Abstract
A gene (mgt) encoding a monofunctional glycosyltransferase (MGT) from Staphylococcus aureus has been identified. This first reported gram-positive MGT shared significant homology with several MGTs from gram-negative bacteria and the N-terminal glycosyltransferase domain of class A high-molecular-mass penicillin-binding proteins from different species. S. aureus MGT contained an N-terminal hydrophobic domain perhaps involved with membrane association. It was expressed in Escherichia coli cells as a truncated protein lacking the hydrophobic domain and purified to homogeneity. Analysis by circular dichroism revealed that secondary structural elements of purified truncated S. aureus MGT were consistent with predicted structural elements, indicating that the protein might exhibit the expected folding. In addition, purified S. aureus MGT catalyzed incorporation of UDP-N-acetylglucosamine into peptidoglycan, proving that it was enzymatically active. MGT activity was inhibited by moenomycin A, and the reaction product was sensitive to lysozyme treatment. Moreover, a protein matching the calculated molecular weight of S. aureus MGT was identified from an S. aureus cell lysate using antibodies developed against purified MGT. Taken together, our results suggest that this enzyme is natively present in S. aureus cells and that it may play a role in bacterial cell wall biosynthesis.
Bacterial cell wall biosynthesis is a complex dynamic event involving participation of a variety of enzymes (12). Peptidoglycan, a key component of bacterial cell walls, is composed of glycan strands cross-linked by peptide bridges (4, 11). Two cell wall biosynthetic enzymes that have received much attention, transpeptidase and glycosyltransferase (1, 3, 21), play critical roles in the terminal stages of peptidoglycan formation. Glycosyltransferase is responsible for elongation of the glycan strands using lipid-linked disaccharide-pentapeptide as the substrate. Transpeptidase cross-links the peptide chains attached to the glycan strands (3). A group of bifunctional high-molecular-weight (HMW) penicillin-binding proteins (PBPs) possessing both glycosyltransferase and transpeptidase activity have been identified in both gram-positive and gram-negative bacteria. The N-terminal domains of these HMW PBPs contain glycosyltransferase activity, while the C-terminal domains possess transpeptidase and PBP activities (3, 20). In addition, these bifunctional enzymes contain an N-terminal hydrophobic region responsible for membrane association (5, 7, 18).
Monofunctional enzymes possessing only glycosyltransferase or transpeptidase activity have also been identified (2, 15). For example, low-molecular weight PBPs in several bacterial species have been shown to carry only dd-carboxypeptidase activity (8, 13, 14, 19). Monofunctional and/or non-PBP-related glycosyltransferase (MGT) activities have been reported for Escherichia coli (10) and gram-positive species such as Staphylococcus aureus and Micrococcus luteus (17). Several laboratories have reported molecular cloning of mgt genes from E. coli, Hemophilus influenzae, Ralstonia eutropha, and other gram-negative bacterial species (2, 15, 18). The proteins encoded by these mgt genes show a high degree of similarity to the N-terminal glycosyltransferase domain of HMW PBPs (15, 18). In addition, purified recombinant E. coli MGT is capable of catalyzing peptidoglycan synthesis in vitro (2).
MGT might also play a key role in peptidoglycan biosynthesis in pathogenic gram-positive bacteria such as S. aureus and Streptococcus pneumoniae (16, 17). In this report, we describe the identification of a complete DNA sequence with an open reading frame (ORF) encoding an ∼31-kDa MGT from the S. aureus genome. A genetically engineered soluble form of S. aureus MGT was expressed in E. coli cells and purified to homogeneity. The purified MGT was characterized with regard to protein structure and enzymatic activity. Using the antibodies developed against purified MGT protein, we demonstrated that MGT was natively expressed in S. aureus cells as a membrane-associated protein.
MATERIALS AND METHODS
Identification and cloning of S. aureus mgt
Genomic DNAs isolated from S. aureus strains ST446 (S. aureus 27S, a methicillin-sensitive strain obtained from Richard Novick) and ST430 (a methicillin-resistant strain obtained from Henry Chambers) were used for identification of the mgt gene reported previously (2). Briefly, S. aureus genomic DNA was digested with a variety of restriction enzymes and subjected to Southern analysis using standard protocols. The hybridization probes used for these analyses were produced by PCR and covered the region encoding a portion of MGT from S. aureus (2).
Inverse PCR was used to obtain the entire S. aureus mgt coding region. To prepare the templates for inverse-PCR analysis, samples of S. aureus ST446 genomic DNA, digested to completion with the restriction enzymes AccI, ClaI, and NdeI, were circularized by self-ligation and then used as templates. PCR primers (primer 1, 5′-CAACGATTAGCGACAGAGATG-3′, and primer 2, 5′-TGCCAACTGGTTGATAATACG-3′) were designed on the basis of the published DNA sequence of ORF2 (GenBank accession number L19300 [2]). Inverse PCR was performed using Taq DNA polymerase for a total of 30 cycles with the following cycling pattern: melting at 94°C for 30 s, annealing at 55°C for 30 s, and polymerization at 72°C for 1 min. The amplified DNA fragments were then sequenced. Based on the DNA sequence obtained, the 3′ end of S. aureus mgt was identified and the entire mgt DNA sequence was established. The nucleotide and the predicted amino acid sequences of S. aureus mgt can be obtained from GenBank (accession number AF287468).
Expression of recombinant S. aureus MGT proteins in E. coli
DNA sequence analysis of S. aureus mgt revealed two in-frame translational start sites 16 amino acids apart at the N terminus of mgt. Considering that previously characterized MGTs had an average length of approximately 240 amino acids, we selected the second translational start codon as the initiation of translation because it produced a protein of 253 amino acids, close to the average MGT amino acid length. A DNA fragment containing S. aureus mgt was generated by PCR using the primers 5′-GAACATGGATCCCATATGAAACACGAACCTCAC-3′ (primer 3) and 5′-TGCGGATCCTTAACGATTTAATTGTGACATAG-3′ (primer 4) with S. aureus genomic DNA as the template. For cloning purposes, these two primers were designed to incorporate a BamHI site at the 3′ end and an NdeI site at the 5′ end of S. aureus mgt. PCR conditions were as described above except only 25 cycles were used. The DNA fragment generated by PCR was gel purified and subcloned into the E. coli expression vector pET-16b (Novagen, Madison, Wis.). The resulting construct (pRBP1) introduced an N-terminal His tag (10 histidine residues) to aid in protein purification.
To express a truncated form of MGT lacking the hydrophobic N-terminal domain, another expression vector was prepared. The resultant mgt gene encoded a truncated protein with a deletion of 67 amino acids from its N terminus. PCR primer 4 and primer 5 (5′-TATTTTGGATCCCATATGGATAATGTGGATGAACTAAG-3′) were then used to amplify the desired coding region under the PCR conditions described above. Again, these primers incorporated 5′ NdeI and 3′ BamHI sites in the amplified DNA product. After digestion with NdeI and BamHI, the gel-purified DNA fragment encoding the truncated version of MGT was ligated into plasmid pET-16b (Novagen), forming plasmid pRBP2. The accuracy of the mgt coding regions in these plasmids was confirmed by DNA sequence analysis.
For expression of MGT proteins, E. coli BL-21(DE3) pLysS cells (Novagen), transformed with either pRBP1 or pRBP2, were grown overnight in TY broth (10 mg of tryptone per ml, 5 mg of yeast extract per ml, and 5 mg of NaCl per ml) containing 0.1 mg of ampicillin per ml on a shaker platform at 37°C. To induce protein expression, the overnight broth culture was diluted 1:100 in fresh TY medium with antibiotic, isopropyl-β-d-thiogalactopyranoside (final concentration, 1 mM) was added for induction, and incubation was continued at 30°C for an additional 4 to 5 h. Cells were then harvested by centrifugation.
Purification of recombinant His-ΔMGT
E. coli cells (40 g) transformed with plasmid pRBP2 were resuspended in 400 ml of buffer A (25 mM HEPES [pH 7.5], 0.15 M NaCl, protease inhibitor tablets [Roche], and 200 U of DNase). Cells were then lysed by sonication. The lysate was clarified by centrifugation at 100,000 × g for 40 min at 4°C. The supernatant was loaded onto a 5-ml Pharmacia HiTrap chelating Sepharose column (Ni2+ charged), preequilibrated with buffer B (25 mM HEPES [pH 7.5] and 0.5 M NaCl). After the column was washed with 0.1 M imidazole in buffer B to the baseline absorbance, the bound proteins were eluted using a linear gradient of 0.1 to 0.7 M imidazole in buffer B. Fractions containing the 26-kDa histidine-tagged MGT protein (His-ΔMGT) were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Gel filtration was performed on a size exclusion Superdex-75 column (Pharmacia) preequilibrated with buffer B containing 10% glycerol and 0.5% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}. Protein elution from the Superdex-75 column was monitored by absorbance at 280 nm, and elution fractions were analyzed by SDS-PAGE. Fractions containing highly purified His-ΔMGT were harvested and stored for further analysis.
Generation of antibodies and Western blot analysis
To develop polyclonal antibodies for S. aureus MGT, purified His-ΔMGT was first heat denatured and then subjected to SDS-PAGE analysis. Gel slices containing the ∼26-kDa His-ΔMGT protein were used for immunization of New Zealand White rabbits. Antisera were collected after two booster injections. The quality of the harvested antisera was analyzed by enzyme-linked immunosorbent assays. For Western blot analysis, protein samples were subjected to SDS-PAGE and transferred to nitrocellulose membrane filters. The filters were first incubated with 5% milk in phosphate-buffered saline buffer, then with either the preimmune serum or the polyclonal antibodies against His-ΔMGT (diluted 1:2,000), and finally with the peroxidase-conjugated secondary antibodies (diluted 1:2,000). The cross-linked proteins were detected using ECL immunodetection reagents (Amersham).
Analyses of purified His-ΔMGT
Circular dichroism (CD) spectra of purified S. aureus His-ΔMGT were recorded with an Aviv 62DS spectropolarimeter. Denatured His-ΔMGT was generated by adding guanidine hydrochloride (6 M final concentration) directly to the protein sample. Both native and denatured His-ΔMGT proteins were analyzed at a concentration of 0.5 mg/ml. The displayed spectra were obtained from the averages of values for five scans as described previously (22), with the signal being corrected for background using the enzyme buffer solution.
N-terminal amino acid sequencing of purified proteins was performed on a Procise protein sequencer equipped with a model 140C microgradient delivery system (Applied Biosystems). Purified His-ΔMGT was subjected to SDS-PAGE and then transferred to a polyvinylidene difluoride membrane filter. The protein band, visualized by Coomassie blue staining, was cut from the filter and used for sequence analysis. Electrospray ionization mass spectrometry analysis was conducted using a PESciex API III triple-stage quadrupole mass spectrometer equipped with a pneumatically assisted electrospray (IonSpray) interface as described previously (23).
Enzymatic activity of purified recombinant S. aureus MGT
Cell membranes of Aerococcus viridans (ATCC 10400) were prepared as described previously (9) and used to measure glycan polymerization by monitoring incorporation of 14C-N-acetylglucosamine into trichloroacetic acid (TCA)-precipitable material. To ensure measurement of glycan synthesis, we slightly modified the published procedure (2) as specified below. A typical MGT enzymatic reaction was performed at either pH 6.1 or pH 8.0 as required in a total volume of 70 μl containing the following components: the A. viridans membrane fraction (50 μg), 0.38 mM [12C]- or [14C]UDP-N-acetylglucosamine (specific activity, ∼4,000 cpm/nmol), 0.33 mM UDP-N-acetylmuramylpentapeptide, 50 mM MgCl2, 0.21 mM KCl, 0.83 mM NH4Cl, 250 μg of penicillin G per ml, 50 mM Tris-HCl, 50 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 6.7 mM ATP, and 16 μg of purified S. aureus His-ΔMGT. Reaction mixtures were incubated at 23°C for 1 h and quenched with 1 ml of ice-cold 10% TCA. Precipitates were collected on Whatman GF/C glass fiber filter papers and washed with 5% TCA. Samples were then counted in a liquid scintillation counter after the addition of a toluene-based cocktail as described previously (2). The data shown are averages of values from three separate experiments. The sensitivity of MGT activity to moenomycin A or lysozyme was determined under the same assay conditions.
RESULTS
Identification and cloning of S. aureus mgt
An ORF (ORF2) encoding a portion of a putative MGT was identified in S. aureus previously (2). However, two motifs (RKXXE and KXXXLXXYXN, where X is any amino acid) typically associated with glycosyltransferase were absent in the deduced amino acid sequence of ORF2 (2). A close examination of the DNA sequence revealed the presence of a Sau3A restriction site 6 nucleotides upstream from the stop codon at the end of ORF2, raising the possibility that the missing motifs noted above may have been due to a cloning artifact, which frequently happens in genomic library constructions.
To investigate this possibility, genomic DNA isolated from S. aureus strain ST446 was digested with a variety of restriction enzymes and subjected to Southern analysis using a hybridization probe that covered the region encoding the reported portion of MGT from S. aureus (2). The hybridization pattern obtained was found to be incompatible with the reported DNA sequence. For example, with ClaI digestion one anticipates a 1.3-kb hybridizing fragment; our results indicated a 2.7-kb hybridizing DNA fragment. Similarly, digestion with NdeI should have produced hybridizing DNA fragments of 1.7 and 1.8 kb; our data could confirm only one of the two fragments, with the other one being replaced by a different fragment of ∼2.5 kb. In addition to using Southern analysis, we also investigated this possibility by PCR amplification of regions up- or downstream of the suspected Sau3A site. No amplification products were obtained if PCR primers were positioned on either side of this Sau3A restriction site. To further confirm this conclusion, we performed similar analyses using a different S. aureus strain, ST430, a methicillin-resistant strain. Additional hybridizing bands were not found in genomic DNA from either strain. Thus, duplication and a subsequent partial deletion of mtgA were unlikely. Results from both strains were the same and strongly suggested that an entire mgt gene encoding a putative MGT is present in the S. aureus genome.
Inverse PCR was conducted to clone the entire S. aureus mgt gene as described in Materials and Methods. The newly acquired complete mgt gene from S. aureus encoded all four conserved motifs in the proper spatial orientation observed in other bacterial glycosyltransferases. Amino acid residues 152 to 156 (RKVKE) represented the previously missing RKXXE motif, and residues 170 to 179 (KNEILSFYLN) contained the KXXXLXXYXN motif. Examination of the native DNA sequence downstream of S. aureus mgt did not reveal a coding region homologous to a transpeptidase domain, confirming that the MGT encoded was not the N-terminal domain of an HMW PBP. In addition, hydropathy analysis revealed that this MGT contained a cluster of hydrophobic amino acids (residues 46 to 64) near its N terminus, indicating that this region might traverse the cell membrane in a manner similar to that of other MGTs identified thus far (18). Amino acid sequence comparisons indicated that S. aureus MGT shared significant homology to the N-terminal transglycosylase domain of HMW PBPs from gram-negative and gram-positive bacteria in addition to other characterized MGTs (Fig. 1).
FIG. 1.
Amino acid sequence comparisons of MGTs and the glycosyltransferase domain of PBPs. Numbers in boxes are percentages of amino acid identity between the two indicated proteins. Amino acid sequence comparisons were done with the GAP program from the Genetics Computer Group sequence analysis software package from the University of Wisconsin Department of Genetics. Database accession numbers are provided at the end of each protein description. Only the N-terminal (transglycosylase) domains of PBPs were used in these comparisons. Abbreviations are as follows: Sau, S. aureus; Eco, E. coli; Hin, H. influenzae; Reu, R. eutropha; Kpn, Klebsiella pneumoniae; Ngo, Neisseria gonorrhoeae; Spn, Streptococcus pneumoniae; and Bsu, Bacillus subtilis.
Expression and purification of recombinant MGT in E. coli
A full-length MGT encoded by S. aureus mgt was expressed in E. coli cells. Although full-length MGT, with an anticipated molecular mass of ∼31 kDa, could be overproduced in transformed E. coli cells, it was present mainly in the particulate fraction of the cell lysate (data not shown). These results indicated that S. aureus MGT might be a putative transmembrane or membrane-associated protein, consistent with the results of hydropathy analysis mentioned above.
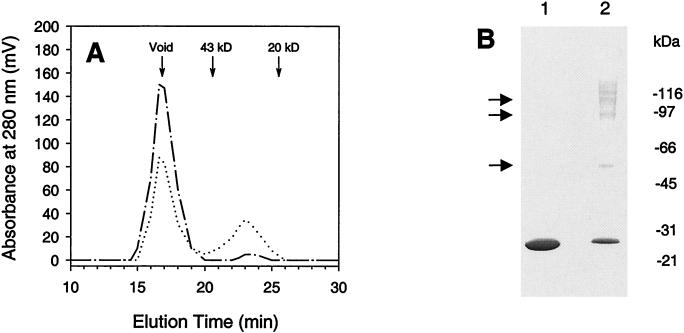
Initial attempts to solubilize membrane-associated full-length S. aureus MGT using various nonionic detergents failed. Therefore, we designed another expression vector, pRBP2, in which the first 67 amino acids were deleted. As expected, E. coli cells transformed with pRBP2 produced a truncated MGT (His-ΔMGT) that was present in the soluble fraction of the cell crude extracts (Fig. 2, lanes 1 and 2). This overexpressed MGT was not found in the control cells transformed with the vector alone. Purification of soluble His-ΔMGT was achieved by one-step Ni2+-affinity column chromatography as described in Materials and Methods (Fig. 2). Using this system, we could purify ∼0.7 mg of His-ΔMGT, with a purity of >95%, from 3.0 g of transformed E. coli cells.
FIG. 2.
Purification of recombinant His-ΔMGT. An aliquot containing His-ΔMGT was loaded onto a 4-to-20% gradient gel for separation, followed by Coomassie blue staining. Lane 1, whole-cell lysate; lane 2, solubilized fraction; lane 3, Ni2+ column pools; lanes 4 and 5, monomer and oligomer pools after Superdex-75 column chromatography, respectively. Lane 6 reflects the results of the Western blot analysis of purified His-ΔMGT (50 ng) using the polyclonal anti-His-ΔMGT antibodies as described in Materials and Methods.
Purified His-ΔMGT exhibited the expected molecular mass of ∼26 kDa on denaturing SDS-polyacrylamide gels. Using ion spray mass spectrometric analysis, we confirmed the molecular weight of the recombinant His-ΔMGT. Western blot analysis indicated that His-ΔMGT was recognized by the polyclonal antibodies raised against gel-purified His-ΔMGT (Fig. 2, lane 6). The identity of His-ΔMGT was further confirmed by N-terminal amino acid sequencing; the first 15 amino acids were identical to those predicted from the DNA sequence of the His-tagged truncated form of S. aureus mgt.
Structural analysis of S. aureus MGT
Although most of the overproduced His-ΔMGT was present in the soluble fraction (Fig. 2), gel filtration analysis showed that nearly all His-ΔMGT appeared in the void volume of the size exclusion column (Fig. 3A), suggesting that His-ΔMGT might aggregate in solution. Addition of 0.5% CHAPS into the protein preparation decreased the aggregation level, which generated a portion of His-ΔMGT (∼35%) that eluted at the monomer position (Fig. 3A).
FIG. 3.
Self-interaction of S. aureus His-ΔMGT. (A) Gel filtration analysis of His-ΔMGT. Shown are the elution profiles of purified His-ΔMGT proteins from a Superdex-75 column in the absence (dashed and dotted line) and presence (dotted line) of 0.5% CHAPS detergent. Elution positions for protein standards are marked. (B) Chemical cross-linking of purified His-ΔMGT. Purified His-ΔMGT (5 μg) was incubated with either the solvent dimethyl sulfonate (lane 1) or the linker disuccinimidyl suberate (lane 2) for 2 h at 4°C as described in the text. The samples were then subjected to separation by SDS-PAGE followed by Coomassie blue staining. Cross-linked complexes are indicated by arrows.
To verify the oligomerization of purified S. aureus His-ΔMGT, we analyzed the protein on a native gel under nondenaturing conditions. Purified His-ΔMGT appeared on the native gel as a smeared band close to the top of the gel (data not shown), consistent with His-ΔMGT forming complexes. Self-interaction was further confirmed by chemical cross-linking experiments. As seen in Fig. 3B, different oligomers, including a dimer, trimer, and tetramer of His-ΔMGT, could be identified using the primary-amine-reactive linker disuccinimidyl suberate. In addition, the cross-linked protein bands were recognized by the polyclonal antibodies against His-ΔMGT as revealed by Western blot analysis (data not shown), confirming that these protein bands contained S. aureus MGT.
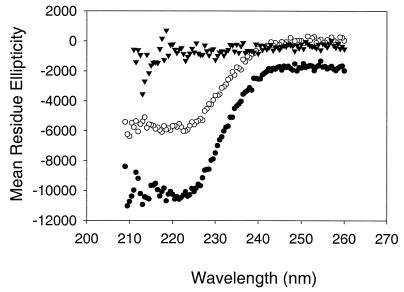
Using the monomeric and oligomeric fractions collected from the size exclusion column, we next conducted structural analysis of these MGT protein samples in order to gain a better understanding of the protein folding. CD spectroscopy, responsive to the contribution of protein secondary structural elements, was used to evaluate the conformation of the monomeric and oligomeric forms of the purified His-ΔMGT. Their CD spectra are shown in Fig. 4, and the compositions of the structural features of the purified His-ΔMGT are summarized in Table 1. A significant difference in the components of the α-helix and β-sheet between the monomer and oligomeric protein was observed (Table 1). The secondary-element features of the monomeric protein were closer to those predicted on the basis of its primary amino acid sequence using the Garnier-Osguthorge-Robson method (6). In addition, the CD spectra of the denatured His-ΔMGT monomer and oligomers, which were different from those generated using the nondenatured protein samples, were also obtained (Fig. 4).
FIG. 4.
CD spectrometric analysis of purified S. aureus His-ΔMGT samples. The CD spectra were collected as described in the text using highly purified His-ΔMGT samples. Filled circles, monomer; open circles, oligomer; triangles, monomer denatured by 6 M guanidine hydrochloride.
TABLE 1.
Structural analysis of His-ΔMGT protein derived from S. aureus
Glycosyltransferase activity of purified S. aureus His-MGT
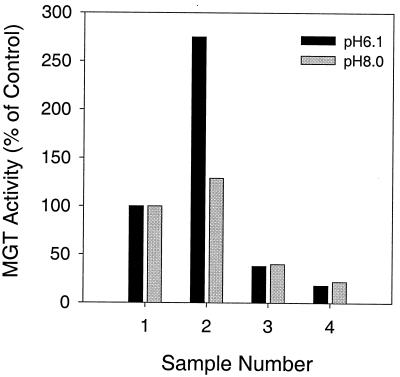
To test if the purified S. aureus His-ΔMGT was enzymatically active, a glycosyltransferase assay was performed. A previous report suggested that MGT from E. coli had a pH optimum between 6.0 and 6.5 for its catalytic activity and that the glycosyltransferase activity of HMW PBPs from E. coli preferred a pH between pH 7.5 and 8.0 (10). Accordingly, we tested the ability of a cell membrane preparation from A. viridans to incorporate a radiolabeled nucleotide-linked sugar precursor, UDP-N-acetylglucosamine, into a glycan polymer at pHs 6.1 and 8.0. These results are presented in Fig. 5. When purified S. aureus His-ΔMGT was tested at pH 6.1, the incorporation of UDP-N-acetylglucosamine increased ∼3-fold, but when it was tested at pH 8.0, only a 25% increase was seen in the reactions (Fig. 5). Addition of moenomycin A, a known glycosyltransferase inhibitor, or lysozyme, an enzyme that cleaves specific disaccharide bonds in peptidoglycan, reduced the formation of the radioactive product (Fig. 5). These results confirmed that purified His-ΔMGT catalyzed the incorporation of the radioactive substrate into peptidoglycan.
FIG. 5.
Peptidoglycan synthesis catalyzed by purified S. aureus His-ΔMGT. Peptidoglycan synthesis reactions were performed as described in the text. Bar 1, control reaction; bar 2, control reaction plus S. aureus His-ΔMGT; bar 3, control reaction plus His-ΔMGT and 10 μg of moenomycin A per ml; bar 4, control reaction plus S. aureus His-ΔMGT and 300 μg of lysozyme per ml.
Presence of MGT in actively growing S. aureus cells
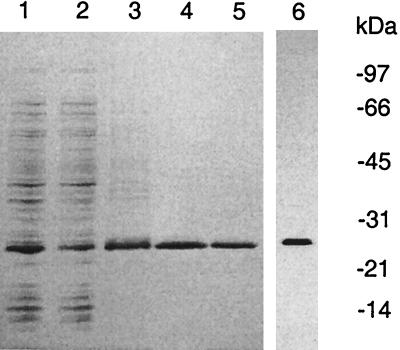
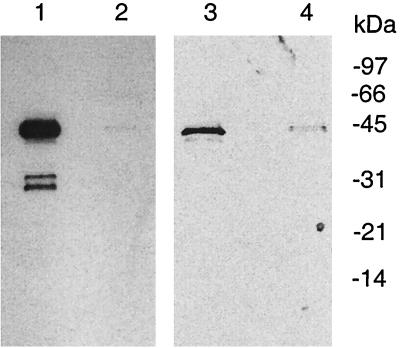
Western blot analysis was used to detect the expression of MGT in actively growing S. aureus cells. Crude cell lysates of S. aureus grown to log phase were probed with polyclonal antibodies specifically developed against purified His-ΔMGT. As seen in Fig. 6, three major proteins, with molecular masses of approximately 42, 32, and 30 kDa, reacted with the antibodies against S. aureus MGT. The 42-kDa protein was assumed to be well-known membrane-bound protein A because this protein was reactive to the preimmune serum (Fig. 6, lanes 3 and 4) as well as the antibodies developed against other unrelated proteins (data not shown). The second protein had a molecular mass of ∼32 kDa, matching the predicted size of a full-length S. aureus MGT protein (Fig. 6, lane 1). Our data also revealed that these proteins were found mainly in the particulate fraction of cell lysates (Fig. 6), consistent with the expected localization for both protein A and MGT. The identity of the ∼30-kDa protein reacting with the antibodies was not clear; it might represent a proteolytic fragment of MGT.
FIG. 6.
Detection of MGT in S. aureus cells. S. aureus cells (0.7 g) grown to log phase were collected and lysed by sonication. The whole-cell lysate was centrifuged at 75,000 × g for 30 min to generate a supernatant fraction (lanes 2 and 4) and particulate fraction (lanes 1 and 3). After separation by SDS-PAGE, Western blotting was performed using either polyclonal antibodies against purified His-ΔMGT (lanes 1 and 2) or preimmune serum (lanes 3 and 4) as described in the text.
DISCUSSION
Glycosyltransferase activity derived from either bifunctional PBPs or the more recently identified MGTs may represent a useful target for the development of potent antibiotics. One of our efforts in searching for novel glycosyltransferase enzymes from pathogenic bacteria included the identification of a complete DNA sequence (mgt) encoding a putative MGT in S. aureus. To the best of our knowledge, this is the first fully characterized mgt reported for gram-positive bacteria.
Our results suggested that S. aureus MGT possessed glycan synthesis activity that was sensitive to assay conditions such as pH, in agreement with results published previously for E. coli MGT (2). Consistent with our expectations, the glycan-synthesizing activity of purified S. aureus MGT was inhibited by moenomycin A and the product was sensitive to lysozyme treatment. The observed MGT activity was low under the conditions tested; this may have resulted from the use of a truncated enzyme, a lack of cofactors, and/or simply the measurement methods used (2, 17).
It has been reported by Park and Matsuhashi that peptidoglycan synthesis in S. aureus is catalyzed mainly by non-PBP-associated transglycosylases (17). To date, it is not clear how much this MGT contributes to the non-PBP-related transglycosylase activity reported for S. aureus cells (17). Final conclusions on this point, as well as its essentiality in S. aureus cells, remain to be determined in future experiments. It is worth noting that gene disruption of an H. influenzae MGT was not a lethal event (data not shown). The knockout results obtained from H. influenzae were not surprising because the glycosyltransferase activity involved in cell wall biosynthesis might be contributed in a redundant fashion by several HMW PBPs plus MGTs. Therefore, one might anticipate that S. aureus MGT is not essential to S. aureus. However, since the purified MGT had catalytic activity and this activity is sensitive to moenomycin A, which also inhibits other cell wall biosynthetic glycosyltransferases, it is reasonable to think that this enzyme can be used in in vitro assays to find inhibitors of glycosyltransferases in general.
Purified S. aureus His-ΔMGT demonstrated self-interaction in solution even though the hydrophobic N-terminal domain had been deleted. This self-interaction was not the result of covalent disulfide (S—S) bridges because this protein did not contain cysteine residues. Oligomerized His-ΔMGT possessed percentages of secondary structural elements, such as the α-helix and β-sheet, different from those of the monomeric form. However, other major elements such as β-turns, important for establishing the overall three-dimensional structure of a protein, and a random coil, which provides dynamic flexibility to the protein, were present to similar degrees in the His-ΔMGT monomer and oligomers. This observation may explain the similar enzymatic activities of these different enzyme preparations. Apparently, His-ΔMGT's self-interaction did not significantly affect the active-site conformation. We have not examined the state of the full-length MGT protein in S. aureus with regard to this phenomenon. Thus, the significance of MGT self-interaction is not clear at this time.
The mere presence of an ORF within an organism does not constitute proof that the encoded protein is expressed. With S. aureus mgt, we were able to detect a membrane-associated protein of the appropriate molecular weight with polyclonal antibodies exhibiting specificity for S. aureus MGT (Fig. 6). These results suggested that S. aureus mgt is not a silent gene and that MGT is actively expressed in S. aureus, most likely as a membrane-associated protein. These data, together with its measurable enzymatic activity, support the notion that this protein may play a role in bacterial cell wall biosynthesis. In this regard, the S. aureus MGT may represent a model for further characterization of non-PBP-related transglycosylases as well as for development of novel antibiotics.
ACKNOWLEDGMENTS
We thank Genshi Zhao, Larry Blaszczak, Melissa Clage, and Angelika Kraft for critical reading of the manuscript and Deborah Mullen and Rohn Millican for technical assistance.
REFERENCES
- 1.Anderson J S, Meadow P M, Haskin M A, Strominger J L. Biosynthesis of the peptidoglycan of bacterial cell walls. Utilization of uridine diphosphate acetylmuramyl pentapeptide and uridine diphosphate acetylglucosamine for peptidoglycan synthesis by particulate enzymes from Staphylococcus aureus and Micrococcus lysodeikticus. Arch Biochem Biophys. 1966;116:487–515. doi: 10.1016/0003-9861(66)90056-7. [DOI] [PubMed] [Google Scholar]
- 2.Di Beradino M, Dijkstra A, Stuber D, Keck W, Gubler M. The monofunctional glycosyltransferase of Escherichia coli is a member of a new class of peptidoglycan-synthesizing enzymes. FEBS Lett. 1996;392:184–188. doi: 10.1016/0014-5793(96)00809-5. [DOI] [PubMed] [Google Scholar]
- 3.Di Guilmi A M, Mouz N, Andrieu J-P, Hoskins J, Jaskunas S R, Gagnon J, Dideberg O, Vernet T. Identification, purification and characterization of transpeptidase and glycosyltransferase domains of Streptococcus pneumoniae penicillin-binding protein 1a. J Bacteriol. 1998;180:5652–5659. doi: 10.1128/jb.180.21.5652-5659.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dmitriev B A, Ehlers S, Rietschel E T. Layered murein revisited: a fundamentally new concept of bacterial cell wall structure, biogenesis and function. Med Microbiol Immunol. 1999;187:173–181. doi: 10.1007/s004300050090. [DOI] [PubMed] [Google Scholar]
- 5.El Kharroubi A, Jaques P, Piras G, Van Beeumen J, Coyette J, Ghuysen J-M. The Enterococcus hirae R40 penicillin-binding protein 5 and the methicillin-resistant Staphylococcus aureus penicillin-binding protein 2 are similar. Biochem J. 1991;280:463–469. [PMC free article] [PubMed] [Google Scholar]
- 6.Garnier J, Gibrat J-F, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 7.Ghuysen J-M. Molecular structures of penicillin-binding proteins and β-lactamases. Trends Microbiol. 1994;10:372–380. doi: 10.1016/0966-842x(94)90614-9. [DOI] [PubMed] [Google Scholar]
- 8.Ghuysen J-M. Penicillin-binding proteins. Wall peptidoglycan assembly and resistance to penicillin: facts, doubts and hopes. Int J Antimicrob Agents. 1997;8:45–60. doi: 10.1016/s0924-8579(96)00358-5. [DOI] [PubMed] [Google Scholar]
- 9.Hammes W P, Neuhaus F. Biosynthesis of peptidoglycan in Gaffkya homari: role of the peptide subunit of uridine diphosphate-N-acetylmuramyl-pentapeptide. J Bacteriol. 1974;120:210–218. doi: 10.1128/jb.120.1.210-218.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hara H, Suzuki H. A novel glycan polymerase that synthesizes uncross-linked peptidoglycan in Escherichia coli. FEBS Lett. 1984;168:155–160. doi: 10.1016/0014-5793(84)80226-4. [DOI] [PubMed] [Google Scholar]
- 11.Holtje J-V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labischinski H, L. a J. Cell wall targets in methicillin-resistant staphylococci. Drug Res Updates. 1999;2:319–325. doi: 10.1054/drup.1999.0101. [DOI] [PubMed] [Google Scholar]
- 13.Markiewicz Z, Broome-Smith J K, Schwarz U, Spratt B G. Spherical E. coli due to elevated levels of d-alanine carboxypeptidase. Nature. 1982;297:702–704. doi: 10.1038/297702a0. [DOI] [PubMed] [Google Scholar]
- 14.Matsuhashi M, Wachi M, Ishino F. Machinery for cell growth and division: penicillin-binding proteins and other proteins. Res Microbiol. 1990;141:89–103. doi: 10.1016/0923-2508(90)90101-u. [DOI] [PubMed] [Google Scholar]
- 15.Paik J, Jendrossek D, Hakenbeck R. A putative monofunctional glycosyltransferase is expressed in Ralstonia eutropha. J Bacteriol. 1997;179:4061–4065. doi: 10.1128/jb.179.12.4061-4065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park W, Seto H, Hackenbeck R, Matsuhashi M. Major peptidoglycan transglycosylase activity in Streptococcus pneumoniae that is not a penicillin-binding protein. FEMS Microbiol Lett. 1985;27:45–48. [Google Scholar]
- 17.Park W, Matsuhashi M. Staphylococcus aureus and Micrococcus luteus peptidoglycan transglycosylases that are not penicillin-binding proteins. J Bacteriol. 1984;157:538–544. doi: 10.1128/jb.157.2.538-544.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spratt B G. Monofunctional biosynthetic peptidoglycan transglycosylases. Mol Microbiol. 1996;19:639–640. doi: 10.1046/j.1365-2958.1996.442924.x. [DOI] [PubMed] [Google Scholar]
- 19.Spratt B G. Penicillin-binding proteins and the future of β-lactam antibiotics. J Gen Microbiol. 1983;129:1247–1260. doi: 10.1099/00221287-129-5-1247. [DOI] [PubMed] [Google Scholar]
- 20.Terrak M, Ghosh T K, van Heijenoort J, Van Beeumen J, Lampilas M, Aszodi J, Ayala J A, Ghuysen J-M, Nguyen-Disteche M. The catalytic, glycosyl transferase and acyl transferase modules of the cell wall peptidoglycan-polymerizing penicillin-binding protein 1b of Escherichia coli. Mol Microbiol. 1999;34:350–364. doi: 10.1046/j.1365-2958.1999.01612.x. [DOI] [PubMed] [Google Scholar]
- 21.van Heijenoort J. Assembly of the monomer unit of bacterial peptidoglycan. Cell Mol Life Sci. 1998;54:300–304. doi: 10.1007/s000180050155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q M, Johnson R B. Activation of human rhinovirus 14 3C protease. Virology. 2001;280:80–86. doi: 10.1006/viro.2000.0760. [DOI] [PubMed] [Google Scholar]
- 23.Wang Q M, Johnson R B, Cohen J D, Voy G T, Richardson J M, Jungheim L N. Development of a continuous fluorescence assay for rhinovirus 14 3C protease using synthetic peptides. Antivir Chem Chemother. 1997;4:303–310. [Google Scholar]