Summary
Background
Brazil is a unique and understudied setting for malaria, with complex foci of transmission associated with human and environmental conditions. An understanding of the population genomic diversity of P. vivax parasites across Brazil can support malaria control strategies.
Methods
Through whole genome sequencing of P. vivax isolates across 7 Brazilian states, we use population genomic approaches to compare genetic diversity within country (n = 123), continent (6 countries, n = 315) and globally (26 countries, n = 885).
Findings
We confirm that South American isolates are distinct, have more ancestral populations than the other global regions, with differentiating mutations in genes under selective pressure linked to antimalarial drugs (pvmdr1, pvdhfr-ts) and mosquito vectors (pvcrmp3, pvP45/48, pvP47). We demonstrate Brazil as a distinct parasite population, with signals of selection including ABC transporter (PvABCI3) and PHIST exported proteins.
Interpretation
Brazil has a complex population structure, with evidence of P. simium infections and Amazonian parasites separating into multiple clusters. Overall, our work provides the first Brazil-wide analysis of P. vivax population structure and identifies important mutations, which can inform future research and control measures.
Funding
AI is funded by an MRC LiD PhD studentship. TGC is funded by the Medical Research Council (Grant no. MR/M01360X/1, MR/N010469/1, MR/R025576/1, MR/R020973/1 and MR/X005895/1). SC is funded by Medical Research Council UK grants (MR/M01360X/1, MR/R025576/1, MR/R020973/1 and MR/X005895/1) and Bloomsbury SET (ref. CCF17-7779). FN is funded by The Shloklo Malaria Research Unit - part of the Mahidol Oxford Research Unit, supported by the Wellcome Trust (Grant no. 220211). ARSB is funded by São Paulo Research Foundation - FAPESP (Grant no. 2002/09546–1). RLDM is funded by Brazilian National Council for Scientific and Technological Development - CNPq (Grant no. 302353/2003–8 and 471605/2011–5); CRFM is funded by FAPESP (Grant no. 2020/06747–4) and CNPq (Grant no. 302917/2019–5 and 408636/2018–1); JGD is funded by FAPESP fellowships (2016/13465–0 and 2019/12068–5) and CNPq (Grant no. 409216/2018–6).
Keywords: Malaria, Plasmodium, Plasmodium vivax, Non-falciparum malaria, Brazil, South America, Whole genome sequencing, Population genetics, Vector-borne diseases, Drug resistance, Genomics
Research in context.
Evidence before this study
Human malaria caused by infections with P. vivax parasites are a significant threat to global health. Whilst less pathogenic than P. falciparum, P. vivax infections are more widely spread, with one third of the world's population at risk of infection, resulting in a huge impact on morbidity. Prior to this study, whole genome sequencing (WGS) of Brazilian P. vivax isolates has focussed on the two states of Amazonas and Acre, both in the Amazonian region. These studies demonstrated genetically diverse populations of parasites within the Americas, signs of inbreeding in high-transmission sites in Brazil such as Mancio Lima, and evidence of mutations within genes associated with drug susceptibility. There are however other transmission foci within Brazil and the genomic background of these foci has yet to be investigated.
Added value of this study
This study analyses high-quality genome data from 885 clinical isolates of P. vivax sourced globally, including 315 from South America, and 123 from Brazil covering seven states. Using the largest genomic dataset of Brazilian P. vivax isolates, we demonstrate a complex population structure at country level, with a distinct population of isolates from São Paulo that may be P. simium. We find Brazil-specific mutations in genes associated with mosquito life stages and drug susceptibility and suggest potential novel candidates for further investigation.
Implications of all the available evidence
Malaria continues to be an important public health issue in Brazil. To tackle this disease effectively, it is crucial to understand the genetic make-up of the underlying parasites. Through WGS studies, it is possible to identify genetic differences between populations that may enable us to target parasites more effectively. By screening drug resistance markers, it is possible to determine the most effective treatment regimen to use. Here, we investigate the largest genomic dataset of clinical P. vivax isolates in Brazil, and determine mutations within genes associated with drug susceptibility, alongside Brazilian-specific variants in genes associated with mosquito transmission stages, potentially informing an understanding of transmission in the country and the wider region.
Introduction
The Plasmodium vivax parasite causes the highest malaria burden outside of sub-Saharan Africa,1 with more than one third of the global population at risk due to its wide geographical range.2 Complications associated with P. vivax infections can lead to severe, life-threatening syndromes.3 P. vivax infections underly the majority (>80%) of the >700k annual malaria cases in the Americas, including in South America, where countries surrounding the Amazon rain forest areas, such as Brazil, Colombia, and Venezuela, have hotspots of endemic disease.1 In South America, malaria transmission dynamic studies are complicated by P. vivax and P. falciparum co-infections, which have differences in their life cycles and transmission patterns.1 Further, significant challenges continue to thwart P. vivax control, including the ability of parasites to form dormant hypnozoite stages within the liver, leading to relapses of malaria if not treated using a radical cure of primaquine.4 Unfortunately, individuals with glucose-6-phosphate dehydrogenase deficiency are at risk of developing severe haemolysis if treated with primaquine or tafenoquine, and therefore along with pregnant women and infants, are ineligible for radical cure.5 Additionally, control measures are compromised by the presence of sub-microscopic and asymptomatic P. vivax infections, leading to untreated human parasite reservoirs.6 Human settlement and mobility, including through peri-urban expansion, gold mining-related activities, and deforestation in the Amazon, all lead to significantly higher risk of malaria infections.5,7 In addition, self-medication with poor adherence, reported in these regions, can contribute to relapses of P. vivax infections and may contribute to selection of mutations leading to parasite drug resistance.8,9 There are many gaps in our knowledge of P. vivax infections, including malaria in pregnancy which is understudied, and a lack of knowledge of genetic markers for drug resistance, specifically regarding the first line antimalarial, chloroquine, to which resistance has emerged in many countries including South America,10 calling for a rapid change in P. vivax control strategy.
Brazil has a diverse geographical profile leading to variation in malaria transmission, with foci split into three discrete groups, each with unique settings for transmission. First, the Amazon rainforest in the northeast of Brazil, which accounts for 99% of all malaria cases, and transmission is led by Anopheles darlingi and An. albitarsis complex mosquitoes. Second, the north-western coastal border of Brazil, where transmission is lower and due to the An. aquasalis mosquito species. Third, the Atlantic Rainforest on the south-western coastal border of the country, where transmission is mainly mediated by An. bellator and An. cruzii.11 In this southern region, P. simium is transmitted by An. cruzii, and is genetically highly related to P. vivax. P. simium is found mainly in non-human primates12 but human cases have been recorded in São Paulo and Rio de Janeiro.13 Within Brazil, there is both inter-state transmission and importation of malaria from neighbouring countries. Due to their proximity, countries bordering the Amazonian region (French Guiana, Guyana, Venezuela, and Peru) play an important role in malaria transmission in the Amazon. Importation of malaria, whether between countries, or within country is a threat to elimination, as malaria-free regions neighbouring those with malaria transmission are at a constant risk of importation and resulting outbreaks.11
Human genetics may contribute to P. vivax transmission dynamics in Brazil, with the presence of the Duffy negative (Fy-) blood group phenotype, which hinders P. vivax invasion of human erythrocytes.14 The Fy-phenotype appears to be more common on the north-western coast of Brazil, with some areas demonstrating >50% frequency.15 In contrast, the phenotype appears less common in studies based in the Amazonian region, where vivax-malaria transmission is highest. For example, the Fy-phenotype frequency is only 2.8% in Presidente Figueiredo, where malaria transmission is high (annual parasite index (API) of 301.65 malaria cases per 1000 individuals).16 Whilst Fy-was previously thought to provide complete protection to P. vivax infection, there have been reports,17, 18, 19, 20 including in Brazil,21 that vivax-infections can occur in Fy-individuals. Furthermore, there are concerns that vivax-malaria in Fy-individuals may present as an asymptomatic infection with lower asexual parasitaemias than Duffy positive individuals, which could lead to a large silent parasite reservoir, complicating malaria eradication.22,23
In contrast to the wider transmission of P. vivax in Brazil, P. falciparum infections are restricted to hotspot areas, mostly found within the Amazonian rainforest states of Amazonas and Acre, the two states which account for >45% of all malaria cases.24 Control measures for malaria have been designed against P. falciparum infections and are widely known to be less effective at tackling P. vivax, due to key differences in parasite biology. For example, P. vivax parasites are able to create dormant liver stage parasites, known as hypnozoites, which are not cleared using routine antimalarials, and require additional treatment, known as radical cure.25 Additionally, P. vivax parasites are viable within a wider temperature range than P. falciparum, allowing for their spread into a greater geographical area,26 with transmission further aided by the permissibility of P. vivax to multiple mosquito vector species.27
In Brazil, chloroquine is still used to treat P. vivax infections, even though resistance has already been documented in both P. falciparum and P. vivax parasites.10 Surveillance of drug resistance in P. vivax parasites is a challenge as the underlying genetic markers for resistance are unknown. Whole genome sequencing (WGS) of P. vivax could provide insights into genetic mutations underlying both drug resistance and population structure. Studies have shown that P. vivax parasites within South America display high levels of genetic diversity, comparable to high transmission regions such as Southeast Asia.28 This difference may be due to the complex pattern of human migration in Brazil, including inter-state movement for work opportunities and historical waves such as during the slave trade and colonization; all potentially leading to multiple introductions of genetically different Plasmodium parasites.28, 29, 30, 31
Previous studies have shown that South American P. vivax parasites form a distinct global subpopulation,32, 33, 34 with informative barcoding loci found within orthologs of genes known to be important for mosquito development stages and possible targets to inhibit parasite transmission, including the pvcrmp gene family (with orthologs in P. berghei associated with sporozoite development and onwards transmission35,36), pvs47 and pvs48/45.37 Drug susceptibility loci are also informative for barcoding, including pvmdr1, whose ortholog in P. falciparum is associated with multi drug resistance.33,34,38,39 P. vivax parasites within South America are known to demonstrate general country level separation,28,32 with Brazilian and Peruvian isolates clustering together.28,34 The P. vivax parasite population structure in Brazil remains unclear, with the vast majority of currently available WGS data collected from malaria infections in Acre.32,40,41 Brazil has a complex setting, due to both the three distinct P. vivax transmission foci, and the context of human migration. Here, we perform a population genomic analysis of the largest WGS dataset for P. vivax isolates from 10 regions within Brazil (n = 123) spanning 7 states, position them in a global context using a filtered global database (n = 885), characterise the within country and wider regional ancestral and population structure, and identify loci under selective pressure. We reveal a complex population of parasites within Brazil, with vast genomic diversity in areas of high transmission, and Brazilian specific signals of selection in genes associated with drug susceptibility.
Methods
Whole genome sequence data
A total of 1113 isolates were analysed, including publicly available (n = 1023)32,34,40,42, 43, 44 and novel sequence data from Brazil (n = 89). After quality control (as described in Bioinformatic analysis), the dataset consisted of 885 isolates spanning all regions where P. vivax infections are endemic: (i) South America (n = 315: Brazil 123, Colombia 34, Guyana 3, Mexico 20, Panama 46, Peru 89); (ii) East Africa (n = 84; Eritrea 13, Ethiopia 53, Madagascar 4, Sudan 9, Uganda 5); (iii) South Asia (n = 114; Afghanistan 27, Bangladesh 1, India 48, Pakistan 37, Sri Lanka 1); (iv) South East Asia (SEA; n = 286; Cambodia 71, China 12, Laos 2, Myanmar 28, Thailand 160, Vietnam 13); and (v) the Western Pacific and southern South East Asia (SSEA; n = 86; Indonesia 9, Malaysia 50, Papua New Guinea 26, The Philippines 1) (Table S1). These included newly sequenced isolates (n = 51) and publicly available data (n = 834) in the final filtered dataset. Newly sequenced isolates were obtained from whole blood samples from seven states in Brazil (Acre 4; Amapá, 10; Rondônia, 4; Amazonas 3; São Paulo 12; Mato Grosso 5; Pará 13), leading to a total of 123 high quality WGS data from isolates within Brazil spanning all areas of P. vivax transmission (see Fig. S1 for a map; Table S2).
The whole blood samples were obtained from symptomatic malaria patients. All samples were collected with the appropriate ethical approval from relevant authorities, including from Hospital Universitário Antonio Pedro, Universidade Federal Fluminense (ref. CAAE 06214118.2.0000.5243) and Faculdade de Medicina de São José do Rio Preto (ref. CAAE 01774812.2.0000.5415), Centro de Pesquisa em Medicina Tropical Rondônia (CAAE 61442416.7.0000.0011), and Instituto de Ciências Biomédicas (ICB/USP; ref. CAAE: 03930812.8.0000.5467). Informed consent was obtained from all individuals. DNA was extracted from whole blood samples using the QIAamp DNA Blood Mini Kit (Qiagen), quantified using a Qubit (v2.0) fluorometer, and single-species P. vivax infections were confirmed using qPCR. Selective whole genome amplification (SWGA) using a set of previously described primers45 was used to increase the relative levels of P. vivax DNA within the sample, allowing for whole genome sequencing (WGS).46 Amplified isolates were sequenced using the Illumina MiSeq and HiSeq4000 platforms using paired-150 bp read kits through The Applied Genomics Centre, LSHTM.
Bioinformatic analysis
FASTQ files generated from the Illumina sequencing reads (from both publicly available (n = 1023) and newly sequenced isolates (n = 89), available from the European Nucleotide Archive under project codes PRJEB56411, PRJEB44419, PRJEB36199 and the MalariaGEN P. vivax Genome Variation project,40 were trimmed using TRIMMOMATIC (v0.39) with the following parameters: LEADING:3, TRAILING:3, SLIDINGWINDOW:4-20, MINLEN:36.47 Trimmed reads were aligned to the PVP01 P. vivax reference genome (v1)48 (https://plasmodb.org) using BWA-MEM software (v0.7.12).49 BAM files were processed using samtools (v1.10) functions fixmate and markdup. We used a “training set” of high-quality P. vivax SNPs from previously published work50 to calibrate variant calling (see34). Using the training set, GATK's BaseRecalibrator and ApplyBQSR functions were run within a robust framework51, to produce improved and corrected BAM files for all isolates. SNPs and indels were determined using GATK's HaplotypeCaller (v4.1.4.1; options -ERC GVCF; otherwise default settings) to produce variant call format files, which contain all SNPs and insertions and deletions (indels) identified.51 The GATK ValidateVariants function was used to validate the resulting Genomic Variant Call Format Files (GVCFs), which were subsequently imported into a GenomicDB using the GATK function GenomicsDBImport. GATK's GenotypeGVCFs function was used to create a combined VCF including all isolates. A total of 3,932,759 unfiltered SNPs were identified across the 1113 isolates. Variants within subtelomeric regions and Variant Quality Score Log-Odds (VQSLOD) scores <0 were removed. A total of 228 isolates with more than 40% of SNPs missing genotype data were excluded from downstream analysis. The final dataset consisted of 885 isolates and 454,681 high quality SNPs used for population genetic analysis. SNPs were annotated with their downstream effect using SnpEff software.52
Population genetic analysis
Multiplicity of infection (MOI) was calculated at a country level using the FWS score implemented in the moimix package (https://github.com/bahlolab/moimix), as well as at an individual isolate level using estMOI software.53 Population structure of isolates was investigated using a principal component analysis (PCA) based on pairwise SNP Manhattan distances between isolates. Maximum-likelihood (phylogenetic) trees were created using IQTREE software (v2.1.2)54 on a nucleotide alignment consisting of the high quality isolates SNP positions. Ancestral analysis was performed using the ADMIXTURE (v1.3.0) package on matrices of high-quality SNPs with a linkage disequilibrium correlation coefficient ≤0.1. ADMIXTURE predicts the most likely number of ancestral populations (K) within a dataset using cross-validation error.55 We calculated the (pairwise) fixation indices (FST) for SNPs between population groups (at global regional, country and two grouping levels within the Brazilian population; clade and geographic groupings) to investigate alleles driving the differences between populations using the VCFtools (v0.1.16) function --weir-fst-pop.56 Nucleotide diversity (Nei and Li π) was calculated genome-wide using VCFtools within each Brazilian state (Pará, n = 13; Amapá, n = 10; Mato Grosso, n = 5; Rondônia, n = 5; Acre, n = 74; Amazonas, n = 4; São Paulo, n = 12) using sliding windows of 25 kbp.
Positive and balancing selection and IBD analysis
We screened monoclonal (FWS >95%) isolates for signals of positive selection at both the regional and country level, with a focus on South American, and specifically Brazilian samples, using the REHH package (v3.2.1) in R.57 The integrated haplotype homozygosity score (iHS)58 was calculated to identify signals of within population selection, and the Rsb59 score was calculated to demonstrate signals of selection between two assigned populations. Both measures were calculated at the regional and country level, as well as within Brazil at two different grouping classifications (clade groupings from the phylogenetic tree, and geographical groupings into Group A and Group B (Table S2)). Candidate regions were identified from iHS and Rsb results using default parameters and a p-values of <1 × 10−4 and <1 × 10−5, respectively. Only populations with >10 isolates and genes with >5 SNPs were included in analysis. Where there were >10 isolates per country, monoclonal Isolates (FWS >95%) were screened at the country level for identity-by-descent (IBD) using the hmmIBD package with default parameters. For IBD analysis, a recombination rate of 13.5 kb per centimorgan (cM) was used, based on previous work in P. falciparum60 and commonly used in P. vivax research.28,61 This P. falciparum based recombination rate is the default setting in hmmIBD,62 but despite an absence of an equivalent robust estimate for P. vivax, genome-wide analysis has shown that the rates may be similar between the two species.63 Pairwise comparisons for isolates presenting evidence of IBD were plotted using a sliding window of 50 kbp along the genome location. Signals of selection at the regional level (for populations with >10 isolates), and within Brazil at the gene level (for genes with >5 SNPs), were investigated using the Tajima's D metric, which was calculated using the PEGAS package (v0.14).64
Role of funding source
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Results
P. vivax isolates and sequencing data
WGS data of Brazilian samples (n = 123) includes isolates from human infections spanning 10 regions (Goianésia do Pará, Novo Repartimento, Itaituba (Pará State), Macapá, Oiapoque (Amapá State), Rio Branco (Acre State), Porto Velho (Rondônia State), Barcelos (Amazonas State), and Mato Grosso and São Paulo States), and builds on public sequence data originating from infections in Acre and Rondônia28,33,41 (see Fig. S1 for a map of all Brazilian isolates). WGS data was analysed with 1113 isolates of P. vivax spanning 26 countries, and a total of 3,932,759 SNPs were identified.32, 33, 34,40 After filtering (see Methods), a final combined “high quality” dataset consisted of 885 isolates with a total of 454,681 unique SNP positions in the core genome of P. vivax, excluding the hypervariable regions. The filtered isolates were assigned into regional groups: South America (n = 315, including Brazil (n = 123), Colombia, Guyana, Mexico, Panama, Peru), South Asia (n = 114; Afghanistan, Bangladesh, India, Sri Lanka, Pakistan), East Africa (n = 84; Eritrea, Ethiopia, Madagascar, Sudan, Uganda), South East Asia (SEA; n = 286; Cambodia, China, Laos, Myanmar, Thailand, Vietnam), and Southern SEA (SSEA; n = 86; Malaysia, Papua New Guinea, Indonesia, The Philippines). These regions are based on previous genomics work, which demonstrated they are distinct from each other34 (Table S1, Table S2, Fig. S1). As expected, the overall sequence coverage before quality control (mean 49-fold; median 16.7-fold) was lower than post-filtering (mean 60.6-fold; median 29.5-fold).
Four ancestral populations in South America with a distinct Brazilian parasite population
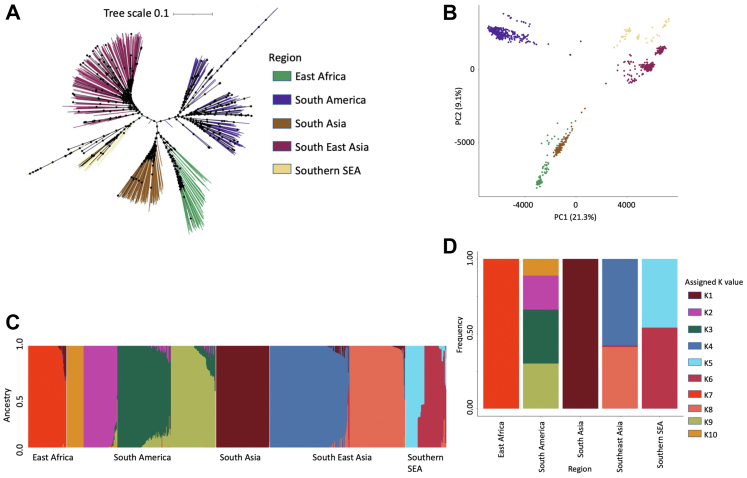
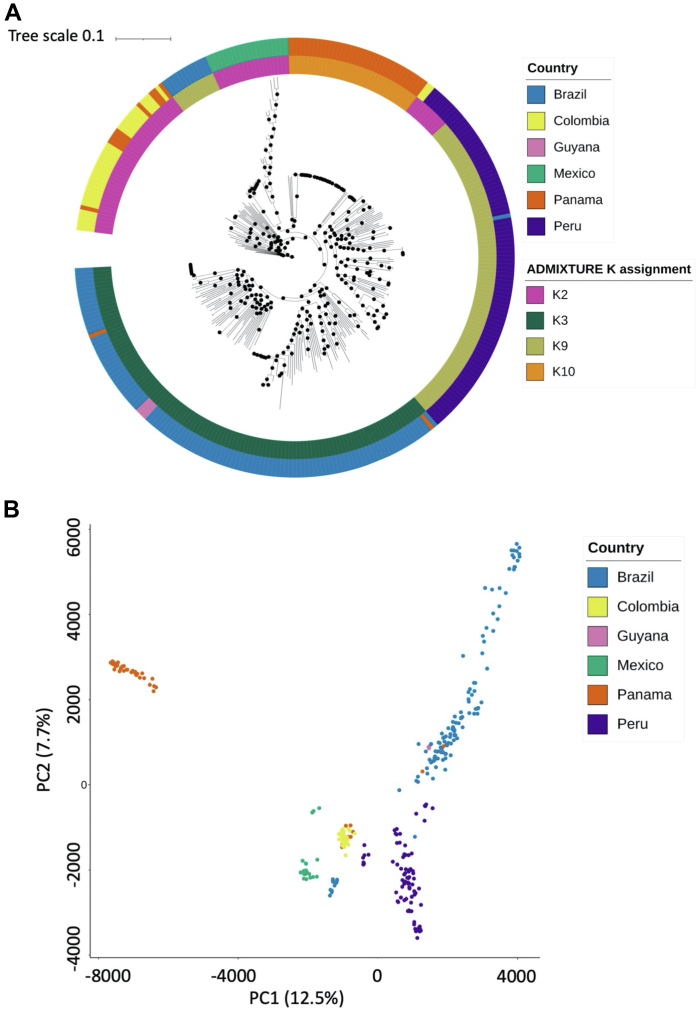
Both SNP-based maximum likelihood tree and principal component analysis (PCA) on the final dataset (n = 885 isolates, unique SNPs = 454,681) revealed the expected regional separation34 of P. vivax parasites with distinct clusters forming for South America, as well as for East Africa, South Asia, SEA, and SSEA (Fig. 1). An ADMIXTURE analysis suggested that there are ten ancestral populations spread across the five global regions, including four within South America (K2, K3, K9 and K10) and six elsewhere (East Africa K7; South Asia K1; SEA K8, K6 and K4; SSEA K6 and K5) (Fig. 1, Fig. S2). Within the South American subset of isolates (n = 315), a maximum-likelihood tree and PCA analysis based on the 102,765 unique SNPs, revealed country-level separation, including for Brazil (except São Paulo samples), with some minor overlap between Panama and Colombia, and both Panama and Guyana with Brazil (Fig. 2). There is a high concordance between ADMIXTURE population and country of origin (K3 Brazil, Guyana; K2 Mexico, Colombia; K9 Peru; K10 Panama; Fig. 2), with highly clonal clusters for Mexico and Panama consistent with previous studies.32,65 The samples from São Paulo (n = 12) cluster together in a clade separated from the remaining Brazilian samples and close to the Mexican clade (Fig. 2 and Fig. 3). These samples could be P. simium, being collected in the geographic region where this parasite has been reported, with the majority of them containing two putative P. simium barcoding mitochondrial SNPs (T4133C, A4467G).13 Deletions in pvdbp1 and pvrbp2a loci reported in P. simium but not in P. vivax66 could not be characterised with high certainty due to poor sequencing coverage at these regions. None of the 12 São Paulo isolates had coverage >5-fold across the length of pvdbp1, pvrbp2a, and their wider flanking regions, leading to uncertainty in deletion calling.
Fig. 1.
Population structure of 885 P. vivax isolates from 26 countries. A) Maximum likelihood phylogenetic tree generated using IQTREE from the pairwise SNP matrix of the complete global dataset of 885 samples and 454,681 SNPs. IQTREE was run using ModelFinder, tree search, ultrafast bootstrap and SH-aLRT test. Bootstrap scores between 50% and 100% are annotated on the tree branches with a black circle. Branches are colouring according to regional grouping (East Africa, n = 84, green branches; South America, n = 315, purple branches; South Asia, n = 114, brown branches; Southeast Asia (SEA), n = 286, dark pink branches; Southern SEA, n = 86, yellow branches). The phylogenetic tree file was visualised in iTOL with midpoint rooting. B) Principal component (PC) analysis displaying principal components 1 and 2 of the distance matrix generated using the SNP matrix. Each point represents and individual sample, coloured according with the region assigned in (A). PC 1 summarises 21.3% of the total variation whilst principal component 2 summarises 9.1% of the total variation. C) ADMIXTURE analysis of the global dataset predicted a total of 10 ancestral populations spread across each region: East Africa, n = 1; South America, n = 4; South Asia, n = 1; SEA, n = 3; SSEA, n = 2. D) Bar plot summarising the number and proportion of each ancestral population within each region.
Fig. 2.
Population structure of South American isolates. A) Maximum likelihood (ML) tree using SNP data (102,765 unique SNPs) from 315 isolates from South America (Brazil, n = 123; Colombia, n = 34; Mexico, n = 20; Panama, n = 46; Peru, n = 89; Guyana, n = 3). The outer circle track is coloured according to the country of each isolate (Brazil - blue, Colombia - yellow, Guyana - pink, Mexico - green, Panama - orange, Peru - purple), and the inner circle track denotes the population assigned to each isolate after ADMIXTURE analysis of the entire global dataset. ADMIXTURE denoted four populations within South America (K2 - pink, K3 - dark green, K9 - light green, K10 - orange). IQTREE was used to generate ML trees using ModelFinder software (which calculated GTR + F + R10 as the model with the best fit), tree search, ultrafast bootstrap and SH-aLRT test. B) Principal component analysis of the pairwise distance matrix generated using the 102,765 SNP matrix from 315 South American isolates. Each point denotes a sample, which is coloured according to the country, as with the tree in A).
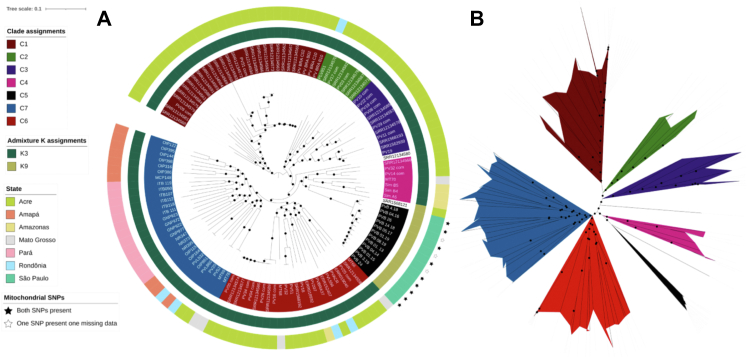
Fig. 3.
Population structure within Brazil isolates. Maximum likelihood tree using SNP data (70,757 unique SNPs) from 123 isolates from seven states within Brazil. A) Circular phylogenetic tree in iTOL, with outer colour strip coloured according to the state (Pará, n = 13; Amapá, n = 10; Mato Grosso, n = 5; Rondônia, n = 5; Acre, n = 74; Amazonas, n = 4; São Paulo, n = 12), the inner colour strip highlighting the ADMIXTURE population assignments from the global analysis (K3, n = 13; K9, n = 110), and the inner label coloured according to the 7 clades assigned based on the tree topology (C1, n = 29; C2, n = 8; C3, n = 12; C4, n = 7; C5, n = 12; C6, n = 24; C7, n = 29). Two isolates, SRR12134580 and SRR1568121 were not assigned a clade grouping. Isolates containing the two investigated mitochondrial SNPs (T4133C and A4467G in PvP01_MIT_v1) are labelled using a black star on the outer perimeter of the colour track; isolates containing both SNPs have a black filled in star, isolates where one SNP is present, but there is missing data for the other SNP are denoted with a white star with a black outline. B) Mid-point rooted visualisation of the same tree in A) to demonstrate clade groupings. The maximum likelihood tree for both plots was generated using IQTREE with ModelFinder software (which assigned TVM + F + R5 as the model with the best fit), tree search, ultrafast bootstrap and SH-aLRT test. Bootstrap values between 50% and 100% are indicated by a black circle midway along each branch length.
Loci informative for South American and Brazilian population differentiation
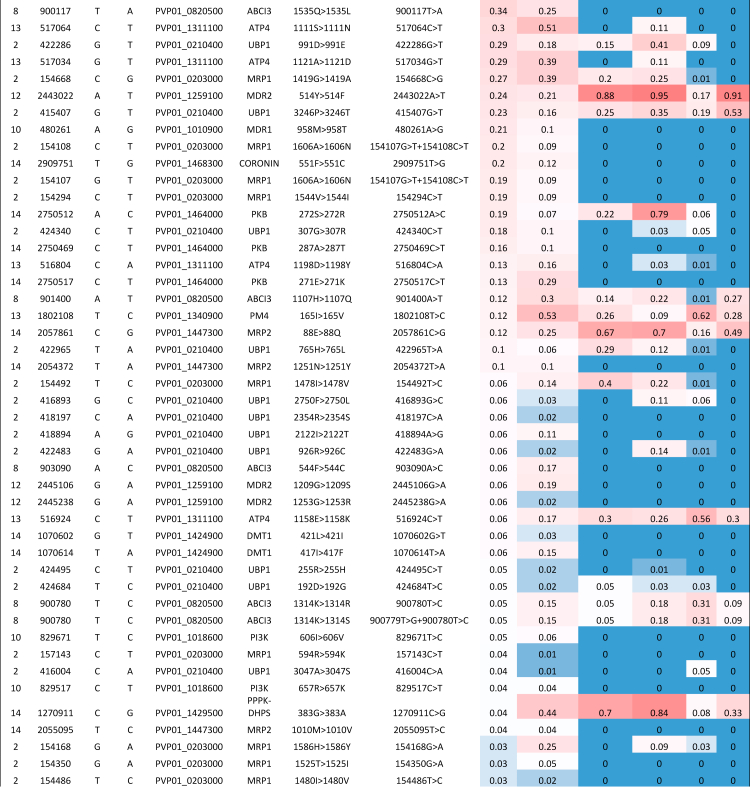
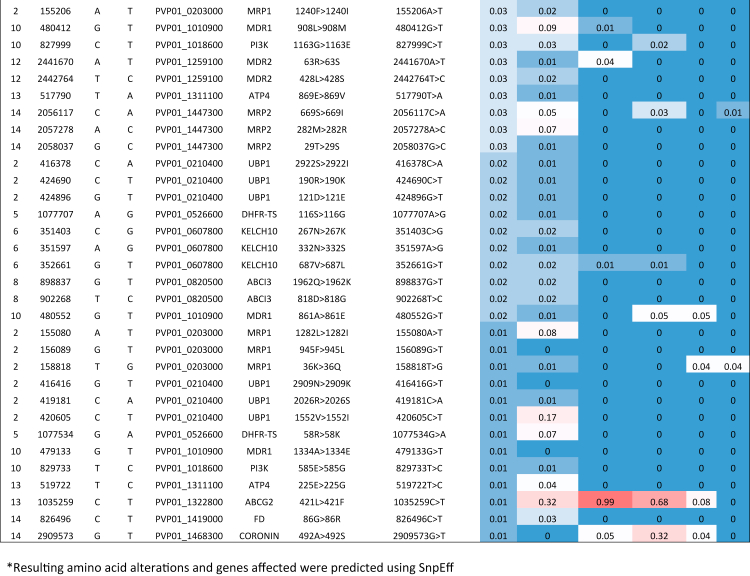
The fixation index (FST) was used to identify genes driving the P. vivax population differentiation for South American and its country-wide isolates. The isolates from São Paulo that may be P. simium infections were removed from further P. vivax population genetic analyses due to inconclusive species identification. When comparing South American isolates (n = 303) to other regions, the greatest number of highly differentiating (FST ≥0.99) SNPs are seen with SSEA (>150 SNPs with FST ≥0.99) (South Asia 44, Southeast Asia 37, East Africa 22) (Table S3). Across all pairwise regional comparisons, highly differentiating (FST ≥ 0.99) SNPs in South America were found in genes potentially involved in gene regulation (pvjmjc167), mosquito life stages (pvcrmp3, pvp28, pvp47, pvp48/4536,68, 69, 70), drug resistance (pvmdr171), gliding motility and cell traversal (pvtrap, pvtlp72,73), and those encoding parasite surface proteins (pvmsp1074) (Table S3). These genes overlapped with South American-specific differentiating SNPs (FST >0.9, vs. non-South American, n = 570; Table S4; Table 1). A nonsynonymous SNP leading to amino acid substitution 698S > 698G in pvmdr1, fixed in both the Brazilian and South American population, differentiated South American parasites from those in SSEA and SEA (FST = 1 and 0.99, respectively) in accordance with previous findings34 (Table 2, Table S3).
Table 1.
Non-synonymous SNPs with top 20 FST scores that differentiate P. vivax isolates from South America and Brazil.
| Region | Chr | Pos | Ref | Alt | Gene name | AA changea | Nucleotide change | Fstb |
|---|---|---|---|---|---|---|---|---|
| South America | 13 | 337753 | A | C | CRMP3 | 1719K > 1719N | 337753A > C | 0.999 |
| South America | 12 | 327858 | G | A | P48/45 | 418R > 418K | 327858G > A | 0.998 |
| South America | 11 | 1265741 | A | T | PVP01_1129500 | 236N > 236F | 1265740A > T+1265741A > T | 0.998 |
| South America | 11 | 1265741 | A | T | PVP01_1129500 | 236N > 236I | 1265741A > T | 0.998 |
| South America | 12 | 323603 | C | T | P47 | 24L > 24F | 323603C > T | 0.996 |
| South America | 11 | 1276259 | A | T | PVP01_1129700 | 2225T > 2225S | 1276259A > T | 0.993 |
| South America | 13 | 336962 | G | A | CRMP3 | 1456V > 1456M | 336962G > A | 0.992 |
| South America | 12 | 424886 | A | T | PVP01_1210400 | 195R > 195W | 424886A > T | 0.990 |
| South America | 14 | 1322275 | C | A | PVP01_1430500 | 1067L > 1067I | 1322275C > A | 0.986 |
| South America | 11 | 1272806 | G | A | PVP01_1129700 | 1074E > 1074K | 1272806G > A | 0.986 |
| South America | 4 | 652108 | G | C | P230p | 158L > 158V | 652108G > C | 0.985 |
| South America | 7 | 747984 | G | A | PVP01_0716800 | 575G > 575S | 747984G > A | 0.982 |
| South America | 9 | 1199491 | C | T | PVP01_0927300 | 572E > 572K | 1199491C > T | 0.980 |
| South America | 11 | 1483830 | A | G | PVP01_1134800 | 579K > 579R | 1483830A > G | 0.975 |
| South America | 14 | 2662867 | T | A | PVP01_1461600 | 403I > 403L | 2662867T > A | 0.973 |
| South America | 9 | 892855 | G | C | PVP01_0920500 | 1053Q > 1053H | 892855G > C | 0.973 |
| South America | 9 | 878674 | C | G | PVP01_0920200 | 517G > 517A | 878674C > G | 0.967 |
| South America | 11 | 1262951 | G | A | PVP01_1129400 | 20P > 20S | 1262951G > A | 0.966 |
| South America | 8 | 556253 | G | A | PVP01_0813100 | 150S > 150N | 556253G > A | 0.966 |
| South America | 11 | 1514101 | T | C | ApiAP2 | 319N > 319D | 1514101T > C | 0.962 |
| Brazil | 10 | 481636 | C | T | MDR1 | 500D > 500N | 481636C > T | 0.921 |
| Brazil | 12 | 1621163 | C | G | ApiAP2 | 869R > 869G | 1621163C > G | 0.895 |
| Brazil | 13 | 818665 | T | C | PVP01_1317400 | 39K > 39E | 818665T > C | 0.876 |
| Brazil | 13 | 809067 | G | A | PVP01_1317200 | 1086R > 1086Q | 809067G > A | 0.876 |
| Brazil | 5 | 440493 | T | C | NT2 | 117F > 117S | 440493T > C | 0.875 |
| Brazil | 2 | 377716 | C | A | PVP01_0209100 | 590G > 590V | 377716C > A | 0.869 |
| Brazil | 12 | 1618925 | A | G | ApiAP2 | 123I > 123V | 1618925A > G | 0.868 |
| Brazil | 4 | 530215 | T | C | PVP01_0412900 | 299E > 299G | 530215T > C | 0.860 |
| Brazil | 1 | 716831 | A | T | PVP01_0116000 | 4344L > 4344M | 716831A > T | 0.859 |
| Brazil | 12 | 1860075 | C | T | PVP01_1245000 | 1553A > 1553T | 1860075C > T | 0.853 |
| Brazil | 13 | 810706 | G | A | PVP01_1317200 | 1578G > 1578D | 810706G > A | 0.849 |
| Brazil | 11 | 915559 | G | T | PK4 | 1694T > 1694N | 915559G > T | 0.839 |
| Brazil | 6 | 179243 | A | T | PVP01_0604500 | 441L > 441M | 179243A > T | 0.835 |
| Brazil | 9 | 1366817 | C | G | SR1 | 236E > 236Q | 1366817C > G | 0.832 |
| Brazil | 14 | 2887017 | C | T | PVP01_1467700 | 33A > 33T | 2887017C > T | 0.830 |
| Brazil | 14 | 2153846 | G | T | PVP01_1449600 | 1581P > 1581T | 2153846G > T | 0.820 |
| Brazil | 10 | 490615 | C | G | PVP01_1011000 | 842G > 842A | 490615C > G | 0.818 |
| Brazil | 14 | 115657 | A | G | RBP2a | 719K > 719E | 115657A > G | 0.815 |
| Brazil | 13 | 336738 | C | T | CRMP3 | 1381P > 1381L | 336738C > T | 0.815 |
| Brazil | 7 | 360367 | A | C | PVP01_0706700 | 544K > 544Q | 360367A > C | 0.815 |
AA amino acid.
Within Region vs. all other isolates.
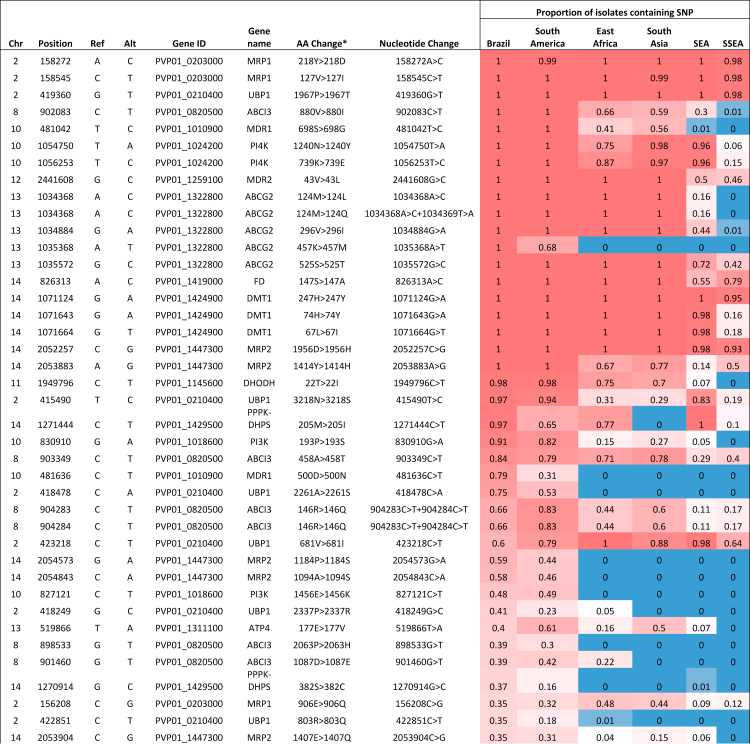
Table 2.
Mutations in putative drug resistance genes in Brazil, with reference to other regions across the globe.
Frequencies from 0 (blue) to 1 (red).
SEA, Southeast Asia; SSEA, Southern Southeast Asia.
aResulting amino acid alterations and genes affected were predicted using SnpEff software.
Within South America, Brazilian P. vivax (n = 111, vs. other South America, n = 192) informative SNPs (FST>0.8) were found within genes associated with drug susceptibility (pvmdr171), gene expression (pvapiap275), mosquito life stages (pvcrmp336) and a gene encoding reticulocyte binding protein, pvrbp2a76 (position 719K > 719E, FST = 0.82) (Table 1, Table S5). By focusing on country-level pairwise comparisons with Brazil, the highest number of differentiating SNPs (FST >0.95) were observed against Mexico (62 SNPs), followed by Panama (29 SNPs), Colombia (14 SNPs) and Peru (4 SNPs) (Table S6), consistent with differences in geographical distance and genetic clustering in the maximum likelihood tree and PCA analysis (Fig. 2). Of note, are putative drug resistance mutations, including within pvpppk-dhps (M205I: Brazil 97% vs. Mexico 0%; FST = 0.95; S. America 65%) previously observed in China77 and Thailand,78 and pvmdr1 (V221L: Panama 92% vs. Brazil 0%; FST = 0.97) previously observed in Peru79 (Table S6).
Distinct populations within Brazil associated with parasite surface proteins and drug resistance loci
All Brazilian regions have similarly low levels of nucleotide diversity (average π across all states, excluding São Paulo, 3.06 x 10–4), with the lowest diversity seen within São Paulo (π = 0.54 x 10−4) (Fig. S3). A SNP-based maximum likelihood tree (n = 70,757 SNPs) of only Brazilian isolates (n= 123) revealed seven distinct clades (C1–C7) (Fig. 3), including a likely P. simium clade with the samples from São Paulo (C5), corresponding to the ADMIXTURE population K9 (Fig. 3, Fig. S4). Clades C1, C2, C3, C4 and C6 mostly cover isolates from Acre and the Amazonas, with a small number of isolates from Rondônia in clades C2 (n = 1) and C6 (n = 2), and a small number of isolates from Mato Grosso in clades C4 (n = 1) and C6 (n = 2), demonstrating the vast genetic variability of isolates in the Amazon basin. Clade C7 covers isolates from Amapá and Pará located in northern Brazil, with a small number of isolates from Rondônia (n = 2), Acre (n = 2), and Mato Grosso (n = 2) (Fig. 3, Fig. S4). Two isolates from Acre did not fall into a clade grouping (Fig. 3). Whilst the population structure within Brazil appears to be complex, it is important to note that excluding those from São Paulo, all other Brazilian isolates clustered together as population K3 in the global ADMIXTURE analysis, which was a distinct population of Brazilian isolates (Fig. 3).
Informed by the population structure observed, subsequent analysis within Brazil compared different clades as well as two regional groups (A: Amazonas, Acre, Mato Grosso and Rondônia states (n = 88); B: Amapá and Pará states (n = 23)) (Fig. S5, Table S2). FST scores are heavily impacted by population size, therefore only clades with >15 isolates were compared to each other (excluding clades C2 to C4 from comparisons) (Table S7). Highly differentiating non-synonymous SNPs (FST >0.85) separating clades C1, C6 and C7 were identified (Table S7), including in many conserved proteins of unknown function and genes associated with reticulocyte binding (merozoite surface protein, pvmsp1),80 liver stages of infection (pvlisp281), and within a Plasmodium-specific ABC transporter (pvabci3) whose ortholog has been linked to a drug resistance mechanism in P. falciparum.82 Clades C6 and C7, which are associated with isolates from Acre and Amapá-Pará states respectively only have 11 highly differentiating mutations (FST >0.85), all synonymous SNPs. When comparing regional groups A and B, most highly differentiating SNPs were observed on chromosome 6 within the Plasmodium interspersed repeat gene family (pvpir). Pvpir genes are the largest gene family within Plasmodium spp (found within P. vivax, as well as simian and rodent malaria parasites), thought to play a role in host red blood cell invasion and immune evasion83 (Table S8).
Multi-clonality and signals of relatedness and homology within parasite populations
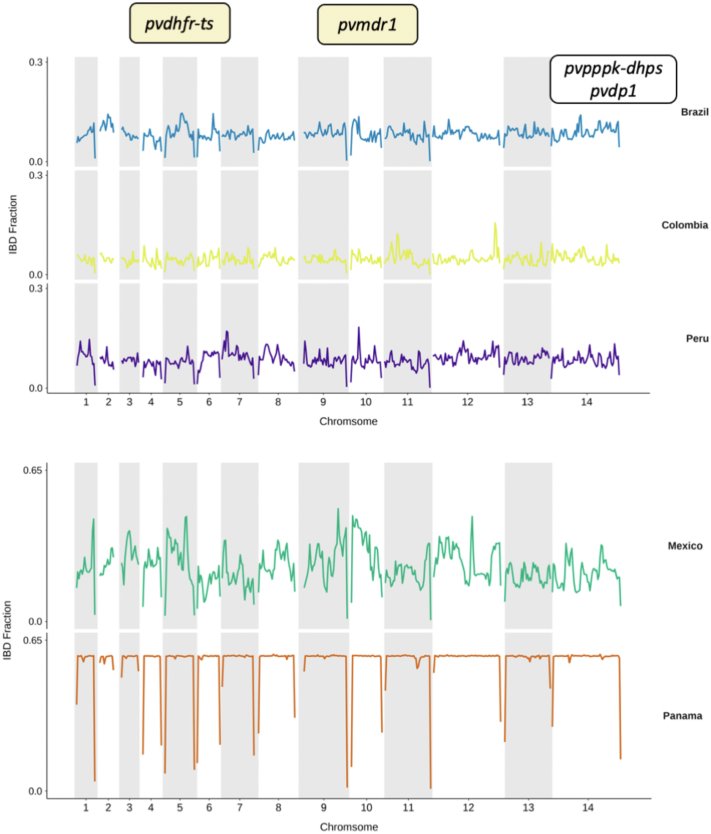
Multi-clonality, as measured by within-sample diversity (FWS metric < 95%), was present in 206 (23.2%) of all isolates, being more common among SEA (35.4%) and SSEA (33.7%), suggesting a higher chance of co-transmission of multiple P. vivax strains in these regions (Table S1, Fig. S6). In the Brazilian isolates, monoclonal infections were common, with FWS >0.95 observed in 87.8% of the 123 isolates. Multiclonality was more common within clades C4, C6 and C7 (28.6%, 20.8% and 24.1%, respectively, of all isolates with FWS <0.95). Multiclonality was also more likely in regional group B (30.4% of all isolates with FWS <0.95) than group A (9.1% isolates FWS <0.95) (Table S2, Fig. S7). Multiclonality appears less common in this analysis than previously presented for isolates from the region of Mancio Lima,44 which is likely due to differences in SNP filtering, where we perform FWS analysis on the already filtered dataset. Analysis of identity-by-descent (IBD), to quantify isolate relatedness, was performed at country level on the global dataset of monoclonal isolates (n = 679) (Table S1), and revealed Malaysia (median IBD 0.335), Panama (0.971) and Mexico (0.232) with the greatest fractions of IBD, with all other populations with fractions less than 0.0561 (Ethiopia 0.0561, Peru 0.0544, Colombia 0.0462, Brazil 0.0426, India 0.0236, Pakistan 0.0137, Cambodia 0.0123, Afghanistan 0.0121, Myanmar 0.00698, Papua New Guinea 0.00607) (Fig. S8, Table S9). Across genome-wide sliding windows of 50kbp, there are several global patterns of signals of high IBD (Table S10). In Brazil, a segment on chromosome 5 encompassing pvdhfr-ts, a gene associated with pyrimethamine resistance, demonstrates a high signal of IBD (0.122) (Fig. 4).39 Additionally, there is a segment of chromosome 10 encompassing pvmdr1, a gene associated with multi-drug resistance (Brazilian IBD = 0.136), which demonstrates a high signal also in East Africa (0.124) and South America (0.276)39 (Fig. 4). Brazil also has a high signal of IBD on chromosome 14, observed in other countries, where both pvdbp1, a gene associated with erythrocyte invasion,84 and pvdhps-pppk, a gene associated with sulfadoxine resistance,39 are found (Brazilian IBD = 0.133) (Fig. 4).
Fig. 4.
Country level comparisons of identity by descent (IBD) across the whole genome of monoclonal P. vivax isolates. IBD fractions along 50 kbp sliding windows across the genome at country level separation. The top 1% of IBD fractions for each country is summarised in Table S10. Genes of interest which demonstrate high signals of IBD are annotated. Where signals of high IBD are conserved across all countries within South America, the gene annotation is at the top of all plots and highlighted in yellow (pvdhfr-ts on chromosome 5 and pvmdr1 on chromosome 10). For country specific signals of high IBD where genes of interest are found, the gene annotation is found above the line graph for each country (pvpppk-dhps/pvdbp1 within chromosome 14 in Brazil).
We investigated patterns of IBD for clades C1, C6 and C7 (all with >15 isolates). Clade C1 specific signals of IBD were found within chromosomes 9 (encompassing pvama1, a potential vaccine candidate85) (IBD = 0.692), and 3 sequential segments within chromosome 14 (positions 2.35Mbp to 2.50Mbp), which included many genes, some of unknown function (Table S11). In this region a gene encoding the clustered-asparagine-repeat-protein (pvCARP) is also found, which is associated with the host immune response to malaria infection.86 For clade C6, signals were identified on chromosome 3, 5, 11, 12, 13 and 14 (IBD >0.3), encompassing a GPI-anchored micronemal antigen (pvGAMA) on chromosome 5 which is an essential invasion protein in P. falciparum infections, suggested as a potential vaccine candidate,87 and loci encoding putative AP2 domain transcription factors associated with gene regulation.75 Clade 7 IBD signals were found in chromosome 14, where both pvdbp1, a gene associated with P. vivax erythrocyte invasion,84 and pvpppk-dhps, a gene associated with sulfadoxine susceptibility,39 are located (IBD = 0.134) (Table S11). High signals of IBD were observed across all three clades (C1, C6 and C7) within chromosome 14 (average IBD = 0.329). Signals of IBD across the two geographical groupings (A, B) were polarizing, with signals in chromosomes 2 and 5 for Group A, including a region encompassing the eukaryotic initiation factor-2α, potentially associated with artemisinin resistance in Plasmodium parasites.88 For Group B, there were within segments of chromosome 14, including among other genes, the pvpppk-dhps, pvdbp1 and pvrbp1a, a gene associated with erythrocyte invasion89 (Table S12).
Regions under selection in South American and Brazilian subpopulations
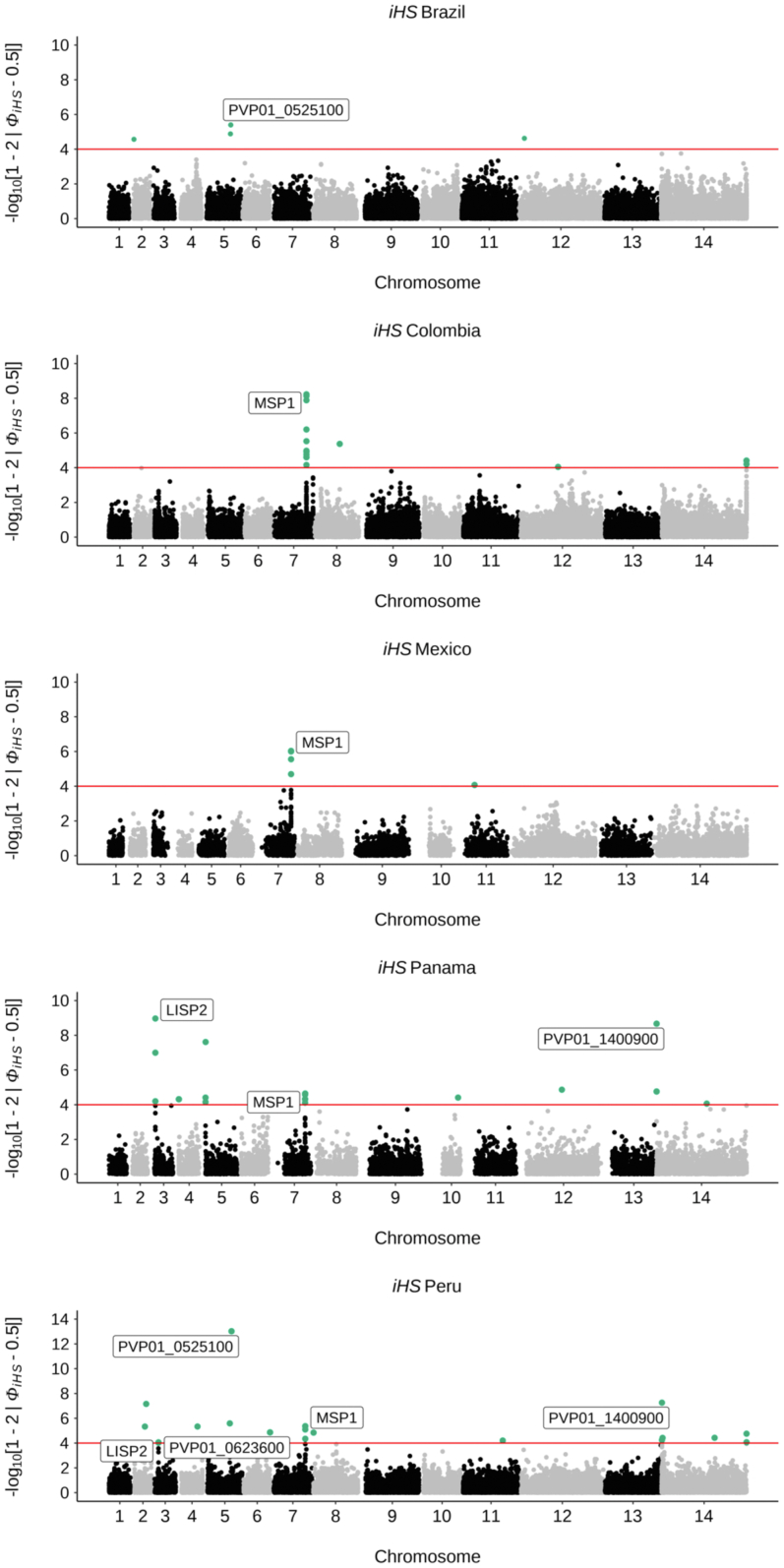
Genome-wide analysis to identify positive selective sweeps was performed using the “single population” integrated haplotype score (iHS) across monoclonal isolates (n = 679). Surface protein genes (pvmsp1, pvmsp4, pvmsp5) were detected in all global regions except for SSEA (Table S13). Within East Africa, South Asia and SEA, signals of positive selection were identified within chromosome 2 in a region that encompasses several genes including the pvmrp1, a gene associated with drug susceptibility.34 In both South Asia and SEA, signals of positive selection were found within chromosome 5, where the pvdhfr-ts gene is located. Loci associated with erythrocyte binding were also identified, including pvdbp1 in SEA (Table S13, Fig. S9). Across East Africa, South Asia, SEA, and SSEA, analyses detected signals of positive selection within chromosome 3 which includes pvlisp2, linked to parasite development in the liver.81 Signals of positive selection within South America include regions of the genome where multiple Plasmodium Poly-Helical Interspersed Sub-Telomeric (PHIST) proteins are encoded on chromosome 5. These proteins peripherally-localised in infected erythrocytes and in P. falciparum are involved in functions such as protein trafficking, membrane rigidity and intercellular signalling.90 Other loci identified included the leucine-rich repeat protein (pvlrr8) and the surface protein pvmsp1 along with a paralog pvmsp1p-1991 (Table S13). Within South America, we looked for signals of positive selection at the country level (for countries with >10 isolates). Signals were detected in pvmsp1 within Colombia, Panama, and Peru, in pvdbp1 within Peru, and in pvlisp2 within both Panama and Peru (Fig. 5, Table S14). There were only 5 SNPs detected within Brazil which demonstrated signals of positive selection, with just 2 SNPs in coding regions (Plasmodium exported protein PVP01_0525100, pvphist) (Fig. 5, Table S15). Within Brazil, signals of positive selection using iHS were detected in chromosomes 8 and 14 in Amazonian clade C6, including the ABC-transporter pvabci3 (PVP01_082050), whose orthologue is associated with drug resistance in P. falciparum.92 In Clade C7 isolates (associated with Amapá and Pará states) candidate regions for positive selection were seen within loci encompassing surface proteins (e.g., pvmsp1, pvmsp4, pvmsp5), pvlisp2, pvlrr8 and pvdbp involved in erythrocyte invasion (Table S16).
Fig. 5.
Evidence of selection (iHS) within South American countries. Genome-wide iHS scores in a Manhattan plot for all countries within South America where there are >10 isolates. SNPs within genes with iHS score of P< 1 × 10−4 are highlighted in green and gene names are annotated for candidate regions of high iHS for genes with validated functions (MSP1, PVP01_0728900; LISP2, PVP01_0304700). For expanded gene families and genes with unknown functions, gene IDs are given (PVP01_0525100, PHIST protein; PVP01_1400900, exported plasmodium protein of unknown function (PUF); PVP01_0623600, PIR protein. Raw outputs of iHS scores, alongside proposed candidate regions for South America and Brazil specifically are summarised in Tables S14–S16.
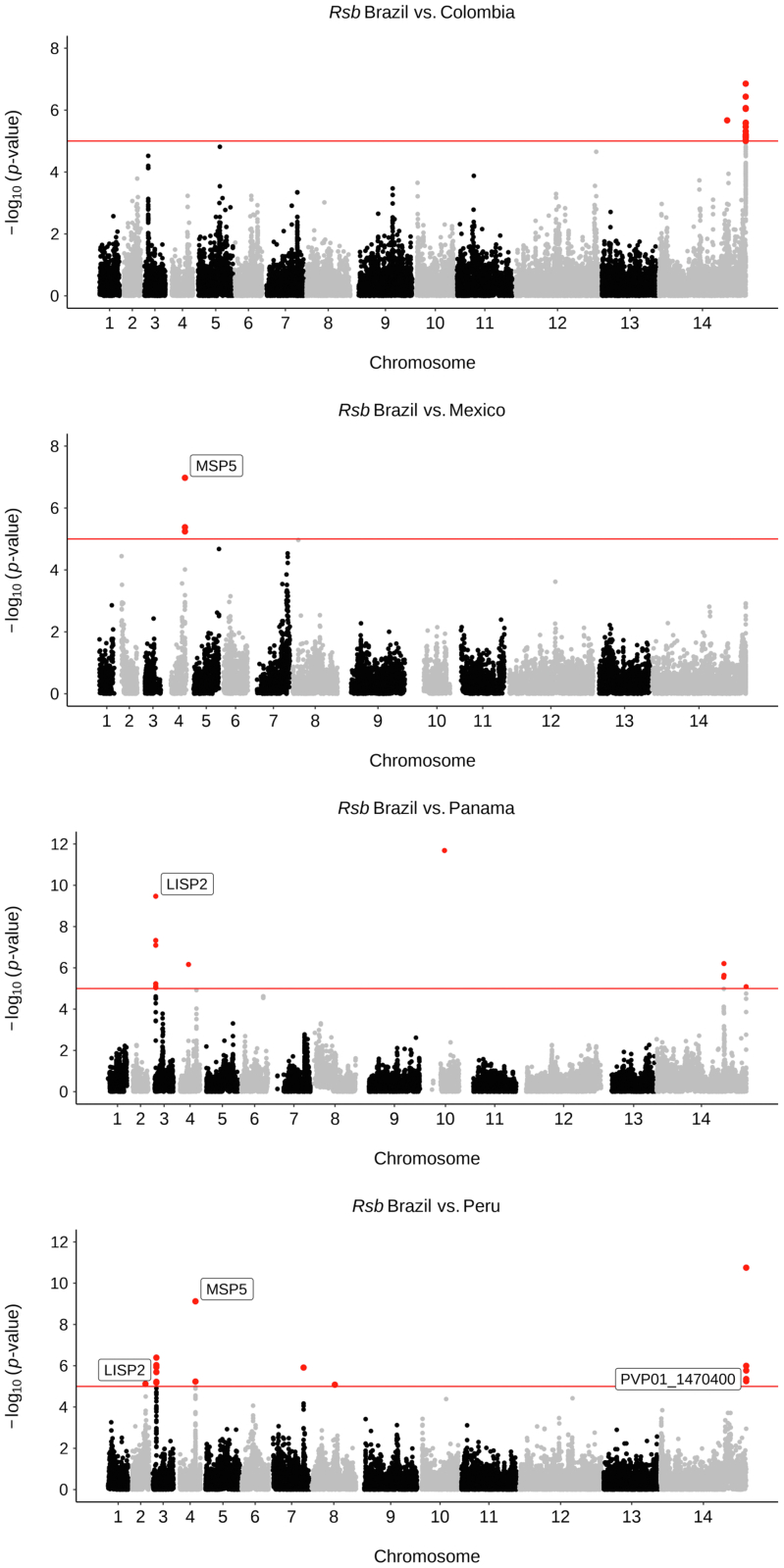
The between population Rsb method was applied to detect positive selection at both the regional and country level (Table S17; Table S18; P < 1 × 10−5). When comparing South America against the other global regions, multiple SNPs within pvmsp1, associated with reticulocyte binding,80 demonstrated signs of positive selection. Similarly, SNPs in the gene encoding PvPHIST exported protein were also detected in all pairwise comparisons, except with SSEA. The surface protein encoding gene pvmsp5 (chromosome 4) was detected between South America and SEA. Comparisons of Brazil to other South American countries revealed multiple SNPs within pvlisp2 (Brazil vs. Panama, Peru), pvmsp5 (Brazil vs. Mexico, Peru) and pvmsp1, pvmsp4 and pvmsp5 (Brazil vs. Peru) (Fig. 6, Table S18, Table S19). Within Brazil, two candidate genomic regions were detected between clades C1 and C6, where surface proteins were found (e.g., pvmsp4, pvmsp5), in addition to two regions between clades C1 and C7 (including pvsmp1, pvmsp1p, and pvlrr8). Five candidate genetic regions were identified when comparing clades C6 and C7, which included the pvlisp2 gene and multiple merozoite surface proteins. Between regional groups A and B, 5 candidate regions were identified, which included the pvlisp2, pvdbp and pvmsp1 genes (Table S20).
Fig. 6.
Evidence of selection between countries in South America (Rsb). Manhattan plots for genome-wide Rsb analysis for P. vivax isolates within South America at the country level. SNPs with P < 1 × 10−5 are highlighted in red, and gene names are annotated for candidate regions of selection. Gene names are given for genes with validated locations and functions (MSP5, PVP01_0418400; LISP2, PVP01_0304700), whereas gene IDs are given for genes within expanded gene families or genes with unknown functions (PVP01_1470400, exported PUF). Rsb results output for South America, in addition to within Brazil analyses are summarised in Tables S17–S20.
In addition to positive selection signals, we investigated genes (with >5 SNPs) under balancing selection by applying the Tajima's D statistic to all monoclonal isolates (n = 679). As expected, most Tajima's D values for genes across global regions were negative (median: South America −0.437, SEA -1.82, South Asia −0.756, East Africa −0.385, SSEA -0.904), with the most negative value globally occurring in SEA, suggesting population expansion in this region (Fig. S10). Within South America, median values for Tajima's D were negative in Brazil (−0.034), Colombia (−0.046) and Panama (−0.330), while positive in Mexico (0.173) and Peru (0.078) indicating a population decrease or a genetic bottleneck (Fig. S11). The top 50 genes with the highest and lowest Tajima's D metric in South America are summarised (Table S21), with the most positive including pvmsp1, pvmsp5, pvlisp2, pvrbp1a, and many genes encoding exported proteins, including PvPHIST, suggesting balancing selection. The findings from the same analysis for Brazil overlapped, and includes genes pvmsp1, pvlisp2, pvrbp1a, and pvcyrpa, in addition to loci encoding PvPHIST and PvPIR proteins (Table S22).
Identification of mutations and allele frequencies in P. vivax drug resistance candidate genes
Treatment failures have been reported with P. vivax infections, however the molecular determinants for reduced drug efficacy are not clearly defined. We investigated the presence of SNPs within orthologs of genes associated with resistance in P. falciparum, alongside loci identified by selection metrics, including hits from previous population genomics studies34,40,50 (Table 2, Table S23). There are similar patterns of frequencies of SNPs within potential resistance-associated genes between Brazil and the other South American isolates likely due to similar drug regimens across this region. Of note are SNPs which appear close to fixation within the Brazilian population, found within pvubp1 (potentially associated with artemisinin resistance in P. falciparum93), multidrug resistance associated proteins MDR1, MDR2, MRP2, phosphatidylinositol 4-kinase pvpi4k (the target of novel antimalarial class imidazopyrazines94), DHODH (a drug target for DSM265, a novel antimalarial in clinical trials, shown to be less effective against P. vivax infections than P. falciparum95), ferredoxin – pvfdx (potentially associated with artemisinin resistance in P. falciparum96), pvpppk-dhps (associated with sulfadoxine resistance39), and genes coding for putative ABC transporters (pvabci3, pvabcg2), whose orthologues may be associated with antimalarial resistance in P. falciparum infections71,92 (Table 2). Some of these mutations are observed in high frequency in South America but have quite different frequencies compared with other global regions, including a missense mutation within pvmdr1 (698S > 689G), which is fixed in all South American isolates, and found in approximately half of the populations in East Africa and South Asia, but rare and non-existent in SEA and SSEA respectively. Another pvmdr1 mutation (500D > 500N) is also present in Brazil with high frequency (80%) but with lower frequency in wider South America (31%) and not identified in any other continents. Similarly, a missense mutation within pvabcg2 (457K > 457M) is fixed within Brazil and present in South America (69%), but not observed elsewhere (Table 2). Pvabcg2 encodes an ATP binding cassette (ABC) transporter, which are commonly known to be associated with multiple drug resistance phenotypes in many organisms 74 and is linked to the gametocyte stages of parasite development.97
Discussion
Whilst P. vivax infections pose a serious risk to global health, genomic analyses of this species, particularly in South America where the parasite is predominant, are scarce in comparison to the more pathogenic P. falciparum. Brazil is a unique setting for malaria transmission, with distinct foci relating to the local environments and resultant vector landscapes. To date, all previously published WGS data from Brazil has originated from isolates obtained mainly from Acre and a few from Rondônia, in the north-western region of the country. Previous population genomic analyses have demonstrated that South American isolates (n = 146) are a distinct population with high genetic diversity,28 with three ancestral populations (Mexico, Peru, Colombia/Brazil),34 in contrast to our study, which reveals four main populations (n = 315; Brazil, Mexico/Colombia, Peru, Panama). Previous work has revealed geographical clustering of isolates from Brazil and Peru,28 but whilst closely related in our analysis, they are distinct. Earlier work focused solely on Mancio Lima, and found high levels of inbreeding.44 Studies of P. vivax from 4 countries (Brazil, Colombia, PNG, India), using microsatellite markers, have demonstrated high similarity between isolates from Brazil (Manaus) and India (Bikaner), and high genetic diversity irrespective of the transmission situation.98 Microsatellite data has also shown high diversity within and between Amazon parasite populations (Manaus, Porto Velho), with Amapa and Para infections being the most divergent,99 consistent with our findings that also suggest these two states are a distinct diverged clade.
Here, we provide the first insight into the genomic diversity of P. vivax isolates from all three malaria endemic regions in Brazil, spanning seven states, to determine the broader population structure within the country, as well as its position within a continental and global resolution. Using 855 global isolates of P. vivax across 26 countries, we placed South America in the global context, demonstrating that they form a distinct population with more ancestral populations than other global regions. The four distinct ancestral South American populations mostly correspond to country groups, in accordance with previous studies demonstrating nation-level separation within this continent.28,34 Using 123 isolates from Brazil, we demonstrated that the population structure is complex, with samples clustering across seven distinct clades, clearly separating the Northern states (Amapá and Pará) and the highly clonal potential P. simium cluster from São Paulo. Isolates from the Amazonian basin fall within five (of the seven) clades, consistent with the high malaria transmission in the large region leading to greater population diversity.
WGS data can reveal genetic differences within and between populations, which may be indicative of signals of differential selection, including those resulting from differences in the implementation of antimalarial drug regimens. For example, artemisinin resistance in P. falciparum isolates was confirmed through detecting signals of selection between populations around the Pfkelch13 gene, agnostic to a resistance phenotype, and mutations in that locus were found to correlate with differences in parasite clearance rates after treatment with artemisinin.100,101 It is therefore possible that genome-wide screens of selection for P. vivax may reveal much needed novel candidates of drug resistance. In this context, the monitoring for signals of selection may inform on the effectiveness of control measures, but can also reveal important insights into patterns of parasite adaptation. Understanding the genetic differences across parasite populations can inform on the origin of parasites, leading to the development of molecular barcodes for both P. falciparum102 and P. vivax parasites33 to accurately predict the source of infections, including importation events. These geographically informative molecular barcodes can be used as an easier alternative to WGS for determining patterns of parasite transmission, and predicting the source of infection outbreaks, which can be extremely useful in countries nearing elimination to determine between native transmission and imported malaria.
Using comparative population genomics, our results highlight many South American-specific SNPs within genes involved in different parasite life stages and associated with drug resistance. Genes involved in mosquito life stages, such as gametocyte proteins PVS48/45 and PVS47, may be reflective of the different mosquito vectors present in South America compared to other regions, and could be potential molecular barcode candidates for identifying parasites originating in South America. Other studies have also identified mosquito-related proteins under selection in other P. vivax endemic regions.34,40,50 Additionally, South American-specific SNPs were also found within genes encoding parasite surface proteins (e.g., pvmsp1/4/5) and drug resistance (e.g., pvmdr1). Several signals of selection and homology were identified in loci associated with drug resistance, specifically within pvdhfr-ts and pvmdr1 across all South American isolates, which may reflect similar selection pressures due to a like drug regimens within continent. In addition, a Brazilian-specific signal was observed within pvdhps-pppk, the determinant of sulfadoxine resistance in P. falciparum. Sulfadoxine resistance in P. vivax has been reported in South America but, in contrast to P. falciparum, the molecular marker has not been confirmed, in part due to the lack of an in vitro culture method for P. vivax. Other possible candidates for further investigation linked to antimalarial drugs included pvmrp1, pvmrp2, and an ABC transporter I family member (pvabci3), revealed as signals of positive selection and/or SNPs fixed in Brazilian samples. The orthologous pfmrp1 gene in P. falciparum is a multidrug-resistance candidate, and has been shown to be under strong selection in across populations, with mutations associated with reduced susceptibility to sulfadoxine-pyrimethamine, chloroquine and mefloquine, and pyronaridine.103 The ABCI3 protein is a Plasmodium-specific ABC family member, and SNP and gene amplification variants in P. falciparum have recently been shown to confer anti-plasmodial drug resistance across a variety of compounds.92 However, no such investigations of pvabci3 have been applied to P. vivax.
Determining the downstream effect of SNPs for P. vivax research is complicated due to the lack of a routine in vitro culture method for this parasite species. It is possible to perform orthologue replacement transgenesis in P. knowlesi104 as this parasite can be cultured in human erythrocytes105 and is the most closely related species to P. vivax. This system allows the functional investigation of the role of genetic variants, such as in drug susceptibility or red blood cell invasion. Brazil-specific SNPs in genes involved in red blood cell invasion (pvrbp2a, pvrbp1, pvcyrpa), and signals of positive selection in PHIST family members were also detected, which may reflect immune selective pressure or regional-specific host factors on erythrocytes. Invasion genes are possible P. vivax vaccine candidates, and understanding the genetic diversity of these loci across global populations can inform on their potential efficacy.
The liver stage pvlisp2 gene, which differentiates between dormant hypnozoites and early developing parasites,81 was identified when investigating signals of selection across South American populations and within Brazilian clades. Genetic markers in pvlisp2 can assist the development of drug discovery assays predictive of anti-relapse activity.81
Overall, our work provides insights into the genomic diversity across all three malaria endemic regions in Brazil, as well as in the broader context of South America and other continents. The results highlight many novel and previously detected genes and mutations, which may reflect ongoing evolutionary interactions with the vector and human hosts in the different regional settings and in response to antimalarial drugs. Our insights will inform functional studies, which can determine the role of the candidate loci during the parasite life cycle and in response to treatment and anti-relapse therapies. Ultimately, this work will assist with the design of much needed tools for infection control, ultimately working towards malaria elimination.
Contributors
TGC and SC conceived and directed the project. SS, AC, ATC, DF, GVT, FN, KS, DN, CJS, JD, MSM, DBP, CM, ARSB, RLDM, and SMS organised sample collection and processing. AI, MC, and SC undertook laboratory work including sequencing. AI and EM performed bioinformatic analysis under the supervision of SC and TGC, and together they interpreted the results. EDB provided software. AI, TGC, and SC wrote the first draft of the manuscript. All authors commented on the results and on the manuscript and approved the final submission.
Data sharing statement
Raw sequence data is available from the European Nucleotide Archive under project code PRJEB56411 (see Table S2 for accession numbers of novel Brazilian isolates). The dataset also includes sequenced isolates from the MalariaGEN P. vivax Genome Variation project and described elsewhere.32,34,40,42, 43, 44
Editor note
The Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
Declaration of interests
The authors have declared that no competing interests exist.
Acknowledgements
AI is funded by an MRC LiD PhD studentship. TGC is funded by the Medical Research Council UK (Grant no. MR/M01360X/1, MR/N010469/1, MR/R025576/1, MR/R020973/1 and MR/X005895/1). SC is funded by Medical Research Council UK grants (MR/M01360X/1, MR/R025576/1, MR/R020973/1 and MR/X005895/1) and Bloomsbury SET (Ref. CCF17-7779). The Shloklo Malaria Research Unit is part of the Mahidol Oxford Research Unit, supported by the Wellcome Trust (Grant no.220211). Further funding support was available by FAPESP (Process 02/9546–1), CNPq (Process 302353/2003–8 and process 471605/2011–5) to ARSB and RLDM. CRFM is funded by São Paulo Research Foundation - FAPESP (Grant no. 2020/06747–4) and Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq (Grant no. 302917/2019–5 and 408636/2018–1). JGD is funded by fellowships from FAPESP (2016/13465–0 and 2019/12068–5) and CNPq (Grant no. 409216/2018–6). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.lana.2022.100420.
Contributor Information
Taane G. Clark, Email: taane.clark@lshtm.ac.uk.
Susana Campino, Email: Susana.campino@lshtm.ac.uk.
Appendix A. Supplementary data
References
- 1.WHO . 2020. World Malaria Report 2020. [Google Scholar]
- 2.Howes R.E., Battle K.E., Mendis K.N., et al. Global epidemiology of Plasmodium vivax. Am J Trop Med Hyg. 2016;95:15–34. doi: 10.4269/ajtmh.16-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandre M.A., Ferreira C.O., Siqueira A.M., et al. Severe Plasmodium vivax malaria, Brazilian Amazon. Emerg Infect Dis. 2010;16:1611–1614. doi: 10.3201/eid1610.100685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization Guidelines for the treatment of malaria. 2015. https://books.google.co.uk/books?hl=en&lr=&id=IVo0DgAAQBAJ&oi=fnd&pg=PP1&ots=9Ukc5pR7eP&sig=ZCxSqmVQZmlgUmQCjHD3tvxL-sc&redir_esc=y#v=onepage&q&f=false 3rd ed. Available at:
- 5.Recht J., Siqueira A., Monteiro W.M., et al. Malaria in Brazil, Colombia, Peru and Venezuela: current challenges in malaria control and elimination. Malar J. 2017;16:1–18. doi: 10.1186/s12936-017-1925-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almeida A.C.G., Kuehn A., Castro A.J.M., et al. High proportions of asymptomatic and submicroscopic Plasmodium vivax infections in a peri-urban area of low transmission in the Brazilian Amazon. Parasites Vectors. 2018;11:1–13. doi: 10.1186/s13071-018-2787-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacDonald A.J., Mordecai E.A. Amazon deforestation drives malaria transmission, and malaria burden reduces forest clearing. Proc Natl Acad Sci USA. 2019;116:22212–22218. doi: 10.1073/pnas.1905315116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz G., Lasso A.M., Murillo C., Montenegro L.M., Echeverry D.F. Evidence of self-medication with chloroquine before consultation for malaria in the southern pacific coast region of Colombia. Am J Trop Med Hyg. 2019;100:66. doi: 10.4269/ajtmh.18-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douine M., Lazrec Y., Blanchet D., et al. Predictors of antimalarial self-medication in illegal gold miners in French Guiana: a pathway towards artemisinin resistance. J Antimicrob Chemother. 2018;73:231–239. doi: 10.1093/jac/dkx343. [DOI] [PubMed] [Google Scholar]
- 10.Gonçalves L.A., Cravo P., Ferreira M.U. Emerging Plasmodium vivax resistance to chloroquine in South America: an overview. Mem Inst Oswaldo Cruz. 2014;109:534–539. doi: 10.1590/0074-0276130579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlos B.C., Rona L.D.P., Christophides G.K., Souza-Neto J.A. A comprehensive analysis of malaria transmission in Brazil. Pathog Glob Health. 2019;113:1–13. doi: 10.1080/20477724.2019.1581463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Alvarenga D.A.M., de Pina-Costa A., de Sousa T.N., et al. Simian malaria in the Brazilian Atlantic forest: first description of natural infection of capuchin monkeys (Cebinae subfamily) by Plasmodium simium. Malar J. 2011;14 doi: 10.1186/s12936-015-0606-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brasil P., Zalia M.G., de Pina-Costa A., et al. Outbreak of human malaria caused by Plasmodium simium in the Atlantic Forest in Rio de Janeiro: a molecular epidemiological investigation. Lancet Glob Heal. 2017;5:e1038–e1046. doi: 10.1016/S2214-109X(17)30333-9. [DOI] [PubMed] [Google Scholar]
- 14.Langhi D.M., Bordin J.O. Duffy blood group and malaria. Hematology. 2006;11:389–398. doi: 10.1080/10245330500469841. [DOI] [PubMed] [Google Scholar]
- 15.Howes R.E., Patil A.P., Piel F.B., et al. The global distribution of the Duffy blood group. Nat Commun. 2011;2(1):1–10. doi: 10.1038/ncomms1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langhi D.M., Jr, Albuquerque S., Serafim R., et al. Serological and molecular study of the duffy blood group among malarial endemic region residents in Brazil. J Brazilian Soc Trop Med. 2022;55 doi: 10.1590/0037-8682-0490-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmerman P.A. Plasmodium vivax infection in duffy-negative people in Africa. Am J Trop Med Hyg. 2017;97:636–638. doi: 10.4269/ajtmh.17-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ménard D., Barnadas C., Bouchier C., et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci USA. 2010;107:5967–5971. doi: 10.1073/pnas.0912496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendes C., Dias F., Figueiredo J., et al. Duffy negative antigen is no longer a barrier to Plasmodium vivax--molecular evidences from the African West Coast (Angola and Equatorial Guinea) PLoS Negl Trop Dis. 2011;5 doi: 10.1371/journal.pntd.0001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niang M., Sane R., Sow A., et al. Asymptomatic plasmodium vivax infections among duffy-negative population in Kedougou, Senegal. Trop Med Health. 2018;46:1–5. doi: 10.1186/s41182-018-0128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavasini C.E., de Mattos L.C., Couto A.U.D., et al. Plasmodium vivax infection among Duffy antigen-negative individuals from the Brazilian Amazon region: an exception? Trans R Soc Trop Med Hyg. 2007;101:1042–1044. doi: 10.1016/j.trstmh.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 22.Abate A., Bouyssou I., Mabilotte S., et al. Vivax malaria in Duffy-negative patients shows invariably low asexual parasitaemia: implication towards malaria control in Ethiopia. Malar J. 2022;21(1):1–10. doi: 10.1186/s12936-022-04250-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abou-Ali R.K., Dhyani A., Terço A.L., et al. Impact of Duffy polymorphisms on parasite density in Brazilian Amazonian patients infected by Plasmodium vivax. Malar J. 2019;18:1–9. doi: 10.1186/s12936-019-2918-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Oliveira Padilha M.A., de Oliveira Melo J., Romano G., et al. Comparison of malaria incidence rates and socioeconomic-environmental factors between the states of Acre and Rondônia: a spatio-Temporal modelling study. Malar J. 2019;18:306. doi: 10.1186/s12936-019-2938-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu C.S., White N.J. The prevention and treatment of Plasmodium vivax malaria. PLoS Med. 2021;18 doi: 10.1371/journal.pmed.1003561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gething P.W., Van Boeckel T.P., Smith D.L., et al. Modelling the global constraints of temperature on transmission of Plasmodium falciparum and P. vivax. Parasit Vectors. 2011;4 doi: 10.1186/1756-3305-4-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Battle K.E., Gething P.W., Elyazar I.R.F., et al. The global public health significance of Plasmodium vivax. Adv Parasitol. 2012;80 doi: 10.1016/B978-0-12-397900-1.00001-3. [DOI] [PubMed] [Google Scholar]
- 28.de Oliveira T.C., Rodrigues P.T., Menezes M.J., et al. Genome-wide diversity and differentiation in New World populations of the human malaria parasite Plasmodium vivax. PLoS Negl Trop Dis. 2017;11 doi: 10.1371/journal.pntd.0005824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cruz Marques A. Human migration and the spread of malaria in Brazil. Parasitol Today. 1987;3:166–170. doi: 10.1016/0169-4758(87)90170-0. [DOI] [PubMed] [Google Scholar]
- 30.Taylor J.E., Pacheco M.A., Bacon D.J., et al. The evolutionary history of Plasmodium vivax as inferred from mitochondrial genomes: parasite genetic diversity in the Americas. Mol Biol Evol. 2013;30:2050. doi: 10.1093/molbev/mst104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues P.T., Valdivia H.O., de Oliveira T.C., et al. Human migration and the spread of malaria parasites to the New World. Sci Rep. 2018;8:1–13. doi: 10.1038/s41598-018-19554-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hupalo D.N., Luo Z., Melnikov A., et al. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat Genet. 2016;48:953–958. doi: 10.1038/ng.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diez Benavente E., Campos M., Phelan J., et al. A molecular barcode to inform the geographical origin and transmission dynamics of Plasmodium vivax malaria. PLoS Genet. 2020:1–19. doi: 10.1371/journal.pgen.1008576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benavente E.D., Manko E., Phelan J., et al. Distinctive genetic structure and selection patterns in Plasmodium vivax from South Asia and East Africa. Nat Commun. 2021;12 doi: 10.1038/s41467-021-23422-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J., Fernandez-Reyes D., Sharling L., et al. Plasmodium cysteine repeat modular proteins 1-4: complex proteins with roles throughout the malaria parasite life cycle. Cell Microbiol. 2007;9:1466–1480. doi: 10.1111/j.1462-5822.2006.00885.x. [DOI] [PubMed] [Google Scholar]
- 36.Douradinha B., Augustijn K.D., Moore S.G., et al. Plasmodium Cysteine Repeat Modular Proteins 3 and 4 are essential for malaria parasite transmission from the mosquito to the host. Malar J. 2011;10 doi: 10.1186/1475-2875-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tachibana M., Suwanabun 2 N., Kaneko O., et al. Plasmodium vivax gametocyte proteins, Pvs48/45 and Pvs47, induce transmission-reducing antibodies by DNA immunization. Vaccine. 2015;33:1901–1908. doi: 10.1016/j.vaccine.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Price R.N., Cassar C., Brockman A., et al. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob Agents Chemother. 1999;43:2943–2949. doi: 10.1128/aac.43.12.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buyon L.E., Elsworth B., Duraisingh M.T. The molecular basis of antimalarial drug resistance in Plasmodium vivax. Int J Parasitol Drugs Drug Resist. 2021;16:23–37. doi: 10.1016/j.ijpddr.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearson R.D., Amato R., Auburn S., et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet. 2016;48:959–964. doi: 10.1038/ng.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Oliveira T.C., Corder R.M., Early A., et al. Population genomics reveals the expansion of highly inbred Plasmodium vivax lineages in the main malaria hotspot of Brazil. PLoS Negl Trop Dis. 2020;14(10) doi: 10.1371/journal.pntd.0008808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Auburn S., Getachew S., Pearson R.D., et al. Genomic analysis of plasmodium vivax in southern Ethiopia reveals selective pressures in multiple parasite mechanisms. J Infect Dis. 2019;220(11) doi: 10.1093/infdis/jiz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Auburn S., Benavente E.D., Miotto O., et al. Genomic analysis of a pre-elimination Malaysian Plasmodium vivax population reveals selective pressures and changing transmission dynamics. Nat Commun. 2018;9(1):1–12. doi: 10.1038/s41467-018-04965-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Oliveira T.C., Corder R.M., Early A., et al. Population genomics reveals the expansion of highly inbred Plasmodium vivax lineages in the main malaria hotspot of Brazil. PLoS Negl Trop Dis. 2020;14:1–16. doi: 10.1371/journal.pntd.0008808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cowell A.N., Loy D.E., Sundararaman S.A., et al. Selective whole-genome amplification is a robust method that enables scalable whole-genome sequencing of Plasmodium vivax from unprocessed clinical samples. mBio. 2017;8:1–15. doi: 10.1128/mBio.02257-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ibrahim A., Benavente E.D., Nolder D., et al. Selective whole genome amplification of Plasmodium malariae DNA from clinical samples reveals insights into population structure. Sci Rep. 2020;10:1–11. doi: 10.1038/s41598-020-67568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Auburn S., Böhme U., Steinbiss S., et al. A new Plasmodium vivax reference sequence with improved assembly of the subtelomeres reveals an abundance of pir genes. Wellcome Open Res. 2016;1:4. doi: 10.12688/wellcomeopenres.9876.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benavente E.D., Ward Z., Chan W., et al. Genomic variation in Plasmodium vivax malaria reveals regions under selective pressure. PLoS One. 2017;12:1–15. doi: 10.1371/journal.pone.0177134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DePristo M.A., Banks E., Poplin R., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cingolani P., Platts A., Wang L.L., et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Assefa S.A., Preston M.D., Campino S., et al. EstMOI: estimating multiplicity of infection using parasite deep sequencing data. Bioinformatics. 2014;30:1292–1294. doi: 10.1093/bioinformatics/btu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen L.T., Schmidt H.A., Von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Danecek P., Auton A., Abecasis G., et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gautier M., Klassmann A., Vitalis R. Rehh 2.0: a reimplementation of the R package rehh to detect positive selection from haplotype structure. Mol Ecol Resour. 2017;17:78–90. doi: 10.1111/1755-0998.12634. [DOI] [PubMed] [Google Scholar]
- 58.Voight B.F., Kudaravalli S., Wen X., Pritchard J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang K., Thornton K.R., Stoneking M. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 2007;5:e171. doi: 10.1371/journal.pbio.0050171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miles A., Iqbal Z., Vauterin P., et al. Indels, structural variation, and recombination drive genomic diversity in Plasmodium falciparum. Genome Res. 2016;26:1288–1299. doi: 10.1101/gr.203711.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daron J., Boissière A., Boundenga L., et al. Population genomic evidence of Plasmodium vivax Southeast Asian origin. Sci Adv. 2021;7:3713–3741. doi: 10.1126/sciadv.abc3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaffner S.F., Taylor A.R., Wong W., Wirth D.F., Neafsey D.E. hmmIBD: software to infer pairwise identity by descent between haploid genotypes. Malar J. 2018;17:196. doi: 10.1186/s12936-018-2349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bright A.T., Manary M.J., Tewhey R., et al. A high resolution case study of a patient with recurrent Plasmodium vivax infections shows that relapses were caused by meiotic siblings. PLoS Negl Trop Dis. 2014;8 doi: 10.1371/journal.pntd.0002882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paradis E. Pegas. An R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- 65.Buyon L.E., Santamaria A.M., Early A.M., et al. Population genomics of Plasmodium vivax in Panama to assess the risk of case importation on malaria elimination. PLoS Negl Trop Dis. 2020;14 doi: 10.1371/journal.pntd.0008962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mourier T., de Alvarenga D.A.M., Kaushik A., et al. The genome of the zoonotic malaria parasite Plasmodium simium reveals adaptations to host-switching. bioRxiv. 2019;19:841171. doi: 10.1186/s12915-021-01139-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cui L., Fan Q., Cui L., Miao J. Histone lysine methyltransferases and demethylases in Plasmodium falciparum. Int J Parasitol. 2008;38:1083–1097. doi: 10.1016/j.ijpara.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tomas A.M., Margos G., Dimopoulos G., et al. P25 and P28 proteins of the malaria ookinete surface have multiple and partially redundant functions. EMBO J. 2001;20:3975. doi: 10.1093/emboj/20.15.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Molina-Cruz A., Canepa G.E., Barillas-Mury C. Plasmodium P47: a key gene for malaria transmission by mosquito vectors. Curr Opin Microbiol. 2017;40:168. doi: 10.1016/j.mib.2017.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Dijk M.R., Janse C.J., Thompson J., et al. A central role for P48/45 in malaria parasite male gamete fertility. Cell. 2001;104:153–164. doi: 10.1016/s0092-8674(01)00199-4. [DOI] [PubMed] [Google Scholar]
- 71.Koenderink J.B., Kavishe R.A., Rijpma S.R., Russel F.G.M. The ABCs of multidrug resistance in malaria. Trends Parasitol. 2010;26:440–446. doi: 10.1016/j.pt.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 72.Sultan A.A., Thathy V., Frevert U., et al. TRAP is necessary for gliding motility and infectivity of plasmodium sporozoites. Cell. 1997;90:511–522. doi: 10.1016/s0092-8674(00)80511-5. [DOI] [PubMed] [Google Scholar]
- 73.Moreira C.K., Templeton T.J., Lavazec C., et al. The Plasmodium TRAP/MIC2 family member, TRAP-Like Protein (TLP), is involved in tissue traversal by sporozoites. Cell Microbiol. 2008;10:1505. doi: 10.1111/j.1462-5822.2008.01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Puentes A., Ocampo M., Rodríguez L.E., et al. Identifying Plasmodium falciparum merozoite surface protein-10 human erythrocyte specific binding regions. Biochimie. 2005;87:461–472. doi: 10.1016/j.biochi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 75.Jeninga M.D., Quinn J.E., Petter M. ApiAP2 transcription factors in apicomplexan parasites. Pathogens. 2019;8 doi: 10.3390/pathogens8020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Malleret B., Sahili A.E., Tay M.Z., et al. Plasmodium vivax binds host CD98hc (SLC3A2) to enter immature red blood cells. Nat Microbiol. 2021;6:991–999. doi: 10.1038/s41564-021-00939-3. [DOI] [PubMed] [Google Scholar]
- 77.Ding S., Ye R., Zhang D., et al. Anti-folate combination therapies and their effect on the development of drug resistance in Plasmodium vivax. Sci Reports. 2013;3:1–6. doi: 10.1038/srep01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pornthanakasem W., Riangrungroj P., Chitnumsubet P., et al. Role of Plasmodium vivax dihydropteroate synthase polymorphisms in sulfa drug resistance. Antimicrob Agents Chemother. 2016;60:4453–4463. doi: 10.1128/AAC.01835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Villena F.E., Maguiña J.L., Santolalla M.L., et al. Molecular surveillance of the Plasmodium vivax multidrug resistance 1 gene in Peru between 2006 and 2015. Malar J. 2020;19:1–10. doi: 10.1186/s12936-020-03519-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rodríguez L.E., Urquiza M., Ocampoet M., et al. Plasmodium vivax MSP-1 peptides have high specific binding activity to human reticulocytes. Vaccine. 2002;20:1331–1339. doi: 10.1016/s0264-410x(01)00472-8. [DOI] [PubMed] [Google Scholar]
- 81.Gupta D.K., Dembele L., Voorberg-van der Wel A., et al. The Plasmodium liver-specific protein 2 (LISP2) is an early marker of liver stage development. Elife. 2019;8 doi: 10.7554/eLife.43362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miguel-Blanco C., Murithi J.M., Benaventeet E.D., et al. The antimalarial efficacy and mechanism of resistance of the novel chemotype DDD01034957. Sci Rep. 2021;11 doi: 10.1038/s41598-021-81343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cunningham D., Lawton J., Jarra W., Preiser P., Langhorne J. The pir multigene family of Plasmodium: antigenic variation and beyond. Mol Biochem Parasitol. 2010;170:65–73. doi: 10.1016/j.molbiopara.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 84.Prajapati S.K., Singh O.P. Insights into the invasion biology of Plasmodium vivax. Front Cell Infect Microbiol. 2013;3 doi: 10.3389/fcimb.2013.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Remarque E.J., Faber B.W., Kocken C.H.M., Thomas A.W. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol. 2008;24:74–84. doi: 10.1016/j.pt.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 86.Wahlgren M., Bejarano M.T., Troye-Blomberget M., et al. Epitopes of the Plasmodium falciparum clustered-asparagine-rich protein (CARP) recognized by human T-cells and antibodies. Parasite Immunol. 1991;13:681–694. doi: 10.1111/j.1365-3024.1991.tb00563.x. [DOI] [PubMed] [Google Scholar]
- 87.Arumugam T.U., Takeo S., Yamasakiet T., et al. Discovery of GAMA, a plasmodium falciparum merozoite micronemal protein, as a novel blood-stage vaccine candidate antigen. Infect Immun. 2011;79:4523. doi: 10.1128/IAI.05412-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang M., Gallego-Delgado J., Fernandez-Ariaset C., et al. Inhibiting the plasmodium eIF2α kinase PK4 prevents artemisinin-induced latency. Cell Host Microbe. 2017;22:766–776.e4. doi: 10.1016/j.chom.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Han J.-H., Lee S.K., Wang B., et al. Identification of a reticulocyte-specific binding domain of Plasmodium vivax reticulocyte-binding protein 1 that is homologous to the PfRh4 erythrocyte-binding domain. Sci Rep. 2016;6 doi: 10.1038/srep26993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tarr S.J., Moon R.W., Hardege I., Osborne A.R. A conserved domain targets exported PHISTb family proteins to the periphery of Plasmodium infected erythrocytes. Mol Biochem Parasitol. 2014;196:29. doi: 10.1016/j.molbiopara.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheng Y., Wang Y., Ito D., et al. The Plasmodium vivax merozoite surface protein 1 paralog is a novel erythrocyte-binding ligand of P. vivax. Infect Immun. 2013;81:1585. doi: 10.1128/IAI.01117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Murithi J.M., Deni I., Pasaje C. F.A., et al. The Plasmodium falciparum ABC transporter ABCI3 confers parasite strain-dependent pleiotropic antimalarial drug resistance. Cell Chem Biol. 2021;29:824–839. doi: 10.1016/j.chembiol.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Henrici R.C., van Schalkwyk D.A., Sutherland C.J. Modification of pfap2μ and pfubp1 markedly reduces ring-stage susceptibility of plasmodium falciparum to artemisinin in vitro. Antimicrob Agents Chemother. 2019;64 doi: 10.1128/AAC.01542-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McNamara C.W., Lee M.C., Lim C.S., et al. Targeting plasmodium phosphatidylinositol 4-kinase to eliminate malaria. Nature. 2013;504:248–253. doi: 10.1038/nature12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Llanos-Cuentas A., Casapia M., Chuquiyauri R., et al. Antimalarial activity of single-dose DSM265, a novel plasmodium dihydroorotate dehydrogenase inhibitor, in patients with uncomplicated Plasmodium falciparum or Plasmodium vivax malaria infection: a proof-of-concept, open-label, phase 2a study. Lancet Infect Dis. 2018;18:874–883. doi: 10.1016/S1473-3099(18)30309-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kimata-Ariga Y., Morihisa R. Functional analyses of plasmodium ferredoxin Asp97Tyr mutant related to artemisinin resistance of human malaria parasites. J Biochem. 2021;170:521–529. doi: 10.1093/jb/mvab070. [DOI] [PubMed] [Google Scholar]
- 97.Tran P.N., Brown S.H.J., Mitchell T.W., et al. A female gametocyte-specific ABC transporter plays a role in lipid metabolism in the malaria parasite. Nat Commun. 2014;5:1–3. doi: 10.1038/ncomms5773. [DOI] [PubMed] [Google Scholar]
- 98.Menegon M., Bardají A., Martínez-Espinosaet F., et al. Microsatellite genotyping of plasmodium vivax isolates from pregnant women in four malaria endemic countries. PLoS One. 2016;11 doi: 10.1371/journal.pone.0152447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rezende A.M., Tarazona-Santos E., Fontes C.J.F., et al. Microsatellite loci: determining the genetic variability of Plasmodium vivax. Trop Med Int Health. 2010;15:718–726. doi: 10.1111/j.1365-3156.2010.02535.x. [DOI] [PubMed] [Google Scholar]
- 100.Cheeseman I.H., Miller B.A., Nair S., et al. A major genome region underlying artemisinin resistance in malaria. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Takala-Harrison S., Clark T.G., Jacobet C.G., et al. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin. Proc Natl Acad Sci USA. 2013;110:240–245. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rodrigues P.T., Alves J.M., Santamaria A.M., et al. Using mitochondrial genome sequences to track the origin of imported Plasmodium vivax infections diagnosed in the United States. Am J Trop Med Hyg. 2014;90:1102–1108. doi: 10.4269/ajtmh.13-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rijpma S.R., van der Velden M., 1, Bilos A., et al. MRP1 mediates folate transport and antifolate sensitivity in Plasmodium falciparum. FEBS Lett. 2016;590:482–492. doi: 10.1002/1873-3468.12079. [DOI] [PubMed] [Google Scholar]
- 104.Mohring F., Hart 1 M.N., Rawlinsonet T.A., et al. Rapid and iterative genome editing in the malaria parasite Plasmodium knowlesi provides new tools for P. vivax research. Elife. 2019;8 doi: 10.7554/eLife.45829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moon R.W., Hall J., Rangkuti F., et al. Adaptation of the genetically tractable malaria pathogen Plasmodium knowlesi to continuous culture in human erythrocytes. Proc Natl Acad Sci USA. 2013;110:531–536. doi: 10.1073/pnas.1216457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.