Abstract
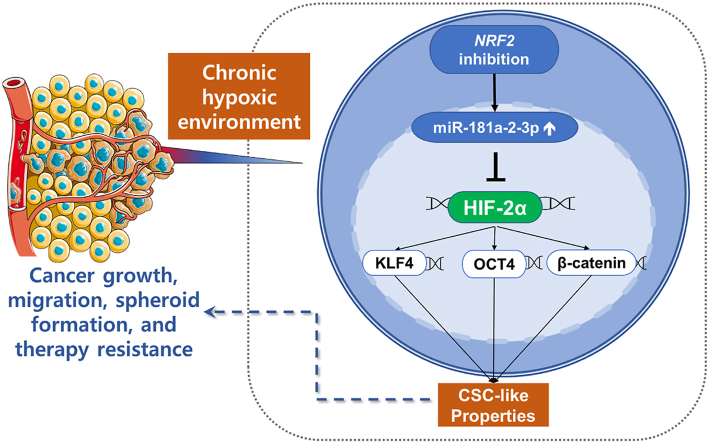
The acquisition of the cancer stem cell (CSC) properties is often mediated by the surrounding microenvironment, and tumor hypoxia is considered an important factor for CSC phenotype development. High levels of NRF2 (Nuclear Factor Erythroid 2-Like 2; NFE2L2), a transcription factor that maintains cellular redox balance, have been associated with facilitated tumor growth and therapy resistance. In this study, we investigated the role of NRF2 in hypoxia-induced CSC phenotypes in colorectal cancer cells. Chronic hypoxia for 72 h resulted in CSC phenotypes, including elevation of krupple-like factor 4 (KLF4) and octamer-binding transcription factor 4 (OCT4), and an increase in cancer migration and spheroid growth with concomitant hypoxia-inducible factor 2α (HIF-2α) accumulation. All these chronic hypoxia-induced CSC properties were attenuated following HIF-2α-specific silencing. In this chronic hypoxia model, NRF2 inhibition by shRNA-based silencing or brusatol treatment blocked HIF-2α accumulation, which consequently resulted in decreased CSC marker expression and inhibition of CSC properties such as spheroid growth. In contrast, NRF2 overactivation by genetic or chemical approach enhanced the chronic hypoxia-induced HIF-2α accumulation and cancer migration. As a molecular mechanism of the NRF2-inhibition-mediated HIF-2α dysregulation, we demonstrated that miR-181a-2-3p, whose expression is elevated in NRF2-silenced cells, targeted the HIF-2α 3′UTR and subsequently suppressed the chronic hypoxia-induced HIF-2α and CSC phenotypes. The miR-181a-2-3p inhibitor treatment in NRF2-silenced cells could restore the levels of HIF-2α and CSC markers, and increased cancer migration and sphere formation under chronic hypoxia. In line with this, the miR-181a-2-3p inhibitor transfection could increase tumorigenicity of NRF2-silenced colorectal cancer cells. Collectively, our study suggests the involvement of NRF2/miR181a-2-3p signaling in the development of HIF-2α-mediated CSC phenotypes in sustained hypoxic environments.
Keywords: Cancer stem cell phenotype, Chronic hypoxia, HIF-2α, NRF2, miR-181a-2-3p
Graphical abstract
Highlights
-
•
HIF-2α mediates CSC phenotypes under chronic hypoxic condition.
-
•
NRF2 inhibition blocks the chronic hypoxia-induced HIF-2α accumulation in colorectal cancers.
-
•
NRF2 inhibition suppresses the chronic hypoxia-induced CSC phenotypes by increasing HIF-2α-targeting miR-181a-2-3p.
1. Introduction
The tumor mass contains a small subpopulation of cells that fuel tumor growth and therapy resistance, which are called tumor-initiating cells or cancer stem cells (CSCs) [1]. Since their first report on acute myeloid leukemia, CSCs have been identified in solid tumors, including breast, lung, and colon cancers [[2], [3], [4]]. The self-renewal and differentiation abilities of CSCs are critical drivers of cancer malignancy, including metastasis, invasion, therapy resistance, and immune evasion [5,6]. Although the characterization of CSCs remains challenging, several markers have been identified that contribute to stemness, including cluster of differentiation 133 (CD133) in glioblastoma [7], CD44 and krupple-like factor (KLF4) in breast cancers [8,9], octamer-binding transcription factor 4 (OCT4) in melanomas [10], and SRY (sex determining region Y)-box 2 (SOX2) and homeobox protein NANOG (NANOG) in colorectal cancers [11]. The signaling pathways such as Wnt/β-catenin and neurogenic locus notch homolog protein (NOTCH) have been reported to increase in CSCs, and these are known to involve in CSC self-renewal and tumorigenicity [12].
The development of CSC phenotypes is supported by the surrounding microenvironment. Tumor hypoxia is considered as an important microenvironment factor facilitating cancer initiation, progression, and acquisition of CSC-like properties [13,14]. Upon hypoxic environment, hypoxia-inducible factors (HIFs) upregulate groups of genes to maintain cancer survival [15]. HIFs consist of an oxygen-reactive α subunit (HIF-α) and stable β subunit {HIF-β, aryl hydrocarbon nuclear translocator 1 (ARNT1)}, which are members of the bHLH-PAS domain proteins [16]. Among the HIF-α proteins, HIF-1α and HIF-2α are the best characterized in mammals. Under sufficient levels of O2, HIF-α proteins are constantly hydroxylated by proline hydroxylase domain (PHD) proteins, which are enzymes that use oxygen as a substrate. Hydroxylated HIF-α proteins are then recognized by the von Hippel Lindau (pVHL) tumor suppressor protein for rapid degradation by the 26S proteasome [17]. In an anaerobic environment, HIF-α proteins are stabilized due to the lack of PHD activity and heterodimerize with ARNT1 to bind to the hypoxic response element (HRE) of their target genes [17].
Despite having similar structures, HIF-1α and HIF-2α are different in their specific functions, and not only share overlapping target genes but also have specific target genes [18]. HIF-1α is mainly activated during acute hypoxia and promotes angiogenesis by activating vascular endothelial growth factor (VEGF) expression [17]. It also increases glycolytic capacity, which is a hallmark of cancer, by elevating glycolysis-related enzymes such as glucose transporter 1, and plays a role in cell survival by targeting the anti-apoptotic protein Bcl-2 [19]. In contrast, HIF-2α is more stable under chronic hypoxic conditions, and growing evidence shows that HIF-2α is likely to possess oncogenic activity by regulating cancer stemness [18]. HIF-2α directly regulates OCT4 expression to support stem cell function, embryo development, and tumor growth [20]. In glioblastoma specimens, HIF-2α and CSC markers were colocalized, and the depletion of HIF-2α in CD133-enriched glioma stem cells reduced their tumorigenic activity in vitro and in vivo [21]. HIF-2α upregulates the expression of the stem cell factor (SCF), a multifunctional cytokine, and consequently promotes angiogenesis and metastasis in hepatocellular carcinoma [22]. Breast cancer stemness is mediated by HIF-2α via the induction of mitochondrial reactive oxygen species (ROS) under hypoxia [23].
NRF2 (Nuclear Factor Erythroid 2-Like 2; NFE2L2) plays an important role as a transcription factor in the expression of multiple cytoprotective genes in response to electrophilic and oxidative stress. NRF2 activation is primarily regulated by Kelch-like ECH-associated protein 1 (KEAP1), an adaptor protein for ubiquitination and continuous degradation of NRF2 by the 26S proteasome under normal conditions [24,25]. In the presence of stress, conformational changes in the KEAP1 protein structure allow NRF2 nuclear translocation to activate gene subsets, including NAD(P)H: quinone oxidoreductase (NQO1) and glutamate-cysteine ligase (GCL). However, constitutive NRF2 activation has been reported in multiple types of cancers, and its association with facilitated cancer growth, survival, and therapy resistance has been firmly established [26].
In our previous studies, knockdown of NRF2 expression in colorectal and breast cancer cells suppressed HIF-1α accumulation under hypoxic conditions for 24 h, thus inhibiting HIF-1α-dependent tumor angiogenesis and metabolic shift [27,28]. Additionally, we provided evidence that the elevation of NRF2 levels is associated with CSC properties, including therapy resistance and enhanced tumor growth, spheroid growth, and migration/invasion capacity in breast, colorectal, and ovarian carcinoma cells [[29], [30], [31], [32]]. Based on these findings, we hypothesized the potential contribution of NRF2 to hypoxia-induced CSC-like properties, and investigated the effect of NRF2 on HIF-2α accumulation and CSC phenotype development in chronic hypoxia-exposed colorectal cancer cells. Additionally, the molecular link between NRF2 and HIF-2α regulation has been explored by focusing on differentially expressed micro RNAs (miRNAs) in NRF2-silenced colorectal cancer cells.
2. Materials and methods
2.1. Reagents
5-Fluorouracil (5-FU), Z-Leu-Leu-Leu-al (MG132), brusatol, doxorubicin, and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich Co. (Saint Louis, MO, USA). Bardoxolone methyl was purchased from Selleckchem (Boston, MA, USA). TB Green real-time polymerase chain reaction (PCR) master mix was purchased from Takara Bio Inc. (Kusatsu, Shiga, Japan). HIF-1α antibody (#610958) was purchased from BD Biosciences (Palo Alto, CA, USA). Antibodies against HIF-2α (#7096), KLF4 (#4038), and pVHL (#68547) were obtained from Cell Signaling Technology Inc. (Danvers, MA, USA). Antibodies against the catalytic subunits of GCL (GCLC; #207777), matrix metallopeptidase 2 (MMP2, #86607), NRF2 (#62352), and OCT4 (#109183) were purchased from Abcam (Trumpington, Cambridge, UK). Antibodies against ARNT1 (#55526), KEAP1 (#15246), lamin B (#6216), NQO1 (#16464), VEGF (#152), and β-actin (#47778) were purchased from Santa Cruz Biotechnology Inc. (Dallas, TX, USA). All primers for PCR analysis (Table 1), silencing RNAs, and miRNA mimics and inhibitors were synthesized by Bioneer Corp. (Daejeon, Republic of Korea).
Table 1.
Primer sequences for qPCR analysis of human genes and miRNAs.
| Gene name | Forward | Reverse |
|---|---|---|
| KLF4 | 5′-ACACTTGTGATTACGCGGGCTGC-3′ | 5′-GGCGAATTTCCATCCACAGCCG-3′ |
| OCT4 | 5′-TGCAGAAGTGGGTGGAGGAAGC-3′ | 5′-TGGCTGATCTGCTGCAGTGTG-3′ |
| MMP2 | 5′-CGTCGCCCATCATCAAGTTCCC-3′ | 5′-CCTTCAGCACAAACAGGTTGCAGC-3′ |
| VEGF | 5′-TTGTACAAGATCCGCAGACG-3′ | 5′-TTCTGTCGATGGTGATGGTG-3′ |
| EPAS1 | 5′-ATCAGCTTCCTGCGAACACA -3′ | 5′-GCTCCACCTGTGTAAGTCCC-3′ |
| PHD1 | 5′-ACGGGCTCGGGTACGTAAG-3′ | 5′-CCCAGTTCTGATTCAGGTAATAGATACA-3′ |
| PHD2 | 5′-ACTGGGATGCCAAGGTAAGTG-3′ | 5′-CTCGTGCTCTCTCATCTGCAT-3′ |
| PHD3 | 5′-GGAGAGGTCTAAGGCAATGG-3′ | 5′-ACTTCGTGTGGGTTCCTACG-3′ |
| ACTB | 5′-CCATGTACGTTGCTATCCAG-3′ | 5′-GGCCATCTCTTGCTCGAAGT-3′ |
| miR-181a-2-3p | 5′-ACCACTGACCGTTGACTGTACC-3′ | Universal primer |
| miR-2278 | 5′-GAGAGCAGTGTGTGTTGCCTGG-3′ | Universal primer |
| U6 | 5′-CGCAAGGATGACACGCAAATTC-3′ | Universal primer |
2.2. Plasmids
The pMir-Target plasmids containing the 3′UTR of human HIF-2α (wild-type, point, and whole sequence mutation in the miR-181a-2-3p binding sites) were purchased from OriGene Technologies Inc. (Rockville, MD, USA). HIF-2α overexpression plasmid with mutation in its proline residues (pcDNA3-HA–HIF–2α-P405A/P531A) was a gift from William Kaelin (Addgene #18956) [33]. Mammalian expression plasmid for human NRF2 (pcDNA3-Myc3-NRF2) was a gift from Yue Xiong (Addgene #21555) [34]. The miArrest™ miR-181a-2-3p inhibitor expression plasmid (pEZX-AM04-miR-181a-2-3p inhibitor, #HmiR-AN0233-AM04) and its backbone vector control (pEZX-AM04, #CmiR-AN0001-AM04) were purchased from Genecopoeia (Rockville, MD, USA).
2.3. Cell culture
HCT116 and HT29 colorectal cancer cell lines were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). HCT116 and HT29 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) and Roswell Park Memorial Institute (RPMI) 1640 medium, respectively. All media were purchased from Welgene Inc. (Daegu, Republic of Korea) and supplemented with 10% fetal bovine serum (FBS; Corning Inc., Corning, NY, USA) and 1% penicillin/streptomycin (Welgene Inc.). Stable NRF2- and KEAP1-silenced cell lines and their corresponding scrambled control cell lines were previously established [27,35]. Cells were grown at 37 °C in a humidified atmosphere containing 5% CO2. Hypoxic incubation was performed in a hypoxic incubator (Astec Co. Ltd., Kasuya, Fukuoka, Japan) humidified with 1% O2 and 5% CO2 at 37 °C as described previously [28].
2.4. Transfection of siRNA, microRNA mimic and inhibitor
The cells were seeded in antibiotic-free complete medium at a density of 5 × 105 cells/well and grown overnight. The next day, cells were transfected using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) with predesigned siRNAs targeting HIF-1α (#1, 5′-GUGGUUGGAUCUAACACUA(dTdT)-3′ and 5′-UAGUGUUAGAUCCAACCAC (dTdT)-3’; #2, 5′-CAUGAAAGCACAGAUGAAU(dTdT)-3′ and 5′-AUUCA UCUGUGCUUUCAUG(dTdT)-3’; #3, 5′-CUCAUCCAUGUGACCAUGA(dTdT)-3′ and 5′-UCAUGGUCACAUGGAUGAG(dTdT)-3′), HIF-2α (#1, 5′-CGUGAG AACCUGAGUCUCA(dTdT)-3′ and 5′-UGAGACUCAGGUUCUCACG(dTdT)-3’; #2, 5′-ACUACGUCCUGAGUGAGAU(dTdT)-3′ and 5′-AUCUCACUCAGG ACGUAGU(dTdT)-3′, #3, 5′-GAUCUUUUCGAAGCUGUUA(dTdT)-3′ and 5′-UAACAGCUUCGAAAA GAU C(dTdT)-3′), or scrambled control siRNA from Bioneer Corp. For miRNA transfection, 10 nmol of mimic or inhibitor of miR-181a-2-3p and miR-2278-5p and the corresponding negative control miRNA were used. For plasmid transfection, 1 μg plasmid or its corresponding control was added to the complex mixture. After 24 h of transfection, the transfection complex-containing medium was removed, and the cells were re-seeded at a density of 2 × 105 cells and incubated for 72 h.
2.5. Cell counting
Cells were seeded at a density of 5 × 104 cells in 6-well plates. After incubation in hypoxic conditions for 24, 48, or 72 h, the cells were harvested and stained with trypan blue. Viable cells were counted using a TC10 automated cell counter (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
2.6. WST assay
Cells were plated at a density of 1.25 × 103 cells/well in a 96-well plate and incubated with 5-FU or doxorubicin for 72 h in a hypoxia chamber. Next, 10 μL of WST solution (EZ-Cytox, DoGenBio, Seoul, Republic of Korea) was added to each well and further incubated for 1 h. Absorbance was measured at 450 nm using a SpectraMax spectrophotometer (Molecular Devices, San Jose, CA, USA) as described previously [36].
2.7. Immunoblot analysis
Hypoxia-incubated cells were washed using phosphate-buffered saline (PBS) supplemented with 100 mM MG132 and rapidly lysed with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 1% nonyl phenoxypolyethoxylethanol-40, 1% sodium dodecyl sulfate, and 10% sodium deoxycholate) containing a protease inhibitor cocktail (Sigma-Aldrich Co.). Protein concentrations were determined using a bicinchoninic acid kit (Thermo Fisher Scientific Inc.,Waltham, MA, USA). Proteins (15 mg/mL) were separated on 6%, 10%, or 15% SDS-polyacrylamide gels and transferred onto nitrocellulose membranes (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) [37]. Membranes were blocked with 5% skim milk (BD Biosciences) for 1 h and incubated overnight with the corresponding primary antibody. The membranes were then incubated with the corresponding secondary antibody combined with horseradish peroxidase (Thermo Fisher Scientific Inc.) for an hour. Chemiluminescent images were captured using an ImageQuant LAS 4000 Mini luminescent image analyzer (GE Healthcare Bio-Sciences). Protein levels from three independent experiments were calculated using ImageJ analysis software [38].
2.8. Total RNA isolation and quantitative PCR (qPCR) analysis
Total RNA was isolated from cells using TRIzol (Ambion, Inc. Austin, TX, USA). For mRNA expression analysis, 200 ng of RNA was transcribed into cDNA using GoScript Reverse Transcriptase (Promega Corp., Madison, WI, USA). qPCR was performed using TB Green qPCR Mix with forward and reverse primer pairs (Table 1) in a total volume of 20 μL. β-Actin was used as the reference gene. miRNA cDNA was synthesized from 500 ng of RNA using a miScript RT kit (Qiagen, Hilden, Germany). qPCR analysis was performed with a miScript SYBR green PCR kit (Qiagen) using miRNA-specific forward primers (Table 1) and universal primers provided by the kit manufacturer. Forward primer U6 was used as the reference gene. All primers were synthesized by Bioneer Corp. Expression quantitation was performed on a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.) as previously described [39]. The data were processed using Bio-Rad CFX Maestro Software 2.3 (Bio-Rad Laboratories, Inc.).
2.9. Isolation of nuclear fraction
After growth under normoxic or hypoxic condition for 72 h, the cells were harvested and washed with ice-cold PBS. The cells were processed using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher Inc.) according to the manufacturer's protocol. Briefly, cytoplasmic extraction reagents I and II were added at 200:11 ratio to separate the cytoplasmic fraction from the cell lysates. After centrifugation, an ice-cold nuclear extraction reagent was added to isolate the nuclear fraction.
2.10. Soft agar colony formation assay
Anchorage-independent growth ability was assessed by suspending 5 × 103 cells in the top soft agar layer (0.35% agarose) on 6-well plates, which were pre-coated with 0.5% base agar. Colonies were allowed to grow at 37 °C in a normoxic or hypoxic (1% O2) incubator for 2–3 weeks and counted using an ECLIPSE Ti inverted microscope by NIS-Elements AR (V.4.0) computer software program (Nikon Instruments Korea, Seoul, Republic of Korea) [29].
2.11. Transwell migration assay
The cells were treated under normoxic or hypoxic condition for 72 h, suspended in serum-free medium, and seeded in a Transwell chamber (8.0 μm pore size, Corning Inc.) at a density of 2 × 104 cells/well. The chambers were placed in complete medium and further incubated under normoxic or hypoxic conditions for 18 h. Migrated cells on the lower surface of the membrane were fixed and stained using Diff-Quick reagent (Sysmex, Kobe, Japan) before capture using ToupView software (ToupTek, Hangzhou, Zhejiang, China) as described previously [36]. The density of migrated cells was calculated using ImageJ software.
2.12. Sphere formation assay
HCT116 cells were grown at a density of 1 × 105 cells/mL in a 1:1 ratio of serum-free DMEM and Nutrient Mixture F-12 (Welgene Inc.) supplemented with 2% B-27 (Life Technologies), 20 ng/mL epithelial growth factor, 20 ng/mL basic fibroblast growth factor (R&D Systems, Minneapolis, MN, USA), 5 μg/mL bovine insulin (Cell Applications Inc., San Diego, CA, USA), 0.5 μg/mL hydrocortisone (Sigma Aldrich, Co.), and 1% penicillin/streptomycin on ultralow attachment 100 mm plates (Corning Inc.). The sphere formation assay was conducted by growing the colonospheres on ultra-low attachment 96-well plates (Corning Inc.) for 3 d following hypoxic incubation for 72 h. The sphere number was then measured using ToupView software (ToupTek) as described previously [30].
2.13. Animal experiments
Six-week-old male BALB/c (nu/nu) mice (Orient Bio Inc., Seongnam, Republic of Korea) were housed in a pathogen-free environment with standard feeding and water ad libitum. One week after the adaptation period, the mice were injected with sc-HCT116 or shNRF2-HCT116 cells (1 × 106 or 2.5 × 106 cells/200 μL) transfected with pEZX-AM04-miR-181a-2-3p inhibitor and its control plasmid in their right flank. For brusatol treatment, a total of 2 × 106 HCT116 cells were injected. Once the average tumor volume reaches 80 mm3, the mice were randomly divided into two groups, and brusatol (2 mg/kg) and vehicle control (5% DMSO in DPBS) were administered intraperitoneally for a total of 15 doses interspersed with a 5-day resting period. Tumor growth was monitored by measuring the diameter of the tumors using calipers twice a week. Tumor volume was calculated using formula V=(a2xb)/2, where a is the width and b is the length of the tumor [27]. This study was approved by the Animal Ethics Committee of the Catholic University of Korea (CUK-IACUC-2020-006; CUK-IACUC-2022-037). The mice were sacrificed, and tumor sections were preserved by snap-freezing using liquid nitrogen until further analysis. The tumor sections were weighed and mixed with RIPA buffer (10 mg tissue/1 mL buffer) supplemented with protease and phosphatase inhibitor cocktail before being homogenized using a tissue homogenizer (IKA, Staufen, Germany). The protein extracts were centrifuged at 12,000 rpm for 20 min and re-centrifuged at 7000×g for another 20 min at 4 °C before being used for quantification and immunoblot analysis.
2.14. Luciferase reporter assay
The cells were seeded in 24-well plates at a density of 2 × 102 cells/well and grown overnight. Three hours before transfection, the medium was replaced by 1X reduced-serum medium (Opti-MEM, Gibco, Waltham, MA, USA). A total of 100 μL of Lipofectamine 2000, 0.1 μg pRL-TK Renilla luciferase, 1 μg pMir Target of HIF-2α 3′UTR, and 100 pmol of miRNA mimic were added and the cells were further incubated overnight. The reporter assay was performed using the Dual Luciferase Reporter Assay System (Promega Corp.) [40]. Transfected cells were washed with PBS and lysed using a passive lysis buffer at room temperature. Luciferase activity was detected following the addition of LARII substrate and Stop&Glo at 10s measurement on a Centro XS3 LB960 microplate luminometer (Berthold Technologies, Baden Württemberg, Germany).
2.15. Clinical analysis of miR-181a-3-5p expression in colorectal cancers using public datasets
Gene expression data from the colon cancer dataset (n = 105) available from the Clinical Proteomic Tumor Analysis Consortium (CPTAC-2 Prospective) [41] and pan-cancer analysis of whole genomes (n = 749) from the International Cancer Genome Consortium and The Cancer Genome Atlas (ICGC/TCGA) [42] were analyzed using cBioPortal (https://www.cbioportal.org/) [43,44]. The correlations between miR-181a-2 or miR-2278 levels and OCT4 mRNA levels were plotted as RNA-seq data using the estimation maximization (RSEM) algorithm on a log2 scale. The correlations were evaluated using both Spearman rank correlation and Pearson correlation coefficients. GSE49246 [45] was used to study the expression of human miRNAs in public colorectal cancer (CRC) datasets. Samples from healthy and colon cancer patients were grouped, and the expression values from both groups were further analyzed using statistical analysis.
2.16. Statistical analysis
The data were analyzed using Student's t-test and one-way or two-way ANOVA followed by Bonferroni's multiple comparison test to determine significantly different groups. These analyses were performed using GraphPad Prism5 (GraphPad Software, Inc., La Jolla, CA, USA). Statistical significance was set at P < 0.05.
3. Results
3.1. Chronic hypoxia induces CSC phenotypes and HIF-2α accumulation in colorectal cancer cells
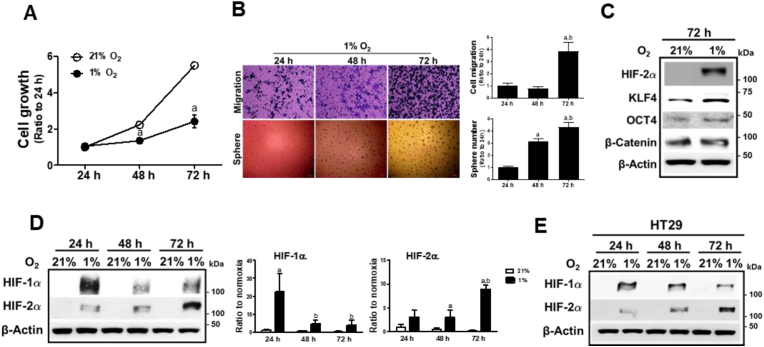
Hypoxia is known to support CSC maintenance and is considered a hallmark of aggressive cancer [1]. To investigate whether chronic hypoxia is responsible for the acquisition of stemness phenotypes of cancer cells, we exposed colorectal cancer HCT116 cells to normoxic (21% O2) or hypoxic condition (1% O2) for 24, 48, and 72 h, and monitored cell growth. Hypoxic condition affected cell growth, and the 72 h-hypoxic incubation suppressed cell growth by 56% compared to the normoxic condition (Fig. 1A). However, when these hypoxia-exposed HCT116 cells were subjected to cancer migration and sphere formation assays, the 72 h hypoxic incubation exhibited substantially increased migration and sphere-forming capacities compared to the 24 h and 48 h hypoxic cells (Fig. 1B). Along with aggressive phenotypes, 72 h-hypoxic incubation elevated CSC transcription factors such as KLF4, OCT4, and β-catenin (Fig. 1C), which suggests that chronic hypoxia induces CSC-like phenotypes in colorectal cells.
Fig. 1.
HIF-2α is increased following prolonged hypoxia. (A) HCT116 cells were incubated in normoxic (21% O2) or hypoxic (1% O2) condition for 24, 48, or 72 h, and viable cell number was determined using an automated cell counter. Values are means ± SD of three experiments, aP <0.05 compared with the normox group. (B) Cell migration and sphere formation were assessed in HCT116 cells following incubation in hypoxic condition for 24, 48, or 72 h. Data are expressed as means ± SD of three experiments. aP <0.05 compared with the 24 h hypoxic group. bP < 0.05 compared with the 48 h hypoxic group. (C) Protein levels of HIF-2α, KLF4, OCT4, and β-catenin were determined following 72 h hypoxic incubation of HCT116. (D) Protein levels of HIF-1α and HIF-2α were monitored in HCT116 cells following normoxic or hypoxic incubation for 24, 48, or 72 h. Data are expressed as means ± SD from three experiments. aP <0.05 compared with each normoxic control. bP < 0.05 compared with the 24 h hypoxic group. (E) Protein levels of HIF-1α and HIF-2α were monitored in HT29 cells following normoxic or hypoxic incubation for 24, 48, or 72 h.
Because HIFα plays an indispensable role in cancer adaptation under low-oxygen conditions, HIFα protein levels were estimated. While HIF-1α levels rapidly increased after 24 h of hypoxia, HIF-2α levels showed a maximal elevation after 72 h of hypoxic incubation (Fig. 1D). Similar results were obtained for HT29 colorectal cancer cells (Fig. 1E). This observation implies that HIF-2α elevation in chronic hypoxic condition may contribute to the development of CSC properties in colorectal cancers.
3.2. HIF-2α mediates CSC-like properties under chronic hypoxic condition
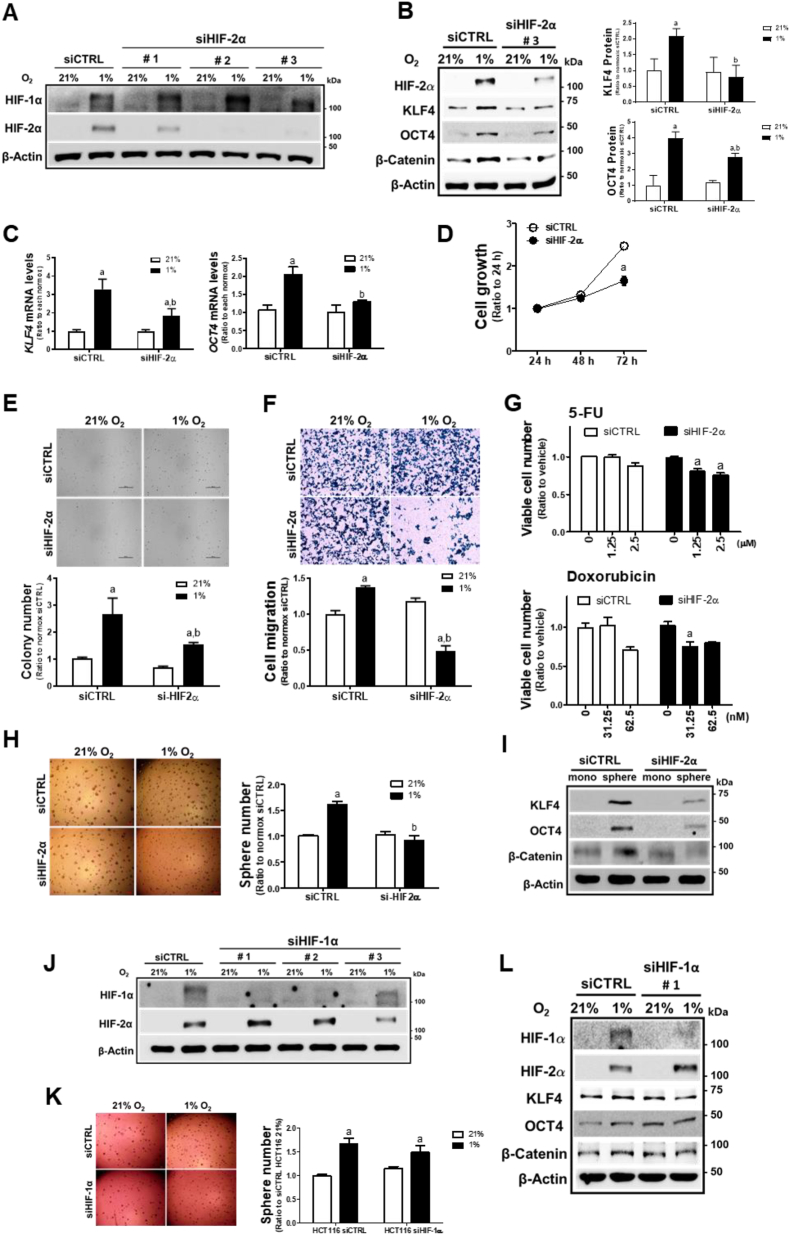
On the basis of the acquisition of stemness phenotypes and HIF-2α accumulation under chronic hypoxic conditions, we investigated the potential role of HIF-2α. For this, different types of HIF-2α-targeting siRNA (siHIF-2α #1, #2, and #3) were transfected in HCT116 cells and HIF-1α and HIF-2α levels were determined following hypoxic incubation. Both siHIF-2α #2 and #3 inhibited HIF-2α but not HIF-1α, which indicates a selective inhibition of HIF-2α by these siRNAs (Fig. 2A). Accordantly, we selected the siHIF-2α #3 for subsequent experiments. Transfection with siHIF-2α #3 blocked the elevation of the CSC markers KLF4, OCT4, and β-catenin after 72 h of hypoxia (Fig. 2B) and suppressed KLF4 and OCT4 mRNA levels (Fig. 2C). These results suggest a specific contribution of HIF-2α to hypoxia-induced CSC marker elevation. Next, we monitored the in vitro cell growth, anchorage-independent growth, migration, resistance to drug therapy, and sphere-forming ability of the 72 h hypoxia-exposed HCT116 to examine the role of HIF-2α in the development of CSC-like properties. Silencing HIF-2α suppressed hypoxia-exposed cell growth (Fig. 2D) and anchorage-independent colony formation (Fig. 2E). Transwell assays showed a substantial reduction in the migration of HIF-2α-silenced HCT116 cells (Fig. 2F). The WST assay indicated that HIF-2α-silenced HCT116 cells were relatively more sensitive to 5-FU- and doxorubicin-induced cytotoxicity under chronic hypoxic conditions (Fig. 2G). Moreover, the colonosphere number, which was increased in cells exposed to hypoxia for 72 h, was completely suppressed by HIF-2α-silencing (Fig. 2H), and the increase in KLF4, OCT4, and β-catenin in spheroid HCT116 was ablated by HIF-2α-silencing (Fig. 2I). Similar results were observed when the siHIF-2α #2 was transfected: levels of hypoxia-induced CSC markers, cell migration, and sphere formation were all suppressed by siHIF-2α #2 (Supplementary Fig. S1). Moreover, specific inhibition of HIF-2α suppressed hypoxia-inducible KLF4 and OCT4 elevation in additional cell line HT29 (Supplementary Fig. S2).
Fig. 2.
HIF-2α mediates CSC-like properties under prolonged hypoxia. (A) Protein levels of HIF-1α and HIF-2α were determined following transfection with HIF-2α-specific siRNAs (siHIF-2α #1, #2, #3) and nonspecific siRNA (siCTRL) in hypoxic HCT116 cells. (B) Protein levels of HIF-2α, KLF4, OCT4, and β-catenin were determined following prolonged hypoxic incubation for 72 h in siHIF-2α (#3) or siCTRL HCT116 cells. Values are means ± SD from three experiments. aP <0.05 compared with normoxic siCTRL. bP < 0.05 compared with the hypoxic siCTRL. (C)Transcript levels of KLF4 and OCT4 were measured in hypoxic siCTRL and siHIF-2α (#3) HCT116 cells (72 h) using RT-PCR analysis. Values are expressed as means ± SD of three experiments, aP <0.05 compared with each normoxic group and bP < 0.05 compared with the hypoxic siCTRL control. (D) Cell growth was assessed in siCTRL and siHIF-2α (#3) HCT116 cells following hypoxic incubation for 24, 48, or 72 h. Data are expressed as means ± SD of three experiments. aP <0.05 compared with the siCTRL control. (E) Soft agar colony formation was determined in siCTRL and siHIF-2α (#3) HCT116 cells following incubation under hypoxic conditions for 72 h, and the number of colonies was counted. Data are expressed as the means ± SD of three experiments. aP <0.05 compared with each normoxic group. bP < 0.05 compared with the hypoxic siCTRL control. (F) Transwell cell migration assay was performed using siCTRL and siHIF-2α (#3) following incubation under 72 h hypoxic condition. Data are expressed as the means ± SD of three experiments. aP <0.05 compared with each normoxic group. bP < 0.05 compared with the hypoxic siCTRL control. (G) Viability of the siCTRL and siHIF-2α (#3) cells was determined following incubation with 5-fluorouracil (5-FU, 1.25 and 2.5 μM) or doxorubicin (31.25 and 62.5 nM) for 72 h under hypoxia. Data are expressed as means ± SD of three experiments, aP <0.05 compared with each untreated group. (H) Sphere formation was assessed in siCTRL and siHIF-2α (#3) following incubation for 72 h of hypoxia, and sphere numbers were counted. Data are expressed as the means ± SD of three experiments. aP <0.05 compared with each normoxic group. bP < 0.05 compared with the hypoxic siCTRL control. (I) Protein levels of KLF4, OCT4, and β-catenin were determined in siCTRL and siHIF-2α spheres. (J) Protein levels of HIF-1α and HIF-2α were determined following transfection of HIF-1α-specific siRNAs (siHIF-1α #1, #2, #3) and nonspecific siCTRL in HCT116 cells. (K) Sphere formation capacity was assessed in the 72 h-hypoxic siHIF-1α (#1) HCT116 cells. Data are expressed as the means ± SD of three experiments. aP <0.05 compared each normox group. (L) HCT116 cells were transfected with HIF-1α-specific siRNA (#1) and the levels of HIF-1α, HIF-2α, KLF4, OCT4, and β-catenin were monitored following hypoxic incubation for 72 h.
In contrast, specific silencing of HIF-1α (Fig. 2J), which was achieved by siHIF-1α #1, did not show a noticeable effect on chronic hypoxia-induced sphere formation when compared to the normox control cells (Fig. 2K), and the inhibitory effect of HIF-1α silencing on the hypoxia-inducible CSC markers was only marginal (Fig. 2L). Together, these results show that HIF-2α mediates CSC-like properties of colorectal cancer cells following prolonged hypoxic incubation.
3.3. NRF2-silencing suppresses HIF-2α elevation under chronic hypoxic condition
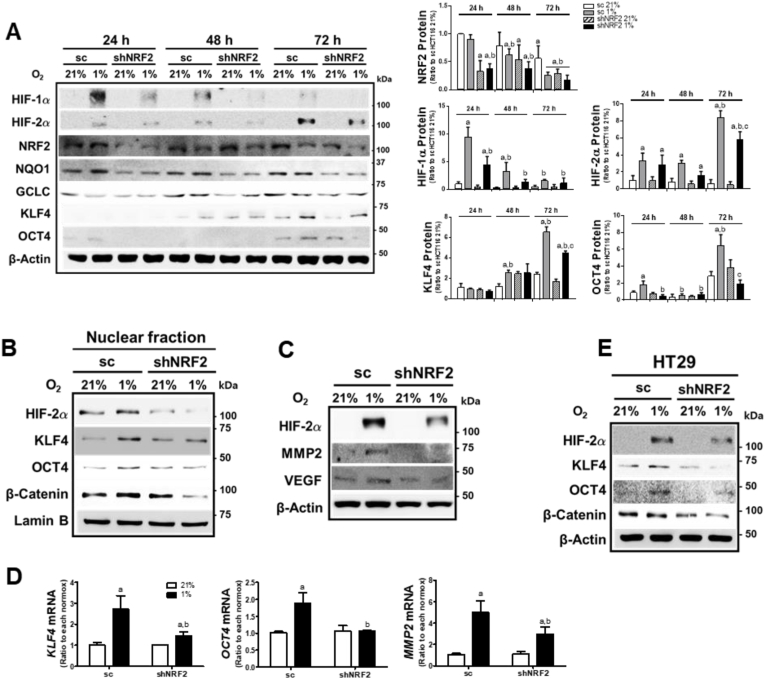
Previously, we observed that the inhibition of NRF2 expression in colorectal cancer cells abrogated HIF-1α accumulation under hypoxic conditions for 24 h [27,28]. In order to explore the potential role of NRF2 in HIF-2α-induced CSC-like properties in a chronic hypoxic environment, stable HCT cell lines with nonspecific shRNA (sc) or NRF2-specific shRNA (shNRF2) were incubated in 1% O2 for 24 h, 48 h, or 72 h. HIF-1α accumulation, which was at the maximal level after 24 h hypoxia, was significantly low in shNRF2 cells (Fig. 3A). HIF-2α accumulation was maximal at 72 h of hypoxia, and NRF2-silencing inhibited HIF-2α elevation. Similarly, KLF4 and OCT4 levels, which increased after 72 h of hypoxic incubation, were attenuated in shNRF2 cells. The levels of NRF2 and its target NQO1 and GCLC were not noticeably changed by chronic hypoxic incubation. Immunoblot analysis of nuclear levels of HIF-2α, KLF4, OCT4, and β-catenin showed similar effect of NRF2-silencing on at 72 h (Fig. 3B). In line with this, prolonged hypoxia-inducible levels of MMP2, a specific target of HIF-2α, and VEGF, a common target of HIF-1α and HIF-2α, were inhibited in NRF2-silenced HCT116 cells (Fig. 3C). These results were confirmed by measuring mRNA levels of MMP2, KLF4, and OCT4 (Fig. 3D). An additional colorectal cell line HT29 showed similar results: levels of HIF-2α and CSC markers diminished in NRF2-silenced HT29 cells after 72 h of hypoxia (Fig. 3E and Supplementary Fig. S3).
Fig. 3.
NRF2 silencing suppresses prolonged hypoxia-induced HIF-2α elevation. (A) The control (sc) and NRF2-silenced HCT116 (shNRF2) cells were incubated in hypoxic condition for 24, 48, or 72 h, and protein levels of HIF-1α, HIF-2α, NRF2, NQO1, GCLC, KLF4, and OCT4 were determined using immunoblot analysis. Values are expressed as means ± SD of three experiments. aP <0.05 compared with each normoxic control. bP < 0.05 compared with the 24 h hypoxic sc control. cP < 0.05 compared with the 72 h hypoxic sc control. (B) Nuclear levels of HIF-2α, KLF4, OCT4, and β-catenin were measured in the nuclear fractions of the sc and shNRF2 cells following 72 h hypoxic incubation. (C) Protein levels of HIF-2α, and MMP2 were determined in the sc and shNRF2 cells following hypoxic incubation for 72 h. (D) Transcript levels of KLF4, OCT4, and MMP2 were assessed in the sc and shNRF2 cells following 72 h hypoxic incubation. Data are expressed as means ± SD of three experiments. aP <0.05 compared with each normoxic group. bP < 0.05 compared with the hypoxic sc control. (E) HT29 cells were transfected with the nonspecific shRNA (sc) or NRF2-specific shRNA (shNRF2), and protein levels of HIF-2α, KLF4, OCT4, and β-catenin were assessed following 72 h hypoxic incubation.
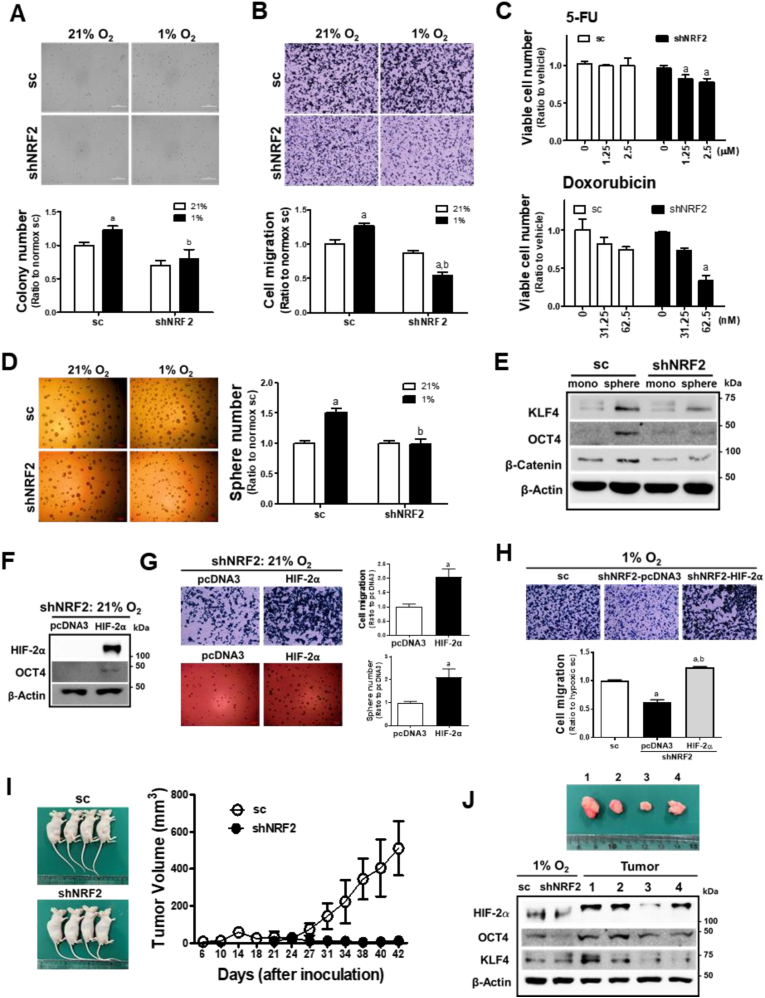
3.4. NRF2-silencing suppresses CSC-like properties in chronic hypoxic environment
To confirm the effect of HIF-2α inhibition in NRF2-silenced cancer cells, CSC-like phenotypes were examined in cells exposed to 72 h of hypoxia. Hypoxia-activated anchorage-independent colony growth (Fig. 4A) and migration (Fig. 4B) were significantly inhibited in NRF2-silenced HCT116 cells. The viability of shNRF2 cells was more suppressed following 5-FU and doxorubicin treatment than that of control cells (Fig. 4C). Chronic hypoxic-stimulated sphere formation was substantially attenuated in shNRF2 cells (Fig. 4D), and CSC markers, which were highly elevated in colonospheres, were diminished in the NRF2-silenced group (Fig. 4E). These results suggest that the development of chronic hypoxia-induced CSC-like properties is dependent on NRF2.
Fig. 4.
NRF2 mediates prolonged hypoxia-induced CSC-like properties. (A) Soft agar colony formation was assessed in the sc and shNRF2 HCT116 cells, which were incubated in hypoxic condition for 72 h, and number of colonies was counted. (B) Transwell migration assay was carried out in the sc and shNRF2 cells following incubation in hypoxic condition for 72 h. Data are expressed as means ± SD of three experiments. aP <0.05 compared with each normoxic control. bP < 0.05 compared with the hypoxic sc control. (C) Viable cell number was determined following incubation with 5-fluorouracil (5-FU) or doxorubicin under hypoxic condition. Data are expressed as means ± SD of three experiments. aP <0.05 compared with each treated sc control. (D) Sphere formation capacity assessed in the sc and shNRF2 cells following incubation in hypoxic condition for 72 h. Data are expressed as means ± SD of three experiments. aP <0.05 compared with each normoxic control. bP < 0.05 compared with the hypoxic sc control. (E) Protein levels of KLF4, OCT4, and β-catenin were determined in the sc and shNRF2 spheres. (F) Protein levels of HIF-2α and OCT4 were measured in normoxic shNRF2 HCT116 cells following transfection with HIF-2α overexpression plasmid (pcDNA3-HA–HIF–2α-P405A/P531A) and blank pcDNA3. (G) Cell migration and sphere formation were assessed in normoxic shNRF2 cells following transfection with HIF-2α overexpression plasmid and blank pcDNA3. Values are means ± SD of three experiments. aP <0.05 compared with pcDNA3 control. (H) Cell migration was determined in shNRF2 HCT116 cells following transfection with HIF-2α overexpression plasmid or blank pcDNA3 in chronic hypoxic condition (72 h). Values are means ± SD of three experiments. aP <0.05 compared with the sc control cells. bP < 0.05 compared with pcDNA3 control. (I) Tumor growth was monitored following the inoculation of the sc and shNRF2 HCT116 cells in BALBc (nu/nu) mice. (J) Levels of HIF-2α, OCT4, and KLF4 were determined in tumors from the sc control group. For the comparison, cell lysates from the sc and shNRF2 cells which were exposed to hypoxic condition for 72 h. Relative tumor size of the sc control groups is shown together.
In an attempt to verify the role of HIF-2α, the HIF-2α plasmid with proline residues mutation (P405A/P531A) was transfected. Overexpression of HIF-2α in normoxic condition promoted migration and sphere formation capacities of the shNRF2 HCT116 cells with concomitant increase in OCT4 (Fig. 4F and G). Moreover, hypoxia-induced shNRF2 migration, which was reduced when compared to the sc control, was restored by the forced expression of HIF-2α (Supplementary Fig. S4 and Fig. 4H).
Consistent with the in vitro results, shNRF2 cells did not develop tumors in the mouse xenograft study (Fig. 4I). In contrast, the sc control HCT116 developed tumors from 21st day after inoculation, and tumoral levels of HIF-2α, OCT4, and KLF4 were found to be high, which are comparable to those in hypoxic HCT116 cells (Fig. 4J).
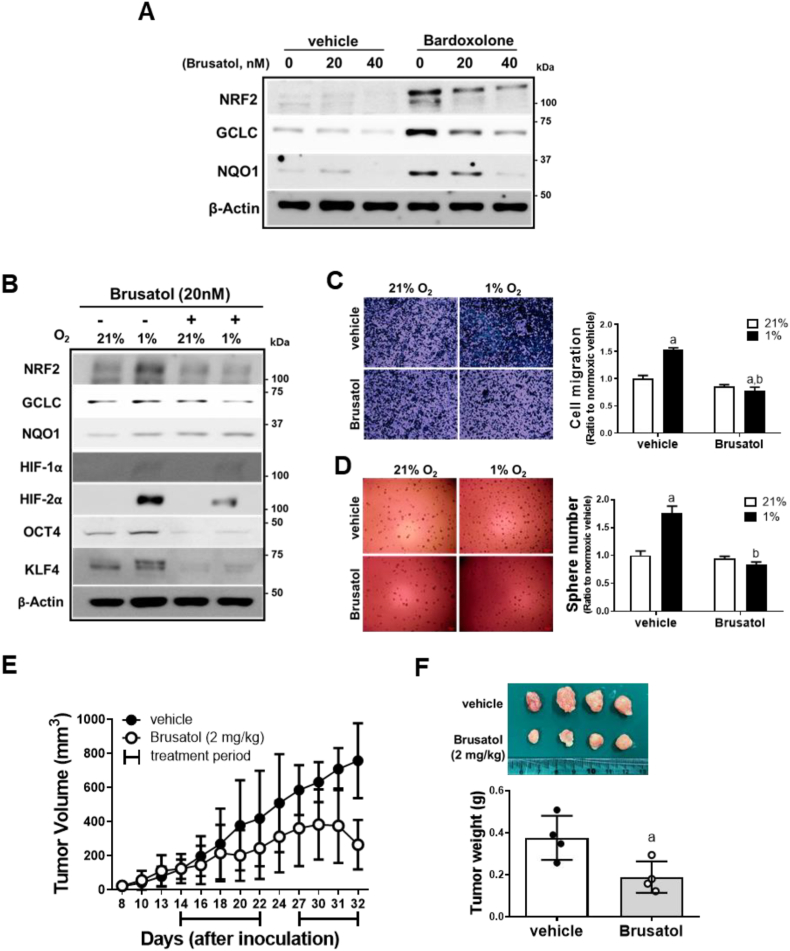
3.5. NRF2 inhibitor brusatol suppresses CSC-like properties in chronic hypoxic environment
Brusatol has been shown to inhibit NRF2 and thereby enhance chemotherapy efficacy in cancers [46]. In HCT116, brusatol (20 and 40 nM) incubation for 24 h effectively reduced the bardoxolone-induced NRF2 accumulation and target gene expression (Fig. 5A). Similarly to NRF2-silencing, brusatol treatment diminished HIF-2α levels and suppressed migration/sphere formation of HCT116 in 72 h-hypoxic condition (Fig. 5B–D). In vivo, brusatol administration (2 mg/kg) for 15 days significantly retarded tumor growth in HCT116-inoculated mice (Fig. 5E and F). These results confirm the effect of NRF2 inhibition on hypoxia-induced CSC-like properties.
Fig. 5.
Brusatol-mediated NRF2 inhibition represses HIF-2α induced-CSC-like properties. (A) Protein levels of NRF2, GCLC, and NQO1 were measured following brusatol (20 and 40 nM) or vehicle incubation (DMSO) for 24 h with or without bardoxolone (0.1 μM) treatment. (B) Levels of NRF2, GCLC, NQO1, HIF-1α, HIF-2α, OCT4, and KLF4 were monitored following brusatol (20 nM) incubation under 72 h hypoxic condition. (C–D) Cell migration and sphere formation were assessed in brusatol or vehicle-incubated HCT116 cells under 72 h hypoxic condition. Values are expressed as means ± SD of three experiments. aP <0.05 compared with normoxic vehicle. bP < 0.05 compared with hypoxic vehicle control. (E) Tumor growth was assessed following brusatol treatment (2 mg/kg) in BALB/C (nu/nu) mice inoculated with HCT116 cells. The treatment was started once the tumor volumes reached ±80 mm3. Brusatol was administered for 15 days, excluding a 5-day break. (F) Tumors were collected and weighed at the end of brusatol administration. Values are means ± SD of four animals. aP <0.05 compared with vehicle group.
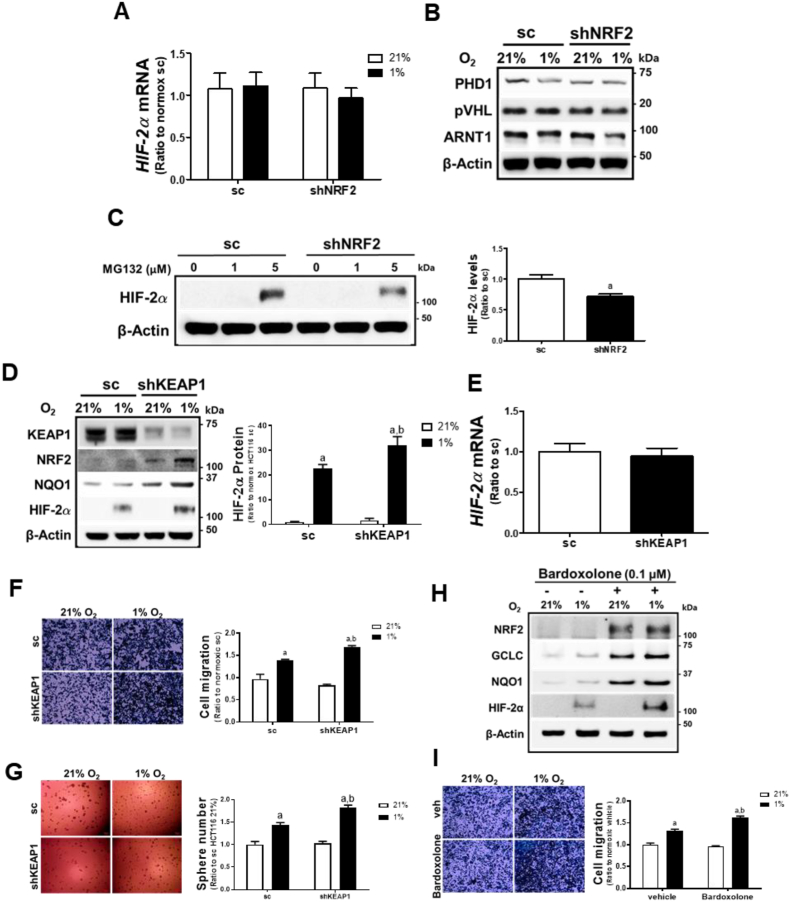
3.6. NRF2 level- is positively correlated with HIF-2α level
To elucidate the mechanism of HIF-2α inhibition in hypoxic NRF2-silenced cancer cells, we first monitored mRNA levels of HIF-2α. The shNRF2 cells did not show a noticeable difference in HIF-2α mRNA levels compared to the sc control, and no changes were observed after 72 h of hypoxia (Fig. 6A), which indicates that reduced HIF-2α transcription is not a cause of HIF-2α inhibition. Additionally, the levels of PHD1, pVHL, and ARNT1 were not altered by NRF2-silencing (Fig. 6B). Measurement of PHD1, PHD2, and PHD3 mRNA levels also showed that differential PHDs expression is not associated with HIF-2α inhibition (Supplementary Fig. S5A). Of note, when proteasome-mediated HIF-2α degradation was blocked by MG132 incubation, HIF-2α levels were still attenuated in NRF2-silenced cells (Fig. 6C).
Fig. 6.
NRF2 level is positively associated with HIF-2α level-. (A) Transcript levels of HIF-2α were measured in the sc and shNRF2 HCT116 cells following 72 h hypoxic incubation. (B) Levels of PHD1, pVHL, and ARNT1 were determined in the hypoxic sc and shNRF2 cells using immunoblot analysis. (C) Protein levels of HIF-2α were assessed following incubation of the sc and shNRF2 HCT116 cells with MG132 (1 and 5 μM). Values are means ± SD of three experiments. aP <0.05 compared with the sc control. (D) Protein levels of KEAP1, NRF2, NQO1, and HIF-2α were monitored in KEAP1-silenced HCT116 cells (shKEAP1) following 72 h hypoxic incubation. HIF-2α protein levels are means ± SD of three experiments. aP <0.05 compared with normoxic sc. bP < 0.05 compared with hypoxic sc control. (E) Transcript levels of HIF-2α were measured in the sc and shKEAP1 HCT116 cells using RT-PCR analysis. (F–G) Cell migration and sphere formation were assessed in sc and shKEAP1 cells following 72 h hypoxia. Values are means ± SD of three experiments. aP <0.05 compared with normoxic sc. bP < 0.05 compared with hypoxic sc control. (H) Protein levels of NRF2, GCLC, NQO1, and HIF-2α were determined after pre-treatment of the sc control cells with bardoxolone (0.1 μM) or vehicle (DMSO) for 24 h followed by hypoxic incubation (72 h). (I) Migration of bardoxolone-treated sc control was determined following hypoxic incubation for 72 h. Data are expressed as means ± SD of three experiments. aP <0.05 compared with normoxic vehicle. bP < 0.05 compared with hypoxic vehicle control.
We also examined the NRF2-dependent HIF-2α regulation using KEAP1-silencing and bardoxolone treatment. NRF2 and its target NQO1 were elevated in KEAP1-silenced HCT116 cells, and chronic hypoxia (72 h)-induced HIF-2α accumulation was further enhanced compared to the sc control (Fig. 6D), although HIF-2α mRNA levels did not show any differences (Fig. 6E). KEAP1-silencing enhanced migration and sphere formation capacities in chronic hypoxic condition (Fig. 6F and G). When shKEAP1 cells were incubated with MG132, the HIF-2α level was higher than that in control cells (Supplementary Fig. S5B). Similarly, bardoxolone treatment increased NRF2, GCLC, and NQO1 levels, and elevated HIF-2α accumulation following a 72 h-hypoxic incubation (Fig. 6H). Chronic hypoxia-stimulated HCT116 migration was also enhanced following bardoxolone treatment (Fig. 6I). Taken together, these results suggest that NRF2 levels are positively correlated with chronic hypoxia-induced HIF-2α levels and that NRF2 appears to contribute to HIF-2α regulation during the translational process.
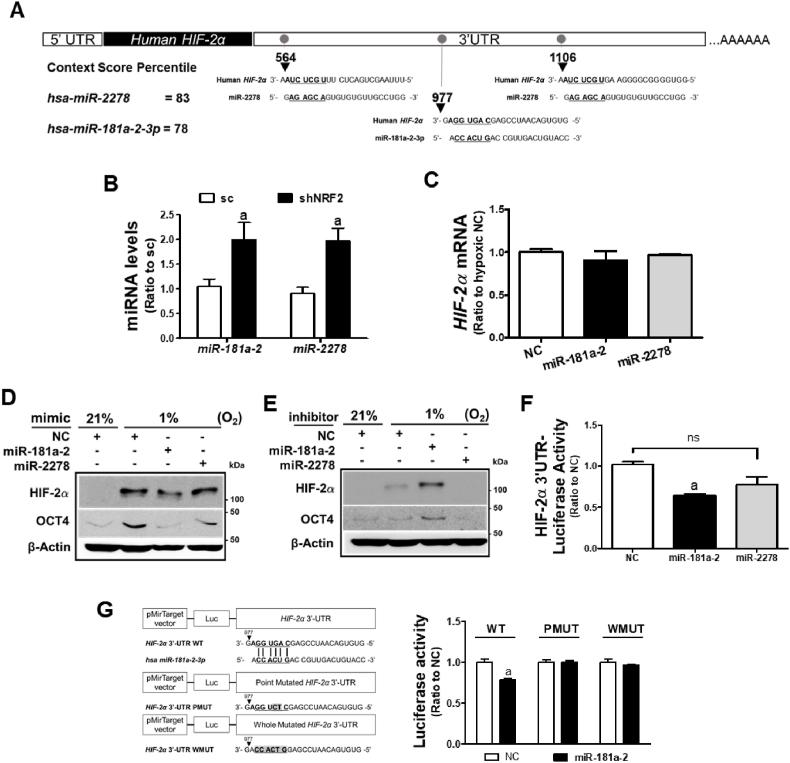
3.7. NRF2-silencing-induced miR-181a-2-3p inhibits HIF-2α elevation
Since it was observed that regulation of transcription and protein stability were not involved in HIF-2α inhibition following NRF2-silencing, we explored the potential association of miRNAs with this change. For this, we selected common miRNAs that were upregulated more than 1.5-fold by NRF2-silecing both in HCT116 and HT29 cells from our prior microarray analysis [39], and searched for miRNAs predictably targeting HIF-2α 3′-untranslated region (UTR) with more than 75% context percentile rank using the TargetScan database (Table 2). Among the 23 miRNAs, miR-181a-2-3p (miR-181a-2) and miR-2278-5p (miR-2278) were found to target the HIF-2α 3′-UTR with context score percentiles of 78% and 83%, respectively (Table 2 and Fig. 7A).
Table 2.
Common microRNAs, which are upregulated (>1.5 fold) both in HCT116 and HT29, following a stable silencing of NRF2. Hypoxia-related target genes were predicted using TargetScan database. n.d., not determined.
| miRNAs | shNRF2/sc > 1.5-fold |
Hypoxia-related target gene prediction | |
|---|---|---|---|
| HCT116 | HT29 | ||
| hp_hsa-mir-1908-5p | 1.505 | 1.511 | HIF1A |
| hsa-miR-181a-2-3p | 1.505 | 1.949 | HIF2A |
| hsa-miR-4787-5p | 1.511 | 1.945 | n.d. |
| hsa-miR-200a-3p | 1.568 | 1.702 | n.d. |
| hp_hsa-mir-1302-10-3p | 1.572 | 1.613 | n.d. |
| hsa-miR-3122-5p | 1.588 | 1.721 | HIF3A |
| hsa-miR-181c-5p | 1.590 | 2.013 | ARNT2 |
| hsa-miR-4466-5p | 1.598 | 1.822 | n.d. |
| hsa-miR-2392-5p | 1.611 | 1.667 | n.d. |
| hsa-miR-3152-3p | 1.638 | 1.868 | HIF1A |
| hp_hsa-mir-3960-5p | 1.640 | 1.604 | n.d. |
| hsa-miR-4763-3p | 1.643 | 1.531 | HIF3A, ARNT2 |
| hp_hsa-mir-548q-5p | 1.645 | 2.112 | HIF3A, ARNT2 |
| hp_hsa-mir-124-2-3p | 1.659 | 1.708 | ARNT1, ARNT2 |
| hsa-miR-3201-5p | 1.674 | 2.754 | HIF1A |
| hsa-miR-146a-5p | 1.898 | 1.713 | HIF1A, HIF3A |
| hsa-miR-93-3p | 1.953 | 1.852 | HIF1A, ARNT1, ARNT2 |
| hsa-miR-2278-5p | 2.034 | 1.615 | HIF2A |
| hsa-miR-4720-5p | 2.131 | 1.990 | HIF1A, ARNT2 |
| hsa-miR-4490-5p | 2.209 | 3.118 | HIF1A, HIF3A, ARNT2 |
| hsa-miR-3921-5p | 2.499 | 2.366 | n.d. |
| hsa-miR-4532-5p | 2.582 | 1.713 | n.d. |
| hsa-miR-3188-5p | 4.568 | 1.938 | HIF1A, HIF3A, ARNT2 |
Fig. 7.
Involvement of miR-181a-2-3p in HIF-2α regulation in NRF2-silenced cancer cells. (A) Schematic diagram of the putative binding sites of miR-181a-2-3p (miR-181a-2) and miR-2278-5p (miR-2278) in the 3′-UTR of the human HIF-2α. (B) Levels of miR-181a-2 and miR-2278 were determined in the sc and shNRF2 HCT116 cells. Data are expressed as means ± SD of three experiments. aP <0.05 compared with the sc control. (C) HIF-2α mRNA levels were evaluated following the transfection of HCT116 cells with miR-181a-2 and miR-2278. (D) HIF-2α and OCT4 levels were determined following transfection of the control HCT116 cells with miR-181a-2 or miR-2278 under prolonged hypoxia. (E) HIF-2α and OCT4 levels were monitored following transfection of the shNRF2 cells with miR-181a-2 inhibitor or miR-2278 inhibitor under prolonged hypoxia. (F) HIF-2α 3′UTR-derived luciferase activities were measured in HCT116 following the transfection with miR-181a-2 or miR-2278. Data are expressed as means ± SD of three experiments. aP <0.05 compared with the negative control miRNA (NC). (G) The putative binding site of miR-181a-2 was mutated at the specific point sequence (HIF-2α 3′UTR PMUT) or at the whole sequence (HIF-2α 3′UTR WMUT) in the 3′UTR of HIF-2α gene. Luciferase activities from the wild-type, the PMUT-, or WMUT-HIF-2α 3′UTR were measured following miR-181a-2. Data are expressed as means ± SD of three experiments. aP <0.05 compared with the negative control miRNA group (NC).
Consistent with the microarray results, qPCR analysis showed that miR-181a-2 and miR-2278 levels increased 2-fold in shNRF2 cells compared to the control (Fig. 7B). Transfection of HCT116 cells with miR-181a-2 and miR-2278 mimic did not affect HIF-2α mRNA levels (Fig. 7C). When these two miRNAs were transfected into the sc cells and further incubated under hypoxic conditions for 72 h, miR-181a-2 reduced HIF-2α and OCT4 elevations, but not miR-2278 (Fig. 7D). Conversely, transfection of shNRF2 cells with inhibitors of two miRNAs, only miR-181a-2 enhanced hypoxia-induced HIF-2α and OCT4 levels (Fig. 7E). In line with these results, transfection of HCT116 cells with miR-181a-2 repressed luciferase activity derived from the 3-UTR of HIF-2α, confirming the inhibitory function of miR-181a-2 (Fig. 7F). Moreover, the specific binding of miR-181a-2 to the HIF-2α 3′UTR was confirmed by mutation analysis. When the putative binding site of miR-181a-2 was mutated in the HIF-2α 3′-UTR, miR-181a-2 lost its inhibitory effect on HIF-2α 3′-UTR-derived luciferase activity (Fig. 7G). These results suggest that miR-181a-2, which is elevated in NRF2-silenced colorectal cancer cells, targets the HIF-2α gene.
3.8. NRF2-silencing-induced miR-181a-2-3p suppresses hypoxia-induced CSC-like properties
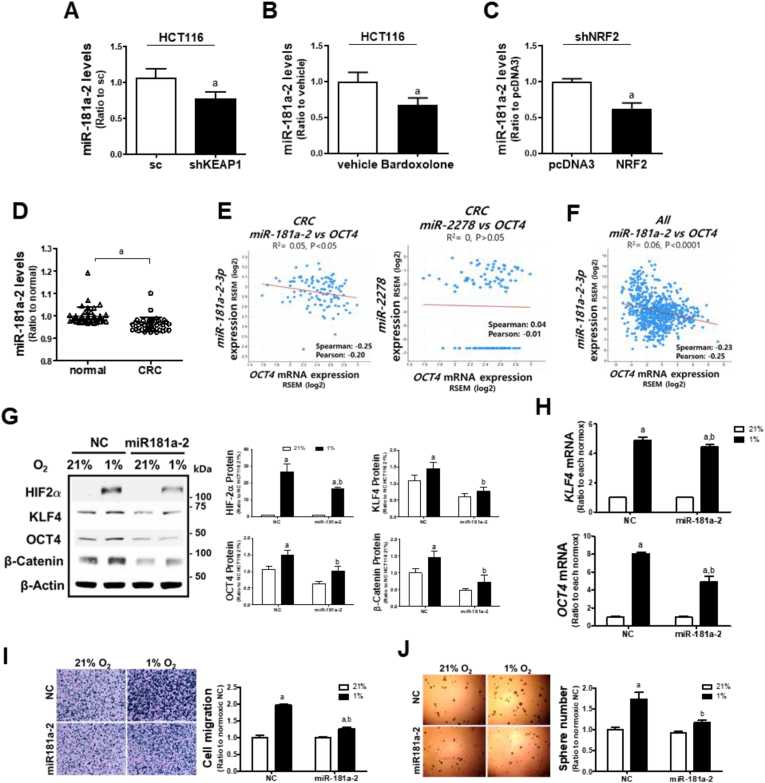
In line with NRF2-silencing effect, the negative correlation between NRF2 and miR-181a-3 levels could be confirmed in additional experiments. Levels of miR-181a-2 were lower in KEAP1-silenced HCT116 cells (Fig. 8A), bardoxolone-treated HCT116 (Fig. 8B), and NRF2-overexpressed shNRF2 HCT116 cells (Fig. 8C).
Fig. 8.
miR-181a-2-3p suppresses chronic hypoxia-induced CSC properties. (A) miR-181a-2 levels were determined in the sc and KEAP-silenced HCT116 (shKEAP1). (B) Levels of miR-181a-2 were measured following bardoxolone (0.1 μM) treatment. (JC) miR-181a-2 levels were assessed in shNRF2 cells following transfection with NRF2 overexpression plasmid. Data are expressed as means ± SD of three experiments. aP <0.05 compared with each control. (D) Relative miR-181a-2 levels in normal and colorectal cancer (CRC) samples were analyzed from GSE49246 public datasets (n = 40). aP <0.05 compared with the normal sample datasets. (E) Clinical correlation between OCT4 mRNA and miR-181a-2 or miR-2278 were evaluated using colorectal cancer (CRC) dataset (n = 105) from the Clinical Proteomic Tumor Analysis Consortium (CPTAC). (F) Clinical correlation between OCT4 mRNA and miR-181a-2 was analyzed using pan-cancer analysis of whole genomes dataset (n = 749) from International Cancer Genome Consortium and The Cancer Genome Atlas (ICGC/TCGA). (G) Protein levels of HIF-2α, KLF4, OCT4, and β-catenin were measured following the transfection of HCT116 with miR-181a-2 and prolonged hypoxic incubation. (H) Transcript levels of KLF4 and OCT4 were assessed in HCT116 cells after the transfection of miR-181a-2 and hypoxic incubation. Data are expressed as means ± SD of three experiments. aP <0.05 compared with each normoxic control, bP < 0.05 compared with the hypoxic NC control. (I–J) Cell migration and sphere formation capacities were evaluated following the transfection of HCT116 with miR-181a-2 and prolonged hypoxic incubation. Data are expressed as means ± SD of three experiments. aP <0.05 compared with each normoxic group, bP < 0.05 compared with the hypoxic NC control.
The association of miR-181a-3 with cancer HIF-2α could be further confirmed using clinical datasets. miR-181a-2 levels were significantly lower in colorectal cancer tissues than in normal tissues (Fig. 8D), and there were negative correlations between miR-181a-2 levels and HIF-2α target OCT4 levels in colorectal cancers (Fig. 8E) and total cancers (Fig. 8F). In the case of miR-2278, no correlation was observed between miR-2278 levels and OCT4 (Fig. 8E), and its expression in colorectal cancers did not show a statistical difference when compared to normal tissues (data not shown).
Next, to verify the effect of miR-181a-2 on HIF-2α-mediated CSC-like properties, we transfected control cells with miR-181a-2 and monitored the CSC marker levels after 72 h of hypoxia. As expected, miR-181a-2 diminished hypoxia-inducible HIF-2α, KLF4, OCT, and β-catenin protein levels (Fig. 8G) and attenuated hypoxia-inducible KLF4 and OCT4 mRNA levels (Fig. 8H). Consistent with the repressed CSC markers, miR-181a-2 inhibited chronic hypoxia-stimulated cell migration (Fig. 8I) and sphere formation (Fig. 8J). Taken together, these results suggest that miR-181a-2, which is negatively correlated with NRF2 level, suppresses CSC-like properties in chronic hypoxic condition.
3.9. miR-181a-2-mediated HIF-2a inhibition suppresses CSC-like properties in chronic hypoxic condition
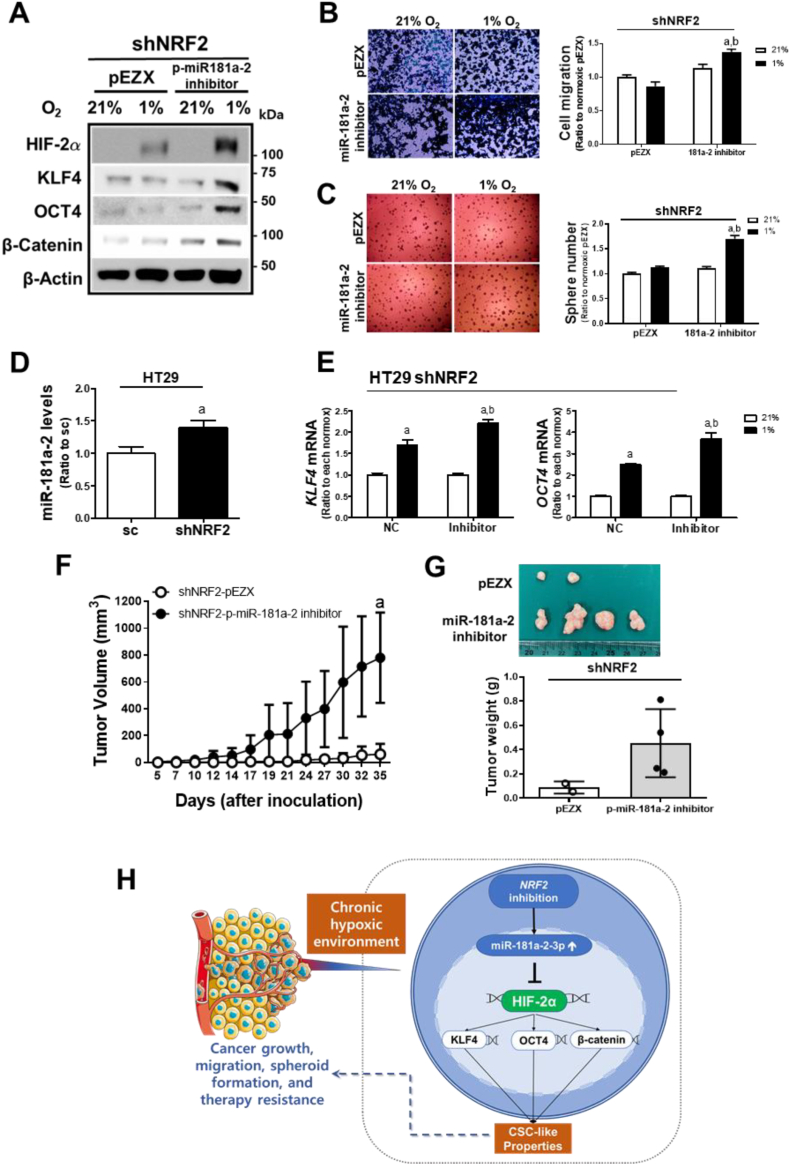
In an attempt to verify the role of miR-181a-2 in CSC properties, effect of the miR-181a-2 inhibitor was examined. Transfection of NRF2-silenced HCT116 cells with the miR-181a-2 inhibitor restored chronic hypoxia-inducible HIF-2α and CSC marker levels (Fig. 9A). Migration and sphere formation of the shNRF2 cells were also increased by the miR-181a-2 inhibitor (Fig. 9B and C). These results were confirmed using an additional colorectal HT29 cell line. Stable silencing of NRF2 in HT29 cells increased miR-181a-2 levels (Fig. 9D), and treatment of NRF2-silenced HT29 with the miR-181a-2 inhibitor elevated hypoxia-inducible KLF4 and OCT4 expression (Fig. 9E). As in vivo evidence, NRF2-silenced HCT116 with the miR-181a-2 inhibitor transfection showed a substantially increased tumor growth when compared to the blank plasmid-transfected shNRF2 cells (Fig. 9F and G). Collectively, our results demonstrate that the effect of NRF2-silencing on CSC-like properties in a chronic hypoxic environment is mediated through the elevation of miR-181a-2.
Fig. 9.
miR-181a-2-3p mediates the suppressive effect of NRF2- silencing on hypoxia-induced CSC properties. (A) Protein levels of HIF-2α, KLF4, OCT4, and β-catenin were measured following the transfection of shNRF2 HCT116 with pEZX-AM04-miR-181a-2-3p inhibitor (p-miR-181a-2 inhibitor) or control pEZX plasmid under 72 h hypoxic incubation. (B–C) Transwell migration assay and sphere formation were performed in shNRF2 cells transfected with p-miR-181a-2 inhibitor or the pEZX control plasmid. Values are means ± SD of three experiments, aP <0.05 compared with normoxic pEZX control, bP < 0.05 compared with the hypoxic pEZX control. (D) Levels of miR-181a-2 were monitored in NRF2-silenced HT29 (shNRF2 HT29) cells. Data are expressed as means ± SD of three experiments. aP <0.05 compared with the sc control. (E) Transcript levels of KLF4 and OCT4 were measured in the shNRF2 HT29 cells following the miR-181a-2 inhibitor transfection. Data are expressed as means ± SD of three experiments, aP <0.05 compared with each normoxic control, bP < 0.05 compared with the hypoxic NC control. (F) Tumor volumes were measured following inoculation of the p-miR-181a-2 inhibitor-transfected shNRF2 HCT116 cells in BALB/C(nu/nu) mice. (G) Tumors were collected and weighed. Tumors were only grown in two mice in the shNRF2 group. Values are means ± SD of four animals. aP <0.05 compared with the pEZX control group. (H) A schematic diagram of the effect of NRF2 levels on the prolonged hypoxia-induced HIF-2α activation and CSC-like properties.
4. Discussion
HIFs signaling leads to the immediate adaptive responses of cancer cells to a low oxygen environment and promotes cancer survival and growth by inhibiting apoptosis, inducing metabolic conversion, and acquiring drug resistance and metastatic potential, which eventually results in poor clinical outcomes [16]. In particular, stabilization of HIFs has been considered a prominent mechanism for regulating the genes that contribute to the maintenance of stemness properties and development of aggressive cancer phenotypes [13]. In neuroblastoma and small-cell lung carcinoma, hypoxia was demonstrated to maintain a highly tumorigenic stem cell population, which showed the capability of migration and invasion in in vitro and in vivo experimental models [47]. Hypoxic exposure of breast cancer cells induced HIFs-dependent demethylation of NANOG mRNA and further increased NANOG activity to enhance tumor initiation capacity [48]. For the relative contribution of HIFs to the development of CSC phenotypes, specific role of HIF-2α has been demonstrated in multiple studies. HIF-2α levels were differentially increased in glioma stem cells when compared to non-stem glioma cells, and HIF-2α-silencing selectively suppressed VEGF expression only in glioma stem cells. Whereas, HIF-1α was found to contribute to VEGF expression in both glioma stem and non-stem cells [21]. Hypoxia promotes stem-like properties in non-stem glioblastoma cells with a concomitant increase in HIF-2α, and overexpression of HIF-2α enhances CSC markers and tumorigenic properties of non-stem glioblastoma cells [49]. Intestine-specific deletion of VHL constitutively upregulated HIFs, thereby increasing colon tumorigenesis in ApcMin/+ mice. This effect was attenuated following the double deletion of VHL and HIF-2α, indicating the critical role of HIF-2α in hypoxia-induced tumorigenic signaling [50]. Consistent with these reports, we observed an association between HIF-2α and CSC phenotype development in colorectal cancer cells. Under a 1% O2 hypoxic environment, the HIFs proteins showed differential accumulation patterns. While HIF-1α rapidly increased under these conditions, HIF-2α protein elevation was maximal after 72 h of hypoxia. In addition, CSC markers and sphere-forming capacity were enhanced in a chronic hypoxic environment (72 h), and these CSC phenotypes were all suppressed by HIF-2α-silencing, but not by HIF-1α-silencing, which suggests the critical contribution of HIF-2α to the development of CSC phenotypes under chronic hypoxic conditions.
Regardless of its protective role in normal healthy cells, NRF2 signaling is often activated in many types of cancer and plays a significant role in cancer survival and progression. Dysregulation of cancer NRF2 signaling seems to be an adaptive strategy to maintain homeostasis under stressful tumor environments and involves various molecular events. Frequent somatic mutations have been found in the binding regions of KEAP1 and NRF2, and these changes lead to constitutive NRF2 activation and malignant properties [51,52]. KEAP1 gene methylation or NRF2 exon 2 deletion has also been reported in lung and colorectal cancers, and resultant NRF2 overactivation is associated with poor prognosis in patients [53,54]. Phosphorylated p62/SQSTM1, an autophagy protein adaptor, can increase NRF2 activity by competing with NRF2 for KEAP1 binding; thus, its accumulation in tumor cells leads to hepatocellular carcinoma malignancies [55]. The key role of NRF in cancer malignancy is further supported by recent reports showing an association between NRF2 signaling and CSC phenotypes [56]. Upregulation of NRF2 has been observed in breast cancer spheroids and this elevation contributed to spheroid growth and survival, and chemotherapy resistance [32,57]. Aldehyde dehydrogenase (ALDH)-positive breast CSC population, which shows high NRF2 levels, is responsible for the radioresistance of breast tumors [58]. NRF2 activation in breast cancer spheroids activates FoxO3a/Bim-1 signaling, which promotes the self-renewal capacity of breast CSCs [59]. In contrast, silencing of NRF2 or pharmacological NRF2 inhibition sensitized breast CSCs to radiotherapy by enhancing ROS and suppressing DNA repair [60]. Similar to these reports, we observed that NRF2 signaling is activated via p62 and GSK-3β signaling in CD44-positive breast cancer, CD133-positive colorectal cancer, and ALDH-high ovarian cancer cells, and NRF2-sileing suppressed CSC phenotypes such as migration, spheroid formation, drug resistance, and tumor growth [[29], [30], [31]]. In addition to the above reports, our current study provides evidence that NRF2 is involved in the development of CSC properties acquired by cancer cells under hypoxic conditions. Xenograft tumors from control HCT116 cells showed high levels of HIF-2α, OCT4, and KLF4, which implies the activation of hypoxia-induced HIF-2α signaling; however, NRF2-silenced cells failed to develop tumors (Fig. 4I and J). The treatment with brusatol, an inhibitor of NRF2, suppressed hypoxia-inducible HIF-2α accumulation, CSC marker elevation, and migration/sphere formation in hypoxic HCT116 cells, and inhibited HCT116 xenograft tumors (Fig. 5). Conversely, chronic hypoxia-induced HIF-2α accumulation and CSC properties were enhanced by NRF2 activation that was achieved by KEAP1-silencing and bardoxolone treatment (Fig. 6D–I). In particular, we showed that NRF2 inhibition can suppress chronic hypoxia-induced CSC phenotypes by elevating miR-181a-2, a miRNA targeting HIF-2α. Introduction of miR-181a-2 into colorectal cells diminished cell proliferation, anchorage-independent colony growth, sphere formation, cell migration, and drug resistance. In contrast, miR-181a-2 inhibition in NRF2-silenced cells restored HIF-2α levels and CSC marker expression, and facilitated cancer migration/sphere formation and tumor growth (Fig. 9).
Although a direct link between NRF2 and HIF-2α activation has not been reported, compelling evidence has been shown for NRF2 and HIF-1α. Continuous hypoxia increased HIF-1α without NRF2 elevation in lung cancer cells, whereas intermittent hypoxia increased both HIF-1α and NRF2, and ROS-mediated NRF2/TRX1 elevation augmented HIF-1α signaling [61]. The PKR-like ER kinase (PERK)-mediated NRF2 phosphorylation is critical for ROS removal, cell proliferation, and radioprotection in hypoxic pancreatic and lung cancers, and NRF2-knockdown led to HIF-1α suppression and subsequent cancer inhibition [62]. Molecular events between NRF2 and HIF-1α have been suggested by several groups. First, the direct regulation of HIF-1α by NRF2 has been suggested by identifying the functional ARE present 30 kb upstream of the HIF-1α gene [63]. Second, a role for NRF2 target proteins has been suggested. NQO1 was found to directly bind to HIF-1α protein and thereby PDH-mediated proteasomal degradation, resulting in HIF-1α stabilization in human cancers overexpressing NQO1 [64]. Third, in our previous study, the relationship between NRF2 and HIF-1α was explained by miR-181c: NRF2-silencing increased miR-181c levels, which subsequently targeted the mitochondria-encoded cytochrome C oxidase subunit-1 and consequently reduced mitochondrial oxygen consumption, leading to the inhibition of the 24 h-hypoxia-induced HIF-1α accumulation in colorectal and breast cancer cells [27,28].
MiRNAs, which play divergent roles in cancers as oncogenes or tumor suppressors, are often dysregulated in human cancers; therefore, miRNAs are considered as promising targets for developing cancer biomarkers and therapeutics [65]. Several miRNAs have been identified to target HIFs. MiR-200c, an miR-200 family member that regulates epithelial-mesenchymal transition, targets HIF-1α, thereby inhibiting the migration of lung cancer cells [66]. Overexpression of miR-519d-3p diminished HIF-2α levels by binding to its 3′-UTR region, thereby suppressing cell proliferation in hypoxic cervical cancer cells [67]. Recently, miR-526b-3p was demonstrated to inhibit HIF-2α/Notch signaling and consequently suppress the sphere-forming capacity and CSC marker expression [68]. Our results demonstrate a novel role of miR-181a-2 in NRF2-associated HIF-2α suppression and CSC phenotype inhibition. It was shown that miR-181a-2 directly binds to the 3′-UTR of the HIF-2α gene, and alterations in the binding sequences abrogated this binding, confirming the specific binding of this miRNA to HIF-2α. MiR-181a-2-3p, together with miR-181b-2, is located on chromosome 9q33.3, while other miR-181 family members, including miR-181c and miR-181a-1, are located on chromosomes 1p32.1 and 19p13.12 [69]. Although the specific function of miR-181a-2-3p in cancer remains unclear, our analyses of public datasets and clinical studies suggest that miR-181a-2 level is low in colorectal cancers. Similarly, miR-181a-2 levels were found to be low in 46% of cervical cancer samples due to the loss of heterozygosity of chromosome 9q33.3 and promoter hypermethylation [70]. The introduction of miR-181a-2 in cervical cancer attenuates tumorigenesis by targeting the EGF/SOX2 pathway with a concomitant decrease in the CSC population [71]. In agreement with our observations, this result suggests an inhibitory role of miR-181a-2 in CSC phenotypes. Moreover, our analysis of clinical data sets confirmed the negative relationship between miR-181a-2 levels and the CSC marker OCT4 levels in colorectal and total cancer samples.
The molecular mechanism of miR-181a-2 increase by NRF2-silencing remains to be clarified in future studies. Since we observed that miR-181a-2 levels were diminished by KEAP1-silencing and bardoxolone treatment (Fig. 8A and B), the link between NRF2 and miR-181a-2 seems to be negative. Considering reports showing a repressive role of NRF2 in gene transcription [72,73], NRF2-mediated direct regulation might be a possible explanation. In addition, several transcription factors, including SMADs, have been reported to directly regulate miR-181a-2-3p expression [74]. As lines of evidence indicate that NRF2 activation suppresses the TGF-β/SMAD3 pathway, which is eventually associated with tissue fibrosis inhibition [75], NRF2 might hypothetically downregulate miR-181a-2 through the suppression of SMAD3/4 signaling.
Collectively, under chronic hypoxic conditions, NRF2 inhibition can suppress HIF-2α-induced CSC phenotypes, including enhanced colony formation, migration, sphere formation, drug resistance, and tumor growth, by elevating the HIF-2α-3′UTR targeting miR-181a-2 (Fig. 9H). These results further suggest that the NRF2/miR-181a-2 pathway may be a novel therapeutic target to prevent chronic hypoxia-induced CSC properties.
Funding
This work was supported by National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIT, Republic of Korea) (2022R1A2C2011866, 2018R1A2A1A05078894, and 2018R1A6A1A03025108). This study was also supported by the Research Fund, 2021 of The Catholic University of Korea (Republic of Korea).
Declaration of competing interest
M − K Kwak has received research grants from National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIT) and The Catholic University of Korea.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102632.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Prager B.C., Xie Q., Bao S., Rich J.N. Cancer stem cells: the architects of the tumor ecosystem. Cell Stem Cell. 2019;24(1):41–53. doi: 10.1016/j.stem.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M.A., Dick J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet D., Dick J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997;3(7):730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien C.A., Pollett A., Gallinger S., Dick J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 5.Batlle E., Clevers H. Cancer stem cells revisited. Nat. Med. 2017;23(10):1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 6.Lytle N.K., Barber A.G., Reya T. Stem cell fate in cancer growth, progression and therapy resistance. Nat. Rev. Cancer. 2018;18(11):669–680. doi: 10.1038/s41568-018-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glumac P.M., LeBeau A.M. The role of CD133 in cancer: a concise review. Clin. Transl. Med. 2018;7(1):e18. doi: 10.1186/s40169-018-0198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu F., Li J., Chen H., Fu J., Ray S., Huang S., Zheng H., Ai W. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30(18):2161–2172. doi: 10.1038/onc.2010.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Hajj M., Wicha Max S., Benito-Hernandez A., Morrison Sean J., Clarke Michael F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S.M., Liu S., Lu H., Zhang H., Zhang P.J., Gimotty P.A., Guerra M., Guo W., Xu X. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene. 2012;31(47):4898–4911. doi: 10.1038/onc.2011.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munro M.J., Wickremesekera S.K., Peng L., Marsh R.W., Itinteang T., Tan S.T. Cancer stem cell subpopulations in primary colon adenocarcinoma. PLoS One. 2019;14(9) doi: 10.1371/journal.pone.0221963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L., Shi P., Zhao G., Xu J., Peng W., Zhang J., Zhang G., Wang X., Dong Z., Chen F., Cui H. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Targeted Ther. 2020;5(1):8. doi: 10.1038/s41392-020-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semenza G.L. The hypoxic tumor microenvironment: a driving force for breast cancer progression. Biochim. Biophys. Acta Mol. Cell Res. 2016;1863(3):382–391. doi: 10.1016/j.bbamcr.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heddleston J.M., Li Z., Lathia J.D., Bao S., Hjelmeland A.B., Rich J.N. Hypoxia inducible factors in cancer stem cells. Br. J. Cancer. 2010;102(5):789–795. doi: 10.1038/sj.bjc.6605551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keith B., Simon M.C. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129(3):465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pouysségur J., Dayan F., Mazure N.M. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441(7092):437–443. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- 17.Lee P., Chandel N.S., Simon M.C. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat. Rev. Mol. Cell Biol. 2020;21(5):268–283. doi: 10.1038/s41580-020-0227-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh M.Y., Powis G. Passing the baton: the HIF switch. Trends Biochem. Sci. 2012;37(9):364–372. doi: 10.1016/j.tibs.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao X., Zhang J., Huang G., Yan J., Xu C., Dou Z., Sun C., Zhang H. The crosstalk between HIFs and mitochondrial dysfunctions in cancer development. Cell Death Dis. 2021;12(2):215. doi: 10.1038/s41419-021-03505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Covello K.L., Kehler J., Yu H., Gordan J.D., Arsham A.M., Hu C.-J., Labosky P.A., Simon M.C., Keith B. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20(5):557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z., Bao S., Wu Q., Wang H., Eyler C., Sathornsumetee S., Shi Q., Cao Y., Lathia J., McLendon R.E., Hjelmeland A.B., Rich J.N. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15(6):501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X., Dong J., Jia L., Zhao T., Lang M., Li Z., Lan C., Li X., Hao J., Wang H., Qin T., Huang C., Yang S., Yu M., Ren H. HIF-2-dependent expression of stem cell factor promotes metastasis in hepatocellular carcinoma. Cancer Lett. 2017;393:113–124. doi: 10.1016/j.canlet.2017.01.032. [DOI] [PubMed] [Google Scholar]
- 23.Yan Y., He M., Zhao L., Wu H., Zhao Y., Han L., Wei B., Ye D., Lv X., Wang Y., Yao W., Zhao H., Chen B., Jin Z., Wen J., Zhu Y., Yu T., Jin F., Wei M. Cell Death & Differentiation; 2022. A Novel HIF-2α Targeted Inhibitor Suppresses Hypoxia-Induced Breast Cancer Stemness via SOD2-mtROS-PDI/GPR78-UPRER axis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baird L., Yamamoto M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell. Biol. 2020;40(13) doi: 10.1128/MCB.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 System: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018;98(3):1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rojo de la Vega M., Chapman E., Zhang D.D. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34(1):21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim T.-H., Hur E.-g., Kang S.-J., Kim J.-A., Thapa D., Lee Y.M., Ku S.K., Jung Y., Kwak M.-K. NRF2 blockade suppresses colon tumor angiogenesis by inhibiting hypoxia-induced activation of HIF-1α. Cancer Res. 2011;71(6):2260–2275. doi: 10.1158/0008-5472.CAN-10-3007. [DOI] [PubMed] [Google Scholar]
- 28.Lee S., Hallis S.P., Jung K.-A., Ryu D., Kwak M.-K. Impairment of HIF-1α-mediated metabolic adaption by NRF2-silencing in breast cancer cells. Redox Biol. 2019;24 doi: 10.1016/j.redox.2019.101210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D., Choi B.-h., Ryoo I.-g., Kwak M.-K. High NRF2 level mediates cancer stem cell-like properties of aldehyde dehydrogenase (ALDH)-high ovarian cancer cells: inhibitory role of all-trans retinoic acid in ALDH/NRF2 signaling. Cell Death Dis. 2018;9(9):896. doi: 10.1038/s41419-018-0903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park J., Kim S.K., Hallis S.P., Choi B.-H., Kwak M.-K. Role of cd133/NRF2 Axis in the development of colon cancer stem cell-like properties. Front. Oncol. 2022;11 doi: 10.3389/fonc.2021.808300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryoo I.-g., Choi B.-h., Ku S.-K., Kwak M.-K. High CD44 expression mediates p62-associated NFE2L2/NRF2 activation in breast cancer stem cell-like cells: implications for cancer stem cell resistance. Redox Biol. 2018;17:246–258. doi: 10.1016/j.redox.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryoo I.-g., Choi B.-h., Kwak M.-K. Activation of NRF2 by p62 and proteasome reduction in sphere-forming breast carcinoma cells. Oncotarget. 2015;6(No 10) doi: 10.18632/oncotarget.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan Q., Bartz S., Mao M., Li L., Kaelin William G. The hypoxia-inducible factor 2α N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol. Cell. Biol. 2007;27(6):2092–2102. doi: 10.1128/MCB.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furukawa M., Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the cullin 3-roc1 ligase. Mol. Cell. Biol. 2005;25(1):162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung K.-A., Kwak M.-K. Enhanced 4-hydroxynonenal resistance in KEAP1 silenced human colon cancer cells, oxidative medicine and cellular. Longevity. 2013;2013 doi: 10.1155/2013/423965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryu D., Lee J.-H., Kwak M.-K. NRF2 level is negatively correlated with TGF-β1-induced lung cancer motility and migration via NOX4-ROS signaling. Arch Pharm. Res. (Seoul) 2020;43(12):1297–1310. doi: 10.1007/s12272-020-01298-z. [DOI] [PubMed] [Google Scholar]
- 37.Komoto T.T., Lee J., Lertpatipanpong P., Ryu J., Marins M., Fachin A.L., Baek S.J. Trans-chalcone suppresses tumor growth mediated at least in part by the induction of heme oxygenase-1 in breast cancer. Toxicol. Res. 2021;37(4):485–493. doi: 10.1007/s43188-021-00089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung K.-A., Lee S., Kwak M.-K. NFE2L2/NRF2 activity is linked to mitochondria and AMP-activated protein kinase signaling in cancers through miR-181c/mitochondria-encoded cytochrome c oxidase regulation. Antioxidants Redox Signal. 2017;27(13):945–961. doi: 10.1089/ars.2016.6797. [DOI] [PubMed] [Google Scholar]
- 40.Kim S.H., Lee D., Lee J., Yang J.Y., Seok J., Jung K., Lee J. Evaluation of the skin sensitization potential of metal oxide nanoparticles using the ARE-Nrf2 Luciferase KeratinoSens(TM) assay. Toxicol. Res. 2021;37(2):277–284. doi: 10.1007/s43188-020-00071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasaikar S., Huang C., Wang X., Petyuk V.A., Savage S.R., Wen B., Dou Y., Zhang Y., Shi Z., Arshad O.A., Gritsenko M.A., Zimmerman L.J., McDermott J.E., Clauss T.R., Moore R.J., Zhao R., Monroe M.E., Wang Y.-T., Chambers M.C., Slebos R.J.C., Lau K.S., Mo Q., Ding L., Ellis M., Thiagarajan M., Kinsinger C.R., Rodriguez H., Smith R.D., Rodland K.D., Liebler D.C., Liu T., Zhang B., Pandey A., Paulovich A., Hoofnagle A., Mani D.R., Chan D.W., Ransohoff D.F., Fenyo D., Tabb D.L., Levine D.A., Boja E.S., Kuhn E., White F.M., Whiteley G.A., Zhu H., Zhang H., Shih I.-M., Bavarva J., Whiteaker J., Ketchum K.A., Clauser K.R., Ruggles K., Elburn K., Hannick L., Watson M., Oberti M., Mesri M., Sanders M.E., Borucki M., Gillette M.A., Snyder M., Edwards N.J., Vatanian N., Rudnick P.A., McGarvey P.B., Mertins P., Townsend R.R., Thangudu R.R., Rivers R.C., Payne S.H., Davies S.R., Cai S., Stein S.E., Carr S.A., Skates S.J., Madhavan S., Hiltke T., Chen X., Zhao Y., Wang Y., Zhang Z. Proteogenomic analysis of human colon cancer reveals new therapeutic opportunities. Cell. 2019;177(4) doi: 10.1016/j.cell.2019.03.030. 1035-1049.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell P.J., Getz G., Korbel J.O., Stuart J.M., Jennings J.L., Stein L.D., Perry M.D., Nahal-Bose H.K., Ouellette B.F.F., Li C.H., Rheinbay E., Nielsen G.P., Sgroi D.C., Wu C.-L., Faquin W.C., Deshpande V., Boutros P.C., Lazar A.J., Hoadley K.A., Louis D.N., Dursi L.J., Yung C.K., Bailey M.H., Saksena G., Raine K.M., Buchhalter I., Kleinheinz K., Schlesner M., Zhang J., Wang W., Wheeler D.A., Ding L., Simpson J.T., O'Connor B.D., Yakneen S., Ellrott K., Miyoshi N., Butler A.P., Royo R., Shorser S.I., Vazquez M., Rausch T., Tiao G., Waszak S.M., Rodriguez-Martin B., Shringarpure S., Wu D.-Y., Demidov G.M., Delaneau O., Hayashi S., Imoto S., Habermann N., Segre A.V., Garrison E., Cafferkey A., Alvarez E.G., Heredia-Genestar J.M., Muyas F., Drechsel O., Bruzos A.L., Temes J., Zamora J., Baez-Ortega A., Kim H.-L., Mashl R.J., Ye K., DiBiase A., Huang K.-l., Letunic I., McLellan M.D., Newhouse S.J., Shmaya T., Kumar S., Wedge D.C., Wright M.H., Yellapantula V.D., Gerstein M., Khurana E., Marques-Bonet T., Navarro A., Bustamante C.D., Siebert R., Nakagawa H., Easton D.F., Ossowski S., Tubio J.M.C., De La Vega F.M., Estivill X., Yuen D., Mihaiescu G.L., Omberg L., Ferretti V., Sabarinathan R., Pich O., Gonzalez-Perez A., Taylor-Weiner A., Fittall M.W., Demeulemeester J., Tarabichi M., Roberts N.D., Van Loo P., Cortés-Ciriano I., Urban L., Park P., Zhu B., Pitkänen E., Li Y., Saini N., Klimczak L.J., Weischenfeldt J., Sidiropoulos N., Alexandrov L.B., Rabionet R., Escaramis G., Bosio M., Holik A.Z., Susak H., Prasad A., Erkek S., Calabrese C., Raeder B., Harrington E., Mayes S., Turner D., Juul S., Roberts S.A., Song L., Koster R., Mirabello L., Hua X., Tanskanen T.J., Tojo M., Chen J., Aaltonen L.A., Rätsch G., Schwarz R.F., Butte A.J., Brazma A., Chanock S.J., Chatterjee N., Stegle O., Harismendy O., Bova G.S., Gordenin D.A., Haan D., Sieverling L., Feuerbach L., Chalmers D., Joly Y., Knoppers B., Molnár-Gábor F., Phillips M., Thorogood A., Townend D., Goldman M., Fonseca N.A., Xiang Q., Craft B., Piñeiro-Yáñez E., Muñoz A., Petryszak R., Füllgrabe A., Al-Shahrour F., Keays M., Haussler D., Weinstein J., Huber W., Valencia A., Papatheodorou I., Zhu J., Fan Y., Torrents D., Bieg M., Chen K., Chong Z., Cibulskis K., Eils R., Fulton R.S., Gelpi J.L., Gonzalez S., Gut I.G., Hach F., Heinold M., Hu T., Huang V., Hutter B., Jäger N., Jung J., Kumar Y., Lalansingh C., Leshchiner I., Livitz D., Ma E.Z., Maruvka Y.E., Milovanovic A., Nielsen M.M., Paramasivam N., Pedersen J.S., Puiggròs M., Sahinalp S.C., Sarrafi I., Stewart C., Stobbe M.D., Wala J.A., Wang J., Wendl M., Werner J., Wu Z., Xue H., Yamaguchi T.N., Yellapantula V., Davis-Dusenbery B.N., Grossman R.L., Kim Y., Heinold M.C., Hinton J., Jones D.R., Menzies A., Stebbings L., Hess J.M., Rosenberg M., Dunford A.J., Gupta M., Imielinski M., Meyerson M., Beroukhim R., Reimand J., Dhingra P., Favero F., Dentro S., Wintersinger J., Rudneva V., Park J.W., Hong E.P., Heo S.G., Kahles A., Lehmann K.-V., Soulette C.M., Shiraishi Y., Liu F., He Y., Demircioğlu D., Davidson N.R., Greger L., Li S., Liu D., Stark S.G., Zhang F., Amin S.B., Bailey P., Chateigner A., Frenkel-Morgenstern M., Hou Y., Huska M.R., Kilpinen H., Lamaze F.C., Li C., Li X., Li X., Liu X., Marin M.G., Markowski J., Nandi T., Ojesina A.I., Pan-Hammarström Q., Park P.J., Pedamallu C.S., Su H., Tan P., Teh B.T., Wang J., Xiong H., Ye C., Yung C., Zhang X., Zheng L., Zhu S., Awadalla P., Creighton C.J., Wu K., Yang H., Göke J., Zhang Z., Brooks A.N., Fittall M.W., Martincorena I., Rubio-Perez C., Juul M., Schumacher S., Shapira O., Tamborero D., Mularoni L., Hornshøj H., Deu-Pons J., Muiños F., Bertl J., Guo Q., Gonzalez-Perez A., Xiang Q., I.T.P.-C.A.o.W.G.C. The Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y., Reva B., Goldberg A.P., Sander C., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao J., Aksoy Bülent A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269) doi: 10.1126/scisignal.2004088. pl1-pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J.-X., Song W., Chen Z.-H., Wei J.-H., Liao Y.-J., Lei J., Hu M., Chen G.-Z., Liao B., Lu J., Zhao H.-W., Chen W., He Y.-L., Wang H.-Y., Xie D., Luo J.-H. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14(13):1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 46.Ren D., Villeneuve N.F., Jiang T., Wu T., Lau A., Toppin H.A., Zhang D.D. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proceed. Nat. Acad. Sci. USA. 2011;108(4):1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das B., Tsuchida R., Malkin D., Koren G., Baruchel S., Yeger H. Hypoxia enhances tumor stemness by increasing the invasive and tumorigenic side population fraction. Stem Cell. 2008;26(7):1818–1830. doi: 10.1634/stemcells.2007-0724. [DOI] [PubMed] [Google Scholar]
- 48.Zhang C., Samanta D., Lu H., Bullen J.W., Zhang H., Chen I., He X., Semenza G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA. 2016;113(14):E2047–E2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heddleston J.M., Li Z., McLendon R.E., Hjelmeland A.B., Rich J.N. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8(20):3274–3284. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xue X., Taylor M., Anderson E., Hao C., Qu A., Greenson J.K., Zimmermann E.M., Gonzalez F.J., Shah Y.M. Hypoxia-inducible factor-2α activation promotes colorectal cancer progression by dysregulating iron homeostasis. Cancer Res. 2012;72(9):2285–2293. doi: 10.1158/0008-5472.CAN-11-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh A., Misra V., Thimmulappa R.K., Lee H., Ames S., Hoque M.O., Herman J.G., Baylin S.B., Sidransky D., Gabrielson E., Brock M.V., Biswal S. Dysfunctional KEAP1–NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3(10) doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shibata T., Ohta T., Tong K.I., Kokubu A., Odogawa R., Tsuta K., Asamura H., Yamamoto M., Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. USA. 2008;105(36):13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldstein Leonard D., Lee J., Gnad F., Klijn C., Schaub A., Reeder J., Daemen A., Bakalarski Corey E., Holcomb T., Shames David S., Hartmaier Ryan J., Chmielecki J., Seshagiri S., Gentleman R., Stokoe D. Recurrent loss of NFE2L2 exon 2 is a mechanism for Nrf2 pathway activation in human cancers. Cell Rep. 2016;16(10):2605–2617. doi: 10.1016/j.celrep.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 54.Wang R., An J., Ji F., Jiao H., Sun H., Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem. Biophys. Res. Commun. 2008;373(1):151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 55.Saito T., Ichimura Y., Taguchi K., Suzuki T., Mizushima T., Takagi K., Hirose Y., Nagahashi M., Iso T., Fukutomi T., Ohishi M., Endo K., Uemura T., Nishito Y., Okuda S., Obata M., Kouno T., Imamura R., Tada Y., Obata R., Yasuda D., Takahashi K., Fujimura T., Pi J., Lee M.-S., Ueno T., Ohe T., Mashino T., Wakai T., Kojima H., Okabe T., Nagano T., Motohashi H., Waguri S., Soga T., Yamamoto M., Tanaka K., Komatsu M. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat. Commun. 2016;7(1) doi: 10.1038/ncomms12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi B.-H., Kim J.M., Kwak M.-K. The multifaceted role of NRF2 in cancer progression and cancer stem cells maintenance. Arch Pharm. Res. (Seoul) 2021;44(3):263–280. doi: 10.1007/s12272-021-01316-8. [DOI] [PubMed] [Google Scholar]
- 57.Wu T., Harder B.G., Wong P.K., Lang J.E., Zhang D.D. Oxidative stress, mammospheres and Nrf2-new implication for breast cancer therapy? Mol. Carcinog. 2015;54(11):1494–1502. doi: 10.1002/mc.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kamble D., Mahajan M., Dhat R., Sitasawad S. Keap1-Nrf2 pathway regulates ALDH and contributes to radioresistance in breast cancer stem cells. Cells. 2021;10(1) doi: 10.3390/cells10010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim D.-H., Jang J.-H., Kwon O.-S., Cha H.-J., Youn H.-J., Chun K.-S., Surh Y.-J. Nuclear factor erythroid-derived 2-like 2-induced reductive stress favors self-renewal of breast cancer stem-like cells via the FoxO3a-bmi-1 Axis. Antioxidants Redox Signal. 2019;32(18):1313–1329. doi: 10.1089/ars.2019.7730. [DOI] [PubMed] [Google Scholar]
- 60.Qin S., He X., Lin H., Schulte B.A., Zhao M., Tew K.D., Wang G.Y. Nrf2 inhibition sensitizes breast cancer stem cells to ionizing radiation via suppressing DNA repair. Free Radic. Biol. Med. 2021;169:238–247. doi: 10.1016/j.freeradbiomed.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malec V., Gottschald O.R., Li S., Rose F., Seeger W., Hänze J. HIF-1α signaling is augmented during intermittent hypoxia by induction of the Nrf2 pathway in NOX1-expressing adenocarcinoma A549 cells. Free Radic. Biol. Med. 2010;48(12):1626–1635. doi: 10.1016/j.freeradbiomed.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 62.Küper A., Baumann J., Göpelt K., Baumann M., Sänger C., Metzen E., Kranz P., Brockmeier U. Overcoming hypoxia-induced resistance of pancreatic and lung tumor cells by disrupting the PERK-NRF2-HIF-axis. Cell Death Dis. 2021;12(1):82. doi: 10.1038/s41419-020-03319-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lacher S.E., Levings D.C., Freeman S., Slattery M. Identification of a functional antioxidant response element at the HIF1A locus. Redox Biol. 2018;19:401–411. doi: 10.1016/j.redox.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oh E.-T., Kim J.-w., Kim J.M., Kim S.J., Lee J.-S., Hong S.-S., Goodwin J., Ruthenborg R.J., Jung M.G., Lee H.-J., Lee C.-H., Park E.S., Kim C., Park H.J. NQO1 inhibits proteasome-mediated degradation of HIF-1α. Nat. Commun. 2016;7(1) doi: 10.1038/ncomms13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peng Y., Croce C.M. The role of MicroRNAs in human cancer. Signal Transduct. Targeted Ther. 2016;1(1) doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Byun Y., Choi Y.-C., Jeong Y., Lee G., Yoon S., Jeong Y., Yoon J., Baek K. MiR-200c downregulates HIF-1α and inhibits migration of lung cancer cells. Cell. Mol. Biol. Lett. 2019;24(1):28. doi: 10.1186/s11658-019-0152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang L., Shi S., Shi Q., Zhang H., Xia Y., Zhong T. MicroRNA-519d-3p inhibits proliferation and promotes apoptosis by targeting HIF-2α in cervical cancer under hypoxic conditions. Oncol. Res. 2018;26(7):1055–1062. doi: 10.3727/096504018X15152056890500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu J.-H., Li W.-T., Yang Y., Qi Y.-B., Cheng Y., Wu J.-H. MiR-526b-3p attenuates breast cancer stem cell properties and chemoresistance by targeting HIF-2α/Notch signaling. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.696269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ji J., Yamashita T., Budhu A., Forgues M., Jia H.-L., Li C., Deng C., Wauthier E., Reid L.M., Ye Q.-H., Qin L.-X., Yang W., Wang H.-Y., Tang Z.-Y., Croce C.M., Wang X.W. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM–positive hepatic cancer stem cells. Hepatology. 2009;50(2):472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mei Q., Li X., Zhang K., Wu Z., Li X., Meng Y., Guo M., Luo G., Fu X., Han W. Genetic and methylation-induced loss of miR-181a2/181b2 within chr9q33.3 facilitates tumor growth of cervical cancer through the PIK3R3/akt/FoxO signaling pathway. Clin. Cancer Res. 2017;23(2):575–586. doi: 10.1158/1078-0432.CCR-16-0303. [DOI] [PubMed] [Google Scholar]
- 71.Chhabra R. let-7i-5p, miR-181a-2-3p and EGF/PI3K/SOX2 axis coordinate to maintain cancer stem cell population in cervical cancer. Sci. Rep. 2018;8(1):7840. doi: 10.1038/s41598-018-26292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bahn G., Jo D.G. Therapeutic approaches to alzheimer's disease through modulation of NRF2. NeuroMolecular Med. 2019;21(1):1–11. doi: 10.1007/s12017-018-08523-5. [DOI] [PubMed] [Google Scholar]
- 73.Liu P., Rojo de la Vega M., Sammani S., Mascarenhas J.B., Kerins M., Dodson M., Sun X., Wang T., Ooi A., Garcia J.G.N., Zhang D.D. RPA1 binding to NRF2 switches ARE-dependent transcriptional activation to ARE-NRE-dependent repression. Proceed. Nat. Acad. Sci. USA. 2018;115(44):E10352–e10361. doi: 10.1073/pnas.1812125115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Presnell S.R., Al-Attar A., Cichocki F., Miller J.S., Lutz C.T. Human natural killer cell microRNA: differential expression of MIR181A1B1 and MIR181A2B2 genes encoding identical mature microRNAs. Gene Immun. 2015;16(1):89–98. doi: 10.1038/gene.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gong Y., Yang Y. Activation of Nrf2/AREs-mediated antioxidant signalling, and suppression of profibrotic TGF-β1/Smad3 pathway: a promising therapeutic strategy for hepatic fibrosis — a review. Life Sci. 2020;256 doi: 10.1016/j.lfs.2020.117909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.