Abstract
We created plasmids for use in insertion-duplication mutagenesis (IDM) of Neisseria gonorrhoeae. This mutagenesis method has the advantage that it requires only a single cloning step prior to transformation into gonococci. Chromosomal DNA cloned into the plasmid directs insertion into the chromosome at the site of homology by a single-crossover (Campbell-type) recombination event. Two of the vectors contain an erythromycin resistance gene, ermC, with a strong promoter and in an orientation such that transcription will proceed into the cloned insert. Thus, these plasmids can be used to create insertions that are effectively nonpolar on the transcription of downstream genes. In addition to the improved ermC, the vector contains two copies of the neisserial DNA uptake sequence to facilitate high-frequency DNA uptake during transformation. Using various chromosomal DNA insert sizes, we have determined that even small inserts can target insertion mutation by this method and that the insertions are stably maintained in the gonococcal chromosome. We have used IDM to create knockouts in two genes in the gonococcal genetic island (GGI) and to clone additional regions of the GGI by a chromosome-walking procedure. Phenotypic characterization of traG and traH mutants suggests a role for the encoded proteins in DNA secretion by a novel type IV secretion system.
Because many standard genetic tools do not function in Neisseria gonorrhoeae, the construction of special tools for manipulating its genome is a necessity. N. gonorrhoeae is highly competent for natural transformation (13, 37). Therefore several mechanisms for mutagenesis of N. gonorrhoeae that rely on transformation have been developed previously, including shuttle mutagenesis (34), direct cloning of antibiotic resistance markers (46), and direct transformation of constructed mutations without selectable markers (14). Shuttle mutagenesis is highly useful in that it offers a method for transposon mutagenesis in an organism that otherwise cannot be transposon mutagenized (34, 35). The procedure, however, requires several steps. The gene of interest must be cloned into an Escherichia coli plasmid, and the plasmid is then subjected to transposon mutagenesis in E. coli. Screening to identify mutants takes place, and finally the vector is transferred to the organism of interest, where homologous recombination results in the incorporation of the mutation into the chromosome. For efficient transformation of gonococci, the transposon must be flanked by a significant amount of gonococcal DNA (∼500 bp). Therefore, the original target must generally contain more than a kilobase of cloned DNA. The method of direct transformation of constructed mutants in gonococcal genes has similar difficulties and requires extensive screening to find a mutant of interest.
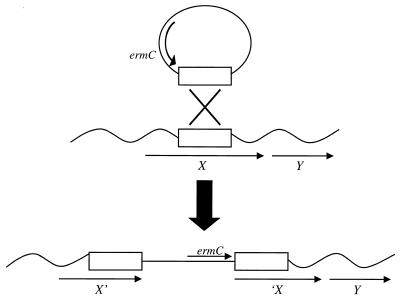
In this paper we describe the development and use of insertion-duplication mutagenesis (IDM) plasmids for producing targeted mutations in N. gonorrhoeae. IDM utilizes chromosomal fragments cloned into plasmids to target insertion into the recipient chromosome by homologous recombination, resulting in the insertion of the vector between duplicated target sequences (25) (see Fig. 1). This method of mutagenesis has been used for Streptococcus pneumoniae for more than 20 years (19), and similar methods have been developed for Streptococcus mutans (41), Bacillus subtilis (9, 24, 27), and Lactobacillus species (20, 21). Additionally, plasmid insertion methods are required for the identification of in vivo-induced genes by the in vivo expression technology (IVET) and differential fluorescence induction (DFI) techniques and have been used for the signature-tagged mutagenesis method in bacteria in which transposons are not functional (22, 29, 42). Wolfgang et al. have reported the use of plasmid insertion in gonococci for purposes of in situ cloning of comP and flanking sequences (45), but the vector was not optimized for this purpose and the mutagenesis process was not characterized. IDM has several advantages over other methods of mutagenesis utilized for N. gonorrhoeae. IDM requires only one cloning step for use in mutagenesis, and every clone of chromosomal DNA can be used to make a mutation. Because the mutation is made from an E. coli plasmid, it is easy to recover the construct that generated a mutation of interest by simply transforming DNA from the mutant into E. coli. Thus, this method has been used to identify plasmid clones capable of restoring spontaneous mutants to the wild-type phenotype (8). In addition, with the IDM vectors that we have created for use in N. gonorrhoeae, it is possible to make both transcriptionally polar and nonpolar mutations.
FIG. 1.
Generation of a nonpolar insertion mutation. IDM results in a single-crossover recombination event, insertion of the plasmid into the chromosome, and duplication of homologous sequences. The strong ermC promoter drives the transcription of downstream genes.
We have recently discovered the presence of a 60- to 70-kb genetic island in N. gonorrhoeae. The gonococcal genetic island (GGI) is variable, and certain versions of the GGI are found significantly more often in N. gonorrhoeae strains isolated from patients with disseminated gonococcal infections (7). Within the GGI, there are multiple genes with homology to those of type IV secretion systems; the greatest similarity is that to the E. coli F-plasmid transfer genes. Type IV secretion systems are characterized by their unique ability to transfer both DNA and protein molecules. Well-studied examples of these systems include the DNA transfer systems of conjugative plasmids, the agrobacterial T-DNA transfer system, and the pertussis toxin secretion system encoded by Bordetella pertussis (4, 18).
Gonococci are naturally competent for transformation during all phases of growth. Although the fine points of gonococcal transformation are not fully understood, it is known that the recipients must produce pilin, PilT, and ComP for transformation in N. gonorrhoeae (2, 37, 45). These data suggest a role for pili and twitching motility in transformation, but their exact role remains unclear. Gonococcal transformation is dependent on RecA for homologous recombination of transforming DNA in the gonococcal chromosome (17). Both single-stranded and double-stranded DNA, as well as supercoiled and linear plasmid DNA, have been shown to transform N. gonorrhoeae (38). However, transforming DNA must contain the 10-bp neisserial DNA uptake sequence (DUS) for efficient uptake into a DNase-resistant state (13). Although significant information is known about the requirements of the recipient in transformation, little is known about mechanism of DNA donation. We have recently shown that gonococci secrete DNA into the medium during growth and that a mutation in a peptidoglycan hydrolase gene in the GGI (atlA) prevents this secretion (7). We have also shown that the GGI contains genes similar to those involved in DNA transfer during conjugation (7). Here we demonstrate that IDM mutations in these genes eliminate DNA secretion by growing gonococci. These data suggest that the GGI encodes a novel type IV secretion system involved in the donation of DNA for natural transformation.
MATERIALS AND METHODS
Bacteria and growth conditions.
Bacterial strains and plasmids used in this study are described in Table 1. E. coli strains were grown on Luria agar plates or in Luria broth (31) at 37°C. N. gonorrhoeae strains were grown either in GCBL (1.5% Proteose Peptone no. 3, 0.4% K2HPO4, 0.1% KH2PO4, 0.1% NaCl) plus Kellogg's supplements (16) and 0.042% NaHCO3 (26) at 37°C with aeration or on GCB agar (Difco) plates under 5% CO2 at 37°C. Erythromycin was used at 10 μg/ml for gonococci and at 500 μg/ml for E. coli (except where otherwise indicated) to select for expression of ermC. Kanamycin was used at 40 μg/ml for E. coli carrying pHSS6 derivatives (35). Chloramphenicol was used at 10 μg/ml for gonococci, and spectinomycin was used at 75 μg/ml alone and at 50 μg/ml when in combination with chloramphenicol.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Properties | Source or reference |
|---|---|---|
| Plasmids | ||
| pUP1 | Template for pIDN plasmids; contains 10-bp neisserial DUS in polylinker; Kmr | 10 |
| pMOB | Source of polylinker for pIDN plasmids; pUC origin; Apr; multicloning site (MCS); T3 and T7 promoters | 39 |
| pHSS23 | Source of original ermC gene | 44 |
| pJD1145 | Source of improved ermC gene for pIDN construction | This work |
| pNH5 | Forward orientation of ermC from pJD1145 in pUP1; Emr Kmr | This work |
| pNH5-1 | Reverse orientation of ermC from pJD1145 in pUP1; Emr Kmr | This work |
| pNH9 | BamHI-PvuII deletion of Kmr in pNH5 | This work |
| pNH9-1 | BamHI-PvuII deletion of Kmr in pNH5-1 | This work |
| pNH9-6 | pMOB polylinker in forward orientation in pNH9 (Emr) | This work |
| pNH9-7 | pMOB polylinker in reverse orientation in pNH9 (Emr) | This work |
| pNH9-8 | pMOB polylinker in forward orientation in pNH9-1 (Emr) | This work |
| pNH9-9 | pMOB polylinker in reverse orientation in pNH9-1 (Emr) | This work |
| pIDN1 | IDM vector: ermC in forward orientation, polylinker in forward orientation | This work |
| pIDN2 | IDM vector: ermC in forward orientation, polylinker in reverse orientation | This work |
| pIDN3 | IDM vector: ermC in reverse orientation, polylinker in forward orientation | This work |
| pIDN4 | IDM vector: ermC in reverse orientation, polylinker in reverse orientation | This work |
| pJD1175 | 642-bp EcoRV-Sau3AI fragment of traH from MS11A cloned into pIDN1 | This work |
| pJD1177 | 642-bp EcoRV-Sau3AI fragment of traH from MS11A cloned into pIDN3 | This work |
| pKS43 | 479-bp Sau3AI fragment of traG from MS11A cloned into pIDN1 | This work |
| pKS53 | 479-bp Sau3AI fragment of traG from MS11A cloned into pIDN3 | This work |
| pNH10-1M | AgeI-NsiI fragment of traG from PCR of MS11A with 23F and EndL6R in XmaI and PstI sites of pNH9-9 | This work |
| pHH1 | HindIII deletion of pNH10-1M | This work |
| pHH2 | ScaI-EcoRV deletion of pNH10-1M | This work |
| pHH3 | ScaI-Ecl136II deletion of pNH10-1M | This work |
| pNH11 | Blunted PCR product of MS11A with 27F and TraH2R in EcoRV site of pNH9-9; contains 5′ end of traH and upstream sequence | This work |
| pGCC6 | Plasmid for complementation in the gonococcal chromosome; lac promoter/operator; lacI Cmr Kmr | 23 |
| pHH15 | PCR product of MS11A with TraH-up and TraH-down, cut with NsiI and EcoRV and cloned into the NsiI and PmeIsites of pGCC6 | This work |
| N. gonorrhoeae | ||
| MS11A | Wild-type N. gonorrhoeae | 32 |
| KS1 | MS11A transformed by IDM with pJD1175, polar mutant of traH (Emr) | This work |
| KS58 | MS11A transformed by IDM with pKS43, polar mutant of traG (Emr) | This work |
| KS16 | MS11A transformed by IDM with pJD1177, nonpolar mutant of traH (Emr) | This work |
| KS59 | MS11A transformed by IDM with pKS53, nonpolar mutant of traG (Emr) | This work |
| HH500 | MS11A transformed by IDM with pNH10-1M (Emr) | This work |
| HH501 | MS11A transformed by IDM with pHH1 (Emr) | This work |
| HH502 | MS11A transformed by IDM with pHH2 (Emr) | This work |
| HH503 | MS11A transformed by IDM with pHH3 (Emr) | This work |
| JD1510 | MS11A atlA::ermC (insertion-deletion mutation) | 6 |
| JD1603 | MS11A transformed by IDM with pNH11 (Emr) | This work |
| JD1545 | MS11A cnp::cat recA6 (Cmr) | 7 |
| MS11-Spc | MS11A Spr | 33 |
| HH507 | JD1545 transformed by IDM with pNH10-1M (traG recA6 Cmr Emr) | This work |
| HH508 | JD1545 transformed by IDM with pNH11 (recA6 Cmr Emr) | This work |
| HH518 | KS16 transformed with pHH15; traH expression in trans (Cmr Emr) | This work |
DNA techniques.
Gel purification of digested DNA fragments was performed using GeneClean (Bio 101). Following all ligations, transformation was performed in chemically competent TOP10 E. coli according to the protocol of the supplier (Invitrogen).
Southern blotting to confirm pIDN insertions was performed according to standard procedures (31). Chromosomal DNA from N. gonorrhoeae was prepared as described by Boyle-Vavra and Seifert (3). DNA digests were separated by gel electrophoresis in a 0.8% agarose Tris-borate-EDTA gel. DNA was transferred to a Stratagene Duralon-UV membrane using a vacuum blotter. Following UV cross-linking, the chromosomal DNA was probed with digoxigenin-labeled pIDN4. Blots were washed at high stringency, and chemiluminescent detection was performed as suggested by the manufacturer (Boehringer Mannheim).
Plasmid screening was performed by whole-cell lysis using the lysis solution of Kado and Liu (3% sodium dodecyl sulfate in 50 mM Tris [pH 12.6]) (15). Patched E. coli colonies were lysed in 100 μl of lysis solution, and 20 μl of the lysate was separated by agarose gel electrophoresis and subsequently stained with ethidium bromide. Following whole-cell lysis, possible positive plasmids were isolated and screened further by restriction enzyme digestion. Plasmid purification was performed as described by Birnboim and Doly (1).
Plasmid constructions.
pUP1 was linearized with HindIII, blunted with T4 DNA polymerase, and ligated to the blunted EcoRI-XbaI fragment from pJD1145 containing the erythromycin resistance gene. Two plasmids carrying kanamycin and erythromycin resistance resulted due to insertion of the ermC fragment in pUP1 in both orientations (pNH5, forward orientation; pNH5-1, reverse orientation). To remove the kanamycin resistance marker, the plasmids were digested with BamHI and PvuII, T4 polymerase blunted, and religated. The product with the forward orientation of ermC was named pNH9, and the product with the reverse orientation was named pNH9-1.
A polylinker for the pIDN vectors was obtained via PCR of pMOB with pMOB-polF and pMOB-polR (Tm, 56°C; extension time, 20 s) (Table 2). The blunted, polynucleotide kinase (PNK)-treated PCR product was ligated to NotI/ClaI-digested, T4 polymerase-blunted pNH9 and pNH9-1 to yield four new plasmids containing all possible orientations of erythromycin and the polylinker: pNH9-6, pNH9-7, pNH9-8, and pNH9-9. To make the plasmids smaller, ∼200 bp of sequence was removed from pNH9-8 and pNH9-9 by PCR with pORI-F and pGCU-R to amplify the desired section of each plasmid (Tm, 55°C; extension time, 90 s). PCR products were blunted, phosphorylated with PNK, and ligated to themselves to form pIDN3 and pIDN4. EcoRI-SphI fragments from pNH9-6 and pNH9-7 containing the polylinker and the ermC gene were cloned into the EcoRI and SphI sites of pNH9-8 and pNH9-9, respectively, to create pIDN1 and pIDN2. The DNA sequence of pIDN4 was determined and was deposited in GenBank under accession number AY034154.
TABLE 2.
PCR primers
| Primer name | Sequence |
|---|---|
| pMOB-polF | 5′-GTA AAA CGA CGG CCA GTG AGC GCG-3′ |
| pMOB-polR | 5′-AAC AGC TAT GAC CAT GAT TAC GCC AAG-3′ |
| pORI-F | 5′-GAA GCT AGC TTA TGC GGT GTG AAA TAC-3′ |
| pGCU-R | 5′-AAC GTT GTT GCC ATT GCT GC-3′ |
| 27F | 5′-CGG TTG ACA ACG CGG ATA TTT CAG G-3′ |
| TraH2R | 5′-GCC AGG TCA AAG GCT ACA CC-3′ |
| 23F | 5′-GAT TCG TTC TGC TGC TGA GGC C-3′ |
| TraH-up | 5′-CGA ATG CAT CTG CTA TAA CCG CTT CAT GG-3′ |
| TraH-down | 5′-CGA GAT ATC GTA CAG CTT TCT CCG TTA CC-3′ |
pNH10-1M was created by PCR amplification of MS11A chromosomal DNA using primers 23F and EndL6R (7), digestion with AgeI and NsiI, and cloning into the PstI and XmaI sites of pNH9-9. Subclones of pNH10-1M were created by deletions of portions of the 890-bp traG insert and religation of the plasmids. pHH1 was created by deleting the 0.6-kb HindIII fragment. pHH2 was created in a similar manner by deleting the ScaI-EcoRV fragment. pHH3 was created by deletion of the ScaI-Ecl136II fragment. pHH1, pHH2, and pHH3 contain 290-, 540-, and 350-bp fragments of the original traG fragment in pNH10-1M, respectively.
pHH15 was constructed in the following manner. The PCR product of TraH-up (containing an NsiI site) and TraH-down (containing a EcoRV site) (Tm, 66°C; extension time, 60 s) was cut with NsiI and EcoRV and cloned into the NsiI and PmeI sites of pGCC6. This plasmid was linearized with NotI and transformed into KS16 (traH mutant) for complementation. pGCC6 targets the inserted gene to a chromosomal location between gonococcal genes lctP and aspC.
Gonococcal transformation.
Spot transformations of gonococci were performed as follows: 1 pmol of each plasmid in a 20-μl volume was spotted in three places on a prewarmed (37°C) GCB agar plate. The spots were allowed to soak into the plate. A P+ colony of MS11A was then streaked across the plate through the DNA spots. After overnight growth at 37°C, colonies were swabbed from the spots with a Dacron swab and resuspended in 600 μl of GCB. This suspension of cells was then diluted and plated on both GCB alone and GCB plus erythromycin, and CFU were counted. Transformation frequencies were determined as the number of Emr CFU per total CFU.
Spot transformation was also used to make IDM strains. After overnight growth of the colonies, the spots containing gonococcal transformants were swabbed onto a GCB-plus-erythromycin plate. These plates were grown for 1 to 2 days, and individual transformants were restreaked.
DNA release assay.
For DNA release experiments, P− transparent gonococcal strains were grown overnight on GCB agar plates. Strains were then inoculated with a sterile Dacron swab into 3 ml of GCBL medium with Kellogg's supplements (16) and 0.042% NaHCO3 (26), with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) added where necessary. These cultures were grown for ∼2 to 2.5 h. Cultures were then diluted to an optical density at 540 nm of 0.2 (t = 0 h) in 3 ml of Cellgro Complete tissue culture medium (Mediatech) with the following supplements: 0.35 mM cysteine, 0.15 mM cystine, 17.9 mM pyruvate, 0.1% soluble cornstarch, 0.042% NaHCO3, Kellogg's supplements, and 1 mM IPTG where necessary. These cultures were grown for an additional 5 h. Culture supernatants were collected at 0 and 5 h and were assayed for DNA release in the following manner. One hundred microliters of a 1:200 dilution of PicoGreen fluorescent dye (Molecular Probes) was added to 100 μl of culture supernatant. DNA release was immediately measured in a fluorometer at an excitation wavelength of 485 nm and an emission wavelength of 535 nm. DNA release per milliliter of culture over 5 h of growth was then calculated.
DNA release was normalized to the total amount of protein in the culture, quantified by the Bio-Rad protein assay. A 500-μl portion of each culture was centrifuged, and the pellet was resuspended in 1 ml of H2O. Cells were sonicated three times for 10 s each time. One milliliter of a 1:5 dilution of Bio-Rad Dye Reagent Concentrate was added to 20 μl of the sonicated cell solution (in duplicate). Only gonococcal cultures that had grown ∼1 log unit in 5 h of growth were used for analysis of DNA release.
Coculture transformation assay.
Coculture transformation assays were performed using the P+ gonococcal strain HH507 or HH508 and MS11-Spc that had been grown on GCB agar overnight. Cells were swabbed into GCBL plus Kellogg's supplements and NaHCO3 and grown for ∼2.5 h at 37°C with aeration. One milliliter of each culture was then centrifuged and resuspended in 4 ml of prewarmed, fresh GCBL plus supplements for inoculation (∼107 CFU/ml). The following cultures were inoculated into 3 ml of prewarmed GCBL plus supplements: 1 ml of donor cells (HH507 or HH508), 1 ml of recipient cells (MS11-Spc), 0.5 ml of donor cells plus 0.5 ml of recipient cells, and 0.5 ml of donor cells plus 0.5 ml of recipient cells plus 25 μg of DNase/ml. These four cultures were diluted and plated for CFU per milliliter (t = 0 h) on GCB, GCB-chloramphenicol, GCB-spectinomycin, and GCB-chloramphenicol-spectinomycin plates. Cultures were then grown at 37°C with aeration for 4 h, after which they were again diluted and plated for CFU per milliliter on plates containing the same antibiotics as above.
Nucleotide sequence accession number.
The DNA sequence of pIDN4 has been deposited in GenBank under accession number AY034154.
RESULTS
Construction of IDM plasmids.
To create an IDM system in gonococci, we considered several criteria, both of other IDM systems and of N. gonorrhoeae. The ideal vector for IDM in gonococci should be nonreplicating in N. gonorrhoeae but should replicate in E. coli for cloning purposes. The vector should also have a high transformation rate; efficient DNA uptake in transformation of N. gonorrhoeae requires the presence of the 10-bp DUS, which is commonly found in gonococcal and meningicoccal DNA (13). The vector should be small to facilitate incorporation into the chromosome, since incorporation of heterologous DNA by gonococci is greatly affected by the length of the heterology (3). In addition, the IDM vector system should have good selection both in gonococci and in E. coli, a useful polylinker for cloning, and the capability to produce both polar and nonpolar insertion-duplication (ID) mutations.
Plasmid pUP1 was used as the starting point for the construction of an insertion-duplication plasmid because it is a relatively small plasmid containing the DUS (10), and like most E. coli plasmids, it does not replicate in gonococci. Additionally, unlike many E. coli cloning vectors, it has no Tn3 resolvase site, and thus DNA cloned into it could be mutagenized with the mini-Tn3 transposons developed for shuttle mutagenesis of gonococci (3, 34). Several factors, however, made pUP1 itself undesirable as an insertional vector for gonococci. The kanamycin resistance marker is not ideal for selection in gonococci due to the inherent kanamycin resistance of nonpiliated variants (12). Also, pUP1 has few sites in the polylinker, and the neisserial DUS is located within the polylinker, making it possible to lose the uptake sequence during cloning.
Erythromycin resistance was chosen as the selectable marker for the IDM plasmids. Erythromycin is preferred over kanamycin because it is a cleaner selection agent in gonococci. However, E. coli is inherently resistant to high levels of erythromycin. To increase our ability to select for transformants containing ermC-carrying plasmids in E. coli, we constructed an improved promoter for the ermC gene. The ermC gene in pHSS23 has a SacI site between the −35 and −10 hexamers, and the vector contained a perfect −35 sequence in the polylinker region. A deletion was made in pHSS23 between the ClaI site in the vector and the SacI site in the ermC promoter to generate a smaller ermC gene with a consensus sigma-70 promoter that has a spacing of 16 bp between the −10 and −35 sequences. Thus, except for the effects of intervening or surrounding sequence, the new ermC promoter should have a strength similar to that of the tac promoter, which also has a perfect consensus sequence and a spacing of 16 bp. This new ermC promoter is strong enough to read through the ermC transcriptional terminator, and for this reason this marker was used by Mehr et al. to make nonpolar mutations upstream of essential genes in N. gonorrhoeae (23). When the enhanced ermC was tested for erythromycin resistance, it was found that E. coli cells carrying the construct grew well in media containing 200 to more than 1,000 μg of erythromycin/ml.
The improved ermC was cloned into pUP1 in both orientations. By creating a plasmid in which transcription will read into the cloned insert, we will be able to create insertions in the gonococcal chromosome that are effectively nonpolar on downstream genes (Fig. 1). The opposite orientation of ermC would be more desirable for creating a polar mutation or when transcription of the insert might result in the production of a detrimental product. Kanamycin resistance was then removed to make the vectors smaller, resulting in plasmids pNH9 and pNH9-1.
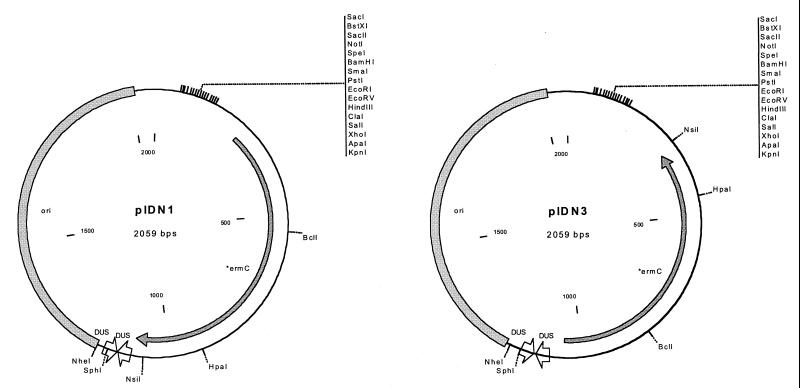
To complete the construction, we inserted a polylinker and decreased the size of the plasmid by removing unnecessary sequence. A 224-bp region containing a polylinker was obtained by PCR amplification of the small sequencing and mutagenesis plasmid pMOB (39). The polylinker was cloned in both orientations to facilitate cloning of fragments in both orientations. This region contains a polylinker with 16 unique sites, as well as binding sites for universal sequencing primers. The region also contains T7 and T3 promoters and could be used for controlled transcription in certain E. coli strains or for in vitro transcription. The final step in the constructions was to eliminate approximately 200 bp of unnecessary DNA between the DUS and the origin. This was accomplished by PCR of the plasmid and religation. This resulted in the final plasmid constructs, which we have named pIDN, for plasmid for IDM of Neisseria (Fig. 2).
FIG. 2.
Plasmid maps of pIDN1 and pIDN3. Plasmids consist only of the ermC gene, a polylinker, an E. coli origin of replication, and two copies of the neisserial DUS. pIDN2 and pIDN4 (not shown) are the same as pIDN1 and pIDN3, respectively, differing only in the orientation of the polylinker.
Testing IDM in N. gonorrhoeae and generation of GGI mutants.
Before the completion of the IDM vectors, we tested our idea that IDM could work efficiently as a method of mutagenesis in N. gonorrhoeae. There are several reasons why IDM might not work well in gonococci. Gonococci appear to undergo a significant degree of illegitimate recombination, which can be seen in attempts to knock out essential genes by insertion of an antibiotic resistance marker. These markers are often incorporated, but the essential gene is duplicated in the transformants, presumably by an illegitimate recombination event (23, 40). Thus, IDM plasmids might be incorporated into arbitrary locations in the chromosome. In addition, N. gonorrhoeae does not efficiently incorporate large regions of heterologous DNA (3). Thus, plasmid insertion frequencies might be too low for IDM to be useful. Gonococci are also known to undergo very high levels of recombination (12, 36). If recombination between the homologous segments of the insertions in the chromosome were to occur at a high frequency, IDM mutations would be lost and the method would not be useful for mutagenesis of N. gonorrhoeae.
During the process of construction of the pIDN vectors, an intermediate plasmid that contained ermC in the reverse orientation and the polylinker in the reverse orientation was created. This plasmid, pNH9-9, was used to test IDM in gonococci. An 890-bp fragment of the GGI gene, traG, from N. gonorrhoeae strain MS11A was cloned into this intermediate plasmid, and the resulting plasmid was named pNH10-1M. Transformation of gonococci with this plasmid resulted in 2,500 Emr transformants/ml. When transformed into gonococci, pNH10-1M inserted into the chromosome, resulting in the traG mutant HH500. Southern analysis confirmed that this mutant contained the expected insertion of the construct interrupting traG.
In order for IDM to be a useful method of mutagenesis in gonococci, it must work with small fragments of genes. It has been found in IDM systems in other organisms that the size of the chromosomal fragment within the IDM vector affects transformation frequency (19). We made deletions in pNH10.1-M, resulting in three plasmids with smaller target regions of DNA: 290-, 540-, and 350-bp fragments of traG. We found that the length of traG sequence affected transformation frequency (Table 3). Frequencies ranged from 1.23 × 10−7 to 4.39 × 10−6 transformants/total CFU (7.21 × 10−8 to 2.09 × 10−6 transformants/μg of plasmid DNA). Plasmids containing smaller fragments resulted in lower frequencies of transformation. However, even the smallest target tested (pHH1, with 290 bp) still attained a transformation frequency higher than 10−7, well within an acceptable range for mutagenesis.
TABLE 3.
Transformation frequencies of plasmid insertion into the gonococcal chromosome
| Plasmid | Insert size (bp) | Transformation frequencya |
|---|---|---|
| pNH10-1M | 890 | 4.39 × 10−6 ± 9.9 × 10−7 |
| pHH1 | 290 | 1.23 × 10−7 ± 3.4 × 10−8 |
| pHH2 | 540 | 8.51 × 10−7 ± 4.2 × 10−8 |
| pHH3 | 350 | 3.52 × 10−7 ± 1.6 × 10−8 |
Reported as the number of Emr transformants per total CFU. All values are significantly different; P < 0.005 by Student's t test.
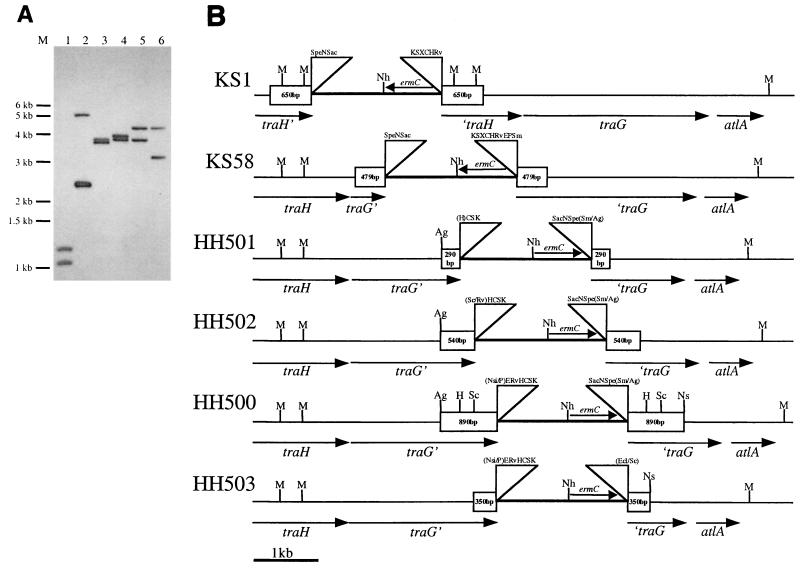
The gonococcal strains HH500, HH501, HH502, and HH503, along with two other IDM strains, KS1 and KS58, were characterized by Southern blotting to confirm their insertion locations (Fig. 3). The blot was probed with pIDN4. The further down the chromosome the plasmid is inserted, the larger the MfeI-NheI fragment is. Except in the case of KS1, which contains a duplication of the MfeI sites in traH, as the insert is moved down the chromosome, the NheI-MfeI fragment grows smaller. In this blot, as well as other Southern blots of these strains, the mutants were found to have correctly incorporated insertion plasmids.
FIG. 3.
(A) Southern blot of IDM strains KS1, KS58, HH501, HH502, HH500, and HH503 (lanes 1 through 6, respectively) digested with MfeI and NheI. This blot was probed with pIDN4, which binds to the plasmid inserted within the N. gonorrhoeae chromosome. (B) Schematic of N. gonorrhoeae IDM insertions.
In a few instances we have identified ID mutants that did not contain the expected construction. In the process of making nearly 80 plasmid insertion mutations in the gonococcal island, we have screened 45 gonococcal ID mutants by Southern analysis. Among these we have obtained only two mutants that did not give the expected restriction map when analyzed by Southern blotting, and in neither case was the unusual insertion the only or the predominant type of insertion. Since the pIDN plasmids contain two copies of the DUS, a sequence repeated many times in the gonococcal chromosome, these unusual plasmid insertions may have resulted from recombination at the DUS. We have also found mutants that contain a duplication of the plasmid insertion. These double insertions appeared as an additional band the size of the plasmid on the Southern blot and may result from insertion of a plasmid dimer formed in E. coli or from a duplication event in the gonococcal chromosome. Dimers have appeared only twice in all of the mutants screened.
To estimate the frequency of excision of IDM plasmids from the gonococcal chromosome, a strain carrying the 890-bp IDM insert in traG was tested for loss of the plasmid insertion in two separate experiments. The strain was grown in broth culture for 20 h without erythromycin and then plated onto GCB plates at various dilutions. After growth overnight, the colonies were replica plated to plates containing erythromycin and to plates containing no antibiotic. The pattern of colonies produced on the plates was recorded using a photodocumentation system, and the images were superimposed. We were unable to detect a single revertant among approximately 105 CFU.
IDM-generated GGI mutants are deficient in DNA release.
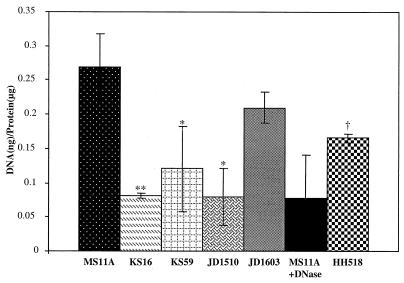
The conventional wisdom is that gonococci donate DNA for transformation by cell death and autolysis (28, 33); however, our data suggest that the putative type IV secretion system in the GGI contributes to DNA donation via specific transport, before autolysis occurs in culture. Strains with mutations in atlA, a peptidoglycan hydrolase gene in the GGI, were found to be deficient in DNA donation. AtlA function may be necessary for assembly of the type IV secretion apparatus (7). Type IV secretion systems characteristically secrete both DNA and protein (4). Therefore, assaying for DNA released into the extracellular medium can be used to evaluate the importance of GGI genes in type IV secretion. Nonpolar ID mutants of the GGI genes, traG and traH (KS59 and KS16, respectively), were tested for the ability to release DNA into the extracellular medium. Both KS16 and KS59 were analyzed by Southern blotting and were found to contain the proper plasmid insertions (data not shown). In addition to these strains, wild-type, atlA mutant, and plasmid control gonococci were tested. JD1603, our positive control, contains a plasmid insert creating a duplication of the beginning of traH (containing the promoter), resulting in a full copy of the gene. This mutation was designed to control for any nonspecific effects of having a pIDN insertion. The traG and traH mutants (KS16 and KS59) exhibited greatly reduced DNA release, with values similar to those of a DNase-treated control (Fig. 4).
FIG. 4.
DNA released into the medium by gonococcal strains MS11A (wild type), KS16 (traH mutant), KS59 (traG mutant), JD1510 (atlA mutant), JD1603 (an intergenic ID mutant), and HH518 (with traH complementation) during 5 h of growth. For comparison, the supernatant of MS11A was treated for 30 min with DNase. DNA release was normalized to the total amount of protein. ∗, P < 0.01; ∗∗, P < 0.05; †, P < 0.01 compared to KS16 (Student's t test).
traH was complemented in trans using a plasmid construct that targeted the inducible complementing gene to an irrelevant location on the chromosome of strain KS16. In this construct traH is under the control of the lac promoter-operator and can be induced using IPTG. Upon induction, the ability of the complemented traH mutant HH518, to release DNA is restored (Fig. 4). Not only does this result demonstrate a requirement for traH in DNA release; it also shows that the original traH mutant is effectively nonpolar as we predicted, i.e., it is not greatly affected with regard to expression of the downstream genes traG and atlA.
Coculture transformation assays.
We examined gonococcal strains HH507 and HH508 for the ability to donate DNA for natural transformation by a coculture transformation assay. In this assay, donor strains have been made incapable of DNA recombination by a mutation in recA. Donor strains (Cmr) were mixed 1:1 with the recipient strain (Spr), and after 4 h of growth, cultures were plated for selection of transformants expressing both antibiotic resistances. The transformation frequency obtained by using HH508 as the donor is ≥50-fold greater than that with HH507 (traG mutant) (Table 4). The addition of DNase abolishes transformation of the recipient strain by HH508. These results support the findings (Fig. 4) that the material released from N. gonorrhoeae is DNA and demonstrate the importance of the type IV secretion of DNA for transformation.
TABLE 4.
Transformation frequencies as determined by coculture assay
| Donor strain | Transformation frequencya |
|---|---|
| HH508 (plasmid control) + MS11-Spc | 2.32 × 10−5 |
| HH508 + MS11-Spc + DNase | <1.06 × 10−7b |
| HH507 (traG mutant) + MS11-Spc | <4.70 × 10−7b |
| HH507 + MS11-Spc + DNase | <3.62 × 10−7b |
| HH508 | <4.50 × 10−8b |
| HH507 | <1.04 × 10−7b |
| MS11-Spc | <1.62 × 10−8b |
Calculated as Cmr Spr CFU/total CFU for HH508, HH507, and MS11-Spc and as Cmr Spr CFU/total recipient (Spr) CFU for mixed cultures. Frequencies are expressed as geometric means from three different experiments.
No transformants were obtained. Values expressed are limits of detection.
Use of IDM for chromosome walking.
In addition to providing a convenient method for creating mutations, IDM also facilitates the cloning of unknown regions of the bacterial chromosome. Morrison et al. have described its use for this purpose in cloning transformation genes of S. pneumoniae (25). More recently, plasmids inserted in the chromosome of Salmonella enterica serovar Typhimurium were used to clone the genes found to be induced during infection in a DFI screen (42). We have used our insertions in the GGI to clone unknown GGI DNA. To clone the region upstream of traH, a strain containing an insertion at traH was mapped by Southern blotting to find a reasonably sized fragment that would contain the plasmid and upstream flanking region. The MfeI fragment was excised from the gel, purified, and ligated. Transformation of E. coli generated 43 transformants. Screening of 18 of the transformants showed that 17 contained a plasmid of the expected size (data not shown). One of the 18 transformants contained a plasmid identical in size to the plasmid used to make the original insertion. This result suggests that at a low frequency the plasmid is excising from the chromosome of N. gonorrhoeae or that E. coli can recombine incompletely digested chromosomal DNA to generate the original plasmid.
DNA sequencing of the region cloned from upstream of traH revealed the presence of two open reading frames. The first showed no significant similarity to any sequence in the databases. The second open reading frame was significantly similar to that encoding TraF of the E. coli F plasmid. The gonococcal TraF homologue showed 45% similarity and 29% identity with the E. coli TraF, with similarity extending over nearly the entire length of the proteins. In E. coli, TraF is a periplasmic protein necessary for F-pilus assembly and conjugative transfer (11).
DISCUSSION
We have characterized the successful use of IDM in N. gonorrhoeae. Using small vectors that we constructed containing as little as 290 bp of homologous DNA, we observed transformation at frequencies that are well within the range for successful mutagenesis and resulted in stable insertion mutations. Using four different-sized fragments of the traG gene, we observed that smaller inserts gave lower gonococcal transformation frequencies. These data are consistent with IDM work performed with S. pneumoniae, where a strong correlation was found between the length of the homologous sequence and the transformation frequency (19). Although there have been a few reports of insertional mutagenesis in Neisseria (30, 45), this is the first report of plasmids created specifically for IDM of Neisseria and the first in-depth analysis of the process.
The pIDN plasmids constructed in this study are not only useful in targeted mutagenesis; they are also useful in chromosome walking, as shown here for the cloning of traF. Additionally, as part of an ongoing GGI cloning and sequencing project, we have used the pIDN plasmids to clone and map approximately 50 kb of the GGI. There is also potential to use IDM for random insertional mutagenesis in N. gonorrhoeae. Lee et al. have shown that by constructing a library of IDM vectors containing random 300-bp fragments of S. pneumoniae chromosomal DNA, it is possible to use IDM in a random mutagenesis method (19). A similar method could be employed for N. gonorrhoeae.
The IDM mutations in traG and traH have provided insight into the functions of these genes in N. gonorrhoeae. traG and traH mutants were shown to release less DNA into the medium than the wild-type parent during growth. Furthermore, a traG mutant was shown to be deficient in the ability to donate chromosomal DNA for natural transformation. This suggests that these genes play a role in a type IV secretion system encoded within the genetic island. E. coli TraH and TraG are necessary for conjugation of the F plasmid. E. coli TraG is involved in mating pair stabilization, and both TraG and TraH function in conjugative pilus assembly (11). However, our DNA transfer in coculture is sensitive to DNase, indicating that the transfer is not occurring by conjugation (Table 4). Furthermore, the DNA can be detected in cell-free medium. Thus, these proteins are not functioning in conjugation. These results raise the question of what the likely roles of these proteins are in gonococci. F-plasmid TraG is an inner membrane protein with multiple membrane-spanning segments. It is essential for DNA transfer and may aid in stabilizing mating contacts at an early stage of the interaction between donor and recipient cell surfaces. It has been speculated that TraG may form part of the channel for DNA transfer during conjugation in E. coli (11). Sequence analysis shows that the GGI traG has 22% identity and 40% similarity with the E. coli F-plasmid traG. The gonococcal TraG may also have a role in the formation of a channel through the membranes by which DNA is exported into the extracellular medium, composing part of the apparatus of a type IV secretion system. TraH of the F plasmid is predicted to be a periplasmic protein involved in F-pilus assembly. It contains an ATP-binding motif (Walker box), suggesting that it may be involved in the energetics of assembly or DNA transfer during conjugation. The GGI TraH also contains a Walker box and has 24% identity and 42% similarity to its F-plasmid homologue. The gonococcal TraH may be performing a similar function, i.e., assembly of a pilus-like apparatus involved in DNA release.
Many questions regarding DNA donation and transformation by N. gonorrhoeae are still unanswered. Do the DNA-donating cells die? We have previously shown that DNA release is observed before autolysis can be detected, in the early stages of growth, when gonococci are growing healthily (7). The great advantage of a DNA donation method would seem to be the ability to release DNA without having to undergo autolysis and death. The DNA donation system described here might operate similar to that of an E. coli Hfr, i.e., the gonococcal chromosome may be nicked or cut and the entire chromosome exported. Alternatively, there may be specific genes or regions of the chromosome that are transferred. We do not yet know the size of the DNA being exported or if the exported DNA molecules are coupled with proteins. Future studies of these questions will enhance the comprehension of the mechanisms and functions of gonococcal DNA donation and transformation.
Currently, type IV secretion systems are known to export three types of substrates: (i)DNA conjugation intermediates (DNA-protein complexes), (ii) the multisubunit pertussis toxin, and (iii) monomeric proteins (5). Agrobacterium tumefaciens, the type IV secretion prototype, transfers T-DNA directly into plant cells and transfers the proteins VirE2 (a single-stranded binding protein) and VirF (a virulence factor) as well. The Legionella pneumophila dot/icm system, also a type IV secretion system, is known to conjugate RSF1010-related plasmids (IncQ) and presumably secretes effector molecules to promote survival within macrophages and free-living amoebae (43). The type IV secretion system of B. pertussis (composed of ptl gene products) is known only to secrete a protein substrate (pertussis toxin). Type IV secretion systems have also been identified in Helicobacter pylori, Rickettsia prowazekii, Brucella spp., and a number of nonpathogenic bacteria (5). With the evidence of both DNA and protein transfer in other type IV secretion systems, further studies of the N. gonorrhoeae GGI and its secretion system may reveal similar findings of effector molecule or virulence factor secretion.
ACKNOWLEDGMENTS
We thank Nathan Heerey for technical work on this project.
We also thank the Cremer Fellowship in the Basic Sciences for financial support of Holly L. Hamilton. This work was supported in part by a grant to the University of Wisconsin Medical School under the Howard Hughes Medical Institute Research Resources Program for Medical Schools.
REFERENCES
- 1.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas G D, Sox T, Blackman E, Sparling P F. Factors affecting genetic transformation of Neisseria gonorrhoeae. J Bacteriol. 1977;129:983–992. doi: 10.1128/jb.129.2.983-992.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle-Vavra S, Seifert H S. Shuttle mutagenesis: two mini-transposons for gene mapping and for lacZ transcriptional fusions in Neisseria gonorrhoeae. Gene. 1993;129:51–57. doi: 10.1016/0378-1119(93)90695-y. [DOI] [PubMed] [Google Scholar]
- 4.Burns D L. Biochemistry of type IV secretion. Curr Opin Microbiol. 1999;2:25–29. doi: 10.1016/s1369-5274(99)80004-6. [DOI] [PubMed] [Google Scholar]
- 5.Christie P J, Vogel J P. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 2000;8:354–360. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dillard J P, Seifert H S. A peptidoglycan hydrolase similar to bacteriophage endolysins acts as an autolysin in Neisseria gonorrhoeae. Mol Microbiol. 1997;25:893–901. doi: 10.1111/j.1365-2958.1997.mmi522.x. [DOI] [PubMed] [Google Scholar]
- 7.Dillard, J. P., and H. S. Seifert. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol. Microbiol., in press. [DOI] [PubMed]
- 8.Dillard J P, Yother J. Genetic and molecular characterization of capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 3. Mol Microbiol. 1994;12:959–972. doi: 10.1111/j.1365-2958.1994.tb01084.x. [DOI] [PubMed] [Google Scholar]
- 9.Duncan C H, Wilson G A, Young F E. Mechanism of integrating foreign DNA during transformation of Bacillus subtilis. Proc Natl Acad Sci USA. 1978;75:3665–3668. doi: 10.1073/pnas.75.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkins C, Thomas C E, Seifert H S, Sparling P F. Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J Bacteriol. 1991;173:3911–3913. doi: 10.1128/jb.173.12.3911-3913.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frost L S, Ippen-Jhler K, Skurray R A. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev. 1994;58:162–210. doi: 10.1128/mr.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbs C P, Meyer T F. Genome plasticity in Neisseria gonorrhoeae. FEMS Microbiol Lett. 1996;145:173–179. doi: 10.1111/j.1574-6968.1996.tb08574.x. [DOI] [PubMed] [Google Scholar]
- 13.Goodman S D, Scocca J J. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1988;85:6982–6986. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunn J S, Stein D C. Use of a nonselective transformation technique to construct a multiply restriction/modification-deficient mutant of Neisseria gonorrhoeae. Mol Gen Genet. 1996;251:509–517. doi: 10.1007/BF02173639. [DOI] [PubMed] [Google Scholar]
- 15.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kellogg D S, Jr, Peacock W L, Deacon W E, Brown L, Pirkle C I. Neisseria gonorrhoeae. Virulence genetically linked to clonal variation. J Bacteriol. 1963;85:1274–1279. doi: 10.1128/jb.85.6.1274-1279.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koomey M, Falkow S. Cloning of the recA gene of Neisseria gonorrhoeae and construction of gonococcal recA mutants. J Bacteriol. 1987;169:790–795. doi: 10.1128/jb.169.2.790-795.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krause S, Pansegau W, Lurz R, de la Cruz F, Lanka E. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J Bacteriol. 2000;182:2761–2770. doi: 10.1128/jb.182.10.2761-2770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee M S, Seok C, Morrison D A. Insertion-duplication mutagenesis in Streptococcus pneumoniae: targeting fragment length is a critical parameter in use as a random insertion tool. Appl Environ Microbiol. 1998;64:4796–4802. doi: 10.1128/aem.64.12.4796-4802.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leenhouts K J, Kok J, Venema G. Campbell-like integration of heterologous plasmid DNA into the chromosome of Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1989;55:394–400. doi: 10.1128/aem.55.2.394-400.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leloup L, Ehrlich S D, Zagorec M, Morel-Deville F. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl Environ Microbiol. 1997;63:2117–2123. doi: 10.1128/aem.63.6.2117-2123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 23.Mehr I J, Long C D, Serkin C D, Seifert H S. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics. 2000;154:523–532. doi: 10.1093/genetics/154.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michel B, Niaudet B, Ehrlich S D. Intermolecular recombination during transformation of Bacillus subtilis competent cells by monomeric and dimeric plasmids. Plasmid. 1982;10:1–10. doi: 10.1016/0147-619x(83)90052-5. [DOI] [PubMed] [Google Scholar]
- 25.Morrison D A, Trombe M-C, Hayden M K, Waszak G A, Chen J-D. Isolation of transformation-deficient Streptococcus pneumoniae mutants defective in control of competence, using insertion-duplication mutagenesis with the erythromycin resistance determinant of pAMβ1. J Bacteriol. 1984;159:870–876. doi: 10.1128/jb.159.3.870-876.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morse S A, Bartenstein L. Factors affecting autolysis of Neisseria gonorrhoeae. Proc Soc Exp Biol Med. 1974;145:1418–1421. doi: 10.3181/00379727-145-38025. [DOI] [PubMed] [Google Scholar]
- 27.Niaudet B, Goze A, Ehrlich S D. Insertional mutagenesis in Bacillus subtilis: mechanism and use in gene cloning. Gene. 1982;19:277–284. doi: 10.1016/0378-1119(82)90017-8. [DOI] [PubMed] [Google Scholar]
- 28.Norlander L, Davies J, Norqvist A, Normark S. Genetic basis for colonial variation in Neisseria gonorrhoeae. J Bacteriol. 1979;138:762–769. doi: 10.1128/jb.138.3.762-769.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polissi A, Pontiggia A, Feger G, Altieri M, Mottl H, Ferrari L, Simon D. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect Immun. 1998;66:5620–5629. doi: 10.1128/iai.66.12.5620-5629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinn M L, Weyer S J, Lewis L A, Dyer D W, Wagner P M. Insertional inactivation of the gene for the meningococcal lactoferrin binding protein. Microb Pathog. 1994;17:227–237. doi: 10.1006/mpat.1994.1068. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Segal G, Russo J J, Shuman H A. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol Microbiol. 1999;34:799–809. doi: 10.1046/j.1365-2958.1999.01642.x. [DOI] [PubMed] [Google Scholar]
- 33.Seifert H S, Ajioka R S, Marchal C, Sparling P F, So M. DNA transformation leads to pilin antigenic variation in Neisseria gonorrhoeae. Nature. 1988;336:392–395. doi: 10.1038/336392a0. [DOI] [PubMed] [Google Scholar]
- 34.Seifert H S, Ajioka R S, Paruchuri D, Heffron F, So M. Shuttle mutagenesis of Neisseria gonorrhoeae: pilin null mutations lower DNA transformation competence. J Bacteriol. 1990;172:40–46. doi: 10.1128/jb.172.1.40-46.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seifert H S, Chen E Y, So M, Heffron F. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1986;83:735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seifert H S, So M. Genetic mechanisms of bacterial antigenic variation. Microbiol Rev. 1988;52:327–336. doi: 10.1128/mr.52.3.327-336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sparling P F. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J Bacteriol. 1966;92:1364–1371. doi: 10.1128/jb.92.5.1364-1371.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stein D C. Transformation of Neisseria gonorrhoeae: physical requirements of the transforming DNA. Can J Microbiol. 1991;37:345–349. doi: 10.1139/m91-056. [DOI] [PubMed] [Google Scholar]
- 39.Strathmann M, Hamilton B A, Mayeda C A, Simon M I, Meyerowitz E M, Palazzolo M J. Transposon-facilitated DNA sequencing. Proc Natl Acad Sci USA. 1991;88:1247–1250. doi: 10.1073/pnas.88.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taha M K, So M, Seifert H S, Billyard E, Marchal C. Pilin expression in Neisseria gonorrhoeae is under both positive and negative transcriptional control. EMBO J. 1988;7:4367–4378. doi: 10.1002/j.1460-2075.1988.tb03335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao L, Tanzer J M, Kuramitsu H K, Das A. Identification of several rod loci and cloning of the rodD locus of Streptococcus mutans. Gene. 1993;126:123–128. doi: 10.1016/0378-1119(93)90600-8. [DOI] [PubMed] [Google Scholar]
- 42.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 43.Vogel J P, Andrews H L, Wong S K, Isberg R R. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–875. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 44.Wainwright L A, Pritchard K H, Seifert H S. A conserved DNA sequence is required for efficient gonococcal pilin antigenic variation. Mol Microbiol. 1994;13:75–87. doi: 10.1111/j.1365-2958.1994.tb00403.x. [DOI] [PubMed] [Google Scholar]
- 45.Wolfgang M, van Putten J P M, Hayes S F, Koomey M. The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol Microbiol. 1999;31:1345–1357. doi: 10.1046/j.1365-2958.1999.01269.x. [DOI] [PubMed] [Google Scholar]
- 46.Zhou D, Apicella M A. Plasmids with erythromycin resistance and catechol 2,3-dioxygenase- or beta-galactosidase-encoding gene cassettes for use in Neisseria spp. Gene. 1996;171:133–134. doi: 10.1016/0378-1119(96)00103-5. [DOI] [PubMed] [Google Scholar]