Abstract
The major role of RecA is thought to be in helping repair and restart stalled replication forks. During exponential growth, Bacillus subtilis recA cells exhibited few microscopically observable nucleoid defects. However, the efficiency of plating was about 12% of that of the parent strain. A substantial and additive defect in viability was also seen for addB and recF mutants, suggesting a role for the corresponding recombination paths during normal growth. Upon entry into stationary phase, a subpopulation (∼15%) of abnormally long cells and nucleoids developed in B. subtilis recA mutants. In addition, recA mutants showed a delay in, and a diminished capacity for, effecting prespore nucleoid condensation.
The RecA protein is ubiquitous among the bacteria studied and possesses one of the most conserved primary amino acid sequences. The function of the RecA protein as a mediator of homologous recombination and as a regulator of the inducible SOS response in both Escherichia coli and Bacillus subtilis has been elucidated in detail (33, 36). The primary role of RecA in E. coli, and perhaps in all bacteria, appears to be its participation in the housekeeping function of repairing and restarting stalled replication forks (5).
The importance of this housekeeping function is illustrated by the fact that E. coli PriA, which is disposable for initiation of DNA replication at oriC, but essential for restarting stalled replication forks, cannot be functionally inactivated without suffering a severe penalty: in growing cultures about 90% of priA mutant cells are dead (16, 25). Similarly, the absence of recombinational repair in E. coli recA mutants results in a 50% loss of viability (4). In recA mutants, the primary cause of death appears to be chromosomal degradation, presumably at sites of stalled DNA replication forks. About 10% of E. coli recA cells are anucleate, and an additional portion show signs of chromosomal degradation (29, 38).
During our analysis of site-specific recombination and chromosome dimerization in B. subtilis, we noted that mutations in recA did not completely suppress the nucleoid segregation defects associated with the inability to resolve dimeric chromosomes in ripX mutants (26). However, the recA mutation by itself had little effect on nucleoid morphology. Subsequently, we noted that about 12% of recA cells were viable during exponential growth and stationary phase (“viable” in this report refers to colony-forming ability). In addition, we have observed that a B. subtilis strain with a recA mutation developed a subpopulation of cells that displayed gross division defects and a loss of nucleoid integrity during the transition from exponential growth to stationary phase. These phenotypes were not seen in E. coli recA mutants. To our knowledge, there have been no previous reports of aberrant nucleoids in recombination-deficient strains of B. subtilis.
The parent strain in this study was BR151 (trpC2 lys-3 metB10). The ΔSPβ phage strain used (YB886) and the ΔSPβ addB72 strain (SL7576) were obtained from the Bacillus Genetic Stock Center and are derivatives of BR151 (37). SL7131, SL7360, and SL7370 have been described previously (26). The B. subtilis recF strain (SL7609) was made by transformation of BR151 with pSAS28, a pBluescript plasmid containing recF::spc (spc between the StyI sites within recF). The ΔSPβ addB72 recF strain (SL7611) was made by transformation of SL7576 with pSAS28. W3110 is an E. coli recA::Tn9 strain (34). The recA-lacZ strain (SL7937) was made by transformation of BR151 with YB3001 DNA (28). comG-lacZ (SL7952) and comK-lacZ (SL7954) strains were made by transformation of BR151 with BD1960 and BD1991 DNA, respectively. Bacterial cultures were grown in Luria-Bertani (LB) medium or modified Schaeffer's sporulation medium (MSSM) (21) at 37°C with shaking at 150 rpm. Media were used fresh or were stored for short periods in the dark before use; glass vessels were used throughout. Medium volume was maintained between 6 and 9% of total flask volume. recA, recF, and addB mutants were grown in glass flasks wrapped in aluminum foil to reduce the effects of ambient light. Mitomycin C (MMC) was used at a concentration of 20 ng ml−1. Nucleoids were stained with 4′, 6-diamidino-2-phenylindole (DAPI) and were observed and photographed as described previously (26). Cell and nucleoid measurements were performed as described earlier (26). β-Galactosidase and sporulation-frequency assays were performed essentially as described previously (19). “T0 ” in these studies refers to the end of exponential growth. “T1, T2, T3, ” etc., are 1, 2, and 3 h, etc., after the end of exponential growth. Plating efficiencies were derived from dilutions of exponential- or stationary-phase cultures. Dilutions were made in Spizizen's minimal salts plus 55 mM glucose, 0.02% casein hydrolysate, 0.1% yeast extract, 20 μg of tryptophan per ml, and 50 μg (each) of methionine and lysine per ml. Plating was performed in duplicate on freshly prepared LB agar, and incubation was at 37°C in the dark. Only plates with between 30 and 400 colonies were used to determine viable counts. In general, Difco components were used in the media. Preliminary experiments indicated that growth rate, cell and nucleoid morphologies, and plating efficiencies of recA mutants were very similar when Oxoid components were substituted for Difco components.
Nucleoid and cell morphology phenotypes of B. subtilis rec mutants during exponential-phase growth.
In Table 1, we present an assessment of nucleoid and cell morphologies for various rec mutant strains and their derivatives during exponential growth. A small proportion of the recA mutant (SL7360) cells showed evidence of partitioning failures and cell wall irregularities. The cell wall defects observed were typically bulges in the lateral wall (Fig. 1E and F). recA cells with cell wall defects almost always contained complex nucleoid structures (Fig. 1E and F). Because the nucleoids in cells with abnormal wall morphologies had a highly variable appearance, we did not attempt to quantify them in our scoring. Anucleate cells in the recA strain were rare (Table 1). In other experiments in which a larger number of cells were observed, the anucleate frequency of two different recA alleles was 0.16% (4 anucleate cells out of 2,500 cells scored). This is notable because a hallmark of E. coli recA strains is their high frequency of anucleate cells (∼10%) (38). Anucleate cells in E. coli recA strains are thought to result from “rec-less” degradation of chromosomal DNA mediated by RecD (29).
TABLE 1.
Nucleoid profiles of various rec strains fixed during mid-exponential growth in LB medium
| Strain (relevant genotype) | % with nucleoid phenotypea
|
Viability
|
||||
|---|---|---|---|---|---|---|
| Parb | Anucleate | Cell wall defectsc | Total defects | CFU mg−1d | Relative plating efficiencye | |
| BR151 (parent) | 1.2 | 0 | 0 | 1.2 | 7.46 × 108 | 1.0 |
| SL7609 (recF::spc) | 8.0 | 0.8 | 0.2 | 9.0 | 5.07 × 108 | 0.61 |
| SL7360 (recA::neo) | 4.2 | 0 | 1.8 | 6.0 | 9.27 × 107 | 0.12f |
| SL7370 (recA::neo ripX::spc) | 16.0 | 2.4 | 6.0 | 24.4 | 4.15 × 107 | 0.056 |
| SL7131 (ripX::spc) | 26.2 | 1.0 | 0 | 27.2 | 4.28 × 108 | 0.57 |
| SL7513 (ΔSPβ parent) | 0.66 | 0 | 0 | 0.66 | 2.55 × 109 | 1.0 |
| SL7576 (ΔSPβ addB72) | 2.6 | 0.4 | 0 | 3.0 | 1.34 × 109 | 0.52 |
| SL7611 (ΔSPβ recF::spc addB72) | 5.3 | 0.83 | 0.17 | 6.3 | 8.03 × 108 | 0.31 |
Minimum of 500 cells scored per strain.
Par nucleoids were of two classes: those that had elongated to > 1/3 of the cell length and those in which partitioning appeared stalled. Typically the stalled nucleoids had an hourglass or dumbbell shape, but had no immediate nucleoid neighbors.
Cell wall defects typically appeared as bulges in the lateral cell wall and normally contained complex nucleoid structures that may be described as “disintegrating.” Examples are provided in Fig. 1E and F.
The numbers of CFU were normalized by bacterial dry weight (in milligrams) with a correction table for spectrophotometric values at an optical density at 600 nm. The average of at least two experiments is shown (see text for details).
Plating efficiencies are relative to the appropriate parent strain: BR151 or SL7513.
The relative plating efficiency value during stationary phase for this strain was 0.16.
FIG. 1.
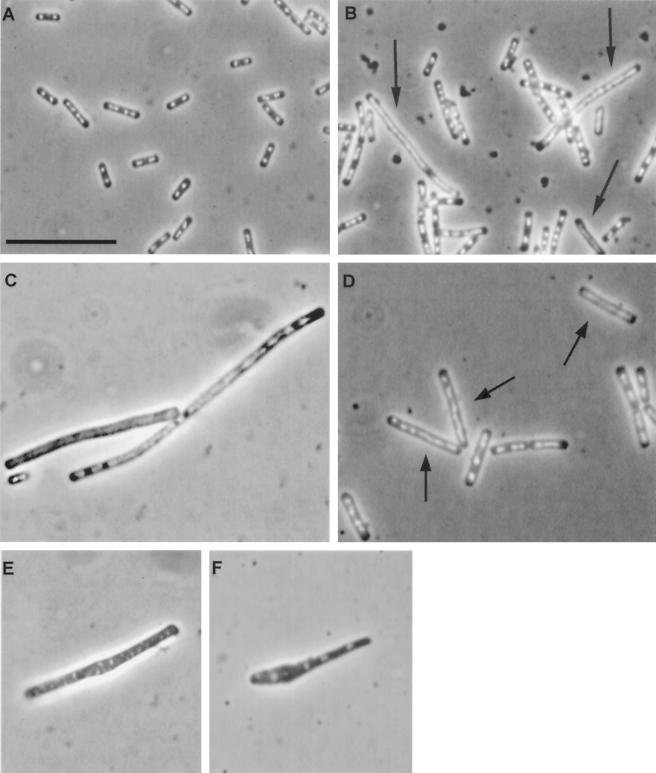
Micrographs of B. subtilis strains treated with DAPI to visualize nucleoids. (A) Parent strain BR151 grown to stationary phase (T1.5) in LB medium. (B) recA mutant SL7360 grown to stationary phase (T1.5) in LB medium. (C) recA mutant SL7360 grown to stationary phase (T1.5) in sporulation medium MSSM. (D) Parent strain BR151 grown in LB medium with MMC (20 ng/ml) added at mid-exponential phase; photo was taken 1.5 h after MMC addition. (E) Example of cell wall defect in recA mutant SL7360 during the exponential phase. (F) Example of cell wall defect in recA ripX mutant SL7370 during the exponential phase. Arrows point to elongated nucleoids. The scale bar in panel A is 10 μm and applies to all panels.
Two major homologous recombination pathways (RecA dependent) are responsible for processing of stalled replication forks in E. coli: the recBCD pathway and the recF pathway (5). The RecD component of RecBCD has been identified as the nuclease responsible for degradation of chromosomes in recA cells of E. coli (20). The counterparts for E. coli recBCD and recF in B. subtilis are addAB and recF (9). Like the recA strain, both the addB and recF strains showed a low frequency of nucleoid partitioning defects. Neither strain, however, showed substantial signs of the cell wall irregularities noted in recA and recA combination mutants (Table 1). Consistent with previous experiments, ripX and ripX recA strains displayed strong evidence of nucleoid partitioning defects. We also note that the frequency of cell wall defects observed with the ripX recA strain was more than triple that seen with the recA strain, suggesting that there is an additive penalty for carrying both mutations (Table 1).
Viability of B. subtilis rec mutants during exponential-phase growth.
The generation time of the recA strain in LB medium was significantly longer than that of the parent strain (recA, 31 min; parent, 22 min). The plating efficiency of an exponentially growing B. subtilis recA strain was about 12% of that seen in the parent strain (Table 1). This score is in agreement with previous results for a recA mutant grown at 30°C (14) and is particularly striking given the mild nucleoid phenotype we have observed. These data are consistent with the concept that recA cells suffer from an inability to repair stalled DNA replication forks. The B. subtilis recA viability as indicated by plating efficiency is less than the 50% viability score reported for E. coli recA mutants (4). The viabilities of the addB and recF mutants were about 50 and 60%, respectively (Table 1). The addB recF double mutant was approximately 30% viable, reinforcing the concept that both recombination paths are utilized during exponential growth. The coparticipation of the B. subtilis AddAB and RecF recombination pathways has previously been shown in transformation and transduction assays (1). In addition, the double-mutant data suggest that a small amount of recombination repair can be mediated independent of AddB and RecF. We note that the presence of a ripX mutation (which impairs chromosome dimer resolution) in a recA strain lowered the plating efficiency twofold (from 12% to 5.6%). This result supports our observation that ripX mutations are not silent in a recA derivative of strain BR151 (26).
Although the frequency of anucleate cells in B. subtilis was substantially lower than that seen in E. coli, the 12% viability of the B. subtilis recA mutant suggests that most of the nucleated recA population is nonviable. Also, the sum of the nucleoid defects in B. subtilis recA cells, recorded here as 6%, presumably underreports the extent of nucleoid defects in a population. Based on these considerations, we speculate that lethal nucleoid damage indeed occurs in B. subtilis, but degradation is not as rapid or complete as it is in E. coli.
Cell division and nucleoid phenotypes of stationary-phase recA cells.
After the end of exponential growth, recA mutants developed a subpopulation of filamented cells with elongated nucleoids (Fig. 1). These unusual cells have been measured as a separate class apart from normal cells to highlight the differences between the two cell types in the recA stationary-phase population. In Table 2, we show that the peak representation of the aberrant subpopulation is about 17% (a similar frequency was observed for a recA4 point mutant strain [data not shown]). The average nucleoid length in the aberrant subpopulation class was found to be two- to fourfold greater than that in the normal-appearing recA cells and in the parent strain. Also, the average cell length of the aberrant subpopulation was two- to threefold larger than that of unaffected recA cells and that of the parent strain (Table 2 and Fig. 1). We observed no signs of septum formation in the areas occupied by these distended recA nucleoids, either by phase-contrast microscopy or by fluorescent microscopy employing the vital membrane stain FM4–64 (22) (Fig. 1) (data not shown).
TABLE 2.
Cell and nucleoid measurements in strains during the transition from exponential growth to the stationary phase
| Time (min)a | Result for strain (relevant genotype):
|
|||||||
|---|---|---|---|---|---|---|---|---|
| BR151 (parent)
|
SL7360 (recA::neo)
|
|||||||
| Frequency of elongation (%)b | Avg nucleoid length (μm)c | Avg cell length (μm)d | Frequency of elongation (%)b | Normal class
|
Elongated class
|
|||
| Avg nucleoid length (μm)c | Avg cell length (μm)d | Avg nucleoid length (μm)c | Avg cell length in μme | |||||
| −20 | 0.7 | 0.93 | 4.9 | 3.8 | 0.83 | 5.5 | 1.3 | 11.4 (2) |
| 0 | 0.2 | 0.84 | 4.9 | 7.5 | 0.76 | 6.3 | 1.6 | 9.1 (5) |
| 20 | 0.5 | 0.88 | 4.4 | 10.5 | 0.89 | 5.6 | 1.5 | 10.2 (6) |
| 40 | 0.2 | 0.8 | 4.3 | 12.9 | 0.82 | 5.3 | 3 | 9.8 (6) |
| 60 | 1.3 | 0.87 | 4.1 | 14.3 | 0.7 | 3.6 | 2.8 | 9.5 (7) |
| 80 | 0.7 | 0.77 | 3.1 | 12.3 | 0.67 | 3.3 | 2.4 | 7.5 (6) |
| 100 | 0.2 | 0.73 | 2.6 | 16.8 | 0.7 | 3.2 | 2.8 | 6.9 (7) |
Time relative to the end of exponential growth.
Elongated in this experiment refers to nucleoids whose length was >50% of the cell length.
A minimum of 150 nucleoids were measured per time point in a minimum of 15 random fields of view.
At least 45 cells were measured per time point in a minimum of 15 random fields of view.
The number of cells measured in the SL7360 elongated class is indicated in parentheses.
Based on the role of RecA in restoration of stalled replication forks, it is plausible that some or all of the cells in stationary-phase cultures of the B. subtilis recA strains that contained elongated nucleoids had accumulated DNA damage that prevented replication fork progression. This supposition is strengthened by our observation that treatment of B. subtilis RecA+ cells with MMC resulted in the development of filamented cells containing elongated nucleoids, a phenotype similar to that seen in recA stationary-phase cells (compare Fig. 1B and D). Additionally, recF and addB mutants showed the same distinctively elongated cells and nucleoids during stationary phase, although not as frequently (50 and 10% relative to the recA mutant [data not shown]). We discuss the growth-phase-specific nature of the aberrant subpopulation in B. subtilis recA cells later.
The reduced frequency of aberrant nucleoids in addB cells was unexpected. A possible explanation is that in the absence of RecA or RecF, unprocessed DNA substrates subsequently become targets for the exonuclease activity of AddAB. This combination of DNA degradation without repair might lead to the observed stationary-phase nucleoid phenotype in recA and recF cells. If this proposed scheme is correct, addB mutants would not be expected to present similar frequencies of the aberrant nucleoid phenotype to those seen in recA and recF mutants, because chromosomal DNA would not be efficiently degraded in the absence of a fully functional AddAB complex.
In an E. coli recA mutant (W3110), we observed no comparable signs of nucleoid elongation during the transition from exponential growth into the stationary phase (data not shown). Furthermore, the amount of cell filamentation in E. coli recA cells actually decreased during this time (exponential, ∼13%; stationary, ∼4%). It is also worth noting in this context that even when treated with MMC, E. coli recA cells continued growing and dividing at stages where plating yielded no CFU (13). Therefore, a second distinction can now be made between E. coli recA and B. subtilis recA cells. Whereas chromosome degradation is extensive and cell division occurs frequently in E. coli recA cells, the more stable nucleoids appear to persist as barriers to septation during the transition into the stationary phase in B. subtilis recA cells (31, 35). A similar absence of septation in cells harboring aberrant nucleoids has been seen in B. subtilis ripX mutants (26).
Why should E. coli and B. subtilis nucleoids appear so different upon entry into stationary phase? E. coli H-NS and integration host factor (IHF) are DNA-binding proteins involved in maintaining nucleoid structure and have been shown to be transcriptional regulators of gene expression. The IHF protein is up-regulated upon entry into the stationary phase (7). While several groups have shown substantial H-NS up-regulation at the stationary phase (6, 30, 32), another study showed little growth phase variation in H-NS levels (10). The B. subtilis DNA-binding proteins with the strongest similarity to these E. coli proteins are HBsu (IHF) and Smc (H-NS) (2, 8, 17, 18). Interestingly, B. subtilis transcription of hbs (encoding HBsu) and the Smc level are both reduced in stationary phase (8, 11), although the HBsu/DNA ratio in spores is similar to that in vegetative cells (24). It is possible, therefore, that E. coli has evolved a more comprehensive system of stationary-phase DNA compaction and protection than B. subtilis (with the exception of the highly protective stationary-phase sporulation program).
recA phenotypes during sporulation.
As was the case in LB medium, only about 10% of sporulating recA cells were viable (Table 3). The same morphological phenotypes found in recA cells making the transition from the exponential phase into the stationary phase under nonsporulation conditions were also seen in postexponential recA cells in the sporulation-inducing medium MSSM (Fig. 1C). The elongated nucleoid structures seen in postexponential recA cells were distinguishable from sporulation-dependent axial filaments that form during stage I of sporulation in B. subtilis (3) as follows. (i) Axial filaments were not formed by the parent strain in LB medium. (ii) The cell and nucleoid lengths were both noticeably larger in affected recA cells than those of the parent strain in sporulation medium at stage I of development. (iii) Unlike stage I sporulation axial filaments, whose appearance is temporary, the phenotypes associated with recA cells that became apparent at the end of exponential growth were maintained for at least 7 h (data not shown).
TABLE 3.
Cell density and sporulation frequencies of parent, recA, and recF strains
| Strain (relevant genotype) | Absolute no. of CFUa
|
Sporulation (%)b | Frequency relative to parent
|
|||
|---|---|---|---|---|---|---|
| No heat | Heat | Sporulation | Viable count | Heat survival | ||
| BR151 (parent) | 8.3 × 107 | 5.5 × 107 | 66 | 1.0 | 1.0 | 1.0 |
| SL7360 (recA::neo) | 8.1 × 106 | 3.6 × 106 | 45 | 0.68 | 0.097 | 0.067 |
| SL7609 (recF::spc) | 5.9 × 107 | 2.9 × 107 | 49 | 0.74 | 0.71 | 0.52 |
Numbers refer to the number of CFU normalized to bacterial dry weight (in milligrams) observed on LB agar. See text for details.
Sporulation was assessed 21 h after the cessation of exponential growth. The percentages shown here were within 2% of those obtained by phase-contrast microscopic examination just prior to plating. Heat treatment was for 20 min at 80°C.
In addition to the cell division and nucleoid irregularities seen early in the sporulation program, we have observed two separate and unfavorable consequences for sporulating recA mutants. First, the timing of prespore nucleoid condensation (27) is delayed by ∼2 h and occurs at a lower frequency than that of the parent strain (Table 4). Second, recA strains sporulate less frequently than their parent strain (Table 3) (26); the reduced sporulation frequency is distinct from the altered resistance properties of recA spores (28). The impairment in prespore condensation and diminished frequency of sporulation are likely related to the aberrant morphologies discussed previously. Regardless of the cause, it should be remembered that the reduced sporulation frequency in recA mutants is superimposed on a severely reduced plating efficiency.
TABLE 4.
Prespore nucleoid condensation at different times after the start of sporulation
| Strain (relevant genotype) | % of condensed nucleoids in prespore compartmenta
|
||||
|---|---|---|---|---|---|
| T2b | T3 | T4 | T5 | T6 | |
| BR151 (parent) | 2.5 | 27.2 | 43 | 38.8 | 22.5 |
| SL7360 (recA::neo) | 0.2 | 0.3 | 1.6 | 11.7 | 11.4 |
| SL7609 (recF::spc) | 1.1 | 14.4 | 27.8 | 32.4 | 22.3 |
The results shown are combined data from two independent experiments. A minimum of 500 cells were scored per time point per strain.
T, time in hours after the end of exponential growth.
Induction of recA during the stationary phase.
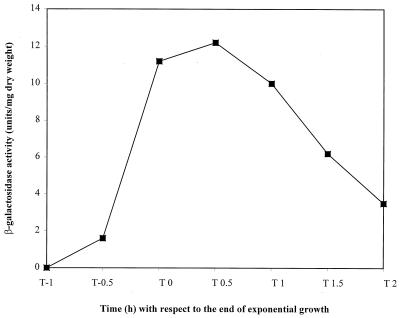
Using a recA-lacZ transcriptional fusion, we have shown that at approximately the same time recA cells begin developing overt cell division and nucleoid irregularities, transcription of the recA gene in BR151 was induced at the end of exponential gowth in LB medium (Fig. 2). (In other experiments, a low, basal level of transcription was observed during exponential growth.) This postexponential induction is in agreement with previous observations of recA transcription in B. subtilis in LB medium (23) and in competence medium (15, 23). RecA is capable of positively regulating gene expression (33, 36). Therefore, we reasoned that the elongated nucleoids in a subpopulation of cells in the stationary phase (Fig. 1B) might be the result of gene silencing in the absence of the normally induced RecA. For example, B. subtilis mecA mutants release comK transcription and the transcription of other ComK-regulated genes, such as comC, -D, -E, and -G, from medium dependence and have been reported to have cell and nucleoid abnormalities similar to those we have seen in stationary-phase recA cells (12). Based on these similarities, we speculated that RecA might be involved in the positive regulation of mecA during the stationary phase in the non-competence-inducing medium used here. Therefore, we evaluated the expression of comK and comG in our parent and recA strains during their transitions into the stationary phase. Neither the comK-lacZ nor the comG-lacZ reporter activity was enhanced in the absence of RecA (data not shown). These data indicate that RecA does not exert a positive regulatory effect on mecA or other genes responsible for regulating comK during the stationary phase in LB medium.
FIG. 2.
recA-lacZ activity during the transition into the stationary phase. Solid diamonds, SL7937 (recA-lacZ). The endogenous activity of the parent strain BR151 has been subtracted from each time point. Results are the average of two experiments.
Acknowledgments
We thank Warren Masker and Robert Britton for helpful discussions. We also thank Peter Setlow for providing YB3001 DNA and David Dubnau for comG- and comK-lacZ fusions.
This work was supported by Public Health Service grant GM-43577 (to P.J.P.) and training grant T32 AI-07101 (to S.A.S.). G.W.B. was supported by a Wellcome Trust Career Development Fellowship (039542/A/98).
REFERENCES
- 1.Alonso J C, Lüder G, Tailor R H. Characterization of Bacillus subtilis recombinational pathways. J Bacteriol. 1991;173:3977–3980. doi: 10.1128/jb.173.13.3977-3980.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Britton R A, Lin D C, Grossman A D. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bylund J E, Haines M A, Piggot P J, Higgins M L. Axial filament formation in Bacillus subtilis: induction of nucleoids of increasing length after addition of chloramphenicol to exponential-phase cultures approaching stationary phase. J Bacteriol. 1993;175:1886–1890. doi: 10.1128/jb.175.7.1886-1890.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capaldo F N, Ramsey G, Barbour S D. Analysis of the growth of recombination-deficient strains of Escherichia coli K-12. J Bacteriol. 1974;118:242–249. doi: 10.1128/jb.118.1.242-249.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox M M, Goodman M F, Kreuzer K N, Sherratt D J, Sandler S J, Marians K J. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 6.Dersch P, Schmidt K, Bremer E. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol Microbiol. 1993;8:875–889. doi: 10.1111/j.1365-2958.1993.tb01634.x. [DOI] [PubMed] [Google Scholar]
- 7.Ditto D M, Roberts D, Weisberg R A. Growth phase variation of integration host factor level in Escherichia coli. J Bacteriol. 1994;176:3738–3748. doi: 10.1128/jb.176.12.3738-3748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez S, Alonso J C. Bacillus subtilis sequence-independent DNA-binding and DNA-bending protein Hbsu negatively controls its own synthesis. Gene. 1999;231:187–193. doi: 10.1016/s0378-1119(99)00105-5. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez S, Ayora S, Alonso J C. Bacillus subtilis homologous recombination: genes and products. Res Microbiol. 2000;151:481–486. doi: 10.1016/s0923-2508(00)00165-0. [DOI] [PubMed] [Google Scholar]
- 10.Free A, Dorman C J. Coupling of Escherichia coli hns mRNA levels to DNA synthesis by autoregulation: implications for growth phase control. Mol Microbiol. 1995;18:101–113. doi: 10.1111/j.1365-2958.1995.mmi_18010101.x. [DOI] [PubMed] [Google Scholar]
- 11.Graumann P L, Losick R, Strunnikov A V. Subcellular localization of Bacillus subtilis SMC, a protein involved in chromosome condensation and segregation. J Bacteriol. 1998;180:5749–5755. doi: 10.1128/jb.180.21.5749-5755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn J, Bylund J, Haines M, Higgins M, Dubnau D. Inactivation of mecA prevents recovery from the competent state and interferes with cell division and the partitioning of nucleoids in Bacillus subtilis. Mol Microbiol. 1995;18:755–767. doi: 10.1111/j.1365-2958.1995.mmi_18040755.x. [DOI] [PubMed] [Google Scholar]
- 13.Hill T M, Sharma B, Valjavec-Gratian M, Smith J. sfi-independent filamentation in Escherichia coli is lexA dependent and requires DNA damage for induction. J Bacteriol. 1997;179:1931–1939. doi: 10.1128/jb.179.6.1931-1939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Love P E, Yasbin R E. Genetic characterization of the inducible SOS-like system of Bacillus subtilis. J Bacteriol. 1984;160:910–920. doi: 10.1128/jb.160.3.910-920.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovett C M, Jr, Love P E, Yasbin R E. Competence-specific induction of the Bacillus subtilis RecA protein analog: evidence for dual regulation of a recombination protein. J Bacteriol. 1989;171:2318–2322. doi: 10.1128/jb.171.5.2318-2322.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marians K J. PriA: at the crossroads of DNA replication and recombination. Prog Nucleic Acid Res Mol Biol. 1999;63:39–67. doi: 10.1016/s0079-6603(08)60719-9. [DOI] [PubMed] [Google Scholar]
- 17.Micka B, Groch N, Heinemann U, Marahiel M A. Molecular cloning, nucleotide sequence, and characterization of the Bacillus subtilis gene encoding the DNA-binding protein HBsu. J Bacteriol. 1991;173:3191–3198. doi: 10.1128/jb.173.10.3191-3198.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moriya S, Tsujikawa E, Hassan A K, Asai K, Kodama T, Ogasawara N. A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol Microbiol. 1998;29:179–187. doi: 10.1046/j.1365-2958.1998.00919.x. [DOI] [PubMed] [Google Scholar]
- 19.Nicholson W L, Setlow P. Sporulation germination and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley and Sons; 1990. pp. 391–429. [Google Scholar]
- 20.Palas K M, Kushner S R. Biochemical and physical characterization of exonuclease V from Escherichia coli. Comparison of the catalytic activities of the RecBC and RecBCD enzymes. J Biol Chem. 1990;265:3447–3454. [PubMed] [Google Scholar]
- 21.Piggot P J, Curtis C A M. Analysis of the regulation of gene expression during Bacillus subtilis sporulation by manipulation of the copy number of spo-lacZ fusions. J Bacteriol. 1987;169:1260–1266. doi: 10.1128/jb.169.3.1260-1266.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pogliano J, Osborne N, Sharp M D, Abanes-De Mello A, Perez A, Sun Y L, Pogliano K. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raymond-Denise A, Guillen N. Expression of the Bacillus subtilis dinR and recA genes after DNA damage and during competence. J Bacteriol. 1992;174:3171–3176. doi: 10.1128/jb.174.10.3171-3176.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross M A, Setlow P. The Bacillus subtilis HBsu protein modifies the effects of α/β-type, small acid-soluble spore proteins on DNA. J Bacteriol. 2000;182:1942–1948. doi: 10.1128/jb.182.7.1942-1948.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandler S J, Samra H S, Clark A J. Differential suppression of priA2::kan phenotypes in Escherichia coli K-12 by mutations in priA, lexA, and dnaC. Genetics. 1996;143:5–13. doi: 10.1093/genetics/143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sciochetti S A, Piggot P J, Sherratt D J, Blakely G. The ripX locus of Bacillus subtilis encodes a site-specific recombinase involved in proper chromosome partitioning. J Bacteriol. 1999;181:6053–6062. doi: 10.1128/jb.181.19.6053-6062.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Setlow B, Magill N, Febbroriello P, Nakhimovsky L, Koppel D E, Setlow P. Condensation of the forespore nucleoid early in sporulation of Bacillus species. J Bacteriol. 1991;173:6270–6278. doi: 10.1128/jb.173.19.6270-6278.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Setlow B, Setlow P. Role of DNA repair in Bacillus subtilis spore resistance. J Bacteriol. 1996;178:3486–3495. doi: 10.1128/jb.178.12.3486-3495.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skarstad K, Boye E. Degradation of individual chromosomes in recA mutants of Escherichia coli. J Bacteriol. 1993;175:5505–5509. doi: 10.1128/jb.175.17.5505-5509.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spassky A, Rimsky S, Garreau H, Buc H. H1a, an E. coli DNA-binding protein which accumulates in stationary phase, strongly compacts DNA in vitro. Nucleic Acids Res. 1984;12:5321–5340. doi: 10.1093/nar/12.13.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Q, Yu X C, Margolin W. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol Microbiol. 1998;29:491–503. doi: 10.1046/j.1365-2958.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- 32.Ueguchi C, Kakeda M, Mizuno T. Autoregulatory expression of the Escherichia coli hns gene encoding a nucleoid protein: H-NS functions as a repressor of its own transcription. Mol Gen Genet. 1993;236:171–178. doi: 10.1007/BF00277109. [DOI] [PubMed] [Google Scholar]
- 33.Walker G C. The SOS response of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1400–1416. [Google Scholar]
- 34.Wertman K F, Wyman A R, Botstein D. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene. 1986;49:253–262. doi: 10.1016/0378-1119(86)90286-6. [DOI] [PubMed] [Google Scholar]
- 35.Woldringh C L, Mulder E, Valkenburg J A, Wientjes F B, Zaritsky A, Nanninga N. Role of the nucleoid in the toporegulation of division. Res Microbiol. 1990;141:39–49. doi: 10.1016/0923-2508(90)90096-9. [DOI] [PubMed] [Google Scholar]
- 36.Yasbin R E, Cheo D, Bol D. DNA repair systems. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: ASM Press; 1993. pp. 529–537. [Google Scholar]
- 37.Yasbin R E, Fields P I, Andersen B J. Properties of Bacillus subtilis 168 derivatives freed of their natural prophages. Gene. 1980;12:155–159. doi: 10.1016/0378-1119(80)90026-8. [DOI] [PubMed] [Google Scholar]
- 38.Zyskind J W, Svitil A L, Stine W B, Biery M C, Smith D W. RecA protein of Escherichia coli and chromosome partitioning. Mol Microbiol. 1992;6:2525–2537. doi: 10.1111/j.1365-2958.1992.tb01429.x. [DOI] [PubMed] [Google Scholar]