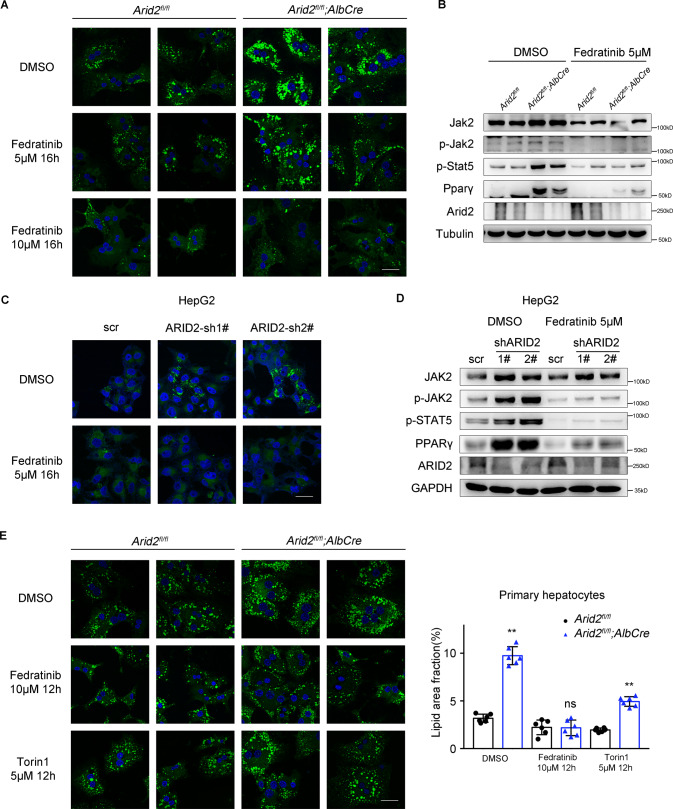
To validate the involvement of JAK2-STAT5 pathway in ARID2-regulated hepatic lipid metabolism, primary hepatocytes isolated from Arid2 LKO mice and their WT littermates were treated with Fedratinib, an FDA approved JAK2 inhibitor. BODIPY staining demonstrated that intracellular lipid deposition gradually decreased with increasing doses of Fedratinib (Fig. 3A and Supplementary Fig. S3A). Decrease in lipid droplets accumulation was more remarkable in Fedratinib-treated Arid2 KO cells, indicating Arid2 deletion renders hepatocytes more sensitive to Fedratinib. In addition, another JAK2 inhibitor Ruxolitinib and Pimozide, an inhibitor of STAT5 phosphorylation were further employed to validate the involvement of JAK2-STAT5 signaling in ARID2-regulated lipid accumulation. Ruxolitinib and Pimozide showed similar but weaker effect when compared with Fedratinib (Supplementary Fig. S3B). The STAT3 inhibitor Cryptotanshinone was used as a negative control. As shown in Fig. 3B, Fedratinib efficiently suppressed the level of phosphorylated Jak2 and Stat5, as well as the expression of Pparγ. The changes caused by Fedratinib in lipid accumulation and gene expression in ARID2 knockdown HepG2 cells were similar to those in Arid2 KO hepatocytes (Fig. 3C, D and Supplementary Fig. S3C). We also confirmed the impact of Pimozide/Fedratinib on ARID2 knockdown HepG2 cells (Supplementary Fig. S3D) and YY-8103 cells for further validation (Supplementary Fig. S3E). mTORC1 plays an significant role in lipid metabolism [23], thus we treated the primary hepatocytes with mTOR inhibitor Torin1 to exclude the potential impact of mTOR on hepatic steatosis induced by Arid2 deficiency. As shown in Fig. 3E, Torin1 only slightly reduced lipid droplets content in Arid2 KO hepatocytes, while Fedratinib showed stronger inhibitory effect on lipid accumulation, which identified JAK2 as the leading cause of hepatic lipid metabolism disorder in Arid2 LKO mice. Therefore, the above data proved that JAK2-STAT5-PPARγ axis mediated the regulation of hepatic lipid metabolism by ARID2.
Fig. 3. Inhibition of JAK2 attenuates lipid droplets accumulation in vitro.
A BODIPY staining of lipid droplets in control and Fedratinib-treated WT and Arid2 KO hepatocytes. DMSO was used as a control. Scale bars, 90 μm. Blue signal, nuclei stained with Hoechst; Green signal, lipid droplets stained with BODIPY. B The inhibitory effect of Fedratinib on Jak2-Stat5 signaling in primary hepatocytes was confirmed by western blot. C BODIPY staining of lipid droplets in control and ARID2 knockdown HepG2 cells treated with DMSO or 5 μM Fedratinib for 16 h. Scale bars, 90 μm. D Inhibition of JAK2-STAT5 signaling by Fedratinib in HepG2 cells. E Primary hepatocytes isolated from WT and Arid2 LKO mice (n = 2 per group) were treated with either 10 μM Fedratinib or 5 μM Torin1 for 12 h. Lipid droplets in the hepatocytes were stained by BODIPY and quantified by Image J (3 images per mouse). Scale bars, 90 μm. All experiments were repeated 3 times independently. Data in E were analyzed with Mann-Whitney test and represented as the mean ± SEM. **p < 0.01, ns not significant.
ARID2 promotes ubiquitination of JAK2 via E3 ligase NEDD4L
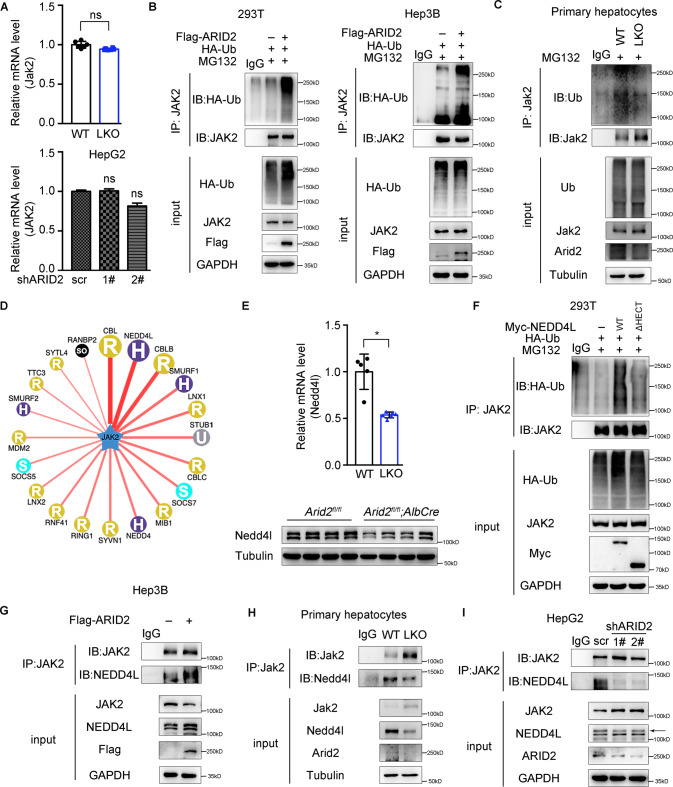
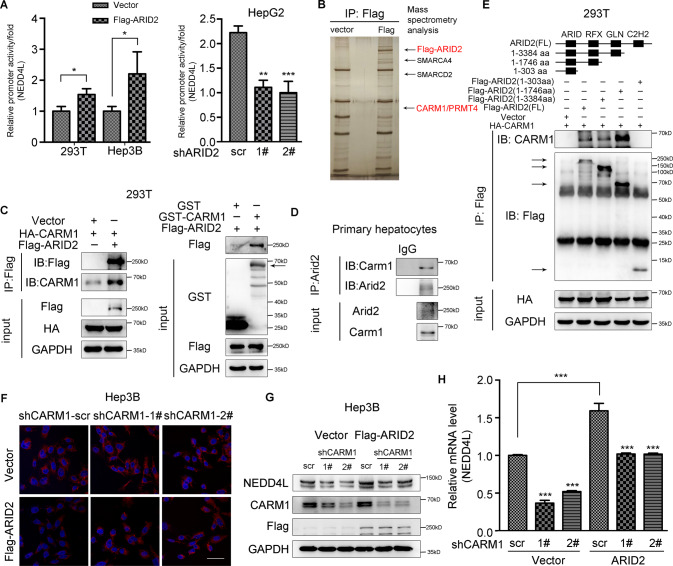
It was worthy to note that expression of JAK2 was highly elevated in the livers of Arid2 LKO mice. Firstly, we wondered whether this change occurred at transcriptional level. However, the mRNA level of hepatic Jak2 in Arid2 LKO mice remained almost the same as that in WT mice, which was further confirmed in ARID2 knockdown HepG2 cells (Fig. 4A). Thus, we speculated that ARID2 may affect JAK2 expression in a post-translational manner. To identify this hypothesis, 293T and Hep3B cells were transfected with Flag-ARID2 plus HA-Ub, and treated with proteasome inhibitor MG132, then the ubiquitination of JAK2 was examined. As shown in Fig. 4B, ARID2 overexpression enhanced JAK2 ubiquitination. Consistently, decreased Jak2 ubiquitination was observed in primary hepatocytes isolated from Arid2 LKO mice (Fig. 4C). Similar observations were confirmed in ARID2 knockdown HepG2 cells (Supplementary Fig. S4A). Then we used Ubi browser website to search for the potential E3 ligase for JAK2. As shown in Fig. 4D, CBL, NEDD4L, CBLB and SMURF1 were the most likely candidates. Notably, only Nedd4l was downregulated in the livers of Arid2 LKO mice at both mRNA and protein level (Fig. 4E), while the mRNA level of Cbl, Cblb and Smurf1 remained unchanged (Supplementary Fig. S4B). NEDD4L overexpression in 293T cells induced robust ubiquitination of JAK2, whereas deletion of the catalytic HECT domain of NEDD4L abrogated this effect (Fig. 4F). To further identify NEDD4L as a bona fide E3 ligase for JAK2, we examined the interaction between NEDD4L and JAK2 in the absence or presence of ARID2 by Co-Immunoprecipitation (Co-IP) assay. As expected, endogenous NEDD4L-JAK2 interaction was enhanced in Hep3B cells by ARID2 overexpression (Fig. 4G), while Arid2 deficiency markedly attenuated the interaction between endogenous JAK2 and NEDD4L in primary hepatocytes (Fig. 4H). Accordingly, similar results were obtained in ARID2 knockdown HepG2 cells (Fig. 4I). Thus, our data suggested that ARID2 promoted the ubiquitination of JAK2 via transcriptionally upregulating NEDD4L expression.
Fig. 4. ARID2 promotes the ubiquitination of JAK2 via the E3 ligase NEDD4L.
A Relative mRNA levels of JAK2 in the livers of WT and Arid2 LKO mice (upper panel, n = 6 per group) and in control and ARID2 knockdown HepG2 cells (lower panel). The mRNA expression levels were normalized to that of Gapdh/GAPDH. B, C Ubiquitination of JAK2 was examined in 293T, Hep3B (B) and primary hepatocytes (C). Cells were treated with 10 μM MG132 for 8 h. IgG was used as a negative control. D Network view of the E3 ligases for JAK2 predicted by Ubi browser website. E Relative mRNA (n = 6 per group) and protein levels of Nedd4l in the liver tissues of WT and Arid2 LKO mice. The mRNA expression level was normalized to that of Gapdh. F Ubiquitination of JAK2 was examined in 293T cells transfected with vector, WT and mutant NEDD4L, respectively. Cells were treated with 10 μM MG132 for 8 h. IgG was used as a negative control. ΔHECT, deletion of HECT domain. G–I Co-IP was performed to examine the interaction between endogenous JAK2 and NEDD4L in Hep3B cells, primary hepatocytes and HepG2 cells. IgG was used as a negative control. All experiments were repeated 3 times independently. All data were analyzed by two-tailed unpaired Student’s t test and represented as the mean ± SEM. *p < 0.05, ns not significant.
ARID2 recruits CARM1 to activate the transcription of NEDD4L
As ARID2 is a DNA-binding component of SWI/SNF chromatin remodeling complex, we analyzed whether ARID2 could bind to the promoter of NEDD4L based on a publicly available ARID2 ChIP-seq data (GSE105731) [24] from ENCODE website, and significant peaks were found in the promoter of NEDD4L as expected (Supplementary Fig. S5A). Luciferase reporter assay revealed that the promoter activity of NEDD4L was increased by ARID2 overexpression in 293T and Hep3B cells and inhibited by ARID2 knockdown in HepG2 cells (Fig. 5A). ARID2 was previously reported to interact with epigenetic regulators to modulate gene transcription [25]. Therefore, an unbiased mass spectrometry analysis was employed to find ARID2 partner. The results showed that ARID2 interacted with several SWI/SNF components, including SMARCA4 and SMARCD2, which validated the experiment. Coactivator associated arginine methyltransferase 1 (CARM1), also known as PRMT4, was chosen for further investigation due to the consistency between its function and the observed phenotype (Fig. 5B). Interaction between exogenous ARID2 and CARM1 was verified by Co-IP, which was further confirmed by Glutathione-S-transferase (GST) pull-down assay (Fig. 5C and Supplementary Fig. S5B). Interaction between endogenous proteins was also confirmed in primary hepatocytes (Fig. 5D and Supplementary Fig. S5C). Next, we sought to figure out the critical amino acid residues within ARID2 responsible for this interaction. Various truncated mutants of ARID2 were constructed according to the ARID2 domain structure [26]. Domain mapping assay showed that the 304-1746aa of ARID2 mediated its interaction with CARM1 (Fig. 5E).
Fig. 5. ARID2 activates the transcription of NEDD4L by recruiting CARM1.
A The influence of ARID2 on the promoter activity of NEDD4L was examined by luciferase reporter assay. B Silver-stained gel showing differential bands between control and ARID2-overexpressing samples. C The interaction between ARID2 and CARM1 was examined by Co-IP assay (left panel) and GST pull-down assay (right panel) in 293T cells co-transfected with the indicated plasmids. Purified GST was used as a control. D Co-IP assay conducted in primary hepatocytes verified the endogenous interaction between ARID2 and CARM1. IgG was used as a negative control. E The interactions between exogenous CARM1 and truncated ARID2 mutants were examined in 293T cells by Co-IP. F The lipid droplets in control and ARID2-overexpressing Hep3B cells with reduced expression of CARM1 were examined by Nile Red staining. Scale bars, 90 μm. Blue signal, nuclei stained with Hoechst; Red signal, lipid droplets stained with Nile Red. G, H Protein and mRNA level of NEDD4L were measured in control and ARID2-overexpressing Hep3B cells with CARM1 knockdown. All experiments were repeated 3 times independently. Relative promoter activity of NEDD4L in panel A was analyzed by two-tailed unpaired Student’s t test, other data were analyzed with one-way ANOVA with Bonferroni post hoc multiple comparison test. All results were represented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
To further clarify the involvement of CARM1 in the upregulation of NEDD4L modulated by ARID2, CAMR1 was knocked down by shRNA in control and ARID2-overexpressing Hep3B cells. As expected, both protein and mRNA levels of NEDD4L were reduced in Hep3B cells after CARM1 knockdown (Fig. 5G, H). In addition, lipid accumulation was decreased upon ARID2 overexpression, but recovered after CARM1 knockdown, associated with the alteration of NEDD4L expression (Fig. 5F and Supplementary Fig. S5D). Similar phenotype was observed in Huh7 cells (Supplementary Fig. S5F). CARM-IN-1, a CARM1 inhibitor which could decrease the protein level of CARM1 also reduced NEDD4L expression in ARID2-overexpressing Hep3B cells (Supplementary Fig. S5E).
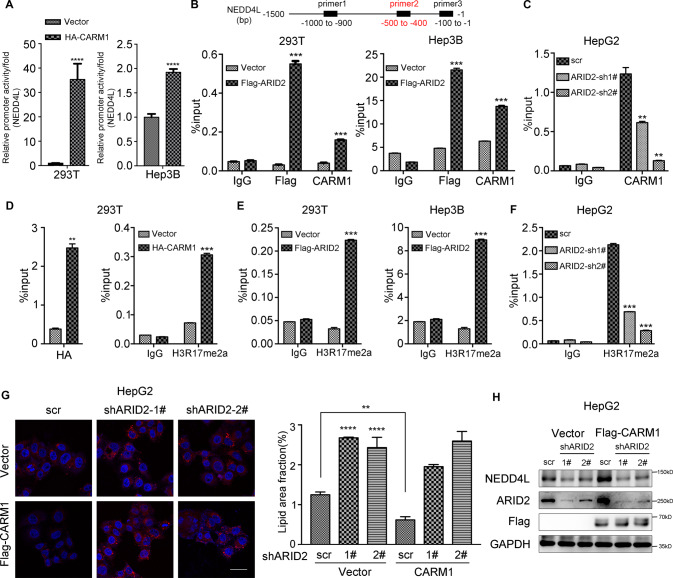
Then, we explored the exact mechanism underlying the transcriptional activation of NEDD4L by ARID2 and CARM1. First, Luciferase reporter assay was performed to confirm the activation of NEDD4L promoter by overexpression of HA-tagged CARM1 in 293T and Hep3B cells (Fig. 6A). Then reporters with different deletions within the NEDD4L promoter region were constructed, and the transcriptional activity of truncated promoters was examined (Supplementary Fig. S6A). We found that three sites were potentially involved in the regulation of NEDD4L by CARM1. Thus, we designed three different primers based on the indicated binding sites of CARM1 and performed ChIP-qPCR assay. We chose binding site 2 (−500 to −400) for subsequent assays, because of the most dramatic changes in ChIP assay with primer 2. Both ARID2 and CARM1 bound to the −500 to −400 bp region upstream of the transcriptional start site of NEDD4L in ARID2-overexpressing 293T and Hep3B cells (Fig. 6B), while ARID2 knockdown significantly reduced CARM1 binding in HepG2 cells (Fig. 6C). CARM1 was reported to asymmetrically dimethylate arginine 17 of histone H3 (H3R17me2a), a marker associated with transcriptional activation [17, 27]. Thus, we examined CARM1 and H3R17me2a occupancy at the promoter region of NEDD4L by ChIP assay, and the enrichment of HA-CARM1 and H3R17me2a was detected (Fig. 6D). Furthermore, H3R17me2a level at the NEDD4L promoter was increased in ARID2-overexpressing 293T and Hep3B cells (Fig. 6E), while decreased in ARID2 knockdown HepG2 cells (Fig. 6F). We have also detected CARM1 and H3R17me2a enrichment at the other two sites within NEDD4L promoter and obtained similar results with primer 1 rather than primer 3 (Supplementary Fig. S6B–F). Therefore, these data suggested that ARID2 promoted the transcription of NEDD4L by recruiting CARM1 to NEDD4L promoter, which led to elevated level of H3R17me2a and subsequent transcriptional activation.
Fig. 6. CARM1 increases H3R17me2a level at the NEDD4L promoter and its recruitment is mediated by ARID2.
A Luciferase reporter assay revealed the activation of NEDD4L promoter by CARM1 overexpression in both 293T and Hep3B cells. B The binding of Flag-tagged ARID2 and CARM1 to specific sites within NEDD4L promoter was examined by ChIP-qPCR using the corresponding Flag and CARM1 antibodies in 293T and Hep3B cells. IgG was used as a negative control. C Binding of CARM1 to the promoter regions of NEDD4L is measured by ChIP-qPCR in control and ARID2 knockdown HepG2 cells. IgG was used as a negative control. D Overexpression of CARM1 resulted in increased H3R17me2a level at the NEDD4L promoter. The ChIP-qPCR was performed with indicated Chip-grade antibodies. IgG was used as a negative control. E, F The levels of H3R17me2a at the NEDD4L promoter in control and ARID2-overexpressing 293T and Hep3B cells as well as ARID2 knockdown HepG2 cells were examined by ChIP-qPCR. IgG was used as a negative control. G, H Nile Red staining (left panel), the corresponding quantification (middle panel, 3 images per group) and western blot (right panel) indicated that CARM1 overexpression in ARID2 knockdown HepG2 cells could not rescue the phenotype and the protein level of NEDD4L. Blue signal, nuclei stained with Hoechst; Red signal, lipid droplets stained with Nile Red. All experiments were repeated 3 times independently. Quantification of lipid droplets in panel G was analyzed with one-way ANOVA with Bonferroni post hoc multiple comparison test, other data were analyzed by two-tailed unpaired Student’s t test. All results were represented as the mean ± SEM. **p < 0.01, ***p < 0.001, ****p < 0.0001.
To further elucidate the critical role of ARID2 in recruiting CARM1, we investigated whether CARM1 overexpression in ARID2 knockdown HepG2 cells could revert the phenotype and NEDD4L expression. As shown in Fig. 6G, ARID2 knockdown increased lipid droplets accumulation even when CARM1 was overexpressed. Moreover, CARM1 overexpression could not rescue NEDD4L expression in ARID2 knockdown cells (Fig. 6H). Here we verified the necessity of ARID2 in recruiting CARM1 to the promoter of NEDD4L.
Inhibition of JAK2 alleviates HFD-induced hepatic steatosis in Arid2 LKO mice
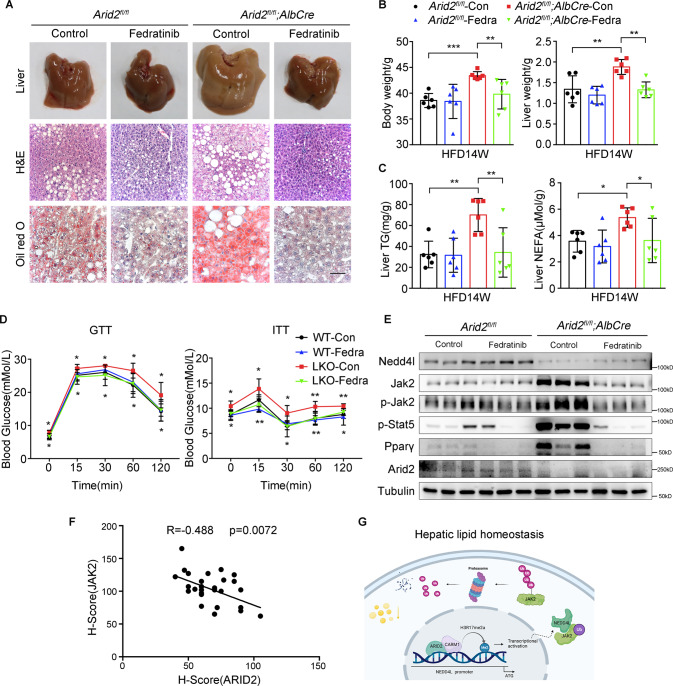
Considering that inhibitors of JAK2, especially Fedratinib, exerted a dramatical inhibition on lipid deposition in Arid2 KO hepatocytes (Supplementary Fig. S3B), we evaluated its therapeutic potential in the treatment of NAFLD induced by ARID2 deficiency. The 6-week-old WT and Arid2 LKO mice were fed with HFD, followed by intragastrical administration of Fedratinib from 16 to 20 weeks (Supplementary Fig. S7A). At the end of the treatment, hepatic steatosis of Arid2 LKO mice was efficiently ameliorated by Fedratinib as indicated by the liver pictures, H&E staining Oil Red O staining (Fig. 7A and Supplementary Fig. S7B). Body weight, liver weight, hepatic TG contents and NEFA levels were significantly decreased in Arid2 LKO mice treated with Fedratinib (Fig. 7B, C), as well as body fat, serum TG contents and serum NEFA levels (Supplementary Fig. S7C). In addition, Fedratinib improved glucose intolerance and insulin resistance induced by HFD in Arid2 LKO mice (Fig. 7D). Expression of lipid metabolism-related genes were rescued as well (Supplementary Fig. S7D). Besides, we confirmed the inhibitory effect of Fedratinib on Jak2-Stat5-Pparγ axis in liver tissues, especially in Arid2 KO tissues (Fig. 7E). To further evaluate the potential application of Fedratinib in NAFLD patients with decreased ARID2 expression, we examined the protein expression of ARID2 and JAK2 and analyzed their correlation in human NAFLD by immunohistochemistry. ARID2 expression was remarkably reduced in the livers from NAFLD patients and showed significantly negative correlation with JAK2 (Fig. 7F and Supplementary Fig. S7E), indicating that Fedratinib may be applicable to NAFLD treatment.
Fig. 7. Fedratinib, a small molecular inhibitor of JAK2, ameliorates HFD-induced hepatic steatosis in Arid2 LKO mice.
A Liver pictures, H&E and Oil Red O staining of liver sections from HFD-fed WT and Arid2 LKO mice treated with Fedratinib or control vehicle (n = 6 per group), respectively. Scale bars, 45 μm. B Body weight and liver weight of WT and Arid2 LKO mice in the indicated groups fed with HFD for 14 weeks (n = 6 per group). C Hepatic TG and NEFA levels were measured in WT and Arid2 LKO mice receiving the indicated treatment (n = 6 per group). D GTTs and ITTs in the treated WT and Arid2 LKO mice at 19 weeks (n = 6 per group). Upper asterisks: significant difference between WT and Arid2 LKO mice. Lower asterisks: significant difference between control group and Fedratinib-treated group in Arid2 LKO mice. E Protein levels of Nedd4l and Jak2-Stat5 signaling pathway in the livers from the mice of indicated groups (n = 3 per group). F The correlation between ARID2 and JAK2 expression in the liver sections from NAFLD patients (n = 29). G Schematic representation demonstrating the molecular mechanism of ARID2 in maintaining hepatic lipid homeostasis. Created with BioRender.com. All experiments were repeated 3 times independently. H-scores were analyzed by Pearson correlation analysis. Other data were analyzed with one-way ANOVA with Bonferroni post hoc multiple comparison test. All results were represented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
Due to its frequent mutation in human cancers, ARID2 has been largely studied in a variety of cancers, including liver cancer [26], lung cancer [28], melanoma [29] and colorectal cancer [30], etc. Among these cancers, liver cancer is the most studied malignancy. ARID2 was reported to modulate DNA damage in HCC cells [8]. In another study, Duan et al revealed that ARID2 targeted cyclin D1 and cyclin E1 to suppress HCC progression [7]. And our lab previously reported that ARID2 suppressed hepatocellular carcinoma metastasis via Snail-DNMT1 axis [31]. However, the role of ARID2 in other liver diseases remains elusive. In the past decades, NAFLD has become a leading cause of chronic liver disease and cirrhosis is the main risk factor for HCC development. Therefore, in the current study, we investigated the function of ARID2 in the development of NAFLD. Our study demonstrated that Arid2 LKO mice are more prone to the development of hepatic steatosis under both normal and over-nourished condition. It is worthy to note that Arid2 LKO mice showed lipid deposition at very early stage and the expression of inflammation- and fibrosis-related genes were increased only in Arid2 LKO mice under HFD conditions. In addition, significant downregulation of ARID2 was detected in the steatotic liver tissues from NAFLD patients compared with nonsteatotic livers. Moreover, in the HFD plus N-nitrosodiethylamine (DEN; 40 mg/kg)–induced HCC mouse model, Oil Red O staining demonstrated that liver-specific deletion of Arid2 promoted lipid accumulation in the tumor section (Supplementary Fig. S8A). These findings indicated that ARID2 deficiency may contribute to more severe NAFLD, even HCC, which required further confirmation in cohort study.
JAK-STAT pathway plays a crucial role in carcinogenesis and inflammation [32, 33]. Emerging evidence has shown that this signaling also affects lipid metabolism in adipose tissue by regulating the expression of PPARγ [13, 14]. The role of JAK-STAT signaling in hepatic metabolism is quite complicated, and conflicting functions were reported by different groups [34–36], which may be due to the activation of this signaling by different factors, and its crosstalk with other signalings. Here, we identified that hepatic lipid accumulation was increased by activation of JAK2-STAT5-PPARγ axis in Arid2 LKO mice livers. As mentioned before, ARID1A could regulate hepatic lipid metabolism in mice [12]. To examine whether other ARID subunits in SWI/SNF complex could affect hepatic lipid metabolism via JAK2-STAT5-PPARγ axis, we analyzed the bulk RNA sequencing (RNA-seq) data from GEO datasets (GSE69567), provided by Jesse et al. [37]. ARID1A, ARID1B and ARID2 were separately knocked down in HepG2 cells to investigate their functional relationships. Gene set enrichment analysis (GSEA) showed significant activation of JAK-STAT signaling pathway in ARID2 knockdown cells (Supplementary Fig. S8B), which is consistent with our RNA-seq data in mice. Although knockdown of ARID1A in HepG2 cells significantly affected fatty acid metabolism, it didn’t influence JAK-STAT signaling pathway (Supplementary Fig. S8C). As for ARID1B knockdown group, there were few alterations in both JAK-STAT signaling and fatty acid metabolism (Supplementary Fig. S8D). These results indicate that only ARID2 deficiency can activate JAK-STAT signaling pathway.
JAK/STAT pathway is mainly regulated by phosphorylation and ubiquitination [38]. Previous studies have revealed that CBL family and SOCS family are the two main E3 ligases that ubiquitylate JAK2 [39, 40]. Here, we disclosed a novel mechanism underlying the regulation of JAK2 by ARID2. We identified NEDD4L as a new E3 ligase for JAK2 based on the following evidence: NEDD4L could interact with JAK2, and WT NEDD4L increased JAK2 ubiquitination while loss-of-function mutant couldn’t. Furthermore, our study revealed that ARID2 recruited epigenetic regulator CARM1 to the promoter of NEDD4L, which increased H3R17me2a level and enhanced NEDD4L transcription. Therefore, our study has disclosed a novel regulatory mechanism of JAK2-STAT5 signaling.
Decrease of ARID2 in steatotic liver tissues and the negative regulation of JAK2-STAT5 signaling by ARID2 promoted us to explore the therapeutic potential of targeting JAK2-STAT5 axis for the treatment of NAFLD with ARID2 deficiency. Interestingly, two JAK2 inhibitors as well as STAT5 inhibitor effectively decreased intracellular lipid accumulation in vitro. Fedratinib was approved by FDA to treat patients with myelofibrosis. We chose this drug to conduct animal study because of its stronger effect than the other two. Fedratinib modestly suppressed hepatic lipid droplets accumulation in WT mice, while significantly alleviated hepatic steatosis in Arid2 LKO mice, which may be attributed to hyper activation of JAK2 upon ARID2 loss. More importantly, the reverse correlation between JAK2 and ARID2 was observed in the steatotic liver tissues from NAFLD patients, indicating that Fedratinib may effectively reduce liver steatosis and improve insulin sensitivity in ARID2low-JAK2high NAFLD patients.
Taken together, we have revealed the important role of ARID2 in maintaining homeostasis of hepatic lipid metabolism and disclosed the underlying mechanism: ARID2 recruits CARM1 to activate NEDD4L transcription, and the increased NEDD4L promotes ubiquitination of JAK2. Subsequently, the decreased JAK2-STAT5 signaling leads to reduced hepatic lipid accumulation (Fig. 7G). Furthermore, our study identified negative correlation between ARID2 and JAK2 in NAFLD patients and indicated the therapeutic potential of Fedratinib in the treatment of NAFLD patients with ARID2 deficiency.
Materials and methods
Human liver samples
The paraffin sections of fatty liver and normal liver tissues were purchased from Shanxi Avilabio Company. NAFLD is histologically diagnosed when lipid accumulation occurs in >5% of hepatocytes. Our study has been approved by the Biomedical Research Ethics Committee, Shanghai Institute for Biological Science, CAS in 2019 (ER-SIBS-251902).
Animals
The Arid2fl/fl mice were provided from Bin Zhou’s lab (Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Science, Shanghai, China). The hepatocyte specific Arid2 knockout mice were generated as follows. The Arid2-flox mice were crossbred with Albumin-cre mice (Jackson Laboratory), successful Arid2 knockout and Arid2 expression could be verified by western blot. The primer pairs used for mouse identification were described in Table S6. All mice were male in C57BL/6 background and all experiments were performed under the approval of the Institutional Animal Care and Use Committee of Shanghai Institute of Nutrition and Health, CAS (IACUC, SINH, CAS). Mice in each group were selected randomly and there was no blinding study for different animal groups.
High-fat diet (HFD)
HFD models were established by feeding mice on a HFD (fat, 60 Kcal%; protein, 20 Kcal%; carbohydrates, 20 Kcal%; Research Diet) continuously for 12 weeks and 14 weeks.
Fedratinib treatment
Six weeks old male mice were fed with HFD for 14 weeks, mice were given intragastric administration every other day with a dose of 120 mg/kg starting from 16 weeks and the mice were sacrificed at the age of 20 weeks.
Primary hepatocytes separation and treatment
Primary hepatocytes were isolated from aged 8–12 weeks mice by liver perfusion as previously described [41]. In short, mice were anesthetized with Tribromoethanol. Liver perfusion buffer was perfused through the portal vein, followed by the perfusion of liver digest buffer for 15 min. After this, the liver was excised, minced and filtered through a cell sieve (70 μm). Hepatocytes were separated via centrifugation at 800 rpm/min for 3 min, purified on 50% percoll. The cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin in a 5% CO2 incubator at 37 °C.
Mouse serum and hepatic lipid analyses
To measure the serum TG, animals were starved for 6 h before tail vein blood collection. Blood serum was further collected after centrifugation at 6000 rpm for 20 min at 4 °C. For lipid tests in liver tissues, we homogenized 30 mg of liver tissues in 1xPBS, and then methanol and chloroform (1:2) were added for extraction. Total TG were measured with a total triglyceride kit (Shanghai Shensuo). NEFA tests were performed according to the instructions of the kit (Wako, Japan).
Nile red staining
Lipid droplets in control and ARID2-overexpressing (GFP-positive) cells were detected by Nile Red, a lipophilic fluorescent dye. The Nile red (ThermoFisher) was dissolved in acetone and diluted 1:1000 in 1xPBS for Nile Red staining. Briefly, cells on slides were fixed in 4% formaldehyde for 15 min and then were washed 3 times with 1xPBS, 5 min per time. Then slides were stained with prepared Nile red for 30 min at 37 °C, after that, slides were washed 3 times, 5 min per time. Next, they were stained with the Hoechst for 3 min, followed by being washed 3 times, 5 min per time. Finally, slides were covered with the film and photographed by confocal microscope (Zeiss, LSM 510 NLO).
BODIPY staining
BODIPY (4,4-difluoro-3a,4a-diaza-s-indacene) is a highly lipophilic neutral fluorophore. Staining was performed similarly to Nile Red. The intensely fluorescent BODIPY (ThermoFisher) was dissolved in dimethylsulfoxide (DMSO) and diluted 1:2000 in 1xPBS for BODIPY staining. Quantification of lipid deposition was determined by Image J.
Plasmid constructs
ARID2 coding sequence was inserted into p23-3×Flag-GFP lentiviral vector and cells transfected with ARID2 gene were screened using flow cytometry by green fluorescence. Truncations of ARID2 were constructed according to its structure [26] using 3×Flag-cmv-14 vector and Jak2 was cloned into the Tag2b-Myc vector. pcDNA3-HA-NEDD4L was thankfully given from Ying Yu’s Lab (Tianjin Medical University, Tianjin, China). NEDD4L and its truncation were cloned into the Tag2b-Myc vector and CARM1 was cloned into the p23-3×Flag-GFP or PCDNA3-HA vector. shRNAs, targeted ARID2 and CARM1, were designed using software provided by Qiagen (Valencia, CA, USA) and inserted into pLKO.1-TRC vector, positive cells were selected by puromycin (4 μg/mL) for 4 days. The sequences of cloned genes and shRNA were listed in Table S1 and Table S2, respectively.
qPCR
qPCR was performed according to the protocol of our laboratory [42]. Briefly, total mRNA was isolated from cultured cells or liver samples using TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. 2 μg cDNA was reverse transcribed into cDNA using Promega Kit (Promega, Madison) according to the manufacturer’s protocol. SYBR Green (YEASEN Biotech) was applied to quantify PCR amplification. Expression levels were calculated using the ΔCT-method. The primer pairs used in our study are described in Table S3.
ChIP-qPCR
The ChIP assay was performed by kit purchased from Cell Signaling Technology (CST) and qPCR was performed as previously described. Results were analyzed using ΔΔCT method. Three paired primers were employed to assess the enrichment of the indicated genes at the NEDD4L promoter and sequences of primers were provided in Table S4. The ChIP signals were normalized to the input samples from the same chromatin extract.
Cell lines
The Human Embryonic Kidney 293T cell line and other human HCC cells were purchased from the Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). All cell lines were cultured in DMEM, 10% FBS and 1% penicillin-streptomycin, all were maintained in a humidified chamber with 5% CO2 at 37 °C. Palmitate (50 μM; Sigma) was added to the medium for 24 h. Stable monoclonal ARID2 gene overexpressed HCC cell lines were created using the flow sorting technology by lentivirus-mediated gene transfection. Briefly, we firstly cloned the ARID2 gene into lentivirus expression vector (P23-3xFlag-GFP). Next, HCC cell lines were infected with packaged lentivirus for 8 h, followed by GFP sorting.
Western blot
Western blot was performed as described previously [42]. Proteins were separated using 6 and 8% SDS-PAGE gels and then transferred to PVDF membranes (0.45 μm). The membranes were blocked in 5% BSA, they were incubated overnight at 4 °C with primary antibodies and then for 70 min at room temperature with the mouse and rabbit secondary antibodies. The protein levels were quantified using Tanon-5200 multifunctional chemiluminescence instrument and normalized to the levels of GAPDH or TUBULIN. The antibodies used in our study were described in in Table S5. Horseradish peroxidase-conjugated secondary antibodies (1:2000) were purchased from Cell Signaling Technology. Full-length original western blots for the results are provided in Supplementary materials.
GST fusion protein purification and GST pulldown
GST-CARM1 was purified from BL21. Briefly, coding sequences were subcloned into pGEX-4T-1 vector. Vectors were transformed into BL21 bacteria. Bacteria was cultured in LB medium with Ampicillin until the culture reached an optical density of 0.8 at 600 nm (OD600). Then IPTG was added to a final concentration of 0.3 mM, afterwards, the incubation was continued for additional 10 h at 30 °C. Bradford reagent was used to determine the concentration of the purified protein. The lysis of cultured cells was incubated with 10 μg purified protein overnight at 4 °C. The second day, lysis was incubated with GST beads (GE) for 1 h at 4 °C and then was analyzed with western blotting using the indicated antibodies.
Immunoprecipitation
Experiments were performed as previously described [31].
Luciferase reporter assay
HEK293T were placed in 24-well plate, 100 ng ARID2 expression vector or empty vector, 50 ng reporter plasmid and 20 ng renilla luciferase were co-transfected into cells for 48 h. However, only 50 ng reporter plasmid and 20 ng Renilla luciferase were co-transfected into stably transfected HCC cell lines and its control cells for 24 h. The experiments were performed in triplicate. Then, cells were lysed by passive lysis buffer (Promega, Madison, WI) and reporter activities were investigated by the Dual-Luciferase Reporter Assay System (Promega).
Immunohistochemistry (IHC)
IHC experiments were performed as described previously [31]. Sections were incubated with primary antibody against ARID2 (1:300, Abcam), and JAK2 (1:50, Cell Signaling Technology) overnight at 4 °C.
Oil Red O staining
Liver tissues were imbedded into Tissue-Tek OCT compound and frozen for oil red o staining. Oil Red O solution was mixed with distilled water at a ratio of 3:2, then the mixture was filtered by 0.45 μm filters. Frozen sections of liver (5 μm) were fixed in 10% buffered formalin for 15 min, rinsed with 60% isopropanol, then stained with Oil Red O working solution for 15 min. After washing with 60% isopropanol, hematoxylin was used to stain the nuclei. Finally, the stained sections were covered and microscopically examined. Lipid accumulation in the livers was quantified by Image J.
RNA sequencing and processing
Total RNA was isolated from liver tissues of WT and Arid2 LKO mice. A total amount of 1 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Differential expression analysis of two groups was performed using the DESeq2 R package (1.16.1). The resulting p-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted p-value < 0.05 found by DESeq2 were assigned as differentially expressed. Statistical enrichment of differential expression genes in KEGG pathways were tested using clusterProfiler R package. The whole procedure was performed by Novogene Co. Ltd (Beijing, China).
Mass spectrometry analysis
Flag-tagged ARID2-overexpressing 293T cells were lysed followed by immunoprecipitation with the Flag antibody according to the immunoprecipitation assay protocol. The supernatants were separated by SDS–PAGE gels and the protein bands were visualized by silver staining (Beyotime Biotechnology) according to the manufacturer’s instructions. The subsequent Mass spectrometry analysis was performed by Shanghai Applied Protein Technology Co. Ltd (Shanghai, China).
Statistical analyses
Sample sizes of all experiments were predetermined based on our experience for this kind of study. No sample was excluded from the analyses. Investigators were not blinded to the group allocation during the experiment and outcome assessment. Results from independent animals, experiments or separately generated samples were treated as biological replicates. Unless otherwise stated, experiments were performed independently in triplicate with the sample size “n” indicated in the figure legend.
All data were analyzed using GraphPad Prism 6.0(Macintosh). Quantitative values are presented as the mean ± S.E.M. Statistical differences between two experimental groups with normally distributed data were analyzed by two-tailed unpaired Student’s t-test, data without normal distribution were analyzed by Mann-Whitney test and analysis between multiple groups was performed using one-way ANOVA with Bonferroni post hoc multiple comparison test. Correlation was analyzed by Pearson correlation analysis. A P value < 0.05 was considered significant. Statistical analysis used in each panel was described in the figure legends.
Supplementary information
Acknowledgements
We thank Zhonghui Weng, Yifan Bu and Lin Qiu from Institutional Center for Shared Technologies and Facilities of SINH, CAS for technical assistance. We appreciate the New World Group for their Charitable Foundation to establish the Institute for Nutritional Sciences, SIBS, CAS-New World Joint laboratory, which have given full support to this study.
Author contributions
HC, JL and DX conceived the project and designed the research studies. HC performed most of the experiments described. HJ, KD, XQ, NM, FZ, YW, QZ and JX provided help with animal and technical assistance in the mouse experiments. QN, SX and BZ assisted with the in vitro assays. BZ provided conceptual advice and helpful discussion. HC analyzed data. HC, JL and DX wrote the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81972757 and 82172950), Youth Innovation Promotion Association of Chinese Academy of Sciences grant (2017324) and Sanofi-SIBS 2018 Young Faculty Award to Jing-Jing Li and the National Natural Science Foundation of China (82030084 and 81730083) to Dong Xie.
Data availability
RNA-seq data supporting the results of this study have been deposited in the NCBI GEO database under accession number GSE216171. All the other data supporting the findings of this study are available within the article and its Supplementary Information files and from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethics statement
Experiments about human tissues have been approved by the Biomedical Research Ethics Committee, Shanghai Institute for Biological Science, CAS in 2019 (ER-SIBS-251902). All animal studies were performed under the approval of the Institutional Animal Care and Use Committee of Shanghai Institute of Nutrition and Health, CAS (IACUC, SINH, CAS).
Footnotes
Edited by M Bianchi
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dong Xie, Email: dxie@sibs.ac.cn.
Jing-Jing Li, Email: tide7@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-01090-0.
References
- 1.Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol. 2013;10:656–65. doi: 10.1038/nrgastro.2013.183. [DOI] [PubMed] [Google Scholar]
- 2.Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol. 2013;10:686–90. doi: 10.1038/nrgastro.2013.171. [DOI] [PubMed] [Google Scholar]
- 3.Adams LA, Lymp JF, Sauver JS, Sanderson SO, Lindor KD, Feldstein A, et al. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 2005;129:113–21. doi: 10.1053/j.gastro.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Matteoni CA, Younossi ZM, Gramlich T, Boparai N, Liu YC, McCullough AJ. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology. 1999;116:1413–9. doi: 10.1016/S0016-5085(99)70506-8. [DOI] [PubMed] [Google Scholar]
- 5.Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908–22. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, Wang W, Li X, He S, Yao J, Wang X, et al. MicroRNA-155 promotes tumor growth of human hepatocellular carcinoma by targeting ARID2. Int J Oncol. 2016;48:2425–34. doi: 10.3892/ijo.2016.3465. [DOI] [PubMed] [Google Scholar]
- 7.Duan Y, Tian L, Gao Q, Liang L, Zhang W, Yang Y, et al. Chromatin remodeling gene ARID2 targets cyclin D1 and cyclin E1 to suppress hepatoma cell progression. Oncotarget. 2016;7:45863–75. doi: 10.18632/oncotarget.10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oba A, Shimada S, Akiyama Y, Nishikawaji T, Mogushi K, Ito H, et al. ARID2 modulates DNA damage response in human hepatocellular carcinoma cells. J Hepatol. 2017;66:942–51. doi: 10.1016/j.jhep.2016.12.026. [DOI] [PubMed] [Google Scholar]
- 9.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muchardt C, Yaniv M. ATP-dependent chromatin remodelling: SWI/SNF and Co. are on the job. J Mol Biol. 1999;293:187–98. doi: 10.1006/jmbi.1999.2999. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Liu C, Li N, Hao T, Han T, Hill DE, et al. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 2008;8:105–17. doi: 10.1016/j.cmet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore A, Wu L, Chuang JC, Sun X, Luo X, Gopal P, et al. Arid1a loss drives nonalcoholic steatohepatitis in mice through epigenetic dysregulation of hepatic lipogenesis and fatty acid oxidation. Hepatology. 2019;69:1931–45. doi: 10.1002/hep.30487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JB, Yoon SJ, Lee SH, Lee MS, Jung H, Kim TD, et al. Ginsenoside Rg3 ameliorated HFD-induced hepatic steatosis through downregulation of STAT5-PPARgamma. J Endocrinol. 2017;235:223–35. doi: 10.1530/JOE-17-0233. [DOI] [PubMed] [Google Scholar]
- 14.Murtaza M, Khan G, Aftab MF, Afridi SK, Ghaffar S, Ahmed A, et al. Cucurbitacin E reduces obesity and related metabolic dysfunction in mice by targeting JAK-STAT5 signaling pathway. PloS One. 2017;12:e0178910. doi: 10.1371/journal.pone.0178910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, et al. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol. 2003;23:7475–87. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci USA. 2005;102:3611–6. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karakashev S, Fukumoto T, Zhao B, Lin J, Wu S, Fatkhutdinov N, et al. EZH2 inhibition sensitizes CARM1-high, homologous recombination proficient ovarian cancers to PARP inhibition. Cancer Cell. 2020;37:157–67.e6. doi: 10.1016/j.ccell.2019.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu F, Ma F, Wang Y, Hao L, Zeng H, Jia C, et al. PKM2 methylation by CARM1 activates aerobic glycolysis to promote tumorigenesis. Nat Cell Biol. 2017;19:1358–70. doi: 10.1038/ncb3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawai M, Namba N, Mushiake S, Etani Y, Nishimura R, Makishima M, et al. Growth hormone stimulates adipogenesis of 3T3-L1 cells through activation of the Stat5A/5B-PPARgamma pathway. J Mol Endocrinol. 2007;38:19–34. doi: 10.1677/jme.1.02154. [DOI] [PubMed] [Google Scholar]
- 21.Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117:1281–3. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 22.Li WX. Canonical and non-canonical JAK–STAT signaling. Trends Cell Biol. 2008;18:545–51. doi: 10.1016/j.tcb.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han J, Wang Y. mTORC1 signaling in hepatic lipid metabolism. Protein Cell. 2018;9:145–51. doi: 10.1007/s13238-017-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. [DOI] [PMC free article] [PubMed]
- 25.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–92. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 26.Loesch R, Chenane L, Colnot S. ARID2 Chromatin Remodeler in Hepatocellular Carcinoma. Cells. 2020;9:2152. [DOI] [PMC free article] [PubMed]
- 27.Coughlan N, Thillainadesan G, Andrews J, Isovic M, Torchia J. β-Estradiol-dependent activation of the JAK/STAT pathway requires p/CIP and CARM1. Biochimica et Biophysica Acta. 2013;1833:1463–75. doi: 10.1016/j.bbamcr.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Moreno T, Monterde B, González-Silva L, Betancor-Fernández I, Revilla C, Agraz-Doblas A, et al. ARID2 deficiency promotes tumor progression and is associated with higher sensitivity to chemotherapy in lung cancer. Oncogene. 2021;40:2923–35. doi: 10.1038/s41388-021-01748-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukumoto T, Lin J, Fatkhutdinov N, Liu P, Somasundaram R, Herlyn M, et al. ARID2 deficiency correlates with the response to immune checkpoint blockade in melanoma. J Investigative Dermatol. 2021;141:1564–72.e4. doi: 10.1016/j.jid.2020.11.026. [DOI] [PubMed] [Google Scholar]
- 30.Bala P, Singh AK, Kavadipula P, Kotapalli V, Sabarinathan R, Bashyam MD. Exome sequencing identifies ARID2 as a novel tumor suppressor in early-onset sporadic rectal cancer. Oncogene. 2021;40:863–74. doi: 10.1038/s41388-020-01537-z. [DOI] [PubMed] [Google Scholar]
- 31.Jiang H, Cao H-J, Ma N, Bao W-D, Wang J-J, Chen T-W, et al. Chromatin remodeling factor ARID2 suppresses hepatocellular carcinoma metastasis via DNMT1-Snail axis. Proc Natl Acad Sci. 2020;117:4770–80. doi: 10.1073/pnas.1914937117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Shea JJ, Holland SM, Staudt LM. JAKs and STATs in immunity, immunodeficiency, and cancer. N. Engl J Med. 2013;368:161–70. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stepkowski SM, Chen W, Ross JA, Nagy ZS, Kirken RA. STAT3: an important regulator of multiple cytokine functions. Transplantation. 2008;85:1372–7. doi: 10.1097/TP.0b013e3181739d25. [DOI] [PubMed] [Google Scholar]
- 34.Li T, Weng J, Zhang Y, Liang K, Fu G, Li Y, et al. mTOR direct crosstalk with STAT5 promotes de novo lipid synthesis and induces hepatocellular carcinoma. Cell Death Dis. 2019;10:619. doi: 10.1038/s41419-019-1828-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J-B, Yoon S-J, Lee S-H, Lee M-S, Jung H, Kim T-D, et al. Ginsenoside Rg3 ameliorated HFD-induced hepatic steatosis through downregulation of STAT5-PPARγ. J Endocrinol. 2017;235:223–35. doi: 10.1530/JOE-17-0233. [DOI] [PubMed] [Google Scholar]
- 36.Barclay JL, Nelson CN, Ishikawa M, Murray LA, Kerr LM, McPhee TR, et al. GH-dependent STAT5 signaling plays an important role in hepatic lipid metabolism. Endocrinology. 2011;152:181–92. doi: 10.1210/en.2010-0537. [DOI] [PubMed] [Google Scholar]
- 37.Raab JR, Resnick S, Magnuson T. Genome-wide transcriptional regulation mediated by biochemically distinct SWI/SNF complexes. PLoS Genet. 2015;11:e1005748. doi: 10.1371/journal.pgen.1005748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yasukawa H, Sasaki A, Yoshimura A. Negative regulation of cytokine signaling pathways. Annu Rev Immunol. 2000;18:143–64. doi: 10.1146/annurev.immunol.18.1.143. [DOI] [PubMed] [Google Scholar]
- 39.Liu CS, Yang-Yen HF, Suen CS, Hwang MJ, Yen JJ. Cbl-mediated K63-linked ubiquitination of JAK2 enhances JAK2 phosphorylation and signal transduction. Sci Rep. 2017;7:4613. doi: 10.1038/s41598-017-04078-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ungureanu D, Saharinen P, Junttila I, Hilton DJ, Silvennoinen O. Regulation of Jak2 through the ubiquitin-proteasome pathway involves phosphorylation of Jak2 on Y1007 and interaction with SOCS-1. Mol Cell Biol. 2002;22:3316–26. doi: 10.1128/MCB.22.10.3316-3326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang FK, Ni QZ, Wang K, Cao HJ, Guan DX, Zhang EB, et al. Targeting USP9X-AMPK Axis in ARID1A-deficient hepatocellular carcinoma. Cell Mol Gastroenterol Hepatol. 2022;14:101–27. doi: 10.1016/j.jcmgh.2022.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng Y-Z, Chen P-P, Wang Y, Yin D, Koeffler HP, Li B, et al. Connective tissue growth factor is overexpressed in esophageal squamous cell carcinoma and promotes tumorigenicity through beta-catenin-T-cell factor/Lef signaling. J Biol Chem. 2007;282:36571–81. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data supporting the results of this study have been deposited in the NCBI GEO database under accession number GSE216171. All the other data supporting the findings of this study are available within the article and its Supplementary Information files and from the corresponding author upon reasonable request.