Abstract
A new gene from Bordetella bronchiseptica, bfrZ encoding a putative siderophore receptor, was identified in a Fur-repressor titration assay. A bfrZ null mutant was constructed by allelic exchange. The protein profile of this mutant is similar to that of the wild-type parent strain. The BfrZ−-BfrZ+ isogenic pair was tested for utilization of 132 different siderophores as iron sources. None of these iron sources acted as a ligand for BfrZ. Translational bfrZ::phoA and transcriptional bfrZ::lacZ fusions were introduced into the B. bronchiseptica bfrZ locus. No alkaline phosphatase or β-galactosidase activity was detected. Sequence analysis of the bfrZ upstream region revealed the presence of two tightly linked genes, bupI and bupR. Both of these genes are located downstream from a Fur-binding sequence. BupI is homologous to Escherichia coli FecI and Pseudomonas putida PupI and belongs to the family of extracytoplasmic-function sigma factors involved in transcription of genes with extracytoplasmic functions. BupR is homologous to the FecR and PupR antisigma factors and is predicted to be localized in the inner membrane. Similar to the surface signaling receptors FecA and PupB, BfrZ bears an N-terminal extension. We found that bfrZ is not transcribed when bupI and bupR are expressed at the same level. However, overexpression of bupI from a multicopy plasmid triggers bfrZ transcription, and under these conditions BfrZ was detected in membrane fractions. By analogy with the FecI-FecR-FecA and PupI-PupR-PupB systems, our data suggest that bfrZ expression is inducible by binding of the cognate ligand to BfrZ and transduction of a signal through the envelope.
Iron is essential for the growth of most microorganisms but is usually not readily accessible. In an oxic environment at neutral pH, the concentration of free Fe3+ in solution is less than 10−18 M, and in the host iron is sequestered by proteins such as transferrin and lactoferrin (for a recent review see reference 9). To fulfill their iron requirement, bacteria have developed very efficient iron uptake systems. Most aerobic and facultatively aerobic bacteria secrete one or two small Fe3+-complexing molecules, named siderophores, and produce specific siderophore receptors at their surfaces. Bacteria usually synthesize multiple receptors to scavenge exogenous iron chelates secreted by other microbial species. However, some pathogens (for instance, Neisseria spp. and Haemophilus influenzae) use heme and the ferriproteins of their hosts as iron sources without producing siderophores (20, 27, 42). Serratia marcescens and Pseudomonas aeruginosa secrete a small protein which can release heme from hemoglobin and then bind to a receptor at the cell surface (28).
Ferrisiderophore transport systems and their regulation have been well characterized in Escherichia coli and several other gram-negative bacteria. The TonB-ExbB-ExbD envelope complex (Ton system) enables transfer of all iron-loaded siderophores to the periplasm, and then siderophore-specific ABC-type systems transport the iron chelates across the inner membrane (9, 32). Genes encoding iron uptake systems are repressed by the Fur protein under high-iron growth conditions (see reference 13 for a recent review). The Fur-Fe2+ complex binds to promoters containing target sequences named Fur-binding sequences (FBS) and thus blocks transcription. The FBS consensus sequence was recently reexamined and is thought to result from combination of at least three adjacent 6-bp NATA-TAT motifs (18) instead of a previously proposed 19-bp palindromic AT-rich sequence (13). In the absence of Fe2+, Fur does not bind to the FBS, and promoters are derepressed. Recently, positive Fur-Fe2+ regulation of the E. coli iron superoxide dismutase promoter was reported. The mechanism of this activation has not been elucidated yet, but it does not involve an FBS (16).
Several iron transport genes are also positively regulated by their cognate ligands via a surface signaling mechanism (8, 13). The ferric dicitrate uptake system is the only ligand-inducible iron uptake system known in E. coli. It has been extensively studied, and the following regulation model has been proposed and refined (8, 17, 22). Binding of ferric dicitrate to the outer membrane FecA receptor generates transmission of a signal from the periplasmic N-terminal extension of FecA to the C-terminal periplasmic domain of inner membrane protein FecR via the Ton system. FecR then transduces the signal across the cytoplasmic membrane and activates the sigma factor FecI. FecI binds to the RNA polymerase core enzyme, and the complex initiates transcription of the fecABCDE operon. The fecIR and fecABCDE operons are linked on the chromosome, and both are Fur repressed (1). A similar surface signaling mechanism has been identified in Pseudomonas putida WCS358 for pupB regulation (24). PupB is a receptor for pseudobactins BN7 and BN8. Upon binding to PupB, these siderophores induce transduction of a signal from the receptor to the cytoplasmic PupI sigma factor via the Ton system and the PupR antisigma factor localized in the envelope. The pupIR operon contains an FBS and is located upstream from pupB (24).
Iron uptake systems and their regulation in Bordetella have not been completely characterized yet. Bordetella bronchiseptica, the etiologic agent of swine atrophic rhinitis and kennel cough, and Bordetella pertussis, the agent of whooping cough, both secrete the siderophore alcaligin (34). The Bordetella fur gene has been identified and has been shown to mediate iron regulation in these species (6, 10; Pradel and Locht, Abstr. 95th Gen. Meet. Am. Soc. Microbiol. 1995, abstr. B-357, 1995). The alcABCDE operon encodes enzymes of the alcaligin biosynthesis pathway (19, 21, 39). AlcR is an AraC type of activator of the alc operon and of the alcaligin receptor gene fauA (7, 11, 39). The alcABCDE, alcR, and fauA promoters contain FBS.
In addition to alcaligin, B. bronchiseptica and B. pertussis have been shown to use enterobactin, heme, hemoglobin, ferrichrome, and desferal as iron sources (4). Four exogenous siderophore receptors have been identified in B. bronchiseptica: BfeA, BfrA, BfrB, and BfrC (3–5). B. pertussis produces only three of these, and BfrA is specific to B. bronchiseptica (4). BfeA binds enterobactin, but the ligands of the Bfr receptors have not been identified yet. The Ton system has been shown to be required for utilization of siderophores, heme, and hemoglobin (36, 40).
To gain more insight into the iron regulatory network in Bordetella, we analyzed a B. bronchiseptica Fur-repressed gene that we had previously isolated in a genetic screen analysis by a Fur titration assay (FURTA) (39, 46). This gene, named bfrZ, encodes a new TonB-dependent receptor for an unidentified ligand, probably an exogenous siderophore. We show here that bfrZ expression is controlled by BupI and BupR, a pair of sigma-antisigma transcription factors. We suggest that upon binding of the cognate siderophore to BfrZ, a signal is transduced through the envelope to BupI present in the cytoplasm to induce bfrZ transcription.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this work are listed in Table 1. E. coli strains were grown at 37°C in Luria-Bertani (LB) medium (33) or on solid media obtained by adding 1.5% (wt/vol) Bacto Agar. In the FURTA (46), the Lac phenotype of E. coli H1717 transformants was tested on MacConkey lactose agar plates containing 50 μM FeCl3. Bordetella strains were grown at 37°C on Bordet-Gengou agar plates supplemented with 1% glycerol and 15% sheep blood. PhoA and β-galactosidase activities of E. coli or B. bronchiseptica strains were assayed on LB medium plates containing 40 μg of bromo-4-chloro-3-indolyl-phosphate (XP) (Sigma, St. Quentin Fallavier, France) per ml and 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Sigma), respectively. Liquid cultures of Bordetella spp. were grown in modified Stainer-Scholte (SS) medium containing 10 mg of FeSO4 · 7H2O per liter (40). The low-iron medium used was SS medium without added FeSO4 · 7H2O (SS-Fe). To ensure that the conditions were iron limiting for B. bronchiseptica, the Chrome Azurol S assay was used to assess alcaligin production by Bordetella cells grown in SS-Fe as described previously (40). Modulation conditions were obtained by adding 50 mM MgSO4 to SS medium or to LB medium plates. When necessary, antibiotics were included in the growth media at the following final concentrations: ampicillin, 150 μg/ml; chloramphenicol, 30 μg/ml; gentamicin, 10 μg/ml; kanamycin, 30 or 300 μg/ml for pEP453 TnphoA mutagenesis; and streptomycin, 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid(s) | Relevant featuresa | Source or reference |
|---|---|---|
| E. coli strains | ||
| CC102 | CC118 with F42 lacI3 zzf-2::TnphoA; Kmr | 30 |
| CC118 | recA1 ΔphoA20 | 30 |
| GM2163 | dam-13::Tn9 dcm-6; Cmr | This laboratory |
| H1717 | aroB fhuF::λplacMu; Kmr | 46 |
| SM10 | Mobilizing strain; Kmr | 45 |
| XL1-Blue | High-efficiency transformation; Tcr | Stratagene |
| B. bronchiseptica strains | ||
| BB1015 | Smr but not rpsL | 39 |
| BBEP173 | BB1015 fur173 | This laboratory |
| BBEP185 | BB1015 bfrZ::lacZ (pEP589 integration); Gnr | This study |
| BBEP186 | BBEP173 bfrZ::lacZ (pEP589 integration); Gnr | This study |
| BBEP187 | BBEP205 bfrZ::lacZ (pEP589 integration); Gnr | This study |
| BBEP205 | BB1015 alcR::Kmr | 39 |
| BBEP231 | BB1015 bfrZ::Kmr | This study |
| BBEP250 | BB1015 bfrZ::phoA (pEP515 integration); BfrZ+ Kmr Gnr | This study |
| BBEP251 | BBEP173 bfrZ::phoA (pEP515 integration); BfrZ+ Kmr Gnr | This study |
| B. pertussis strain | BPSM TohamaI rpsL; Smr | 31 |
| Plasmids | ||
| pBCSK+ | High-copy-number vector; Cmr | Stratagene |
| pUC18, pUC19 | High-copy-number vector; Apr | Roche |
| pUC4K | Source of Kmr cassette | Pharmacia |
| pBBR1MCS | Broad-host-range vector; Cmr | 25 |
| pFus2 | Bordetella suicide vector to generate lacZ transcriptional fusions; Gnr | 2 |
| pJK200SK | Bordetella suicide vector; Gnr | 41 |
| pEP278 | pBCSK+ bearing bfrZ′ on a 2.6-kb PstI fragment | This study |
| pEP410 | pUC19 bearing bfrZ::Kmr | This study |
| pEP416 | pJQ200SK bearing bfrZ::Kmr | This study |
| pEP434 | pUC18 bearing bfrZ::Kmr and bfrZ downstream region | This study |
| pEP453 | pBBR1MCS bearing bfrZ on a PstI-SacI fragment | This study |
| pEP482 | pEP453 bearing bfrZ::TnphoA | This study |
| pEP484 | pEP482 deleted for the transposase gene (ΔBamHI) | This study |
| pEP515 | pJQ200SK bearing bfrZ::phoA | This study |
| pEP589 | pFus2 bearing ′bfrZ′ to generate a bfrZ-lacZ fusion | This study |
| pEP596 | pBCSK+ bearing bfrZ upstream region | This study |
| pEP624 | pBBR1MCS bearing bupI bupR bfrZ′ on an NsiI-ScaI fragment | This study |
| pEP625 | pBBR1MCS bearing bupI bupR′ on an NsiI-SalI fragment | This study |
Apr, Cmr, Gnr, Kmr, Smr, and Tcr, resistance to ampicillin, chloramphenicol, gentamicin, kanamycin, streptomycin, and tetracycline, respectively.
DNA techniques.
Plasmid DNA was isolated by the alkaline lysis method (43). Restriction enzymes and T4 DNA ligase were obtained from Roche (Meylan, France) and were used according to standard procedures (43). Cloned DNA fragments were sequenced by using an ABI PRISM dye terminator cycle sequencing kit and an ABI PRISM 377 sequencer (PE Applied Biosystems, Warrington, United Kingdom) along with a combination of universal, reverse, and custom-synthesized primers. PCRs were carried out with Vent DNA polymerase (New England Biolabs, Inc., Beverly, Mass.).
Computer analysis of sequences.
The nucleotide and protein sequences were analyzed by using the DNA Strider 1.2 software (Service de Biochimie et de Génétique Moléculaire du CEA, Saclay, France). Sequence similarities were identified with the BLASTN, BLASTX, and BLASTP programs (http://www.ncbi.nlm.nih.gov/BLAST/). Sequence alignment was performed with the multalin 5.3.3 software (12). Oligonucleotides were designed with the Oligo 5.0 software (NBI, Plymouth, Minn.).
Construction of pEP278 and pEP416 and cloning of the bfrZ 3′ extremity.
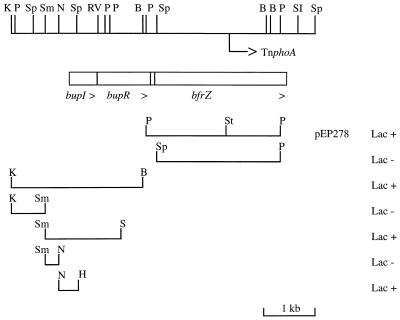
Plasmid pEP278 bearing bfrZ′ on a 2.6-kb PstI fragment was isolated with the FURTA from a partial B. bronchiseptica BB1015 genomic library as described previously (39). The 2.4-kb SphI-PstI fragment bearing ′bfrZ′ was isolated from pEP278 and cloned into pUC19. The resulting plasmid was introduced into E. coli GM2163 dcm (Cmr) and then reisolated from this strain to enable digestion with StuI. The HincII Kmr cassette was isolated from pUC4K and inserted into the unique StuI site of bfrZ (Fig. 1). The resulting plasmid, pEP410, was digested with SphI and XbaI, and the 3.7-kb fragment bearing bfrZ::Kmr was cloned into the Bordetella suicide vector pJQ200SK, digested with the same restriction enzymes, to obtain pEP416. E. coli SM10 was transformed with pEP416 and used as a donor in conjugations with B. bronchiseptica BB1015. B. bronchiseptica BBEP231 was isolated as an Smr Kmr Gns exconjugant, and the correct allelic exchange in this strain was confirmed by Southern blot hybridization (data not shown). To isolate the bfrZ downstream region, genomic DNA of BBEP231 was digested with SphI, and 4- to 5-kb restriction fragments were cloned into pUC18. Recombinant pEP431 Apr Kmr was selected, and the nucleotide sequence of the bfrZ 3′ extremity was determined.
FIG. 1.
Physical map of the B. bronchiseptica bupI bupR bfrZ locus and relevant constructions described in this study. The recombinant plasmid-associated Lac phenotypes in the FURTA are indicated on the right. In this genetic test, a Lac+ phenotype indicates the presence of an FBS on a multicopy plasmid. The boxes representing ORFs are drawn to scale; the arrowheads indicate the direction of transcription. The TnphoA insertion site is indicated by an arrow. Certain restriction sites are indicated as follows: B, BamHI; H, HincII; K, KpnI; N, NsiI; P, PstI; RV, EcoRV; S, SalI; SI, SacI; Sc, ScaI; Sm, SmaI; Sp, SphI; St, StuI.
Siderophore utilization plate assay.
B. bronchiseptica cells and plates were prepared as described previously (40). Filter paper disks impregnated with 10 μl of a siderophore solution (5 mM) were applied to the surfaces. Growth stimulation around the disks was evaluated after 12 h of incubation at 37°C.
Construction of pEP453 and TnphoA mutagenesis.
The complete bfrZ gene was reconstituted in Bordetella replicative plasmid pBBR1MCS as a 3-kb PstI-SacI fragment to obtain pEP453. TnphoA (30) was introduced into E. coli CC118(pEP453) by mating with E. coli CC102. Transpositions of TnphoA onto pEP453 were selected on LB medium containing XP and 300 μg of kanamycin per ml. Plasmids were prepared from pools of bacteria and transformed into E. coli CC118. None of the resulting Cmr Kmr colonies showed a strong PhoA+ phenotype (no dark blue colonies). One clone which exhibited rather low yet detectable PhoA activity (pale blue colonies) was named CC118(pEP482) and was studied further. Sequencing of the fusion joint indicated that TnphoA was inserted in frame into bfrZ, 65 bp downstream from the StuI site (Fig. 1). Plasmid pEP482 was digested with BamHI to delete the transposase gene and was religated to generate pEP484.
Construction of B. bronchiseptica bfrZ::phoA chromosomal fusions.
A 6.6-kb BamHI-EcoRV fragment containing bfrZ::phoAKmr from pEP484 was inserted into the Bordetella Gnr suicide vector pJQ200SK opened with BamHI and SmaI to generate pEP515. As this construct bore the bfrZ promoter region up to the PstI site, pEP515 integration into the chromosome conserved an intact bfrZ copy. E. coli SM10(pEP515) was mated with B. bronchiseptica BB1015 and BBEP173 fur173 to isolate the bfrZ::phoA Smr Kmr exconjugants BBEP250 and BBEP251, respectively.
Construction of B. bronchiseptica bfrZ::lacZ mutants.
An internal bfrZ fragment was amplified from pEP278 by using oligonucleotides HindIII-bfrZ (5′-AAGCTTCGTTGTCGGGCAGCAATCTC-3′) and bfrZ-BamHI (5′-GGATCCGCTCTTGGGCTCCTGGAAG-3′), which hybridized 264 bp downstream from the SphI site and 136 bp upstream from the ScaI site (complementary strand), respectively. The 680-bp PCR product was cloned into the HincII site of pBCSK+. The resulting plasmid was digested with HindIII and BamHI, and the 680-bp fragment obtained was ligated into pFus2 (2) digested with the same enzymes to generate pEP589. This Bordetella Gnr suicide plasmid was introduced into B. bronchiseptica BB1015, BBEP173 fur173, and BBEP205 alcR::Kmr by conjugation with E. coli SM10(pEP589). For each strain, one Smr Gnr exconjugant bearing pEP589 integrated into the chromosome was studied further; these exconjugants were designated BBEP185, BBEP186, and BBEP187. In contrast to the bfrZ::phoA translational mutants, the bfrZ::lacZ transcriptional mutants were defective for BfrZ production, as pEP589 did not contain the bfrZ promoter.
Cloning of the bfrZ upstream region and construction of pEP624 and pEP625.
B. bronchiseptica BBEP185 genomic DNA was digested with NotI, which did not cut pEP589, and ligated. The ligation mixture was used to transform E. coli XL1-Blue to Gnr. A recombinant plasmid resulting from intramolecular ligation of a chromosomal NotI fragment containing pEP589 was isolated. Restriction mapping of this plasmid enabled us to localize the bfrZ upstream region in a 6-kb EcoRI fragment. This fragment was cloned into pBCSK+ to obtain pEP596. The 3-kb region upstream from bfrZ was sequenced up to the KpnI site shown in Fig. 1. Plasmids pEP624 and pEP625 were derivatives of pBBR1MCS containing a 3-kb NsiI-ScaI fragment bearing bupI bupR bfrZ′ cloned into the NsiI and SmaI sites and a 1.2-kb NsiI-SalI bupI bupR′ fragment inserted into the NsiI and SalI sites, respectively.
Cell fractionation and protein analysis.
Cells from 100-ml B. bronchiseptica cultures grown in SS-Fe to an optical density at 600 nm of 3 were harvested by centrifugation, resuspended in 10 ml of 10 mM HEPES (pH 7.4) containing DNase (10 μg/ml) and disrupted with a French pressure cell (SLM-Aminco, Rochester, N.Y.). The pressates were centrifuged at 2,065 × g for 10 min at 4°C to sediment cellular debris and unbroken cells. Whole-cell lysates were then centrifuged at 111,000 × g for 1 h at 4°C to separate soluble and insoluble cell fractions. Whole-membrane pellets were resuspended in 3 ml of 1% Sarkosyl in 10 mM HEPES buffer (pH 7.4) and incubated for 30 min at room temperature. The suspensions were centrifuged at 111,000 × g for 1 h at 4°C to pellet outer-membrane-enriched fractions. The pellets were resuspended in 300 μl of 10 mM HEPES buffer (pH 7.4). After solubilization at 95°C for 5 min in Laemmli buffer (26), proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by using a 5% stacking gel and a 10% separating gel.
Nucleotide sequence accession number.
The nucleotide sequence of the B. bronchiseptica 5,909-bp KpnI-SphI DNA fragment has been assigned EMBL accession no. AJ251793.
RESULTS
Identification of bfrZ.
The FURTA (46) was used to detect potential Fur-binding fragments in a B. bronchiseptica genomic DNA library as described previously (39). In this genetic screen analysis, recombinant high-copy-number plasmids were introduced into an E. coli Δlac strain bearing a Fur-repressible fhuF::lacZ fusion (Lac− phenotype in high-iron growth conditions) and transformants were selected on iron-rich MacConkey agar plates containing antibiotics. Cloned sequences containing FBS titrated out the Fur repressor, resulting in derepression of the chromosomal lacZ fusion. Such transformants formed red colonies on MacConkey agar plates (Lac+ phenotype). Plasmid pEP278 was isolated and studied further since it gave a strong Lac+ phenotype in the assay. Restriction mapping showed that pEP278 contained a 2.6-kb PstI DNA fragment (Fig. 1). Deletion of a 200-bp PstI-SphI fragment at one end of the insert conferred a Lac− phenotype in the FURTA, suggesting that this region contained an FBS (Fig. 1). The nucleotide sequence of the PstI fragment was determined. Consistent with the genetic data, a sequence homologous to the recently reexamined E. coli Fur-binding consensus sequence (18) was identified 123 bp upstream from the SphI site (aATAAcGAatcTCAaTAT; 12 of 18 matches). Sequence analysis revealed a large open reading frame (ORF) starting 80 bp downstream from the putative FBS and extending to the opposite PstI site. Of three potential in-frame initiation codons, only the first ATG was preceded by a sequence resembling the canonical AAGAGG E. coli ribosome binding site. Thus, translation most probably starts at this ATG, which is located 4 bp downstream from a putative AAGGGAGAA ribosome binding site.
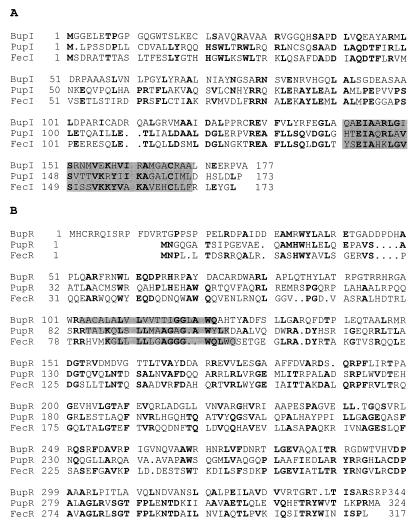
Scanning of the Swiss Protein Data Base revealed that the deduced amino acid sequence of the 821-residue ORF is homologous to that of TonB-dependent receptors. The closest homologue whose ligand has been identified is the ferripyoverdin receptor FpvA of P. aeruginosa PAO1 (GenBank accession no. P48632), with 22% identity, 38% similarity, and 11% gaps in a 699-amino-acid (aa) overlap. The ORF was named bfrZ by analogy with the previously characterized putative B. bronchiseptica siderophore receptor genes bfrA, bfrB, and bfrC (3, 4). The N-terminal region of BfrZ was predicted to contain a 41-residue cleavable signal peptide. BLASTP searches in the GenBank database indicated that the N-terminal sequence of the proposed mature BfrZ protein is similar to that of a class of Pseudomonas sp. siderophore receptors whose production is induced in the presence of their cognate siderophores. A sequence alignment of this region with the corresponding regions of the pseudobactin M114 receptor PbuA of Pseudomonas sp. strain M114 (GenBank accession no. Q08017), the ferripyoverdin receptor FpvA of P. aeruginosa (GenBank accession no. P48632), and the pseudobactin BN7-BN8 receptor PupB of P. putida (GenBank accession no. P38047) is presented in Fig. 2. The N-terminal periplasmic extension of PupB has been shown to be involved in signal transduction and transcriptional control of pupB and other iron transport genes (24). A similar role is suspected for the N terminus of FpvA (44).
FIG. 2.
Sequence alignment for the deduced mature N terminus of BfrZ and the N-terminal periplasmic extensions of three inducible Pseudomonas siderophore receptors. PbuA, pseudobactin M114 receptor of Pseudomonas sp. strain M114; FpvA, ferripyoverdin receptor of P. aeruginosa; PupB, pseudobactin BN7-BN8 receptor of P. putida. Conserved residues are indicated by boldface type.
The presence of bfrZ in other Bordetella genomes was tested by Southern blot hybridization using the 2.6-kb PstI fragment of pEP278 as a probe. No hybridization signal was detected with B. pertussis, Bordetella parapertussis, or Bordetella avium genomic DNA (data not shown). Thus, similar to bfrA, bfrZ is probably specific for B. bronchiseptica.
Characterization of a bfrZ mutant and cloning of the bfrZ 3′ terminus.
B. bronchiseptica BBEP231, a bfrZ::Kmr mutant, was constructed by allelic exchange as described in Materials and Methods. Whole-membrane and outer-membrane-enriched fractions of BBEP231 and the BB1015 parent strain grown in low-iron medium were analyzed by SDS-PAGE. No difference in the protein profiles was observed, suggesting that BfrZ is not abundantly present in the outer membrane (data not shown). BBEP231 exogenous siderophore utilization and BB1015 exogenous siderophore utilization were compared in a plate bioassay. More than 110 pyoverdins and 22 other siderophores from the Jean-Marie Meyer collection (Strasbourg, France) were tested. The two strains had identical iron source utilization profiles (data not shown). Thus, none of the siderophores tested proved to be the BfrZ ligand.
To isolate the bfrZ 3′ extremity, BBEP231 genomic DNA was digested with SphI and cloned into pUC18 to obtain pEP431. Sequencing of the 900-bp BamHI-SphI fragment indicated that the bfrZ stop codon is located 123 bp downstream from the PstI site (Fig. 1). The deduced mature BfrZ protein is a 819-residue molecule with a calculated molecular mass (MM) of 91.3 kDa. Its C-terminal sequence contains a TonB-dependent receptor signature, TIVWGNERRAMLNAQLSF (PROSITE accession no. PDOC00354). A truncated ORF starting about 200 bp downstream from bfrZ and having the same orientation was detected. In a 112-residue overlap, the translation product exhibited 59% similarity with a 127-aa putative inner membrane protein encoded by a Sphingomonas aromaticivorans catabolic plasmid (GenBank accession no. O58848).
Expression of bfrZ::phoA and bfrZ::lacZ fusions.
A translational bfrZ::phoA fusion was isolated by TnphoA mutagenesis of pEP453, an E. coli-Bordetella shuttle vector containing the whole bfrZ gene on a PstI-SphI fragment. The fusion junction of the TnphoA insertion is indicated in Fig. 1. The mutagenized plasmid, pEP482, and its derivative deleted for the transposase gene, pEP484, conferred low but detectable levels of PhoA activity to E. coli ΔphoA cells (data not shown). However, B. bronchiseptica BB1015(pEP484) grown in low-iron conditions expressed no detectable PhoA activity, although these conditions were sufficient to induce alcaligin production. In addition, no PhoA activity was detected in B. bronchiseptica BBEP205 alcR::Kmr bearing pEP484, suggesting that bfrZ expression is not repressed by AlcR, the regulator of alcaligin biosynthesis and alcaligin receptor genes (data not shown). B. pertussis BPSM(pEP484) did not produce any detectable PhoA activity either (data not shown). The same bfrZ::phoA fusion was introduced into the bfrZ locus of B. bronchiseptica BB1015 and its fur173 derivative, BBEP173. During growth in low-iron medium the resulting strains, BBEP250 and BBEP251, expressed no PhoA activity. Taken together, these observations suggest that either the hybrid BfrZ-PhoA protein is highly unstable in Bordetella or additional sequences not present in pEP484 are required for bfrZ expression in Bordetella.
To bypass potential hybrid protein instability, a transcriptional bfrZ::lacZ fusion was constructed and introduced into B. bronchiseptica BB1015, BBEP173 fur173, and BBEP205 alcR::Kmr. None of the strains obtained expressed β-galactosidase activity in low-iron growth conditions. These data suggest that bfrZ transcription is tightly regulated, as has been shown for several siderophore receptor genes in Pseudomonas spp. (13).
Identification of the bupI and bupR genes upstream from bfrZ.
The bfrZ upstream region was subcloned from pEP595, a large recombinant plasmid generated by digestion of BBEP185 bfrZ::lacZ genomic DNA with NotI followed by intramolecular ligation. Sequence analysis of the 2.6-kb KpnI-PstI fragment upstream from bfrZ revealed the presence of two tightly linked ORFs separated from bfrZ by 100 bp and having the same orientation (Fig. 1). BLASTP searches performed with sequences in the GenBank database indicated that the first ORF translates into a product homologous to P. putida PupI and E. coli FecI ECF sigma factors (GenBank accession no. S46355 and JV0111, respectively). The second ORF encodes a protein similar to the P. putida PupR and E. coli FecR antisigma factors (GenBank accession no. S46356 and B37804, respectively). By analogy with the P. putida system, the ORFs were designated bupI and bupR.
The deduced BupI protein is a 177-aa molecule with a calculated MM of 18.8 kDa. It exhibits 29% identity and 49% similarity with PupI in a 130-aa overlap and 26% identity and 50% similarity with FecI in a 129-aa overlap. An alignment of the sequences of these proteins is shown in Fig. 3A. Similar to FecI and PupI, BupI is predicted to possess a C-terminal helix-turn-helix motif. The deduced BupR protein is a 344-aa molecule with a calculated MM of 38.2 kDa. It exhibits 24% identity and 42% similarity with FecR in a 306-aa overlap and 22% identity and 37% similarity with PupR in a 310-aa overlap (Fig. 3B). FecR contains a unique hydrophobic transmembrane segment (aa 85 to 105) which anchors it in the inner membrane, while its N- and C-terminal regions are cytosolic and periplasmic, respectively (37, 48). Protein structure prediction programs suggested that PupR and BupR both have this topology and that they possess a transmembrane region formed by aa 85 to 105 and aa 103 to 123, respectively (http://www.bmm.icnet.uk/∼prof).
FIG. 3.
Alignment of the deduced sequences of the B. bronchiseptica BupI (A) and BupR (B) proteins with the sequences of their E. coli FecI and FecR and P. putida PupI and PupR homologues. Conserved residues are indicated by boldface type. The highlighted motifs are helix-turn-helix C-terminal regions (A) and predicted transmembrane segments (B).
Sequence analysis of the DNA region upstream from bupI did not reveal any homology with sequences in the database. The 2.5-kb KpnI-BamHI fragment was tested with the FURTA and found to be positive (Fig. 1). Several subclones were constructed and tested with the FURTA in order to localize the FBS more precisely. As shown in Fig. 1, the 380-bp NsiI-HincII fragment conferred a Lac+ phenotype in the assay. Examination of the corresponding nucleotide sequence indicated that a putative FBS was present 21 bp upstream from the predicted bupI initiation codon (atTAATGAgAtTtgTTAT; 12 of 18 matches). A GC-rich 10-bp inverted repeat, CCGCCAGGACAGGCCGTCCTGGCGG, located 245 bp upstream from bupI, could form a transcriptional termination signal. Along with the fact that only 1 bp separates bupI and bupR, these observations suggest that bupI bupR is a Fur-repressed operon.
bupI overexpression induces bfrZ expression.
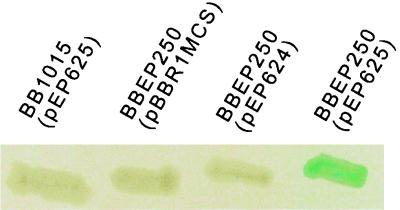
To test whether BupI and BupR are involved in bfrZ expression, we constructed pEP624 and pEP625, which were derivatives of Bordetella replicative plasmid pBBR1MCS bearing the intact bupI bupR operon and bupI and a truncated bupR gene, respectively. These plasmids were introduced into B. bronchiseptica BBEP250 bfrZ::phoA. BBEP250(pBBR1MCS) and BBEP250(pEP624) were found to form white patches on plates containing XP (PhoA− phenotype), while the BBEP250(pEP625) patches were blue (PhoA+) (Fig. 4). The BB1015(pEP625) control strain was PhoA−, which showed that pEP625 did not induce expression of a resident B. bronchiseptica phosphatase activity. In addition, transfer of pEP625 into BBEP185 induced bfrZ::lacZ expression (data not shown). These results indicated that an excess of BupI over BupR induces bfrZ transcription. BBEP250(pBBR1MCS), BBEP250(pEP624), BBEP250(pEP625), and BBEP185(pEP625) were grown in the presence of 50 mM MgSO4 to modulate virulence gene expression. Expression of the bfrZ fusions was not affected (data not shown), suggesting that bfrZ is not controlled by the BvgA-BvgS virulence factor regulatory system (29).
FIG. 4.
Alkaline phosphatase activities of B. bronchiseptica BB1015(pEP625), BBEP250(pBBR1MCS), BBEP250(pEP624), and BBEP250(pEP625) on LB medium plates containing XP and chloramphonicol. pEP624 and pEP625 are pBBR1MCS derivatives bearing bupI bupR and bupI, respectively. White colonies are PhoA−, and blue colonies are PhoA+.
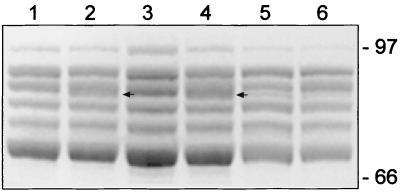
Outer membrane protein-enriched fractions were prepared from BB1015(pBBR1MCS), BB1015(pEP625), BBEP250(pBBR1MCS), BBEP250(pEP625), BBEP231(pEP625), and BBEP185(pEP625) grown in low-iron medium and were analyzed by SDS-PAGE (Fig. 5). As shown in Fig. 5, a protein with an apparent MM of approximately 90 kDa was detected in the extracts from cells bearing an intact bfrZ chromosomal gene and bupI on a multicopy plasmid (lanes 2 and 4). As the calculated MM of BfrZ is 91 kDa, the protein present in Fig. 5, lanes 2 and 4, was most probably BfrZ. This protein was not visible in extracts from cells carrying an intact bfrZ gene but no multicopy bupI gene (Fig. 5, lanes 1 and 3) or in extracts from cells harboring bupI on a plasmid but having an interrupted bfrZ gene on the chromosome (lanes 5 and 6). However, in strains bearing an inactivated bfrZ gene (lanes 5 and 6), bupI overexpression seemed to induce production of a protein that migrated slightly faster than would be predicted from the MM of BfrZ. This membrane protein may be another siderophore receptor whose synthesis requires BupI. Alternatively, BupI overproduction could generate cross-talk and activate expression of a gene normally transcribed via another extracytoplasmic-function (ECF) sigma factor.
FIG. 5.
Effect of bupI overexpression on outer membrane protein composition as determined by SDS-PAGE analysis. Cells were grown in low-iron medium. Lane 1, B. bronchiseptica BB1015 bfrZ+ (pBBR1MCS); lane 2, BB1015 bfrZ+ (pEP625); lane 3, BBEP250 bfrZ+ bfrZ::phoA (pBBR1MCS); lane 4, BBEP250 bfrZ+ bfrZ::phoA (pEP625); lane 5, BBEP231 bfrZ::Kmr (pEP625); lane 6, BBEP185 bfrZ::lacZ (pEP625). The arrows indicate the position of BfrZ. The positions of the MM standards are indicated on the right.
DISCUSSION
We isolated and characterized the bupI bupR bfrZ locus, the first example of a putative inducible exogenous siderophore uptake system in B. bronchiseptica. The bupI bupR genes seem to be transcribed as a single unit from a Fur-repressed promoter, while bfrZ expression requires an excess of BupI over BupR in the absence of the cognate ligand. By analogy with the well-characterized E. coli FecI-FecR-FecA (1, 8) and P. putida PupI-PupR-PupB (24) systems, our data suggest that BupI is an ECF sigma factor localized in the cytoplasm, BupR is an antisigma factor anchored in the inner membrane, and BfrZ is an outer membrane siderophore receptor. However, in the absence of the phenotype of a bfrZ mutant with respect to siderophore uptake, we cannot rule out the possibility that BfrZ is involved in transport of a noniron ligand. Nicholson and Beall previously isolated another Fur-repressed B. bronchiseptica putative ECF sigma-antisigma pair, BtfI-BtfR, but its target gene(s) has not been identified yet (M. L. Nicholson and B. Beall, Abstr. 98th Gen. Meet. Am. Soc. Microbiol., abstr. B-60, 1998). Other observations have suggested that expression of the bfeA enterobactin receptor gene is inducible, as uptake experiments have indicated that the ability of B. bronchiseptica to bind enterobactin is increased when cells are precultured with this exogenous siderophore (47). However, the regulation mechanism remains to be elucidated. In P. aeruginosa, induction of the enterobactin uptake system involves phosphorylation of a two-component regulatory system (14, 15).
The G+C content of the bupI bupR bfrZ locus is 67%, which is similar to those of other Bordetella genes. Southern blot experiments indicated that bfrZ is not present in the strictly human pathogen B. pertussis. We recently used the Bordetella BLAST server of the Sanger Centre to scan the available B. pertussis and B. bronchiseptica genomic DNA sequences for homology with bfrZ (http://www.sanger.ac.uk). No bfrZ sequence was detected in the 105 assembled contigs, which covered most of the B. pertussis genome. A unique bfrZ gene was identified in the 1,777 contigs, which covered part of the B. bronchiseptica genome. The B. pertussis genome is about 1 Mb smaller than that of B. bronchiseptica (http://www.sanger.ac.uk). The latter species has a much wider ecological niche as it can survive and grow even in lakewater (38). Another siderophore receptor, BfrA, has also been shown to be specific to B. bronchiseptica (4). Considering the importance of iron to sustaining life, it is not surprising that B. bronchiseptica has evolved or conserved a significant iron-scavenging potential in addition to other survival strategies.
So far, about 200 siderophores secreted by bacteria or fungi have been identified, but many more remain to be isolated (35). We were able to test a wide range of pyoverdins and other siderophores for their ability to stimulate the growth of B. bronchiseptica in iron-depleted conditions. In addition to the previously identified siderophores ferrichrome and desferal, we observed that coprogen, schizokinen, ferricrocin, vicibactin, ferrichrysin, ferrirubin, aerobactin, protochelin, and several pyoverdins are iron sources for B. bronchiseptica (data not shown). However, we were unable to identify the BfrZ ligand, either because it is absent from the siderophore library or because it is transported via a second receptor in a B. bronchiseptica bfrZ null mutant. Koster et al. reported that a P. putida pupI::Tn5 mutant still uses pseudobactin BN8 as an iron source although it does not produce the cognate receptor PupB, suggesting that an additional receptor for the BN8 siderophore is produced in this strain (23). Pseudobactins BN7, BN8, and M114 are not included in the collection that we tested; thus, we cannot comment on their utilization by B. bronchiseptica. We used the B. bronchiseptica bfrZ+ bfrZ::phoA mutant as a reporter strain to assay bfrZ induction in the presence of siderophores from the collection. No increase in PhoA activity was detected in a plate test, suggesting that the BfrZ ligand is missing from the siderophore library (data not shown).
In the E. coli ferric dicitrate transport system, the N-terminal cytoplasmic region of FecR is required and is sufficient for fecA expression (48). An E. coli strain bearing a nonsense mutation in codon 19 of fecR does not express fecA, but the production of a 56-residue FecR is sufficient to activate the FecI sigma factor to transcribe fecA in the absence of the citrate inducer. In P. putida, disruption of pupR by deletion of its 5′ region and insertion of an Smr interposon triggers constitutive pupB expression (24). Thus, PupR is not necessary for pupB expression. Overproduction of PupI from a plasmid in conjunction with chromosomal expression of pupR leads to pupB expression. PupR appears to be more like a stoichiometric repressor of PupI than an enzymatic activator (24). However, intact PupR is required for optimal pupB transcription in the presence of the pseudobactin BN8 inducer (24). The Fec and Pup systems are related but may differ with respect to the role of the antisigma factor. In B. bronchiseptica, construction of bupR mutants and additional bfrZ expression studies are required to evaluate the function of BupR. As the cognate siderophore of BfrZ has not been identified yet, construction of a chimeric receptor can also be envisioned. By analogy with the PupB-PupA chimera constructed by Koster et al. (24), a hybrid protein formed by the N-terminal extension of BfrZ and the mature FauA alcaligin receptor could be tested for its ability to transduce a signal to the BupI-BupR system and initiate bfrZ transcription upon binding of alcaligin to the FauA moiety.
ACKNOWLEDGMENTS
We thank Jean-Marie Meyer for kindly welcoming E.P. in his lab to test his siderophore collection. We are grateful to Eve Willery for technical assistance with automatic sequencing, to Emmanuelle Fort for photographic work, and to Dominique Raze for friendly computer assistance.
This work was supported by the INSERM, the Institut Pasteur de Lille, the Région Nord Pas-de-Calais, and the Ministère de l'Enseignement Supérieur et de la Recherche.
REFERENCES
- 1.Angerer A, Braun V. Iron regulates transcription of the Escherichia coli ferric citrate transport genes directly and through the transcription initiation proteins. Arch Microbiol. 1998;169:483–490. doi: 10.1007/s002030050600. [DOI] [PubMed] [Google Scholar]
- 2.Antoine R, Alonso S, Raze D, Coutte L, Lesjean S, Willery E, Locht C, Jacob-Dubuisson F. New virulence-activated and virulence-repressed genes identified by systematic gene inactivation and generation of transcriptional fusions in Bordetella pertussis. J Bacteriol. 2000;182:5902–5905. doi: 10.1128/jb.182.20.5902-5905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beall B. Two iron-regulated putative ferric siderophore receptor genes in Bordetella bronchiseptica and Bordetella pertussis. Res Microbiol. 1998;149:189–201. doi: 10.1016/s0923-2508(98)80079-x. [DOI] [PubMed] [Google Scholar]
- 4.Beall B, Hoenes T. An iron-regulated outer-membrane protein specific to Bordetella bronchiseptica and homologous to ferric siderophore receptors. Microbiology. 1997;143:135–145. doi: 10.1099/00221287-143-1-135. [DOI] [PubMed] [Google Scholar]
- 5.Beall B, Sanden G N. A Bordetella pertussis fepA homologue required for utilization of exogenous ferric enterobactin. Microbiology. 1995;141:3193–3205. doi: 10.1099/13500872-141-12-3193. [DOI] [PubMed] [Google Scholar]
- 6.Beall B W, Sanden G N. Cloning and initial characterization of the Bordetella pertussis fur gene. Curr Microbiol. 1995;30:223–226. doi: 10.1007/BF00293637. [DOI] [PubMed] [Google Scholar]
- 7.Beaumont F C, Kang H Y, Brickman T J, Armstrong S K. Identification and characterization of alcR, a gene encoding an AraC-like regulator of alcaligin siderophore biosynthesis and transport in Bordetella pertussis and Bordetella bronchiseptica. J Bacteriol. 1998;180:862–870. doi: 10.1128/jb.180.4.862-870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun V. Surface signaling: novel transcription initiation mechanism starting from the cell surface. Arch Microbiol. 1997;167:325–331. doi: 10.1007/s002030050451. [DOI] [PubMed] [Google Scholar]
- 9.Braun V, Killmann H. Bacterial solutions to the iron-supply problem. Trends Biochem Sci. 1999;24:104–109. doi: 10.1016/s0968-0004(99)01359-6. [DOI] [PubMed] [Google Scholar]
- 10.Brickman T J, Armstrong S K. Bordetella pertussis fur gene restores iron repressibility of siderophore and protein expression to deregulated Bordetella bronchiseptica mutants. J Bacteriol. 1995;177:268–270. doi: 10.1128/jb.177.1.268-270.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brickman T J, Armstrong S K. Essential role of the iron-regulated outer membrane receptor FauA in alcaligin siderophore-mediated iron uptake in Bordetella species. J Bacteriol. 1999;181:5958–5966. doi: 10.1128/jb.181.19.5958-5966.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean C R, Neshat S, Poole K. PfeR, an enterobactin-responsive activator of ferric enterobactin receptor gene expression in Pseudomonas aeruginosa. J Bacteriol. 1996;178:5361–5369. doi: 10.1128/jb.178.18.5361-5369.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean C R, Poole K. Expression of the ferric enterobactin receptor (PfeA) of Pseudomonas aeruginosa: involvement of a two-component regulatory system. Mol Microbiol. 1993;8:1095–1103. doi: 10.1111/j.1365-2958.1993.tb01654.x. [DOI] [PubMed] [Google Scholar]
- 16.Dubrac S, Touati D. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J Bacteriol. 2000;182:3802–3808. doi: 10.1128/jb.182.13.3802-3808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enz S, Mahren S, Stroeher U H, Braun V. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR, and FecI regulatory proteins. J Bacteriol. 2000;182:637–646. doi: 10.1128/jb.182.3.637-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giardina P C, Foster L A, Toth S I, Roe B A, Dyer D W. Identification of alcA, a Bordetella bronchiseptica gene necessary for alcaligin production. Gene. 1995;167:133–136. doi: 10.1016/0378-1119(95)00659-1. [DOI] [PubMed] [Google Scholar]
- 20.Gray-Owen S D, Loosmore S, Schryvers A B. Identification and characterization of genes encoding the human transferrin-binding proteins from Haemophilus influenzae. Infect Immun. 1995;63:1201–1210. doi: 10.1128/iai.63.4.1201-1210.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang H Y, Brickman T J, Beaumont F C, Armstrong S K. Identification and characterization of iron-regulated Bordetella pertussis alcaligin siderophore biosynthesis genes. J Bacteriol. 1996;178:4877–4884. doi: 10.1128/jb.178.16.4877-4884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim I, Stiefel A, Plantor S, Angerer A, Braun V. Transcription induction of the ferric citrate transport genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol Microbiol. 1997;23:333–344. doi: 10.1046/j.1365-2958.1997.2401593.x. [DOI] [PubMed] [Google Scholar]
- 23.Koster M, van de Vossenberg J, Leong J, Weisbeek P J. Identification and characterization of the pupB gene encoding an inducible ferric-pseudobactin receptor of Pseudomonas putida WCS358. Mol Microbiol. 1993;8:591–601. doi: 10.1111/j.1365-2958.1993.tb01603.x. [DOI] [PubMed] [Google Scholar]
- 24.Koster M, van Klompenburg W, Bitter W, Leong J, Weisbeek P. Role for the outer membrane ferric siderophore receptor PupB in signal transduction across the bacterial cell envelope. EMBO J. 1994;13:2805–2813. doi: 10.1002/j.1460-2075.1994.tb06574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovac M E, Phillips R W, Elzer P H, Roop II R M, Peterson K M. pBBR1MCS: a broad host range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Lee B C. Quelling the red menace: haem capture by bacteria. Mol Microbiol. 1995;18:383–390. doi: 10.1111/j.1365-2958.1995.mmi_18030383.x. [DOI] [PubMed] [Google Scholar]
- 28.Letoffe S, Redeker V, Wandersman C. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA haemophore. Mol Microbiol. 1998;28:1223–1234. doi: 10.1046/j.1365-2958.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 29.Locht C. Molecular aspects of Bordetella pertussis pathogenesis. Int Microbiol. 1999;2:137–144. [PubMed] [Google Scholar]
- 30.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menozzi F D, Boucher P E, Riveau G, Gantiez C, Locht C. Surface-associated filamentous hemagglutinin induces autoagglutination of Bordetella pertussis. Infect Immun. 1994;62:4261–4269. doi: 10.1128/iai.62.10.4261-4269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mietzner T A, Tencza S B, Adhikari P, Vaughan K G, Nowalk A J. Fe(III) periplasm-to-cytosol transporters of gram-negative pathogens. Curr Top Microbiol Immunol. 1998;225:113–135. doi: 10.1007/978-3-642-80451-9_7. [DOI] [PubMed] [Google Scholar]
- 33.Miller J H. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 34.Moore C H, Foster L A, Gerbig D G, Jr, Dyer D W, Gibson B W. Identification of alcaligin as the siderophore produced by Bordetella pertussis and B. bronchiseptica. J Bacteriol. 1995;177:1116–1118. doi: 10.1128/jb.177.4.1116-1118.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neilands J B, Nakamura K. Detection, determination, isolation, characterization and regulation of microbial iron chelates. In: Winkelmann G, editor. Handbook of microbial iron chelates. Boca Raton, Fla: CRC Press; 1991. pp. 1–15. [Google Scholar]
- 36.Nicholson M L, Beall B. Disruption of tonB in Bordetella bronchiseptica and Bordetella pertussis prevents utilization of ferric siderophores, haemin and haemoglobin as iron sources. Microbiology. 1999;145:2453–2461. doi: 10.1099/00221287-145-9-2453. [DOI] [PubMed] [Google Scholar]
- 37.Ochs M, Veitinger S, Kim I, Welz D, Angerer A, Braun V. Regulation of citrate-dependent iron transport of Escherichia coli: fecR is required for transcription activation by FecI. Mol Microbiol. 1995;15:119–132. doi: 10.1111/j.1365-2958.1995.tb02226.x. [DOI] [PubMed] [Google Scholar]
- 38.Porter J F, Wardlaw A C. Long-term survival of Bordetella bronchiseptica in lakewater and in buffered saline without added nutrients. FEMS Microbiol Lett. 1993;110:33–36. doi: 10.1111/j.1574-6968.1993.tb06291.x. [DOI] [PubMed] [Google Scholar]
- 39.Pradel E, Guiso N, Locht C. Identification of AlcR, an AraC-type regulator of alcaligin siderophore synthesis in Bordetella bronchiseptica and Bordetella pertussis. J Bacteriol. 1998;180:871–880. doi: 10.1128/jb.180.4.871-880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pradel E, Guiso N, Menozzi F D, Locht C. Bordetella pertussis TonB, a Bvg-independent virulence determinant. Infect Immun. 2000;68:1919–1927. doi: 10.1128/iai.68.4.1919-1927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 42.Ren Z, Jin H, Morton D J, Stull T L. hgpB, a gene encoding a second Haemophilus influenzae hemoglobin- and hemoglobin-haptoglobin-binding protein. Infect Immun. 1998;66:4733–4741. doi: 10.1128/iai.66.10.4733-4741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Schalk I J, Kyslik P, Prome D, van Dorsselaer A, Poole K, Abdallah M A, Pattus F. Copurification of the FpvA ferric pyoverdin receptor of Pseudomonas aeruginosa with its iron-free ligand: implications for siderophore-mediated iron transport. Biochemistry. 1999;38:9357–9365. doi: 10.1021/bi990421x. [DOI] [PubMed] [Google Scholar]
- 45.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–790. [Google Scholar]
- 46.Stojiljkovic I, Baumler A J, Hantke K. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J Mol Biol. 1994;236:531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- 47.Thulasiraman P, Newton S M, Xu J, Raymond K N, Mai C, Hall A, Montague M A, Klebba P E. Selectivity of ferric enterobactin binding and cooperativity of transport in gram-negative bacteria. J Bacteriol. 1998;180:6689–6696. doi: 10.1128/jb.180.24.6689-6696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welz D, Braun V. Ferric citrate transport of Escherichia coli: functional regions of the FecR transmembrane regulatory protein. J Bacteriol. 1998;180:2387–2394. doi: 10.1128/jb.180.9.2387-2394.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]