Abstract
Background/objectives
Admixed populations are a resource to study the global genetic architecture of complex phenotypes, which is critical, considering that non-European populations are severely underrepresented in genomic studies. Here, we study the genetic architecture of BMI in children, young adults, and elderly individuals from the admixed population of Brazil.
Subjects/methods
Leveraging admixture in Brazilians, whose chromosomes are mosaics of fragments of Native American, European, and African origins, we used genome-wide data to perform admixture mapping/fine-mapping of body mass index (BMI) in three Brazilian population-based cohorts from Northeast (Salvador), Southeast (Bambuí), and South (Pelotas).
Results
We found significant associations with African-associated alleles in children from Salvador (PALD1 and ZMIZ1 genes), and in young adults from Pelotas (NOD2 and MTUS2 genes). More importantly, in Pelotas, rs114066381, mapped in a potential regulatory region, is significantly associated only in females (p = 2.76e−06). This variant is rare in Europeans but with frequencies of ~3% in West Africa and has a strong female-specific effect (95% CI: 2.32–5.65 kg/m2 per each A allele). We confirmed this sex-specific association and replicated its strong effect for an adjusted fat mass index in the same Pelotas cohort, and for BMI in another Brazilian cohort from São Paulo (Southeast Brazil). A meta-analysis confirmed the significant association. Remarkably, we observed that while the frequency of rs114066381-A allele ranges from 0.8 to 2.1% in the studied populations, it attains ~9% among women with morbid obesity from Pelotas, São Paulo, and Bambuí. The effect size of rs114066381 is at least five times higher than the FTO SNPs rs9939609 and rs1558902, already emblematic for their high effects.
Conclusions
We identified six candidate SNPs associated with BMI. rs114066381 stands out for its high effect that was replicated and its high frequency in women with morbid obesity. We demonstrate how admixed populations are a source of new relevant phenotype-associated genetic variants.
Introduction
Overweight and obesity are risk factors for noncommunicable diseases, which are responsible for 63% of deaths worldwide [1] and 72% in Brazil [2]. Interindividual differences in BMI result from the effects of multiple genetic variants, environmental factors, and their interactions [3, 4]. Most of BMI heritability, estimated to be ~40%, is attributable to unknown genetic factors [5, 6]. Indeed, a meta-analysis of genome-wide association studies (GWAS) of BMI estimated that 97 loci explain ~2.7% of its variance [3]. The GWAS Catalog [7] and the DANCE web tool [8] report 389 SNPs associated with BMI, with a mean effect size of 0.054 kg/m2 (Fig. S1). Thus, BMI genetic architecture is characterized by a high number of loci with small effect sizes [5].
Our knowledge of the genetic architecture of complex phenotypes is biased [9] because only 22% of individuals included in GWAS are non-Europeans/non-US whites, 2.4% are from Africa, and 1.3% from Latin America [10]. The BMI meta-GWAS by Locke et al. [3] included only 5% of individuals of non-European ancestry among 339,226 individuals. Thus, expanding GWAS-based strategies beyond non-European populations is critical to discover differences in the genetic architecture of BMI among populations. This is especially important for phenotypes such as obesity, whose prevalence is higher in US African Americans, Hispanics, and Native Americans than in European Americans [11, 12], and in Brazil, higher in black women than in white women [13].
Few studies consider the influences of age- and sex-associated genetic factors on BMI. Despite the high correlation of intraindividual measurements of BMI at diverse ages, some genetic variants do have distinct effects depending on age [14–16]. For example, a meta-analysis of 14 GWAS [17] found that variants near to PRKD1, TNNI3K, SEC16B, and CADM2 genes had larger effects on BMI during adolescence/young adulthood than later in the lifespan, while a variant near SH2B1 had the opposite trend. Regarding sex, variants in SEC16B and ZFP64 were identified with stronger effects in women ([3] and see “Discussion”).
Here we study the genetic architecture of BMI in the admixed population of Brazil, the largest and most populous Latin-American country, with more than 200 million inhabitants. Brazilians are the product of about 500 years of admixture between Africans, Europeans, and Native Americans [18] and therefore, are suitable for admixture mapping. This method uses an admixed population to map genomic regions associated both with a specific ancestry and the phenotype of interest. Admixture mapping, by performing less statistical tests respect to classical GWAS, results in higher statistical power. Thus, for medium-sized studies (more feasible in limited resources setting environments hosting non-European populations), admixture mapping improves the power to detect an association when compared to GWAS that include only a few thousands of individuals. So far, admixture mapping has identified seven loci associated with BMI at chromosomes 2 (2p23.3), 3 (3q29), 5 (5q13.3 and 5q14), 15 (15q26), and X (Xq25, Xq13.1) [11, 19, 20], but these studies were restricted to US African American populations.
We performed admixture mapping (followed by fine-mapping) of BMI using data of ~2.3 million SNPs for three Brazilian population-based cohorts, from Northeast (Salvador), Southeast (Bambuí), and South (Pelotas), with distinct admixture and socio-demographic backgrounds, studied by the EPIGEN-Brazil Initiative (https://epigen.grude.ufmg.br, [18]). Salvador has 51% of African ancestry, while Pelotas and Bambuí have predominant European ancestry (76% and 79%, respectively) (Table S1 and Fig. 1). As these cohorts include individuals of three different epochs of life—children, young adults, and older adults—we performed three separate admixture mapping to avoid confounding effects of age. Additionally, we performed a replication by testing the association between 216 BMI GWAS Catalog hits in our three Brazilian cohorts.
Fig. 1. Admixture in the Brazilian cohorts, BMI distributions, and admixture mapping (AM) Manhattan plots with significant peaks.
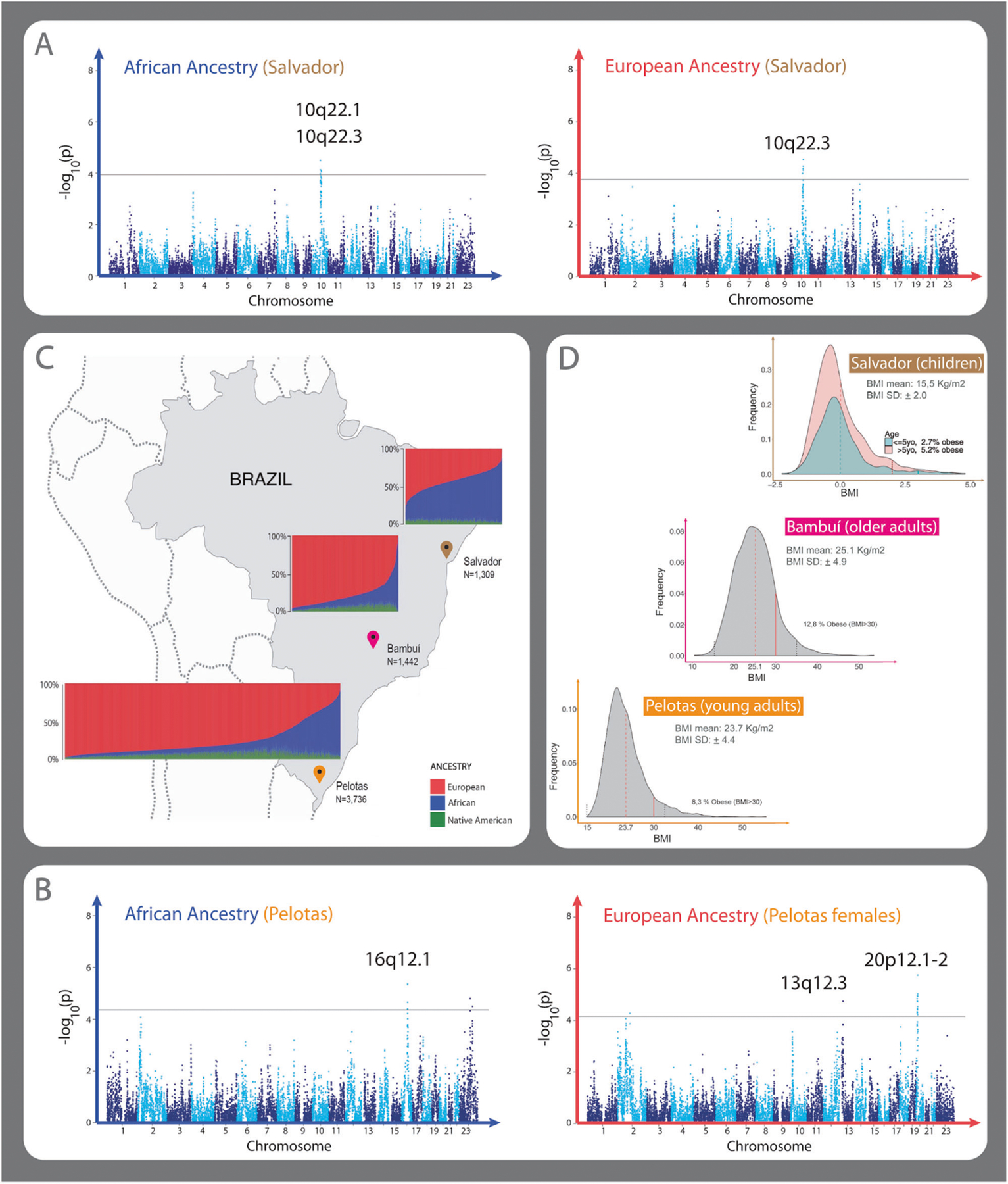
A, B Manhattan plots showing AM peaks using linear regressions with PCAdmix local ancestry inferences. Consensus significant AM peaks for PCAdmix and RFMix local ancestry inferences are specified on each plot. A Manhattan Plot showing the AM results of African (left) and European (right) ancestry in Salvador cohort. African ancestry AM shows two positive significant peaks 10q22.1 (β = 0.36, p value = 3.21e−05) and 10q22.3 (β = 0.36, p value = 7.87e−05). European AM ancestry analysis shows one negative associated peak 10q22.3 (β = −0.36, p value = 2.92e−05). B Manhattan plot showing the AM results of African (left) and European (right) ancestry in the Pelotas cohort. One peak in 16q12.1 (β = −0.80, p value = 4.30e−06) was observed associated with African ancestry, and two associated peaks, 13q12.3 (β = −0.95, p value = 1.84e−05) and 20p12.1–2 (β = −1.05, p value = 1.79–06), with European ancestry in females. Results are presented as log10(p value) to the given ancestry of each window of 100 SNPs along the genome. Black line in the Manhattan plots correspond to the genome-wide threshold p value estimated for the given ancestry and dataset (Table S5). The linear regression coefficient (β) and p values for all peaks correspond to the lead window, the genomic window with the most significant p value in the linear regression result. C Brazilian regions and continental individual ancestry bar plots for each cohort. D Histogram of Z-score adjusted by sex and age according to WHO guidelines in Salvador (top), histogram of BMI in Bambui (center) and Pelotas (bottom) cohorts.
Materials and methods
Study populations and genotyping
The Salvador-SCAALA cohort comprises 1445 children aged 4–11 years in 2005, when BMI was measured. Salvador (Fig. 1) is a city of 2.8 million inhabitants in Northeast Brazil [21]. This population is part of an earlier observational study and represents the population without sanitation in Salvador.
The 1982 Pelotas birth cohort study was conducted in Pelotas, a city in southern Brazil, with 340,000 inhabitants (Fig. 1). Throughout 1982, 99.2% of all births in the city were enrolled. Of these, the 5914 liveborn infants whose families lived in the urban area constituted the original cohort [22]. BMI was measured in 2004/2005, when individuals were 23 years old. The 2012–2013 follow-up measured participants’ body fat, lean, and bone mineral masses using dual-energy X-ray absorptiometry (DXA; GE Lunar Prodigy densitometer) in a full-body scan. We calculated fat mass index by dividing the adjusted fat mass (kg) by squared height (meters).
The Bambuí cohort study of ageing is ongoing in Bambuí, a city of ~15,000 inhabitants, in Minas Gerais State in Southeast of Brazil (Fig. 1). This cohort consisted of all residents aged 60 years and over on January 1997. From 1742 eligible residents, 1606 constituted the original cohort [23–25]. BMI was measured in 1997, when individuals were between 60 and 93 years old.
The EPIGEN-Brazil initiative genotyped individuals from Salvador, Pelotas, and Bambuí cohorts using the Illumina (San Diego, CA, USA) HumanOmni2.5–8v1 and the HumanOmni5–4v1 arrays. We used the consensus working datasets described in Kehdy et al. [18] that went through extensive quality control of SNPs and samples, as detailed in Kehdy et al. [18] and briefly explained in the Supplementary Material. For the three cohorts, measurements were taken by trained research staff and BMI was calculated as weight (kg) divided by squared height (meters). Potential confounding variables are sex, age, and different socioeconomic status (SES) (Table S1). In the end, we had genotyped, BMI, and covariables data for 1222, 3628, and 1342 individuals from the Salvador, Pelotas, and Bambuí cohorts, respectively. The EPIGEN protocol was approved by Brazil’s national research ethics committee (CONEP, resolution number 15895, Brasília). Informed consent was obtained from all subjects. In addition, we used African, European, and Native American individual ancestries estimated in Kehdy et al. [18] and performed on the same set of individuals using the software ADMIXTURE [26] (Fig. 1 and Table S1).
Kinship coefficients
Relatives were identified and removed from Salvador (63 individuals) and Pelotas (83 individuals) cohorts using a network-based approach that aims to eliminate the smallest possible number of individuals [18]. The Bambuí cohort has 516 (36%) individuals with relatives in the cohort. Thus, for Bambuí, we identified families with a categorical variable.
Phasing and local ancestry inference by PCAdmix and RFMix software
We phased our datasets using the software SHAPEIT2 [27], as detailed in ref. [18]. As inferences of continental local chromosome ancestry based on genome-wide data are more uncertain than inferences about individual or population ancestry, we used two methods for local ancestry inference, implemented in the software PCAdmix [28] and RFMix [29]. PCAdmix inferences were performed as in ref. [18]. For RFmix inferences, we used Europeans, Africans, and Native American as parental populations and fixed parameters as described in the Supplementary Methods. For PCAdmix and RFMix results, we considered only the windows which ancestry was inferred with a posterior probability > 0.90.
Admixture mapping
We tested the association between BMI and each local ancestry (African, European, and Native American) across the genome using linear regression models. The regressions were adjusted by age (Salvador and Bambuí), sex, SES, and genome-wide African ancestry. For Bambuí, we corrected for family structure. We used an additive model that considers the number of inferred African, European, or Native American ancestry copies (0, 1, or 2) carried by an individual for each window. Because we found an association between individual African ancestry and BMI in females in Pelotas (Supplementary Methods, Tables S2–S4), we performed a stratified analysis for each sex in this cohort. While for Salvador and Pelotas we used simple linear regression [30], for Bambuí we used robust variance estimators to correct results by family structure [31].
To establish a significance threshold for the admixture mapping, accounting both for multiple testing and linkage disequilibrium (LD) due to admixture, we estimated the effective number of tests (ENT) for each chromosome for each individual [32], and obtained an equivalent Bonferroni p value threshold for significance dividing 0.05 divided by ENT (Table S5). We conservatively used the same genome-wide thresholds to identify admixture mapping hits for X-chromosome. We compared the significant admixture mapping peaks of each chromosome obtained using PCAdmix and RFMix local ancestry inferences (using p values thresholds from Table S5 multiplied by 10), and, considered a consensus significant peak the ones mapped to the same chromosomal region (not only the same chromosome bands) using both inferences. Only these consensus significant admixture mapping peaks were followed-up for fine-mapping.
Imputation, fine-mapping, and annotation
Fine-mapping was performed using both genotyped and imputed SNPs. We imputed our dataset focusing on ±1 Mb centered in the most significant window of each admixture mapping hit (based on PCAdmix). To this, we used IMPUTE2 [33] with a reference panel that merged the public reference panel data from 1000G and 270 individuals from EPIGEN (90 of each cohort) genotyped for 4.3 million SNPs, and considered only SNPs imputed with an info score quality metric > 0.9 [34].
Genotyped and imputed SNPs were tested for association with BMI using the same linear regressions models used for admixture mapping. We excluded SNPs with minor allele frequency < 0.005 for these analyses. We considered significant, the associations with p values less than or equal to the ones obtained for the admixture mapping peaks and suggestively significant those SNPs with a p value higher than the ones obtained for the admixture mapping peaks but not more than one unit of −log (p value). Fine-mapping results were plotted using the LocusZoom tool [35] and annotated using ANNOVAR [36]. We estimated the LD statistics (r2, [37]) on phased data using the software Haploview [38]. A flowchart summarizing the study design is shown in Fig. S12.
Replication cohorts and meta-analysis
We tested for replication the association of rs114066381-A with BMI in other four cohorts: (1) whole-genome data with a mean target coverage of 30x from 651 unrelated females from São Paulo, Brazil, the SABE (Health, Wellbeing, and Aging) study [39–41]. Linear regression was adjusted for age, education level, SES, and individual African ancestry proportion; (2) 1082 women from Puerto Rico (547 non-cancer controls and 535 cases) genotyped on the Affymetrix Axio UK biobank array. rs114066381 was imputed with IMPUTE2 using samples from 1000G as reference. Linear regression was adjusted for age, education level, individual African ancestry proportion, and breast cancer diagnosis; (3) 1103 adult women (age ≥18 years) from Nigeria, Cameroon, Sudan, Ethiopia, Kenya, Tanzania, and Botswana [42]. Individuals were genotyped on either the Illumina 5M-Omni array or the Illumina 1M-Duo BeadChip array. For individuals typed on the last one, rs114066381 was imputed using MiniMac, based on a reference panel of 180 whole-genome sequences from eastern and southern Africa as well as the African populations from the 1000G. Association tests were performed using a linear-mixed model in which age was modeled as a fixed effect and the kinship matrix was used for the random effects term; (4) 859 women from Soweto, South Africa [43] genotyped on the 2,3M H3-Africa array. rs114066381 was imputed using the African reference panel at Sanger Imputation facility. Linear regression was adjusted for age and SES. We used the package metafor [44] to perform the meta-analysis using a random effects model with Hedges method.
Statistical power estimation
Power estimation was performed separately for each EPI-GEN cohort, according to the specific BMI distribution (Table S1). SNPs associated with BMI in the GWAS Catalog were extracted using the keyword “body mass index”, and those with p values > 9e−05 were filtered out. From the 2205 SNPs reported on 61 published studies, we kept 216 SNPs with effect size unambiguously associated to a specific allele and reported as a regression coefficient expressed in kg/m2 from cross-sectional studies, genotyped or imputed in our database. We calculated the statistical power using the latter effect size (regression coefficient) values, and allele frequency and number of individuals from each EPIGEN cohort (Table S1). The type I error rate was set at α = 0.00023. All power estimates were calculated with QUANTO v1.2.4 program [45], assuming an additive genetic model with independent individuals.
Replication analysis of previous GWAS hits
To test the association of previous BMI GWAS hits, we used the regression model used in fine-mapping for all the selected 216 SNPs in the three Brazilian cohorts. p values were adjusted considering 216 independent tests using the Benjamini–Hochberg correction [46].
Genomic in silico analyses
The search for candidate regulatory SNPs was performed using HaploReg v4.1 database (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php, [47]), Ensembl (https://grch37.ensembl.org/, [48]), and RegulomeDB (http://www.regulomedb.org/, [49]). ChIP-seq data were provided by [50], available at HaploReg v4.1 database.
Results
Admixture mapping and fine-mapping
We performed an admixture mapping analysis for the three continental ancestries (African, European, and Native American) in the three cohorts using an additive model considering the number of inferred African, European, or Native American ancestry copies (0, 1, or 2) carried by an individual for each chromosome fragment.
Table 1 shows the five consensus significant admixture mapping peaks found in Salvador and Pelotas. The distribution of BMI for each allele of African or European ancestry for the five peaks are shown in Supplementary Figs. S2–S4. No consensus significant peak was found in older adults from Bambuí.
Table 1.
Admixture mapping peaks obtained both with RFMix and PCAdmix local ancestry inferences.
| Cohort | Genomic region (length in base pairs) | Local ancestry (sub-dataset) | Regression coefficienta | p valuea | Associated region (length in base pairs)b |
|---|---|---|---|---|---|
| Children from Salvador | 10q22.1 (4,300,000) | African | 0.36 | 3.21e–05 | 445,166 |
| 10q22.3 (4,400,000) | African | 0.36 | 7.87e–05 | 255,990 | |
| European | −0.36 | 2.92e–05 | 577,546 | ||
| Young adults from Pelotas | 16q12.1 (5,600,000) | African | −0.80 | 4.30e–06 | 600,961 |
| 13q12.3 (3,300,000) | European (female) | −0.95 | 1.84e–05 | 118,909 | |
| 20p12.1–2 (8,700,000) | European (female) | −1.05 | 1.79e–06 | 3,481,320 |
Regression coefficients, p values, and windows length associated in the linear regressions using PCAdmix local ancestry inferences.
Regression coefficients and p values of the lead genomic window of 100 SNPs, using PCAdmix inference. The lead window is the genomic window with the most significant regression coefficient in the admixture mapping among those windows below the significance cut-off. Regression coefficients are the change in units of BMI (Kg/m2) for each additional copy of a specific ancestry.
Length of the continuous genomic region including significant admixture mapping results. Each window of PCAdmix inference has 100 SNPs.
Fine-mapping on Salvador children
The high African ancestry (51%) in children from Salvador allowed us to identify two genomic regions where this ancestry is positively associated with BMI and within these regions, we identified three significant variants (Tables S8 and S9 and Fig. S5): within 10q22.1, rs1334909357-CTTT in an intron in the PALD1 gene and, within 10q22.3, the linked SNPs rs79947827-A and rs141274185-T (LD: r2 = 0.86) in the ZMIZ1 gene, that encodes a protein that regulates the activity of many transcription factors [51]. Other SNPs in ZMIZ1 are associated with 19 complex disorders, and this gene is among the 21 human genes most associated with complex phenotypes, including not only BMI-related phenotypes such as height and sitting height ratio, but also psychiatric disorders, breast cancer and autoimmune diseases (http://gilderlanio.pythonanywhere.com/home, Fig. S6).
Fine-mapping in young adults from Pelotas
While the low non-European admixture reduces the power to detect non-European associated variants in Pelotas, this is compensated by its larger size (n = 3628) in respect to the Salvador cohort. Also, as Pelotas is a birth cohort, all individuals have the same age, which limits nongenetic variance for BMI. For the entire Pelotas cohort, we identified one genomic region, 16q12.1, where African ancestry is negatively associated with BMI and, within this region, we found one significant SNP, rs76416629-G, 2 kb upstream of NOD2 gene (Tables S6 and S7 and Fig. S5).
Furthermore, we identified a genomic region, 13q12.3, for which European ancestry is associated with lower values of BMI in females and, within this region, two significant SNPs (not in LD, r2 < 0.001). rs113214936-G in the intron of MTUS2, a gene previously associated with obesity-related traits [52]. Our most striking result is the association of the SNP rs114066381-A with a strong effect on BMI in females (beta = 3.99 ± 0.84 kg/m2 per allele, 95% CI: 2.32–5.65, p = 2.76 × 10−6, Table 2 and Fig. 2). This SNP is present in 31 unrelated females (all heterozygous) that have a mean BMI of 27.99 kg/m2, which is larger than the mean BMI for the cohort females (23.61 kg/m2, p = 0.0008, Fig. 3). These 31 females have a mean African ancestry of 35%, while the mean in unrelated females is 16%. Remarkably, the BMI of 25 males carrying the rs114066381-A allele (mean: 23.72 kg/m2) does not differ from the general population (mean: 23.81 kg/m2, p = 0.5397, Fig. 3).
Table 2.
Association of rs114066381-A with body mass index and fat mass index in unrelated females of the Pelotas cohort and replications.
| Phenotype | Cohort | N | Age (years old) | Frequency | Regression coefficient | SE | p value | Power |
|---|---|---|---|---|---|---|---|---|
| Fat mass index | Pelotas | 1417 | 30 | 0.008 | 2.21 | 0.84 | 9.00e–03 | – |
| Body mass index | Pelotas | 1795 | 23 | 0.008 | 3.99 | 0.84 | 2.76e–06 | – |
| Bambui | 516 | 60–93 | 0.010 | 2.93 | 2.65 | 0.26 | 39% | |
| Bambui (all femalesa) | 821 | 60–93 | 0.009 | 2.64 | 2.33 | 0.25 | 53% | |
| Sao Paulo | 651 | 59–99 | 0.019 | 3.34 | 1.12 | 3.26e–03 | 89% | |
| Bambui + Sao Paulo | 1173 | 59–99 | 0.015 | 3.55 | 0.94 | 1.91e–04 | 99% | |
| Salvador | 664 | 4–11 | 0.021 | −0.24 | 0.38 | 0.53 | 9% | |
| Puerto Rico | 1082 | 21–89 | 0.005 | 1.30 | 1.92 | 0.50 | 13% | |
| African populationsb (all femalesa) | 1103 | 17–97 | 0.013 | −0.57 | 0.80 | 0.47 | 9% | |
| Soweto | 859 | 39–60 | 0.020 | 2.39 | 7.63 | 0.75 | 69% |
Using genetic relationship matrix.
Nigeria, Cameroon, Sudan, Ethiopia, Kenya, Tanzania, and Botswana populations.
Fig. 2. LocusZoom plot of the fine-mapping of consensus significant admixture mapping peak in young adults from Pelotas at 13q12.3 associated with European ancestry in females performed using both genotyped and imputed SNPs ±1 Mb from target region (lead windows).
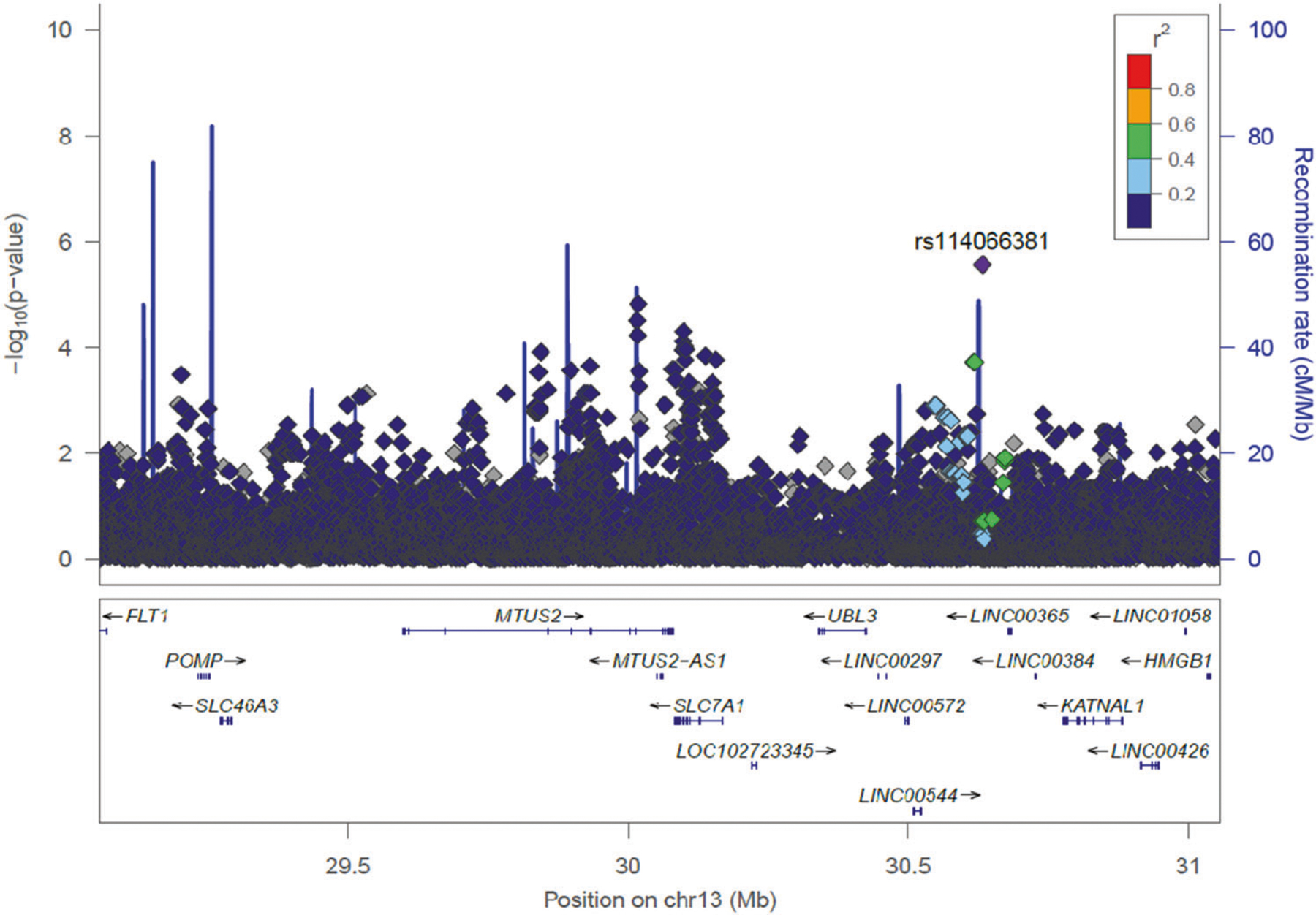
The SNP with the lowest p value is color coded in purple and labeled. The linkage disequilibrium between this SNP and the remaining nearby SNPs is indicated by the color coding according to r2 values based on Africans from 1000 Genomes Project (Color figure online).
Fig. 3. Body mass index (BMI) in females and males’ adults from Pelotas cohort, according to their genotypes in the SNP rs114066381.
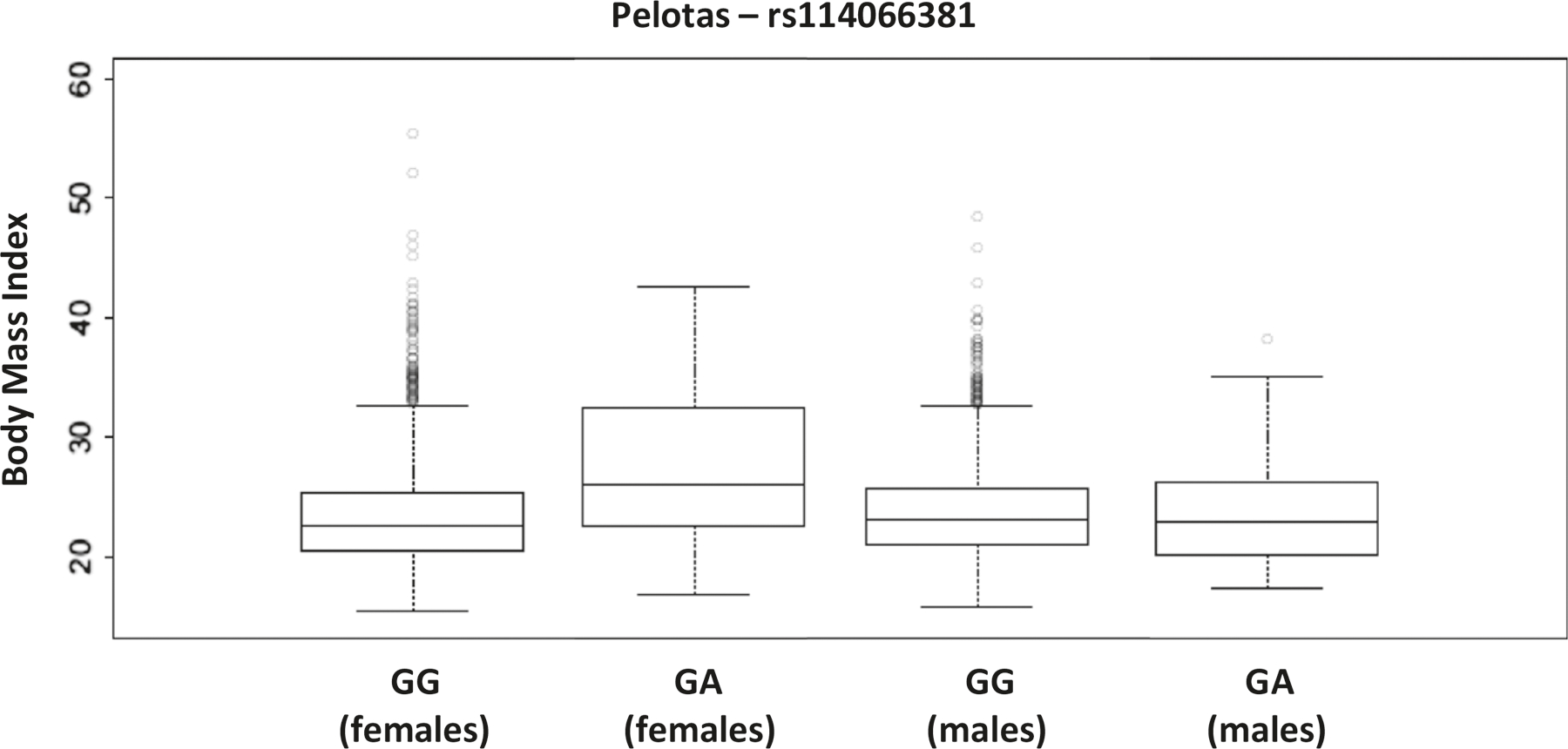
The increase of BMI associated with the rs114066381-A is observed in females (p value = 0.0008), but not in males (p value = 0.5397).
rs114066381 is 2 kb from a CTCF-binding site [48], but no evidence of transcription regulation is shown by RNA-seq [50]. Besides, this genomic region contains binding sites for the histone-interacting proteins KAP1 and SETDB1, as reported by ChIP-seq data (HaploReg v4.1, [47], [50], Fig. S7). However, there is no evidence in the literature that the region acts as an enhancer in vivo. This genomic region is primate-specific, being absent from the genome of other vertebrates (Supplementary Methods, UCSC Genome Browser 2013, [53], Figs. S8 and S9). The derived allele A is very rare in Europeans, but has frequencies of ~3% in West Africans (Table S6).
We confirmed the rs114066381 female-specific association using the fat mass index (a more direct measurement of adiposity), measured by DXA, 7 years after the measurement of BMI on the same individuals (beta = 2.21 kg of fat/m2 per allele, 95% CI: 0.55–3.88, p = 9× 10−3, Table 2). We replicated with 89% of power the association in older adult females from São Paulo (Brazil) (SABE cohort, [40]) and in the merged dataset from São Paulo and Bambuí (power = 99%, Table 2). We tested but did not observe significant association for the other cohorts tested (Table 2). However, a meta-analysis synthesizing the seven effect sizes showed a positive association between rs114066381-A allele and BMI in females (Fig. 4) both considering all effects together or only the effects obtained with admixed populations. Remarkably, while in Pelotas and São Paulo the frequency of rs114066381-A allele is 1.1%, its frequency increases in women with overweight (1.2%) and obesity (1.98%), and attains 9% among women with morbid obesity. The same pattern is also observed in Bambuí and Soweto populations but not in Puerto Rico, in which the frequency of rs114066381-A allele is higher only for women with obesity (Table S10). We observed that the BMI distribution of Salvador and rural Africa are very different compared to the other populations, while in Pelotas, Bambuí, São Paulo, Puerto Rico, and Soweto, 27% of individuals have BMI greater than 25 kg/m2, in Salvador and rural Africa, 11% of individuals fall in this category (and only 0.2% of individuals are morbidly obese in both cohorts, contrasting to more than 1% in the other populations). Moreover, 23% of the individuals of rural Africa are underweight (BMI < 18.5 kg/m2).
Fig. 4. Forest plots from the meta-analysis synthesizing association results between rs114066381 and BMI from seven populations.
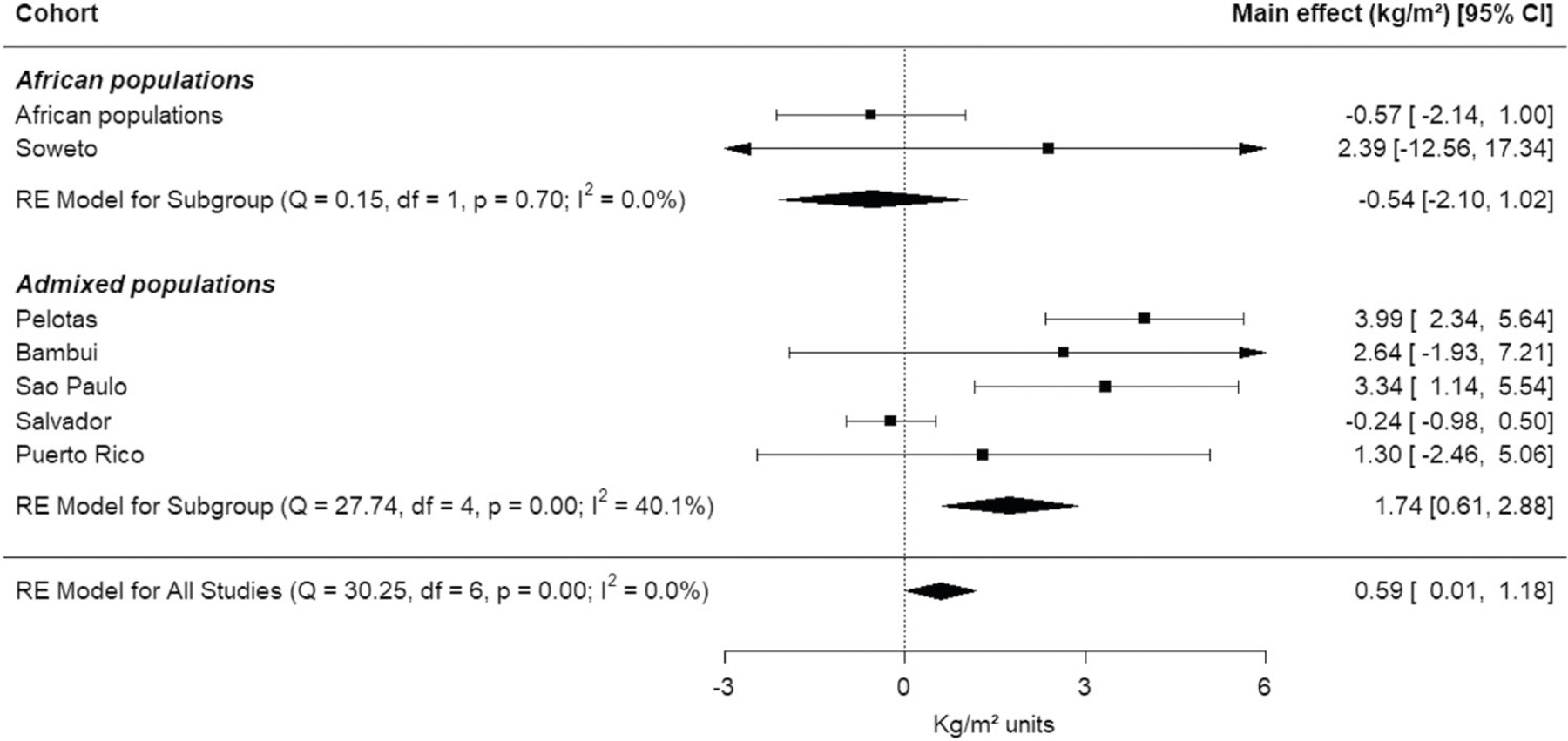
Effect size [95% confidence interval (CI)] in each individual study, subgroups of African populations and admixed populations, and combining all populations.
Power estimation and replication of previous GWAS hits
We estimated the statistical power to detect associations for 216 BMI GWAS catalog hits on the three EPIGEN cohorts, conditioning on the effect sizes reported in kg/m2 units on the GWAS Catalog (took as a population parameter), the BMI distribution, as well as the number of studied individuals and the allele frequencies in each of the EPIGEN cohorts. These 216 GWAS Catalog hits were selected because, in the context of the high heterogeneity of data stored in the GWAS Catalog, their effect sizes (linear regression coefficient) were unambiguously associated with a specific allele and were consistently reported in units of kg/m2. Out of the 216 hits, 189 were observed in adults, 4 in children, and 23 in both, in individuals with predominant European ancestry.
Based on the mean statistical power of the 216 GWAS Catalog hits, and assuming that these SNPs (and their regression coefficient) are part of the genetic architecture of BMI in the Brazilian cohorts, we would expect to observe 24 SNPs out of 216 (average power = 11%, but we observed 0 replications) associated in Salvador children, 22 SNPs (average power = 10%, we observed 20) in Pelotas young adults, and 15 SNPs (average power = 7%, and we observed 8) in Bambuí older adults (Fig. S10 and Table S11). Specifically, in Pelotas, we confirmed the association for the six FTO SNPs included in our analysis (rs9930333, rs62033400, rs8050136, rs3751812, rs1558902, and rs9939609).
Discussion
Leveraging admixture in Brazil, we used genome-wide data of three population-based cohorts to find loci associated with BMI through admixture mapping followed by fine-mapping. We acknowledge that a limitation of our study is the relatively small number of individuals respect to GWAS standards in European or US populations; but this is a limitation shared by several studies, considering the difficulties of achieving larger sample sizes in admixed populations of low-medium income countries. Besides, the present study has important characteristics absent in most studies. First, it relies on population-based cohorts that better capture the phenotypic variation of populations, but are rarely considered in genetic studies [54, 55]. Second, it is one of the few studies that explore the genetic architecture of BMI in three different age stages: children, young adults, and older adults. Third, in the context of under-representation of non-European populations in GWAS, we analyzed populations of South America with African and Native American ancestry. Because none of the six new candidate SNPs to influence BMI reported in this study (Tables S7 and S9) are in LD with previous 389 GWAS Catalog hits of BMI (r2 < 0.022), we conclude that we have contributed to expand the catalog of SNPs of the global genetic architecture of BMI.
Factors supporting the female-specific effect of rs114066381 on adiposity
First, we replicated the female-specific association in an independent admixed cohort from São Paulo and confirmed its association by a meta-analysis. Second, while our initial female-specific association with BMI and fat mass index is based on imputed genotypes, replication in the São Paulo cohort is based on whole-genome data. Third, in the discovery cohort, we not only observed a strong association with BMI, but also with fat mass index measured with DXA, a more direct measure of adiposity collected 7 years later than BMI in the same individuals. Fourth, rs114066381 is located in a potential regulatory region, which makes a biological role for this genomic region plausible. With respect to the pattern of LD, rs114066381 presents an r2 = 0.5 with three SNPs in LD with each other (r2 = 1) mapped in near regulatory regions (in sensu HaploREG v4.1, [47], Fig. S11). In Bambuí the r2 of rs114066381 with the same three SNPs varies between 0.3 and 0.7 (Fig. S11). rs114066381 is ~300 kb from rs7335631 associated with “Fat distribution in HIV” [56], but there is no (r2 < 0.001) LD between these two SNPs in any of our three Brazilian populations. Thus, our results suggest a specific role for rs114066381.
The discovery size effect for rs114066381 (beta = 3.99 ± 0.847 kg/m2) is one of the highest observed for BMI, considering both sexes. The size effects suggested by meta-analyses (Fig. 4) are also high considering the distribution of BMI effect sizes. According with GWAS Catalog (October/2019), the range of estimated beta in kg/m2 for BMI hits is 0.013–4.119 with an average of 0.054 kg/m2.
The female-specific association for rs114066381 is observed in the following context: out of 833 hits reported in GWAS Catalog as associated with BMI with beta reported in kg/m2 (October/2019) independently of sex, 229 are female-specific associations (beta range: 0.009–0.484, beta mean: 0.025) and 134 male-specific [beta range: 0.013–0.095, beta mean: 0.025]. Even if mean effect sizes are similar in men and women, the effect size distribution of women shows a tail of higher beta values, which suggests that higher effect sizes are more common in women than in men. Our finding is paradigmatic of this context: in adult females of Southern Brazil, rs114066381 alone explains a similar portion of the variance of BMI (r2 range for Pelotas, Bambuí, and São Paulo cohorts: 0.008–0.044) as the entire set of 97 GWAS hits recently reported [3]. Also, we can speculate that rs114066381 could be an example of a thrifty genotype [57, 58] associated with energy storage in females and pregnancy (but see [59] for a counterpoint of the thrifty theory).
Replication of other GWAS hits
We replicated 28 of the 216 associations reported for SNPs in previous GWAS, mostly performed in adults of European ancestry, with the Pelotas cohort presenting not only the largest rate of replication (20/216) (Table S11), but also a very good concordance between the observed (20) and expected (22) number of replications. This is consistent with: (1) the larger size of Pelotas cohort; (2) the relative lower SES of the Salvador cohort adds a layer of complexity to the definition of the genetic architecture of BMI, respect to GWAS in predominantly European populations with different socioeconomic background; (3) the age dependence of the genetic architecture of BMI, and the fact that most GWAS of BMI were performed in adults and in Pelotas BMI was also measured in young adults, while in the Salvador and Bambuí cohorts, BMI was measured in children and older adults, respectively. Lasky-Su et al. [54] showed how age-dependent effects can be an important and misjudged cause of non-replication. These results exemplify how differences in age, SES and ancestry contribute to differences in the genetic architecture of BMI in particular and complex traits in general.
In conclusion, we performed three admixture mapping/fine-mapping for BMI and tested the association of GWAS Catalog hits in three Brazilian population-based cohorts. We provide six candidate SNPs associated with African or European ancestry that are associated with BMI. More importantly, our admixture/fine-mapping in Brazilians reveals a West African associated potential regulatory variant (rs114066381), with a female-specific effect on BMI, which seems to be particularly important for the development of morbid obesity. Altogether, our results show that the study of South American admixed populations, as well as other populations worldwide [60–62] are a source of novel non-European associated variants with considerable effect size that may explain in non-European populations an important portion of the current “missing heritability”. This statement can be generalized by the observation that ~25% of the variants discovered in GWASs of BMI were identified by studies with Latin Americans, although they represent only 11% of such studies, indicating the importance of increase the number and size of studies with these populations.
Code availability
Used bioinformatics pipelines are available in the EPIGEN-Brazil Project Scientific Workflow (http://www.ldgh.com.br/scientificworkflow, [34]).
Supplementary Material
Acknowledgements
For analyses, we used the Sagarana cluster (from Centro de Laboratórios Multiusuários do Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais). We thank Miguel Ortega for help in the use of Sagarana, Ms. Evelyn Tay at University of Ghana Medical School (Accra, Ghana) for managing the study, and Ms. Marcelle Bartholomeu and Ms. Àlex Teixeira for technical support. The EPIGEN-Brazil Initiative is funded by the Brazilian Ministry of Health (Department of Science and Technology from the Secretaria de Ciência, Tecnologia e Insumos Estratégicos) through Financiadora de Estudos e Projetos. The EPIGEN-Brazil investigators received funding from the Brazilian Ministry of Education (CAPES Agency), Brazilian National Research Council (CNPq), the Minas Gerais State Agency for Support of Research (FAPEMIG), Rede Mineira de Genômica Populacional e Medicina de Precisão (FAPEMIG-RED-00314–16), and TWAS-CNPq Full PhD fellow, and grant 2019/19998–8, São Paulo Research Foundation (FAPESP).
Footnotes
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41366-021-00761-1.
Compliance with ethical standards
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.WHO. Global status report on noncommunicable diseases 2010 2011. https://www.who.int/nmh/publications/ncd_report2010/en/.
- 2.Schmidt MI, Duncan BB, e Silva GA, Menezes AM, Monteiro CA, Barreto SM. et al. Chronic non-communicable diseases in Brazil: burden and current challenges. Lancet 2011;377:1949–61. http://linkinghub.elsevier.com/retrieve/pii/S0140673611601359. [DOI] [PubMed] [Google Scholar]
- 3.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197–206. 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rask-Andersen M, Karlsson T, Ek WE, Johansson A. Gene-environment interaction study for BMI reveals interactions between genetic factors and physical activity, alcohol consumption and socioeconomic status. PLoS Gene 2017;13:e1006977. 10.1371/journal.pgen.1006977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson MR, English G, Moser G, Lloyd-Jones LR, Triplett MA, Zhu Z. et al. Genotype–covariate interaction effects and the heritability of adult body mass index. Nat Genet 2017;49:1174–81. 10.1038/ng.3912. [DOI] [PubMed] [Google Scholar]
- 6.Wainschtein P Recovery of trait heritability from whole genome sequence data. bioRxiv 2019. 10.1101/588020. [DOI]
- 7.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H. et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 2014;42:D1001–6. http://www.ncbi.nlm.nih.gov/pubmed/24316577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Araújo GS, Lima LHC, Schneider S, Leal TP, Da Silva APC, Vaz De Melo POS. et al. Integrating, summarizing and visualizing GWAS-hits and human diversity with DANCE (Disease-ANCEstry networks). Bioinformatics 2016;32:1247–9. [DOI] [PubMed] [Google Scholar]
- 9.Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell 2019;177:26–31. http://www.ncbi.nlm.nih.gov/pubmed/30901543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morales J, Welter D, Bowler EH, Cerezo M, Harris LW, McMahon AC. et al. A standardized framework for representation of ancestry data in genomics studies, with application to the NHGRI-EBI GWAS Catalog. Genome Biol 2018;19:21. 10.1186/s13059-018-1396-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng CY, Kao WHL, Patterson N, Tandon A, Haiman CA, Harris TB. et al. Admixture mapping of 15,280 African Americans identifies obesity susceptibility loci on chromosomes 5 and X. PLoS Genet 2009;5:e1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nassir R, Qi L, Kosoy R, Garcia L, Allison M, Ochs-Balcom HM. et al. Relationship between adiposity and admixture in African-American and Hispanic-American women. Int J Obes 2012;36:304–13. 10.1038/ijo.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gigante DP, Moura EC, Sardinha LMV. Prevalência de excesso de peso e obesidade e fatores associados, Brasil, 2006. Rev Saude Publica 2009;43:83–9. [DOI] [PubMed] [Google Scholar]
- 14.Bradfield J, Taal H, Timpson N, Scherag A, Lecoeur C, Warrington N. et al. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat Genet 2012;44:526–31. http://www.nature.com/articles/ng.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardy R, Wills AK, Wong A, Elks CE, Wareham NJ, Loos RJF. et al. Life course variations in the associations between FTO and MC4R gene variants and body size. Hum Mol Genet 2010;19:545–52. 10.1093/hmg/ddp504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warrington NM, Howe LD, Paternoster L, Kaakinen M, Herrala S, Huikari V. et al. A genome-wide association study of body mass index across early life and childhood. Int J Epidemiol 2015;44:700–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graff M, Ngwa JS, Workalemahu T, Homuth G, Schipf S, Teumer A. et al. Genome-wide analysis of BMI in adolescents and young adults reveals additional insight into the effects of genetic loci over the life course. Hum Mol Genet 2013. P. 3597–607. https://academic.oup.com/hmg/article/22/17/3597/572524. [DOI] [PMC free article] [PubMed]
- 18.Kehdy FSG, Gouveia MH, Machado M, Magalhães WCS, Horimoto AR, Horta BL. et al. Origin and dynamics of admixture in Brazilians and its effect on the pattern of deleterious mutations. Proc Natl Acad Sci 2015;112:8696–701. 10.1073/pnas.1504447112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basu A, Tang H, Arnett D, Gu CC, Mosley T, Kardia S. et al. Admixture mapping of quantitative trait loci for BMI in African Americans: evidence for loci on chromosomes 3q, 5q, and 15q. Obesity (Silver Spring) 2009;17:1226–31. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2929755&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng C-Y, Reich D, Coresh J, Boerwinkle E, Patterson N, Li M. et al. Admixture mapping of obesity-related traits in African Americans: the Atherosclerosis Risk in Communities (ARIC) Study. Obesity (Silver Spring) 2010;18:563–72. 10.1038/oby.2009.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barreto ML, Cunha SS, Alcântara-Neves N, Carvalho LP, Cruz AA, Stein RT. et al. Risk factors and immunological pathways for asthma and other allergic diseases in children: background and methodology of a longitudinal study in a large urban center in Northeastern Brazil (Salvador-SCAALA study). BMC Pulm Med 2006;6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Victora CG, Barros FC. Cohort profile: the 1982 Pelotas (Brazil) birth cohort study. Int J Epidemiol 2006;35:237–42. http://www.ncbi.nlm.nih.gov/pubmed/16373375. [DOI] [PubMed] [Google Scholar]
- 23.Lima-Costa MF, Firmo JO, Uchoa E. Cohort profile: the Bambui (Brazil) cohort study of ageing. Int J Epidemiol 2011;40:862–7. 10.1093/ije/dyq143. [DOI] [PubMed] [Google Scholar]
- 24.Lima-Costa MF, Rodrigues LC, Barreto ML, Gouveia M, Horta BL, Mambrini J. et al. Genomic ancestry and ethnoracial self-classification based on 5,871 community-dwelling Brazilians (The Epigen Initiative). Sci Rep 2015;5:9812. http://www.ncbi.nlm.nih.gov/pubmed/25913126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lima-Costa MF, de Mello Mambrini JV, Leite MLC, Peixoto SV, Firmo JOA, de Loyola Filho AI. et al. Socioeconomic position, but not African genomic ancestry, is associated with blood pressure in the Bambui-Epigen (Brazil) cohort study of aging. Hypertension 2015. 10.1161/HYPERTENSIONAHA.115.06609. [DOI] [PubMed]
- 26.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 2003;164:1567–87. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1462648&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods 2011;9:179–81. [DOI] [PubMed] [Google Scholar]
- 28.Brisbin A, Bryc K, Byrnes J, Zakharia F, Omberg L, Degenhardt J. et al. PCAdmix: principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum Biol 2012;84:343–64. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3740525&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet 2013;93:278–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1950838&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeileis A Econometric computing with HC and HAC covariance matrix estimators. J Stat Softw 2004;11. http://www.jstatsoft.org/v11/i10/. [Google Scholar]
- 32.Shriner D, Adeyemo A, Rotimi CN. Joint ancestry and association testing in admixed individuals. PLoS Comput Biol 20117: e1002325. 10.1371/journal.pcbi.1002325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009;5:e1000529. 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magalhães WCS, Araujo NM, Leal TP, Araujo GS, Viriato PJS, Kehdy FS. et al. EPIGEN-Brazil Initiative resources: a Latin American imputation panel and the Scientific Workflow. Genome Res 2018;28:1090–5. http://www.ncbi.nlm.nih.gov/pubmed/29903722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010;26:2336–7. http://www.ncbi.nlm.nih.gov/pubmed/20634204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. http://www.ncbi.nlm.nih.gov/pubmed/20601685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theor Appl Genet 1968;38:226–31. 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- 38.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–5. http://www.ncbi.nlm.nih.gov/pubmed/15297300. [DOI] [PubMed] [Google Scholar]
- 39.Barbosa AR, Souza JMP, Lebrão ML, Laurenti R, Marucci M, de FN. Functional limitations of Brazilian elderly by age and gender differences: data from SABE Survey. Cad Saude Publica 2005;21:1177–85. http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0102-311X2005000400020&lng=en&tlng=en. [DOI] [PubMed] [Google Scholar]
- 40.Naslavsky MS, Scliar MO, Yamamoto GL, Wang JYT, Zverinova S, Karp T. et al. Whole-genome sequencing of 1,171 elderly admixed individuals from the largest Latin American metropolis (São Paulo, Brazil). bioRxiv 2020. 10.1101/2020.09.15.298026. [DOI]
- 41.Telenti A, Pierce LCT, Biggs WH, di Iulio J, Wong EHM, Fabani MM. Deep sequencing of 10,000 human genomes. Proc Natl Acad Sci USA 2016. 10.1073/pnas.1613365113. [DOI] [PMC free article] [PubMed]
- 42.Crawford NG, Kelly DE, Hansen MEB, Beltrame MH, Fan S, Bowman SL. et al. Loci associated with skin pigmentation identified in African populations. Science 2017;358:eaan8433. 10.1126/science.aan8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ali SA, Soo C, Agongo G, Alberts M, Amenga-Etego L, Boua RP. et al. Genomic and environmental risk factors for cardiometabolic diseases in Africa: methods used for Phase 1 of the AWI-Gen population cross-sectional study. Glob Health Action 2018;11:1507133. 10.1080/16549716.2018.1507133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viechtbauer W Conducting meta-analyses in R with the metafor Package. J Stat Softw 2010;36. http://www.jstatsoft.org/v36/i03/.
- 45.Gauderman W, Morrison J. QUANTO documentation. (Technical report no. 157) Los Angeles, CA: Department of Preventive Medicine, Universityof Southern California, 2001. 2006. Available at https://preventivemedicine.usc.edu/download-quanto/. [Google Scholar]
- 46.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 1995;57:289–300. [Google Scholar]
- 47.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res 2016;44:D877–81. 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J. et al. Ensembl 2018. Nucleic Acids Res 2018;46:D754–61. http://academic.oup.com/nar/article/46/D1/D754/4634002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012;22:1790–7. 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.The ENCODE Project Consortium. A user’s guide to the Encyclopedia of DNA elements (ENCODE). PLoS Biol 20119: e1001046. 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogers LM, Riordan JD, Swick BL, Meyerholz DK, Dupuy AJ. Ectopic expression of Zmiz1 induces cutaneous squamous cell malignancies in a mouse model of cancer. J Invest Dermatol 2013;133:1863–9. http://linkinghub.elsevier.com/retrieve/pii/S0022202X15363260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA. et al. Novel genetic loci identified for the pathophysiology of childhood obesity in the hispanic population. PLoS ONE 2012;7:e51954. 10.1371/journal.pone.0051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM. et al. The human genome browser at UCSC. Genome Res 2002;12:996–1006. 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lasky-Su J, Lyon HN, Emilsson V, Heid IM, Molony C, Raby BA. et al. On the replication of genetic associations: timing can be everything!. Am J Hum Genet 2008;82:849–58. http://linkinghub.elsevier.com/retrieve/pii/S0002929708001742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manolio TA, Bailey-Wilson JE, Collins FS. Genes, environment and the value of prospective cohort studies. Nat Rev Genet 2006;7:812–20. 10.1038/nrg1919. [DOI] [PubMed] [Google Scholar]
- 56.Irvin MR, Shrestha S, Chen Y- DI, Wiener HW, Haritunians T, Vaughan LK. et al. Genes linked to energy metabolism and immunoregulatory mechanisms are associated with subcutaneous adipose tissue distribution in HIV-infected men. Pharmacogenet Genomics 2011;21:798–807. http://www.ncbi.nlm.nih.gov/pubmed/21897333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neel JV. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”?. Am J Hum Genet 1962;14:353–62. http://www.ncbi.nlm.nih.gov/pubmed/13937884. [PMC free article] [PubMed] [Google Scholar]
- 58.Prentice AM, Hennig BJ, Fulford AJ. Evolutionary origins of the obesity epidemic: natural selection of thrifty genes or genetic drift following predation release?. Int J Obes 2008;32:1607–10. http://www.nature.com/articles/ijo2008147. [DOI] [PubMed] [Google Scholar]
- 59.Wang G, Speakman JR. Analysis of positive selection at single nucleotide polymorphisms associated with body mass index does not support the “Thrifty Gene” hypothesis. Cell Metab 2016;24:531–41. https://linkinghub.elsevier.com/retrieve/pii/S1550413116304302. [DOI] [PubMed] [Google Scholar]
- 60.Chen G, Doumatey AP, Zhou J, Lei L, Bentley AR, Tekola-Ayele F. et al. Genome-wide analysis identifies an african-specific variant in SEMA4D associated with body mass index. Obesity 2017;25:794–800. 10.1002/oby.21804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Granot-Hershkovitz E, Karasik D, Friedlander Y, Rodriguez-Murillo L, Dorajoo R, Liu J. et al. A study of Kibbutzim in Israel reveals risk factors for cardiometabolic traits and subtle population structure. Eur J Hum Genet 2018;26:1848–58. http://www.ncbi.nlm.nih.gov/pubmed/30108283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salinas YD, Wang L, DeWan AT. Multiethnic genome-wide association study identifies ethnic-specific associations with body mass index in Hispanics and African Americans. BMC Genet 2016;17:78. 10.1186/s12863-016-0387-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


