Abstract
The DAN/TIR genes encode nine cell wall mannoproteins in Saccharomyces cerevisiae which are expressed during anaerobiosis (DAN1, DAN2, DAN3, DAN4, TIR1, TIR2, TIR3, TIR4, and TIP1). Most are expressed within an hour of an anaerobic shift, but DAN2 and DAN3 are expressed after about 3 h. At the same time, CWP1 and CWP2, the genes encoding the major mannoproteins, are down-regulated, suggesting that there is a programmed remodeling of the cell wall in which Cwp1 and Cwp2 are replaced by nine anaerobic counterparts. TIP1, TIR1, TIR2, and TIR4 are also induced during cold shock. Correspondingly, CWP1 is down-regulated during cold shock. As reported elsewhere, Mox4 is a heme-inhibited activator, and Mot3 is a heme-induced repressor of the DAN/TIR genes (but not of TIP1). We show that CWP2 (but not CWP1) is controlled by the same factors, but in reverse fashion—primarily by Mot3 (which can function as either an activator or repressor) but also by Mox4, accounting for the reciprocal regulation of the two groups of genes. Disruptions of TIR1, TIR3, or TIR4 prevent anaerobic growth, indicating that each protein is essential for anaerobic adaptation. The Dan/Tir and Cwp proteins are homologous, with the greatest similarities shown within three subgroups: the Dan proteins, the Tip and Tir proteins, and, more distantly, the Cwp proteins. The clustering of homology corresponds to differences in expression: the Tip and Tir proteins are expressed during hypoxia and cold shock, the Dan proteins are more stringently repressed by oxygen and insensitive to cold shock, and the Cwp proteins are oppositely regulated by oxygen and temperature.
The cell wall of Saccharomyces cerevisiae is a rigid structure which determines cell morphology and also serves as a protective barrier, providing mechanical protection and enabling selective uptake of macromolecules (1a, 4, 7). A major component of the cell wall is mannoprotein, comprising about 40% of its mass. Mannoproteins are believed to be determinants of cell wall permeability (31), and certain ones are also essential for developmental events such as mating and transition to hyphal growth (18, 24). Some mannoproteins can be extracted from the cell wall with detergent; others are covalently bound but can be released with glucanase (7). Proteins in the latter category have common features, including a region rich in serine and threonine, a glycosylphosphatidylinositol (GPI) anchor attachment signal at C termini (3, 8), and an endoplasmic reticulum localization sequence; several also have a PAU domain. This segment of about 100 amino acids is shared among a group of proteins known as “seripauperins” (29).
The Cwp2 mannoprotein (28) is one of the most abundant proteins of the cell wall and is believed to play a role in its stabilization, along with another homologous constituent, Cwp1 (23, 27, 28). While Cwp1 and Cwp2 are expressed under normal growth conditions, other mannoproteins are expressed in response to environmental stress. The most extensive response is the induction of several homologous mannoproteins during anaerobic growth: Dan1 (26) (for “delayed anaerobic”), Tip1, Tir1, and Tir2 (9, 16, 17, 22). We refer to the genes encoding these proteins as the DAN/TIR genes. We find that the DAN/TIR group includes genes for five other mannoproteins, designated Dan2, Dan3, Dan4, Tir3, and Tir4 (open reading frames [ORFs] YLR037c, YBR301w, YJR151c, YIL011w, and YOR009w), and that Tir1, Tir3, and Tir4 are required for anaerobic growth. At the same time we show that expression of CWP1 and CWP2 is turned off during anaerobiosis. Hence, one group of proteins is replaced by another group, some of which are essential for anaerobic adaptation. In addition to being oxygen regulated a subset of these genes (TIP1, TIR1, TIR2, and TIR4) are induced during cold shock, while CWP1 is correspondingly down-regulated. CWP1, CWP2, and TIP1 are also differentially regulated during the cell cycle (2), possibly reflecting an increased demand for cell wall proteins during bud formation. There is also suggestive evidence that expression of CWP1 and other cell wall components is coregulated (23).
Until recently, little was known about the regulation of mannoprotein expression, beyond expression patterns in response to various signals. We had earlier found that the regulatory coeffector controlling expression of DAN1 is heme, which functions as an inhibitor of expression in aerobic cells (26). We report elsewhere on a system of regulators which control anaerobic induction and heme repression of DAN1 and the other DAN/TIR genes through a recently identified group of promoter sites (4a). These include principally the Mox4 (or Upc2) activator (1, 5) as well as a group of repressors, Mox1, Mox2, Mot3, and Rox1. We show here that expression of CWP2 is under the control of some of the same factors, acting in an opposite fashion to induce expression during aerobic growth and to block expression in anaerobic cells.
MATERIALS AND METHODS
Plasmids and gene disruptions. (i) pBSTIR1:URA3.
The region containing the TIR1 gene (−1034 to + 996) was amplified using the primers GAGTCGACAAGTATCCAACAGACAGTAGTGCC and GAGAGAATTCATATCTACAAATATCCCGGC. The PCR product was digested with EcoRI and SalI and ligated to pBS-SK [pBluescript SK(+) (Stratagene)] which had been digested with the same enzymes, generating pBSTIR1. To generate the disruption construct, the URA3 gene was inserted into pBSTIR1. For this construction, the URA3 gene from −938 to +1772 was amplified using the primers GAGAGAATTCATCGATCAACTAACATCACACTTGCTGG and GAGAGTCGACACGCGTGGAACACAGTGGAGCCTTG and digested with BspDI (at −938) and SmaI (at +882) for insertion into the BstBI (−490) and EcoRV(+186) sites in pBSTIR1, generating pBSTIR1:URA3. The disruption fragment used to transform FY23 cells was excised with EcoRI and XhoI.
(ii) pBSTIR3:URA3.
The region containing the TIR3 gene (−1084 to +728) was amplified using the primers GAGAGTCGACTGCGGAAAATACTTCGTACC and GAGAGGATCCGCGTTCTTGGAGGTAGCAG. The PCR product was digested with BamHI and SalI and ligated to pBS-SK which had been digested with the same enzymes, generating pBSTIR3. To generate the disruption construct, the URA3 gene was inserted into pBSTIR3. For this construction an NdeI/BspDI linker (TAATCGATATCGAT) was inserted into the NdeI site (at −149) in pBSTIR3. The resulting plasmid was digested with NcoI (at +226), end filled with Klenow polymerase, digested with BspDI, and ligated to the BspDI/SmaI fragment containing URA3 described above, generating pBSTIR3:URA3. The disruption fragment was excised with EcoRI and XhoI.
(iii) pBSTIR4:URA3.
The region containing the TIR4 gene (−993 to +822) was amplified using the primers GAGAGAATTCGTCGACACACGATAAAGTTCTTGAAGAAAG and TGTGACAGCAGAAGAACTAGTAGC. The PCR product was digested with SpeI and SalI and ligated to pBS-SK which had been digested with SpeI and SalI, generating pBSTIR4. This plasmid was digested with NcoI (at +290), end filled with Klenow polymerase, digested with BstBI (at −131), and ligated to the ClaI-SmaI-digested URA3 fragment, generating pBSTIR4:URA3. The disruption fragment was excised with NotI and XhoI.
Centromeric plasmids containing the TIR1, TIR3, and TIR4 genes and their respective native promoters were constructed as follows. For YCpTIR1, a fragment containing the TIR1 gene up to −1034 was excised from pBSTIR1 with EcoRI and SalI and ligated to YCplac22 (12) which had been digested with the same enzymes. For YCpTIR3, a region containing the TIR3 gene (−1084 to +1052) was amplified using the primers GAGAGGATCCGCGTTCTTGGAGGTAGCAG and TGTGGGATCCTTTTCTCGACGGCTGCTAC. The product was digested with SalI and BamHI and ligated to YCplac22 which had been digested with the same enzymes. For YCpTIR4, a region containing the TIR4 gene (−993 to +1651) was amplified using the primers GAGAGAATTCAGTCGACACACGATAAAGTTCTTGAAGAAAG and TCTCGGATCCTCTTCCTGCCCACATTCTG. The product was digested with SalI and BamHI and ligated to YCplac22 which had been digested with the same enzymes.
Strains and growth conditions.
Cells of strain FY23 (30) and derivatives or RZ53 (20) were used for all experiments. Aerobic and anaerobic growth conditions in liquid media (yeast-peptone-dextrose [YPD] or synthetic complete medium lacking uracil [SC-ura]) were as described previously (19). Cells were also grown on SDET-ura agar (SC-ura medium containing 0.5% Tween 80 and 10 μg of ergosterol/ml) at 30o under anaerobic conditions in an anaerobic jar containing a BBL anaerobic GasPak. Cells grown to mid-log phase in YPD were subjected to cold shock by shifting to 13°C for 90 min. Cells were harvested and RNA was extracted as described previously (19). For treatment under anaerobic conditions with heme, RZ53 cells were grown as described previously (26). For induction of expression of the MOX4 gene under the control of galactose, cells carrying YCpGAL1/MOX4 or the YCp33 (12) vector were grown as described previously (1).
Electron microscopy.
For electron microscopy, cells of strains FY23, FY23tir1Δ, FY23tir3Δ, and FY23tir4Δ were grown in anaerobic jars as described above, washed from plates in SC medium, pelleted in microcentrifuge tubes. The pellets were high-pressure frozen and fixed by freeze substitution in 1% osmium in acetone at −90°C (72 h), −60°C (48 h), and 4°C (18 h). Cells were washed twice in acetone and embedded in Epon/Araldite. Thin sections were stained with uranyl acetate and lead.
RESULTS
Homology patterns defining the DAN/TIR gene family.
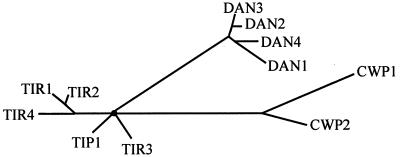
Several mannoprotein genes have been reported to be induced during anaerobiosis: TIR1, TIR2, TIP1, and DAN1. To identify more genes in this group, we searched for ORFs encoding homologous proteins and found five with significant homology to TIR1 and DAN1: YIL011w, YOR009w, YLR037c, YBR301w, and YJR151c. The hypothetical proteins were subjected to analysis with Puzzle_4 software to deduce a possible lineage tree (Fig. 1). According to this set of comparisons, two of the open reading frames, YIL011w and YOR009w, were relatively more homologous to TIR1, and they were designated, respectively, TIR3 and TIR4 (we refer here to TIR1, -2, -3, and -4 and TIP1 as “TIR” genes, for “tip-related” [9]). Three other genes were clustered with DAN1: YLR037c, designated DAN2; YBR301w, designated DAN3; and YJR151c, designated DAN4. The DAN and TIR genes share homology with the seripauperin family of genes in the PAU domain, although unlike most of these proteins, Dan1, Dan4, Tip1, Tir1, Tir3, and Tir4 contain extensive serine-rich domains, presumed to be the site of mannosylation. DAN2 and DAN3 are more typical of the PAU group proteins with PAU domains and short serine-threonine rich regions. In addition most of DAN/TIR group share homology in GPI anchor domains at the C-terminal end, as well as in N-terminal endoplasmic reticulum localization domains. We noted that the CWP1 and CWP2 genes, which encode the major mannoproteins under normal growth conditions, also show weak homology to the PAU domains of the DAN/TIR genes.
FIG. 1.
Homology tree for mannoprotein genes. Puzzle_4 software was used to deduce homology relationships among the DAN/TIR and CWP genes.
Differential responses of the DAN/TIR genes to hypoxia and temperature, including asynchrony.
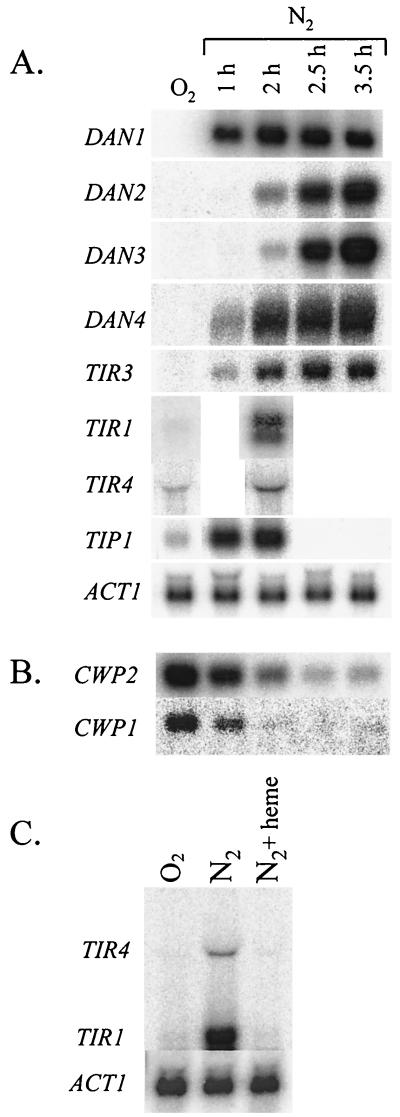
We found that the mRNAs for all the DAN/TIR genes identified in a homology search were induced during anaerobic growth, while under aerobic conditions expression was either weak or undetectable (Fig. 2A). There are differences in expression patterns among subgroups of the DAN/TIR genes. First, the TIR genes are less stringently repressed by oxygen than the DAN genes. Second, as discussed below, four of the TIR genes are induced by cold shock while the DAN genes are not. Finally, there is asynchrony of expression: DAN1, DAN4, and the TIR genes were maximally induced within 1 h of anaerobiosis, but the appearance of DAN2 and DAN3 mRNA was consistently delayed for about two more hours, being maximally induced after 3.5 h (Fig. 2A). This indicated that the genes encoding anaerobic cell wall proteins are programmed for asynchronous expression. As observed earlier for DAN1, expression of all of these genes was negatively regulated by heme, e.g., TIR1 and TIR4 (Fig. 2C).
FIG. 2.
Expression of DAN/TIR and CWP genes during anaerobic growth. (A) Cells of strain FY23 were grown under aerobic or anaerobic conditions as described in the text and harvested for RNA extraction at the intervals indicated. Northern blots were probed with DAN1, DAN2, DAN3, DAN4, TIR1, TIR3, TIR4, TIP1, and ACT1 as a loading control. (B) The RNA samples used for panel A were probed with CWP1 and CWP2. (C) Cells of strain RZ53 were grown in YPD under aerobic conditions or under anaerobic conditions with and without supplementation by heme (25 μg/ml). The Northern blot was probed with TIR4 and TIR1.
Since expression of the DAN/TIR genes depends on the Mox4 activator, we tested the role of Ecm22 protein (ORF YLR228c), which has a high degree of homology to Mox4, especially in a region containing a domain critical to heme regulation. Ecm22 is thought to play a role in cell wall synthesis, since an ecm22:Tn5 mutation causes sensitivity to calcofluor white (21). We found that this allele caused reduced expression of DAN2 and DAN3 (data not shown) but not of the other DAN/TIR genes, suggesting that Ecm22 is a factor in the induction of these two genes.
Down-regulation of the CWP genes during anaerobiosis.
During anaerobiosis, expression of CWP1 and CWP2 was down-regulated more than 10-fold within 2.5 h (Fig. 2B). In effect, the CWP1 and CWP2 mRNAs were replaced by the DAN/TIR mRNAs, suggesting that anaerobic adaptation includes extensive remodeling of the cell surface, where a large fraction of mannoproteins are thought to reside. The loss of CWP1 and CWP2 mRNAs was not instantaneous, showing a half-life of more than half an hour, after the anaerobic shift. This contrasts with other more rapidly degraded mRNAs subject to oxygen induction (20, 25). It is unknown whether the slow decrease is due to mRNA stability or to slow cessation of transcription.
Cold shock induction of TIR1, TIR2, and TIR4.
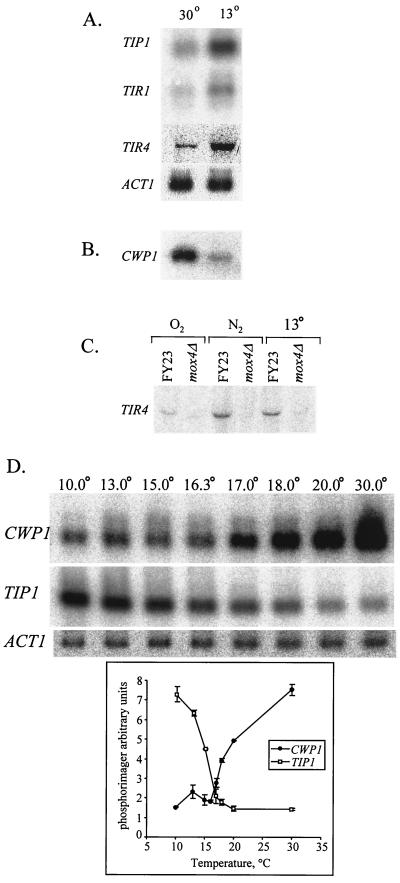
The TIR4 gene was induced during growth at low temperatures (Fig. 3A), as observed earlier for TIP1, TIR1, and TIR2, while the DAN genes and TIR3 remained uninduced (data not shown). Cold shock induction of TIR4 (and of TIR1 [data not shown]) depended on the Mox4 activator (Fig. 3C), suggesting that the hypoxic and hypothermic signal pathways converge through this factor. We also observed that expression of CWP1 mRNA decreased at low temperatures (Fig. 3B) (CWP2 was unaffected). Hence, the reciprocal expression of the CWP and DAN/TIR genes in response to oxygen is echoed in response to hypothermia, among subsets of the two gene groups.
FIG. 3.
Regulation of mannoprotein gene expression during cold shock. Cells of strain FY23 were subjected to cold shock as described in Materials and Methods and harvested for RNA extraction. (A) Northern blots were probed with TIP1, TIR1, and TIR4. (B) Cells of strain FY23 and FY23mox4Δ were grown under aerobic conditions, under anaerobic conditions, or at 13°C. A Northern blot was probed with CWP1. (C) The same blot was probed with TIR4. (D) Cells subjected to cold shock at different temperatures were harvested for RNA extraction. Northern blots were probed with TIP1 and CWP1.
To explore further the effect of temperature on induction of TIP1 and down-regulation of CWP1, we examined the expression at different temperatures and found that induction follows a sigmoidal curve, with an inflection at about 16°C (Fig. 3D). Expression of CWP1 over the same temperature range mirrors that of TIP1 in reverse, although the temperatures of half-maximal induction (∼16°C) and down-regulation (∼18°C) of TIP1 and CWP1 are not identical.
TIR1, TIR3, and TIR4 are required for anaerobic growth.
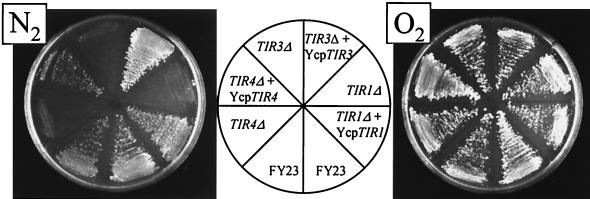
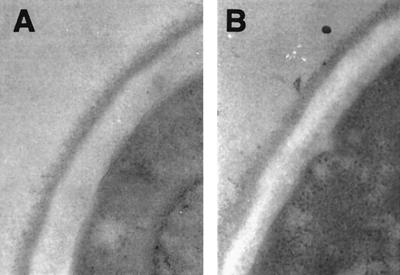
We found earlier that expression of the DAN1 gene is not needed for anaerobic growth (26), and it has been reported that disruption of TIR1 and TIR2 was also without effect (9). In order to assess the importance of some of the other DAN/TIR genes for growth, we generated disruptions of TIR1, TIR3, and TIR4. The tir1Δ, tir3Δ, and tir1Δ strains grew normally on plates under aerobic conditions but not under anaerobic conditions (Fig. 4). When the same strains were transformed with the missing gene carried on a centromeric plasmid, anaerobic growth was restored. These observations indicated that Tir1, Tir3, and Tir4 are necessary for growth without oxygen. Light microscopy of tir1Δ tir3Δ and tir1Δ cells scraped from anaerobic plates showed that most of the cells were unbudded; a small number (less than 1%) had very small buds. In contrast, a majority of wild-type anaerobic cells were budded. This indicated that anaerobic tir knockout cells were able to finish division but not able to restart the cycle, presumably being arrested at G1. Electron microscopy revealed no difference in the density of the outer layer of the cell wall, indicating that there is no gross disruption at least of the pre-existing protein-rich layer caused by loss of the Tir proteins (Fig. 5). None of the three mutant strains showed any growth defect at 15°C.
FIG. 4.
Growth of strains carrying disruptions of TIR genes. Strains FY23, FY23tir1Δ, FY23tir3Δ, and FY23tir4Δ transformed with the indicated plasmids were grown under aerobic and anaerobic conditions on SDET-ura plates.
FIG. 5.
Electron microscopy images of cell walls in anaerobic cells. Cells of strain FY23 (A) and FY23tir3Δ (B) were grown in an anaerobic jar for 36 h and processed for electron microscopy as described in Materials and Methods.
Mot3 is a heme-induced activator of CWP2 expression.
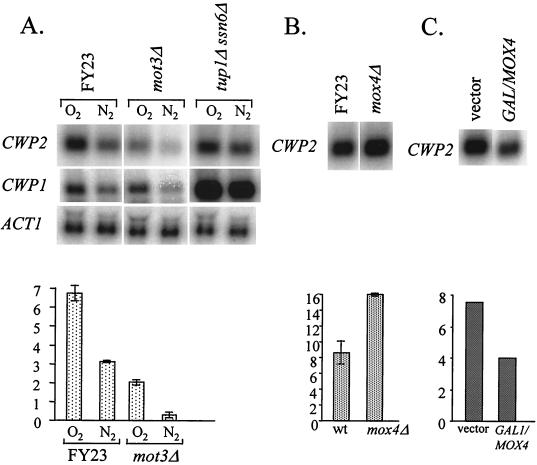
We report elsewhere that each of the DAN/TIR genes is activated by Mox 4 and repressed by a mechanism which includes Mox1 and Mox2 (1). We also found that Mot3 (11, 13) mediates heme repression of several of the DAN/TIR genes (1) and that it is induced by heme in aerobic cells (O. Sertil et al., submitted for publication). Mot3 has been found to act either positively or negatively at a number of different promoters (13), though it is not known why some promoters are activated and others repressed. We tested for a role of the DAN/TIR regulators in control of CWP expression and observed a surprisingly large decrease of expression of CWP2 gene in mot3Δ cells (Fig. 6A), indicating that Mot3 activates CWP2 expression during aerobic growth. Presumably Mot3 acts through three binding sites in the CWP2 promoter (−668, −500, and −479), which would be expected to be in the high-affinity range, based on an earlier report (13). We found expression of CWP1 to be unaffected by the mot3Δ allele (Fig. 6A).
FIG. 6.
Regulatory factors controlling expression of CWP1 and CWP2. (A) Cells of strains FY23, FY23mot3Δ, and FY23tup1Δssn6Δ were grown in YPD under aerobic and anaerobic conditions and harvested for RNA extraction. Northern blots were probed with CWP1, CWP2, and ACT1. Phosphorimager data (in arbitrary units) for relative intensities are shown at the bottom. (B) Cells of strains FY23 and FY23mox4Δ were grown under anaerobic conditions in YPD. Phosphorimager data are shown. (C) Cells of strain FY23 carrying YCpGAL1/MOX4 or the YCplac33 vector were grown at 30°C for 4 h under aerobic conditions in SC-galactose-raffinose medium (1) and harvested for RNA extraction. Data from an autoradiographic scan are shown.
Regulation of CWP2 by Mox4.
Although expression of CWP2 was reduced by 70% in a mot3Δ strain, there was still a difference between aerobic and anaerobic expression, indicating that other factors might be involved in mediating induction. One such factor may be Mox4 or a factor associated with it, since overexpression of MOX4 in aerobic cells under the control of the GAL1 promoter caused a decrease in CWP2 expression (Fig. 6B), along with an increase in DAN/TIR gene expression (data not shown), as observed earlier (1). In addition, mox4Δ cells consistently showed a significant increase in anaerobic CWP2 expression (Fig. 6C) along with the expected loss of expression of the DAN/TIR genes. Hence, Mot3 and Mox4 exert opposite effects on the DAN/TIR and CWP2 promoters. MOT3 expression was not affected by the mox4Δ phenotype (data not shown). Neither the loss nor the overexpression of MOX4 had any effect on the expression of CWP1.
Effect of other transcriptional regulators on the expression of CWP genes.
We also tested the effect of the global repressors Tup1 and Ssn6 on the expression of CWP1 and CWP2. Anaerobic expression of CWP2 was increased modestly—about twofold—by the loss of Tup1 and Ssn6. CWP2 expression may increase in anaerobic tupΔ ssn6Δ cells because these factors normally help repress MOT3 expression in anaerobic cells (Sertil et al., submitted). In contrast, expression of CWP1 was strongly increased in tupΔ ssn6Δ cells both aerobically and anaerobically, reaching levels far higher than the maximum observed during aerobic growth. This suggests that Tup1 and Ssn6 regulate CWP1 expression, by assisting an unidentified promoter-specific repressor. Since this superinduction is observed in aerobic cells, the hypothetical repressor must exert some effect even in the presence of oxygen, and it may be the target of an unknown derepressing signal. The existence of such signals is implied by increased expression of CWP1 in response to cell cycle signals (2) and to loss of cell wall integrity (23). Given the complexity of the inducing signals for CWP1 it is possible that the effect of the tupΔ ssn6Δ mutations is indirect, i.e., that increased expression is caused by the same alterations in cell wall structure which cause the flocculence characteristic of tup1 and ssn6 mutants.
DISCUSSION
We have shown that TIR1, TIR2, TIP1, and DAN1 are members of a larger group of homologous genes encoding mannoproteins involved in anaerobic adaptation. DAN2, DAN3, DAN4, TIR3, and TIR4 are all induced during hypoxia, but their expression is asynchronous, with expression of DAN2 and DAN3 being delayed for about 2 h after expression of the other DAN/TIR genes. We also found that the genes encoding the two major cell wall mannoproteins, CWP2 and CWP1, are down-regulated during anaerobic adaptation. In effect, it appears that oxygen deprivation results in extensive programmed remodeling of the cell wall, with Cwp1 and Cwp2 being replaced by the Dan/Tir proteins. Whether the latter proteins substitute functionally for Cwp1 and Cwp2 remains to be determined.
An analogous substitution process seems to occur when cells are subjected to cold shock, as TIR1, TIR2, TIR4, and TIP1 are induced and CWP1 is down-regulated. In attempting to perceive a rationale for the replacement of one group of cell wall proteins by another during hypoxia and cold shock, one possibility is that the adaptation event is related to membrane fluidity. Cells subjected to these two seemingly unrelated environmental stresses both experience reduction in membrane fluidity—during hypoxia as a result of depletion of unsaturated fatty acids and during hypothermia as a result of membrane phase transition. The cell wall proteins transit the membrane during cell wall assembly; this process may be affected by membrane properties, and conceivably, alternate protein forms or variations in the mechanisms of secretion may accommodate differences in fluidity. Another possibility is that some of these proteins play a role in the transport of sterols, as suggested by the fact that the Mox4 regulator, which controls DAN/TIR gene expression, also controls expression of factors involved in this process (5). Clearly, the special function of the anaerobic cell wall proteins will be more obvious when the function of cell wall proteins and the mechanisms of cell wall assembly are better understood.
We observed that the critical induction temperature both for expression of TIP1 and down-regulation of CWP1 was 16 to 18°C. Hence, induction and down-regulation are half-maximal at a temperature in the range within which phase transition might be expected to occur (10). Another observation suggesting that membrane fluidity is a factor in hypoxic adaptation was that unsaturated fatty acids repress hypoxic and cold shock-induced expression of TIP1. Whether changes in membrane fluidity actually signal low-temperature induction of any of several genes showing this response remains to be determined. Clearly, not all cold shock-induced genes are activated by the same factors. We showed here that cold shock induction of TIR1 and TIR4 requires Mox4, whereas induction of OLE1 and TIP1 was not affected by the mox4Δ allele (data not shown). Hence, if there is a common signal pathway it must diverge at the level of transcriptional regulators.
We noted a correspondence of homology and expression patterns, suggesting that there might be a functional basis for the difference in the patterns of regulation of these genes: (i) TIP1 and the TIR genes are clustered in a homology tree and are less stringently repressed by oxygen than the DAN genes; four TIR genes—TIP1, TIR1, TIR2, and TIR4—are induced by cold shock; (ii) the four DAN genes are also clustered and are stringently regulated and not induced by cold shock; and (iii) the two CWP genes, weakly homologous to the DAN/TIR genes, are quite similar and are induced by oxygen.
In general, the function of mannoproteins is not well understood, though there is evidence that Cwp1 and Cwp2 help maintain cell wall integrity, and a fatty acid esterase activity has been tentatively attributed to Tip1 (14). As a first step in deducing the role of the Dan/Tir proteins, we tested the effect of deleting TIR1, TIR2, or TIR4 and found that each is essential for anaerobic growth. This observation was at variance with an earlier report for TIR1 and TIR2 (9), possibly because of strain differences or differences in the degree of hypoxia achieved in the growth chamber. Anaerobically grown tirΔ cells were unbudded, presumably becoming arrested in G1 after onset of anaerobiosis. The clear growth phenotype of these mutants will facilitate structure-function analysis of the Tir proteins, including the role of the PAU domain.
We have reported elsewhere that a network of regulators is responsible for the coordinate regulation of the DAN/TIR genes. This includes anaerobic induction by the Mox 4 transcriptional activator and aerobic repression by the Mox1 and Mox2 repressors. Some of the DAN/TIR genes are also repressed during aerobic growth by Mot3, a repressor which is induced by heme and repressed in anaerobic cells by Hap1 (Sertil et al., submitted), in parallel with the Rox1 repressor (6, 15, 20). Surprisingly, mutations affecting Mot3 and Mox4 also affected expression of CWP2 but in a manner opposite to their effect on DAN/TIR expression, helping to account for induction of CWP2 in aerobic cells. We have concluded that Mot3, which is known to function as an activator or repressor of different genes through the same binding sites (13), is an important activator of CWP2, presumably through the three Mot3 sites in the promoter. It is interesting that the versatility of Mot3 allows it to mediate both positive and negative regulation by heme. Conversely, Mox4, the activator of the DAN/TIR genes, plays a significant role in repressing CWP2, though the mechanistic basis of this reciprocal effect is unknown. We also observed that the Tup1-Ssn6 repressor complex contributes to repression of anaerobic CWP2 expression, possibly by virtue of its role in repression of MOT3 (Sertil et al., submitted). CWP2 was earlier observed to be regulated during the cell cycle, as would be expected for a gene product associated with bud formation. Interestingly, analysis of genes regulated during the cell cycle revealed that expression of MOT3 is also cyclical, suggesting that fluctuations in Mot3 may account for cell cycle regulation of CWP2.
Regulation of CWP1 is still not well understood, except for a descriptive list of signals affecting its expression, either positively (induction during the cell cycle or when cell wall synthesis is disrupted) or negatively, as shown here during hypoxic or hypothermal stress. Although CWP1 appears to be regulated by oxygen in parallel with CWP2, mot3 and mox4 mutations do not affect its expression, indicating that it is controlled by a different regulatory pathway. However, we did observe constitutive expression of CWP1 in a tup1Δ ssn6Δ strain during hypoxia, indicating that expression in that state is normally blocked by a repressor associated with Tup1-Ssn6. It is worth noting that even though TIP1 is regulated in parallel with the TIR genes, it is also controlled by a different pathway, showing no dependence on Mox4 or Mox1, Mox2, or Mot3 for repression by oxygen or induction by cold shock (data not shown). Work in this area has demonstrated that several distinct but interconnected mechanisms are deployed in yeast to achieve essentially the same effect, i.e., regulation by oxygen, targeting genes in several regulons.
ACKNOWLEDGMENTS
We thank Robert Trimble for useful discussions. We are grateful to Jeff Ault at the EM Core facility at the Wadsworth Center Laboratory of the New York State Department of Health for providing electron microscopy. Light microscopy was carried out with the help of Joseph Mazurkievicz in the Albany Medical College Imaging Core Facility. We are grateful to H. Bussey and M. Lussier for generously providing the ecm22 mutant strain.
This work was supported by a grant from the National Science Foundation (MCB-9723565).
REFERENCES
- 1.Abramova N, Cohen B D, Sertil O, Davies K J A, Lowry C V. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics. 2001;157:1169–1177. doi: 10.1093/genetics/157.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Cabib E, Drgon T, Drgonova J, Ford R A, Kollar R. The yeast cell wall, a dynamic structure engaged in growth and morphogenesis. Biochem Soc Trans. 1997;25:200–204. doi: 10.1042/bst0250200. [DOI] [PubMed] [Google Scholar]
- 2.Caro L H P, Smits G J, Van Egmond P, Chapman J W, Klis F M. Transcription of multiple cell wall protein-encoding genes in Saccharomyces cerevisiae is differentially regulated during the cell cycle. FEMS Microbiol Lett. 1998;161:345–349. doi: 10.1111/j.1574-6968.1998.tb12967.x. [DOI] [PubMed] [Google Scholar]
- 3.Caro L H P, Tettelin H, Vossen J H, Ram A F J, Van den Ende H, Klis F M. In silico identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast. 1997;13:1477–1489. doi: 10.1002/(SICI)1097-0061(199712)13:15<1477::AID-YEA184>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 4.Cid V J, Duran A, Del Rey F, Snyder M P, Nombela C, Sanchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Cohen B D, Sertil O, Abramova N E, Davies K J A, Lowry CV. Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res. 2001;29:799–808. doi: 10.1093/nar/29.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crowley J H, Leak F W, Shianna K V, Tove S, Parks L W. A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J Bacteriol. 1998;180:4177–4183. doi: 10.1128/jb.180.16.4177-4183.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deckert J, Perini R, Balasubramanian B, Zitomer R S. Multiple elements and auto-repression regulate Rox1, a repressor of hypoxic genes in Saccharomyces cerevisiae. Genetics. 1995;139:1149–1158. doi: 10.1093/genetics/139.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Nobel H, Klis F M, Priem J, Munnik T, Van den Ende H. The glucanase-soluble mannoproteins limit cell wall porosity in Saccharomyces cerevisiae. Yeast. 1990;6:491–499. doi: 10.1002/yea.320060606. [DOI] [PubMed] [Google Scholar]
- 8.De Nobel H, Lipke P N. Is there a role for GPIs in yeast cell-wall assembly? Trends Cell Biol. 1994;4:42–45. doi: 10.1016/0962-8924(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 9.Donzeau M, Bourdineaud J P, Lauquin G J. Regulation by low temperature and anaerobiosis of a yeast gene specifying a putative GPI-anchored plasma membrane protein. Mol Microbiol. 1996;20:449–459. doi: 10.1111/j.1365-2958.1996.tb02631.x. [DOI] [PubMed] [Google Scholar]
- 10.Dorfler H, Fabian B. Effect of changes in the lipid composition on the plasma membrane of Saccharomyces cerevisiae through mutation of the phase transition and mixing behavior of the lipid fraction. J Basic Microbiol. 1995;35:207–215. doi: 10.1002/jobm.3620350402. . (In German.) [DOI] [PubMed] [Google Scholar]
- 11.Fujiwara D U, Yoshimoto H, Sone H, Harashima S, Tamai Y. Transcriptional co-regulation of Saccharomyces cerevisiae alcohol acetyltransferase gene, ATF1 and delta-9 fatty acid desaturase gene, OLE1 by unsaturated fatty acids. Yeast. 1998;14:711–721. doi: 10.1002/(SICI)1097-0061(19980615)14:8<711::AID-YEA263>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 12.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 13.Grishin A V, Rothenberg M, Downs M A, Blumer K J. Mot3, a Zn finger transcription factor that modulates gene expression and attenuates mating pheromone signalling in Saccharomyces cerevisiae. Genetics. 1998;149:879–892. doi: 10.1093/genetics/149.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horsted W M, Dey E S, Holmberg S, Kielland-Brandt M C. A novel esterase from Saccharomyces carlsbergensis, a possible function for the yeast TIP1 gene. Yeast. 1998;14:793–803. doi: 10.1002/(SICI)1097-0061(19980630)14:9<793::AID-YEA277>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 15.Keng T. HAP1 and ROX1 form a regulatory pathway in the repression of HEM13 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2616–2623. doi: 10.1128/mcb.12.6.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo K, Inouye M. TIP1, a cold shock inducible gene of Saccharomyces cerevisiae. J Biol Chem. 1991;266:17537–17544. [PubMed] [Google Scholar]
- 17.Kowalski L R Z, Kondo K, Inouye M. Cold-shock induction of a family of TIP1-related proteins associated with the membrane in Saccharomyces cerevisiae. Mol Microbiol. 1995;15:341–353. doi: 10.1111/j.1365-2958.1995.tb02248.x. [DOI] [PubMed] [Google Scholar]
- 18.Lambrechts G M, Bauer F F, Marmur J, Pretorius I S. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc Natl Acad Sci USA. 1996;93:8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowry C V, Zitomer R S. Oxygen regulation of anaerobic and aerobic genes mediated by a common factor in yeast. Proc Natl Acad Sci USA. 1984;81:6129–6133. doi: 10.1073/pnas.81.19.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowry C V, Zitomer R S. ROX1 encodes a heme-induced repression factor regulating ANB1 and CYC7 of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4651–4658. doi: 10.1128/mcb.8.11.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lussier M, White A M, Sheraton J, Di Paolo T, Treadwell J, Southard S B, Horenstein C I, Chen-Weiner J, Ram A F J, Kapteyn J C, Roemer T W, Vo D H, Bondoc D C, Hall J, Zhong W W, Sdicu A M, Davies J, Klis F M, Robbins P W, Bussey H. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marguet D, Lauquin G J. The yeast SRP gene: positive modulation by glucose of its transcriptional expression. Biochem Biophys Res Commun. 1986;138:297–303. doi: 10.1016/0006-291x(86)90279-2. [DOI] [PubMed] [Google Scholar]
- 23.Ram A F J, Kapteyn J C, Montijn R C, Caro L H P, Douwes J E, Baginsky W, Mazur P, Van den Ende H, Klis F M. Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in the release of β-1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J Bacteriol. 1998;180:1418–1424. doi: 10.1128/jb.180.6.1418-1424.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy A, Lu C F, Marykwas D L, Lipke P N, Kurjan J. The AGA1 product is involved in cell surface attachment of the Saccharomyces cerevisiae cell adhesion glycoprotein a-agglutinin. Mol Cell Biol. 1991;11:4196–4206. doi: 10.1128/mcb.11.8.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schjerling C K, Hummel R, Hansen J K, Borsting C, Mikkelsen J M, Kristiansen K, Knudsen J. Disruption of the gene encoding the acyl-CoA-binding protein (ACB1) perturbs acyl-CoA metabolism in Saccharomyces cerevisiae. J Biol Chem. 1996;271:22514–22521. doi: 10.1074/jbc.271.37.22514. [DOI] [PubMed] [Google Scholar]
- 26.Sertil O, Cohen B D, Davies K J D, Lowry C V. The DAN1 gene of S. cerevisiae is regulated in parallel with the hypoxic genes, but by a different mechanism. Gene. 1997;192:199–205. doi: 10.1016/s0378-1119(97)00028-0. [DOI] [PubMed] [Google Scholar]
- 27.Shimoi H, Iimura Y, Obata T. Molecular cloning of CWP1: a gene encoding a Saccharomyces cerevisiae cell wall protein solubilized with Rarobacter faecitabidus protease I. J Biochem (Tokyo) 1995;118:302–311. doi: 10.1093/oxfordjournals.jbchem.a124907. [DOI] [PubMed] [Google Scholar]
- 28.Van der Vaart J M, Caro L H P, Chapman J W, Klis F M, Verrips C T. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J Bacteriol. 1995;177:3104–3110. doi: 10.1128/jb.177.11.3104-3110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viswanathan M, Muthukumar G, Cong Y S, Lenard J. Seripauperins of Saccharomyces cerevisiae: a new multigene family encoding serine-poor relatives of serine-rich proteins. Gene. 1994;148:149–153. doi: 10.1016/0378-1119(94)90249-6. [DOI] [PubMed] [Google Scholar]
- 30.Winston F, Dollard C, Ricupero-Hovasse S L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 31.Zlotnik H, Fernandez M P, Bowers B, Cabib E. Saccharomyces cerevisiae mannoproteins form an external cell wall layer that determines wall porosity. J Bacteriol. 1984;159:1018–1026. doi: 10.1128/jb.159.3.1018-1026.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]