Abstract
Background
Ovarian cancer is the deadliest type of malignant gynecological tumor. Polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs) are involved ovarian cancer and are closely related to adverse outcomes. However, the immunosuppressive mechanism of PMN-MDSCs remains elusive.
Methods
The types and numbers of ANKRD22-expressing cells were investigated by bioinformatics analysis and immunohistochemical staining. Ankrd22-/- C57BL/6 mice were constructed with CRISPR-Cas9 technology. Mouse PMN-MDSCs were obtained from bone marrow (BM)-derived CD11b+Ly6G+Ly6Clow cells sorted by fluorescence-activated cell sorting with treatment of GM-CSF and IL-6, and the immunosuppressive activity of PMN-MDSCs was evaluated by flow cytometry (FCM) and ELISA. The expression level of CCR2 and the exogenous glucose uptake capacity were determined by FCM. RT-qPCR was used to detect ANKRD22 expression in CD11b+HLA-DR-CD14-CD15+ cells from human ovarian cancer tissues, and the correlations of ANKRD22 expression with the clinical characteristics and prognosis of patients were evaluated by the χ2 test.
Results
We identified a novel protein involved in regulating the immunosuppressive ability of PMN-MDSCs, ANKRD22. Ankrd22 expression was high in mouse CD11b+Ly6G+Ly6Clow cells and could be significantly downregulated after exposure to a simulated microenvironmental stimulus. Knockout of Ankrd22 increased the expression level of CCR2 of CD11b+Ly6G+Ly6Clow cells and the immunosuppressive activity of PMN-MDSCs. BM-derived CD11b+Ly6G+Ly6Clow cells of Ankrd22-/- mice significantly promoted the proliferation of ovarian cancer cells in tumor xenograft mouse models. Mechanistically, RNA sequencing showed that Wdfy1 expression was obviously increased in Ankrd22-knockout BM-derived CD11b+Ly6G+ Ly6Clow cells and that ectopic expression of Wdfy1 increased the levels of Arg1, Inos, Ido and Pdl1 in Ankrd22+/+ PMN-MDSCs derived from BM-derived CD11b+Ly6G+Ly6Clow cells. Surprisingly, an ANKRD22-activating candidate small-molecule compound attenuated the immunosuppressive activity of Ankrd22+/+ PMN-MDSCs. Finally, we found that low ANKRD22 levels in CD11b+HLA-DR-CD14-CD15+ cells derived from primary ovarian tissues were associated with a more advanced International Federation of Gynecology and Obstetrics stage, a higher recurrence rate, and a higher neutrophil-to-lymphocyte ratio.
Conclusions
These results suggest that ANKRD22 is a potential novel target for reversing the immunosuppressive effects of PMN-MDSCs.
Keywords: Myeloid-Derived Suppressor Cells, Tumor Microenvironment
WHAT IS ALREADY KNOWN ON THIS TOPIC
PMN-MDSC is one of the common immunosuppressive cells in tumor. How to block or reverse its immunosuppressive ability is a key step to improve the effect of anti-tumor immunotherapy.
WHAT THIS STUDY ADDS
Tumor microenvironment enhanced the immunosuppressive ability of PMN-MDSCs by down-regulating ANKRD22; increasing the expression of ANKRD22 could weaken the immuno -suppressive activity of PMN-MDSCs.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
ANKRD22 is a potential new therapeutic target, laying a solid foundation for development of drugs targeting PMN-MDSCs.
Introduction
Ovarian cancer is the deadliest gynecological cancer, and it seriously threatens the life and health of women.1 Cytoreductive surgery and chemotherapy are the most common treatments for ovarian cancer. The 5-year survival rate of patients with early-stage ovarian cancer can reach 90%, but the 3-year recurrence rate after routine treatment for advanced ovarian cancer is nearly 70%.2 Tumor reduction surgery is the cornerstone of ovarian cancer treatment, and the optimal goal of tumor reduction surgery for advanced ovarian cancer is to achieve no visible tumor cell residue. Additionally, chemotherapy combined with surgery is an effective treatment for ovarian cancer.3 However, recurrent and refractory ovarian cancer does not respond well to this strategy. Notably, targeted therapies such as antivascular endothelial growth factor (VEGF) antibodies, including bevacizumab, and poly-polymerase inhibitors, including olaparib, can be used for maintenance therapy in patients with ovarian cancer.3 Some studies have shown that the efficacy of single-agent immunotherapy targeting programmed cell death protein 1/programmed cell death ligand 1 (PD-1/PD-L1) or cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) can reach 10%–15%.4 5 Similar to that of other types of tumors, the ovarian tumor immune microenvironment (TIME) is formed by ovarian cancer cells, immune cells, mesenchymal cells, cytokines and their related metabolites, and disorder of the TIME accompanies the occurrence, progression and metastasis of ovarian cancer. Several studies have shown that the presence of an immunosuppressive microenvironment weakens the effect of immunotherapy on ovarian cancer,6–8 which suggests that the local immune microenvironment varies greatly across patients and that eliminating or reversing the inhibitory immune microenvironment may be the key to improve curative effects in patients with ovarian cancer. However, the mechanism underlying the formation of the immunosuppressive tumor microenvironment remains poorly understood. Elucidating the mechanism underlying the formation of the immunosuppressive microenvironment in ovarian cancer is necessary to remodel the tumor microenvironment to effectively prevent and treat tumors.
Immune cells are the core cells in the TIME and play roles throughout the process of tumor evolution. Immunosuppressive cells represent an important category of immune cells, and they play a key role in mediating the immune escape and malignant proliferation of tumor cells.4 Myeloid-derived suppressor cells (MDSCs) are the main immunosuppressive cells that mediate tumor immune evasion in the TIME.6 One study showed that TIME-related cytokines, such as tumor necrosis factor (TNF)-α, S100A8/9, granulocyte-macrophage/macrophage colony-stimulating factor (GM/M-CSF), VEGF, prostaglandin E2 (PGE2) and IL (interleukin)-6, have the ability to activate the JAK/STAT3, PI3K/AKT/mTOR, RAS/MAPK and NF-κB pathways in bone marrow (BM) progenitors to promote their proliferation and differentiation into MDSCs.6–8 MDSCs have strong immunosuppressive potential by secreting immunosuppressive molecules, such as arginase-1 (Arg-1), inducible nitric oxide synthase (iNOS) and indoleamine 2,3-dioxygenase (IDO), which competitively inhibit the activity of T cells and deplete these cells.7 Furthermore, MDSCs can mediate T-cell apoptosis or improve T-cell transformation into regulatory T cells (Tregs) by directly interacting with the T-cell receptor (TCR) expressed on T cells.9 Polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs) are closely related to the occurrence and development of ovarian cancer. Cui et al10 found that MDSCs inhibited T-cell activation and enhanced gene expression, ovarian cancer stem cell sphere formation and metastasis. MDSCs localized in the TIME secrete PGE2 to activate intracellular miRNA101 or the CSF2/STAT3 pathway in ovarian cancer cells, resulting in the acquisition of stemness characteristics and increased expression of PD-L1 in ovarian cancer cells and supporting immune escape by ovarian cancer cells.9 10 Targeting PMN-MDSCs can improve the effect of immunotherapy on ovarian cancer, but the mechanism regulating the immunosuppressive activity of PMN-MDSCs in ovarian cancer needs to be further clarified.
MDSCs can be divided into PMN-MDSCs and monocytic myeloid-derived suppressor cells (M-MDSCs) according to morphology. PMN-MDSCs are the main subset of MDSCs that infiltrate the TIME, and mouse PMN-MDSCs can be identified by the phenotype CD11b+Ly6G+Ly6Clow, while human PMN-MDSCs can be identified by the phenotype CD11b+CD14-CD15+/CD66b+11; these surface markers are similar to those of neutrophils. PMN-MDSCs and M-MDSCs have different functional and biological characteristics under various pathological conditions.12 However, while PMN-MDSCs, not M-MDSCs, are the main population that downregulates T-cell immune activity, the mechanism by which the immunosuppressive activity of PMN-MDSCs in the ovarian TIME is regulated needs to be further clarified.
In our previous studies, we identified a novel mitochondrial membrane protein, ankyrin-repeat domain-containing protein 22 (ANKRD22),13 and found that ANKRD22 could decrease macrophage activity in a non-polarized manner.14 The BioGPS database also showed that MDSCs express ANKRD22. However, the function of ANKRD22 in MDSCs remains unclear. Therefore, in this study, we investigated the effect of ANKRD22 expression on the immunosuppressive activity of PMN-MDSCs and found that the TIME could downregulate the expression level of ANKRD22 in CD11b+Ly6G+Ly6Clow cells and that the immunosuppressive activity of PMN-MDSCs with downregulated ANKRD22 was significantly enhanced. Our study suggests that ANKRD22 may be a potential target for reversing the immunosuppressive activity of PMN-MDSCs in the ovarian cancer TIME.
Materials and methods
Animals
The research group commissioned Cyagen Biosciences (Guangzhou, China) to construct Ankrd22 -/- C57BL/6 mice using CRISPR-Cas9 technology. The efficiency of Ankrd22 gene knockout in C57BL/6 mice was verified, and the primers used to verify Ankrd22 knockout were as follows: 5'-GCTGCC CTAAAGTCTTTCCTTCCCC-3' (forward) and 5'-GGGAGTATCGCCATTGAAGCTATCT-3' (reverse). The PCR conditions included annealing at 57°C to produce a fragment with a size of 383 bp. Matched Ankrd22+/+ C57BL/6 mice were purchased from the Shanghai Experimental Animal Resource Center of the Chinese Academy of Sciences (Shanghai, China). BM cells were harvested from female mice aged 6–8 weeks, and tumors were transplanted from female mice aged 4–5 weeks. All animals were bred and raised at the Experimental Animal Research Center of Zhejiang Chinese Medicine University. The ambient temperature of specific pathogen free (SPF) facility in animal center was maintained at 25°C±3°C, a humidity of 30%–70%, and a 12-hour alternating light and dark cycle. The mice were fed standard laboratory food and water.
Study population
This study included 20 newly diagnosed patients with ovarian cancer and 7 patients without ovarian cancer.
The inclusion criteria were as follows: (1) Ovarian cancer patients group: female with stable vital signs, aged ≥18 years old, voluntarily signed informed consent, initially diagnosed and confirmed with ovarian cancer by the gynecological oncologist and pathologist. (2) Non-ovarian cancer control group: female with stable vital signs, aged ≥18 years old, voluntarily signed informed consent, diagnosed by gynecological oncologists as ovarian disease requiring surgery and confirmed by pathologists as benign ovarian lesions, patients with non-ovarian cancer.
The exclusion criteria were as follows: Female with unstable vital signs (eg, concomitant myocarditis, diabetes or autoimmune disease), aged<18 years old, unwilling to sign the informed consent, diagnosed by gynecological oncologists as ovarian disease but not requiring surgery.
Clinical characteristics
The clinical and pathological data of the patients included in this study mainly included age, tumor type, histological type, International Federation of Gynecology and Obstetrics (FIGO) stage, grade, lymph node metastasis, ascites, and recurrence.
Patients with ovarian cancer were 42–74 years old, with a median age of 58 years old. Patients with non-ovarian cancer control were 30–64 years old, with a median age of 48 years old.
Tumor type (ovarian cancer and non-ovarian cancer): We classified patients with ovarian cancer diagnosed by the gynecological oncologist and pathologist according to the patient’s clinical manifestations and postoperative pathology into the ovarian cancer group. Because of the difficulty in obtaining adjacent tissues of ovarian cancer, we included patients diagnosed by gynecological oncologists with ovarian disease (such as ovarian cysts or chocolate cysts) and confirmed by pathologists that the postoperative pathological tissue type was benign lesion not ovarian tumor type in the non-ovarian cancer group.
Histological type (epithelial and nonepithelial): Epithelial ovarian carcinoma, including serous carcinoma, mucinous carcinoma and endometrioid carcinoma, is the most common histological type. Nonepithelial ovarian cancers include germ cell tumors and sex cord stromal tumors.
The clinicopathological features of the patients were listed in online supplemental table 1, and the correlation between different ANKRD22 expression levels and clinicopathological features of patients with ovarian cancer was analyzed according to the expression level of ANKRD22.
jitc-2022-005527supp001.pdf (982.3KB, pdf)
Specimens
Human specimens: Peripheral blood and ovarian tissue were collected from patients. The collected blood was anticoagulated with EDTA-K2 before surgery, and the tumor tissue was divided into three parts: one-third was stored in the fresh tissue preservation solution, which is a special organ preservation fluid for flow cytometry (FCM) (Lianke Biotechnology, Hangzhou, China), one-third was stored in 4% paraformaldehyde/formalin (Haoyang Biological Manufacture, Tianjin, China), and one-third was stored in the −80°C freezer.
Animal specimens: The peripheral blood, tumor tissue, BM and spleen were collected from mice. Blood collected from the mice via the ocular vein was placed in 4% sodium citrate anticoagulant. The tumor tissue was divided into three parts: one-third was stored in a fresh tissue preservation solution, one-third was stored in 4% paraformaldehyde/formalin, and one-third was stored in a −80°C freezer. Single-cell suspensions were obtained by rinsing the BM with 2 mL of Dulbecco’s Modified Eagle’s medium (DMEM) (Corning, New York, USA).
Preparation of single-cell suspensions from mouse peripheral blood and tumor tissues
(1) CD11b+Ly6G+Ly6Clow cells obtained from mouse BM: The femurs and tibias of female C57BL/6 mice were harvested under aseptic conditions, and DMEM medium was used to wash the mouse BM with a 2 mL aseptic syringe. After passing the BM cells through a 40 µm filter membrane, a single-cell suspension was prepared. (2) CD11b+Ly6G+Ly6Clow cells obtained from mouse tumor tissues: Subcutaneous tumor tissues were harvested from mice and washed with phosphate-buffered saline (PBS, Jinuo Biotech, Hangzhou, China), and the surrounding necrotic tissues were trimmed. The tumor tissue was cut and placed in a 5 ml volume of a tissue digestion solution (0.5 mg/mL of type IV collagenase (Sigma, Saint Louis, Missouri, USA) and 1 mg/mL hyaluronidase in serum-free DMEM (Sigma)). The mouse tissue was shaken at 37°C and 180 rpm for 1–2 hours, and the tissue was mixed every 60 min. The digested tissue mixture was filtered through a 40 µm filter membrane to prepare a single-cell suspension from the mouse tissue. (3) CD11b+Ly6G+Ly6Clow cells obtained from mouse peripheral blood: The peripheral blood of C57BL/6 female mice was collected via the ocular vein under sterile conditions and placed in 4% sodium citrate anticoagulant, and each anticoagulant tube was slowly moved up and down several times. According to the instructions of a peripheral blood monocyte isolation kit for mice (Haoyang Biological Manufacture), Ficoll-Hypaque density gradient centrifugation was performed at 2000 rpm for 10 min, and the lower white cell layer was collected after centrifugation to prepare a single-cell suspension of mouse peripheral blood mononuclear cells (PBMCs). (4) Neutrophils obtained from mouse peripheral blood: Peripheral blood was obtained from Ankrd22+/+ and Ankrd22-/- C57BL/6 mice in 4% sodium citrate anticoagulant. According to the instructions of a mouse peripheral blood neutrophil isolation kit (Haoyang Biological Manufacture), neutrophils were isolated from the peripheral blood of Ankrd22+/+ and Ankrd22-/- C57BL/6 mice.
Preparation of single-cell suspensions from human peripheral blood and tumor tissues
(1) Acquisition of CD11b+HLA-DR-CD14-CD15+ cells from human peripheral blood: 5–10 mL peripheral blood from 20 patients with ovarian cancer and 7 female volunteers aged 18 years or older without ovarian cancer was collected into EDTA-K2 anticoagulant tubes, and the anticoagulant tubes were slowly rolled up and down. According to the instructions of a human PBMC isolation kit (Haoyang Biological Manufacture), Ficoll-Hypaque density gradient centrifugation was performed at 2000 rpm for 10 min to extract the lower white cell layer, which was a single-cell suspension of human PBMCs. (2) CD11b+HLA-DR- CD14-CD15+ cells obtained from human ovarian cancer and non-ovarian cancer tissues: Ovarian cancer or non-ovarian cancer tissue was collected and washed with PBS, and the necrotic tissue was trimmed. Tumor tissues were cut and placed in a 5 ml volume of a tissue digestion solution containing 0.5 mg/mL type IV collagenase (Sigma) and 1 mg/mL hyaluronidase (Sigma) in serum-free DMEM. A single-cell suspension was prepared from human ovarian tissue via shaking and digestion at 180 rpm and 37°C for 90 min, followed by filtration through a 40 µm filter membrane.
Fluorescence-activated cell sorting or FCM
Single-cell suspensions were obtained from human ovarian tissue or peripheral blood and from mouse BM, tumor tissue or peripheral blood obtained as described above. Then, the suspensions were centrifuged at 2000 rpm for 10 min. The cell pellets were blocked on ice with PBS buffer containing 2% FBS for 15 min. After centrifugation at 2000 rpm for 10 min, the cells were suspended in a buffer containing primary antibodies and incubated on ice for 30 min in the dark. After removal of any unbound antibodies with a 5 ml volume of complete DMEM, the cell pellets were collected by centrifugation at 2000 rpm for 10 min. Adding 5 mL of red blood cell lysis buffer (Haoyang Biological Manufacture) to the cell pellet and leave it for 10 min at room temperature in the dark, and then the cells were centrifuged at 2000 rpm for 10 min. After the cells were washed with PBS, they were counted and resuspended in a 100 µL flow tube with a filter for FCM (DxFLEX flow cytometry, Beckman Coulter, Brea, California, USA) analysis or fluorescence-activated cell sorting (FACS) (Aria SORP flow sorter, BD Biosciences, Franklin Lakes, New Jersey, USA) sorting.
The antibodies used for mouse BM-derived cells and their subpopulations were anti-human/mouse CD11b-Allophycocyanin (APC) (BioLegend, San Diego, California, USA), anti-mouse Ly6G-fluorescein isothiocyanate (FITC) (BioLegend), anti-mouse Ly6C-Phycoerythrin (PE) (BioLegend) and anti-mouse Gr-1-APC-cyanine 7 (Cy7) (BioLegend). Antibodies that activate T cells are anti-mouse CD3 antibody (BioLegend) and anti-mouse CD28 antibody (BioLegend). Mouse MDSCs were identified as CD11b+GR-1+ cells, mouse PMN-MDSCs were identified as CD11b+Ly6G+Ly6Clow cells, and M-MDSCs were identified as CD11b+Ly6G-Ly6Chigh cells. The purity of CD11b+Ly6G+Ly6Clow cells is up to 97.7% and the CD11b+Ly6G-Ly6Chigh cells is up to 98.1% (online supplemental file 1).
The antibodies used for human CD11b+HLA-DR-CD14-CD15+ cells and their subpopulations were anti-human/mouse CD11b-APC (BioLegend), anti-human HLA-DR-PE (BioLegend), anti-human CD14-APC-Cy7 (BioLegend) and anti-human CD15-peridinin chlorophyll protein (PerCP) (BioLegend). Human PMN-MDSCs were identified as CD11b+HLA-DR-CD14-CD15+ cells. The surface marker phenotype of human M-MDSCs was CD11b+HLA-DR- CD14+CD15-.
The antibodies used for sorting B cells, macrophages and plasma cells were anti-mouse CD19-PerCP/Cy5.5 (BioLegend), anti-mouse F4/80-PE (BioLegend) and anti-mouse CD138-APC (BioLegend). Cells were sorted for subsequent experiments.
Preparation and labeling of single-cell suspensions of the mouse spleen
Mouse spleens were harvested under aseptic conditions, washed with PBS and then cut into pieces in DMEM. After the cell suspensions were filtered through a 40 µm membrane, mouse spleen single-cell suspensions were prepared. Spleen lymphocytes were obtained according to the instructions of the mouse spleen lymphocyte isolation kit (Haoyang Biological Manufacture) and centrifuged at 2000 rpm for 10 min. The cell pellets were washed twice with PBS. Mouse splenic lymphocytes were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE) (BioLegend). 1.0–10×107 cells splenic lymphocytes were resuspended in 5 µM concentration CFSE working solution. The suspension was protected from light and incubated at 37°C for 20 min. DMEM with 10% FBS was added to terminate staining. After centrifugation at 2000 rpm for 10 min, the cells were resuspended in preheated cell culture medium and incubated in the cell incubator for subsequent experiments. Splenic lymphocytes labeled with CFSE were used in subsequent experiments.
Cell culture
(1) Cell lines. ID8 mouse ovarian cancer cells were donated by Professor Dong Chen (Institute of Immunology, Tsinghua University), and B16F10 mouse melanoma cells were purchased from the Cell Bank of the Shanghai Branch of the Chinese Academy of Sciences. Cells were cultured in high-glucose DMEM (Corning) supplemented with 10% FBS (Corning) and 1% penicillin-streptomycin (Haoyang Biological Manufacture). All cells were cultured in a 37°C, 5% CO2 cell incubator. (2) Activated T cells. We collected WT mouse spleen-derived T cells and labeled with CFSE. Then, these T cells were activated by anti-CD3 and anti-CD28 antibodies (BioLegend). (3) Assessment of the immunosuppressive ability of BM-derived CD11b+Ly6G+Ly6 Clow cells. WT mice BM-derived CD11b+Ly6G+Ly6Clow cells with or without 100 ng/mL mouse GM-CSF (Novoprotein, Shanghai, China) and 100 ng/mL mouse IL-6 (Novoprotein) stimulation cocultured with activated T cells for 96 hours. The FITC signal intensity of T cells was detected by FCM and the concentration of IL-2 in the supernatants was determined by a commercial ELISA kit (Proteintech, Wuhan, China). (4) Establishment of the primary PMN-MDSCs culture system in vitro. Primary mouse BM-derived CD11b+Ly6G+Ly6Clow cells obtained by the method described above were cultured with 100 ng/mL GM-CSF (Novoprotein) and 100 ng/mL IL-6 (Novoprotein) and used in subsequent experiments.
Construction of a subcutaneous transplanted tumor mouse model
(1) A subcutaneous tumor model was established in C57BL/6 mice with ID8 cells and primary BM-derived CD11b+Ly6G+Ly6Clow cells. 5×106 mouse ID8 cells were mixed with 10×106 cells CD11b+Ly6G+Ly6Clow cells, which were extracted from the BM of Ankrd22+/+ or Ankrd22-/- C57BL/6 mice, respectively. Total 200 µL/mouse cells were mixed in Matrigel (Corning). ID8 cells and CD11b+Ly6G+Ly6Clow cells with or without treatment of ANKRD22-activating compound in vitro were mixed with Matrigel and injected into the axilla of C57BL/6 mice. (2) A subcutaneous tumor induction model was established with a mixed suspension of mouse peripheral blood derived neutrophils and ID8 cells. We obtain the neutrophils according to the instructions of a mouse peripheral blood neutrophil isolation kit, mixing ID8 cells and mouse neutrophils from C57BL/6 periphral blood as the above method in C57BL/6 mice.
After the above subcutaneous tumor mouse models were established, the formation of subcutaneous tumors in mice was observed every week from the date of tumorigenesis. After the subcutaneous tumors formed, tumor growth was recorded, and a tumor growth curve was generated. When the subcutaneous tumors grew to approximately 1.5 cm or grew until the 8th week, the tumors were collected and weighed. The tumor tissue was preserved in a special organ preservation fluid for FCM (Lianke Biotechnology) and then prepared as a single-cell suspension immediately.
Construction, production and infection procedures for the recombinant lentivirus
Construction and production of the recombinant lentivirus expressing Ankrd22 and Mlixp was performed by GenePharma Corporation (Shanghai, China). Cell infection with recombinant lentivirus was carried out according to the company’s operating procedures. Before lentiviral infection, primary PMN-MDSCs were cultured in six-well plates. 5 µg/mL polybrene (Sigma) and an appropriate amount of virus were added to the cells. After incubation for 48 hours, the cells were collected, washed once and resuspended in PBS. The FITC signal was detected for Ankrd22-overexpressing cells, while the PE signal was detected for MLIXP-overexpressing PMN-MDSCs. The infection efficiency was also verified by fluorescence microscopy.
Detect the immunosuppressive ability of PMN-MDSCs
(1) Detection of cytokines in PMN-MDSCs: After the above-labeled PMN-MDSCs were immobilized with surface antibodies, the cells were cultured in DMEM containing 100 ng/mL GM-CSF+100 ng/mL IL-6 and 81.0 nM phorbol 12-myristate13-acetate (PMA, Sigma) at 37°C and 5% CO2 for 4–6 hours. The cell pellets were collected after centrifugation at 2000 rpm for 10 min. The cells were resuspended in a buffer containing antibodies specific for intracellular FCM and incubated on ice for 30 min in the dark. After neutralizing with 5 mL of complete DMEM and centrifuging at 2000 rpm for 10 min to remove any unbound antibody, the cell pellets were collected. After the cells were washed with PBS, they were counted and resuspended in a 300 µL flow tube for analysis. The antibodies used for the detection were anti-Arg-1 antibody (CST, Danvers, Massachusetts, USA), FITC-conjugated antibody (Jackson ImmunoResearch Laboratories, West Grove, Pennsylvania, USA), PE-conjugated antibody (CST), PE anti-NOS2 (iNOS) antibody (BioLegend), and Alexa Fluor 647 anti-IDO1 (BioLegend). (2) Detecting the effect of PMN-MDSCs on T cells: Mouse BM-derived CD11b+Ly6G+Ly6Clow cells treated with 100 ng/mL GM-CSF and 100 ng/mL IL-6 were cocultured with CFSE-labeled mouse splenic lymphocytes stimulated by CD3 and CD28 (Biolegend) antibodies for 96 hours, and collecting cell and the supernatant for (1) detection of the proliferation number of T cells: Splenic lymphocytes labeled with CFSE (Biolegend) were counted and resuspended in a 300 µL flow tube. The FITC signal was detected to determine T cells proliferating; (2) detection of the immune activity of T cells: The supernatants of splenic T-cell and PMN-MDSCs cocultures were collected for subsequent ELISA to determine the concentration of IL-2 (Proteintech) secreted by T cells.
Effects of a simulated tumor microenvironment on Ankrd22 expression
According to the above method, BM-derived CD11b+Ly6G+Ly6Clow cells of Ankrd22+/+ C57BL/6 mice were plated in 24-well plates and exposed to various simulated tumor microenvironments. (1) Hypoxia: cells were exposed to 0.5% O2 for 24 hours, and then the cells were collected. (2) Glucose deficiency: cells were cultured in glucose deprivation environment for 24 hours, and then the cells were collected. (3) Reactive oxygen species (ROS): Different concentrations of H2O2 (Sangon Bioengineering, Shanghai, China) were added to the medium to achieve final concentrations of 100 µM, 200 µM and 500 µM for 24 hours, and a well without H2O2 was used as the control group. Then, the cells were collected. (4) Transforming growth factor-β (TGF-β): Mouse TGF-β (Novoprotein) was added to the medium to achieve a final concentration of 5 ng/mL or 10 ng/mL, and cells not treated with TGF-β were used as the control group. The cells were cultured for 48 hours and then collected. (5) Interferon-γ (IFN-γ): Mouse IFN-γ (Novoprotein) was added to the medium to achieve a final concentration of 50 ng/mL or 100 ng/mL, and cells not treated with IFN-γ were used as the control group. The cells were cultured for 48 hours and then collected. (6) Senescence microenvironment: In brief, 5 µg/mL or 10 µg/mL Irinotecan hydrochloride trihydrate (CPT-11) (Pfizer, New York, USA) was added to the culture medium, and cells not treated with CPT-11 were used as the control group. The cells were cultured for 24 hours and then collected. (7) Lactic acid: Lactic acid was added to the culture medium at 10 mM, 20 mM or 30 mM, and cells not treated with lactic acid were used as the control group. The cells were cultured for 24 hours and then collected. Cells were collected by centrifugation, and RNA was extracted for subsequent experiments.
RNA extraction, reverse transcription and RT-qPCR
(1) Total RNA was extracted from cells using TRIzol reagent (Macherey-Nagel, Dueren, Germany). The extracted RNA was reverse transcribed into cDNA using the PrimeScript RT Reagent Kit (TaKaRa Bio, Kyoto, Japan) according to the manufacturer’s instructions. (1) RT-qPCR was performed on a CFX96 Connect system (Bio-Rad, Hercules, California, USA) using the TaqMan probe method and a Premix Ex Taq kit (TaKaRa Bio, Kyoto, Japan). The primers and probes were synthesized by Shanghai Sangon Bioengineering Company (Shanghai, China).
The primer and probe sequences were designed as follows:
| Name of primer or probe | Primer or probe sequence (5’−3’) |
| Human-ANKRD22 upstream primer | 5’-CCAGCTTGGACTTCTAGGGA-3’ |
| Human-ANKRD22 downstream primer | 5’-GGCAGATGGGCTCAGAGTAT-3’ |
| Human-ANKRD22 probe | 5’-Fam-TCCCATGCTGGTCCTTCACAGG-Tamra-3’ |
| Human-TUBB upstream primer | 5’-AGGAGGTCGATGAGCAGATG-3’ |
| Human-TUBB downstream primer | 5’-TTGCCAATGAAGGTGACTGC-3’ |
| Human-TUBB probe | 5’-Fam-TGTGACATCCCACCTCGTGGCC-Tamra-3’ |
| Mouse-Ankrd22 upstream primer | 5’-CCAAGGCCTTCATCTCTCCA-3’ |
| Mouse-Ankrd22 downstream primer | 5’-GGCCAAGTCTTCAGAGGGAT-3’ |
| Mouse-Ankrd22 probe | 5’-Fam-AGGACCCATTGCTCACGTTGGAATCT-Tamra-3’ |
| Mouse-Tubb upstream primer | 5’-CAGCAGATGTTCGATGCCAA-3’ |
| Mouse-Tubb downstream primer | 5’-GCTCGTCCACCTCCTTCATA-3’ |
| Mouse-Tubb probe | 5’-Fam-CTGCCTGTGACCCGCGCCAC-Tamra-3’ |
Fam: 5 (6)-Carboxyfluorescein; Tamra: 5 (6)-Carboxytetramethylrhodamine.
(2) Mouse genes were identified by standard PCR. Amplification was carried out on a Life Express TC-96 system (Bio-ER, Hangzhou, China) using a Prime STARHS DNA polymerase kit (TaKaRa Bio, Kyoto, Japan). The annealing temperature was 57°C, and a 383 bp fragment was synthesized. The products were used for subsequent DNA electrophoresis.
The primer sequences were as follows:
| Name of primer | Primer sequence (5’−3’) |
| Mouse-Ankrd22 upstream primer | 5’-GGGAGTATCGCCATTGAAGCTATCT-3’ |
| Mouse-Ankrd22 downstream primer | 5’-GCTGCCCTAAAGTCTTTCCTTCC-3’ |
Luminex chip cytokine assay
CD11b+Ly6G+Ly6Clow cells derived from the BM of Ankrd22+/+ or Ankrd22-/- mice were isolated by FACS and cultured in complete DMEM containing 100 ng/mL GM-CSF and 100 ng/mL IL-6 in a 0.5% O2 anoxic environment for 24 hours, with normoxic culture used as a control. The cell supernatants were collected, stored on dry ice, and sent to Shanghai Huaying Biological Company (Shanghai, China) for detection. According to the instructions of a Luminex Array detection kit (Bioplex Pro Mouse Cytokine; Bio-Rad), which is a standard assay for detecting 385 mouse cytokines, the beads and samples were added to a 96-well plate and incubated at room temperature in the dark for 30 min. After washing three times, 25 µL samples of the mixtures were added to each well for antibody detection, and the plate was incubated at room temperature protected from light for 2 hours. After washing three times, a 50 µL streptavidin-phyoglobinin was added to each well and incubated at room temperature for 10 min. Finally, after washing, the signal was detected with a Bioplex MAGPIX system (Bio-Rad), and the concentration of each cytokine was calculated.
RNA sequencing of CD11b+Ly6G+Ly6Clow cells
Three samples of BM-derived CD11b+Ly6G+Ly6Clow cells from Ankrd22-/- or Ankrd22+/+ mice were obtained by FACS, and the cell pellets contained more than 1×107 cells. After two washes with PBS, 1 mL TRIzol reagent (Macherey-Nagel, Dueren, Germany) was added to extract total RNA from the cells. The extracted RNA was sent to Hangzhou Lianchuan Biotechnology (Hangzhou, China) for RNA sequencing, and the sequencing data were analyzed in R software.
Western blotting
Cell pellets were collected, washed twice with PBS, and treated with a mixture of protease inhibitors Cocktail (Sigma), 100×proteinase inhibitor (Servicebio, Wuhan, China) and RIPA lysis buffer (Yeasen, Shanghai, China). The cells were lysed on ice for 30 min and centrifuged at 13 000 rpm for 5 min. The supernatant was collected, and a 1/2 vol of loading buffer was added and boiled at 100°C for 5 min. SDS-PAGE was performed, and the proteins were transferred to a nitrocellulose membrane (Sartorius Stedim, Gottingen, Germany) at a current of 260 mA for 120 min. At room temperature, the membrane was blocked in a fast protein-free protein blocking solution (Boster, Wuhan, China) for 15 min and then washed twice with 50 mM Tris-HCl saline buffer containing 0.5% Tween-20 (TBST, pH 7.5) (Sangon Bioengineering). The original antibody was diluted to the appropriate concentration of Western blotting (Boster). The membrane was incubated with primary antibody and gently shaken at 4°C overnight. TBST was used to wash the membrane 5 times for 3 min each time, and the secondary antibody was diluted to the appropriate dilution (Boster). Then, the secondary antibody was incubated for 1 hour in TBST with gentle shaking at room temperature. After incubation, the membrane was washed with TBST 5 times for 3 min each and incubated in an enhanced chemiluminescence solution (Yeasen) for 1–3 min. Images were captured on a western blot scanner (Li-Cor Biosciences, Lincoln, Nebraska, USA). The primary antibodies used were as follows: rabbit anti-human β-actin (CST), rabbit anti-human β-tubulin (CST), and house-made mouse anti-human ANKRD22 (clone number: 1A8).13 The secondary antibody used was a horseradish peroxidase-labeled goat secondary antibody (Jackson ImmunoResearch Laboratories).
Histological examination
Immunohistochemical (IHC) staining was performed according to standard protocols. Sections of mouse or human ovarian tissue were incubated at 60°C for 30 min, dewaxed in xylene and hydrated with a series of graded alcohol solutions. The tissue sections were incubated in pH9.0, 10 mM EDTA-Tris buffer (Maxim, Fuzhou, China) at 95°C for 15 min for antigen repair. After cooling to room temperature, the tissue sections were blocked with 3% H2O2 for 10 min. After washing with TBST, the sections were incubated with anti-ANKRD22 (1:150) at room temperature for 1 hour. The tissue sections were washed with TBST 3 times and incubated with biotinylated streptavidin-horseradish peroxidase (SAP) second antibody complex (Maxim) for 30 min at room temperature. After being washed with TBST 3 times, 1–2 drops of diaminobenzidine (DAB) (Maxim) substrate solution were added and incubated for 5 min at room temperature. The reaction was terminated with running water, and the sections were counterstained with hematoxylin (Maxim).
Statistical analysis
All outcomes for continuous variables are expressed as the mean±SD. Unpaired two-sided Student’s t-tests were used to compare two independent samples. Data were analyzed using SPSS V.23.0 statistical software (IBM). A p<0.05 was defined as statistically significant. Results of statistical analysis were plotted using GraphPad Prism V.7.0 software (San Diego, California, USA).
Results
ANKRD22 was highly expressed in CD11b+Ly6G+Ly6Clow cells
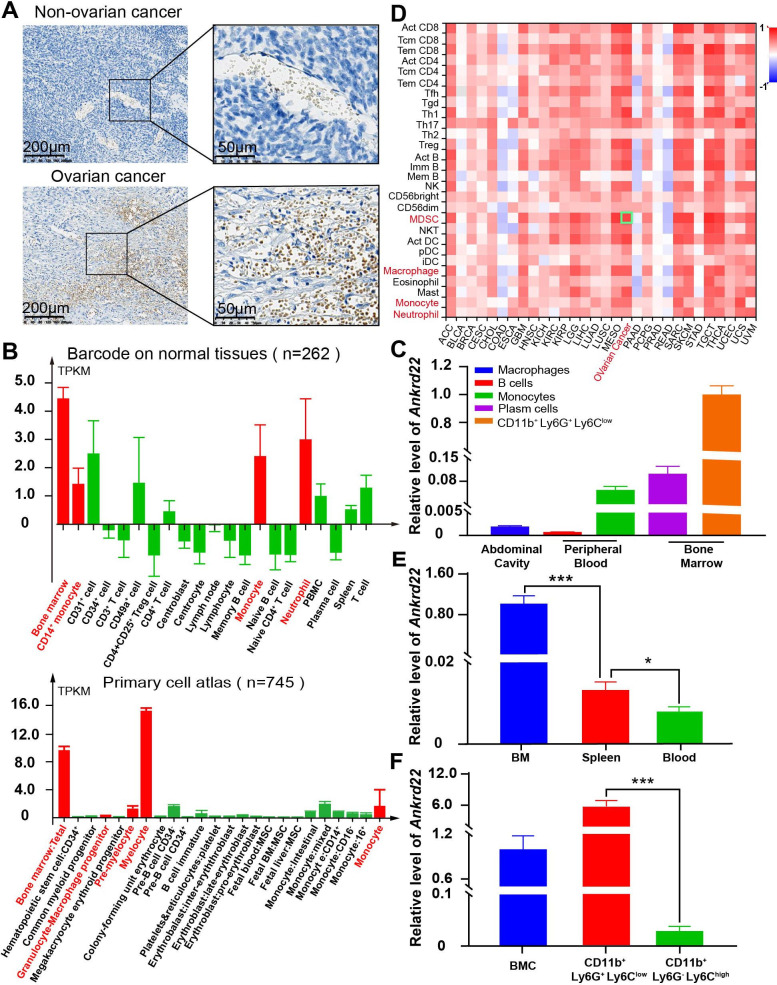
To explore the cell types expressing ANKRD22 and the levels of ANKRD22 expression in ovarian cancer tissues, we used IHC to examine ANKRD22 expression. We found that ovarian cancer cells did not express ANKRD22, but tumor stromal cells did (figure 1A). The Human Protein Atlas (HPA) database showed similar changes; the expression of ANKRD22 in normal ovarian tissue was very low, approximately 0.3 normalized transcripts per million (nTPM), and ovarian cancer cells indeed showed no ANKRD22 expression (www.proteinatlas.org). Since immune-associated cells in the tumor stroma are a major component during the formation of the immunosuppressive TIME, we investigated the expression levels of ANKRD22 in different immune cells. First, the BioGPS database showed that the expression level of ANKRD22 in human myeloid cells and their precursors, neutrophils and monocytes was high (figure 1B) (www.biogps.org/%23goto=genereport&id=52024). Second, RT-qPCR showed that Ankrd22 expression was the highest in mouse BM-derived CD11b+Ly6G+Ly6Clow cells than monocytes and macrophages, while Ankrd22 was low in B cells and plasma cells (figure 1C). Third, the tumor-immune system interaction database showed that the expression level of ANKRD22 in the ovarian cancer microenvironment was proportional to the abundance of invading MDSCs (figure 1D) (http://cis.hku.hk/TISIDB/browse.php?gene=ANKRD22), which implied that MDSCs might express ANKRD22. To this end, we isolated BM-derived CD11b+Ly6G+Ly6Clow cells from C57BL/6 mice, and RT-qPCR results showed that the expression of Ankrd22 in these cells was significantly higher than that derived from the normal spleen or peripheral blood (figure 1E). Finally, we separated BM-derived CD11b+Ly6G+Ly6Clow and CD11b+Ly6G-Ly6Chigh cells by FACS and found that BM-derived CD11b+ Ly6G+Ly6Clow cells expressed higher levels of Ankrd22 than CD11b+Ly6G-Ly6Chigh cells and BM cells by RT-qPCR (figure 1F). These studies suggest that CD11b+Ly6G+Ly6Clow cells have high expression of ANKRD22. However, how the TIME impacts the expression of ANKRD22 in CD11b+Ly6G+Ly6Clow cells and how CD11b+Ly6G+Ly6Clow cells with different levels of ANKRD22 affect the malignant biological behavior of ovarian cancer remain unknown.
Figure 1.
ANKRD22 was highly expressed in CD11b+Ly6G+Ly6 Clow cells. (A) IHC staining of ovarian cancer and normal ovarian tissues from our center showed that no ANKRD22 expression was observed in normal ovarian cells or ovarian tumor cells, while ANKRD22 was expressed in the normal stroma and cancerous ovarian stromal tissues. Scale bars are 200 µm and 50 µm. (B). The BioGPS database showed the expression level of ANKRD22 in different immune cells (n=262) and human bone marrow cells at different stages (n=745); myeloid cells and their precursors, neutrophils, monocytes may express ANKRD22 at a high level. (C). FACS combined with RT-qPCR showed that Ankrd22 expression was the highest in mouse BM-derived CD11b+Ly6G+Ly6Clow cells than monocytes and macrophages, while Ankrd22 was almost undetectable in B cells and plasma cells. (D). The proportion of infiltrating immune cells in human ovarian cancer tissue increased to different degrees. The above data were provided by the lymphocyte infiltration module of the TISIDB. (E). The expression level of Ankrd22 in CD11b+Ly6G+Ly6Clow cells from the bone marrow, spleen or peripheral blood of the wild-type C57BL/6 mice was detected by RT-qPCR. n=3, *p<0.05, ***p<0.001. Two-sample Student’s t-test. (F). BM-derived CD11b+Ly6G+Ly6Clow cells and BM-derived CD11b+Ly6G-Ly6Chigh cells were sorted by FACS, and the expression level of Ankrd22 in these cells was detected by RT-qPCR, respectively. n=3, ***p<0.001. Two-sample Student’s t-test. BM, bone marrow; BMC, bone marrow cells; FACS, fluorescenceactivated cell sorting; IHC, immunohistochemical; TISIDB, Tumor-Immune System Interaction Database.
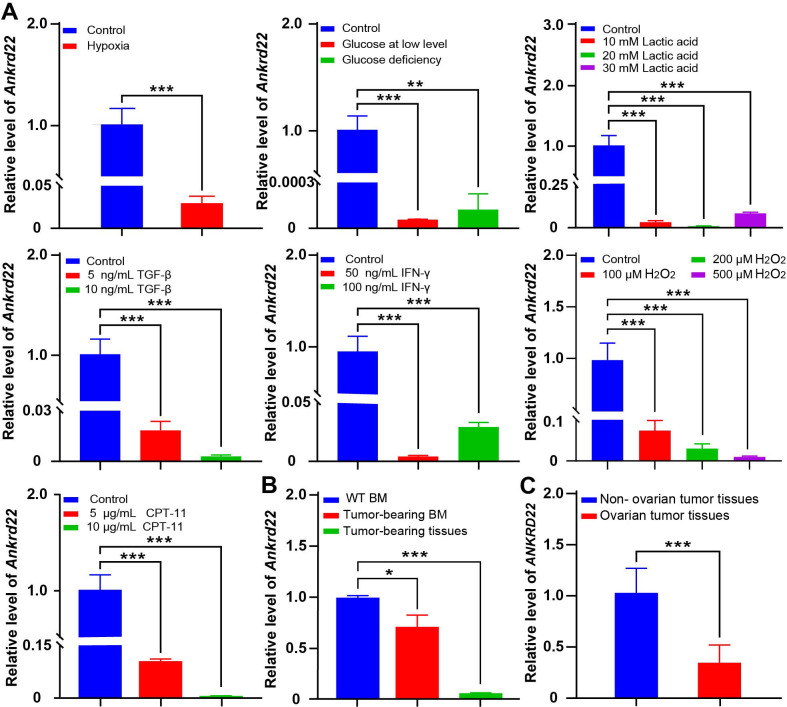
The tumor microenvironment affected the expression of Ankrd22 in BM-derived CD11b+Ly6G+Ly6Clow cells
To verify the effect of the tumor microenvironment on Ankrd22 expression in CD11b+Ly6G+ Ly6Clow cells, we simulated the tumor microenvironment in vitro and examined changes in the Ankrd22 expression level in CD11b+Ly6G+Ly6Clow cells derived from the BM of wild-type (WT) Ankrd22+/+ C57BL/6 mice under different stimuli. FACS was performed to sort mouse BM-derived CD11b+Ly6G+Ly6Clow cells, and FCM showed the sorting efficiency of CD11b+Ly6G+Ly6Clow cells (online supplemental figure 1). RT-qPCR showed that hypoxia, glucose deficiency, lactic acid accumulation, TGF-β, IFN-γ, ROS and induced senescence significantly downregulated the expression of Ankrd22 in CD11b+Ly6G+Ly6Clow cells (figure 2A). Evaluation of CD11b+Ly6G+Ly6Clow cells from the BM also showed that the expression level of Ankrd22 in tumor-bearing mice was significantly lower than those cells from control mice, and local tumor-derived CD11b+Ly6G+Ly6Clow cells have lower levels of Ankrd22 expression than BM-derived CD11b+Ly6G+Ly6Clow cells (figure 2B). Similar changes were also observed in CD11b+HLA-DR-CD14-CD15+ cells derived from fresh tumor tissue of patients with ovarian cancer (figure 2C). These studies suggest that the tumor microenvironment can downregulate Ankrd22 expression in CD11b+Ly6G+Ly6Clow cells.
Figure 2.
The tumor microenvironment affected the expression of Ankrd22 in BM-derived CD11b+Ly6G+Ly6 Clow cells. (A) RT-qPCR was used to detect the expression level of Ankrd22 in CD11b+Ly6G+Ly6Clow cells cultured under hypoxia, glucose deprivation, lactic acid accumulation, TGF-β, IFN-γ, ROS and CPT-11 inducing senescence,n=3, **p<0.01, ***p<0.001. (B) A subcutaneous ovarian cancer model was established in C57BL/6 mice. BM-derived CD11b+Ly6G+Ly6Clow cells and subcutaneous tumor tissues derived CD11b+Ly6G+Ly6Clow cells were sorted by FACS from tumor-bearing mice after 8 weeks, and the Ankrd22 expression level in CD11b+Ly6G+Ly6Clow cells was detected by RT-qPCR. CD11b+Ly6G+Ly6Clow cells from the bone marrow of wild-type mice were used as a control. *p<0.05, ***p<0.001. Two-sample Student’s t-test. (C) Ovarian tissues were collected from patients with and without ovarian cancer undergoing ovarian tissue excision. CD11b+HLA-DR-CD14-CD15+ cells were sorted by FACS after digestion, and the expression level of ANKRD22 were detected by RT-qPCR (n=6 for non-ovarian cancer and n=20 for ovarian cancer). ***p<0.001, two-sample Student’s t-test. BM, bone marrow; FACS, fluorescenceactivated cell sorting.
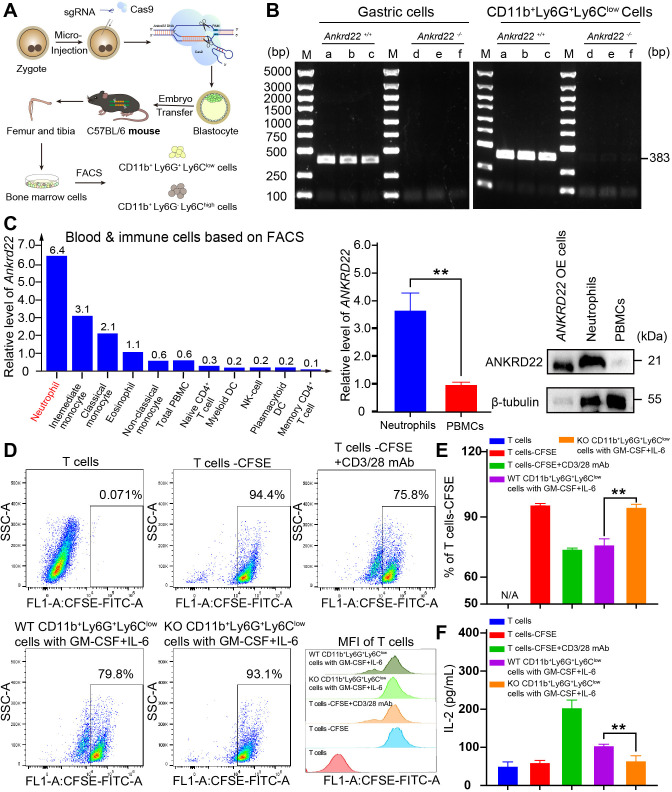
Knockout of Ankrd22 enhanced the immunosuppressive activity of PMN-MDSCs
To investigate the effect of Ankrd22 expression on the immunosuppressive activity of PMN-MDSCs, we constructed Ankrd22-knockout C57BL/6 mice (KO mice) with CRISPR-Cas9 technology (figure 3A). FACS was used to isolate BM-derived CD11b+Ly6G+Ly6Clow cells from KO mice, and PCR analysis showed that Ankrd22 was completely knocked out in CD11b+Ly6G+Ly6Clow cells (figure 3B). These results suggested that this mouse model could be used for subsequent experimental studies.
Figure 3.
Knockout of Ankrd22 enhanced the immunosuppressive activity of PMN-MDSCs. (A) Diagram of the construction of the Ankrd22-knockout C57BL/6 mouse model. (B). Bone marrow was collected from KO mice and WT mice to prepare single-cell suspensions, and then CD11b+Ly6G+Ly6Clow cells were sorted by FACS. PCR and agarose gel electrophoresis were used to detect Ankrd22. M: marker. n=3. Ankrd22 in gastric epithelial cells was used as a positive control. (C). The HPA database showed the expression level of ANKRD22 among human blood and immune cells based on flow sorted. Human peripheral blood neutrophils and mononuclear cells were extracted following the instruction of human peripheral blood neutrophil/mononuclear cell isolation kit, and the expression of ANKRD22 was measured by RT-qPCR and western blotting. The lysate of the ANKRD22-overexpressing (OE) 293 T cells as a positive control. n=3, **p<0.01. Two-sample Student’s t-test. (D, E) FCM was used to detect the percentage of proliferating CFSE-labeled T cells after coculture with CD11b+Ly6G+Ly6Clow cells which were treated with 100 ng/mL GM-CSF and 100 ng/mL IL-6 for 96 hours. CFSE fluorescence intensity of T cells were showed. n=3, **p<0.01. Two-sample Student’s t-test. (F). The content of IL-2 in the supernatant of cocultured cells was detected by ELISA. n=3, **p<0.01. Two-sample Student’s t-test. CFSE, carboxyfluorescein diacetate succinimidyl ester; FACS, fluorescenceactivated cell sorting; FCM, flow cytometry; HPA, Human Protein Atlas; PMN-MDSCs, polymorphonuclear myeloid-derived suppressor cells; WT, wild type.
Since PMN-MDSCs and neutrophils both have a CD11b+Ly6G+Ly6Clow phenotype, there is no characteristic molecular marker to distinguish them, and in accordance with the HPA database, mature neutrophils in human peripheral blood also express ANKRD22 at a high level (figure 3C). To determine whether the BM-derived CD11b+Ly6G+Ly6Clow cells could activate their immunosuppressive activity, CD11b+Ly6G+Ly6Clow cells derived from the BM of WT or KO mice were treated with GM-CSF and IL-6 and then cocultured with CFSE-labeled T cells derived from splenic immune cells. FCM and ELISA were used to detect the inhibitory effects of these BM-derived cells on T-cell proliferation and IL-2 secretion. The results showed that these BM-derived CD11b+Ly6G+Ly6Clow cells from WT or KO mice under GM-CSF and IL-6 stimulation significantly inhibited the proliferation and IL-2 secretion of T cells and that the effects of CD11b+Ly6G+Ly6Clow cells derived from KO mouse BM on T cells were more obvious (figure 3D–F, online supplemental figure 2). These studies suggested that BM-derived CD11b+Ly6G+ Ly6Clow cells under GM-CSF and IL-6 stimulation exhibited characteristics consistent with PMN-MDSCs and that silencing Ankrd22 enhanced the immunosuppressive activity of these PMN-MDSCs.
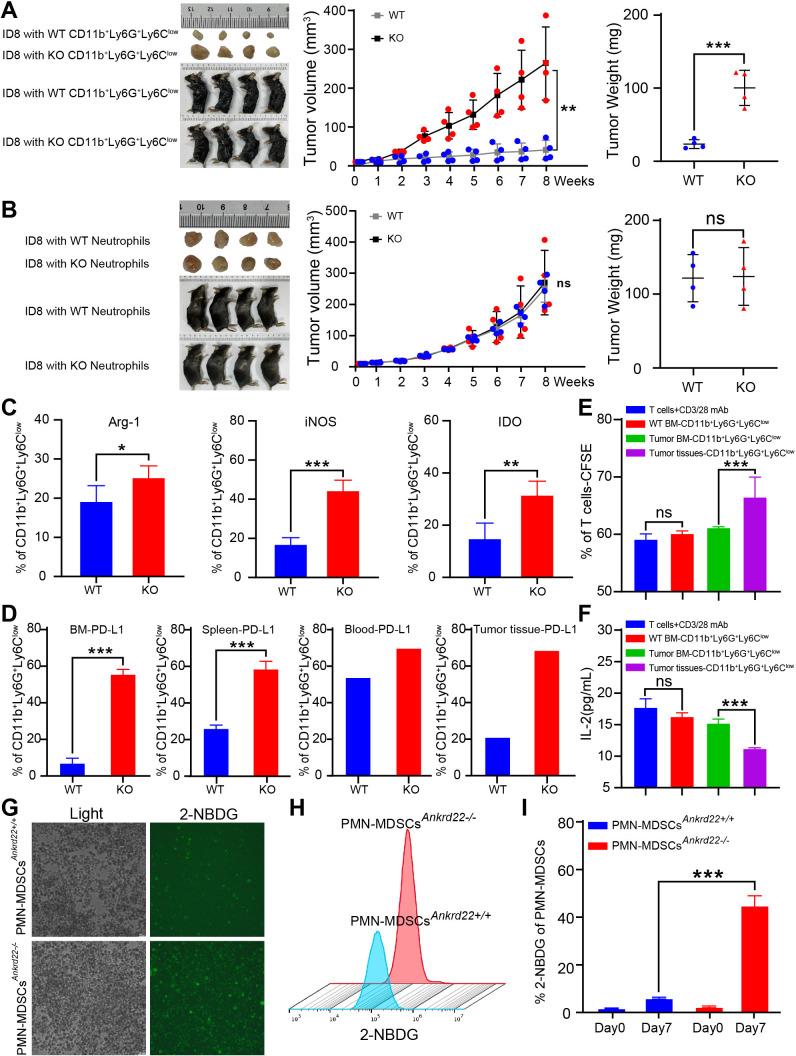
Knockout of Ankrd22 promoted the proliferation of ovarian cancer cells through enhanced immunosuppressive activity of PMN-MDSCs in transplanted tumor mouse models
The above study implied that BM-derived CD11b+Ly6G+Ly6Clow cells under GM-CSF and IL-6 stimulation had PMN-MDSCs characteristics and relatively strong immunosuppressive activity in vitro. We wondered whether similar phenomena could be observed in a transplanted tumor mouse model. To investigate this, CD11b+Ly6G+Ly6Clow cells were isolated from the BM of KO and WT C57BL/6 mice via FACS, and then these cells were mixed with mouse ID8 ovarian cancer cells in Matrigel for subcutaneous transplanted tumor experiments in C57BL/6 mice. The results showed that CD11b+Ly6G+ Ly6Clow cells from Ankrd22-/- mice promoted ovarian cancer cell proliferation more strongly than their WT counterparts (figure 4A). Similar results were observed in a transplanted melanoma tumor mouse model (data not shown). However, no significant difference in subcutaneous oncogenicity was found when mouse ID8 ovarian cancer cells were mixed with neutrophils from the peripheral blood of KO and WT mice (figure 4B). These results suggested that BM-derived CD11b+Ly6G+Ly6Clow cells from Ankrd22-/- mice could promote the proliferation of ovarian cancer cells in vivo.
Figure 4.
Knockout (KO) of Ankrd22 promoted the proliferation of ovarian cancer cells by enhancing the immunosuppressive ability of polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs) in transplanted tumor mouse models. (A) 5×106 ID8 cells/mouse mixed with 10×106 BM-derived CD11b+Ly6G+Ly6Clow cells from Ankrd22+/+ or Ankrd22-/- C57BL/6 mice, were suspended in Matrigel and injected into the axilla of WT C57BL/6 mice. Tumor diameters were measured with a Vernier caliper once a week for 8 weeks. The growth of subcutaneous tumors in mice was recorded. At the end of the experiment, the subcutaneous tumor weight was determined. n=4, **p<0.01, ***p<0.001. Two-sample Student’s t-test. (B). 5×106 ID8 cells/ mouse were mixed with 10×106 neutrophils from mouse peripheral blood and injected into the axilla of WT C57BL/6 mice. Tumor diameters were measured with a Vernier caliper once a week for 8 weeks. The growth of subcutaneous tumors in mice was recorded. At the end of the experiment, the subcutaneous tumor weight was determined. n=4, NS: p>0.05. Two-sample Student’s t-test. (C). Subcutaneous tumor tissues were collected from KO and WT mice, and intracellular Arg-1, iNOS and IDO on CD11b+ Ly6G+Ly6Clow cells were detected by flow cytometry (FCM) after stimulation with 81.0 nM PMA and 1.34 µM ionomycin. n=4, *p<0.05, **p<0.01, ***p<0.001. (D) FCM was used to detect PD-L1 on CD11b+Ly6G+Ly6Clow cells derived from the bone marrow, spleen, peripheral blood or subcutaneous tumor tissues of the tumor-bearing mice in WT and KO groups. ***p<0.001. Two-sample Student’s t-test. (E.) FCM was used to detect the percentage of proliferating carboxyfluorescein diacetate succinimidyl este (CFSE)-labeled T cells after coculture with CD11b+Ly6G+Ly6Clow cells from mice tumor tissues without treatment of 100 ng/mL GM-CSF and 100 ng/mL IL-6 for 96 hours. CFSE fluorescence intensity of T cells were showed. n=3, **p<0.01. Two-sample Student’s t-test. (F). The content of IL-2 in the supernatant of cocultured cells was detected by ELISA. n=3, **p<0.01. Two-sample Student’s t-test. (G–I). CD11b+Ly6G+ Ly6Clow cells from the tumor-bearing mice were cultured with DMEM containing 2-NBDG under 100 ng/mL GM-CSF and 100 ng/mL IL-6 stimulation for 7 days. The intensity of green fluorescence in the PMN-MDSCs was detected by immunofluorescence microscopy. The FCM results show the percentage of PMN-MDSCs with FITC fluorescence. n=3, ***p<0.001. BM, bone marrow; FITC, fluorescein isothiocyanate.
To investigate the mechanism underlying the promotion of ovarian cancer cell proliferation by CD11b+Ly6G+Ly6Clow cells from Ankrd22-/- mice, we detected the expression levels of PMN-MDSCs-related immunosuppressive molecules, such as Arg-1, iNOS and IDO, in subcutaneous tumor tissues from tumor-bearing C57BL/6 mice. Compared with the control group, the Ankrd22-knockout group showed significantly increased expression levels of Arg-1, iNOS and IDO in CD11b+Ly6G+Ly6Clow cells from the tumor tissues (figure 4C). Moreover, the expression levels of PD-L1 in CD11b+Ly6G+Ly6Clow cells derived from the BM, spleen, peripheral blood and subcutaneous tumor tissues of C57BL/6 mice were also significantly increased (figure 4D). Furthermore, we found that CD11b+Ly6G+Ly6Clow cells from mice tumor tissues could suppress T cell proliferation without additional stimulation with GM-CSF and IL-6, while BM-derived CD11b+Ly6G+Ly6Clow cells did not show any suppressive activity without previous treatment with GM-CSF and IL-6 (figure 4E, F, online supplemental figure 3). Glucose uptake ability was also significantly enhanced in PMN-MDSCsAnkrd22-/- induced by GM-CSF and IL-6 (figure 4G–I). These results suggested that TIME promoted the transition from CD11b+Ly6G+Ly6Clow cells to PMN-MDSCs and silencing Ankrd22 could enhance the immunosuppressive activity of PMN-MDSCs by increasing the expression of immunosuppressive molecules.
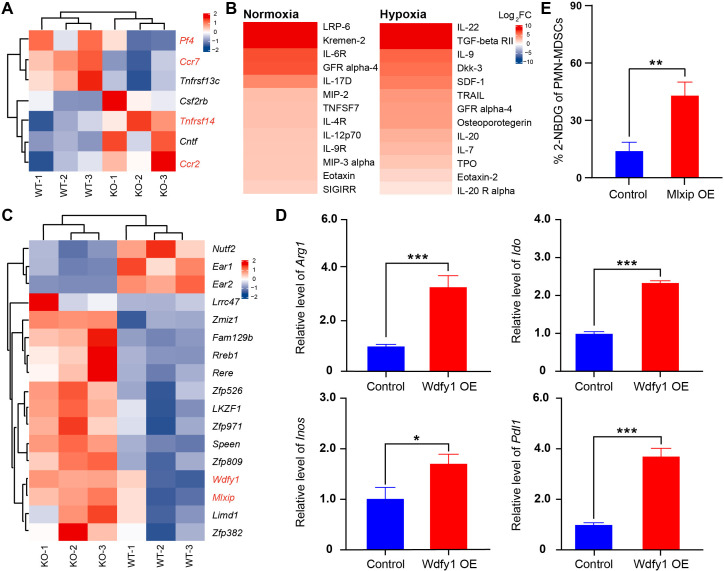
The immunosuppressive activity of PMN-MDSCsAnkrd22-/- was enhanced by WDFY1
To explore the molecular mechanism by which Ankrd22 deletion regulates the function of PMN-MDSCs, we compared changes in RNA transcription levels between BM-derived CD11b+Ly6G+ Ly6Clow cells between Ankrd22-/- and Ankrd22+/+ mice by RNA sequencing. The results showed that Ccr2 and Tnfrsf14 expression was significantly upregulated in the KO group compared with the WT group, while Pf4 and Ccr7 expression was significantly downregulated (figure 5A). Additionally, we detected the cytokines secreted by PMN-MDSCs under hypoxia with a cytokine chip assay. We found that MIP-2 and MIP-3α expression was significantly upregulated in the culture supernatant of PMN-MDSCsAnkrd22-/- compared with that of PMN-MDSCsAnkrd22+/+ under normoxia and that IL-22, TGF-β and DKK-3 expression was significantly upregulated under hypoxia (figure 5B). Furthermore, to explore the affect of ANKRD22 deficiency on the expression of Arg-1, iNOS, IDO and PD-L1 in PMN-MDSCs, we compared differential mRNA expression in BM-derived CD11b+Ly6G+ Ly6Clow cells from Ankrd22-/- mice with that from Ankrd22+/+ mice, and RNA sequencing showed that the expressions of Wdfy1 and Mlxip were significantly increased in BM-derived CD11b+Ly6G+Ly6Clow cells from Ankrd22-/-mice than that from Ankrd22+/+ mice (figure 5C). RT-qPCR indicated that the expression of Arg1, Inos, Ido and Pdl1 was significantly increased when PMN-MDSCs cells were successfully transfected with a Wdfy1-overexpressing plasmid (figure 5D). Notably, overexpression of MLXIP promoted glycolysis in PMN-MDSCs (figure 5E). These studies suggested that the immunosuppressive activity of PMN-MDSCsAnkrd22-/- was enhanced by WDFY1 and MLXIP.
Figure 5.
The immunosuppressive activity of polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs)Ankrd22-/- was enhanced by Wdfy1. (A) RNA sequencing. Bone marrow (BM)-derived CD11b+Ly6G+Ly6Clow cells from Ankrd22-/- and Ankrd22+/+ mice which sorted by fluorescenceactivated cell sorting (FACS) were checked. The heatmap shows mainly changed genes between these two groups, n=3. (B). BM-derived CD11b+Ly6G+Ly6Clow cells from Ankrd22-/- and Ankrd22+/+ mice were cultured with 100 ng/mL GM-CSF and 100 ng/mL IL-6, and the cell supernatants were collected for cytokine chip analysis after normoxic or hypoxic culture for 24 hours. The heatmap shows changes in the expression of cytokines in the supernatants of PMN-MDSCsAnkrd22-/- and PMN-MDSCsAnkrd22+/+. n=3. (C). Heatmap analysis. RNA sequencing showed that the expression of the transcription factors Wdfy1 and Mlxip was significantly upregulated in BM-derived CD11b+Ly6G+Ly6Clow cells from Ankrd22-/- mice. (D). PMN-MDSCs from wild-type (WT) mice were transfected with Wdfy1 and maintained with 100 ng/mL GM-CSF and 100 ng/mL IL-6, and then immunosuppressive molecules, such as Arg1, Ido, Inos and Pdl1, in the PMN-MDSCs were detected by RT-qPCR. n=3, *p<0.05,***p<0.001. Two-sample Student’s t-test. (E). BM-derived CD11b+Ly6G+ Ly6Clow cells from WT mice cultured with 100 ng/mL GM-CSF and 100 ng/mL IL-6 and 2-NBDG was infected with lentivirus overexpressing Mlxip. The ratio of 2-NBDG in the PMN-MDSCs was detected by flow cytometry (FCM) and the graph showed the exogenous glucose uptake ability of PMN-MDSCs. n=3, **p<0.01. Two-sample Student’s t-test. OE, overexpression.
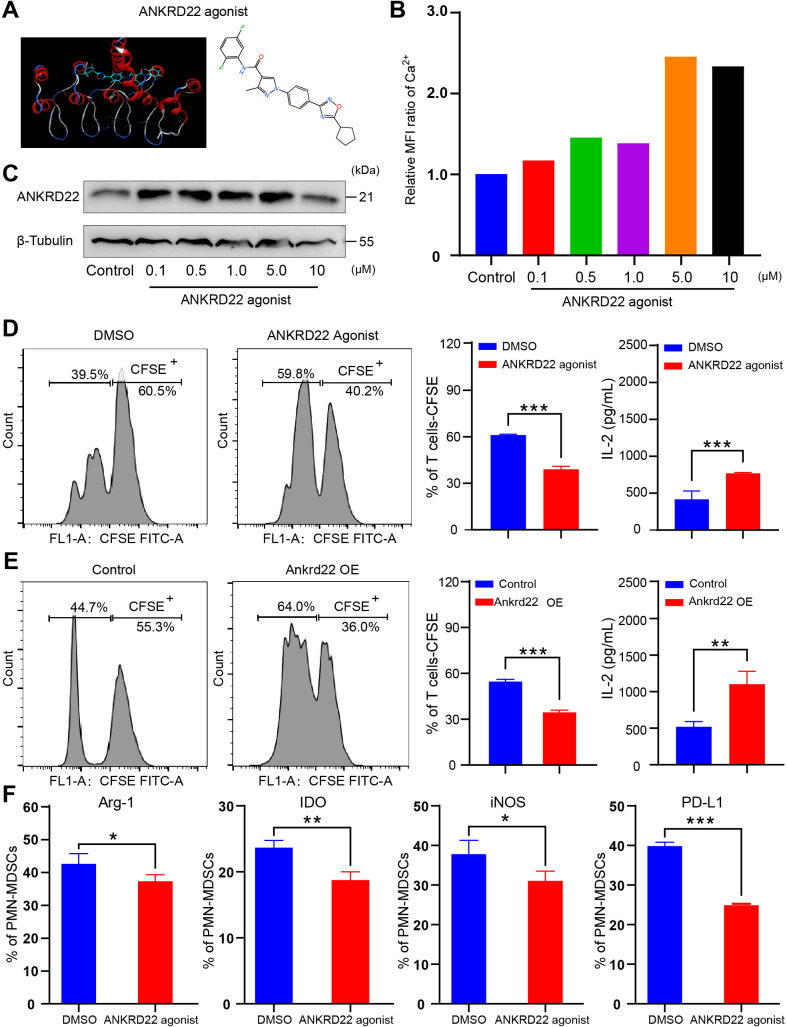
An ANKRD22 agonist reversed the immunosuppressive activity of PMN-MDSCs
The above studies confirmed that silencing ANKRD22 could promote the immunosuppressive activity of PMN-MDSCs, so we wondered whether upregulation of ANKRD22 in PMN-MDSCs could reverse this effect. To this end, we first identified a candidate small-molecule compound that could interact with ANKRD22 through 3D modeling and virtual screening (figure 6A). In vitro experiments showed that the lead compound could enhance the expression of ANKRD22 and increase the intracellular Ca2+ concentration (figure 6B, C). Furthermore, WT mice BM-derived CD11b+Ly6G+Ly6Clow cells stimulated with GM-CSF and IL-6 were cocultured with CFSE-labeled splenic T cells. FCM showed that either addition of candidate ANKRD22-activating compounds or overexpression of Ankrd22 in these CD11b+Ly6G+Ly6Clow cells significantly reduced the abilities of these cells to inhibit T-cell proliferation and IL-2 secretion (figure 6D, E). The FCM results showed that Arg-1, iNOS, IDO and PD-L1 levels were reduced after PMN-MDSCs were treated with the ANKRD22 agonist (figure 6F, online supplemental figure 4). Further, the ANKRD22 agonist weakened the inhibitory function of PMN-MDSCs on ID8 ovarian cancer cell proliferation in WT C57BL/6 mice (online supplemental figure 4). These results suggest that the ANKRD22 agonist can reverse the immunosuppressive function of PMN-MDSCs.
Figure 6.
An ANKRD22 agonist reversed the immunosuppressive activity of polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs). (A) Schematic diagram of ANKRD22-activating small-molecule compounds. (B) Human THP-1 cells were cocultured with agonist at concentrations of 0, 0.1, 0.5, 1, 5 and 10 µM for 24 hours. The mean fluorescence intensity of Ca2+ was measured to evaluate whether the small-molecule compound was effective. (C) Colorectal cancer LS-174T cells were cocultured with agonists at concentrations of 0, 0.1, 0.5, 1, 5 and 10 µM for 24 hours. The expression level of ANKRD22 was detected by western blotting. (D) BM-derived CD11b+Ly6G+Ly6Clow cells from wild-type (WT) mice, stimulated with 100 ng/mL GM-CSF and 100 ng/mL IL-6 (PMN-MDSCsAnkrd22+/+), were cocultured with actived carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled spleen-derived T cells in the presence of 1.0 µM ANKRD22-activating small-molecule compound for 96 hours. DMSO was added to the FCM culture medium as a control. Flow cytometry (FCM) showed the effect of PMN-MDSCs on T-cell proliferation after addition of the activating small-molecule compound. ELISA was used to detect IL-2 concentration (pg/mL) in the supernatant of PMN-MDSCs cocultured with T cells. n=3, ***p<0.001. Two-sample Student’s t-test. (E) PMN-MDSCsAnkrd22+/+ were infected with recombinant lentivirus overexpressing Ankrd22. FCM shows the effect of PMN-MDSCs on T-cell proliferation after infection. ELISA was used to detect the IL-2 concentration in the cell supernatant of PMN-MDSCs cocultured with T cells. n=3, **p<0.01, ***p<0.001. Two-sample Student’s t-test. (F) PMN-MDSCs were treated with an ANKRD22 agonist for 72 hours in vitro, and the results showed that the levels of Arg-1, iNOS, IDO and PD-L1 were reduced in PMN-MDSCs which were with ANKRD22 agonist. n=3, *p<0.05, **p<0.01, ***p<0.001. Two-sample Student’s t-test. BM, bone marrow.
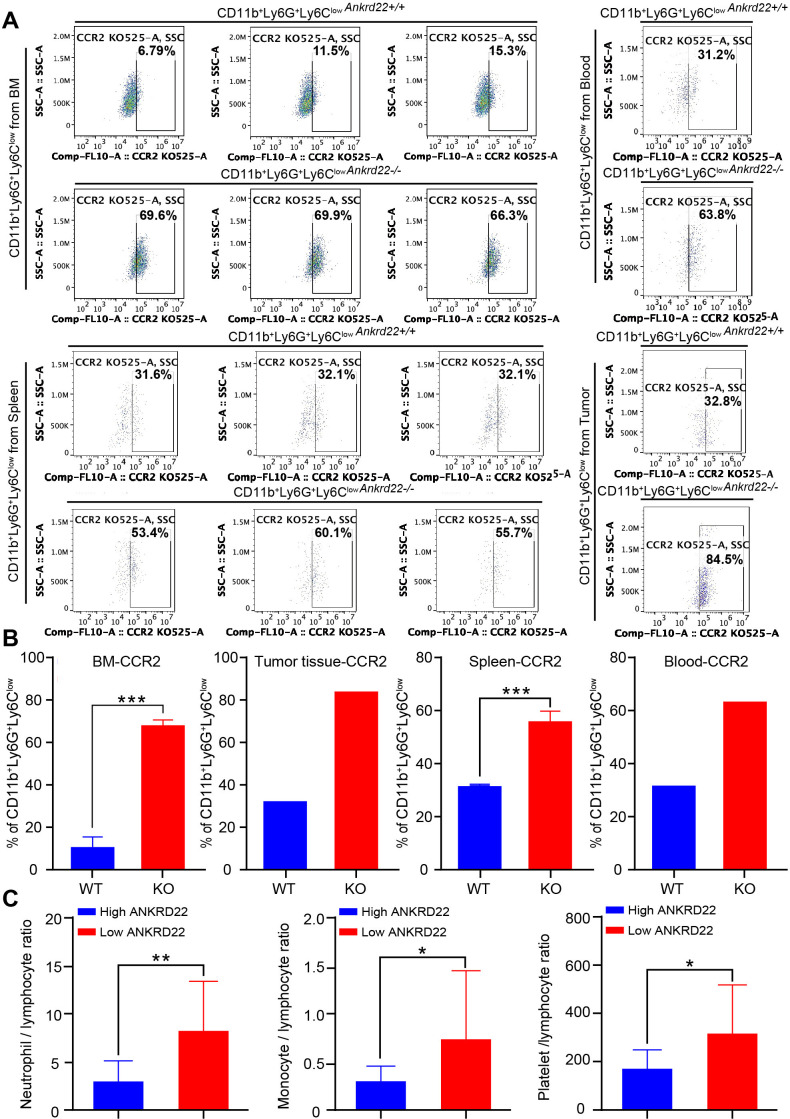
Ankrd22 knockout increased the expression of CCR2 to promote the recruitment of CD11b+Ly6G+Ly6Clow cells to ovarian cancer tissue and were linked to the clinical prognosis of ovarian cancer
Studies have shown that CCR2 plays an important role in the chemotaxis of immune cells. Does ANKRD22 play a role in promoting the chemotaxis of PMN-MDSCs to ovarian cancer tissues? To explore whether the expression of Ankrd22 affects the expression level of CCR2 on CD11b+Ly6G+Ly6Clow cells, BM-derived CD11b+Ly6G+Ly6Clow cells of Ankrd22+/+ and Ankrd22-/- mice were sorted. After being mixed with ID8 ovarian cancer cells, the cells were subcutaneously transplanted into C57BL/6 mice and then collected 8 weeks later. FCM detection showed that the expression of CCR2 on CD11b+Ly6G+Ly6Clow cells derived from the BM, spleen, peripheral blood or subcutaneous tumor tissue was significantly increased in the KO group (figure 7A, B). These results suggested that silence of Ankrd22 could upregulate the expression of CCR2 on the surface of CD11b+Ly6G+Ly6Clow cells of the tumor-bearing mice and promote the chemotaxis of CD11b+Ly6G+Ly6Clow cells to the local ovarian cancer microenvironment. Finally, to analyze the correlation between the number of PMN-MDSCs and the clinical prognosis of ovarian cancer, CD11b+HLA-DR-CD14-CD15+ cells were sorted from fresh ovarian cancer tissues by FACS, and then RT-qPCR was used to detect ANKRD22 expression. Patients with ovarian cancer were divided into a high-expression group and a low-expression group according to their ANKRD22 expression level, and the clinical characteristics of the patients were collected to conduct a correlation analysis. The results showed that low expression of ANKRD22 in CD11b+HLA-DR-CD14-CD15+ cells derived from ovarian cancer tissue was associated with a more advanced FIGO stage, a higher recurrence rate (table 1), and a higher neutrophil-to-lymphocyte ratio (NLR) (figure 7C). These results suggest that patients with low ANKRD22 expression in CD11b+HLA-DR-CD14-CD15+ cells derived from ovarian tissue have a poorer prognosis.
Figure 7.
ANKRD22 knockout increases CCR2 expression and affects the clinical characteristics and prognosis of ovarian cancer. (A, B) BM-derived CD11b+Ly6G+Ly6Clow cells from Ankrd22-/- and Ankrd22+/+ mice were mixed with ID8 cells to construct the tumor-bearing mouse model on WT mice. After tumor formation in mice, the BM, spleen, peripheral blood and subcutaneous tumor tissues were collected to isolate CD11b+Ly6G+Ly6Clow cells by fluorescenceactivated cell sorting (FACS), and the expression of CCR2 was determined by flow cytometry (FCM). n=3, ***p<0.001. Two-sample Student’s t-test. (C) According to the expression level of ANKRD22, patients with ovarian cancer were divided into a high-expression group and a low-expression group, and the numbers of neutrophils, monocytes, platelets and lymphocytes in the peripheral blood were determined. (A) Neutrophil-to-lymphocyte ratio (NLR), n=20, **p<0.01; (B) Monocyte-to-lymphocyte ratio (MLR), n=20, *p<0.05; (C) Platelet-to-lymphocyte ratio (PLR), n=20, *p<0.05. Two-sample Student’s t-test. BM, bone marrow; WT, wild type.
Table 1.
The correlation between different ANKRD22 expression levels andclinicopathological features of patients with ovarian cancer
| Characteristics | All patients | ANKRD22 Expression | P value | |
| Low | High | |||
| No | 20 | 7 | 13 | |
| Age (years) | 0.630 | |||
| <55 | 9 | 3 | 6 | |
| ≥55 | 11 | 4 | 7 | |
| Pathological type | 0.374 | |||
| Non-epithelial | 9 | 2 | 7 | |
| Epithelial | 11 | 5 | 6 | |
| Diffentiation | 0.651 | |||
| Poor differentiated | 7 | 3 | 4 | |
| Well differentiated | 13 | 4 | 9 | |
| NLR | 0.044 | |||
| <2.14 | 7 | 0 | 7 | |
| ≥2.14 | 13 | 7 | 6 | |
| MLR | ||||
| <0.24 | 5 | 2 | 3 | |
| ≥0.24 | 15 | 5 | 10 | |
| PLR | 0.114 | |||
| <146.11 | 5 | 0 | 5 | |
| ≥146.11 | 15 | 7 | 8 | |
| Recurred | 0.031 | |||
| Yes | 5 | 4 | 1 | |
| No | 15 | 3 | 12 | |
| FIGO stage | 0.044 | |||
| I–III | 7 | 0 | 7 | |
| IV | 13 | 7 | 6 | |
FIGO, International Federation of Gynecology and Obstetrics; MLR, monocyte-to-lymphocyte ratio; NLR, neutrophil-tolymphocyte ratio; PLR, platelet-to-lymphocyte ratio.
Discussion
Our study shows that ANKRD22 is highly expressed in CD11b+Ly6G+Ly6Clow cells. The tumor microenvironment can downregulate the expression of Ankrd22 in BM-derived CD11b+Ly6G+Ly6Clow cells and promote the conversion from CD11b+Ly6G+Ly6Clow cells to PMN-MDSCs. Based on an Ankrd22 knockout mouse model, Ankrd22 knockout enhanced the chemotaxis of CD11b+Ly6G+Ly6Clow cells to ovarian cancer tissues by increasing the expression of CCR2. At the same time, after Ankrd22 was knocked out, the immunosuppressive activity of PMN-MDSCs was increased, leading to the formation of an immunosuppressive tumor microenvironment, which ultimately promoted the progression of ovarian cancer.
ANKRD22 is an ankyrin (ANK) domain repeat protein that is located at chromosome 10q23.31 and is involved in the genesis and development of a variety of tumor cells.13 ANKRD22 expression increased in basal breast cancer, non-small cell lung cancer and colon cancer cells but decreased in prostate cancer, and these changes affect tumor progression and prognosis.13–17 Our previous study found that the TIME promoted the expression of ANKRD22 in colorectal cancer cells through the p38/MAX pathway, and ANKRD22 restored the stem cell characteristics of colorectal cancer cells by promoting glycolysis and lipid metabolism reprogramming.13 To investigate which immune cells expressed ANKRD22, we searched BioGPS and HPA and found that in addition to gastric epithelial cells,14 myeloid cells and their precursors, neutrophils, monocytes, B cells, plasma cells, and other cell types may also express ANKRD22 at a high level. Finally, we sorted the above cells according to the cell surface cluster of differentiation (CD). Through RT-qPCR, we found that the expression level of Ankrd22 was high in CD11b+Ly6G+Ly6Clow cells obtained from the mouse BM, while Ankrd22 in B cells and plasma cells was low.
CD11b+Ly6G+Ly6Clow cells include PMN-MDSCs and neutrophils.18 However, PMN-MDSCs and neutrophils still lack effective molecular markers for identification. Although Lectin-like oxidized low density lipoprotein receptor 1 (LOX-1) can be used as a marker to distinguish human PMN-MDSCs from neutrophils, identification markers for mouse-derived PMN-MDSCs are still lacking.19 The ‘gold standard’ for distinguishing PMN-MDSCs from neutrophils is the ability to inhibit the immune activity of T cells.18 In vitro, GM-CSF and IL-6 can activate mice BM-derived naive CD11b+Ly6G+Ly6Clow cells into PMN-MDSCs to gain the immune inhibitory capacity. To confirm whether mouse BM-derived CD11b+Ly6G+Ly6Clow cells have PMN-MDSC characteristics, we used FACS to separate mouse BM-derived CD11b+Ly6G+Ly6Clow cells cultured with GM-CSF+IL-6 and tested their ability to promote T cell proliferation and induce T-cell release of IL-2. In addition, we evaluated whether mouse BM-derived CD11b+Ly6G+Ly6Clow cells had immunosuppressive activity by inoculating them with ovarian cancer cells for tumorigenesis experiments in immunocompetent mice to observe the tumor growth. The results showed that BM-derived CD11b+Ly6G+Ly6Clow cells could significantly inhibit the proliferation of T cells and the release of IL-2 by T cells under GM-CSF+IL-6 stimulation and could significantly enhance the tumorigenicity of ovarian cancer cells in vivo. However, neutrophils isolated from mouse peripheral blood did not have the above inhibitory capacity, suggesting that CD11b+Ly6G+Ly6Clow cells derived from mouse BM have the characteristics of PMN-MDSCs after GM-CSF and IL-6 stimulation. Furthermore, we observed the effect of tumor microenvironment stimulation on Ankrd22 expression in mouse BM-derived CD11b+Ly6G+Ly6Clow cells. In contrast to observations in colorectal cancer cells, Ankrd22 expression in mouse BM-derived CD11b+Ly6G+Ly6Clow cells was significantly lower than that in the control group after exposure to different tumor microenvironment stimulus, GM-CSF+IL-6 and coculture with ID8 ovarian cancer cells, suggesting that mouse BM-derived CD11b+Ly6G+Ly6Clow cells may not rely on the p38/MAX pathway to regulate the expression of Ankrd22.
An increasing number of studies have shown that PMN-MDSCs exert their immunosuppressive effects by enhancing the expression of Arg-1, iNOS and IDO.20–22 In our study, we found that Ankrd22 knockout increased the expression of immunosuppressive molecules of PMN-MDSCs, such as Arg-1, iNOS, IDO, and PD-L1. The potential ANKRD22 agonist could reverse this increase, suggesting that the expression of immunosuppressive molecules of PMN-MDSCs, Arg-1, iNOS, IDO and PD-L1, was related to Anrkd22. To clarify how ANKRD22 regulates the immunosuppressive function of PMN-MDSCs, we referred to our previous research showing that ANKRD22 is an important molecule involved in cell glycolysis and lipid metabolism.13 In this study, we compared the changes in the transcription factor expression profiles of Ankrd22 knockout and non-Ankrd22 knockout CD11b+Ly6G+Ly6Clow cells. We found that Zfp382, Limd1, Mlxip, Wdfy1, and other factors were significantly upregulated in Ankrd22 knockout CD11b+Ly6G+Ly6Clow cells. A comprehensive analysis of the literature suggests that WDFY1 and MLXIP may be related to the immunosuppressive function and disordered energy metabolism of PMN-MDSCs.23–25 Therefore, we used a specific plasmid targeting Wdfy1 overexpression in non-Ankrd22 knockout PMN-MDSCs derived from mouse BM. RT-qPCR showed that WDFY1 overexpression increased the expression of Arg1, Inos, Ido and Pdl1 in PMN-MDSCs, suggesting that Wdfy1 may be the target of Ankrd22 in the inhibition of PMN-MDSCs. WDFY1 encodes a phosphatidylinositol 3-phosphate binding protein, which contains a FYVE zinc finger domain and multiple WD40 repeat domains.23 WDFY1 can mediate the interaction between Toll-like receptor 3 (TLR3) or Toll-like receptor 4 (TLR4) and TICAM1 and positively regulate signaling pathways mediated by TLR3 and TLR4, thus promoting the activation of the TLR3/4 ligand-induced transcription factors IRF3 and NF-κB. Activation of the TLR3/4-NF-κB pathway has been proven to promote the expression of IDO.26 Stanton et al also found that the WDFY1 protein can participate in autophagy through VEGF-C/NRP-2.23 In addition, MLXIP and MLX form heterodimers, which activate the transcription of target genes by binding DNA and participate in the regulation of cellular glycometabolism and lipid metabolism. MLXIP is also a metabolic sensor of the c-Myc pathway.24 MLXIP restricts oxidative phosphorylation by reducing the activity of pyruvate dehydrogenase, thus sensing metabolic pressure in B-ALL cells.24 Depletion of MLXIP reduces the transformation ability of B-ALL cells in vitro and decreases their malignancy potential in vivo.24 Our study showed that Ankrd22 knockout in PMN-MDSCs enhanced the uptake of exogenous glucose, while overexpression of MLXIP in non-Ankrd22 knockout PMN-MDSCs also promoted the uptake of exogenous glucose, suggesting that MLX transcription factor family members may be targets of ANKRD22. However, how MLXIP-mediated changes in the energy metabolism of PMN-MDSCs enhance their immunosuppressive function, and how ANKRD22 interacts with MLXIP and WDFY1 to mediate the differentiation of PMN-MDSCs and the acquisition of their immunosuppressive function needs further study. Although Ankrd22 knockout enhanced the immunosuppressive activity of PMN-MDSCs, we found that systemic Ankrd22 knockout did not affect the mortality of mice or increase infection rates or tumorigenicity, suggesting that another regulatory mechanism is involved in immune status.
Finally, we also found that Ankrd22 knockout increased the expression of CCR2 in CD11b+Ly6G+Ly6Clow cells, which increased the chemotaxis of CD11b+Ly6G+Ly6Clow cells to local tumor tissue. Studies have shown that multiple chemokines are expressed in ovarian cancer and other cancer TIMEs, and these chemokines can recruit MDSCs to promote the formation of an immunosuppressive microenvironment.27 28 CCL2/CCR2 is an important molecular mechanism underlying the chemotaxis of MDSCs to local tumor sites.29 A CCR2 antagonist inhibited the infiltration of TAMs and MDSCs and delayed the growth of tumors in a mouse tumorigenesis model.30 Incomplete radiofrequency ablation (iRFA) of liver cancer can cause chronic inflammation in the body, and residual liver cancer cells secrete CCL2, which induces TAMs and MDSCs to accumulate in local tumors and weakens the effect of anti-PD-1 treatment.31 Based on the important role of the CCL2/CCR2 axis in the development and differentiation of MDSCs, Flores-Toro et al32 established a mouse model of glioblastoma and found that the combination of anti-CCR2 and anti-PD-1 therapy could reduce the number of MDSCs in the local TIME but increase the number of tumor-infiltrating lymphocytes and IFN-γ expression, which prolonged the median survival time of tumor-bearing mice. Although we observed that Ankrd22 knockout also promotes the chemotaxis of CD11b+Ly6G+Ly6Clow cells to ovarian cancer tissues through CCL2/CCR2, how Ankrd22 knockdown increases CCR2 expression of CD11b+Ly6G+Ly6Clow cells, especially the relationship between MLXIP and WDFY1, remains to be further clarified.
In conclusion, Ankrd22 knockout increases the chemotaxis and immunosuppressive activity of PMN-MDSCs in local tumors and indirectly promotes the growth of ovarian cancer cells by inducing the formation of an immunosuppressive microenvironment. Thus, ANKRD22 is a potential drug target to reverse the immunosuppressive activity of PMN-MDSCs.
Footnotes
HC and KY contributed equally.
Contributors: HC and YZ conceived and designed the study. All authors participated in the experiments and collaboratively in drafting the manuscript. HC and KY contributed to FACS and histological examination. HC and LP contributed to RT-qPCR and Western blot. HC, KY and LP performed animal experiments. YZ and JZ supervised the study. JF, YZ and JZ performed funding acquisition. JZ is responsible for the overall content as guarantor.
Funding: This work was supported by grants from the National Natural Science Foundation of China (81902633) for JF, Zhejiang Provincial Natural Science Foundation of China (LY20H160028) for JZ and Key Research and Development Program of Zhejiang Province of China (2020C03013) for YZ.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. Not applicable.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
All animal experiments were approved by the Second Affiliated Hospital of the Zhejiang University School of Medicine (Reference number: 2020-96) and the Medical Animal Ethics Committee of Zhejiang Chinese Medicine University (Reference number: IACUC-20210628-07). All samples were collected from the Second Affiliated Hospital of the Zhejiang University School of Medicine. This study was approved by the Ethics Committee of the Second Affiliated Hospital of the Zhejiang University School of Medicine (Reference number: 2022–0059).
References
- 1.Kuroki L, Guntupalli SR. Treatment of epithelial ovarian cancer. BMJ 2020;371:m3773. 10.1136/bmj.m3773 [DOI] [PubMed] [Google Scholar]
- 2.du Bois A, Baert T, Vergote I. Role of neoadjuvant chemotherapy in advanced epithelial ovarian cancer. J Clin Oncol 2019;37:2398–405. 10.1200/JCO.19.00022 [DOI] [PubMed] [Google Scholar]
- 3.Demircan NC, Boussios S, Tasci T, et al. Current and future immunotherapy approaches in ovarian cancer. Ann Transl Med 2020;8:1714. 10.21037/atm-20-4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bejarano L, Jordāo MJC, Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Discov 2021;11:933–59. 10.1158/2159-8290.CD-20-1808 [DOI] [PubMed] [Google Scholar]
- 5.Hamanishi J, Mandai M, Ikeda T, et al. Safety and antitumor activity of anti-PD-1 antibody, nivolumab, in patients with platinum-resistant ovarian cancer. J Clin Oncol 2015;33:4015–22. 10.1200/JCO.2015.62.3397 [DOI] [PubMed] [Google Scholar]
- 6.Hegde S, Leader AM, Merad M. Mdsc: markers, development, states, and unaddressed complexity. Immunity 2021;54:875–84. 10.1016/j.immuni.2021.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li K, Shi H, Zhang B, et al. Myeloid-Derived suppressor cells as immunosuppressive regulators and therapeutic targets in cancer. Signal Transduct Target Ther 2021;6:362. 10.1038/s41392-021-00670-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veglia F, Perego M, Gabrilovich D. Myeloid-Derived suppressor cells coming of age. Nat Immunol 2018;19:108–19. 10.1038/s41590-017-0022-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Wang J, Wu W, et al. Myeloid-Derived suppressor cells promote epithelial ovarian cancer cell stemness by inducing the CSF2/p-STAT3 signalling pathway. FEBS J 2020;287:5218–35. 10.1111/febs.15311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui TX, Kryczek I, Zhao L, et al. Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microrna101 and suppressing the corepressor ctbp2. Immunity 2013;39:611–21. 10.1016/j.immuni.2013.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veglia F, Sanseviero E, Gabrilovich DI. Myeloid-Derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol 2021;21:485–98. 10.1038/s41577-020-00490-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veglia F, Hashimoto A, Dweep H, et al. Analysis of classical neutrophils and polymorphonuclear myeloid-derived suppressor cells in cancer patients and tumor-bearing mice. J Exp Med 2021;218:e20201803. 10.1084/jem.20201803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan T, Liu J, Xu S, et al. ANKRD22, a novel tumor microenvironment-induced mitochondrial protein promotes metabolic reprogramming of colorectal cancer cells. Theranostics 2020;10:516–36. 10.7150/thno.37472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Wu J, Wang R, et al. ANKRD22 drives rapid proliferation of lgr5+ cells and acts as a promising therapeutic target in gastric mucosal injury. Cell Mol Gastroenterol Hepatol 2021;12:1433–55. 10.1016/j.jcmgh.2021.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y, Liu H, Gong Y, et al. ANKRD22 enhances breast cancer cell malignancy by activating the Wnt/β-catenin pathway via modulating NUSAP1 expression. Bosn J Basic Med Sci 2021;21:294–304. 10.17305/bjbms.2020.4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin J, Fu W, Dai L, et al. ANKRD22 promotes progression of non-small cell lung cancer through transcriptional up-regulation of E2F1. Sci Rep 2017;7:4430. 10.1038/s41598-017-04818-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu Y, Yang S, Pan T, et al. ANKRD22 is involved in the progression of prostate cancer. Oncol Lett 2019;18:4106–13. 10.3892/ol.2019.10738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giese MA, Hind LE, Huttenlocher A. Neutrophil plasticity in the tumor microenvironment. Blood 2019;133:2159–67. 10.1182/blood-2018-11-844548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Condamine T, Dominguez GA, Youn JI, et al. Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci Immunol 2016;1:aaf8943. 10.1126/sciimmunol.aaf8943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Sun HW, Yang YY, et al. Reprogramming immunosuppressive myeloid cells by activated T cells promotes the response to anti-PD-1 therapy in colorectal cancer. Signal Transduct Target Ther 2021;6:4. 10.1038/s41392-020-00377-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Redd PS, Ibrahim ML, Klement JD, et al. Setd1B activates iNOS expression in myeloid-derived suppressor cells. Cancer Res 2017;77:2834–43. 10.1158/0008-5472.CAN-16-2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okła K, Czerwonka A, Wawruszak A, et al. Clinical relevance and immunosuppressive pattern of circulating and infiltrating subsets of myeloid-derived suppressor cells (mdscs) in epithelial ovarian cancer. Front Immunol 2019;10:691. 10.3389/fimmu.2019.00691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanton MJ, Dutta S, Zhang H, et al. Autophagy control by the VEGF-C/NRP-2 axis in cancer and its implication for treatment resistance. Cancer Res 2013;73:160–71. 10.1158/0008-5472.CAN-11-3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sipol A, Hameister E, Xue B, et al. Mondoa drives malignancy in B-ALL through enhanced adaptation to metabolic stress. Blood 2022;139:1184–97. 10.1182/blood.2020007932 [DOI] [PubMed] [Google Scholar]
- 25.Lu Y, Li Y, Liu Q, et al. MondoA-thioredoxin-interacting protein axis maintains regulatory T-cell identity and function in colorectal cancer microenvironment. Gastroenterology 2021;161:575–91. 10.1053/j.gastro.2021.04.041 [DOI] [PubMed] [Google Scholar]
- 26.Hemmati S, Sadeghi MA, Mohammad Jafari R, et al. The antidepressant effects of GM-CSF are mediated by the reduction of TLR4/NF-ĸB-induced IDO expression. J Neuroinflammation 2019;16:117. 10.1186/s12974-019-1509-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horikawa N, Abiko K, Matsumura N, et al. Expression of vascular endothelial growth factor in ovarian cancer inhibits tumor immunity through the accumulation of myeloid-derived suppressor cells. Clin Cancer Res 2017;23:587–99. 10.1158/1078-0432.CCR-16-0387 [DOI] [PubMed] [Google Scholar]
- 28.Obermajer N, Muthuswamy R, Odunsi K, et al. Pge (2) -induced CXCL12 production and CXCR4 expression controls the accumulation of human mdscs in ovarian cancer environment. Cancer Res 2011;71:7463–70. 10.1158/0008-5472.CAN-11-2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zilio S, Bicciato S, Weed D, et al. Ccr1 and CCR5 mediate cancer-induced myelopoiesis and differentiation of myeloid cells in the tumor. J Immunother Cancer 2022;10:e003131. 10.1136/jitc-2021-003131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang D, Li X, Li J, et al. Apobec3B interaction with PRC2 modulates microenvironment to promote HCC progression. Gut 2019;68:1846–57. 10.1136/gutjnl-2018-317601 [DOI] [PubMed] [Google Scholar]
- 31.Shi L, Wang J, Ding N, et al. Inflammation induced by incomplete radiofrequency ablation accelerates tumor progression and hinders PD-1 immunotherapy. Nat Commun 2019;10:5421. 10.1038/s41467-019-13204-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flores-Toro JA, Luo D, Gopinath A, et al. Ccr2 inhibition reduces tumor myeloid cells and unmasks a checkpoint inhibitor effect to slow progression of resistant murine gliomas. Proc Natl Acad Sci U S A 2020;117:1129–38. 10.1073/pnas.1910856117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-005527supp001.pdf (982.3KB, pdf)
Data Availability Statement
Data are available on reasonable request. Not applicable.