Abstract
Environmental isolates of Vibrio cholerae of eight randomly amplified polymorphic DNA (RAPD) fingerprint types from Calcutta, India, that were unusual in containing toxin-coregulated pilus or cholera toxin genes but not O1 or O139 antigens of epidemic strains were studied by PCR and sequencing to gain insights into V. cholerae evolution. We found that each isolate contained a variant form of the VPI pathogenicity island. Distinguishing features included (i) four new alleles of tcpF (which encodes secreted virulence protein; its exact function is unknown), 20 to 70% divergent (at the protein level) from each other and canonical tcpF; (ii) a new allele of toxT (virulence regulatory gene), 36% divergent (at the protein level) in its 5′ half and nearly identical in its 3′ half to canonical toxT; (iii) a new tcpA (pilin) gene; and (iv) four variant forms of a regulatory sequence upstream of toxT. Also found were transpositions of an IS903-related element and function-unknown genes to sites in VPI. Cholera toxin (ctx) genes were found in isolates of two RAPD types, in each case embedded in CTXφ-like prophages. Fragments that are inferred to contain only putative repressor, replication, and integration genes were present in two other RAPD types. New possible prophage repressor and replication genes were also identified. Our results show marked genetic diversity in the virulence-associated gene clusters found in some nonepidemic V. cholerae strains, suggest that some of these genes contribute to fitness in nature, and emphasize the potential importance of interstrain gene exchange in the evolution of this species.
Vibrio cholerae is a genetically diverse species that lives in warm-water environments, often associated with plankton and other aquatic organisms (9, 15, 31). Strains of just two of the approximately 200 currently known O-antigen serogroups (50; T. Shimada, personal communication), O1 and O139, cause epidemic cholera—the acute, devastating diarrheal disease that afflicts many thousands of people annually, especially in developing countries. Although certain other serogroups cause sporadic diarrheal disease (30, 37), most V. cholerae organisms probably do not infect humans. Epidemic strains are also distinguished from most others by their production of cholera toxin and a toxin-coregulated pilus (TCP) (15, 21).
Epidemic strains of V. cholerae have a complex natural history in areas of cholera endemicity, involving rapid transmission to humans via contaminated food and water during each year's cholera season and persistence or proliferation in aquatic organisms or abiotic niches at other times. Several recent major changes in patterns of epidemic cholera may stem in large part from a combination of human factors and environmental fluctuation that could affect the distribution of aquatic host species. The bacterial changes include (i) the replacement of classical biotype O1 strains beginning in 1961 by strains of the new El Tor O1 biotype (15), (ii) the reemergence of classical biotype strains in parts of Bangladesh in the early 1980s and their disappearance a dozen years later (40; A. K. Siddiqui, personal. communication), (iii) the emergence of the new O139 serogroup in 1992 (38) and its persistence along with El Tor O1 strains in South Asia since then (15, 16), and (iv) the sudden appearance of cholera in Peru in 1991 (46), which coincided with El Niño climate warming and changes in aquatic ecosystems (9, 29, 44).
Genetic recombination between divergent bacterial strains can be advantageous, especially in complex or variable environments, because it generates new genotypes more efficiently than does mutation alone. Its importance in V. cholerae evolution is illustrated by comparison of O139 and El Tor O1 strains, which contain quite different O-antigen biosynthetic genes but are closely related in most other genetic loci. This suggests that O139 antigen genes were transferred from an unknown donor into an El Tor-related strain that was well suited to human infection and the South Asian environment. O139 recombinants may have been selected by their ability to infect adults with anti-O1 immunity, in addition to nonimmune children (3, 4, 43, 51). A history of recombination involving other gene loci has also been detected by DNA sequence and multilocus enzyme electrophoresis analyses (3, 6, 22, 45).
Advantageous genes can also spread efficiently via specialized genetic elements whose establishment in new bacterial lineages does not require homology-based recombination. For example, the cholera toxin genes of epidemic V. cholerae strains are within the genome of a filamentous M13-related phage designated CTXφ (48). Although a CTXφ prophage can be carried as a plasmid, it is usually found integrated into the chromosome, often in a multicopy tandem array, and controlled by a prophage repressor. This repressor is inactivated in the bacterial response to DNA damage, thereby allowing production of progeny phage that can infect (lysogenize) new bacterial hosts (14). CTXφ repressors of three different specificities are known, and strains carrying one prophage can often be infected or lysogenized by CTXφ of other repressor specificities (11, 25). In a second example, the genes for TCP form part of a 40-kb segment that is absent from many nonepidemic strains, that has been designated a pathogenicity island (VPI) (23), and that might also correspond to a temperate filamentous phage (24). As remarkable examples of evolutionary coadaptation, the CTXφ virion uses TCP as a receptor during infection (48), and the VPI-encoded ToxT regulatory protein helps turn on transcription of both TCP genes (also located in VPI) and cholera toxin genes (in CTXφ) in response to particular host or environmental conditions (7, 10, 15, 28).
There have been several reports of unusual nonepidemic strains carrying tcpA (pilin) or ctx (cholera toxin) genes (8, 32–34). Here we present a more detailed analysis of two dozen such isolates from Calcutta. Our results show that these genes are also contained within VPI- and CTXφ-type genetic elements, demonstrate a remarkable pattern of localized diversity in them, and emphasize the potential importance of gene exchange as a force in V. cholerae genome evolution.
MATERIALS AND METHODS
Bacterial strains.
The 24 environmental isolates of V. cholerae studied here were obtained from water samples from Calcutta, India, by enrichment culture (8 of 122 tested) and PCR-based screening for tcpA (classical or El Tor) or ctx genes and O-antigen typing, as described previously (8) (Table 1). Three reference epidemic strains of V. cholerae were also used: O139 (SG24); O1 Ogawa, El Tor biotype (VC20); and O1, Ogawa, classical biotype (O395) (2).
TABLE 1.
Characteristics of environmental isolates of V. cholerae
| RAPD type | SCE isolate(s) | Collectiona | Serogroup | VPI
pathogenicity islandb
|
CTXφ
prophagec
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LJ | tcpA | tcpF | F-T | toxT | ig1 | rstR | rstA | rstC | ctx | ||||
| 1a | 4, 6 | 1 | O8 | +L | cla | II | II | epi | − | − | − | − | − |
| 1b | 359 | 7 | O8 | +L | cla | II | ND | epi | − | − | − | − | − |
| 1a | 5 | 1 | O11 | +L | cla | II | ND | epi | − | − | − | − | − |
| 2 | 225, 226, 228 | 3a | O35 | epi | cla | III | III.IS | env | − | − | − | − | − |
| 2 | 227 | 3b | O35 | epi | cla | III | III.IS | env | − | − | − | − | − |
| 3 | 258 | 5 | O42 | epi | cla | IV | IV | env | − | − | − | − | − |
| 4 | 256, 257, 259, 260, 261, 265 | 5 | O42 | epi | cla | IV | IV | env | − | − | − | − | − |
| 4 | 264 | 5 | O42 | epi | cla | IV | IV | env | + | 5 | 264 | + | − |
| 5 | 263 | 5 | O10 | epi | cla | III | III | env | + | 4∗ | + | + | − |
| 6 | 188, 200, 201 | 2 | O44 | epi | env | V | V | epi | + | 4∗, cal | + | + | + |
| 7 | 223 | 4 | O27 | epi | env | V | V | epi | + | 4∗∗ | + | + | + |
| 7 | 354 | 8 | O27 | epi | env | V | V | epi | + | 4∗∗ | + | + | + |
| 8 | 340, 341 | 6 | O69 | epi | elt | IV | IV | env | − | − | − | − | − |
Collection dates and sources: 1, 7 July 1997, plankton, Lake Subhas Sarobar, Calcutta; 2, 2 December 1997, water from fish pond in village of Polan Heights near Calcutta; 3, 8 January 1998, plankton (a) and water (b) from pond in Mohish Batan region of Calcutta; 4, 8 January 1998, plankton from Polan Heights fish pond; 5, 10 February 1998; water from pond in Mohish Batan region of Calcutta; 6, 5 May 1998, plankton water from pond in Mohish Batan region, Calcutta; 7, 19 May 1998, water from Polan Heights fish pond; and 8, 19 May 1998, plankton water from pond in Mohish Batan region, Calcutta.
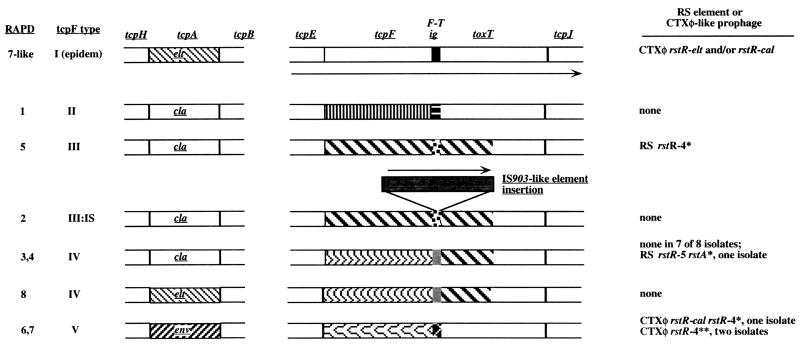
Abbreviations: LJ, left junction (+L, yielded a 2.3-kb PCR product instead of the usual 1-kb product due to gene translocation [Fig. 7]); epi, epidemic; cla, classical; elt; El Tor; env, environmental. The amino acid sequences of the encoded TcpA proteins are shown in Fig. 3. The amino acid sequences of TcpF proteins encoded by alleles II, III, IV, and V, each of which is distinct from TcpF of canonical epidemic strains (designated type I here), are diagrammed in Fig. 5, and their relatedness is summarized in Table 3. Five types of sequence in the ∼200 to 230 bp between tcpF and toxT (designated F-T) were identified (type I, canonical, from strain 569B, but also 99 to 100% matched to those of other epidemic [El Tor and classical biotype] strain sequences in the database. Other types are diagrammed in Fig. 6 and described in the text.).
+, allele very similar to those in epidemic strains; −, no gene detected; ND, not determined. The rstR4∗ and rstR4∗∗ alleles are diagrammed in Fig. 9. rstA264 designates a gene that is unrelated to that in canonical CTXφ.
DNA fingerprinting.
Genomic DNAs prepared by standard cetyltrimethylammonium bromide-phenol extraction from 1.5-ml overnight cultures were used for DNA fingerprinting by the random amplified polymorphic DNA (RAPD) method (1). Reactions were carried out in 25 μl containing 2.5 μl of 10× PCR buffer, 20 ng of V. cholerae genomic DNA, 4 μl of 25 mM MgCl2, 20 pmol of primers 1281 (5′-AACGCGCAAC) or 1283 (5′-GCGATCCCCA), 1 U of AmpliTaq DNA polymerase, and 2.5 μl of 2.5 mM deoxynucleoside triphosphates under a drop of mineral oil for 45 cycles of 94°C for 1 min, 36°C for 1 min, and 72°C for 2 min in a Perkin-Elmer TC480 thermal cycler. After PCR, 8-μl aliquots of product were electrophoresed in 1% agarose gels containing 0.5 mg of ethidium bromide/ml and photographed under UV light. A 1-kb DNA ladder (Gibco BRL, Rockville, Md.) was used as a size marker in all gels.
Characterization of CTXφ and VPI.
PCR tests for VPI pathogenicity island and CTXφ prophage genes were carried out using primers listed in Table 2 (the approximate locations of genes in VPI and in CTXφ that we studied are shown in Fig. 1). Hybridization was carried out with DNA probes generated by PCR from epidemic-strain DNAs. PCR was performed in volumes of 20 μl containing 10 ng of genomic DNA, 10 pmol of primer, and 1 U of Taq DNA polymerase for 30 cycles of 94°C for 40 s, 55°C (or 60°C when higher specificity was needed) for 40 s, and 72°C for a time chosen based on the size of the expected fragment (1 min/kb). Long-distance PCR was performed using the Advantage genomic PCR kit (Clontech Laboratories Inc., Palo Alto, Calif.) when needed (fragments longer than a few kilobases). Each PCR was carried out in a volume of 100 μl containing 8 ng of genomic DNA, 40 pmol of each primer, 2 μl of 50× Advantage genomic polymerase mix, 10 μl of 10× genomic PCR buffer, 2 μl of 50× deoxynucleoside triphosphate mix, 10 μl of MgCl2 (25 mM), and MilliQ water. The amplification conditions were: preincubation at 94°C for 15 or 30 s and then 35 cycles of 94°C for 30 s and 68°C for 6 or 12 min, with final extensions at 68°C for 6 or 12 min for PCRs using primers tcpI-F and tcpA-R (classical variant) and primers tcpA-F (classical) and tcpF-R to generate fragments of 3.9 and 9.2 kb, respectively. All PCRs were carried out in a TC480 thermal cycler (Perkin-Elmer Cetus, Foster City, Calif.).
TABLE 2.
Sequences of primers used in this study
| Primera | Primer sequence (5′-3′) | Coordinateb (5′ end) | Size (bp)c | Use |
|---|---|---|---|---|
| LJ-F | GTGAATCTTGATGAGACGCTCTG | 872910 | 1,011 | PCR |
| LJ-R | GGTGAGCCAGGCTTATTTGGG | 873920 | PCR | |
| SCE4.LJ-F2 | GGATCATCAACCGAAGAGGTAGA | This study | Sequencing | |
| SCE4.LJ-R2 | GCCTGCATCACCACATTCCTCATA | This study | Sequencing | |
| SCE4.LJ-F3 | GGTTAGCTCTTCCATCGACAACA | This study | Sequencing | |
| SCE4.LJ-R3 | GGTCTGGAAATACCACAGCGT | This study | Sequencing | |
| aldA-F | GCATCTGATTATGACCAAAGAATAGG | 875610 | 1,116 | PCR |
| aldA-R | GTCAATGGATGAAGCCACACAGTG | 876725 | PCR | |
| tagA-F | GGTGGTAAGATATTCACTCTTAATG | 877014 | 351 | PCR |
| tagA-R | GAGACATCTATAGAATACTGGCTG | 877364 | PCR | |
| tcpI-F | GCCGTCTCCGCATTAAGCTCTGCAC | 887200 | PCR | |
| tcpI-F2 | GCCAACGTAATGATGCACGCAAGTA | 888350 | 1,212 | PCR |
| tcpH-R2 | CGTTACACCAAGTGCTACAACGAAT | 889561 | PCR | |
| tcpA-F(cla) | CACGATAAGAAAACCGGTCAAGAG | M33514 | 617 | PCR |
| tcpA-R(cla) | ACCAAATGCAACGCCGAATGGAGC | M33514 | PCR | |
| tcpA-F(elt) | GAAGAAGTTTGTAAAAGAAGAACAC | 890480 | 471 | PCR |
| tcpA-R(elt) | GAAAGGACCTTCTTTCACGTTG | 890951 | PCR | |
| tcpQ-R | GAGGACTGTTCTGCAATCTGCTCAT | 892609 | PCR | |
| tcpF-F | GAGTTCCACATGCAGAAACAGGA | 899444 | PCR | |
| tcpF-R | GCCACGAATGTGGCTGTTATCTTATC | 899724 | PCR | |
| toxT-F | ACTGTATAGCAAAGCATATTCAGAGA | 899658 | 1,115 | PCR |
| toxT-R | CAGTGATACAATCGAAAATAGGATC | 900772 | PCR | |
| acfB-R | GAGCGTGCTTTATCATGGTCGAT | 902296 | PCR | |
| int-F | GAAGTAATGAAACCGATAAGTGG | 913253 | 347 | PCR |
| int-R | TGCTTTGTACCAGTCACAGATAG | 912907 | PCR | |
| RJ-F | TCGTTAGCGTGTCGGTTCGCAGG | 914405 | 1,499 | PCR |
| RJ-R | TGCTTTGTACCAGTCACAGATAG | 912907 | PCR | |
| ig-1-F | GAGCCTGTGACACTCACCTTGTAT | 1566438 | 629 | PCR |
| rstA-R | GCATAAGGAACCGACCAAGCAAGAT | 1565810 | PCR | |
| rstC-R | GCTCAGTCAATGCCTTGAGTTG | 1564169 | PCR | |
| orfU-F | GGTGTTATTTGATGGCTGCATG | 1570758 | 3,713 | PCR |
| ctxB-R | GCTTCAGTAAGATATGCAATCCTCAG | 1567046 | PCR | |
| tcpE-F2 | GCTCCTGACAATGGCTGTTTATTCA | 898458 | PCR | |
| tcpF.n-F2 | GCTATTCCTACTCCAGACCAACTA | This study | Sequencing | |
| toxT.n-F2 | GAGTTTGATTGTCGATCTCAGTT | This study | Sequencing | |
| toxT-R2 | CACAGTTCCAGAGGAGCAGAT | This study | Sequencing | |
| tcpJ-R2 | GGGCACATGGAGCGATTTGAAT | 900876 | Sequencing | |
| tcpJ-R | GCTTGGTCCAAGAGGGATAT | 901444 | PCR | |
| 226.tcpF-F | GGATATTCCTACAGCTGAGCAGTT | This study | Sequencing | |
| 226.toxT-R | GGTCATATGTGCTCACTATCTCT | This study | Sequencing |
LJ, left junction; RJ, right junction; aldA, toxT, and int, genes in the V. cholerae pathogenicity island; ig1, RS, orfU, ctxB, and rstA, DNA segments and genes in the CTXφ prophage; tcpI, tcpQ, tcpF, and acfB, genes in the V. cholerae pathogenicity island VPI.
Unless stated otherwise, the coordinates used here correspond to positions in the complete genome sequence of V. cholerae El Tor N16961 (19), available at http://www.tigr.org/tdb/CMR/gvc/htmls/SplashPage.html.
Size of diagnostic PCR product generated with pairs of primers (only those important to this analysis are given).
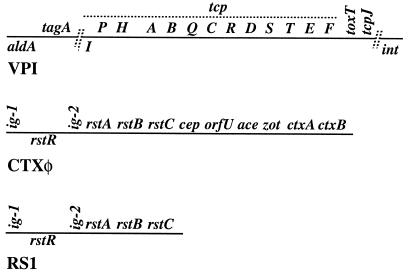
FIG. 1.
Abbreviated maps of VPI, CTXφ, and RS1, including relative positions of genes that were studied here. These maps are not complete or to scale. Genes indicated above the lines are transcribed left to right; those below the lines are transcribed in the opposite orientation These maps are based on references 15, 19, and 48.
Amplified products were electrophoresed in ethidium bromide agarose gels (SeaKem; BMA, Rockland, Maine), and visualized and documented using a video documentation system. PCR products were purified using a PCR product purification kit (Qiagen Inc., Chatsworth, Calif.) and used as probes for dot blot hybridization. For hybridization, 10 to 15 ng of genomic DNA from each V. cholerae isolate was spotted on Hybond N+ membrane (Amersham, Arlington Heights, Ill.) and dried for 30 min. The membrane was then denatured with a solution of 0.5 M NaOH and 1.5 M NaCl for 7 min and neutralized with two washes of 150 ml of 0.5 M Tris-HCl (pH 7.4) and 1.5 M NaCl for 3 min each, after which the membrane was dried for 1 h. DNA was fixed to the membrane by UV cross-linking. Hybridization probes were prepared by PCR from reference strains of V. cholerae O139 (SG24), V. cholerae O1 Ogawa, El Tor biotype (VC20), and V. cholerae O1, Ogawa, classical biotype (O395), using primers listed in Table 1. About 200 ng of each probe DNA was conjugated to horseradish peroxidase, and hybridization to filters was detected with a chemiluminescent substrate (Amersham Pharmacia Biotech, Piscataway, N.J.) on X-ray film, as previously described (47).
DNA sequencing.
DNAs corresponding to parts of the rstR region of CTXφ were PCR amplified using primers ig-1-F and rstA-R or ig-1-F and rstR-R (formerly RS-R) (Table 1); parts of the tcpA region were amplified using primers tcpA-F of the classical variant and tcpQ-R, and PCR products used for sequencing were purified using a Qiagen kit and sequenced directly (without cloning) using a Taq dye terminator sequencing kit (Perkin-Elmer), appropriate primers, and an automated DNA sequencer (ABI Prism 377; ABI, Foster City, Calif.). The sequences were aligned using the DNAsis software program and analyzed using the Basic Local Alignment Search Tool program available on the National Center for Biotechnology Information web site or programs in the Genetics Computer Group (Madison, Wis.) package, PHYLIP of J. Felsenstein (http: //evolution.genetics.washington.edu/phylip.html), and clustal W of T. J. Gibson and colleagues (http://www.csc.fi/molbio/progs/clustalw).
Nucleotide sequence accession numbers.
The nucleotide sequence data for VPI elements reported here have been deposited in GenBank with the following accession numbers: AF133307 (rstR-4** from SCE223), AF133308 (rstR-5 from SCE264), AF13309 (rstR-4* from SCE263), AF13310 (rstR-cal from SCE188), AF319656 (rstA from SCE264), AF208385 (tcpA-env from SCE188), AF306795 (tcpE-tcpJ segment from SCE4), AF306796 (tcpE-tcpJ segment from SCE226, which contains an IS903-related element), AF378526 (tcpE-tcpJ segment from SCE263, which lacks an IS903-related element), AF306797 (tcpE-tcpJ segment from SCE256), AF306798 (tcpE-tcpJ segment from SCE200), AF319954 (left junction region of VPI from SCE4, including a DNA segment translocated from chromosome 2), AF319652 (tcpP from SCE4), AF319653 (tcpP from SCE226), AF319654 (tcpP from SCE256), and AF319655 (tcpP from SCE200). For locations of these segments, see Fig. 1. For a summary of salient features of strains, see Table 1.
RESULTS
RAPD types.
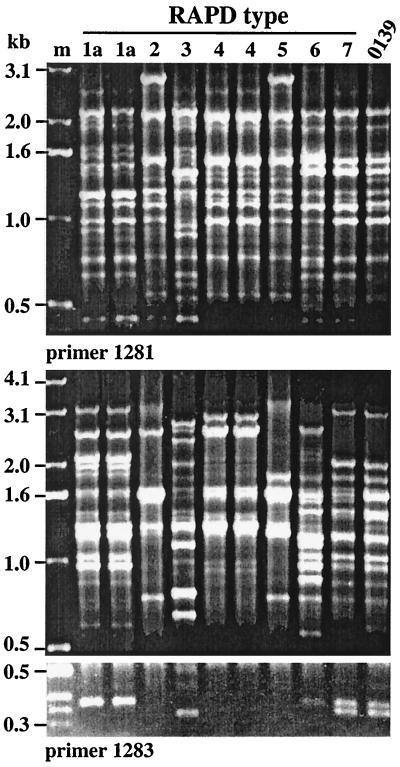
The 24 V. cholerae isolates studied here (Table 1) had been chosen based on positive PCR with primers specific for tcpA genes of classical or El Tor biotype strains (designated tcpA-cla and tcpA-elt, respectively) or ctx genes; they were of eight O antigen types, each of which was distinct from the O1 or O139 epidemic type (8). Our arbitrarily primed PCR (RAPD) fingerprinting identified eight types (Fig. 2), probably each corresponding to a different background genotype. Isolates of the same RAPD type were often from the same collection, suggesting that they might be sibs. However, the same RAPD type was obtained in different collections in two cases (types 1 and 7), and isolates with matched RAPD patterns were distinguishable by other traits or DNA markers in two other cases (types 1a and 4) (Table 1). Although isolates of a given RAPD type tended to be of the same serogroup, but two serogroups (O8 and O11) were represented in isolates of RAPD type 1, and conversely, isolates of two RAPD types (3 and 4) were each of the same serogroup (O42). These exceptions can be ascribed to interstrain gene transfer or to mutation affecting O-antigen biosynthetic genes. In addition, one RAPD pattern (type 7) closely matched that of O139 and El Tor O1 epidemic strains (Fig. 2), but the two isolates of this type (which were collected at different times) differed from epidemic strains in O antigens and several other determinants (Table 1).
FIG. 2.
RAPD profiles of representative V. cholerae environmental isolates generated using primers 1281 and 1283. Lane designations correspond to RAPD types discussed in the text and to SCE isolates defined in Table 1, as follows: 1a, SCE4 and SCE5; 2, SCE226; 3, SCE258; 4, SCE260 and SCE264; 5, SCE263; 6, SCE200; and 7, SCE223. The O139 epidemic strain used is SG24. m, 1-kb ladder (size marker).
VPI pathogenicity islands.
In initial characterizations (8), 19 of the 24 V. cholerae isolates were found to carry tcpA alleles closely matched to either tcpA-cla or tcpA-elt, whereas the other five isolates (two RAPD types) seemed not to contain either of these tcpA alleles, even though only they carried ctx genes (8). In follow-up experiments we observed strong hybridization of DNAs from each of our 24 isolates (the five anomalous, possibly tcpA-deficient isolates included) with probes specific for regions adjacent to tcpA (a 3.9-kb tcpI-tcpA segment on the left and a 9.2-kb tcpA-tcpF segment on the right) (Fig. 1 and data not shown). Further PCR and hybridization tests showed that each of the 24 isolates also carried aldA and tagA sequences near the VPI left end and int near its right end, and that each also yielded PCR products corresponding to the junctions between the left and right ends of VPI and flanking DNA (primers LJ-F and LJ-R and primers RJ-F and RJ-R) (Table 1). The only unusual result from these PCR tests was obtained with isolates of RAPD type 1 (SCE4, -5, -6, and -359), which yielded a 2.3-kb rather than a 1.0-kb PCR product with left-junction-specific primers. These results suggested that each isolate contained an intact or nearly intact VPI pathogenicity island.
New alleles in VPI. (i) tcpA.
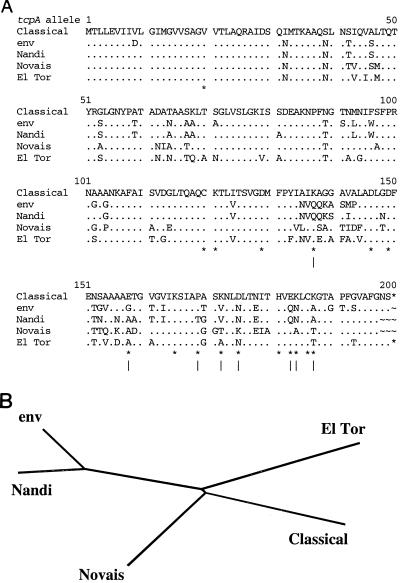
The region near tcpA of isolates that lacked tcpA-cla or tcpA-elt alleles (RAPD types 6 and 7) was studied further. PCR with primers flanking tcpA (tcpI-F and tcpQ-R) yielded a product of the size matching that from epidemic strains (∼5.4 kb), and equivalent PCR using a primer specific for a conserved sequence near the 5′ end of tcpA (tcpA-F) along with tcpQ-R yielded a ∼2.1-kb product in each case. DNA sequencing of two such isolates (SCE188 and SCE354; RAPD types 6 and 7) identified a new tcpA allele (tcpA-env), only ∼74% identical at the DNA level (77 to 78% at the protein level) to corresponding 600-bp sequences from tcpA-elt (GenBank accession no. U09807) and tcpA-cla (accession no. X64098) (36, 39). This new tcpA-env allele was also distinct from tcpA alleles found recently in other nonepidemic isolates (Fig. 3).
FIG. 3.
TcpA alignment and phylogenetic tree. (A) Alignment of inferred amino acid sequences of TcpA-cla with the TcpA-env protein found here (from SCE188) and with other TcpA proteins (Nandi, accession no. AF139626 [33]; Novais, accession no. AF030309 [34]; and El Tor, accession no. M33514). Periods indicate residues identical to those found in TcpA-cla; only amino acid differences (relative to TcpA-cla) are specified. ∗, position whose functional importance in TcpA-cla was tested mutationally (27). (B) Inferred phylogenetic relationships between tcpA alleles.
Much of the divergence among these various tcpA alleles was in the carboxy-terminal half, which is thought to be exposed on the pilus surface (27; R. Chattopadhyaya and A. C. Ghose, personal communication; R. K. Taylor, personal. communication) (Protein Database accession no. 1QT2 [http: //www.rcsb.org/pdb]); the amino-terminal third, which is buried in the mature pilus structure, is relatively better conserved among isolates. Among the many differences between the present tcpA-env allele and the tcpA-cla allele (Fig. 3A) is the K187A substitution, which in the tcpA-cla context increases pilus-mediated autoagglutination 30-fold (27).
(ii) toxT and tcpF.
Initial PCR tests (8) had also suggested that isolates of five RAPD types either lacked the toxT gene or contained an allele that differed significantly from that in epidemic strains (RAPD types 2, 3, 4, 5, and 8 in Table 1). Our additional PCR tests indicated that the adjacent tcpF gene was also either missing or divergent. However, tcpE and tcpJ sequences, which flank the tcpF-toxT segment, were found in every isolate by PCR with primers tcpE-F2 and tcpJ-R. In addition, PCR with these primers yielded products from each of the 24 isolates: the expected 3 kb in 20 of 24 cases and a 1-kb-longer product in the other four, which all belonged to one RAPD type (type 2). DNA sequencing showed that the strains that had not yielded PCR products with standard toxT primers contained a new mosaic toxT allele: 62 to 64% identity (protein level) to that of canonical toxT in its first half (5′-most 148 codons), and 99% identity to canonical toxT in its second (carboxy-terminal) half (Fig. 4), the region that determines specificity of ToxT protein binding to DNA regulatory sites.
FIG. 4.
Alignment of ToxT from strains SCE263 and 569B (classical biotype, epidemic strain) (GenBank accession no. B45247). toxT alleles nearly identical to that of SCE263 were found by sequencing in isolates SCE226 and SCE256.
Further sequencing revealed four markedly different alleles of tcpF, a gene that encodes a secreted protein whose exact role in virulence is not known (Taylor, personal communication). The inferred TcpF proteins were 31 to 79% identical to one another and to canonical TcpF (Fig. 5; Table 3). Two of the new tcpF alleles were associated with the novel toxT allele, and the other two were in isolates carrying toxT alleles that were nearly identical to those of epidemic strains (Fig. 6).
FIG. 5.
TcpF alignments generated with clustal W. Types II, III, IV, and V are from environmental isolates as indicated in Table 1 and in Fig. 6. Reference strain 569B (here designated type I) is a classical biotype O1 serogroup epidemic strain (tcpF sequence from GenBank accession no. L01623).
TABLE 3.
Identities (protein level) among tcpF alleles
| Type | % Identity with
typea:
|
||||
|---|---|---|---|---|---|
| II | III | IV | V | I (epidemic) | |
| II | 100 | 33 | 39 | 79 | 66 |
| III | 100 | 33 | 33 | 31 | |
| IV | 100 | 36 | 33 | ||
| V | 100 | 70 | |||
Identities were inferred from DNA sequences of tcpF genes of SCE4 (type II), SCE263 (type III), SCE258 (type IV), and SCE200 (type V). The epidemic (type I) strain used as a reference is 569B.
FIG. 6.
Allele content of environmental isolates. Each pattern of shading represents a different highly divergent sequence, as shown in Fig. 3, 4, and 5, and described in the text for tcpA, toxT, and tcpF and the F-T intergenic (ig) sequence.
tcpF-toxT intergenic sequence.
Between tcpF and toxT lies a noncoding sequence that in epidemic strains is about 230 bp long and contains binding sites for positive (TcpP and ToxR) and negative (H-NS) regulatory proteins which help link toxT expression to environmental conditions, host signals, and general bacterial physiology (18, 35, 52). A tcpF-toxT intergenic sequence of about 200 to 220 bp was also found in each environmental isolate analyzed, and four types (referred to as F-T types) were distinguished. The sequences F-T II (found in RAPD type 1) and F-T V (in RAPD types 6 and 7) were 88 to 90% identical to one another and to the F-TI segment of epidemic strains. The F-T III (in RAPD type 5) and F-T IV (in RAPD types 3 and 4) sequences were 67% identical to each other in a 169-bp shared segment but did not seem to be related to the corresponding intergenic sequence in epidemic strains. The F-T intergenic sequence in RAPD type 2 isolates contained a transposable element insertion (see below) but otherwise was identical to that in SCE263; therefore, it was designated F-T III:IS.
Conserved sequences in VPI.
The F-T intergenic sequence contains binding sites for multiple regulatory factors, including TcpP protein (18, 35), which is encoded within VPI. Accordingly, 1.1-kb segments containing the 666-bp tcpP gene and upstream sequences were PCR amplified from four isolates representing each of the four different F-T intergenic sequence types (SCE4, -226, -256, and -200). The sequences obtained from them were ≥97% identical to those of canonical epidemic strains in each case. Thus, sequence divergence in the F-T intergenic space in these environmental isolates is probably not associated with major changes in the DNA binding specificity of the TcpP regulatory protein.
Our sequencing of the tcpE-tcpJ segment also yielded about 130 to 180 bp in tcpE, just upstream of tcpF, and another 300 to 400 bp in tcpJ, just downstream of toxT from each of the four strains analyzed. These flanking sequences were, in each case, ≥96% identical to sequences from canonical epidemic strains. This reinforces the view of VPI as a mosaic, containing interspersed regions of conserved and divergent sequence, a feature that is common in temperate phage (20).
Insertion in F-T intergenic space.
The isolates of RAPD type 2, whose tcpE-tcpJ region contained 1 kb of extra DNA, were found to carry an IS903-related element with 98% DNA sequence identity to one that had been found in the V. cholerae genome (containing the transposase open reading frame [ORF] VC0501) (19). In our RAPD type 2 isolates, this element was inserted 60 bp downstream of the 3′ end of the new tcpF allele in an F-T intergenic sequence that was otherwise identical to that in a RAPD type 5 isolate (SCE263). The element was positioned such that if regulatory sites in this segment were arranged as they are in the divergent F-T segment in canonical strains (35), it would separate the binding site for H-NS from those for TcpP and ToxR; its orientation would also allow transcription initiated from upstream genes or the element's transposase promoter to continue into toxT. Thus, the inserted element might affect toxT expression.
The inserted element found here seems likely to contain a complete transposase gene. The closely related element found outside VPI in the fully sequenced genome of an epidemic strain contained a 10-bp deletion (frameshift) at position 327 in the 921-bp transposase gene (gene VCA0501 in reference 19; coordinates based on our sequence). No inverted repeat structure equivalent to the 18-bp terminal inverted repeats of canonical IS903 (12, 17) was found in either inserted V. cholerae element.
Translocation deletion at the left end of VPI.
The basis of the unexpectedly large left junction PCR product in isolates of RAPD type 1 noted above (Table 1) was investigated by sequencing this product from isolate SCE4. Inserted in this left-junction DNA was a 1.6-kb segment containing the ORF VCA0577 and much of VCA0578, in place of 300 bp that includes the left junction of VPI with ancestral DNA in canonical V. cholerae strains (Fig. 7). Since VPI is in chromosome 1 and these genes lie in chromosome 2 in the fully sequenced V. cholerae genome (19), the rearrangement seen in SCE4 probably represents a case of interchromosomal translocation. Comparison of sequences identified a 15-of-17-bp match between chromosomes 1 and 2 at the left end of the translocated segment, but no equivalent match was found at the right end. The VCA0577 and VCA0578 genes are referred to as “function unknown,” with no match to other entries in current databases. Thus, this insertion-deletion rearrangement may have resulted from chance recombination in a segment of short homology and/or illegitimate recombination. This translocation might be adaptive (improve the bacterial phenotype) or reflect an evolutionary accident that has not been eliminated by genetic drift or contraselection if deleterious.
FIG. 7.
Left-junction translocation. The insertion-deletion structure found at the left end of VPI in isolates of RAPD type 1 by PCR and sequencing, as described in the text, is shown. VC and VCA designations refer to genes in V. cholerae chromosomes 1 and 2, respectively,in a V. cholerae reference strain (19). Numbers above the map refer to rearrangement breakpoints, inferred by comparison with the full genome sequence (19).
CTXφ prophage.
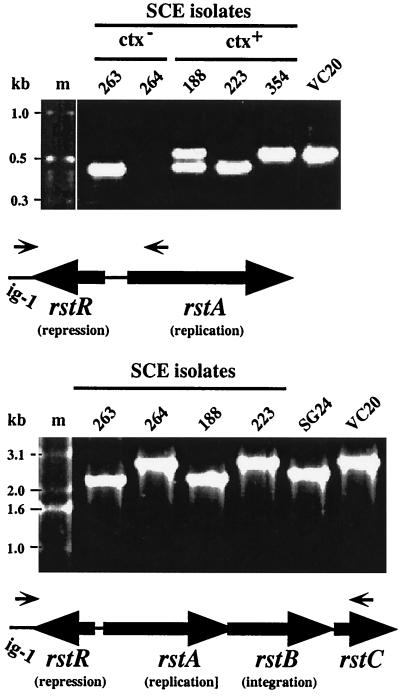
Cholera toxin genes had been found in five isolates (8) belonging to two RAPD types. Our PCR and hybridization tests indicated that these ctx genes were embedded in CTXφ-like prophages. DNA segments that may correspond to RS1 prophage fragments, which contains just genes and sites for phage replication, integration, and immunity (Fig. 1), were found in ctx-negative isolates of two other RAPD types (Table 1). First, PCR of DNAs from the five ctx-positive isolates with primers specific for CTXφ prophage genes (orfU-F and ctxB-R) (Fig. 1) yielded products of about 3.7 kb, which are also obtained using epidemic strains. Second, these five DNAs and also DNAs from two ctx-negative isolates (SCE263 and SCE264) were found to hybridize with an RS1 probe, generated with primers ig-1-F and rstC-R. However, none of the ctx-negative isolates hybridized with a probe specific for sequences between RS1 and ctx (generated with primers orfU-F and ctxB-R). Third, PCR amplification with primers specific for conserved sequences that flank the rstR repressor gene (ig-1-F and rstA-R) generated products from six of the seven RS1 hybridization-positive isolates (Fig. 8) but not from any of the other 17 isolates studied here. A single 450-bp band was obtained from two isolates (SCE223 [ctx positive] and SCE263 [ctx negative]); a single 600-bp band was obtained from another isolate (SCE354 [ctx positive]), suggesting a different rstR allele; and a doublet (450 and 600 bp) was obtained from each of three other isolates (SCE188, SCE200, and SCE201, all ctx positive and RAPD type 6). However, no PCR product was obtained with these primers from the seventh RS1 hybridization-positive isolate (SCE264) (Fig. 8A). Fourth, PCR using the more distal rstC-R primer along with ig-1-F yielded a fragment in the expected size range (2.3 to 2.7 kb) from each of the seven RS1 hybridization-positive isolates (including the anomalous SCE264) (Fig. 8B).
FIG. 8.
Representative results of PCR amplification of rstR and adjacent regions. Lane designations correspond to SCE isolates, except that VC20 and SG24 designate serogroup O1 El Tor and O139 epidemic strains, respectively, used for reference. (Top) Amplification with primers ig-1-F (specific for conserved sequence in the CTXφ intergenic region, just to the left of rstR) and rstA-R. Note that no amplification product was obtained from SCE264 with these primers and that SCE188 yielded two different products, indicating that it is a double lysogen. (Bottom) Amplification with primers ig-1-F and rstC-R. Note that an amplification product was obtained from SCE264 with these primers.
PCR products generated with ig-1-F and rstA-R or rstC-R primers from representative isolates were sequenced, and putative ORF structures were identified (Fig. 9). The 600-bp (larger) fragment from SCE188 was 100% identical in regions of overlap to the sequence of rstR-cal (25). This segment, like rstR-classical and rstR-eltor, was shown by others to have repressor function (11, 26), even though it seemed to contain overlapping ORFs of 59 and 15 codons (6-codon overlap). The sequence of the smaller (450-bp) fragment contained two ORFs (64 and 32 codons; 10-codon overlap) and was a 99% match to that of the corresponding 450 bp from SCE263, a ctx-negative isolate. It did not match other sequences in the current GenBank database. Based on position relative to other CTXφ sequences, we suggest that this sequence also encodes a new prophage repressor, and we provisionally designate it rstR-4*. This sequence was also a 95% match to a 450-bp fragment from SCE223, which, however, contains one continuous ORF of 86 codons [due to one additional T in a poly(T) tract; T7 in SCE223 and T6 in SCE188]; we designate this variant sequence rstR-4**.
FIG. 9.
ORF structures of putative repressor genes (rstR regions) of CTXφ-related prophages inferred from sequences of PCR products. These two new sequences are designated as the fourth and fifth members of a family of CTXφ-related prophage repressor genes and are therefore designated rstR-4 and rstR-5, pending tests for repressor function.
The sequence of the PCR product generated from the unusual SEC264 isolate with primers ig-1-F and rstC-R was a 95% match over 243 bp at one end to canonical rstC (GenBank accession no. U83795), which indicated that this PCR product did indeed come from the targeted genomic region and was not spurious. Three other ORFs were also identified in this 2.5-kb PCR product, none of which closely matched known V. cholerae genes. Immediately adjacent to ig-1 (the location of rstR repressor genes) was a 67-codon ORF; we designate it rstR-5 provisionally, based on similarity in position, orientation, and size to other rstR genes and an inference that it might also encode a repressor. Between rstR-5 and rstC were one ORF with 34% protein-level identity to ORF320 of filamentous pseudomonal phage IF1 (accession no. AAC62160) followed by an ORF with 43% protein-level identity to gene V (single-stranded DNA binding protein) of filamentous coliphages M13 and f1 (accession no. AAA32219.1); these were provisionally designated rstA-264 and rstB-264, respectively, based on their positions. ORF320 and gene V are each involved in DNA replication. Whether their homologs in SCE264 also mediate CTXφ prophage replication and integration, as do rstA and rstB of canonical CTXφ prophages, is uncertain, especially because SCE264 seems to contain only this fragment of a prophage genome, not an intact CTXφ-related prophage.
DISCUSSION
The present study of 24 Calcutta-region environmental isolates of V. cholerae, chosen initially because of their unusual combination of tcpA (pilin) or ctx (cholera toxin) gene sequences and O antigens, gives an intriguing glimpse into microbial genetic diversity. Although the strains were of just eight RAPD fingerprint types, six major variants of the VPI pathogenicity island and several different CTXφ prophages or related RS1-type sequences were found in them. In retrospect, the initial screen (for tcpA and ctx) (8) was rather limited: many more types of VPI and CTXφ elements will probably be found when the screen is repeated using additional, perhaps more highly conserved, genes from these elements. Based on current data, however, it is clear that VPI and CTXφ are mosaics, consisting of blocks of sequence that match those in epidemic strains, adjacent to sequences that had not been seen previously. The boundaries between most such segments coincide with ends of genes or of functional domains, a pattern reminiscent of that in lambdoid phage (20). Such mosaicism implies a history of genetic exchange among these elements and among the V. cholerae strains that carry them.
Each of the five isolates that carried ctx genes also contained a new tcpA allele (tcpA-env) (Table 1; Fig. 3). The pilin protein that it encodes differs from that of classical strains at 22% of positions, including 8 of 17 that had been tested by mutation and identified as functionally important (27). Assuming this pilin to be functional, each of its potentially deleterious motifs is probably compensated by other amino acid motifs that also distinguish this pilin from that of classical biotype strains.
Individual TcpA pilin proteins assume a two-domain ladle-like structure, with many copies assembled like overlapping shingles in the mature bacterial pilus (5, 27). The amino-terminal one-third (“handle”) of each is buried within the pilus, and the carboxy-terminal two-thirds (“blade”) is exposed (5). Most divergence among various TcpA proteins and related pilins is found in this carboxy-terminal domain (Fig. 3) (5), suggesting that much of this diversity may be adaptive. The diversity might reflect, for example, natural selection for (i) immune evasion during infection, (ii) phage susceptibility or resistance in the environment, (iii) efficient aggregation in humans or the environment, (iv) adherence to aquatic hosts or abiotic sites, and/or (v) efficiency in other steps in biofilm formation (5, 27, 49).
Isolates of five RAPD types contained a new mosaic allele of toxT, a gene encoding an AraC-type regulatory protein that stimulates transcription of several virulence-associated gene clusters and that helps coordinate bacterial responses to external stimuli (10, 15). The divergence between the new and canonical ToxT proteins is almost exclusively in their amino-terminal halves (Fig. 4), the domain that, by extrapolation from AraC (42), might bind particular metabolites and/or regulatory proteins. It is attractive to imagine that this new ToxT protein helps mediate responses to intracellular signals and environmental stimuli that are distinct from those to which canonical ToxT responds and that this new toxT allele thereby contributes to colonization of particular hosts or abiotic niches.
Four new alleles of an extended regulatory sequence between tcpF and toxT were found. In epidemic strains this F-T sequence contains binding sites for TcpP and ToxR, which each stimulate toxT transcription, and also for H-NS, a protein that diminishes toxT transcription (35, 52). The interplay of these factors (whose own abundance or activity may vary with physiologic state) affects the level of toxT expression and thereby the expression of many genes in response to external conditions. The four new F-T intergenic sequences found here differ markedly from those of epidemic strains, and direct tests will be needed to learn which regulatory factors act on them. Whether the IS element in one of these F-T sequences affects toxT transcription is also not known.
Four new alleles of tcpF were found, none closely related to tcpF of epidemic strains (Fig. 5 and 6; Tables 1 and 3). Although TcpF is a secreted protein and needed for virulence in epidemic strains, its exact role is not understood (Taylor, personal communication). It is tempting to imagine, however, that each variant TcpF protein found here is also functional and that it might contribute to bacterial survival or growth in particular environmental niches.
The finding of ctx genes in environmental isolates raises the possibility that this toxin contributes to V. cholerae growth in certain environmental niches. The ctx genes were embedded in CTX prophages that were also distinct from those of epidemic strains. Since the lysogens we found also contained a new tcpA-env allele, and canonical CTXφ uses TCP as receptors, the new CTXφ might infect via other structures and enjoy a host range distinct from that of the canonical phage. Several isolates seemed to contain only a fragment of CTXφ, probably equivalent to the 2-kb RS1 element that is found next to full-length (∼7-kb) CTXφ prophages in epidemic strains. The finding of an RS1 element in only one isolate of RAPD type 4 suggests interstrain transfer of an RS1-containing DNA segment or its empty site, or the insertion or excision of RS1 as an autonomous element.
Three putative rstR prophage repressor genes were found: one identical to rstR-calcutta, which was present along with rstR-eltor in a recent O139 strain (11, 25), and two others, designated rstR-4 and rstR-5. These new rstR segments are inferred to also encode prophage repressors, based on similarities in position, orientation, and gene size to other well-documented rstR repressor genes (26) and on the general conservation of arrangements of functionally equivalent genes in divergent phage genomes (20). Given such conservation, perhaps the new genes between rstR-5 and rstC in strain SCE264 also function in replication and integration. Among rstR reading frames, most seem either to be very short or interrupted by frameshift mutations (Fig. 9) (25, 26). The possible occurrence of translational frameshifting in this locus and its possible importance (13, 41) have not been tested.
In conclusion, these studies of the VPI pathogenicity islands and CTXφ prophages reinforce a sense that V. cholerae is extremely diverse genetically. We suggest that the VPI and CTXφ elements, certain of which are important during V. cholerae human infection, can also benefit the bacterium in certain other hosts or abiotic sites. The relatively few V. cholerae strains that cause epidemic cholera may have been created by gene transfers that linked genes that facilitate transmission in human populations with those fostering persistence and proliferation in nearby environmental niches. The specialized CTXφ and VPI elements that figure importantly in this scenario are themselves mosaics of genes from different sources. The potential for scrambling of functional modules within these elements and their transmission between strains may also speed bacterial adaptation to diverse or inconstant environments and contribute to the emergence of new, highly virulent strains of V. cholerae in at-risk human societies.
ACKNOWLEDGMENTS
We are indebted to Ron Taylor and Matt Waldor for stimulating discussions and to T. Shimada and A. K. Siddiqui for permission to cite unpublished results.
This research was supported by grants from the Japan International Cooperation Agency (JICA/NICED Project O54-1061-E-O) at NICED, Calcutta; from the Department of Biotechnology, Government of India (no. BT/MB/VAP/3/2/98); and from the U.S. Public Health Service (AI38166, AI49161, DK53727, and P30 DK52574).
REFERENCES
- 1.Akopyanz N, Bukanov N O, Westblom T U, Kresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pyloridetected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basu A, Mukhopadhyay A K, Sharma C, Jyot J, Gupta N, Ghosh A, Bhattacharya S K, Takeda Y, Faruque A S G, Albert M J, Nair G B. Heterogeneity in the organization of the CTX genetic element in strains of Vibrio choleraeO139 Bengal isolated from Calcutta, India and Dhaka, Bangladesh and its possible link to the dissimilar incidence of O139 cholera in the two locales. Microb Pathog. 1998;24:175–183. doi: 10.1006/mpat.1997.0186. [DOI] [PubMed] [Google Scholar]
- 3.Beltran P, Delgado G, Navarro A, Trujillo F, Selander R K, Cravioto A. Genetic diversity and population structure of Vibrio cholerae. J Clin Microbiol. 1999;37:581–590. doi: 10.1128/jcm.37.3.581-590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bik E M, Bunschoten A E, Gouw R D, Mooi F R. Genesis of the novel epidemic Vibrio choleraeO139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 1995;14:209–216. doi: 10.1002/j.1460-2075.1995.tb06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blank T E, Zhong H, Bell A L, Whittam T S, Donnenberg M S. Molecular variation among type IV pilin (bfpA) genes from diverse enteropathogenic Escherichia colistrains. Infect Immun. 2000;68:7028–7038. doi: 10.1128/iai.68.12.7028-7038.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byun R, Elbourne L D, Lan R, Reeves P R. Evolutionary relationships of pathogenic clones of Vibrio choleraeby sequence analysis of four housekeeping genes. Infect Immun. 1999;67:1116–1124. doi: 10.1128/iai.67.3.1116-1124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll P A, Tashima K T, Rogers M B, DiRita V J, Calderwood S B. Phase variation in tcpH modulates expression of the ToxR regulon of Vibrio cholerae. Mol Microbiol. 1997;25:1099–1111. doi: 10.1046/j.1365-2958.1997.5371901.x. [DOI] [PubMed] [Google Scholar]
- 8.Chakraborty S, Mukhopadhyay A K, Bhadra R K, Ghosh A N, Mitra R, Shimada T, Yamasaki S, Faruque S M, Takeda Y, Colwell R R, Nair G B. Virulence genes in environmental strains of Vibrio cholerae. Appl Environ Microbiol. 2000;66:4022–4028. doi: 10.1128/aem.66.9.4022-4028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colwell R R. Global climate and infectious disease: the cholera paradigm. Science. 1996;274:2025–2031. doi: 10.1126/science.274.5295.2025. [DOI] [PubMed] [Google Scholar]
- 10.Cotter P A, DiRita V J. Bacterial virulence gene regulation: an evolutionary perspective. Annu Rev Microbiol. 2000;54:519–526. doi: 10.1146/annurev.micro.54.1.519. [DOI] [PubMed] [Google Scholar]
- 11.Davis B M, Kimsey H H, Chang W, Waldor M K. The Vibrio choleraeO139 Calcutta bacteriophage CTXφ is infectious and encodes a novel repressor. J Bacteriol. 1999;181:6779–6787. doi: 10.1128/jb.181.21.6779-6787.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derbyshire K M, Hwang L, Grindley N D. Genetic analysis of the interaction of the insertion sequence IS903 transposase with its terminal inverted repeats. Proc Natl Acad Sci USA. 1987;84:8049–8053. doi: 10.1073/pnas.84.22.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farabaugh P J. Translational frameshifting: implications for the mechanism of translational frame maintenance. Prog Nucleic Acid Res Mol Biol. 2000;64:131–170. doi: 10.1016/s0079-6603(00)64004-7. [DOI] [PubMed] [Google Scholar]
- 14.Faruque S, Asadulghani M, Rahman M M, Waldor M K, Sack D A. Sunlight-induced propagation of the lysogenic phage encoding cholera toxin. Infect Immun. 2000;68:4795–4801. doi: 10.1128/iai.68.8.4795-4801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faruque S M, Albert M J, Mekalanos J J. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garg P, Ramamurthy T, Ghosh A, Bhattacharya S K, Takeda Y, Nair G B. Molecular shifts within clonal populations of Vibrio cholerae O1 and O139 and its impact on the persistence and spread of cholera. In: Gupta S, Sood O P, editors. Molecular intervention in diseases. New Delhi, India: Yogesh Pracashan; 1998. pp. 43–49. [Google Scholar]
- 17.Grindley N D, Joyce C M. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc Natl Acad Sci USA. 1980;77:7176–7180. doi: 10.1073/pnas.77.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hase C C, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M D, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White O. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juhala R J, Ford M E, Duda R L, Youlton A, Hatfull G F, Hendrix R W. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 21.Kaper J B, Morris J G, Jr, Levine M M. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karaolis D K, Lan R, Reeves P R. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J Bacteriol. 1995;177:3191–3198. doi: 10.1128/jb.177.11.3191-3198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karaolis D K R, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio choleraepathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karaolis D K R, Somara S, Maneval D R, Jr, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 25.Kimsey H, Nair G B, Ghosh A, Waldor M K. Diverse CTXφs and evolution of new pathogenic Vibrio cholerae. Lancet. 1998;353:457–458. doi: 10.1016/S0140-6736(05)79193-5. [DOI] [PubMed] [Google Scholar]
- 26.Kimsey H H, Waldor M K. CTXφ immunity: application in the development of cholera vaccines. Proc Natl Acad Sci USA. 1998;95:7035–7039. doi: 10.1073/pnas.95.12.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirn T J, Lafferty M J, Sandoe C M, Taylor R K. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol Microbiol. 2000;35:896–910. doi: 10.1046/j.1365-2958.2000.01764.x. [DOI] [PubMed] [Google Scholar]
- 28.Krukonis E S, Yu R R, DiRita V J. The Vibrio choleraeToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol Microbiol. 2000;38:67–84. doi: 10.1046/j.1365-2958.2000.02111.x. [DOI] [PubMed] [Google Scholar]
- 29.Lobitz B, Beck, L. L, Huq A, Wood B, Fuchs G, Faruque A S, Colwell R R. Climate and infectious disease: use of remote sensing for detection of Vibrio choleraeby indirect measurement. Proc Natl Acad Sci USA. 2000;97:1438–1443. doi: 10.1073/pnas.97.4.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris J G., Jr Non-O group 1 Vibrio cholerae: a look at the epidemiology of an occasional pathogen. Epidemiol Rev. 1990;12:179–191. doi: 10.1093/oxfordjournals.epirev.a036052. [DOI] [PubMed] [Google Scholar]
- 31.Nair G B, Sarkar B L, De S P, Chakraborti M K, Bhadra R K, Pal S C. Ecology of Vibrio choleraein the freshwater environs of Calcutta, India. Microb Ecol. 1988;15:203–216. doi: 10.1007/BF02011713. [DOI] [PubMed] [Google Scholar]
- 32.Nair G B, Oku Y, Takeda Y, Ghosh A, Ghosh R K, Chattopadhyay S, Pal S C, Kaper J B, Takeda T. Toxin profiles of Vibrio choleraenon-O1 from environmental sources in Calcutta, India. Appl Environ Microbiol. 1988;54:3180–3182. doi: 10.1128/aem.54.12.3180-3182.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nandi B, Nandy R K, Vicente A C, Ghosh A C. Molecular characterization of a new variant of toxin-coregulated pilus protein (TcpA) in a toxigenic non-O1/non-O139 strain of Vibrio cholerae. Infect Immun. 2000;68:948–952. doi: 10.1128/iai.68.2.948-952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novais R C, Coelho A, Salles C A, Vincente A C P. Toxin co-regulated pilus cluster in non-O1, non-toxigenic Vibrio cholerae: evidence of a third allele of pilin gene. FEMS Microbiol Lett. 1999;171:49–55. doi: 10.1111/j.1574-6968.1999.tb13411.x. [DOI] [PubMed] [Google Scholar]
- 35.Nye M B, Pfau J D, Skorupski K, Taylor R K. Vibrio choleraeH-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J Bacteriol. 2000;182:4295–4303. doi: 10.1128/jb.182.15.4295-4303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogierman M A, Zabihi S, Mourtzios L, Manning P A. Genetic organization and sequence of the promoter-distal region of the tcp gene cluster of Vibrio cholerae. Gene. 1993;126:51–60. doi: 10.1016/0378-1119(93)90589-u. [DOI] [PubMed] [Google Scholar]
- 37.Ramamurthy T, Bag P K, Pal A, Bhattacharya S K, Bhattacharya M K, Sen D, Shimada T, Takeda T, Takeda Y, Nair G B. Virulence patterns of Vibrio choleraenon-O1 strains isolated from hospitalised patients with acute diarrhoea in Calcutta, India. J Med Microbiol. 1993;39:310–317. doi: 10.1099/00222615-39-4-310. [DOI] [PubMed] [Google Scholar]
- 38.Ramamurthy T, Garg S, Sharma R, Bhattacharya S K, Nair G B, Shimada T, Takeda T, Karasawa T, Kurazano H, Pal A, Takeda Y. Emergence of novel strain of Vibrio choleraewith epidemic potential in southern and eastern India. Lancet. 1993;341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 39.Rhine J A, Taylor R K. TcpA pilin sequences and colonization requirements for O1 and O139 Vibrio cholerae. Mol Microbiol. 1994;13:1013–1020. doi: 10.1111/j.1365-2958.1994.tb00492.x. [DOI] [PubMed] [Google Scholar]
- 40.Samadi A R, Shahid N, Eusuf A, Yunus M, Huq M I, Khan M U, Rahman A S M M, Faruque A S G. Classical Vibrio choleraebiotype displaces El Tor in Bangladesh. Lancet. 1983;i:805–807. doi: 10.1016/s0140-6736(83)91860-3. [DOI] [PubMed] [Google Scholar]
- 41.Sekine Y, Ohtsubo E. Frameshifting is required for production of the transposase encoded by insertion sequence 1. Proc Natl Acad Sci USA. 1989;86:4609–4613. doi: 10.1073/pnas.86.12.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soisson S M, MacDougall-Shackleton B, Schleif R, Wolberger C. The 1.6 A crystal structure of the AraC sugar-binding and dimerization domain complexed with d-fucose. J Mol Biol. 1997;273:226–237. doi: 10.1006/jmbi.1997.1314. [DOI] [PubMed] [Google Scholar]
- 43.Sozhamannan S, Deng Y K, Li M, Sulakvelidze A, Kaper J B, Johnson J A, Nair G B, Morris J G., Jr Cloning and sequencing of the genes downstream of the wbf gene cluster of Vibrio choleraeserogroup O139 and analysis of the junction genes in other serogroups. Infect Immun. 1999;67:5033–5040. doi: 10.1128/iai.67.10.5033-5040.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Speelmon E C, Checkley, W. W, Gilman R H, Patz J, Calderon M, Manga S. Cholera incidence and El Nino-related higher ambient temperature. JAMA. 2000;283:3072–3074. [PubMed] [Google Scholar]
- 45.Stine O C, Sozhamannan S, Gou Q, Zheng S, Morris J G, Jr, Johnson J A. Phylogeny of Vibrio cholerae based on recAsequence. Infect Immun. 2000;68:7180–7185. doi: 10.1128/iai.68.12.7180-7185.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tauxe T, Seminario L, Rapia R, Libel M. The Latin American epidemic, p 321–344. In: Wachsmuth K, Blake P A, Olsvik O, editors. Vibrio cholerae and cholera: molecular to global perspectives. Washington, D.C.: ASM Press; 1994. [Google Scholar]
- 47.Waldor M K, Mekalanos J J. toxR regulates virulence gene expression in non-O1 strains of Vibrio choleraethat cause epidemic cholera. Infect Immun. 1994;62:72–78. doi: 10.1128/iai.62.1.72-78.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 49.Watnick P, Kolter R. Biofilm, city of microbes. J Bacteriol. 2000;182:2675–2679. doi: 10.1128/jb.182.10.2675-2679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamai S, Okitsu T, Shimada T, Katsube Y. Distribution of serogroups of Vibrio choleraenon-O1 non-O139 with specific reference to their ability to produce cholera toxin and additional of novel serogroups. Jpn J Assoc Infect Dis. 1997;71:1037–1045. doi: 10.11150/kansenshogakuzasshi1970.71.1037. [DOI] [PubMed] [Google Scholar]
- 51.Yamasaki S, Shimizu T, Hoshino K, Ho S T, Shimada T, Nair G B, Takeda Y. The genes responsible for O-antigen synthesis of Vibrio cholerae O139 are closely related to those of Vibrio choleraeO22. Gene. 1999;237:321–332. doi: 10.1016/s0378-1119(99)00344-3. [DOI] [PubMed] [Google Scholar]
- 52.Yu R R, DiRita V J. Analysis of an autoregulatory loop controlling ToxT, cholera toxin, and toxin-coregulated pilus production in Vibrio cholerae. J Bacteriol. 1999;181:2584–2592. doi: 10.1128/jb.181.8.2584-2592.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]