Key Points
Question
Can a grand challenge be used to facilitate the advancement of automated digital breast tomosynthesis (DBT) cancer detection technology?
Findings
This diagnostic study, in which 8 challenge teams developed algorithms to detect lesions on 22 032 DBT volumes, resulted in tumor detection performances as high as a mean biopsied lesion sensitivity of 0.957, which arose from the development of several novel approaches.
Meaning
The variety of approaches that participants used in this study, alongside their released code and our released tumor detection benchmarking platform, present a starting point for future research in this area.
This diagnostic study describes a grand challenge for using artificial intelligence (AI) to detect cancer in digital breast tomosynthesis images.
Abstract
Importance
An accurate and robust artificial intelligence (AI) algorithm for detecting cancer in digital breast tomosynthesis (DBT) could significantly improve detection accuracy and reduce health care costs worldwide.
Objectives
To make training and evaluation data for the development of AI algorithms for DBT analysis available, to develop well-defined benchmarks, and to create publicly available code for existing methods.
Design, Setting, and Participants
This diagnostic study is based on a multi-institutional international grand challenge in which research teams developed algorithms to detect lesions in DBT. A data set of 22 032 reconstructed DBT volumes was made available to research teams. Phase 1, in which teams were provided 700 scans from the training set, 120 from the validation set, and 180 from the test set, took place from December 2020 to January 2021, and phase 2, in which teams were given the full data set, took place from May to July 2021.
Main Outcomes and Measures
The overall performance was evaluated by mean sensitivity for biopsied lesions using only DBT volumes with biopsied lesions; ties were broken by including all DBT volumes.
Results
A total of 8 teams participated in the challenge. The team with the highest mean sensitivity for biopsied lesions was the NYU B-Team, with 0.957 (95% CI, 0.924-0.984), and the second-place team, ZeDuS, had a mean sensitivity of 0.926 (95% CI, 0.881-0.964). When the results were aggregated, the mean sensitivity for all submitted algorithms was 0.879; for only those who participated in phase 2, it was 0.926.
Conclusions and Relevance
In this diagnostic study, an international competition produced algorithms with high sensitivity for using AI to detect lesions on DBT images. A standardized performance benchmark for the detection task using publicly available clinical imaging data was released, with detailed descriptions and analyses of submitted algorithms accompanied by a public release of their predictions and code for selected methods. These resources will serve as a foundation for future research on computer-assisted diagnosis methods for DBT, significantly lowering the barrier of entry for new researchers.
Introduction
Breast cancer is the leading cause of cancer death for women worldwide,1 and detection is a challenging process that requires the involvement of experienced radiologists. Digital breast tomosynthesis (DBT) creates high-resolution quasi–3-dimension (3D) scans consisting of multiple adjacent reconstruction slices, which reduces the effect of overlapping tissues seen in 2D mammography. This improves cancer detection rates but at the cost of increased reading time.2 An AI-based DBT cancer detection tool with radiologist-level performance could significantly reduce cancer screening costs and time and improve detection performance, which would be particularly helpful at sites that do not have access to fellowship-trained radiologists.
The most common AI method for image analysis is deep learning, which involves the training of nonlinear hierarchical models with many parameters (known as neural networks) to perform difficult tasks, such as image classification,3 object detection,4 and semantic segmentation,5,6 enabled by large data sets and specialized computing power.7 Deep learning detection algorithms have even surpassed radiologist performance8,9 due to their ability to learn far more complex features than earlier computer-assisted diagnosis systems, which had limited clinical applicability.10,11 In fact, algorithms with high sensitivity may even detect cancers missed by radiologists, serving as a second independent reader.12 However, developing deep learning algorithms for medical image analysis faces significant challenges, including a lack of sufficient, well-organized, and labeled training data; a lack of benchmark and test data as well as clearly defined rules for comparing algorithms, especially important because systems with significant false-positive rates can reduce radiologist sensitivity12; and limited access to previously developed algorithms for comparison. Moreover, DBT lesion detection introduces further difficulties for deep learning, including high scan resolution, high anatomical variability of both normal and abnormal breast tissue, and a very high class-imbalance of normal to cancerous cases for screening DBT.
In this article, we provide a practical foundation for the future open development and evaluation of algorithms for DBT lesion detection by providing a collection of analyses and resources for researchers, based on a new publicly available data set. Namely, we created a well-defined benchmark for evaluating future DBT lesion detection algorithms13; a description of several state-of-the-art algorithms for the task; a public release and comparative analysis of the predictions made by these algorithms, allowing for comparison with future approaches; and code for several of the algorithms, where possible. To generate these resources, we hosted a grand challenge, DBTex, for the automated detection of lesions in screening DBT scans. DBTex was divided into 2 phases, from December 14, 2020, to January 25, 2021, and May 24 to July 26, 2021, respectively. Challenges such as BraTS,14 ImageNet,15 other Kaggle competitions,16 and others have long been used to move the field forward by motivating intense and competitive research.
Several recent works have used deep learning to either classify DBT scans for the presence of lesions17,18,19,20,21,22,23,24,25,26,27,28,29 or localize lesion(s) within DBT scans. Localization tasks include determining the exact shape of these lesions, known as segmentation,12,30,31 or drawing bounding boxes around them, known as detection.32,33,34,35,36,37,38,39,40 Our challenge task was the detection of masses and architectural distortions in DBT scans.
Challenge teams developed and trained their detection methods on a large data set of healthy participants, with limited scans containing lesions, from a recently released large, public radiologist-labeled data set of DBT volumes from 5060 patients. After the training phase, participants were provided with a smaller validation data set to fine-tune their methods. At the end of the challenge, teams applied their methods to a previously unseen test set of scans with normal and cancerous tissue, which was used to obtain final rankings. While pathology and lesion locations of the training set were shared with participants as a reference standard, they were made unavailable for the validation and test sets.
Methods
This study was approved by the Duke University Health System institutional review board with a waiver of informed consent due to its retrospective nature. The Duke University Breast Cancer Screening DBT (BCS-DBT) data set, which was provided by the challenge organizers, was publicly available data. Three teams used additional data: the NYU B-Team used an internal data set approved by the NYU Langone Health institutional review board, ZeDuS used an internal institutional review board–approved data set, and VICOROB used the OPTIMAM data set (OMI-DB), whose ethical approval is publicly available.41 This study followed the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) reporting guideline.
Data Set
DBTex was built on Duke University’s BCS-DBT data set,32 which was designed to be the first large, curated and labeled, and publicly available DBT data set, hosted on the Cancer Imaging Archive.42,43 It includes 22 032 reconstructed DBT volumes (a stack of spatially adjacent 2D scan slices) from scans of 5060 participants, with annotations for biopsied lesions provided by 2 trained radiologists. A given DBT scan has separate volumes corresponding to at least 1 and as many as 4 of the anatomical views of the breasts: left craniocaudal (LCC), right craniocaudal (RCC), left mediolateral oblique (LMLO), and right mediolateral oblique (RMLO).
Each of the radiologists who completed the annotations had at least 18 years of experience with breast imaging. Scans were classified as normal, actionable (further imaging requested), benign (lesion found, negative biopsy), or cancerous (lesion found, positive biopsy). Additionally, for benign and cancerous cases, radiologists provided annotations in the form of a tight bounding box around each lesion. If a lesion annotation were present in a volume, the annotation was assigned to the central slice of the volume. (There are approximately 70 slices for each scan volume.) Annotations for microcalcifications were not included. For the challenge, the data set was stratified by participant into training, validation, and test sets, as outlined in Table 1. Lesion boxes and volume class labels were only provided to challenge teams for the training set. In phase 1 of the challenge, teams were provided with 700 scans from the training set, 120 from the validation set, and 180 from the test set, while the second phase used the entire data set. All lesion cases were included in both phases. We provide further logistical details for the challenge in eAppendix 2 in Supplement 1.
Table 1. Statistics of the Data Sets Used for the Challenge.
| Characteristics | No. (%) | ||
|---|---|---|---|
| Training set | Validation set | Test set | |
| Participants | |||
| Total | 4362 (100) | 280 (100 | 418 (100) |
| Normal | 4109 (94.2) | 200 (71.4) | 300 (71.8) |
| Actionable | 178 (4.1) | 40 (14.3) | 60 (14.4) |
| Benign | 62 (1.4) | 20 (7.1) | 30 (7.2) |
| Cancer | 39 (0.9) | 20 (7.1) | 30 (7.2) |
| Total DBT volumes, No. | 19 148 | 1163 | 1721 |
| Bounding boxes for biopsied lesions, No. | 224 | 75 | 136 |
Abbreviation: DBT, digital breast tomosynthesis.
Statistical Analysis
Teams were tasked with developing algorithms that take a DBT volume as input and detect any biopsy-proven (cancerous or benign) lesions found within by generating proposed bounding boxes that enclose the lesion(s). To evaluate this task on the validation and test sets of scans with class labels and lesion bounding boxes unknown to participants, teams were asked to provide bounding box locations (horizontal and vertical pixel coordinates and slice index) and sizes, accompanied by prediction scores indicating a level of certainty for each box for any lesions detected by their models. These scores could be on any scale, but the scale had to be unified across the evaluation of the data set and were used by challenge organizers (along with the bounding boxes submitted by participants and reference standard) to evaluate the overall detection performance of an algorithm.
The overall performance evaluation for an algorithm was based on free-response receiver operating characteristic (FROC) curves, which examine the sensitivity of each model with respect to the number of false-positive (FP) predictions created by the model for each view in the test set. Details of how a prediction was deemed a true positive appear in eAppendix 2 in Supplement 1. The primary performance metric was calculated only on DBT volumes with biopsied lesions (benign or malignant) and was the mean sensitivity (ie, the true-positive rate) over 1, 2, 3, and 4 FPs per volume. We average over the different FP counts to comprehensively reflect the overall performance curve across different sensitivities and specificities. This metric is similar to the competition performance metric for assessing lung nodule detection.44
The secondary performance metric used to break ties, if any, was the sensitivity at 2 FPs per DBT volume calculated using all DBT volumes (eTable in Supplement 1). To win the challenge, a team’s performance did not need to demonstrate a statistically significant improvement over that of the runner-up. The number of views in the test set with biopsied lesions was the same in both challenges, meaning that the primary performance metric is identical. All evaluation code is publicly available.45 Finally, we created a webpage for future evaluations of the DBTex performance metric (on both the validation and test sets).13 This will allow algorithms developed in the future to have a standardized tool for model selection (by the validation set) and performance metric (on the test set).
Results
Grand Challenge Results
In Table 2, we present the ranked roster of participating teams: their affiliations, method names, final scores for all challenge phases that they participated in, and a reference to their code when possible.45,46,47,48,49,50,51,52,53,54,55,56 We also provide the performance results of 2 simple baseline models on the task, both with code: the model that accompanied the BCS-DBT data set release32 and a basic faster region-based convolutional neural network (R-CNN) model (eAppendix 2 in Supplement 1). We provide detailed summaries of all algorithms in eAppendix 2 in Supplement 1, with a link to all prediction results. The teams that participated in both challenge phases had the same final ranking order, so we display the results of both phases in the same table. eAppendix 1 in Supplement 1 presents secondary metric results.
Table 2. Challenge Resultsa.
| Ranking | Team name | Affiliations | Methods | Training set | Mean sensitivity for biopsied lesions (95% CI) | Phase where team achieved best performance | Code available |
|---|---|---|---|---|---|---|---|
| 1 | NYU B-Team | New York University—Langone Health | Phase 1: EfficientDet, Max-Slice-Selection, and Augmentation and Ensembled Perturbations; phase 2: phase 1 methods with cancer cell prediction head and multilocation crop | Phases 1 and 2: DBTex1 and internal data set | 0.957 (0.924-0.984) | 2 | No |
| 2 | ZeDuS | IBM Research—Haifa | Phase 1: RetinaNet ensemble with heatmap NMS; phase 2: phase 1 methods with SWIN46 and NFNet47 | Phases 1 and 2: DBTex1 with internal data set | 0.926 (0.881-0.964) | 2 | Yes, both phases48 |
| 3 | VICOROB | VICOROB—University of Girona | Phase 1: Fast R-CNN, ensembled; phase 2: phase 1 methods with FP reduction (no ensemble) | Phases 1 and 2: DBTex1 with OPTIMAM/OMI-DB | 0.886 (0.836-0.930) | 2 | Yes, both phases49,50 |
| 4 | Prarit | Queen Mary University of London—CRST and School of Physics and Astronomy | Unknown | Unknown | 0.822 (0.754-0.884) | 1 | No |
| 5 | UCLA-MII | UCLA Medical & Imaging Informatics | Phase 1: Faster R-CNN, FPN,51 IoSIB, and Blob Detector | Phase 1: DBTex1 | 0.814 (0.751-0.875) | 1 | Yes, phase 152 |
| 6 | Pranjalsahu | Stony Brook—Department of Computer Science | Phase 1: Faster R-CNN with Confidence Peak Finder | Phase 1: DBTex1 | 0.790 (0.717-0.854) | 1 | Yes; phase 153 |
| 7 | Team-PittRad | University of Pittsburgh—Department of Radiology | Phase 1: YOLOv554 and Cross Stage Partial Networks | Phase 1: DBTex1 | 0.786 (0.720-0.852) | 1 | Yes, phase 155 |
| 8 | Coolwulf | Unknown | Unknown | Unknown | 0.390 (0.301-0.475) | 1 | No |
| NA | Baseline modelb | NA | Faster R-CNN | DBTex1 | 0.379 (0.304-0.456) | NA | Yes56 |
| NA | Data set baseline modelb | NA | DenseNet32 | DBTex1 | 0.444 (0.366-0.523) | NA | Yes45 |
Abbreviations: FP, false positive; FPN, feature pyramind network; IoSIB, intersection over the smaller intersecting box; NA, not applicable; NFNet, Normalizer-Free-ResNets; NMS, nonmaximal suppression; R-CNN, region-based convolutional neural network; SWIN, shifted window transformer.
95% confidence intervals (CI) were computed using bootstrapping, with 5000 bootstraps.
Not submitted for challenge.
Analysis of Grand Challenge Results
Beyond the individual performances of each participant algorithm, we analyzed the collective results of the challenge to obtain a holistic measure for the capability of state-of-the-art DBT lesion detection algorithms. First, we examined how lesion detection difficulty varied between different cases. Next, we analyzed the success of the submitted algorithms by aggregating their predictions.
Lesion Detection Difficulty Ranking
We considered 2 extremes: the easiest lesions to detect and the most difficult. Figure 1A and 1B show the 2 lesions that were easiest to detect according to our difficulty measures (eAppendix 2 in Supplement 1), alongside the algorithms’ corresponding predictions. We see that most submitted algorithms made similar predictions for these lesions. Figure 1C and D show the 2 most challenging lesions, which resulted in disagreeing predictions. Overall, we found that within 4 FPs per DBT volume all algorithms detected 16 of 136 lesion annotations (12%), and there was only 1 lesion (<1%) that was not detected by any algorithm.
Figure 1. The Least and Most Difficult Lesions to Detect.
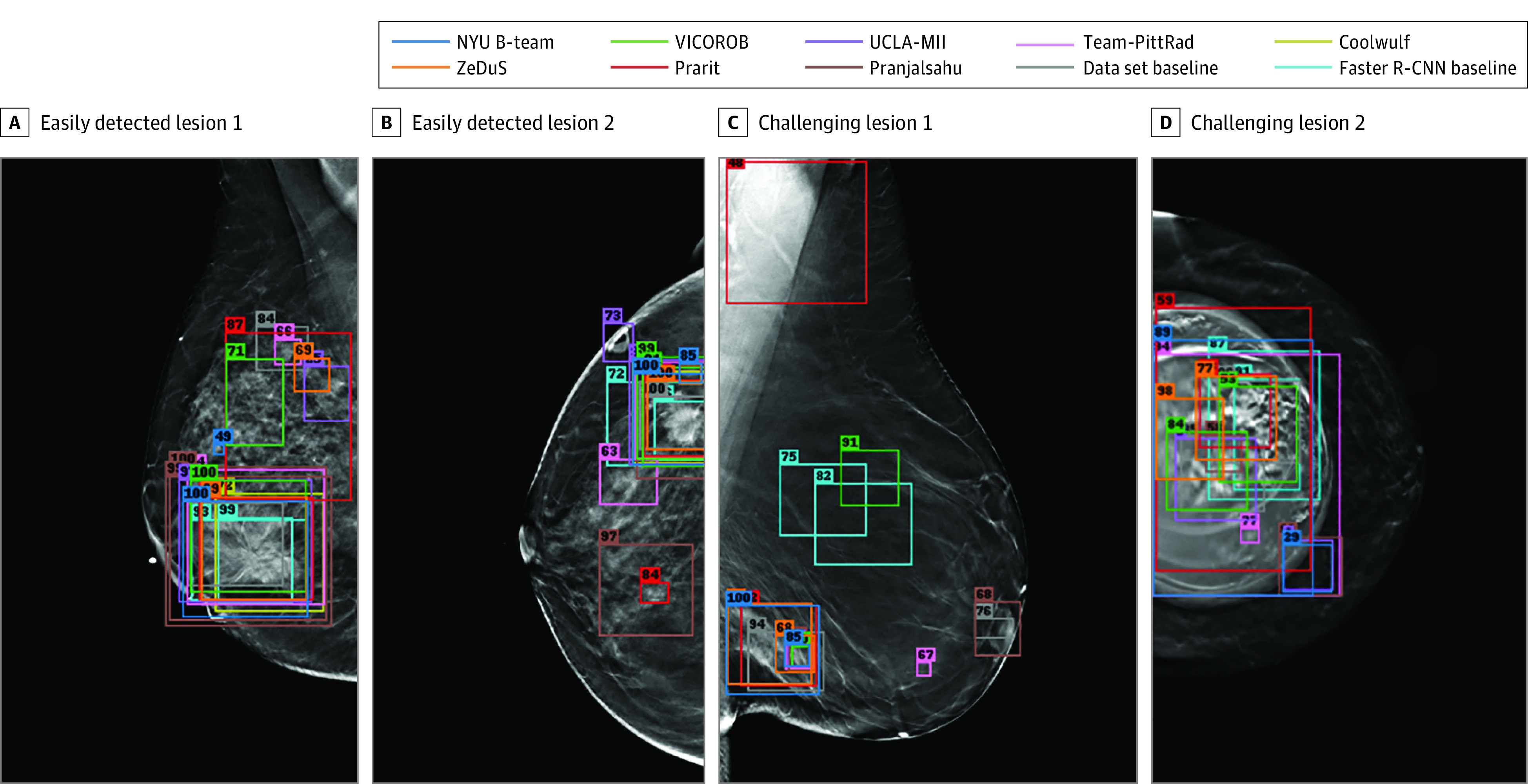
A and B, Examples of digital breast tomosynthesis volumes containing annotated lesions that were the easiest to detect. On average, all 10 algorithms detected lesions in A and with 0.13 and 0.16 false positives, respectively. C and D, Examples of digital breast tomosynthesis volumes containing annotated lesions that were the most difficult to detect. The lesion in panel C was not detected by any algorithm, and the lesion in panel D was detected by only 2 of 10 algorithms with 1.34 false positives on average (due to the presence of a breast implant). Detection bounding boxes indicate submitted algorithm predictions. The number in the upper-left corner of each box indicates the percentile of the corresponding algorithm’s score with respect to the distribution of all algorithm scores for the volume. At most, 2 boxes per algorithm are shown, and the colors of each algorithm’s boxes correspond to the free-response receiver operating characteristic curves shown in Figure 2.
Next, we analyzed the association of lesion detection difficulty with (1) the classification of the lesion being cancerous or benign and (2) lesion type (mass or architectural distortion). We compare these lesion characteristics with our measures of lesion difficulty in Table 3. We also measured the correlation between the number of algorithms that detected a lesion and the lesion size (bounding box diagonal length) to be 0.28, using Spearman rank correlation coefficient.
Table 3. Comparison of Lesion Detection Difficulty Metrics and Lesion Characteristics for the Test Set.
| Metric | Mean (SD) | |||
|---|---|---|---|---|
| Lesion diagnosis | Lesion type | |||
| Benign | Cancer | Mass | Architectural distortion | |
| Total count, No. | 70 | 66 | 121 | 15 |
| No. of algorithms that detected lesion within 4 FPs per volume | 7.47 (1.73) | 7.82 (1.76) | 7.63 (1.78) | 7.73 (1.44) |
| FPs corresponding to correct prediction considering teams that detected it within 4 FPs per volume | 0.77 (0.49) | 0.55 (0.43) | 0.64 (0.47) | 0.91 (0.47) |
Abbreviation: FP, false positive.
Combining Predictions
To aggregate lesion detection results from all methods (not including the baseline models), ie, to merge lesion bounding box predictions that were made by different algorithms, we normalized the detection confidence scores assigned by each algorithm to its predicted bounding boxes, across all algorithms, by transforming all scores for each algorithm to a percentile range. Next, we merged lesion box predictions across the width, height, and slice dimensions using the weighted boxes fusion algorithm57 (eAppendix 2 in Supplement 1).
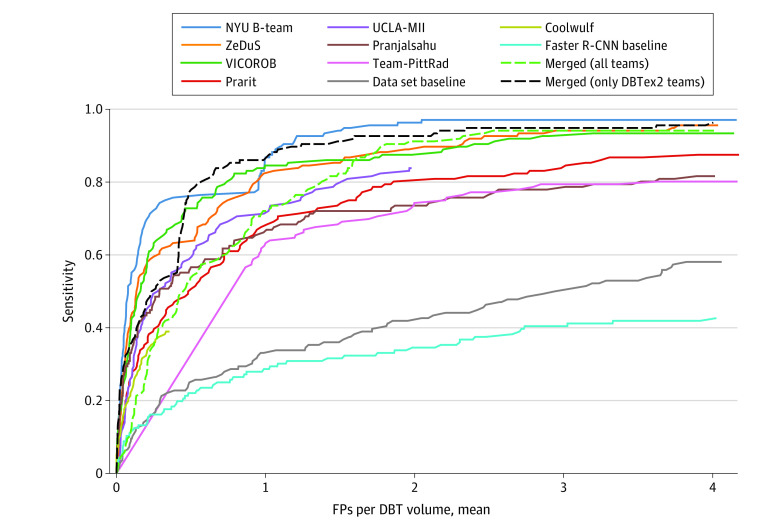
After this merging procedure, we obtained a set of merged predicted lesion bounding boxes with accompanying prediction scores aggregated from each model. We computed merged results for both (1) all submitted algorithms and (2) only phase 2 submissions. For the 3 teams that participated in both phases, we use the results from phase 2, as they were superior to phase 1 submissions in all cases. The mean sensitivity at 1, 2, 3, and 4 FPs per volume was 87.9% (95% CI, 83.6%-91.8%)for the former group and 92.6% (95% CI, 88.8%-95.9%) for the latter. On the same metric, for phase 2, NYU B-Team achieved 94.3% (95% CI, 90.7%-97.2%); ZeDuS, 90.4% (95% CI, 86.1%-94.3%); and VICOROB, 89.6% (95% CI, 85.0%-93.7%). We show these results as FROC curves in Figure 2, including the Faster R-CNN baseline and the baseline model from Buda et al.32
Figure 2. Free-Response Receiver Operating Characteristic Detection Curves for All Methods.
Includes all participants, baseline models, and merged predictions from all algorithms and only the top 3 models from phase 2.
Discussion
The purpose of this challenge was to facilitate the development of state-of-the-art machine learning algorithms for the challenging task of DBT lesion detection. Our goal was to address the current lack of public, standardized resources for research in this field, as prior works have often relied on private data sets, detection models, or both. We approached this by (1) establishing a standardized, publicly accessible testing benchmark, evaluation pipeline, and training data set for this task; (2) hosting a grand challenge to encourage the concentrated development of algorithms; and (3) encouraging the release of publicly available algorithm code.
By merging the results from all submissions, we found that most submitted algorithms achieved strong performance on this task. All submitted algorithms that we analyzed (eAppendix 2 in Supplement 1) relied on some object detection neural network. The key properties that led some submissions to perform better than others were (1) the specific model used, (2) any novel refinements that teams made to their methods, and (3) the training data used. Leading teams used more recent detection architectures, such as EfficientDet58 (NYU B-Team), highly optimized for modern object detection tasks, or RetinaNet37 (ZeDuS), which is especially well-suited for small objects (eg, small lesions) and class-imbalanced data sets like the BCS-DBT training set. The top 3 teams also used model ensembling, the aggregation of multiple models’ predictions to improve overall robustness. This method is well-suited for improving the generalizability of models,59 which is especially applicable to this task because breast tissue and breast lesions have high morphological variability. Finally, winning teams also used additional internal training data that provided more lesion examples to learn from. This is important because the detection model itself can only carry performance so far; the training of deep models is data driven, so the variety and quantity of lesion examples to learn from will significantly affect an algorithm’s ability to generalize to new data.
The importance of training data, model choice, and novel methods is especially apparent when submitted results are compared with the 2 baseline models (Table 2), which were trained only on the provided BCS-DBT training set and had poorer performance than most submissions. However, some submissions (eg, UCLA-MII, pranjalsahu, Team-PittRad) that also only used the provided data performed almost as well as algorithms that used additional data, while vastly outperforming the baseline models (Table 2). As such, the usage of supplementary data appears not to be the only necessary factor for achieving good performance, but also the development of specialized techniques for the unique characteristics of the data set or domain, which the baseline models did not have. This shows that DBT tumor detection is a challenging problem for typical detection models, but large performance improvements are possible if the model development is targeted for this modality. Finally, an additional tactic that some teams used (eg, VICOROB and UCLA-MII) was pretraining their detection models on common universal natural image data sets, such as COCO,60 before training on the target domain of DBT data, giving the models a starting point for visual feature recognition.
Limitations
This study has limitations. All teams relied on supervised training of their detection algorithms, ie, directly recognizing visual features that discriminate between healthy and cancerous examples. One potential obstacle for this approach is the presence of anomalous objects that may mislead detection. The case shown in Figure 1D is an example of this, where an implant distracted most of the models from the mass present in the image. This behavior is due to the data-driven nature of training deep learning models; if an object appears in a test image that is rare within the training data, this may interfere with model predictions. Despite the strong overall results of the algorithms, the presence of these rare cases cannot be ignored in the clinical setting; this could be mitigated by the use of anomaly detection methods.61,62
Another limitation is that each of the top 3 finalists used additional training data including lesions outside of the provided data set, so no conclusion can be drawn about which method would be superior given the same training data set. However, submitted algorithms were still directly compared in their effectiveness at detecting lesions in the test set. An additional limitation of our study is that the benchmark was computed only on true-positive cases. This was because the data set has missing views but only for biopsied cases, which could be (intentionally or not) taken advantage of by participants by prioritizing predictions for these missing-view cases or determining which cases are biopsied by the presence of missing views. However, by comparing the competition results (Table 2) with the FROC curves in Figure 2, the latter of which were computed on all cases, we found there to be no notable difference between (1) the metric computed on all cases and (2) only true-positive cases, so the effect of this factor is minimal.
The scope of the test set was somewhat limited because it included only 136 lesions (due to the natural screening rarity of breast cancer) and because microcalcification annotations are not included in the data set.32 The ability of our benchmark to measure clinical detection performance across a range of institutions is also limited because the data originated only from the Duke University Health System. However, the leading algorithms’ success of using supplementary training data indicates that DBT scans created at different sites still possesses common features to learn from and implies that the algorithms were able to generalize across multiple data domains.
Conclusions
In this diagnostic study of AI for DBT, submitted algorithms for the DBTex challenge gave promising breast lesion detection performance over a range of difficult cases, improving over existing baseline models by a wide margin. To accompany these results, we presented a benchmark evaluation platform for assessing detection algorithms, a large public data set, and code for certain submitted algorithms. The success of this challenge marks a large improvement in DBT tumor detection methods, and the public resources we provide lay the groundwork for the development of clinically relevant computer-assisted diagnosis systems.
eAppendix 1. Additional Challenge Results
eTable. Secondary Metric Challenge Results
eAppendix 2. Additional Methods Details
Data Sharing Statement
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Gao Y, Moy L, Heller SL. Digital breast tomosynthesis: update on technology, evidence, and clinical practice. Radiographics. 2021;41(2):321-337. doi: 10.1148/rg.2021200101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In: Pereira F, Burges CJ, Bottou L, Weinberger KQ. Advances in Neural Information Processing Systems 25 (NIPS 2012). Curran Associates Inc; 2012:1097-1105. Accessed January 19, 2023. https://papers.nips.cc/paper/2012/hash/c399862d3b9d6b76c8436e924a68c45b-Abstract.html [Google Scholar]
- 4.Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. In: Cortes C, Lawrence N, Lee D, Sugiyama M, Garnett R. Advances in Neural Information Processing Systems 28 (NIPS2015). Curran Associates Inc; 2015:91-99. Accessed January 19, 2023. https://papers.nips.cc/paper/2015/hash/14bfa6bb14875e45bba028a21ed38046-Abstract.html [Google Scholar]
- 5.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, eds. Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015. Springer International Publishing; 2015:234-241, doi: 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 6.Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18(2):203-211. doi: 10.1038/s41592-020-01008-z [DOI] [PubMed] [Google Scholar]
- 7.O’Mahony N, Campbell S, Carvalho A, et al. Deep learning vs. traditional computer vision. In: Arai K, Kapoor S, eds. Advances in Computer Vision. Springer International Publishing; 2020:128-144, doi: 10.1007/978-3-030-17795-9_10 [DOI] [Google Scholar]
- 8.Shen Y, Wu N, Phang J, et al. An interpretable classifier for high-resolution breast cancer screening images utilizing weakly supervised localization. Med Image Anal. 2021;68:101908. doi: 10.1016/j.media.2020.101908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez-Ruiz A, Lång K, Gubern-Merida A, et al. Stand-alone artificial intelligence for breast cancer detection in mammography: comparison with 101 radiologists. J Natl Cancer Inst. 2019;111(9):916-922. doi: 10.1093/jnci/djy222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le EPV, Wang Y, Huang Y, Hickman S, Gilbert FJ. Artificial intelligence in breast imaging. Clin Radiol. 2019;74(5):357-366. doi: 10.1016/j.crad.2019.02.006 [DOI] [PubMed] [Google Scholar]
- 11.Oliver A, Freixenet J, Martí J, et al. A review of automatic mass detection and segmentation in mammographic images. Med Image Anal. 2010;14(2):87-110. doi: 10.1016/j.media.2009.12.005 [DOI] [PubMed] [Google Scholar]
- 12.Geras KJ, Mann RM, Moy L. Artificial intelligence for mammography and digital breast tomosynthesis: current concepts and future perspectives. Radiology. 2019;293(2):246-259. doi: 10.1148/radiol.2019182627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.CodaLab. SPIE-AAPM-NCI-DAIR Digital Breast Tomosynthesis Cancer Detection Challenge (DBTex): open benchmark. Accessed January 24, 2023. https://spie-aapm-nci-dair.westus2.cloudapp.azure.com/competitions/9
- 14.Menze BH, Jakab A, Bauer S, et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging. 2015;34(10):1993-2024. doi: 10.1109/TMI.2014.2377694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russakovsky O, Deng J, Su H, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis. 2015;115:211-252. doi: 10.1007/s11263-015-0816-y [DOI] [Google Scholar]
- 16.Kaggle Inc . Kaggle. Accessed January 19, 2023. https://www.kaggle.com/
- 17.Fotin SV, Yin Y, Haldankar H, Hoffmeister JW, Periaswamy S. Detection of soft tissue densities from digital breast tomosynthesis: comparison of conventional and deep learning approaches. Proceedings of SPIE. 2016;9785. doi: 10.1117/12.2217045 [DOI] [Google Scholar]
- 18.Kim DH, Kim ST, Ro YM. Latent feature representation with 3-D multi-view deep convolutional neural network for bilateral analysis in digital breast tomosynthesis. Proc IEEE Int Conf Acoust Speech Signal Process. 2016:927-931. doi: 10.1109/ICASSP.2016.7471811 [DOI] [Google Scholar]
- 19.Geras KJ, Wolfson S, Shen Y, et al. High-resolution breast cancer screening with multi-view deep convolutional neural networks. ArXiv. Preprint posted online June 28, 2018. doi: 10.48550/arXiv.1703.07047 [DOI]
- 20.Yousefi M, Krzyżak A, Suen CY. Mass detection in digital breast tomosynthesis data using convolutional neural networks and multiple instance learning. Comput Biol Med. 2018;96:283-293. doi: 10.1016/j.compbiomed.2018.04.004 [DOI] [PubMed] [Google Scholar]
- 21.Samala RK, Chan HP, Hadjiiski LM, Helvie MA, Richter C, Cha K. Evolutionary pruning of transfer learned deep convolutional neural network for breast cancer diagnosis in digital breast tomosynthesis. Phys Med Biol. 2018;63(9):095005. doi: 10.1088/1361-6560/aabb5b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Zhang Y, Han EY, et al. Classification of whole mammogram and tomosynthesis images using deep convolutional neural networks. IEEE Trans Nanobioscience. 2018;17(3):237-242. doi: 10.1109/TNB.2018.2845103 [DOI] [PubMed] [Google Scholar]
- 23.Liang G, Wang X, Zhang Y, et al. Joint 2D-3D Breast Cancer Classification. 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). doi: 10.1109/BIBM47256.2019.8983048 [DOI] [Google Scholar]
- 24.Zhang Y, Wang X, Blanton H, et al. 2D convolutional neural networks for 3D digital breast tomosynthesis classification. 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). doi: 10.1109/BIBM47256.2019.8983097 [DOI] [Google Scholar]
- 25.Mendel K, Li H, Sheth D, Giger M. Transfer learning from convolutional neural networks for computer-aided diagnosis: a comparison of digital breast tomosynthesis and full-field digital mammography. Acad Radiol. 2019;26(6):735-743. doi: 10.1016/j.acra.2018.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh S, Matthews TP, Shah M, et al. Adaptation of a deep learning malignancy model from full-field digital mammography to digital breast tomosynthesis. ArXiv. Preprint posted online January 23, 2020. doi: 10.48550/arXiv.2001.08381 [DOI]
- 27.Li X, Qin G, He Q, et al. Digital breast tomosynthesis versus digital mammography: integration of image modalities enhances deep learning-based breast mass classification. Eur Radiol. 2020;30(2):778-788. doi: 10.1007/s00330-019-06457-5 [DOI] [PubMed] [Google Scholar]
- 28.Matthews TP, Singh S, Mombourquette B, et al. A multisite study of a breast density deep learning model for full-field digital mammography and synthetic mammography. Radiol Artif Intell. 2020;3(1):e200015. doi: 10.1148/ryai.2020200015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez-Ruiz A, Teuwen J, Vreemann S, et al. New reconstruction algorithm for digital breast tomosynthesis: better image quality for humans and computers. Acta Radiol. 2018;59(9):1051-1059. doi: 10.1177/0284185117748487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai X, Yang W, Li R. DBT masses automatic segmentation using U-Net neural networks. Comput Math Methods Med. 2020;2020:7156165. doi: 10.1155/2020/7156165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bai J, Posner R, Wang T, Yang C, Nabavi S. Applying deep learning in digital breast tomosynthesis for automatic breast cancer detection: a review. Med Image Anal. 2021;71:102049. doi: 10.1016/j.media.2021.102049 [DOI] [PubMed] [Google Scholar]
- 32.Buda M, Saha A, Walsh R, et al. A data set and deep learning algorithm for the detection of masses and architectural distortions in digital breast tomosynthesis images. JAMA Netw Open. 2021;4(8):e2119100. doi: 10.1001/jamanetworkopen.2021.19100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan M, Li Y, Zheng S, Peng W, Tang W, Li L. Computer-aided detection of mass in digital breast tomosynthesis using a faster region-based convolutional neural network. Methods. 2019;166:103-111. doi: 10.1016/j.ymeth.2019.02.010 [DOI] [PubMed] [Google Scholar]
- 34.He K, Gkioxari G, Dollár P, Girshick R. Mask R-CNN. ArXiv. Preprint posted online January 24, 2018. doi: 10.48550/arXiv.1703.06870 [DOI]
- 35.Fan M, Zheng H, Zheng S, et al. Mass detection and segmentation in digital breast tomosynthesis using 3D-mask region-based convolutional neural network: a comparative analysis. Front Mol Biosci. 2020;7:599333. doi: 10.3389/fmolb.2020.599333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. ArXiv. Preprint posted online December 10, 2015. doi: 10.48550/arXiv.1512.03385 [DOI]
- 37.Lin T-Y, Goyal P, Girshick RB, He K, Dollár P. Focal loss for dense object detection. 2017 IEEE Int Conf Comput Vis ICCV. doi: 10.1109/ICCV.2017.324 [DOI] [PubMed] [Google Scholar]
- 38.Lotter W, Diab AR, Haslam B, et al. Robust breast cancer detection in mammography and digital breast tomosynthesis using an annotation-efficient deep learning approach. Nat Med. 2021;27(2):244-249. doi: 10.1038/s41591-020-01174-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Redmon J, Divvala S, Girshick R, Farhadi A. You only look once: unified, real-time object detection. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). doi: 10.1109/CVPR.2016.91 [DOI] [Google Scholar]
- 40.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). doi: 10.1109/CVPR.2017.243 [DOI] [Google Scholar]
- 41.Optimam mammography imaging. Approval and ethics. Accessed January 24, 2023. https://medphys.royalsurrey.nhs.uk/omidb/project-information/approval-ethics/
- 42.Buda M, et al. Breast cancer screening—digital breast tomosynthesis (BCS-DBT). doi: 10.7937/E4WT-CD02 [DOI]
- 43.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045-1057. doi: 10.1007/s10278-013-9622-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niemeijer M, Loog M, Abramoff MD, Viergever MA, Prokop M, van Ginneken B. On combining computer-aided detection systems. IEEE Trans Med Imaging. 2011;30(2):215-223. doi: 10.1109/TMI.2010.2072789 [DOI] [PubMed] [Google Scholar]
- 45.GitHub. Duke DBT data. Accessed January 24, 2023. https://github.com/mazurowski-lab/duke-dbt-data
- 46.Liu Z, Lin Y, Hu H, et al. Swin transformer: hierarchical vision transformer using shifted windows. 2021 IEEE/CVF International Conference on Computer Vision (ICCV). doi: 10.1109/ICCV48922.2021.00986 [DOI] [Google Scholar]
- 47.Brock A, De S, Smith SL, Simonyan K. High-performance large-scale image recognition without normalization. Proceedings of the 38th International Conference on Machine Learning. Accessed January 23, 2023. http://proceedings.mlr.press/v139/brock21a/brock21a.pdf [Google Scholar]
- 48.GitHub. IBM: work reduction DBT. Accessed January 24, 2023. https://github.com/IBM/work-reduction-dbt
- 49.GitHub. VICOROB DBT challenge. Accessed January 24, 2023. https://github.com/ICEBERG-VICOROB/vicorob_DBT_Challenge
- 50.GitHub. DBT phase 2. Accessed January 24, 2023. https://github.com/ICEBERG-VICOROB/DBT_phase2
- 51.Lin T-Y, Dollár P, Girshick R, et al. Feature pyramid networks for object detection. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). doi: 10.1109/CVPR.2017.106. [DOI] [Google Scholar]
- 52.GitHub. DBTex. Accessed January 24, 2023. https://github.com/aguron/DBTex
- 53.GitHub. DBTNet. Accessed January 24, 2023. https://github.com/PranjalSahu/DBTNet
- 54.Jocher G, Stoken A, Borovec J, et al. Ultralytics/yolov5: v3.1—bug fixes and performance improvements. Accessed January 26, 2023. https://zenodo.org/record/4154370#.Y9K4kHbMI2w
- 55.GitHub. Team Pitt-Rad-DBTex 1. Accessed January 24, 2023. https://github.com/IRL-UP/TeamPittRad-DBTex1
- 56.GitHub. DBTex-baseline. Accessed January 24, 2023. https://github.com/mazurowski-lab/DBTex-baseline
- 57.Solovyev R, Wang W, Gabruseva T. Weighted boxes fusion: ensembling boxes from different object detection models. Image Vis Comput. 2021;107:104117. doi: 10.1016/j.imavis.2021.104117 [DOI] [Google Scholar]
- 58.Tan M, Pang R, Le QV. EfficientDet: scalable and efficient object detection. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition. doi: 10.1109/CVPR42600.2020.01079 [DOI] [Google Scholar]
- 59.Ganaie MA, Hu M, Malik AK, Tanveer M, Suganthan PN. Ensemble deep learning: a review. ArXiv. Preprint posted online August 8, 2022. doi: 10.48550/arXiv.2104.02395 [DOI]
- 60.Lin T-Y, Maire M, Belongie S, et al. Microsoft COCO: common objects in context. In Computer Vision—ECCV 2014. Springer International Publishing, 2014: 740-755. doi: 10.1007/978-3-319-10602-1_48 [DOI] [Google Scholar]
- 61.Tack J, Mo S, Jeong J, Shin J. CSI: novelty detection via contrastive learning on distributionally shifted instances. Accessed January 23, 2023. https://proceedings.neurips.cc/paper/2020/file/8965f76632d7672e7d3cf29c87ecaa0c-Paper.pdf
- 62.Swiecicki A, Konz N, Buda M, Mazurowski MA. A generative adversarial network-based abnormality detection using only normal images for model training with application to digital breast tomosynthesis. Sci Rep. 2021;11(1):10276. doi: 10.1038/s41598-021-89626-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eAppendix 1. Additional Challenge Results
eTable. Secondary Metric Challenge Results
eAppendix 2. Additional Methods Details
Data Sharing Statement