Abstract
Introduction
We tried to show the effect of sinomenine (SIN) in diabetic peripheral neuropathic pain (DPNP) and the related underlying mechanism.
Methods
Network pharmacological analysis and bioinformatics analysis were carried out for identification of the active ingredients of Sinomenium acutum and the related genes. The DPNP rat model was constructed and primary rat spinal cord microglial cells were isolated for in vitro cell experiments. The therapeutic role of SIN in DPNP was determined in vivo and in vitro through analysis of microglial cell activation and inflammatory response.
Results
Therapeutic role of S. acutum in DPNP was mainly achieved by regulating 14 key genes, among which the target gene prostaglandin‐endoperoxide synthase 2 (PTGS2) of SIN might be the key gene. An in vivo experiment showed that SIN inactivated the inositol‐requiring enzyme 1 alpha–X‐box binding protein 1 pathway by downregulating PTGS2, which relieved pain symptoms in DPNP rats. It was confirmed in vivo that SIN inhibited the pathway through PTGS2 to alleviate the activation of spinal cord microglial cells and inflammatory response.
Conclusion
SIN decreases the expression of PTGS2 to inactivate the inositol‐requiring enzyme 1 alpha–X‐box binding protein 1 signaling pathway, which inhibits microglial activation, as well as the release of inflammatory factors, thus alleviating DPNP.
Keywords: Diabetic peripheral neuropathic pain, Sinomenine, Spinal cord microglia
Molecular mechanism diagram of the role of SIN in DPNP by regulating PTGS2‐mediated IRE1α‐XBP1s pathway. SIN down‐regulates the expression of PTGS2 to inactivate IRE1α‐XBP1s signaling pathway, which inhibits microglial cell activation and the release of inflammatory factors, finally alleviating DPNP.
![]()
INTRODUCTION
Diabetic peripheral neuropathic pain (DPNP) is a kind of pain directly resulting from the aberrant somatosensory system in diabetes patients in light of the International Association for the Study of Pain 1 . Inflammation plays an important role in the onset, as well as the maintenance, of DPNP 2 . Notably, inhibited generation and activation of microglial cells in the dorsal spinal cord have been found to share an association with the neuropathic pain state, including that in DPNP 3 .
The Sinomenium acutum stem is a kind of popular traditional Chinese medicine applied for treatment of bone and joint diseases; sinomenine (SIN) is the only chemical marker in mainstream pharmacopeias that controls the quality of the S. acutum stem 4 . SIN plays an antinociceptive role in rats with neuropathic pain 5 . Treatment with SIN can partially inhibit prostaglandin‐endoperoxide synthase 2 (PTGS2) expression to exert a protective function in inflammatory pain in a rat model 6 . The PTGS2 inhibitor, meloxicam, can exert antiallodynic effects on DPNP in a mouse model 7 . Of note, PTGS2 can bind to endoplasmic reticulum stress sensor inositol‐requiring enzyme 1 alpha (IRE1α) and thus activate its expression, resulting in IRE1α splicing of X‐box binding protein 1 (XBP1) mRNA 8 . RE1α and XBP1s are activated in neuropathic pain triggered by chronic constriction injury 9 . Notably, inhibition of IRE1α–XBP1 ameliorates the progression of hyperalgesia induced by opioids 10 . Based on the aforementioned results, we proposed a hypothesis here that SIN might affect the development of DPNP by regulating the PTGS2‐mediated IRE1α–XBP1s signaling pathway.
MATERIALS AND METHODS
Ethical approval
This experimental procedure and animal use protocol were approved by the Animal Ethics Committee of Affiliated Huaihua Hospital, Hengyang Medical School, University of South China, and the animal experiments followed the Animal Welfare Act and Public Health Service guidelines for the management and use of experimental animals specified by the National Institutes of Health.
Network pharmacological analysis
The compound components of S. acutum were searched from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform database and screened with the condition of oral bioavailability ≥30% and drug likeness ≥0.18 11 , and the corresponding targets were downloaded. Perl is a programming language suitable for writing simple scripts and complex applications. We used Strawberry Perl 5.30.1.1 combined with the UniProtKB search function in the UniProt database to correct the target name to the corresponding official gene symbol (limiting the species condition as “Homo sapiens”).
Bioinformatics analysis
Human DPNP‐related microarray GSE95849 was obtained through the Gene Expression Omnibus database. The microarray was equipped with the platform annotation file GPL22448, and contained six normal control samples and six DPNP samples (Table 1). The differential gene expression profiles were obtained using the R language “limma” package https://bioconductor.org/packages/limma/, with setting of |logFC| > 1, and P < 0.05 reviewed as the screening criteria of differential genes. The Pearson correlation coefficient of gene expression profiles in microarray GSE95849 was calculated by R language, setting P < 0.05 as the screening criteria for obtaining differential genes of the relevant gene.
Table 1.
Clinical and demographic characteristics of the participants from microarray dataset
| Normal control | DPN | |
|---|---|---|
| n | 6 | 6 |
| Sex | Female | Female |
| Age (years) | 51.17 ± 6.08 | 57.33 ± 7.92 |
| Fasting glucose (mmol/L) | 5.10 ± 0.21 | 9.95 ± 2.11* |
P < 0.05 versus normal control.
DPN, diabetic peripheral neuropathy.
Venn map delineation online
Through the Xiantao Scholar website (https://www.xiantao.love/), the target genes of active ingredients of S. acutum were intersected with differentially expressed genes from the Gene Expression Omnibus microarray data to obtain candidate genes.
Pharmacological network visualization and protein–protein interaction network construction
Cytoscape software (Free Software Foundation, Boston, MA, USA) was used to draw the network relationship diagram of 14 candidate genes and their corresponding components of S. acutum components. Through the database STRING, the interaction of candidate genes was analyzed, where the species condition was limited to “Homo sapiens”. Cytoscape software was applied to construct a regulatory relationship network, with the degree and combine score values represented by circle size and color, and the top 10 key genes were selected according to the degree value.
ClusterProfiler enrichment analysis
Using the R language “ClusterProfiler” package, the gene ontology and Kyoto Encyclopedia of Genes and Genomes functional enrichment analyses of 14 key genes were carried out, including biological processes, cellular components, molecular function and signaling pathways. The color of the dot showed the P‐value, and the dot size showed the number of targets enriched.
DPNP rat model construction and blood glucose monitoring
A total of 75 male Sprague–Dawley rats aged 6–8 weeks (150–200 g; Vital River, Beijing, China) were selected.
Rats were randomly used as control rats, or subjected to model establishment without further treatment (DPNP rats) or further treated with SIN (intraperitoneal injection of SIN), SIN + overexpression (oe)‐negative control (NC; intraperitoneal injection of SIN and tail vein injection of NC of overexpression lentivirus), SIN + oe‐PTGS2 (intraperitoneal injection of SIN and tail vein injection of PTGS2 overexpression lentivirus), SIN + oe‐PTGS2 + MKC8866 (intraperitoneal injection of SIN, tail vein injection of PTGS2 overexpression lentivirus and oral administration of MKC8866), with 12 rats in each group. DPNP rats received intraperitoneal injection of 50 mg/kg of streptozotocin (STZ; S0130; Sigma‐Aldrich, St. Louis, MO, USA) to induce diabetes. A total of 12 rats in the control group were injected with the same dose of normal saline. One week after STZ injection, blood was drawn from the rat orbit, and fasting blood glucose level was measured using a glucose meter (OneTouch Ultra Mini; Johnson & Johnson, New Brunswick, NJ, USA). The rats with blood glucose levels >250 mg/dL were considered to have diabetes, and rats with increased behavioral sensitivity to pain were identified as successful DPNP model rats. The success rate of DPNP modeling was 95%, and the successfully modeled rats were randomly divided into five groups.
All DNPN rats were administered since successfully modeled, namely, on the first day of the second week after STZ injection. SIN ([+]‐4‐hydroxy‐3,7‐dimethoxy‐17‐methylmorphin‐7‐en‐6‐one) was purchased from Aladdin (115‐53‐7; Shanghai, China) and dissolved in 0.9% normal saline. The drug was administered intraperitoneally daily at a dose of 1 mL/kg on the first day of successful DPNP model construction, namely, on the first day of the second week after STZ injection 5 . Behavioral sensitivity testing was carried out 3 h after each administration of SIN.
Next, 10 μL lentivirus was injected into the tail vein of STZ rats on the first day of the successful DPNP model construction. The viral titer of lentivirus was 5 × 108 TU. Meanwhile, 300 mg/kg MKC8866 (HY‐104040; MedChemExpress, Monmouth Junction, MJ, USA) was orally administered every other day. MKC8866 was an inhibitor of the IRE1α–XBP1s pathway 12 . Rats were killed under anesthesia with 1% pentobarbital sodium 4 weeks after STZ injection. The spine of the rat was dissected, and the L4–L6 segment of the spine was obtained from the rat and the nucleus pulposus tissue was isolated for subsequent experiments. Blood glucose levels and bodyweight of the rats were determined before they were killed. A schematic diagram for model establishment, treatment and sampling is shown in Figure 1.
Figure 1.
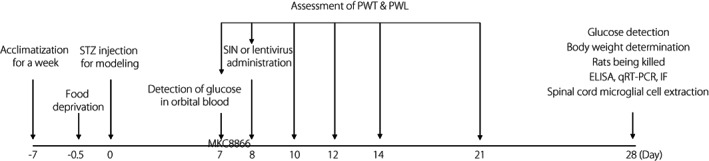
Schematic diagram of time points in the animal experiment. ELISA, enzyme‐linked immunosorbent assay; IF, immunofluorescence; PWL, paw withdrawal latency; PWT, paw withdrawal threshold; qRT–PCR, reverse transcription quantitative polymerase chain reaction; SIN, sinomenine; STZ, streptozotocin.
Neural pain behavioral test
Mechanical allodynia and thermal hyperalgesia were assayed for evaluating the response of rats to neural pain, as previously described 13 . During the test, pain hypersensitivity thresholds were tested using an electronic mechanical pain gauge (BME‐404; Boerni, Tianjin, China) and a 50% mechanical paw withdrawal threshold (PWT) was recorded. In addition, an automatic pain stimulator (BME‐410C; Boerni, Tianjin, China) was used with the paw withdrawal latency (PWL) recorded. Behavioral analysis was processed in a quiet room between 09.00 and 12.00 hours.
Primary microglial cell extraction and culture
Rats were euthanized. The spinal cord tissue was extracted from the L4 to L6 segment of the spine, immersed in 4 mL pre‐cooled Hank's solution (H1387; Sigma, Shanghai, China) containing 15 mmol/L HEPES (Gibco, Carlsbad, CA, USA) and 0.5% glucose (Sigma‐Aldrich), ground and filtered. Cells were collected by centrifugation at 400 g for 10 min. Cell supernatants were collected and subjected to gradient‐density centrifugation using Percoll (P4937; Sigma, Shanghai, China), followed by collection of cells at the junction of the 50%/75% Percoll interface. Expression of the microglial cell marker OX42 was measured by immunofluorescence to identify the purity of microglial cells. The collected cells were rinsed with pre‐cooled phosphate‐buffered saline, and then resuspended in Dulbecco's modified Eagle's medium supplemented with 1% bovine serum albumin (11965092; Gibco, Grand Island, NY, USA), 10% fetal bovine serum (10099141; Gibco) and 1% penicillin/streptomycin (15070063; Thermo Fisher Scientific Inc., Waltham, MA, USA) for the subsequent experiments.
Microglial cells were seeded in six‐well plates routinely cultured for 24 h to induce inflammation. Some cells without any treatment were used as controls, and the others were supplemented with 25 mmol/L glucose (G8270; Sigma) for a 24‐h intervention. High glucose (HG)‐stimulated cells were treated with SIN (12 μmol/L SIN for a 24‐h intervention), with SIN and further infected with oe‐NC or oe‐PTGS2 lentivirus for 48 h, or with SIN + oe‐PTGS2 (for 48 h) + 0.3 μmol/L MKC8866 (an IRE1a–XBP1s pathway inhibitor; for 24 h). All the lentiviruses were purchased from Genechem (Shanghai, China).
Enzyme‐linked immunosorbent assay
Expression of PTGS2, IER1α, XBP1s, tumor necrosis factor (TNF)‐α, interleukin (IL)‐1β and IL‐6 in rat spinal cords and microglial cell supernatants was determined using the enzyme‐linked immunosorbent assay (ELISA) kit: rat PTGS2 (125–8,000 pg/mL, EKE62250; Biomatik, Wilmington, DE, USA), rat IER1α (46.9–3,000 pg/mL, EKF59786; Biomatik), rat XBP1s (156–10,000 pg/mL, EKN49348; Biomatik), rat TNF‐α (78.13–5,000 pg/mL, EKE61994; Biomatik), rat IL‐1β (31.25–2000 pg/mL, EKE61988; Biomatik) and rat IL‐6 (62.5–4,000 pg/mL, EKE61991; Biomatik).
Immunofluorescence
Paraffin sections of the L4–L6 segment of the spine were subjected to antigen retrieval. Next, the sections were immunostained with mouse anti‐Ox42 (ab1211, 10 μg/mL; Abcam Inc., Cambridge, MA, USA), mouse anti‐neuronal nuclear antigen (NeuN; ab104224, 1:1000; Abcam), rabbit anti‐NeuN (ab177487, 1:100; Abcam), rat anti‐glial fibrillary acidic protein (GFAP; ab4648, 1:50; Abcam), rabbit anti‐GFAP (ab7260, 1:50) and rabbit anti‐PTGS2 (ab179800, 1:50; Abcam) at 4°C overnight, as well as fluorescent secondary antibodies including goat anti‐mouse (ab150115, 1:500; Abcam Inc.) and goat anti‐rabbit (ab150077, 1:500; Abcam Inc.) at room temperature for 60 min. An Olympus BX51 fluorescence microscope (Olympus, Tokyo, Japan) or laser scanning confocal microscope (FV500; Olympus) was used to blindly observe all the sections, followed by photographing. Analysis was carried out using the Image Pro‐Plus program and fluorescence intensity was recorded.
For cell samples, when spinal cord microglia reached 40–50%, they were fixed with 1 mL of precooled 95% ethanol for 30 min at −20°C. The ethanol was discarded, and the cells were incubated with 5% bovine serum albumin at room temperature for 60 min. Excess serum was discarded and the cells were incubated with the corresponding primary antibody. The subsequent operations were as same as the tissue samples.
Reverse transcription quantitative polymerase chain reaction
Total ribonucleic acid (RNA) was extracted from cells and frozen tissue samples using TRIZOL reagent (15596‐018; Solarbio, Beijing, China). Total RNA was reversely transcribed into complementary deoxyribonucleic acid using the PrimeScript™ reverse transcription polymerase chain reaction kit (Takara, Dalian, China). Quantitative polymerase chain reaction was carried out on a LightCycler 480 system (Roche Diagnostics GmbH, Mannheim, Germany) using the SYBR Premix Ex Taq™ (Takara Biotechnology Ltd., Dalian, China). Glyceraldehyde‐phosphate dehydrogenase was used as an internal reference to standardize the messenger RNA expression. The primers were designed and synthesized by Anhui General Biotechnology Co., Ltd (Anhui, China). Primer sequences for PTGS2 were forward: 5′‐GTGGAAAAGCCTCGTCCAGA‐3′ and reverse: 5′‐TCCTCCGAAGGTGCTAGGTT‐3′; for glyceraldehyde‐phosphate dehydrogenase were forward: 5′‐AGACAGCCGCATCTTCTTGT‐3′ and reverse: 5′‐TACGGCCAAATCCGTTCACA‐3′. The relative quantitative method ( method) was used to calculate the relative transcript level of the target gene.
Statistical analysis
SPSS 21.0 (IBM Corp., Armonk, NY, USA) was applied to analyze data. Measurement data from three independent experiments were expressed as the mean ± standard deviation. Comparisons on data between two groups were carried out using the independent samples t‐test. Data comparisons among multiple groups were carried out by one‐way analysis of variance (anova). Comparisons on data between two groups that obeyed normal distribution were carried out using t‐tests. P < 0.05 showed statistical significance.
RESULTS
Network pharmacological analysis screened 115 effective target genes of S. acutum
Six active ingredients (Table S1) were obtained after screening 14 , including MOL000358: β‐sitosterol, MOL000621: 16‐epi‐isositsirikine, MOL000622: magnograndiolide, MOL000623: michelenolide, MOL000625: SIN and MOL00027: stepholidine.
As shown in Table S2, a total of 115 target proteins were obtained. Furthermore, the target proteins were converted into target genes through Perl language. As shown in Table S3, there were 38 target genes of β‐sitosterol, 28 target genes of 16‐epi‐isositsirikine, four target genes of magnograndiolide, one target gene of michelenolide, 15 target genes of SIN and 29 target genes of stephanine.
A total of 14 differential genes that S. acutum might act on to treat DPN were screened
The microarray GSE95849 was finally obtained, which contained 6 control samples and 6 DPNP samples. Analysis of the GSE95849 microarray dataset identified 4,676 differential genes, including 2,195 downregulated genes and 2,481 upregulated genes (Figure 2a,b).
Figure 2.
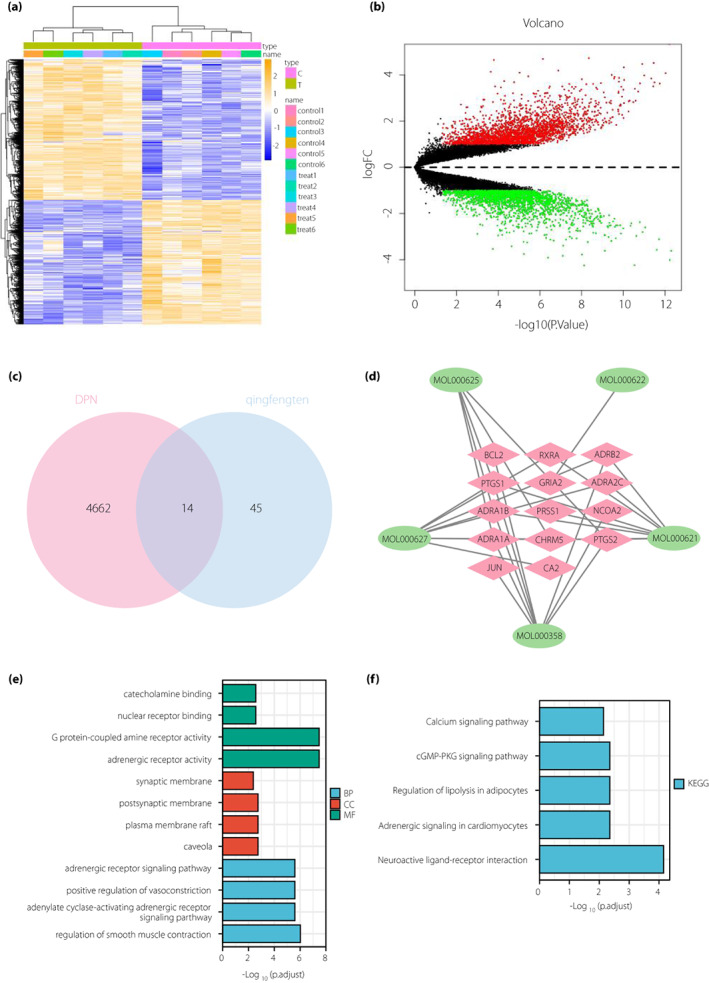
A total of 14 differential genes that Sinomenium acutum might act on to treat diabetic peripheral neuropathy (DPN) are screened. (a) Heat map of the differential genes in the GSE95849 microarray (C represents control group [n = 6], and T represents diabetic peripheral neuropathic pain [DPNP] group [n = 6]. Blue indicates down‐regulation and yellow indicates upregulation). (b) Volcanic map of the differential genes in the GSE95849 microarray (n = 6 for the control and DPNP groups, respectively. Black dots represent genes that are not differentially expressed, red dots represent the upregulated genes and green dots represent downregulated genes). (c) Venn diagram for the intersection of differential genes in GSE95849 microarray and effective target genes obtained from network pharmacological analysis (the bar chart below shows the number of differential genes in DPNP and S. acutum). (d) Network diagram of “Sinomenium acutum‐ingredients‐target” (green circles represent the active ingredients of S. acutum and red diamonds represent the target genes which the active ingredients act on). (e) Bar chart of gene ontology function analysis of the 14 candidate genes at the biological process (BP), cellular component (CC) and molecular function (MF) levels. (f) Bar chart of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the 14 candidate genes.
Differential genes in GSE95849 microarray dataset were intersected with target genes of active ingredients of S. acutum obtained from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform database. The results showed that 59 genes were left after the intersection. There were 14 intersection genes between Gene Expression Omnibus and target genes of active ingredients of S. acutum (Figure 2c, Table S4), which were RXRA, ADRB2, ADRA2C, BCL2, ADRA1B, PRSS1, PTGS1, PTGS2, ADRA1A, CHRM5, GRIA2, NCOA2, CA2 and JUN. Five of them (PTGS1, CHRM5, PTGS2, ADRA1A and ADRA1B) were target genes of SIN.
Cytoscape software (Figure 2d) showed that the main active ingredients of S. acutum in DPNP were MOL000621: 16‐epi‐isositsirikine, MOL000622: magnogradiolide, MOL000625: SIN, MOL000627: stephanine and MOL000358: β‐sitosterol, corresponding to eight, one, five, eight and four target genes, respectively.
Furthermore, gene ontology and Kyoto Encyclopedia of Genes and Genomes function enrichment analyses of 14 key genes were carried out, the results (Figure 2e,f) showed that candidate genes were mainly involved in the main biological processes, including vascular smooth muscle contraction, adrenergic receptor signaling pathway and adenylate cyclase activation adrenergic receptor signaling pathway. The main cell components included synaptic membrane and postsynaptic membrane. The main molecular functions included G protein coupled amine receptor activity, adrenergic receptor activity, nuclear receptor binding and catecholamine binding. Neuroactive ligand receptor interaction was the main Kyoto Encyclopedia of Genes and Genomes pathway enriched. It has been documented that neuroactive ligand receptor interactions are associated with neuroprotection and inflammatory responses 15 , 16 . In addition, adrenergic signaling pathway in cardiomyocytes, regulation of adipocyte lipolysis and CGMP‐PKG signaling pathway have been reported to be closely related to endoplasmic reticulum stress 17 , 18 , 19 .
SIN in S. acutum might alleviate DPNP by regulating the IRE1α–XBP1s pathway through PTGS2
To further screen out the effective ingredients and target genes, 14 candidate genes were imported into the STRING database for construction of the protein–protein interaction diagram (Figure 3a). Next, the data were imported into Cytoscape software to construct the protein–protein interaction network (Figure 3b,c). Furthermore, according to the order of degree value in the protein–protein interaction network from large to small, 10 key targets were obtained, which were GRIA2, JUN, ADRA1B, PTGS2, ADRA2C, ADRA1A, ADRB2, PTGS1, NCOA2 and RXRA in turn (Figure 3d).
Figure 3.
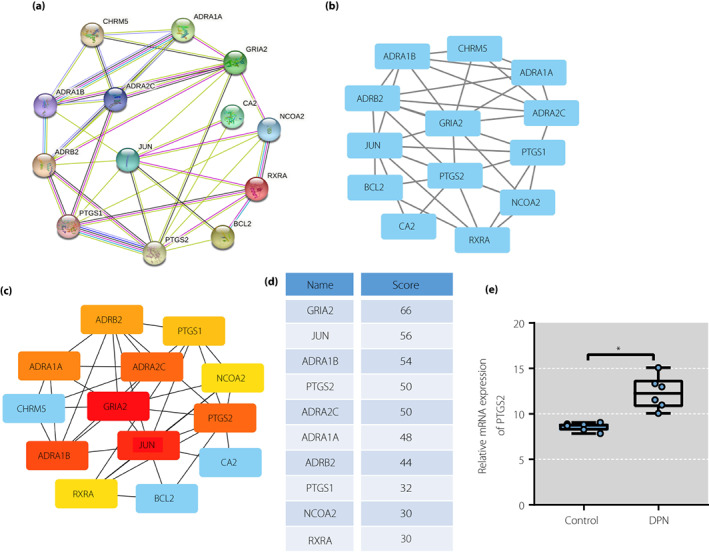
Sinomenine (SIN) in Sinomenium acutum might alleviate diabetic peripheral neuropathic pain by regulating inositol‐requiring enzyme 1 alpha–X‐box binding protein 1 pathway through prostaglandin‐endoperoxide synthase 2 (PTGS2). (a) Protein–protein interaction network of candidate genes analyzed using String database. (b) Protein–protein interaction network constructed using Cytoscape. (c) Protein–protein interaction network of top 10 core genes screened out using Cytoscape. (d) Top 10 core genes scored using Cytoscape. (e) Bar chart of the expression of PTGS2 in GSE95849 microarray (n = 6 for the control and diabetic peripheral neuropathic pain groups, respectively). DPN, diabetic peripheral neuropathy. mRNA, messenger ribonucleic acid.
SIN is an inhibitor of PTGS2 in vitro and suppresses the expression of PTGS2 20 , 21 . Inhibition of PTGS2 can relieve peripheral neuropathic pain in rats 22 . Therefore, SIN was selected as a key active ingredient in S. acutum for analysis, and PTGS2 was selected as the target gene.
It has been documented that the activation of IRE1α–XBP1s pathway can promote the release of pro‐inflammatory factors, and regulation of IRE1α–XBP1s can relieve pain 23 . PTGS2 can activate the IRE1α–XBP1s pathway 8 . In addition, to verify that SIN alleviated DPNP by regulating IRE1 through the IRE1α–XBP1s pathway, GSE95849 microarray dataset was analyzed. The results showed that PTGS2 expression was increased in the DPNP human samples compared with the control human samples (Figure 3e).
SIN blocked the IRE1α–XBP1s pathway by downregulating PTGS2 to relieve pain in DPNP rats
After measurement of blood glucose level and bodyweight of the rats, we identified higher blood glucose levels and lower bodyweight in DPNP rats relative to sham‐operated rats. In contrast to DPNP rats, DPNP rats treated with SIN had lower blood glucose levels and higher bodyweight; whereas both of oe‐PTGS2 and SIN treatment led to an increase in blood glucose level and a reduction in bodyweight, the effect of which could be reversed by further administration of MKC8866 (Figure 4a,b). The results of pain behavior analysis (Figure 4c) showed that PWT decreased in the DPNP rats at day 3 after STZ injection. The PWL decreased at day 1 and the lowest value was achieved at day 7 after STZ injection, and maintained until the day 14 after STZ injection, suggesting that stable mechanical and thermal pain sensitivity was formed in DPNP rats. In contrast to the DPNP rats, DPNP rats treated with SIN had increased PWT and PWL; whereas both of oe‐PTGS2 and SIN treatment reduced PWT and PWL, the effect of which could be reversed by further administration of MKC8866.
Figure 4.
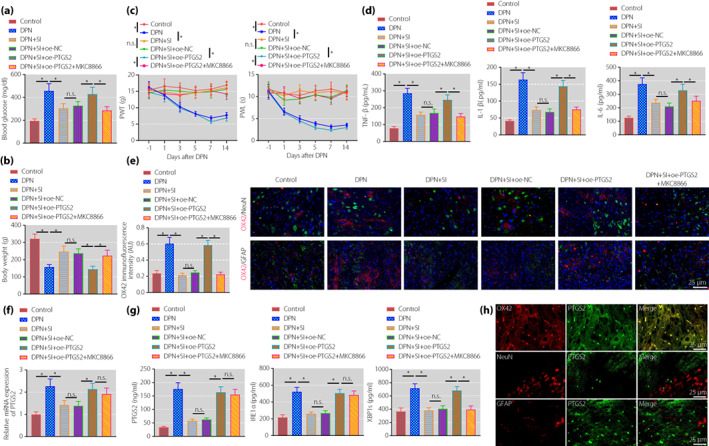
Sinomenine inhibits the inositol‐requiring enzyme 1 alpha (IRE1α)–X‐box binding protein 1 (XBP1s) pathway activation by downregulating prostaglandin‐endoperoxide synthase 2 (PTGS2) to relieve pain symptoms in diabetic peripheral neuropathic pain (DPNP) rats. (a) Detection of blood glucose level in DPNP rats. (b) Detection of bodyweight of DPNP rats. (c) Evaluation of the paw withdrawal threshold and paw withdrawal latency of rats before and after streptozotocin injection. (d) The expression of tumor necrosis factor‐α (TNF‐α), interleukin (IL)‐1β and IL‐6 in rat spinal cord tissue measured by enzyme‐linked immunosorbent assay. (e) Microglia cell activation in the spinal cord of rats measured by immunofluorescence (scale bar: 25 μm). (f) PTGS2 expression in the spinal cord of rats determined by quantitative reverse transcription polymerase chain reaction. (g) The expression of PTGS2, IRE1α and XBP1s in the spinal cord of rats measured by enzyme‐linked immunosorbent assay. (h) PTGS2‐positive cells in the spinal cord of DPNP rats measured using immunofluorescence (scale bar: 25 μm). *P < 0.05 versus the control group. n.s. indicates that the data between the two groups are not statistically significant (n = 10 in each group). DPN, diabetic peripheral neuropathy; NC, normal control; oe, overexpression; SI, sinomenine.
As shown by ELISA (Figure 4d), the expression of TNF‐α, IL‐1β and IL‐6 was elevated in DPNP rats, which was decreased after SIN treatment. Compared with DPNP rats treated with SIN and oe‐NC, the above‐mentioned inflammatory factors increased in DPNP rats treated with SIN and oe‐PTGS2. Compared with DPNP rats treated with SIN and oe‐PTGS2, inflammatory factors decreased in DPNP rats treated with SIN, oe‐PTGS2 and MKC8866. Immunofluorescence results (Figure 4e) showed that the microglial marker, OX42, in the spinal dorsal horn tissue was not co‐expressed with the neuronal marker, NeuN, and the astrocyte marker, GFAP. Furthermore, the expression of OX42 was higher in the DPNP rats than that in the sham‐operated rats, indicating higher microglial activity in the spinal cord in DPNP rats. In contrast to the DPNP rats, microglial activity of the DPNP rats treated with SIN was weakened. In contrast to the DPNP rats treated with SIN and oe‐NC, microglial activity was increased in the DPNP rats treated with SIN and oe‐PTGS2. Relative to the DPNP rats treated with SIN and oe‐PTGS2, opposite microglial activity occurred in the DPNP rats treated with SIN, oe‐PTGS2 and MKC8866.
In addition, we identified elevated expression of PTGS2, IRE1 and XBP1s in the DPNP rats, which was decreased after SIN treatment. In contrast to the DPNP rats treated with SIN and oe‐NC, elevated expression of PTGS2, IRE1 and XBP1s was witnessed in the DPNP rats treated with SIN and oe‐PTGS2. Relative to the DPNP rats treated with SIN and oe‐PTGS2, the expression of PTGS2 and IRE1 was unchanged, and the expression of XBP1s was reduced in the DPNP rats treated with SIN, oe‐PTGS2 and MKC8866 (Figure 4f,g). PTGS2‐positive cells in the spinal cord of DPNP rats were detected by immunofluorescence (Figure 4h), and the results showed that PTGS2 was mainly co‐expressed with OX‐42, but not with NeuN and GFAP.
SIN inhibited the IRE1α–XBP1s pathway through PTGS2 to suppress the activation of spinal cord microglial cells and inflammatory response
Next, we isolated rat spinal cord microglial cells for further exploration. As shown by immunofluorescence, >98% of cells were OX‐42‐positive, showing that microglial cells with high purity were isolated from rat spinal cord (Figure 5a). PTGS2 was overexpressed in microglial cells, validating the successful transfection (Figure 5b).
Figure 5.
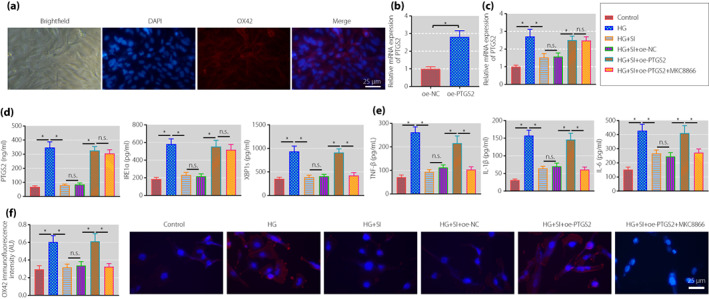
Sinomenine inhibits the inositol‐requiring enzyme 1 alpha (IRE1α)–X‐box binding protein 1 (XBP1s) pathway through prostaglandin‐endoperoxide synthase 2 (PTGS2) to suppress the activation of spinal cord microglia and inflammatory response. (a) The expression of OX‐42 was detected by immunofluorescence to identify the purity of the isolate (scale bar: 25 μm). (b) The transfection efficiency of PTGS2 in spinal cord microglia measured by quantitative reverse transcription polymerase chain reaction. (c) PTGS2 expression in rat spinal cord microglia determined by quantitative reverse transcription polymerase chain reaction. (d) The expression of PTGS2, IRE1α and XBP1s in rat spinal cord microglia measured by enzyme‐linked immunosorbent assay. (e) The expression of tumor necrosis factor‐α (TNF‐α), interleukin (IL)‐1β and IL‐6 in supernatants of rat spinal cord microglia measured using enzyme‐linked immunosorbent assay. (f) The activation of rat spinal cord microglia determined by immunofluorescence (scale bar: 25 μm). *P < 0.05 versus the control group. n.s. indicates that the data between the two groups are not statistically significant. The cell experiments were repeated three times. HG, high glucose; NC, normal control; oe, overexpression; SI, sinomenine.
Further, microglial cell inflammatory response was stimulated with HG. After reverse transcription quantitative polymerase chain reaction and ELISA, we found that the expression of PTGS2, IRE1 and XBP1s was increased in the HG‐stimulated cells. In contrast to the HG‐stimulated cells, the expression of the above factors was reduced in those treated with SIN. In comparison with the HG‐stimulated cells treated with SIN and oe‐NC, opposite results were obtained in those treated with SIN and oe‐PTGS2. In contrast to the HG‐stimulated cells treated with SIN and oe‐PTGS2, the expression of PTGS2 and IRE1 was unchanged, and the expression of XBP1s was reduced in those treated with SIN, oe‐PTGS2 and MKC8866 (Figure 5c,d).
The expression of inflammatory factors in cell supernatant was detected by ELISA, and the results (Figure 5e) showed that the expression of TNF‐α, IL‐1β and IL‐6 was increased in the HG‐stimulated cells, which was reduced after SIN treatment. Relative to SIN + oe‐NC treatment, the above expression was elevated after further treatment with oe‐PTGS2. The trend in response to SIN and oe‐PTGS2 in the HG‐stimulated cells was reversed by co‐treatment with SIN, oe‐PTGS2 and MKC8866. Immunofluorescence results (Figure 5f) showed that compared with the control cells, the expression of OX42 was increased in the HG‐stimulated cells. In contrast to the HG‐stimulated cells, the expression of OX42 was decreased in those treated with SIN. Relative to the HG‐stimulated cells treated with SIN and oe‐NC, increased OX42 expression was shown in those treated with SIN and oe‐PTGS2, and co‐treatment with SIN, oe‐PTGS2 and MKC8866 could reverse the trend.
DISCUSSION
In the current study, we found that SIN could alleviate DPNP through regulation of PTGS2‐mediated IRE1α–XBP1s signaling pathway.
The network pharmacological analysis combined with bioinformatics analysis carried out in the present study showed that SIN was an important active ingredient of S. acutum to exert efficacy on DPNP, and that PTGS2, the target gene of SIN, might be the key gene involved in treating DPNP. Consistent with the present results, the therapeutic effect of SIN in reducing diabetic neuropathic pain has been also previously documented 24 . In addition, SIN was shown to enhance the efficacy of gabapentin or ligustrazine hydrochloride in neuropathic pain in rodent animals 25 . Additionally, SIN could alleviate microglial mobilization and inhibit neuroinflammation to suppress experimental autoimmune encephalomyelitis 26 . A previous study showed that SIN can mediate anesthesia in the peripheral nervous system through modulation of voltage‐gated sodium channels 27 . Similarly, it has also been shown that SIN might exert a modulatory effect on the TRPV signaling pathway through p38MAPK and, thus, on the peripheral nervous system response 24 , 28 , 29 . SIN could relieve cancer‐induced bone pain partially by inhibiting microglial activation and repressing microglial JAK2/STAT3 in a rat model 30 . As previously reported, treatment with SIN could partially inhibit the expression of PTGS2 to exert protective function on inflammatory pain in a rat model 6 . Furthermore, SIN could diminish the expression of PTGS2 in a mouse model of osteoarthritis, resulting in repression of the inflammatory response 31 . Notably, the role of PTGS2 in neuropathic pain and microglia has also been previously unveiled. Inhibition of PTGS2 using meloxicam could play an antiallodynic role in diabetic mice with neuropathic pain, with the action site on the periphery 7 . In addition, highly expressed PTGS2 could aggravate inflammatory neuronal damage induced by microglial activation 32 . As one recent study showed, SIN can inhibit inflammation and related pro‐inflammatory factors, including IL‐1β, IL‐6 and TNF‐α, by inhibiting NLRP3 inflammasome 26 , which is highly consistent with the present study.
Mechanistically, it was shown in the present study that PTGS2 inhibited the IRE1α–XBP1s signaling pathway to alleviate DPNP. Oxidative stress is one of the neuropathic factors in diabetes patients 33 . The linkage between oxidative stress with COX‐2 has been previously documented 34 . Accumulating evidence has documented the interaction between PTGS2 and IRE1α–XBP1s. PTGS2 can upregulate IRE1α expression by binding to it, resulting in IRE1α splicing of XBP1 messenger RNA 8 . Intriguingly, it was previously shown that the treatment with the PTGS2 inhibitor, celecoxib, could contribute to a decline in the expression of IRE1 35 . Elevated expression of IRE1 could cause endoplasmic reticulum stress and aid in increasing paclitaxel‐induced peripheral neuropathy in a rat model 36 . Intriguingly, the activation of IRE1α and XBP1s was reported in chronic constriction injury‐triggered neuropathic pain 9 .
In conclusion, SIN downregulates the expression of PTGS2 to inactivate IRE1α–XBP1s signaling pathway, which inhibits microglial cell activation and the release of inflammatory factors, finally alleviating DPNP (Figure 6). Nevertheless, further study should focus on other neurological performance measurements, such as motor and/or sensory nerve conduction velocities and nerve blood flow, as well as the density of myelinated and unmyelinated nerve fibers or the intraepidermal nerve fiber density, for further validation of the present findings.
Figure 6.
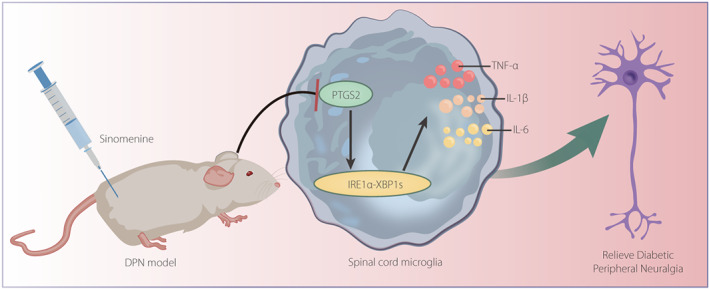
Molecular mechanism diagram of the role of sinomenine in diabetic peripheral neuropathic pain by regulating prostaglandin‐endoperoxide synthase 2 (PTGS2)‐mediated the inositol‐requiring enzyme 1 alpha (IRE1α)–X‐box binding protein 1 (XBP1s) pathway. Sinomenine downregulates the expression of PTGS2 to inactivate the IRE1α–XBP1s signaling pathway, which inhibits microglial cell activation and the release of inflammatory factors, finally alleviating diabetic peripheral neuropathic pain. DPN, diabetic peripheral neuropathy; IL, interleukin; TNF‐α, tumor necrosis factor‐α.
DISCLOSURE
The authors declare no conflict of interest.
Approval of the research protocol: This experimental procedure and animal use protocol were approved by the Animal Ethics Committee of Affiliated Huaihua Hospital, Hengyang Medical School, University of South China.
Informed consent: N/A.
Registry and the registration no. of the study/trial: 2021.09.23; HHSYY‐EC‐202109‐K6.
Animal studies: The animal experiments followed the Animal Welfare Act and Public Health Service guidelines for the management and use of experimental animals specified by the National Institutes of Health.
Supporting information
Table S1 | Network pharmacological analysis screens 115 effective target genes of Sinomenium acutum.
Table S2 | The target proteins corresponding to the six active ingredients.
Table S3 | The related target genes.
Table S4 | Information about 14 intersection genes between Gene Expression Omnibus and target genes of active ingredients of Sinomenium acutum.
ACKNOWLEDGMENTS
This work was supported by Excellent Youth Foundation of Hunan Provincial Department of Education (19B477), Project of Hunan Provincial Health Commission (20200018, 20200037) and Natural Science Foundation of Hunan Province (2021JJ70043).
REFERENCES
- 1. Wei KS, Gu MZ, Zhu JW, et al. Current views of diabetic peripheral neuropathic pain comorbid depression – a review. Eur Rev Med Pharmacol Sci 2020; 24: 10663–10670. [DOI] [PubMed] [Google Scholar]
- 2. Zhang B, Yu Y, Aori G, et al. Tanshinone IIA attenuates diabetic peripheral neuropathic pain in experimental rats via inhibiting inflammation. Evid Based Complement Alternat Med 2018; 2018: 2789847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Toth CC, Jedrzejewski NM, Ellis CL, et al. Cannabinoid‐mediated modulation of neuropathic pain and microglial accumulation in a model of murine type I diabetic peripheral neuropathic pain. Mol Pain 2010; 6: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang YF, He F, Wang CJ, et al. Discovery of chemical markers for improving the quality and safety control of Sinomenium acutum stem by the simultaneous determination of multiple alkaloids using UHPLC‐QQQ‐MS/MS. Sci Rep 2020; 10: 14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhu Q, Sun Y, Zhu J, et al. Antinociceptive effects of sinomenine in a rat model of neuropathic pain. Sci Rep 2014; 4: 7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yuan Y, Zhang Y, He X, et al. Protective effects of sinomenine on CFA‐induced inflammatory pain in rats. Med Sci Monit 2018; 24: 2018–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kimura S, Kontani H. Demonstration of antiallodynic effects of the cyclooxygenase‐2 inhibitor meloxicam on established diabetic neuropathic pain in mice. J Pharmacol Sci 2009; 110: 213–217. [DOI] [PubMed] [Google Scholar]
- 8. Groenendyk J, Paskevicius T, Urra H, et al. Cyclosporine A binding to COX‐2 reveals a novel signaling pathway that activates the IRE1alpha unfolded protein response sensor. Sci Rep 2018; 8: 16678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gui Y, Li A, Zhang J, et al. alpha‐Asarone alleviated chronic constriction injury‐induced neuropathic pain through inhibition of spinal endoplasmic reticulum stress in an liver X receptor‐dependent manner. Anesth Analg 2018; 127: 775–783. [DOI] [PubMed] [Google Scholar]
- 10. Lin TT, Qu J, Wang CY, et al. Rescue of HSP70 in spinal neurons alleviates opioids‐induced hyperalgesia via the suppression of endoplasmic reticulum stress in rodents. Front Cell Dev Biol 2020; 8: 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li X, Xiang L, Lin Y, et al. Computational analysis illustrates the mechanism of Qingfei Paidu decoction in blocking the transition of COVID‐19 patients from mild to severe stage. Curr Gene Ther 2022; 22: 277–289. [DOI] [PubMed] [Google Scholar]
- 12. Sheng X, Nenseth HZ, Qu S, et al. IRE1alpha‐XBP1s pathway promotes prostate cancer by activating c‐MYC signaling. Nat Commun 2019; 10: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miao J, Zhou X, Ji T, et al. NF‐kappaB p65‐dependent transcriptional regulation of histone deacetylase 2 contributes to the chronic constriction injury‐induced neuropathic pain via the microRNA‐183/TXNIP/NLRP3 axis. J Neuroinflammation 2020; 17: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang P, He H, Xu S, et al. Potential molecular target prediction and docking verification of Hua‐Feng‐Dan in stroke based on network pharmacology. Evid Based Complement Alternat Med 2020; 2020: 8872593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang Y, Chen M, Zhai Z, et al. Long non‐coding RNAs Gabarapl2 and Chrnb2 positively regulate inflammatory signaling in a mouse model of dry eye. Front Med (Lausanne) 2021; 8: 808940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qian J, Zhao X, Wang W, et al. Transcriptomic study reveals recovery of impaired astrocytes contribute to neuroprotective effects of danhong injection against cerebral ischemia/reperfusion‐induced injury. Front Pharmacol 2018; 9: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang B, Zhang P, Tan Y, et al. C1q‐TNF‐related protein‐3 attenuates pressure overload‐induced cardiac hypertrophy by suppressing the p38/CREB pathway and p38‐induced ER stress. Cell Death Dis 2019; 10: 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang L, Zhang B, Huang F, et al. Curcumin inhibits lipolysis via suppression of ER stress in adipose tissue and prevents hepatic insulin resistance. J Lipid Res 2016; 57: 1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang F, Ma H, Butler MR, et al. Potential contribution of ryanodine receptor 2 upregulation to cGMP/PKG signaling‐induced cone degeneration in cyclic nucleotide‐gated channel deficiency. FASEB J 2020; 34: 6335–6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang H, Yin P, Shi Z, et al. Sinomenine, a COX‐2 inhibitor, induces cell cycle arrest and inhibits growth of human colon carcinoma cells in vitro and in vivo. Oncol Lett 2016; 11: 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lv Y, Li C, Li S, et al. Sinomenine inhibits proliferation of SGC‐7901 gastric adenocarcinoma cells via suppression of cyclooxygenase‐2 expression. Oncol Lett 2011; 2: 741–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stevens AM, Saleem M, Deal B, et al. Targeted cyclooxygenase‐2 inhibiting nanomedicine results in pain‐relief and differential expression of the RNA transcriptome in the dorsal root ganglia of injured male rats. Mol Pain 2020; 16: 1744806920943309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chopra S, Giovanelli P, Alvarado‐Vazquez PA, et al. IRE1alpha‐XBP1 signaling in leukocytes controls prostaglandin biosynthesis and pain. Science 2019; 365: eaau6499. [DOI] [PubMed] [Google Scholar]
- 24. Rao S, Liu S, Zou L, et al. The effect of sinomenine in diabetic neuropathic pain mediated by the P2X3 receptor in dorsal root ganglia. Purinergic Signal 2017; 13: 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao T, Shi T, Wiesenfeld‐Hallin Z, et al. Sinomenine facilitates the efficacy of gabapentin or ligustrazine hydrochloride in animal models of neuropathic pain. Eur J Pharmacol 2019; 854: 101–108. [DOI] [PubMed] [Google Scholar]
- 26. Kiasalari Z, Afshin‐Majd S, Baluchnejadmojarad T, et al. Sinomenine alleviates murine experimental autoimmune encephalomyelitis model of multiple sclerosis through inhibiting NLRP3 inflammasome. J Mol Neurosci 2021; 71: 215–224. [DOI] [PubMed] [Google Scholar]
- 27. Lee JY, Yoon SY, Won J, et al. Sinomenine produces peripheral analgesic effects via inhibition of voltage‐gated sodium currents. Neuroscience 2017; 358: 28–36. [DOI] [PubMed] [Google Scholar]
- 28. Li G, Gong J, Lei H, et al. Promotion of behavior and neuronal function by reactive oxygen species in C. elegans . Nat Commun 2016; 7: 13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Joksimovic SL, Jevtovic‐Todorovic V, Todorovic SM. The mechanisms of plasticity of nociceptive ion channels in painful diabetic neuropathy. Front Pain Res (Lausanne) 2022; 3: 869735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen SP, Sun J, Zhou YQ, et al. Sinomenine attenuates cancer‐induced bone pain via suppressing microglial JAK2/STAT3 and neuronal CAMKII/CREB cascades in rat models. Mol Pain 2018; 14: 1744806918793232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu Y, Lin Z, Yan Z, et al. Sinomenine contributes to the inhibition of the inflammatory response and the improvement of osteoarthritis in mouse‐cartilage cells by acting on the Nrf2/HO‐1 and NF‐kappaB signaling pathways. Int Immunopharmacol 2019; 75: 105715. [DOI] [PubMed] [Google Scholar]
- 32. Yang L, Zhou R, Tong Y, et al. Neuroprotection by dihydrotestosterone in LPS‐induced neuroinflammation. Neurobiol Dis 2020; 140: 104814. [DOI] [PubMed] [Google Scholar]
- 33. Kato A, Tatsumi Y, Yako H, et al. Recurrent short‐term hypoglycemia and hyperglycemia induce apoptosis and oxidative stress via the ER stress response in immortalized adult mouse Schwann (IMS32) cells. Neurosci Res 2019; 147: 26–32. [DOI] [PubMed] [Google Scholar]
- 34. Pathak SK, Sharma RA, Steward WP, et al. Oxidative stress and cyclooxygenase activity in prostate carcinogenesis: targets for chemopreventive strategies. Eur J Cancer 2005; 41: 61–70. [DOI] [PubMed] [Google Scholar]
- 35. Su W, Tai Y, Tang SH, et al. Celecoxib attenuates hepatocyte apoptosis by inhibiting endoplasmic reticulum stress in thioacetamide‐induced cirrhotic rats. World J Gastroenterol 2020; 26: 4094–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Semis HS, Kandemir FM, Kaynar O, et al. The protective effects of hesperidin against paclitaxel‐induced peripheral neuropathy in rats. Life Sci 2021; 287: 120104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 | Network pharmacological analysis screens 115 effective target genes of Sinomenium acutum.
Table S2 | The target proteins corresponding to the six active ingredients.
Table S3 | The related target genes.
Table S4 | Information about 14 intersection genes between Gene Expression Omnibus and target genes of active ingredients of Sinomenium acutum.


