Abstract
Regulation of gene expression can be analyzed by a number of different techniques. Some techniques monitor the level of specific mRNA directly, and others monitor indirectly by determining the level of enzymes encoded by the mRNA. Each method has its own inherent way of normalization. When results obtained by these techniques are compared between experiments in which differences in growth rates, strains, or stress treatments occur, the normalization procedure may have a significant impact on the results. In this report we present a solution to the normalization problem in RNA slot blotting experiments, in which mRNA levels routinely are normalized to a fixed amount of extracted total RNA. The cellular levels of specific mRNA species were estimated using a renormalization with the total RNA content per cell. By a combination of fluorescence in situ rRNA hybridization, which estimates the relative level of rRNA per cell, and slot blotting to rRNA probes, which estimates the level of rRNA per extracted total RNA, the amount of RNA per cell was calculated in a series of heat shock experiments with the gram-positive bacterium Lactococcus lactis. It was found that the level of rRNA per cell decreased to 30% in the course of the heat shock. This lowered ribosome level led to a decrease in the total RNA content, resulting in a gradually increasing overestimation of the mRNA levels throughout the experiment. Using renormalized cellular mRNA levels, the HrcA-mediated regulation of the genes in the hrcA-grpE-dnaK operon was analyzed. The hybridization data suggested a complex heat shock regulation indicating that the mRNA levels continued to rise after 30 min, but after renormalization the calculated average cellular levels exhibited a much simpler induction pattern, eventually attaining a moderately increased value.
RNA blotting techniques such as Northern blotting and dot or slot blotting are widely recognized and used as means of estimating differential gene expression in bacteria. Through determinations of hybridization signals with specific probes, the amount of specific mRNA can be determined. Although RNA blotting often is found to be well suited for estimating levels of specific mRNA molecules from populations of cells, there are conditions in which this technique might be compromised. This is due to the standard way of normalization, which is performed by applying equal amounts of RNA from all samples to be investigated on a filter or gel (e.g., 0.5 μg of total RNA). rRNA constitutes up to 80% of the total RNA in a cell, and the actual cellular content of rRNA therefore represents a dominant part of the total RNA applied on the filter in a slot blot analysis. Thus, RNA blotting monitors not only mRNA regulation but also carries information about rRNA levels.
It has previously been shown that some rRNA species are degraded during a heat shock. In 1970, Rosenthal and Iandolo (15) described a heat-induced dissociation of the 30S particle and degradation of 16S rRNA in Staphylococcus aureus. Interestingly, neither the 50S particle nor the 23S rRNA was degraded under these conditions. Similar degradation patterns have been found for Bacillus subtilis and Salmonella enterica serovar Typhimurium (7, 13, 22). These studies were performed using sucrose gradient centrifugation of ribosomes and agarose gel electrophoresis of rRNA. Recently, the heat shock-dependent degradation pattern for S. enterica serovar Typhimurium 16S and 23S rRNA was confirmed using fluorescence in situ hybridization (FISH) (20, 21), which clearly demonstrated the effect of heat shock on the rRNA content at the single-cell level.
Variations in the cellular content of rRNA may occur over a range of conditions. Besides heat shock, nutrient starvation and altered growth rates result in changes in the rRNA content. Studies on Escherichia coli starved for a carbon source (9, 10, 12) or inorganic ions (2, 9, 19) showed degradation of rRNA as well. The almost linear relationship between growth rate and rRNA content was demonstrated as early as 1958 by Schaechter et al. (18) in S. enterica serovar Typhimurium, and later this growth dependency was confirmed by FISH analysis of E. coli (3) and Pseudomonas putida (14). Bacteria may thus have various contents of rRNA, and perturbations of the conditions for the cells, e.g., the application of heat shock, shifts in growth rate, or the introduction of certain mutations, may cause changes in the rRNA content. This may have further impact on the determinations of specific mRNA molecules in RNA slot blot experiments and may introduce a source of errors. A decrease of the total cellular RNA (rRNA) to 50% during an experiment would lead to a twofold overestimate of the mRNA level in a standard hybridization analysis due to the application of RNA from twice the number of cells.
These problems of RNA slot blotting due to the standard normalization procedure have been assessed here for cultures of Lactococcus lactis subsp. cremoris MG1363 subjected to heat shock. Through the combination of RNA slot blot analysis and FISH, improved normalization data were obtained. The FISH analysis provided information about the cellular content of rRNA, which were used to renormalize the data obtained from the RNA slot blot analysis.
The presented focus on the normalization procedure not only is relevant in relation to techniques like Northern blotting analysis but it might also prove to be of substantial significance in future technologies as well. When using DNA microarrays for determinations of specific mRNA levels, two experimental situations are often compared, i.e., RNA is extracted from two systems and applied to the DNA microarrays (5). As for Northern blotting, most often the data are normalized to total RNA, since fixed amounts of total RNA are applied to the arrays.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
Wild-type Lactococcus lactis subsp. cremoris MG1363 (6) was grown in the SA medium of Jensen and Hammer (8) supplemented by 1% glucose (GSA). Cultures used for heat shock experiments were grown exponentially without shaking for at least 10 h at 30°C in 200 ml of GSA without exceeding an optical density at 450 nm (OD450) of 0.5. Before transfer to 43°C, aliquots of the cultures were put into 50-ml plastic tubes, each containing 50 ml of culture, which results in a gradual increase in temperature to 43°C over 5 min (11). Samples (20 ml) were taken 0, 5, 10, 20, 35, and 60 min after the transfer to 43°C.
Oligonucleotides.
The oligonucleotide probes used in the present study are shown in Table 1. Detection of 16S rRNA by FISH and by slot blotting was performed using the sequence-identical oligonucleotide probes LLC68Cy3 and AN64 specific for L. lactis subsp. cremoris (16), while the sequence-identical probes for detection of 23S rRNA were EUB1933 and AN65, specific for the bacterial domain (1). Both LLC68Cy3 and EUB1933 were monolabeled with the indocarbocyanine fluorescent dye Cy3 for use in FISH and were purchased from Hobolth DNA Syntese (Hillerød, Denmark). Primers for slot blotting were purchased from DNA Technology (Århus, Denmark).
TABLE 1.
Oligonucleotides
| Primer | Sequencea | Targetb | Reference |
|---|---|---|---|
| AN47F | GTTACGCACTGAAATTCCAC | L. lactis hrcA (828) | 4 |
| AN48rT7 | TAATACGACTCACTATAGCTACAGGTCCTACAACTG | L. lactis hrcA (1174) | 4 |
| AN49F | GCAAAATGTGCAACGTCGCG | L. lactis grpE (1496) | 4 |
| AN50rT7 | TAATACGACTCACTATAGCTGTCAGCTGGATGTTCG | L. lactis grpE (1781) | 4 |
| AN51F | GCAGTTATCACTGTTCCTGC | L. lactis dnaK (2264) | 4 |
| AN52rT7 | TAATACGACTCACTATAGCTTGACCAAGGTCGATTCCG | L. lactis dnaK (2603) | 4 |
| AN64 | TGCAAGCACCAATCTTCATC | L. lactis subsp. cremoris 16S rRNA | 16 |
| AN65 | ACCCGACAAGGAATTTCGC | Eubacterial 23S rRNA | 1 |
| AN66F | TTGGCTTATTGGCAGTTGC | L. lactis dnaK (∼4336) | 4 |
| AN67rT7 | TAATACGACTCACTATAGCTGACAGCCCAATTGCCG | L. lactis dnaK (∼4675) | 4 |
| AN68R | AGCACTCTTTACCTCTAAG | L. lactis hrcA | 4 |
| LLC68Cy3 | Cy3-TGCAAGCACCAATCTTCATC | L. lactis subsp. cremoris 16S rRNA | 16 |
| EUB1933 | Cy3-ACCCGACAAGGAATTTCGC | Eubacterial 23S rRNA | 1 |
The nucleotide sequences are shown with the 5′ end to the left. T7 polymerase tail sequences are in bold.
Numbers in parentheses refer to the starting position according to the nucleotide sequence of the dnaK operon (4).
RNA isolation.
RNA was extracted from approximately 109 cells by use of the FastRNA Kit-BLUE (Bio 101) according to the manufacturer's instructions. All RNA preparations were treated with RNase-free DNase (Roche), extracted once with phenol and once with phenol-CHCl3 (1:1), and resuspended in water.
In vitro RNA synthesis.
Gene-specific riboprobes were synthesized in vitro by transcription of PCR-generated DNA templates. The T7 promoter sequence (TAATACGACTCACTATA) was incorporated in the DNA templates via the downstream PCR primers. In vitro transcription was performed at 37°C for 1 h in a transcription mixture containing 2 μg of the PCR product, 20 U of T7 RNA polymerase (Promega), 1× transcription buffer (Promega), 10 μM dithiothreitol, 20 U of RNasin (Promega), 0.5 mM (each) ATP, UTP, and GTP, 12 μM CTP, 2 μl of [α-32P]CTP (10 μCi/μl, >400 Ci/mmol), and water to 20 μl. One unit of RNase-free DNase (Roche) was added and the mixture was incubated for 15 min at 37°C before it was added directly to the hybridization buffer.
Slot blotting.
Immediately before use, 500 ng of DNase-treated RNA was lyophilized and resuspended in 0.5 ml of a denaturing solution containing 10 mM NaOH and 1 mM EDTA. The RNA samples were blotted onto Zeta-probe nylon membranes (Bio-Rad) by use of a Bio-Dot SF slot blot apparatus (Bio-Rad). After a brief rinse in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (17) plus 0.1% sodium dodecyl sulfate (SDS) for 1 min at room temperature, the membrane was air dried for 10 min and fixed by microwave heating for 2 min at 950 W. The membranes were prehybridized for 2 h in a hybridization buffer containing 1 mM NaCl, 4 mM Na2P4O7, 5× Denhardt's solution (17), 1% SDS, 10% (wt/vol) polyethylene glycol 6000, 50 mM Tris-HCl (pH 7.5), 250 μg of yeast tRNA per ml, and 50% formamide, after which the 32P-labeled riboprobe was added. After an overnight hybridization at 42°C, the membranes were washed twice at room temperature in 2× SSC for 5 min, twice at 65°C in 0.2× SSC plus 1% SDS for 30 min, and twice at 65°C in 0.1× SSC for 30 min. After being washed, the membrane was wrapped in plastic wrap. The amount of radioactivity retained by the riboprobes on the membranes was measured in a Packard Instant Imager.
Cell fixation.
For cell fixation, 300 μl of bacterial culture at an OD450 of approximately 0.7 (according to the time points of sampling as described above) was added to 900 μl of freshly prepared 4% paraformaldehyde in 130 mM NaCl in 10 mM phosphate buffer, pH 7.2 (PBS). The suspension was vortexed for 2 min and incubated for 20 h at 4°C. After incubation, cells were pelleted by centrifugation at 15,000 × g for 5 min and the supernatant was discarded. The cellular pellet was then resuspended in 900 μl of PBS, and after the addition of 100 μl of 0.1% Nonidet P-40 (NP-40) (Sigma) the cell suspension was vortexed for 2 min. Cells were harvested by centrifugation at 15,000 × g for 5 min, and the supernatant was discarded. Five hundred microliters of 0.1% NP-40 was added, and the cells were vortexed for 2 min. Cells were harvested by centrifugation at 6,000 × g for 5 min and resuspended in 100 μl of 2× storage buffer (0.2% NP-40 in 40 mM Tris [pH 7.5]) by vortexing for 2 min. Finally, 100 μl of 96% ethanol was added and the suspension was vortexed before storage at −20°C.
Cell wall permeabilization.
Prior to in situ hybridization, the fixed cells were permeabilized with lysozyme. An amount of cells optimal for subsequent microscopic examination (approximately 3 μl) was applied on a 6-well, heavy Teflon-coated, acid alcohol-washed, and poly-l-lysin-coated slide (14) and dried at 37°C for 30 min. Twenty microliters of lysozyme solution (1 mg of lysozyme/ml in 100 mM Tris [pH 7.5], 50 mM EDTA) was added to each well, and the slides were incubated at 0°C for 20 min in a sealed container (50-ml centrifuge tube). Except for the RNase-treated negative control, the slides were gently rinsed with distilled water (dH2O) and dehydrated by sequential washes in 50, 80, and 96% ethanol (3 min each) and air dried. For the RNase-treated control, 20 μl of RNase (0.5 mg of RNase/ml [Sigma] in 100 mM Tris [pH 7.5]–50 mM EDTA) was added to each well. The slide was incubated at 20°C for 30 min, gently rinsed with dH2O, and dehydrated as described above.
In situ hybridization.
The permeabilized and dried cells were hybridized as follows. Ten microliters of hybridization mixture (15% formamide, 0.9 M NaCl, 100 mM Tris [pH 7.2], 0.1% SDS) containing 50 ng of the probe was added to each hybridization well. Cells were incubated with hybridization solution for 16 h at 37°C in a moisture chamber (50-ml centrifuge tube containing 1 ml of the hybridization mixture). For washing, the slides were submersed in 100 ml of the following washing solutions. First, the slides were washed in washing solution I (15% formamide, 0.9 M NaCl, 100 mM Tris [pH 7.2], 0.1% SDS) for 20 min at 37°C. Then they were washed for 15 min in washing solution II (0.1 M Tris [pH 7.2], 0.9 M NaCl) at 37°C, before a final rinsing in 100 ml of dH2O. For the initial titration of the probe, formamide concentrations of 10, 15, 20, 30, and 40% were used.
Microscopy.
Hybridized samples were analyzed by use of a Carl Zeiss Axioplan epifluorescence microscope. The excitation source was a 100-W HBO bulb, and digital images were captured with a 12-bit cooled slow-scan charge-coupled device camera (KAF 1400 chip; Photometrics, Tucson, Ariz.). The charge-coupled device camera was controlled by PMIS software (Photometrics). Cy3 was visualized by use of filter sets XF40 (BP 560/40 excitation filter, 590-nm dichroic filter, and LP 600 emission filter; Omega Opticals, Brattleboro, Vt.). For each sample, five images (arbitrarily chosen areas) were captured, each containing approximately 200 cells, yielding a total of approximately 1,000 cells for statistical analysis of each sample.
Image processing (Cellstat).
The amount of fluorescence from Cy3-labeled cells was quantified by use of the Cellstat image analysis program (http://www.im.dtu.dk/cellstat/index.html) (14). The bases for Cellstat object recognition are cell size (area in pixels) and various shape parameters. Settings were chosen to allow identification of single cells as well as small chains of cells. The fluorescence intensity for each cell was calculated per unit of area, thus compensating for the increased fluorescence from chains of cells.
RESULTS AND DISCUSSION
Investigation of mRNA synthesis rates.
In order to document specific gene regulation it would be desirable to determine the frequency of successfully transcribing RNA polymerases that initiate from a certain promoter. However, since the normalization of mRNA synthesis to a promoter or a gene is very difficult to obtain, the general normalization to total cellular mass, dry weight, total protein content, or total RNA content is more widely used. In the standard Northern blot, or any other RNA blotting procedure, a constant amount of total extracted RNA from each sample is usually applied to the membrane. When the resulting mRNA levels calculated from hybridization signals are compared between samples, e.g., between cells grown under different conditions, the fold of regulation is usually calculated directly from these total RNA normalized levels. This direct way of documenting genetic regulation may be appropriate under some conditions where the total amount of RNA does not vary significantly in the two compared situations. If, however, the total cellular RNA concentration varies or is unknown, the standard representation could be misleading.
rRNA constitutes the largest fraction of RNA in the cell (up to approximately 80%). Thus, any fluctuation of the cellular rRNA content will influence the total RNA content, thereby causing normalization errors. By FISH analysis of rRNA, estimates of the rRNA content per cell (rRNA/cell) can be obtained. Accordingly, rRNA slot blot analysis determines the amount of rRNA per total RNA (rRNA/total RNA). A combination (division) of these data yields estimates of the cellular total RNA (total RNA/cell), which is an important parameter for interpretation of RNA slot blots. In order to demonstrate this, the amounts of rRNA and specific mRNA species were assessed during a heat shock by application of both RNA slot blotting and FISH.
Estimation by FISH of 16S and 23S rRNA levels in single cells during heat shock.
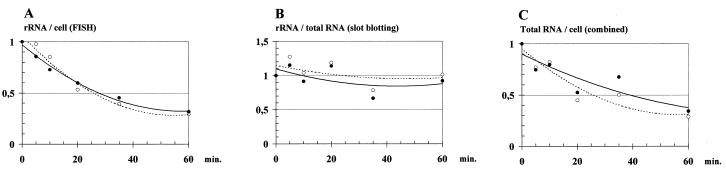
The level of rRNA in L. lactis subsp. cremoris MG1363 before and after a heat shock from 30 to 43°C was investigated by the use of FISH. A culture was grown exponentially for at least eight generations at 30°C, and samples were taken at 0, 5, 10, 20, 35, and 60 min relative to the transfer of the culture to 43°C. The fluorescent probes used in the experiment were specific for either 16S or 23S rRNA. The 16S rRNA probe recognized only L. lactis subsp. cremoris rRNA, while the 23S rRNA probe was specific for all eubacterial 23S rRNA sequences (see Materials and Methods). The FISH analysis demonstrated that the cellular level of both rRNA species decreased during the heat shock (Fig. 1A). After 60 min at 43°C the amount of rRNA was reduced to approximately 30% of the value before the heat shock. Thus, at this time point individual L. lactis cells contained less than one-third of the preshift amount of rRNA.
FIG. 1.
RNA levels in L. lactis during heat shock. MG1363 was grown exponentially at 30°C for at least eight generations. At time zero, the culture was transferred to the sublethal temperature, 43°C. Cells were harvested at 0, 5, 10, 20, 35, and 60 min. (A) FISH analysis of the cellular levels of 16S and 23S rRNA. (B) Slot blot analysis of the levels of 16S and 23S rRNA in samples containing 0.5 μg of total RNA. (C) Average cellular RNA levels (total RNA/cell), calculated by dividing levels from panel A (rRNA/cell) by levels from panel B (rRNA/total RNA). Open and filled circles indicate 16S and 23S rRNA, respectively. The levels are given relative to the level at time zero.
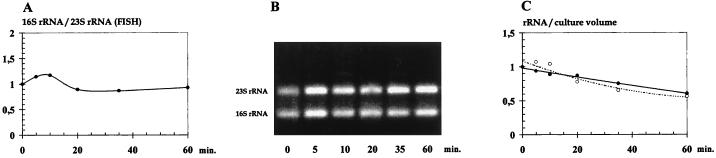
The data presented above (and in Fig. 1) show that the 16S and 23S rRNA molecules are degraded at similar rates during heat shock. This finding contradicts previous findings for B. subtilis, S. enterica serovar Typhimurium, and S. aureus (7, 13, 15, 20, 21), where the 23S rRNA was found to be stable and the 16S rRNA rapidly degraded during heat shock. In Fig. 2A it is clear that the relative amount of the two rRNA species remains constant during the heat shock. This result was confirmed by agarose gel electrophoresis of total RNA (Fig. 2B) where the relative intensities of the bands corresponding to 16S and 23S rRNA remained constant during the heat shock.
FIG. 2.
Equal degradation of 16S and 23S rRNA during heat shock. MG1363 was grown exponentially at 30°C for at least eight generations. At time zero, the culture was transferred to the sublethal temperature, 43°C. Cells were harvested at 0, 5, 10, 20, 35, and 60 min. (A) Relative cellular levels of 16S and 23S rRNA, calculated from the data shown in Fig. 1A by division of the 16S levels by the 23S levels. (B) Agarose gel analysis showing the relative abundance of 16S and 23S rRNA in (0.5 μg) of total RNA extracts used in the slot blotting experiments. (C) Absolute levels of 16S and 23S rRNA per culture volume during heat shock. The values were calculated by multiplying the cellular levels of rRNA shown in Fig. 1A with the relative OD450 of the culture at the corresponding time. The combined value ([rRNA/cell] × [cells/ml] = rRNA/ml) shows whether rRNA is distributed to daughter cells (constant values) or whether it is degraded (decreasing values).
In order to investigate the fate of surplus ribosomes during a heat shock treatment of a culture of L. lactis, the absolute levels of rRNA and total RNA were determined. If ribosome synthesis stops completely, the total amount of ribosomes in the culture will remain constant for a period of time. If ribosomes are degraded, however, the absolute level of ribosomes will decrease. The amount of rRNA per cell, determined by FISH, was multiplied by the OD450 of the culture at the corresponding time point, and the combined value was taken as a measure of the absolute amount of rRNA per volume of culture (i.e., the total concentration of rRNA). The data from this investigation are shown in Fig. 2C, from which it is clear that the absolute rRNA level slowly decreases during the heat shock to reach an rRNA level after 60 min at approximately 60% of the unstressed level. This indicated that the loss of 40% of the ribosomes was due to degradation rather than dilution.
A combination of FISH and RNA slot blot yields the average total RNA per cell.
RNA slot blotting was used to estimate the amounts of 16S and 23S rRNA during heat shock by using a standard procedure. The probes used in this population analysis and the single-cell analysis described above were identical. Apparently, the levels of both 16S and 23S rRNA (Fig. 1B) were constant during the heat shock, in contrast to the findings on the single-cell level (Fig. 1A). This contradiction can be explained by the normalization of the slot blot technique, according to which a fixed amount of total RNA (0.5 μg) was applied to the filter for each sample.
As described above, the FISH analysis gave an estimate of the rRNA per cell and the slot blot analysis gave an estimate of the rRNA level per total RNA. A combination (division) of the two sets of results would yield the average total RNA level per cell. Not surprisingly, the results presented in Fig. 1C showed, through this manipulation, that the average total RNA per cell decreased during a heat shock. After 60 min of heat shock the total RNA level was decreased to approximately 30%. This finding is of great importance for mRNA blotting experiments, since RNA slot blots, as discussed above, usually are normalized to the amount of total RNA by following the standard procedure.
Normalization to cellular RNA is a prerequisite for estimation of mRNA levels.
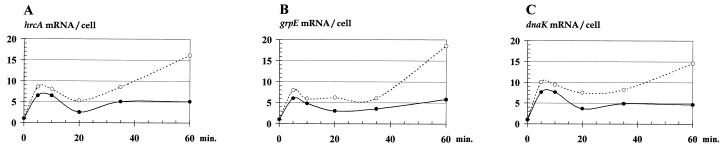
The need for renormalization is, thus, obvious when RNA slot blots are used to estimate amounts of single specific mRNA species during a heat shock, since the degradation of rRNA seriously affects the calculations in slot blot mRNA quantifications. Determination of the cellular content of RNA during heat shock provides a solution to this problem. In order to demonstrate this, the amounts of mRNA expressed for all genes in the L. lactis dnaK operon (4) were quantified by hybridization to slot blots identical to those used for rRNA slot blotting (Fig. 1B). In Fig. 3, the temporal expression patterns for the hrcA (Fig. 3A), grpE (Fig. 3B), and dnaK (Fig. 3C) genes are shown, both normalized to the amount of total RNA (dotted lines) and renormalized, using data for total cellular RNA levels, to yield average cellular mRNA levels.
FIG. 3.
hrcA, grpE, and dnaK mRNA levels during heat shock analyzed by slot blotting. MG1363 was grown exponentially at 30°C for at least eight generations. At time zero, the culture was transferred to the sublethal temperature, 43°C. Cells were harvested at 0, 5, 10, 20, 35, and 60 min. Levels of mRNA in RNA samples containing 0.5 μg of total RNA were analyzed by slot blotting using riboprobes specific for the hrcA (A), grpE (B), or dnaK (C) genes. Values for each mRNA species are given relative to the levels at time zero. Open circles represent values solely from slot blot hybridization experiments. Filled circles represent renormalized average cellular mRNA levels. The latter values were calculated by multiplication of the slot blotting levels with the cellular RNA levels (from Fig. 1C): mRNA/cell = (mRNA/total RNA) × (total RNA/cell).
Based on the uncorrected data, the temporal pattern of hrcA, grpE, and dnaK mRNA levels during heat shock appears to be biphasic. After a fast increase and decrease during the first 30 min after heat shock, transcription continues to constantly increase. When renormalized, another (and more comprehensible) pattern is revealed, showing no increase after 30 min. This latter pattern is in close agreement with the temporal pattern for DnaK synthesis, as analyzed by two-dimensional polyacrylamide gel electrophoresis (11). The application of the conventional RNA slot blot analysis thus leads to a vast overestimation of the contents of the specific mRNA due to the degradation of rRNA, resulting in a distorted temporal pattern of regulation. Therefore, knowledge of the cellular RNA concentration is a prerequisite for meaningful mRNA estimations.
Conclusion.
By combination of two common hybridization techniques, RNA slot blotting and fluorescence in situ rRNA hybridization, estimates of the amounts of total RNA per cell in the course of a heat shock treatment of a culture of L. lactis were obtained (Fig. 1C). The close agreement between the values of total RNA per cell calculated from either the 16S or 23S rRNA data lends credit to the procedure. The use of this information to renormalize the slot blot data provided more precise estimates of the temporal pattern of the levels of specific mRNA species, as illustrated in Fig. 3. It could be argued that the procedure is unnecessarily laborious, since the rRNA constitutes the major part of the total RNA, and the quantification of rRNA by slot blotting, therefore, would maximally result in a 5 to 10% change during renormalization. Without sacrificing much accuracy, the rRNA FISH data could represent the relative amount of total RNA per cell and thus could be used for renormalization. This was clearly the case when data from Fig. 1A was compared to data from Fig. 1C, but we recommend the inclusion of the extra rRNA slot blot, because errors in RNA quantification would be neutralized in the subsequent division. In Fig. 1B, the rRNA levels per amount of total RNA vary within 80 and 120% of the unstressed level. We believe that this variation is due to errors in RNA quantification by UV absorbency. If this is the case, the mRNA slot blotting experiments would have the same problem. By renormalization, however, the errors are neutralized when estimating the average cellular mRNA levels.
In studies of gene regulation of heat shock-induced genes in different strains (e.g., regulatory mutants), renormalization also makes it possible to compare amounts of specific mRNA species between strains. Some mutations may influence and alter the behavior of ribosome degradation, and preliminary data from a heat shock experiment using an hrcA mutant (data not shown) suggests a severe degradation of rRNA resulting in a 500% overestimation of specific mRNA expression if not renormalized to total cellular RNA. The demonstrated fate of rRNA in L. lactis during heat shock focuses the attention to the normalization procedure used in techniques like Northern analysis, at least in heat shock experiments exceeding 10 min, but it is also clear that when studying other stress forms, or when introducing an altered growth rate, the standard normalization could pose a serious problem.
ACKNOWLEDGMENTS
This work was supported by a grant from the Danish Research and Development Program for Food Technology (FØTEK3) through the Center for Advanced Food Studies (LMC) and grants from the Danish Biotechnology Program.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis B D, Luger S M, Tai P C. Role of ribosome degradation in the death of starved Escherichia coli cells. J Bacteriol. 1986;166:439–445. doi: 10.1128/jb.166.2.439-445.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. . (Erratum, 245:1312.) [DOI] [PubMed] [Google Scholar]
- 4.Eaton T, Shearman C, Gasson M. Cloning and sequence analysis of the dnaK gene region of Lactococcus lactis subsp. lactis. J Gen Microbiol. 1993;139:3253–3264. doi: 10.1099/00221287-139-12-3253. [DOI] [PubMed] [Google Scholar]
- 5.Eisen M B, Brown P O. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- 6.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurst A, Hughes A. Stability of ribosomes of Staphylococcus aureus S6 sublethally heated in different buffers. J Bacteriol. 1978;133:564–568. doi: 10.1128/jb.133.2.564-568.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen P R, Hammer K. Minimal requirements for exponential growth of Lactococcus lactis. Appl Environ Microbiol. 1993;59:4363–4366. doi: 10.1128/aem.59.12.4363-4366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan R, Apirion D. Decay of ribosomal ribonucleic acid in Escherichia coli cells starved for various nutrients. J Biol Chem. 1975;250:3174–3178. [PubMed] [Google Scholar]
- 10.Kaplan R, Apirion D. The fate of ribosomes in Escherichia coli cells starved for a carbon source. J Biol Chem. 1975;250:1854–1863. [PubMed] [Google Scholar]
- 11.Kilstrup M, Jacobsen S, Hammer K, Vogensen F K. Induction of heat shock proteins DnaK, GroEL, and GroES by salt stress in Lactococcus lactis. Appl Environ Microbiol. 1997;63:1826–1837. doi: 10.1128/aem.63.5.1826-1837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maruyama H B, Okamura S. Ribosome degradation and the degradation products in starved Escherichia coli. V. Ribonucleoprotein particles from glucose-starved cells. J Bacteriol. 1972;110:442–446. doi: 10.1128/jb.110.1.442-446.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller L L, Ordal Z J. Thermal injury and recovery of Bacillus subtilis. Appl Microbiol. 1972;24:878–884. doi: 10.1128/am.24.6.878-884.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Møller S, Kristensen C S, Poulsen L K, Carstensen J M, Molin S. Bacterial growth on surfaces: automated image analysis for quantification of growth rate-related parameters. Appl Environ Microbiol. 1995;61:741–748. doi: 10.1128/aem.61.2.741-748.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenthal L J, Iandolo J J. Thermally induced intracellular alteration of ribosomal ribonucleic acid. J Bacteriol. 1970;103:833–835. doi: 10.1128/jb.103.3.833-835.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salama M, Sandine W, Giovannoni S. Development and application of oligonucleotide probes for identification of Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1991;57:1313–1318. doi: 10.1128/aem.57.5.1313-1318.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Schaechter M, Maaloe O, Kjeldgaard N O. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 19.St. John A C, Goldberg A L. Effects of starvation for potassium and other inorganic ions on protein degradation and ribonucleic acid synthesis in Escherichia coli. J Bacteriol. 1980;143:1223–1233. doi: 10.1128/jb.143.3.1223-1233.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tolker-Nielsen T, Larsen M H, Kyed H, Molin S. Effects of stress treatments on the detection of Salmonella typhimurium by in situ hybridization. Int J Food Microbiol. 1997;35:251–258. doi: 10.1016/s0168-1605(97)01242-7. [DOI] [PubMed] [Google Scholar]
- 21.Tolker-Nielsen T, Molin S. Role of ribosome degradation in the death of heat-stressed Salmonella typhimurium. FEMS Microbiol Lett. 1996;142:155–160. doi: 10.1111/j.1574-6968.1996.tb08423.x. [DOI] [PubMed] [Google Scholar]
- 22.Tomlins R I, Ordal Z J. Precursor ribosomal ribonucleic acid and ribosome accumulation in vivo during the recovery of Salmonella typhimurium from thermal injury. J Bacteriol. 1971;107:134–142. doi: 10.1128/jb.107.1.134-142.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]