Abstract
The Candida Genome Database provides access to biological information about genes and proteins of several medically important Candida species. The website is organized into easily navigable pages that enable data retrieval and analysis. This chapter shows how to explore the CGD Home page and Locus Summary pages, which are the main access points to the database. It also provides a description of how to use the GO analysis tools, GO Term Finder and GO Slim Mapper, and how to browse large scale datasets using the JBrowse genome browser. Finally, it shows how to search and retrieve data for user-defined sets of genes using the Advanced Search and Batch Download tools.
Keywords: Candida, genome database, expression analysis, gene ontology, GO Slim, JBrowse
1. Introduction
The Candida Genome Database (CGD; http://www.candidagenome.org) started in 2004 as an online resource that provides free access to genomic, genetic and molecular biology information about Candida albicans. At that time, CGD was based on the newly assembled genomic sequence of strain SC5314 (1). The primary goal was to link the sequence data with the literature-derived experimental data in a single, easy to navigate web-based resource. Since then, the expanding scope of Candida research, facilitated by the progress in sequencing of genomes from other strains and species, has prompted the incorporation of similar data for C. glabrata CBS138, C. parapsilosis CDC317, and C. dublinensis CD36. Most recently, the genome of Candida auris B8441 was added to represent a newly emerging pathogen of great medical importance (2). CGD also serves as an archive that provides access to genomic sequences for other related strains and species, including C. albicans WO-1, C. guilliermondii ATCC_6260, C. lusitaniae ATCC_42720, C. orthopsilosis Co 90-125, C. tropicalis MYA-3404, Debaryomyces hansenii CBS767, and Lodderomyces elongisporus NRLL YB-4239 (3).
At the core of CGD lies human curation, a process that involves manually extracting gene-specific experimental information from the published, peer-reviewed literature and associating those annotations with the relevant genomic features (4). Genes and their annotations are organized in such a fashion that the information is easily browsable, searchable, and retrievable for further analysis and perusal. CGD curators also make sure that every curated piece of information is traceable to its original source, usually a publication in a scientific journal, thus providing access to all available experimental details and their interpretations. CGD also includes a rigorous catalog of orthologs between species (5) and of protein domain structure, which allows consistent predictions of functions for genes that have not been experimentally characterized (6) Thus, CGD provides a structured, unbiased and continuously updated collection of a large variety of experimental results and computational predictions that has become indispensable for Candida researchers
To ensure a uniform representation of biological information across different organisms, most genome databases use controlled vocabularies to annotate various attributes of genes and gene products. The most widely used vocabulary for capturing the key aspects of gene product biology is the Gene Ontology (GO; http://www.geneontology.org/; (7). GO is a system of standardized terms with defined relationships that describe a primary activity of the gene product (Molecular Function), a broader cellular role the gene product is involved in (Biological Process), and the predominant localization, such as a protein complex, a subcellular structure, or an organelle (Cellular Component). CGD uses GO as the main vocabulary to annotate genes. Another data type that CGD captures, mutant phenotypes, is curated using Ascomycete Phenotype Ontology (APO), a vocabulary developed at the Saccharomyces Genome Database (SGD) (8) that we adapted to the specific needs of Candida biology.
The information in CGD is organized in a system of interlinked web pages, designed with the goal of making them intuitive, easily navigable and user-friendly. However, the sheer complexity of the data presented in CGD can make it difficult for a newcomer to find the right information. This chapter provides help for navigating the site and highlights some new features of CGD. We present an overview of the main entry point, the Home Page, and the central organizing principle of the database, the Locus Summary Page. We show how to perform some of the most common types of analysis that utilize GO annotations. We also explain how to use many features of the JBrowse genome browser. Finally, we show how to retrieve various types of data for multiple genes in a format amenable for further analyses.
2. Methods
Exploring CGD Home and Locus Summary pages
The CGD home page (http://www.candidagenome.org) serves as a place for database-related announcements, community news, and upcoming meetings of interest to the Candida community. It also provides a starting point for many of the features and tools available at CGD. The search box is the primary gateway to a variety of data types available in the database and is present on most CGD pages (see Note 1). The Locus Summary page (Figure 1) offers an up-to-date summary of what is known about a particular gene (see Note 2).
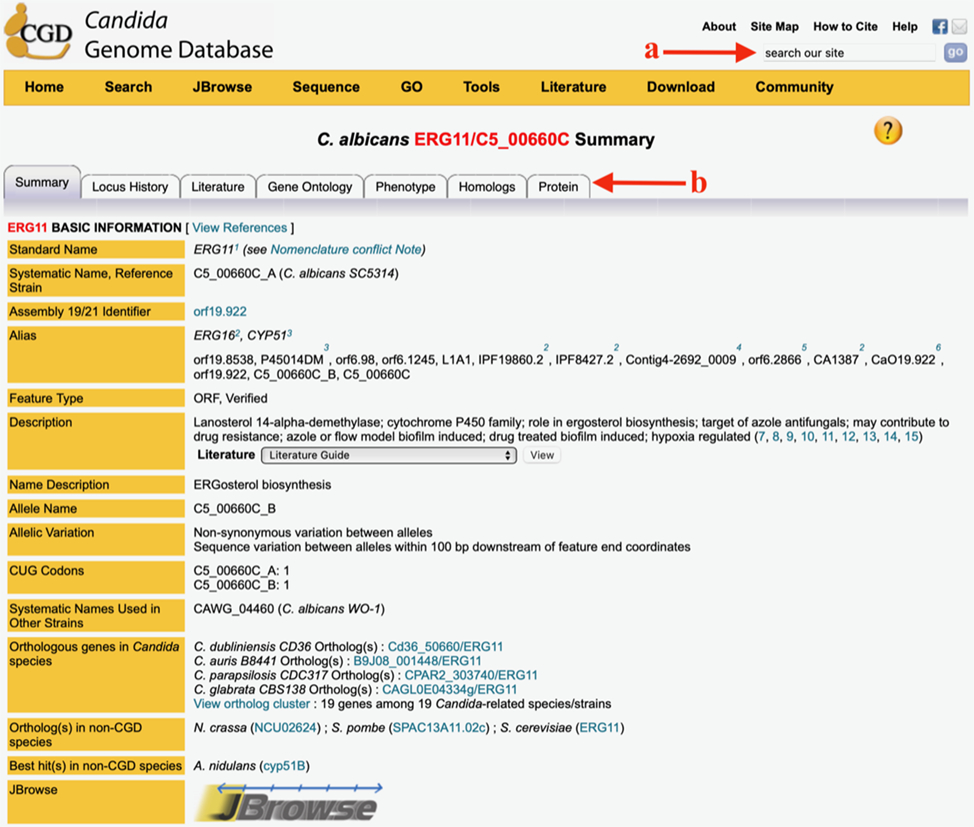
Figure 1.
Basic Information section of the of the Summary Tab of the Locus Summary page for C. albicans gene ERG11. The red arrows point to the “search our site” box (a) and the tabs that lead to pages with more detailed information (b). Other sections of the Summary Tab are available by scrolling down.
-
2.1
Open the home page (http://www.candidagenome.org) and explore the Banner at the top of the page (see Figure 1A). This Banner, present on most CGD pages, provides quick links to multiple data search and analysis tools, literature tools, as well as bulk download tools and various community-related information. Hover the mouse over each item to reveal a drop-down menu with available options.
-
2.2
Enter your query into the “Search our site” box above the Banner. If you enter a unique gene name, such as a systematic name, clicking on the result will jump directly to the Locus Summary page for that gene (see Note 3). If your query produces multiple hits, for instance, a gene name that is used in several Candida species represented in CGD, you will get a “CGD Quick Search Result” page that lists the type and number of hits, from both general search categories (such as GO) and broken down by species specific matches. Positive hits are hyperlinked to either their respective Locus Summary pages, or to an intermediate list of individual hits. Select an individual hit to open its Locus Summary page.
-
2.3
The Locus Summary page contains several tabs, with the Summary tab open by default. Typically, for a protein-coding gene, the other tabs are Locus History, Literature, Gene Ontology, Phenotype, Homologs, and Protein. Flip back and forth through the tabs to see specific types of information they include.
-
2.4
The Basic Information section at the top of the Summary tab (see Figure 1) contains a description that summarizes the most significant features of the locus in a concise, headline-like format. It also lists all the names associated with the locus including the standard name - typically the genetic name under which the gene was first published. If there are other names by which the gene has been referred to in the literature, they are listed as aliases. The identifier assigned during genome sequencing is listed as Systematic Name along with the name of the reference strain used in the sequencing project (see Note 4). There is also a note that indicates whether there are any synonymous or non-synonymous variations between the two alleles in SC5314 and if there are any non-standard CUG codons in either allele. The Basic Information section allows easy navigation to genes in other organisms. Click on any hyperlinked name listed among “Orthologous genes in Candida species” to open its Locus Summary page in CGD. You can also click on the “View ortholog cluster” link, which will show the orthologs from 15 Candida species in their genomic context, a report produced by the Candida Gene Order Browser (5,9). To explore orthologs from other fungal species, select a gene from “Ortholog(s) in non-CGD species” and you will access information at other resources: AspGD, Broad Institute, PomBase, and SGD, for genes from A. nidulans, N. crassa, S. pombe, S. cerevisiae, respectively (see Note 5). At the bottom of this section, there is a thumbnail showing the chromosomal location of the gene, hyperlinked to GBrowse (10), which provides a graphical interface to inspect the genomic context of the gene.
-
2.5
The GO Annotations and Mutant Phenotypes sections show current information about the function of the gene. For more detailed information, including the references on which these annotations are based, open the Gene Ontology or Phenotype tab, respectively. GO annotations on the Summary tab are divided by the GO aspect (Molecular Function, Biological Process, Cellular Component) and by the type of annotation evidence. Manually curated annotations are assigned by a CGD curator on the basis of published, experimental (in most cases) results. Computational annotations are produced by transferring experiment-based GO annotations from orthologous genes in other species, or by predictions based on domain structure. Each annotation is accompanied by an evidence code that indicates the reason behind the annotation, for instance, IMP means Inferred from Mutant Phenotype. Click on any evidence code to see a table of all the evidence codes and their definitions. Annotations based on sequence similarity, genetic or physical interactions also list the source gene(s) and the organism. Click on any gene name to see a report for that gene in CGD or in an external database. Each GO term itself is hyperlinked to another CGD page that provides more information, including term definitions, a diagram depicting the relevant segment of the ontology, and a list of other genes in CGD that are also annotated with that term. Similarly, each mutant phenotype is hyperlinked to a page that lists all other genes in CGD that display the same phenotype.
-
2.6
The Sequence Information section shows the basic data about the gene (chromosomal coordinates, intron-exon structure), but also provides easy access to sequences and sequence analysis tools. Open the drop-down menu next to Retrieve Sequences to see available options that include retrieval of DNA sequence in several configurations and, for protein-coding genes, for the predicted protein sequence as well. Similarly, open the Sequence Analysis Tools drop-down menu to start BLAST searches, restriction analysis or primer design tools. More analysis tools are available from the Banner on top of the page; click on Search, Sequence, Tools, or Download to see the options. In addition, at the bottom of the Sequence Information section there are links to sequence data available from external resources, such as GenBank, UniProt and others.
3. GO Term finder and GO Slim Mapper, tools for GO analysis
The GO vocabularies are constructed as hierarchies, where more general terms, so-called “parents”, encompass more specific “child” terms. When making annotations, curators are required to assign the most specific (granular) term that is supported by the evidence presented in the publication. Within the hierarchical structure of the ontology, annotation to a term also implies annotation to its parent term(s), to the top of the hierarchy. This feature of GO is taken into account during analysis of large-scale data, when identifying common biological features in a set of genes that are, for instance, co-regulated in a genome-wide expression experiment. To find statistically significant similarities in GO annotations for multiple genes, CGD uses GO Term Finder (11) that is able to determine if there are any GO terms that annotate genes in a list (either directly, or indirectly via the GO hierarchy) at a rate greater than would be expected by chance. Given the complexity of GO, it can also be desirable to group genes into broad categories using only high-level terms. The GO Slims are such lists of high-level terms from each ontology branch (Molecular Function, Biological Process, and Cellular Component), carefully selected to cover most of the curated GO information in CGD. The tool, GO Slim Mapper, is able to categorize (map) a large set of genes to user-selected GO Slim terms.
3.1. Using GO Term Finder
-
3.1.1
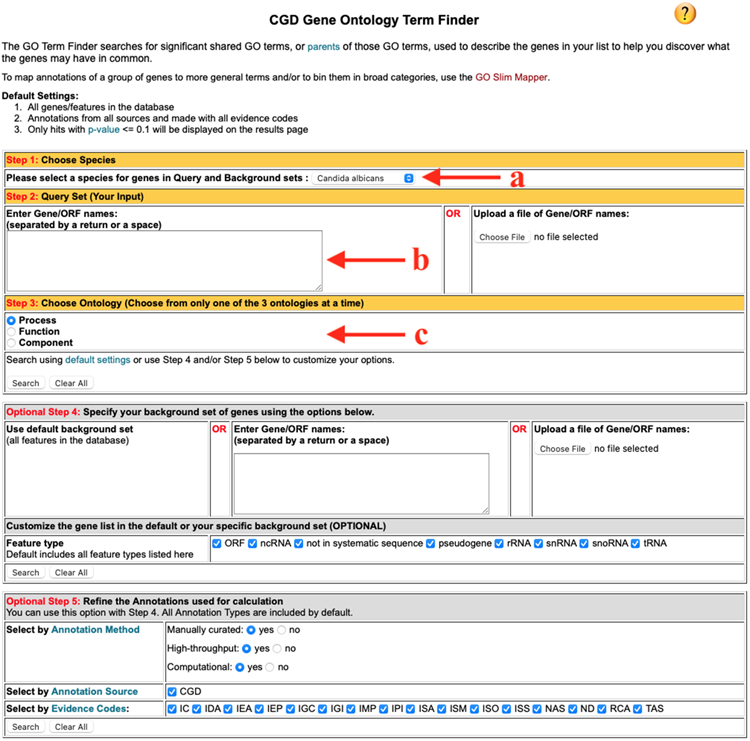
Open the GO Term Finder page (Figure 2) by selecting it from the options in the Search or GO pull-down menus in the Banner (see Note 6).
-
3.1.2
Select the species in Step 1; the default species is Candida albicans, but you can run this analysis for other CGD-curated species: C. glabrata, C. auris, C. dublinensis, or C. parapsilosis
-
3.1.3
In Step 2, enter a list of gene names. You can either type or paste the names of the genes in the input box or upload a file that contains the list. Either genetic names (CGD Standard Names, e.g., AAF1) or systematic names (C3_06470W_A, or orf19 identifiers, e.g., orf19.7436) may be used (see Note 7)
-
3.1.4
In Step 3, select one of the three aspects of GO (biological process, molecular function, or cellular component) by checking the boxes. The tool only searches one of the three branches at a time.
-
3.1.5
Click the Search button after Step 3 to use the default settings or go further down to Steps 4 and 5 to specify and customize your background set and/or refine the types of annotations in your background set.
-
3.1.6
You may change your background set in Step 4. The default background set includes all the genes in the database that have at least one GO annotation. You can also customize the background set by choosing which feature type(s) it should include.
-
3.1.7
In Step 5 you can deselect specific types of GO annotations that should not be used for calculations. By default, annotations collected by all methods and with all types of evidence are included,
-
3.1.8
The results page displays the significant shared GO terms (or their parents) in both graphic and table form, within the set of genes entered on the previous page.
-
3.1.9
The graphic shows the GO tree that includes terms used directly or indirectly in annotations for the genes in your list. The terms are color-coded to indicate their statistical significance (p-value). Genes associated with the GO terms are shown in gray boxes, with links to their respective Locus Summary pages.
-
3.1.10
The table below the graph lists each significant GO term, the number of times the GO term is used to annotate genes in the list and the number of times that the term is used to annotate genes in the background set (see Note 8).
-
3.1.11
Additional columns list the p-value, the False Discovery Rate (FDR), and a list of all the genes annotated, either directly or indirectly, to the term. FDR is an estimate of the percent chance that a particular GO term might actually be a false positive. It represents the fraction of the nodes with p-values as good or better than the node with this FDR that would be expected to be false positives.
-
3.1.12
The statistical significance of the association of a particular GO term with a group of genes in the list is indicated by the p-value: the probability of seeing at least x number of genes out of the total n genes in the list annotated to a particular GO term, given the proportion of genes in the whole genome that are annotated to that GO Term. The smaller the p-value, the more significant the particular GO term association with the group of genes is (i.e., the less likely to occur by chance).
Figure 2.
GO Term Finder query building window. The red arrows point to the pull-down menu for selecting the species (a); the input box where your list of genes is entered (b); and the radio buttons for selecting the GO aspect (c).
3.2. Using GO Slim Mapper
-
3.2.1
Open the GO Slim Mapper window by selecting the options in the Search or GO pull-down menus in the Banner. Select the species for your query in Step 1. In Step 2 you can type or paste your list of genes or upload them as a file.
-
3.2.2
In Step 3 use the pull-down menu to select the GO Set Name: GO Slim Component, GO Slim Function, or GO Slim Process. The list of terms from the selected set appears in the window in Step 4.
-
3.2.3
In Step 4 you can specify which particular term, or terms, you want to use. The default setting is “Select ALL Terms.”, but you can highlight only terms you are interested in.
-
3.2.4
In optional Step 5 you can exclude certain types of annotations, e.g. Computational and High-throughput.
-
3.2.5
Click Search to start the process; note that long lists of genes will take significant time to analyze.
-
3.2.6
Results appear in a table with three columns: the GO Slim terms chosen, with a link to graphical depiction of that branch of GO, the percentage of genes in your list annotated to each term, and the genes from your list that are annotated to that term. You can also download the results in a tab-delimited file.
4. Analyze the genome using JBrowse
Genome browsers allow visualization of large amounts of genomic data in an intuitive graphical format. The results of genome-wide studies are overlaid on the genomic sequence, providing a broad overview of entire regions and, at the same time, a quick access to individual features of interest. In CGD, genome analysis datasets, ranging from gene expression to sequence variants and conservation, are available in JavaScript-based Genome Browser JBrowse (12).
-
4.1
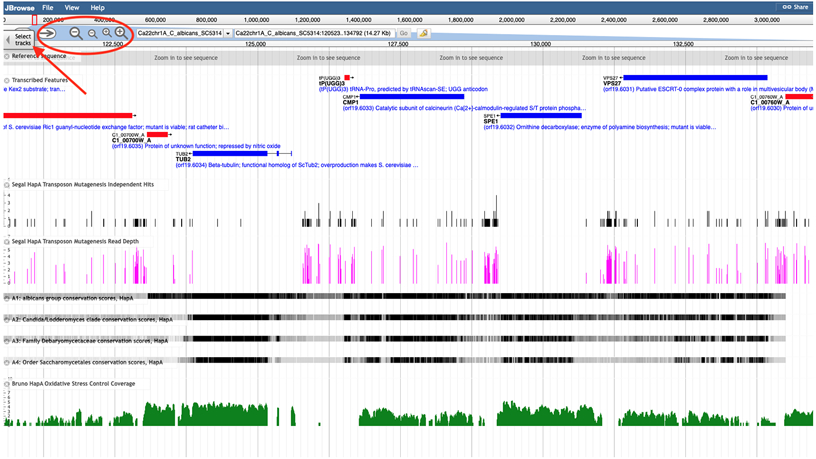
Open the JBrowse window from any Locus Summary page by clicking on the JBrowse thumbnail at the bottom of the Basic Information section. The browser window that opens is centered on the feature of the Locus Summary page, from which it was launched (Figure 3A). Alternatively, you can select JBrowse from the top Banner, select the species you are interested in, and the browser window will be centered on an arbitrary location in the genome (see Note 9).
-
4.2
The navigation bar at the top of the browser window provides controls for browsing. You can shift the display left or right using the left or right arrow, respectively. You can also zoom in or out by clicking on the “+” or “−” icons. The top line, with chromosomal coordinates, has a red box that depicts the currently displayed region of the chromosome; the box can be dragged or resized to display a different region. The two pull-down menus allow selection of a different chromosome and a different chromosomal location. The second box also allows entering a systematic name of a feature to bring into the browser window.
-
4.3
The browser opens with several datasets (“tracks”) preloaded. You can remove a track from the display by clicking on “x” to the left of the track label, or by selecting an option from the pull-down menu attached to the track label. The menu gives access to several other options, including more detailed information about the track, adjustments to the way the track is displayed, and an option to save the track data.
-
4.4
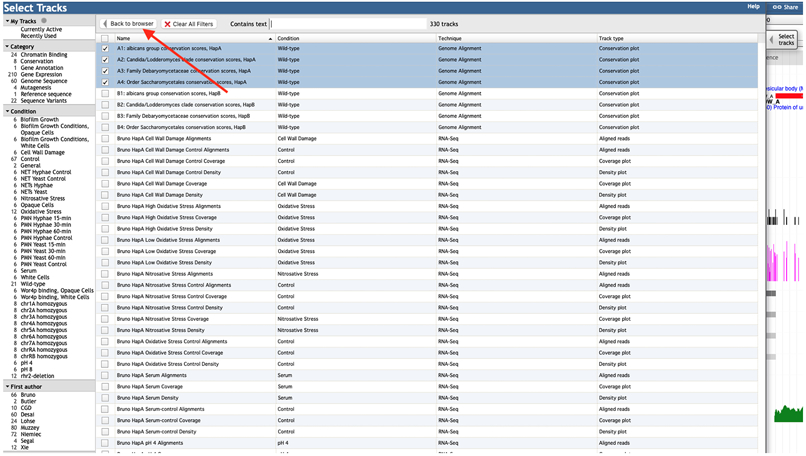
To add a track to the display, click on “Select tracks” in the upper left corner. This will open the table with the available tracks (Figure 3B). To select a track, click on the checkbox next to its description. The panel on the left contains a list of categories, including techniques, the scope and experimental conditions under study, first author names, that allow filtering the tracks. Once the tracks are selected, click on the “Back to browser” button in upper left corner.
Figure 3A.
JBrowse browser window. The display shows a fragment of C. albicans Chromosome 1, Haplotype A. The bars depict transcribed features, color-coded for the + or − strand, and a small arrow indicating the direction of transcription. Several tracks are selected by default. The red oval indicates the navigation tools, and the red arrow next to it points to the “Select tracks” button.
Figure 3B.
JBrowse Select Tracks window (truncated). The main panel shows the available tracks with their Conditions, Techniques and Types. The left panel shows categories that allow filtering the tracks in the main panel. The red arrow points the “Back to browser” button.
5. Using the Advanced Search and Batch Download tools
In conducting research today, it is often necessary to analyze entire sets of genes. The Advanced Search tool allows selecting a group of genes based on several user-defined criteria, and the Batch Download tool facilitates downloading the sequences and other information for a user-provided list of genes.
5.1. Using the Advanced Search tool
-
5.1.1
Open the Advanced Search tool from the Banner, under the Search pulldown menu.
-
5.2.1
In Step 1, select one of the five Candida species; the default option is Candida albicans SC5314.
-
5.2.3
In the required Step 2, select one, several, or all types of chromosomal features by clicking on appropriate checkboxes. Note that no feature type is selected by default.
-
5.1.4
The optional Step 3 allows selecting additional annotated properties, and provides a list of chromosomes you can choose if you want to restrict the search to a particular chromosome or mitochondrial genome. For C. albicans, you can also select either haplotype A or B chromosome. It also allows restricting the search to genes annotated with particular GO-Slim terms, or excluding some annotation methods, such as computational and certain evidence codes (see Note 10).
-
5.1.5
Finish building your query by clicking on Search at the bottom of the page. Depending on the complexity of the query and the number of hits, the search can take a while.
-
5.1.6
The Advanced Search Results page presents a table with the names of the genes, along with their description and relevant GO terms. The systematic names are hyperlinked to their respective Locus Summary pages. At the bottom of the table, there are links to Further Analysis and Download that allow retrieving the search results in an Excel spreadsheet, viewing the GO Annotation Summary, or sending the list directly to the GO Term Finder, GO Slim Mapper or Batch Download tool.
5.2. Using the Batch Download tool
-
5.2.1
Start the Batch Download tool from the Banner, under the Download menu. Or use the Search Results page that opens the Batch Download tool (Figure 4).
-
5.2.2
In Step 1, you can type in, or copy and paste the gene names, separated by return. You can also upload a text file containing the names or use the list already pre-loaded into the input box from your previous search. You also need to specify the species and the genome Assembly (currently Assembly 19, 21 and 22 are available, with the latest Assembly 22 selected by default).
-
5.2.3
The other option in Step 1 is to specify a chromosome or contig and enter the coordinates for the features you want. You can also upload a text file with the coordinates. The required format of the file is explained below the Choose file button.
-
5.2.4
Step 2 allows you to specify what types of data you want to retrieve. You can retrieve several types of sequence data (genomic DNA, or ORF proteins, etc.). You can also select other types of information about the genes. The chromosomal Feature information, for instance, will give you, among other data, all names and aliases associated with the genes on in your input list.
-
5.2.5
Clicking on Submit at the bottom of the page will start the search. Please note that with many hits, the search may take a while.
-
5.2.6
The results are saved in a tab-delimited text file that remains available for download for six hours afterwards.
Figure 4.
Batch Download query building window. The arrows indicate the input box (a); the pull-down menus for species/assembly selection (b); the chromosome/contig/coordinates input boxes (c); and checkboxes for selecting the data type(s) to retrieve (d).
5. Notes
The query entered into the search box may be a Candida gene or protein name (standard or systematic name, or an alias), author or colleague name, PubMed ID, or any keyword (such as a functional term or phenotype). It can even be a name of an ortholog from one of the non-CGD species. When there are multiple hits, a list of matches is displayed.
In addition to protein-coding genes, several other entities have their Locus Summary pages in CGD, including various RNA genes (tRNA, rRNA, snRNA, etc.), as well as a variety of chromosomal features, such as centromeres, telomeres, repeated sequences, and others.
“Search our site” box accepts a wildcard character *. For example, enter “act*” to retrieve any piece of data starting with “act”. Also, the search box has an autocomplete feature, which provides suggestions when you start typing your query.
For C. albicans, the systematic name shown on the Locus Summary page is always the name of the haplotype A allele, as denoted by an “_A” suffix, with the corresponding haplotype B allele listed below. Since the systematic names from previous assemblies of C. albicans genome (so called “orf19” names) continue to be widely used, the Assembly 19/21 identifier is also shown.
The ortholog mappings among Candida strains, and between Candida strains and S. cerevisiae, are derived from the curated syntenic groupings at the Candida Gene Order Browser (CGOB) (13). The ortholog mappings between Candida strains and S. pombe, A. nidulans, and N. crassa are made by pairwise comparisons using the InParanoid software (14).
Links to both GO Term Finder and GO Slim Mapper appear at the bottom of many pages showing results of searches that produce a list of genes. Selecting those links open the respective tool with the gene list already pre-loaded into the input box.
When a name is entered that is an alias for one gene or feature, the program will map the name to that gene. If the name is an alias for more than one gene but not a Standard or Systematic name for any genes, the program will present a list of possible mappings. The user can decide which gene was intended and edit the input.
Because the frequency of any given annotation within the background set is compared against the frequency of the annotation within the query set (input), the choice of background set affects the significance of the results that are returned by the tool. Please note that the specific background set of genes that was used in the absence of any user-defined set (the default background set) has changed over time.
In JBrowse, the two haplotypes in C. albicans SC5314 are displayed separately. The JBrowse window opens the “A” haplotype by default. You can select the other haplotype by selecting a chromosome with the “B” suffix from the pull-down menu.
The Evidence Codes are intended to indicate the confidence level. Annotations based on experimental evidence, such as “Inferred from Direct Assay” (IDA), or “Inferred from Mutant Phenotype” (IMP), are usually considered more trustworthy than annotation based on sequence comparisons (“Inferred from Sequence Similarity” (ISS), for instance), or on large-scale electronic analyses (“Inferred from Electronic Annotation” (IEA)). You should always pay attention to the Evidence Codes and select those that best match the context of your research.
References
- 1.Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, Davis RW, Scherer S (2004) The diploid genome sequence of Candida albicans. Proc Natl Acad Sci U S A 101 (19):7329–7334. doi: 10.1073/pnas.0401648101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP (2017) Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin Infect Dis 64 (2):134–140. doi: 10.1093/cid/ciw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inglis DO, Arnaud MB, Binkley J, Shah P, Skrzypek MS, Wymore F, Binkley G, Miyasato SR, Simison M, Sherlock G (2012) The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res 40 (Database issue):D667–674. doi: 10.1093/nar/gkr945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skrzypek MS, Nash RS (2015) Biocuration at the Saccharomyces genome database. Genesis 53 (8):450–457. doi: 10.1002/dvg.22862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, Cuomo CA (2009) Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459 (7247):657–662. doi: 10.1038/nature08064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skrzypek MS, Binkley J, Binkley G, Miyasato SR, Simison M, Sherlock G (2017) The Candida Genome Database (CGD): incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res 45 (D1):D592–D596. doi: 10.1093/nar/gkw924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Gene Ontology C (2019) The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47 (D1):D330–D338. doi: 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engel SR, Balakrishnan R, Binkley G, Christie KR, Costanzo MC, Dwight SS, Fisk DG, Hirschman JE, Hitz BC, Hong EL, Krieger CJ, Livstone MS, Miyasato SR, Nash R, Oughtred R, Park J, Skrzypek MS, Weng S, Wong ED, Dolinski K, Botstein D, Cherry JM (2010) Saccharomyces Genome Database provides mutant phenotype data. Nucleic Acids Res 38 (Database issue):D433–436. doi: 10.1093/nar/gkp917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzpatrick DA, O'Gaora P, Byrne KP, Butler G (2010) Analysis of gene evolution and metabolic pathways using the Candida Gene Order Browser. BMC genomics 11:290. doi: 10.1186/1471-2164-11-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stein LD (2013) Using GBrowse 2.0 to visualize and share next-generation sequence data. Brief Bioinform 14 (2):162–171. doi: 10.1093/bib/bbt001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G (2004) GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20 (18):3710–3715. doi: 10.1093/bioinformatics/bth456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buels R, Yao E, Diesh CM, Hayes RD, Munoz-Torres M, Helt G, Goodstein DM, Elsik CG, Lewis SE, Stein L, Holmes IH (2016) JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol 17:66. doi: 10.1186/s13059-016-0924-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maguire SL, OhEigeartaigh SS, Byrne KP, Schroder MS, O'Gaora P, Wolfe KH, Butler G (2013) Comparative genome analysis and gene finding in Candida species using CGOB. Mol Biol Evol 30 (6):1281–1291. doi: 10.1093/molbev/mst042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostlund G, Schmitt T, Forslund K, Kostler T, Messina DN, Roopra S, Frings O, Sonnhammer EL (2010) InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res 38 (Database issue):D196–203. doi: 10.1093/nar/gkp931 [DOI] [PMC free article] [PubMed] [Google Scholar]