Abstract

With the aim to improve the design of metal complexes as stabilizers of noncanonical DNA secondary structures, namely, G-quadruplexes (G4s), a series of cyclic dinuclear Au(I) N-heterocyclic carbene complexes based on xanthine and benzimidazole ligands has been synthesized and characterized by various methods, including X-ray diffraction. Fluorescence resonance energy transfer (FRET) and CD DNA melting assays unraveled the compounds’ stabilization properties toward G4s of different topologies of physiological relevance. Initial structure–activity relationships have been identified and recognize the family of xanthine derivatives as those more selective toward G4s versus duplex DNA. The binding modes and free-energy landscape of the most active xanthine derivative (featuring a propyl linker) with the promoter sequence cKIT1 have been studied by metadynamics. The atomistic simulations evidenced that the Au(I) compound interacts noncovalently with the top G4 tetrad. The theoretical results on the Au(I) complex/DNA Gibbs free energy of binding were experimentally validated by FRET DNA melting assays. The compounds have also been tested for their antiproliferative properties in human cancer cells in vitro, showing generally moderate activity. This study provides further insights into the biological activity of Au(I) organometallics acting via noncovalent interactions and underlines their promise for tunable targeted applications by appropriate chemical modifications.
Short abstract
Cyclic dinuclear Au(I) NHCs as selective G4-stabilizers.
Introduction
In the last few decades, gold-based drugs have been added to the medicinal chemistry toolbox as new therapeutic agents featuring promising antiproliferative effects against cancer cells, bacteria, and protozoa and endowed with different modes of action with respect to classical organic drugs.1−4 Interest in antimicrobial gold complexes originated at the end of the 19th century from the work of Koch, reporting on the bacteriostatic activity of K[Au(CN)2].5 Later on, gold therapy (chrysotherapy) has been widely used against rheumatoid arthritis, including in the form of the oral drug auranofin [triethylphosphine(2,3,4,6-tetra-O-acetyl-β-1-d-(thiopyranosato-S)Au(I)] (Ridaura). Early studies on auranofin’s anticancer effects revealed activity levels in vitro comparable to those of the Pt(II) complex cisplatin, leading to a vast number of Au(I) complexes with phosphane ligands being evaluated for their antiproliferative effects in vitro and in vivo.6 Recently, auranofin itself has been actively repurposed in pilot trials and clinical studies for anticancer therapy against chronic lymphocytic leukemia and ovarian cancer among others.7,8
Unfortunately, coordination Au(I) complexes can be easily reduced to colloidal gold in a physiological environment, causing either metallodrug inactivation or toxicity in vivo. To achieve an improved control over metal speciation in biological systems, organogold compounds, featuring a stable Au–carbon bond, have been widely explored in medicinal chemistry.9 Among the possible organometallic ligands, N-heterocyclic carbenes (NHCs) perfectly fulfill the prerequisites for an optimal drug design and tuning of the metal complex’s reactivity and physicochemical properties, due to their high structural versatility and ease of derivatization. Thus, numerous cytotoxic Au(I) NHC complexes have been synthesized and characterized for their mechanisms of pharmacological action.10−12 These studies showed the influence of the NHC scaffold on the anticancer activity of the resulting metal compounds—with, for example, imidazol-2-ylidene carbenes being generally more active than benzimidazol-2-ylidene ones—as well as the role of the functionalization of the NHC backbone, of the wingtip substituents and of the ancillary ligands (Figure 1A) on the resulting metallodrug’s bioactivity.13−15 Most of the reported Au(I) NHCs exert their antiproliferative effects via direct binding to protein/enzyme targets, following ligand exchange reactions with thiols or selenol groups in amino acids’ side chains.16,17 Positively charged Au(I) NHCs endowed with a high lipophilic character were also observed to selectively target mitochondria in cancer cells,18 inducing calcium-sensitive mitochondrial membrane permeabilization accompanied by mitochondrial swelling, as well as by inhibition of mitochondrial enzymes.
Figure 1.
(A) General structure of Au(I) NHC complexes and possible modifications and (B) structure of the cationic bis-NHC Au(I) complex [Au(9-methylcaffeine-8-ylidene)2]+ (AuTMX2) as a selective G4 stabilizer.19
In 2014, in collaboration with the group of Le Gendre, some of us reported on the synthesis and antiproliferative properties of xanthine-derived Au(I) NHC complexes as anticancer agents.19 Unexpectedly, the compounds were generally inactive as protein binders and enzyme inhibitors, and electrospray ionization mass spectrometry studies (ESI-MS) showed scarce reactivity with amino acids and model proteins.20 This observation led to the hypothesis that the compounds’ bioactivity may be based on stable noncovalent adducts with biomolecules. Notably, the lead compound in this series, the cationic caffeine-based bis-NHC Au(I) complex [Au(9-methylcaffeine-8-ylidene)2]+ (AuTMX2, Figure 1B), emerged as a very effective stabilizer of noncanonical nucleic acid structures, namely, G-quadruplex (G4) DNA. G4s are secondary DNA structures formed in guanine-rich sequences self-assembled by Hoogsteen-type hydrogen bonds and have been identified in human telomeres and promoter regions of many genes, where they regulate telomere homeostasis, gene transcription, and DNA replication.21 Stabilization of G4s by small molecules may, therefore, induce anticancer effects due to the resulting inhibition of telomere extensions or oncogene expression.22 Metal complexes of different transition metals have been widely investigated as G4 stabilizers and hold promise in drug discovery.23,24
Interestingly, AuTMX2 was also endowed with selectivity for G4 structures over duplex B-DNA, at variance with the bis-benzimidazolylidene Au(I) analogue.19 The observed selectivity could be rationalized by the ability of the caffeine-ylidene scaffold to mimic the guanines of the G4 framework. Moreover, relativistic Au···π-interactions may also play a role in the interaction of the Au(I) compound with guanines.25 According to initial structure–activity relationships19,26 and X-ray diffraction (XRD) studies of the adduct formed by AuTMX2 with a model of human telomeric G4 (hTel23),27 the compound binds noncovalently between neighboring quadruplexes in the crystal lattice, and the presence of two NHC ligands is essential to achieve the highest possible stabilization of the G4 structure. Further structural characterization of the binding modes of AuTMX2 with different G4s was achieved by advanced atomistic simulations, evidencing the importance of π–π stacking and possibly electrostatic interactions in stabilizing the Au(I) compound/G4 adducts.28
In cells, AuTMX2 shows selectivity for cancerous over nontumorigenic cells and against the ovarian cancer A2780 cells with respect to other human cancer cell lines.19 Recently, to shed light into the mechanisms of anticancer action of AuTMX2, shotgun proteomics was applied enabling analysis of the global protein expression changes of AuTMX2-treated A2780 cancer cells.20 The obtained results, combined with various pharmacological methods, evidenced a multi-modal activity of AuTMX2 based on noncovalent interactions with intracellular targets, and involving alterations in the nucleolus, telomeres, actin stress-fibers, as well as activation of redox-related stress-response.20 Considering that, beside humans, putative G4-forming sequences have been found in other mammalian genomes, as well as in yeasts, protozoa,29,30 bacteria, and viruses,31 this type of organogold complexes, active via noncovalent interactions and targeting secondary nucleic acid structures, could be valuable in other research disease areas. In this context, AuTMX2 and selected derivatives were tested against Leishmania amazonensis and Leishmania braziliensis in vitro, showing promising effects in both promastigote and amastigote cells.32
Intrigued by the peculiar reactivity of AuTMX2 with nucleic acid structures with respect to other cytotoxic metallodrugs, we attempted to improve its G4-stabilizing properties by different strategies. Unfortunately, xanthine-derived Au(I) NHC complexes with different substituents at the N-7 position of the ligand scaffold markedly reduced affinity toward G4 stabilization compared to AuTMX2.19 Similarly, functionalization of the N-1 position, as well as substitution of one of the two NHC ligands with alkynyl moieties to achieve neutral complexes, resulted in loss of G4s′ stabilizing effects.26 These findings were justified by the occurrence of steric effects in the substituted AuTMX2 analogues, eventually interfering with the noncovalent G4 adduct formation and relative stability; while lack of suitable interactions, either electrostatic or involving the alkynyl ligands with the G-tetrads occurred for the neutral heteroleptic Au(I) derivatives.
Therefore, to further improve the G4 stabilization properties, we envisioned to “double” the AuTMX2 unit in the compound’s scaffold, designing cyclic dinuclear Au(I) bis-NHC complexes AuB1–3 in which two purine-derived NHC ligands are coupled by an aliphatic bridge (Scheme 1A). Linkers connecting the two Au(I) bis-NHCs via the wing-tip nitrogens are varied in length to find a balance between steric hindrance and the flexibility required for planarization and efficient π-interaction with G4s. Flexible linkers affect the conformation of the gold macrocycles also with respect to the formation of intramolecular aurophilic interactions.33 Moreover, dinuclear gold complexes in which two Au(I) centers are in close proximity, or can at least approach each other through low energy conformational changes, can potentially develop intramolecular aurophilic interactions between the two metal centers, leading to further tuning of the electronic structure and overall compound’s stability and affecting other supramolecular interactions in the proximity of the gold centers.34 Unfortunately, only two (AuB2–3) out of the three derivatives could be fully isolated and characterized. In addition, three benzimidazole-derived analogues AuC1–3 (Scheme 1B) were also synthesized for comparison purposes. The five Au(I) compounds have been characterized by different methods, including nuclear magnetic resonance (NMR) spectroscopy, high-resolution electrospray mass spectrometry (HR-ESI-MS), and elemental analysis, as well as by XRD. Moreover, the compounds were studied for their stability in solution and in the presence of N-acetyl cysteine as a model biological nucleophile by 1H NMR spectroscopy and HR-ESI-MS. The most stable derivatives were then investigated for their G4 DNA stabilizing properties by fluorescence resonance energy transfer (FRET) DNA melting, as well as by circular dichroism (CD), elucidating differences among the two families of Au(I) compounds, particularly in terms of selectivity between G4s and duplex DNA binding affinity in comparison to the benchmark AuTMX2. The binding of the most stabilizing compound within the purine-based cyclic Au(I) bis-NHC family with a promoter G4 sequence (cKIT1) was also studied by atomistic simulations, namely, metadynamics (metaD) free energy calculations.35 The in silico results confirm and complement the experimental data, providing further structural and energetics information on the ligand binding mechanism, including a quantitatively well-defined free-energy landscape. Finally, preliminary antiproliferative activity assays were conducted in a small panel of human cancer cell lines, as well as in nontumorigenic cells in vitro.
Scheme 1. Reaction Scheme Leading to the Metallacyclic Au(I) Purine- (A) and Benzimidazole-Based (B) NHC Complexes.

Results and Discussion
Synthesis of Cyclic Dinuclear Au(I) NHC Complexes
A detailed path for the synthesis of the cyclic dinuclear Au(I) bis-NHC complexes is reported in Scheme 1 and features three main steps: (i) alkyl-bridging of the NHC ligands, (ii) ligand methylation, and (iii) Au(I) complexation. While imidazole and benzimidazole-based cyclic Au(I) compounds have been already reported in the literature,36−38 to the best of our knowledge, the purine-based ligands H2B1–3PF6 have never been synthesized so far. In general, the functionalization of this type of scaffolds is less straightforward with respect to the analogue benzimidazole ligands due to the lower N9 nucleophilicity, requiring an excess of strong electrophiles for substitution reactions.
Starting with the caffeine-based NHC ligands, two theophylline units have been first alkyl-bridged via nucleophilic substitution using dibromo-alkane precursors, leading to A1–3 in good yields (up to 79%, Scheme 1A). Notwithstanding, the low solubility in polar-aprotic solvents of these compounds hampered their methylation with Meerwein’s salt via previously reported routes.37 Inspired by reactions reported by Hori et al. for the solvent-free N9-methylation of xanthines,39,40 compounds A2 and A3 were finally methylated in molten methyl tosylate at 160 °C (Figures S1 and S2) prior to the anion exchange reaction, which allowed to afford H2B2PF6 and H2B3PF6 in good overall yields of 41 and 49%, respectively (Scheme 1A). The successful theophyllinium ion achievement was also confirmed by 1H and 13C NMR spectroscopy in CD3CN (Figures S3–S9), showing a downfield shift of the C8H and C8 signals up to 8.35 and 8.52 ppm (1H) and 140.00 and 139.56 ppm (13C), respectively, for H2B2PF6 and H2B3PF6.
These harsh conditions were regrettably not suitable for A1 methylation, as 1H NMR spectroscopy on the crude reaction mixture showed the prevalence of the 9-methylcaffeinium ion, likely forming via fragmentation of the monomethylated intermediate. Following another procedure by Hori et al.,41 appropriate A1-methylation conditions were achieved using 30 equiv of dimethyl sulfate in dry nitrobenzene at 90 °C (Figure S10). Afterward, the mixture was used without further purification to afford the ligand H2B1PF6 via the counterion exchange reaction in a methanol/water mixture. Complete characterization of the product was carried out using elemental analysis, as well as 1H, 13C, 31P, and 1H–13C HSQC NMR spectroscopy, the latter showing downfield shifts of the imidazolium proton and carbon signals compared to reagent A1 (Figures S11–S14).
Concerning benzimidazole-based scaffolds, the imidazolium ligands H2C1–3 (Scheme 1B) have already been reported by Kühn’s group in 2010, with their synthesis involving harsh conditions such as a pressure tube at 120 °C.42 While the methylene-bridged ligand H2C1Br could only be synthesized via this methodology, a diverse approach was used to afford H2C2Br and H2C3BF4. While H2C2Br could be obtained by refluxing 1-methyl benzimidazole with the linking agent in acetonitrile, the pathway to the propyl-bridged ligand (H2C3BF4) had to be inverted, whereby the linking of the two benzimidazole units had to occur prior to their methylation (Figures S15–S17). As for the caffeine-based scaffolds, a further counterion exchange reaction allowed one to isolate the PF6 salts of the ligands (H2C1–3PF6) in moderate to good yields, with their analytical data matching the literature.42
To afford most of the cyclic Au(I) bis-NHC complexes, except for derivatives AuB1 and AuC2, a so-called “weak-base approach” was applied (Scheme 1).43 A protocol by Nolan and coworkers43 was slightly modified by replacing the use of acetone with acetonitrile to circumvent solubility issues of the NHC ligands. In the presence of K2CO3, the reactions to AuB2, AuB3, AuC1, and AuC3 proceeded to completion at ca. 60 °C within a day. Of note, while AuC3 was previously synthesized by Tubaro and coworkers from the bromine salt of the ligand (H2C3Br),36 in our case milder temperature conditions were required using the hexafluorophosphate salt.
As this approach failed to afford the other two complexes, AuB1 and AuC2, in situ transmetalation of silver carbene was attempted, following a procedure recently reported by us.26 Notwithstanding, only the benzimidazole derivative AuC2 was successfully isolated upon reaction, while several unidentified side products emerged during the reaction to afford AuB1, unfortunately impeding its isolation. Complete characterization of the cyclic Au(I) bis-NHC complexes was achieved by high-resolution mass spectrometry, elemental analysis, and NMR spectroscopy, the latter clearly showing the disappearance of the carbenic proton upon Au(I) complexation (Figures S18–S35 in the Supporting Information).
X-ray Structural Characterization
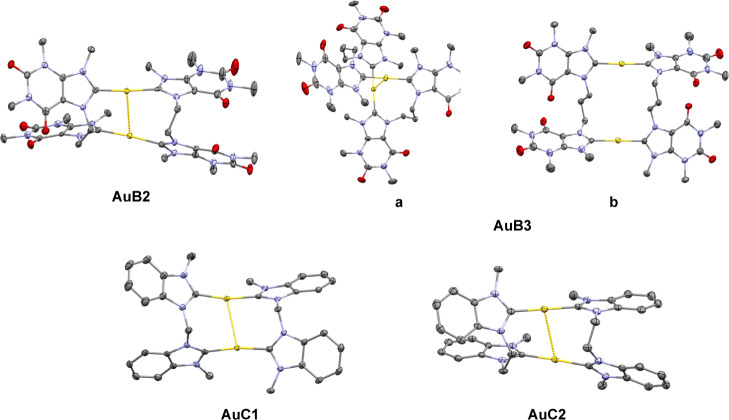
Crystals of AuB2, AuB3, and AuC1 (Figure 2) were grown by slow diffusion of diethyl ether in saturated dimethylformamide (AuB2 and AuB3) or acetonitrile (AuC1) solutions, while AuC2 crystals (Figure 2) grew upon slow evaporation of saturated acetonitrile solutions. All of their structures were determined by XRD (Figure 2, Table 1 and S1–S5 in the Supporting Information), whereas the structure of AuC3 was already reported in the literature.36 The Au–carbene bond lengths (see Table 1 for selected bond lengths and angles) are between 2.016(5) and 2.028(4) Å, in the typical range of NHC–Au(I) complexes found in the Cambridge Structural Database (CSD).44,45 The C–Au–C angles deviate from perfect linearity by different degrees [range from 168.73(9) to 177.68(14)°] but are within literature-reported values for dinuclear Au(I) complexes of bis-NHC ligands. A survey of CSD data shows that deviations from 180° by around 8° or more do basically not occur when monodentate NHC ligands are involved but instead are frequently observed with bidentate or multidentate NHC ligands, which can be attributed to the strain of the ligands and intramolecular aurophilic interactions.44,45 With intramolecular Au···Au distances between 2.9188(7) and 3.2118(4) Å, strong aurophilic interactions can be observed in all compounds with a fold back conformation.46 Intriguingly, the AuB3 crystal structure features two independent molecules with different conformations (Figure 2). Only the first one (Figure 2a), with a twisted conformation resembling a cutout from a helix, features an aurophilic interaction—the shortest one [2.9188(7) Å] among all our compounds. A similar arrangement was reported for a macrocyclic benzimidazole-based tetra-NHC complex of Ag.47 This structure is the only one in our study in which the two NHCs binding one Au(I) center are not strongly rotated around the C–Au–C axis; instead, they are bent outward with angles of 23.11 and 30.68° between each other, while the two NHC planes of the same ligand are roughly parallel (angles of 3.57 and 13.00°).
Figure 2.
Molecular structures of the cyclic dinuclear Au(I) bis-NHC complexes AuB2, AuB3, AuC1, and AuC2. Compound AuB3 features two independent molecules (a,b) with different conformations. Ellipsoids are displayed at the 50% probability level. Hydrogen atoms as well as co-crystallized solvent molecules and counterions are omitted for clarity. Color code: Au (yellow), C (gray), N (blue), and O (red).
Table 1. Selected Distances [Å] and Angles [deg] for the Complexes Depicted in Figure 2 (AuB3 Has Two Independent Molecules in the Asymmetric Unit, a and b as Shown in Figure 2)a.
| AuB2 | AuB3 (a) | AuB3 (b) | AuC1 | AuC2 | |
|---|---|---|---|---|---|
| Au–C | 2.023(4) | 2.021(5) | 2.016(5) | 2.013(2) | 2.027(3) |
| 2.028(4) | 2.020(5) | 2.019(2) | 2.022(3) | ||
| 2.024(4) | 2.023(5) | 2.024(2) | 2.022(3) | ||
| 2.020(4) | 2.021(5) | 2.021(2) | 2.027(3) | ||
| Au–Au | 3.1307(5) | 2.9188(7) | 5.7990(8) | 3.027(1) | 3.2118(4) |
| C–Au–C | 170.58(18) | 171.38(19) | 176.10(19) | 170.03(9) | 177.68(14) |
| 175.59(19) | 172.76(19) | 168.73(9) | 176.22(14) | ||
| NHC–NHC (via Au) | 40.90 | 23.11 | 63.10 | 55.84 | 46.49 |
| 53.12 | 30.68 | 53.46 | 59.25 | ||
| NHC–NHC (via bridge) | 69.76 | 13.00 | 63.10 | 80.13 | 41.58 |
| 47.83 | 3.57 | 72.28 | 54.16 | ||
| Ccarbene–Nbridge | 1.336(5) | 1.331(6) | 1.337(6) | 1.365(3) | 1.371(4) |
| 1.342(6) | 1.336(6) | 1.337(6) | 1.358(3) | 1.355(5) | |
| 1.338(6) | 1.332(6) | 1.358(3) | 1.355(5) | ||
| 1.346(5) | 1.341(7) | 1.360(3) | 1.365(5) | ||
| Ccarbene–Nterminal | 1.366(5) | 1.373(6) | 1.361(6) | 1.347(3) | 1.345(4) |
| 1.377(5) | 1.369(6) | 1.366(6) | 1.343(3) | 1.354(5) | |
| 1.367(5) | 1.371(6) | 1.344(3) | 1.358(5) | ||
| 1.369(5) | 1.374(7) | 1.343(3) | 1.344(4) |
NHC–NHC angles are based on mean planes through the five atoms of the respective imidazole rings and the smaller angle between the planes is always chosen.
In the second structure of AuB3 (Figure 2b), the Au atoms are apart by 5.7990(8) Å and the conformation can be described as outstretched with a slightly folded bridge, whereas the literature-reported AuC3 has a fully outstretched bridge with a Au···Au distance of 6.827(2) Å.36 Moreover, in the second molecule of AuB3 and in the other complexes, the angles between the NHC planes of the same ligand are much larger (>40°, Table 1). Particularly in the ethylene-bridged complexes, the NHC moieties are heavily folded toward each other. Compared to the out-of-plane bending in the first molecule of AuB3; in the other cases, the NHCs bound to the same Au atom are rotated by 40.90–63.10° (Table 1) around the C–Au–C axis. This is probably imposed by the limited flexibility of the bidentate ligands as di-NHC Au complexes of monodentate NHC ligands usually favor a coplanar arrangement (within tolerance and in the absence of bulky wingtips), as reported, for example, in AuTMX2 or [Au(dimethylbenzimidazolylidene)2]+ with different counterions.48,49
It has been previously shown that the Ccarbene–N bonds in AuTMX2 and related compounds are of different lengths, which was rationalized by consideration of the mesomeric structures of disubstituted theophyllinium ions, suggesting that the bond order of the C–N bond closer to the adjacent carbonyl group of the pyrimidine ring is higher and corresponding to a shorter bond length by ca. 0.03 Å.19 In accordance with this observation, very similar differences in the Ccarbene–N bond lengths of AuB2 and AuB3 were noticed (Table 1). The benzimidazole-derived complexes AuC1 and AuC2 as well as literature-reported AuC3,36 however, do not feature such systematic differences but instead display smaller variations or no differences within standard deviations at all.
Reactivity with Model Thiols
The stability of the newly synthesized Au(I) NHC compounds was assessed over 24 h in a mixture DMSO-d6/D2O (80:20) by 1H NMR spectroscopy, showing no spectral changes in this timeframe (Figures S36–S40 in the Supporting Information). To further characterize the compounds’ reactivity toward model biological nucleophiles, each complex was dissolved in a mixture DMSO-d6/D2O (80:20) and exposed for 24 h to an equimolar amount of N-acetyl-l-cysteine (NAC), with the reaction monitored via 1H NMR spectroscopy. It should be noted that, in the applied experimental conditions, NAC undergoes auto-oxidation to cystine over time with the appearance of new peaks at around 4.45, 3.09, and 1.87 ppm (see reference spectrum in Figure 3A), as already reported in the literature.26
Figure 3.
(A) 1H NMR spectra in DMSO-d6/D2O (80:20) of AuB3 alone and in the presence of an equimolar amount of NAC; the NAC spectrum is also reported as reference. Newly formed peaks are labeled with *. (B) Evolution of the 4.10 (blue) and 1.83 ppm 1H NMR signals (green) over 24 h reaction. (C) HR-ESI-MS of the reaction mixture after 24 h.
While AuB2, AuC2, and AuC3 (Figures S41, S44 and S45 in the Supporting Information) did not show any reactivity toward the model nucleophile, significant changes in the 1H NMR spectra were recorded for AuB3 over time (Figure 3). The compound is stable over ca. 3 h; afterward, a new multiplet is detected at around 4.5 ppm (blue star in Figure 3A, close to the signal belonging to the aliphatic linking chain of the unreacted complex), which reaches its maximum at 8 h (Figure 3B) and stays stable over 24 h, while additional new peaks appear in the range 3.0–4.1 ppm. Similarly, a new signal is detected at 1.83 ppm, in the region where NAC resonates (green star in Figure 3A), whereas no further peaks appear in the aromatic range, suggesting that no protonation of the carbenic carbon occurs. Concerning complex AuC1, new peaks can be detected after NAC addition whose intensity increases over time (Figure S42). After 24 h, the methylene-bridged benzimidazole-based Au(I) complex, AuC1, showed a new multiplet of low intensity between 8.16 and 8.23 ppm and in the high-field range, two more peaks appeared at 1.79 and 1.56 ppm, respectively, while NAC signals varied their intensity over time (Figure S42). Such variations are likely to correspond to NAC–Au adduct formation; nevertheless, the main species present corresponds to the intact Au(I) complex even after 24 h.
To further elucidate the nature of the new species formed, the AuB3–NAC reaction mixture was analyzed via HR-ESI-MS in comparison to the sample of the unreactive analogue AuC3 (Figures 3C and S43). While the latter, as expected, displayed only the species corresponding to the intact gold compound, the AuB3–NAC sample spectrum shows both the signal of the intact complex and an additional monocharged peak at 791.2044 m/z, which could not be identified. Overall, these results indicate that the five cyclic compounds are sufficiently stable in solution and even when forming adducts with biological nucleophiles, their reactivity is markedly slower with respect to other Au(I) NHC systems.
DNA G4-Stabilization Properties
The G4-stabilizing properties of the isolated bis-NHC Au(I) complexes, AuB2, AuB3, and AuC1–3, were assessed via the FRET DNA melting assay using a panel of five G4s in a 5:1 stoichiometry [Au(I) NHC complex: G4, see the Experimental Section for details]. The G4s were chosen to represent both the telomeric (hTelo) and promoter (cKIT1, cKIT2, BCL2, and hTERT) regions of the polynucleotide, as well as different G4 strand orientations such as hybrid-mixed (3 + 1) (hTelo and BCL2) and parallel (cKIT1, cKIT2, and hTERT).50−52 The bis-caffeine complex AuTMX2 was tested as a benchmark, also because its cKIT2 and BCL2 G4-stabilizing properties have never been reported so far.26,28
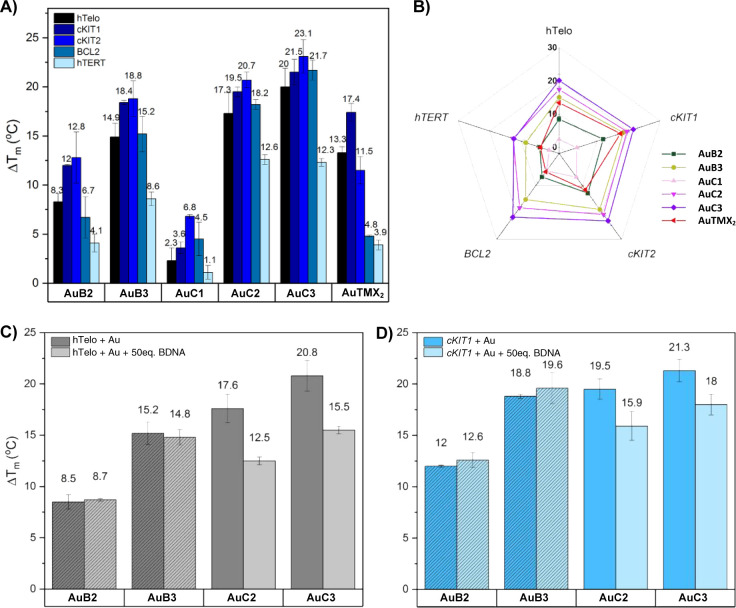
As shown in Figure 4A,B, all the Au(I) bis-NHC complexes can stabilize the selected panel of G4s, with their ΔTm increasing with the length of the linker chain. This trend is particularly evident within the xanthine-based complex series, with the propyl-bridged Au(I) NHC complex AuB3 outperforming the shorter bridged AuB2 by ca. 6 °C per G4 structure. Similarly, in the benzimidazole series, AuC3 displays a ca. 2–3 °C higher ΔTm than AuC2, while the methylene-bridged complex AuC1 barely stabilizes the selected G4s (with ΔTm ranging from 1.1 to a max of 6.8 °C depending on the G4). In the xanthine series, AuB3 shows a comparable stabilizing capability compared to the benchmark AuTMX2 toward hTelo and cKIT1 but higher ΔTm compared to the other G4s. Specifically, AuB3 features larger stabilization effects than AuTMX2 for cKIT2 and hTERT (2-fold increase) and BCL2 (3-fold increase). In the benzimidazole series, both AuC2 and AuC3 can stabilize each G4 structure more prominently than the AuTMX2, with an average increase in ΔTm (between 12.3 and 23.1 °C depending on the G4).
Figure 4.
G-quadruplex stabilization effects induced by the Au(I) NHC complexes studied by the FRET DNA melting assay. (a) ΔTm (°C) of selected G4s solutions in 60 mM potassium cacodylate (pH = 7.4) in the presence of Au(I) NHC complexes (5 equiv); (b) representation of the ΔTm (°C) in a radial plot (from 0 to 30 °C). ΔTm (°C) of hTelo (c) and cKIT1 (d) samples in 60 mM potassium cacodylate (pH = 7.4) in the presence of 5 equiv of selected Au(I) NHC complexes and 50 equiv of CT-DNA. A pattern is shown to differentiate the purine series from the benzimidazole one. Data are shown as mean ± SEM of at least three independent experiments.
In general, a trend of stabilization capability toward the different G4s could be observed. Figure 4B shows that, while AuTMX2 features a certain degree of selectivity, poorly stabilizing BCL2 and hTERT with respect to other G4s, in the bimetallic complexes a comparable stabilization of the different secondary structures occurs. Noteworthy, the stabilization enhancement toward BCL2 by AuB3 compared to AuTMX2 represents an interesting element of novelty and deserves further investigation due to its role in cancer: BCL2 indeed encodes for an antiapoptotic protein, whose overexpression greatly contributes to cancer development and resistance to chemotherapeutic drugs.53 Additionally, while the benzimidazole-based complexes show no preference between the different G4s, AuB2 and AuB3 induce a 4 °C higher stabilization of cKIT1 and cKIT2 compared to the investigated hybrid-mixed conformations, highlighting a mild preference toward parallel G4 structures. To further validate the trend of stabilization via another method, to exclude the influence of the FRET fluorochromes in the binding, we decided to perform additional CD DNA melting studies on selected compounds, namely, the most active AuB2, AuB3, and AuC3 derivatives. The results are reported in Figure S47 and show that the compounds feature the same range and trend of ΔTm values as assessed by FRET with AuB2 < AuB3 < AuC3.
Afterward, the two series of cyclic Au(I) bis-NHC complexes were challenged with an excess (50 equiv) of duplex calf-thymus DNA (CT-DNA) in the presence of hTelo or cKIT1, chosen as models for hybrid and parallel mimicking G4s, respectively. Both the benzimidazole-based compounds, AuC2 and AuC3, experienced a loss in stabilization by ca. 28%, while neither AuB2 nor AuB3 were affected by the presence of the CT-DNA (Figure 4C,D). The results are in line with the high G4 selectivity observed in the case of AuTMX2 and of previously reported mononuclear N1-substituted caffeine-based Au(I) NHC complexes.26 Therefore, although the two benzimidazole-based complexes could stabilize the G4s to a greater extent than AuTMX2, they are less selective DNA binders as their mononuclear Au(I) bis-(N,N-dimethyl-benzimidazole) derivative.19 Further studies are necessary to establish selectivity for G4 over duplex DNA in more physiologically relevant conditions (e.g., higher excess of duplex DNA).
Additional FRET DNA melting assays were performed with increasing equivalents (from 1 to 5 equiv) of the two most potent representative Au(I) complexes in both families, AuB3 and AuC3, using hTelo and cKIT1 as models. The obtained results are depicted in Figure 5 and show that the system reaches a maximum in stabilization at an Au(I) complex/G4 ratio of 2:1 in each case. Moreover, the overall trend of stabilization is always more pronounced for the benzimidazole derivatives with respect to the caffeine-based analogues.
Figure 5.

Stabilization effects induced by 1–5 equiv of AuB3 (sky blue dashed lines) or AuC3 (dark blue dashed lines) complexes toward hTelo (squares) or cKIT1 (circles) studied by FRET DNA melting in 60 mM potassium cacodylate (pH = 7.4).
CD Titrations
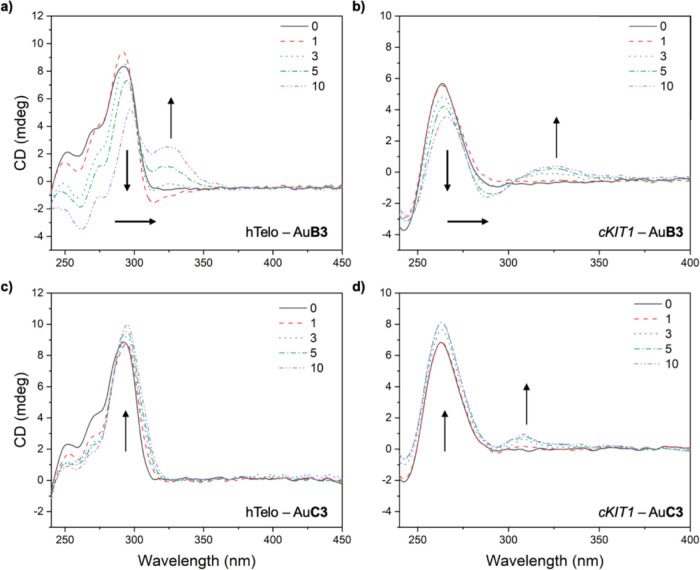
To gain insights into the structural modifications occurring to the G4s upon interaction with the cyclic Au(I) bis-NHC complexes, CD titrations of solution of hTelo and cKIT1 in the presence of increasing amounts of the most active G4 stabilizers of the two families, AuB3 and AuC3, were performed (Figure 6). As expected, the two G4s have a different conformation resulting in markedly different CD spectra (black traces, Figure 6). The hybrid (3 + 1) hTelo profile is characterized by a positive band at 290 nm, a shoulder at 270 nm, and a negative band at 240 nm (black solid lines in Figure 6A,C), while the parallel G4 folding of cKIT1 produces an intense positive and a weaker negative band at 263 and 240 nm, respectively (black solid lines in Figure 6B,D).54
Figure 6.
CD spectra of hTelo (a,c) and cKIT1 solutions (b,d) in the presence of increasing amounts (1–10 equiv) of AuB3 (a,b) and AuC3 (c,d) recorded in Tris–HCl/KCl (10/50 mM, pH = 7.4) buffer. Directions of spectral changes are shown by the arrows.
Increasing amounts (1–10 equiv) of gold complexes result in the appearance of weak-induced CD bands (ICD) above 300 nm in most cases (made exception of the hTelo-AuC3 adduct), diagnostics of the interaction of the achiral cyclic Au(I) bis-NHC complexes with the chiral G4 backbone, giving rise to new chromophores.55 These bands, reported also for other G4-interacting metal complexes,56,57 suggest the tight binding of the gold compounds to the DNA secondary structures. Moreover, while groove-binders commonly produce intense ICD signals, DNA intercalators and G4 end-stackers cause weak or absent induced bands.55 Considering the moderate magnitude of these bands in our experiments, it is reasonable to assume that the AuB3 and AuC3 likely interact with the G-quadruplexes by π-stacking onto the G4-tetrad.
Additionally, the presence of the purine-based representative complex AuB3 also results in a bathochromic effect and intensity loss of the positive bands for both G4s (Figure 6A,B). Conversely, AuC3 leads to a moderate increase in the absorbance of the positive peaks of both hTelo and cKIT1, located at 290 or 263 nm, respectively (Figure 6C,D). It should be noted that these latter effects and spectral changes in the region <300 nm may also be due to an additive effect of further ICD bands of the complexes. Intriguingly, the hTelo CD spectrum in the presence of AuC3 does not show any ICD signal and yet experiences the loss of the DNA “shoulder” band located at 270 nm (Figure 6C), which is not the case when AuB3 is concerned (Figure 6A). This event, together with the increase of the 290 nm band, suggests a likely transition from a hybrid mixed to an antiparallel hTelo conformation in the presence of AuC3, as already described for other metal complexes in the literature.56,58
MetaD Study of Noncovalent Adduct Formation
Afterwards, metadymanics was applied to investigate the interactions between di-nuclear AuB3 and cKIT1 in comparison with the mono-nuclear AuTMX2 complex previously reported.28 MetaD broadens the scope of molecular dynamics simulations by accelerating rare events along selected reaction coordinates, named collective variables (CV), from which the free energy of complex molecular systems59 can be integrated. It has been successfully applied to calculate the free-energy surface (FES) of noncovalent substrate/drug adducts with biomolecules,60,61 including DNA secondary structures.28
Well-tempered metaD calculations consisted of manifold 1000 ns (1 ms) of simulation time. Distances between each AuB3 gold atom and the upper K+ ion at the center of cKIT1, respectively, were chosen as CVs (CV1 and CV2 in Figure 7A). A FES was obtained for each run. A representative FES heat map in shown Figure 7B, see the Experimental Section for details.
Figure 7.
(A) Position of AuB3 and cKIT1 at the beginning of the metaD simulation, showing in stick the upper tetrad and upper K+ ion (purple sphere) and in ball-and-stick the cyclic dinuclear Au(I) bis-NHC complex. CVs (CV1 and CV2) correspond to distances of Au+ ions from the K+ ion (Å). (B) Representative FES depicted as a heat map showing two energy minima based on the two CV distances. (C) Comparison of averaged ΔG for both metaD and FRET DNA melting assays for AuTMX2(28) and AuB3, defined as a bar graph with standard deviation added (state I and state II refer to CV1 < CV2 and CV2 < CV1 respectively).
Gibbs free energy differences and distances were defined with respect to the lowest ΔG value of the FES at the corresponding complex-target distance, for each run (values are detailed in Table S6). Figure 7C reports the ΔG values calculated for AuB3 in comparison to those previously reported for the caffeine-based benchmark AuTMX2.28 In the CV representation (Figure 7B, heatmap), two free energy minima are visible (see dark blue minima in Figure 7B), which correspond to two equally probable binding states (state I and state II) of comparable energy, namely, −39 ± 7.3 and −36.8 ± 3.8 kJ/mol–1, respectively (Figure 7C). Of note, the results are in line with the experimentally calculated ΔG values from FRET DNA melting experiments (see Figure 7C and experimental for details). Moreover, they are also comparable to those previously reported for AuTMX2(28) and in line with the experimental observation that the two complexes feature similar G4 stabilizing activity toward cKIT1 (Figure 4). Overall, from detailed inspection of the simulations, only one caffeine unit (TMX) in AuB3 interacts closely with guanine bases (G6 and G10) of the upper tetrad region (Table S6, Movie S1 and Figure 8) despite the “doubling” of the AuTMX2 scaffold. More specifically, hydrogen bonding and π–π stacking/π-alkyl interactions with G6 and G10 of the tetrad and the loop base A1 were the most prevalent (79% of the trajectories) (Figure 8), confirming the top stacking of the complex as surmised from the ICD band appearance. To a lesser extent, interactions were also observed with the other two guanine bases of the top tetrad, G2 (58%) and G13 (42%), respectively.
Figure 8.
Positions and interactions of the upper G-tetrad nucleobases of cKIT1 with AuB3 (top) and AuTMX2 (bottom) as studied by metaD. (A) G4-adducts of AuB3 and AuTMX2 with cKIT1 in their lowest energy conformations (state I in the case of AuB3). (B) Zoom into the interactions of AuB3 and AuTMX2 with the upper G4 tetrad with π–π stacking interactions shown as an orange surface. (C) 2D interaction diagram shows increased π–π stacking with AuTMX2 when compared to AuB3 with nucleobases G6 (DG6) and G10 (DG10).
Concerning the two binding states (I and II) of AuB3 with cKIT1, a “parallel mode” to base A1, with π–π stacking between AuB3 and bases G6 and G10 (Figure 9A), and a “perpendicular mode” to base A1 with π–π stacking between AuB3 and bases G6 and G10 (Figure 9B) can be identified. These two modes are equivalent (equal occurrence of both parallel and perpendicular binding modes) and shape the symmetric appearance of the FES heatmap in Figure 7B.
Figure 9.
Positions of AuB3 on the upper G-tetrad of cKIT1 in the “parallel” orientation with respect to A1 (A) observed in 21% of the simulation and in the “perpendicular” orientation (B) observed in 47% of the simulations.
Interestingly, residue A1 is observed to switch between an “open” and “closed” conformation (Figure 10) in 9 out of 19 trajectories. While the “closed” arrangement hinders the binding of AuB3 to G2 and G13, the “open” conformation in principle enables the compound to gain access to the nucleobases. However, as mentioned above, only one of the four caffeine units of AuB3 consistently interacts with the upper tetrad (Figure 8B,C), with a second adjacent TMX forming π–π stacking interactions with A1, in either the “open” or “closed” conformation (Figure 9). This lack of further interactions with the upper tetrad residues G2 and G13, even while in the “open” position, is due to the inability of AuB3 to maintain an overall planar conformation. Accordingly, no significant increase in binding affinity is observed in the ΔG values between these two states (closed position = −37.6 ± 4.3 kJ/mol–1, open position = −39.3 ± 8.1 kJ/mol–1).
Figure 10.
Position of AuB3 with the upper G-tetrad in both the “open” (A) and “closed” (B) conformations of A1 (AuB3 and A1 shown in ball and stick representation, cKIT1 shown as a ribbon with bases as a wire. A1 is colored in green for clarity).
On comparing the binding position between the dinuclear Au(I) complex with AuTMX2, the latter interacts with both G6 and G10 guanines located at the G-tetrad top (Figure 8B,C), whereas AuB3 prefers to interact with only one guanine at a time (Figure 8B,C). Therefore, because AuB3 cannot outperform AuTMX2 in noncovalent interactions, key to its ΔG value is a broader positional variety in molecule-to-G4 interaction poses, which is not observed in AuTMX2 nor in other G4-stabilizing metal complexes.62
In conclusion, specific interactions with bases G6 and G10 appear to be key for ligand affinity and cKIT1 stabilization. The shallow saddle point in the FES (about 5 kJ mol–1 above either minimum, Figure 7B) implies a scenario of AuB3 oscillating between the two energy minima. This dynamic behavior fully compensates for the “imperfect” match between the semi-rigid, sterically demanding AuB3 molecule and the G-tetrad motif (as illustrated above), which results into a sizable free energy. We, therefore, speculate that FRET DNA melting experiments detect an average value resulting from AuB3 dynamical (oscillating) and statistical (parallel and perpendicular) orientations. We dub this “shaky lid” behavior of AuB3 on G-tetrad “dynamical docking”.
Antiproliferative Activity
The gold compounds were further tested for their antiproliferative activity in three human cancer cell lines, namely, A2780 ovarian cancer, MDA-MB-231, and MCF-7 breast cancer cells lines. The selected breast cancer cells differ in the expression of estrogen (ER)/progesterone (PR) hormone receptors and human epidermal growth factor receptor type 2. While MDA-MB-231 cells are considered triple negative, because they lack all three receptor types, MCF-7 cells are hormone-dependent (ER+/PR+). Moreover, the compounds were also tested against the nontumorigenic VERO E6 (healthy kidney) cell line. The obtained EC50 values are reported in Table 2 in comparison to the respective ligands and to the benchmark cisplatin and auranofin.13 All ligands were inactive up to a concentration of 100 μM, which highlights the metal contribution in terms of activity enhancement. All the Au(I) complexes were inactive or poorly active in MCF-7 and VERO E6 cells but showed EC50 values ranging from ca. 0.44 to ca. 55 μM against A2780 and MDA-MB-231 cells. While in depth investigation of the modes of action should be further addressed in future studies, preliminary structure–activity relationships could be derived: (a) the xanthine-derived Au(I) complexes are moderately active in all cell lines, with EC50 values in line with those previously reported for AuTMX2,28 (b) ethyl linkers seem beneficial in both series, resulting in the highest activities and following the activity trend: ethylene > propylene > methylene. The only gold complex that outperformed the reference metallodrugs was AuC2 in A2780 ovarian cancer cells (EC50 ca. 0.44 μM, selectivity index >200) and additionally provided the highest selectivity indexes for each cancer cell line compared to both references auranofin and cisplatin.
Table 2. EC50 Values (μM, n ≥ 3) and Selectivity Indexes Calculated as EC50(VERO E6)/EC50(Cancer Cell Line) of the Au(I) NHC Complexes in Human Cancer and Nontumorigenic Cellsa.
| A2780 (ovarian) | MDA-MB-231 (breast) | MCF-7 (breast) | Vero E6 (noncancerogenic kidney) | |
|---|---|---|---|---|
| cisplatin | 0.80 ± 0.08 (15.30) | 11.7 ± 1.5 (1.0) | 27.9 ± 2.3 (0.4) | 12.2 ± 1.2 |
| auranofin | 0.27 ± 0.07 (20.44) | 3.1 ± 0.1 (1.8) | 3.0 ± 0.4 (1.8) | 5.5 ± 0.3 |
| AuTMX2 | 16.2 ± 2.1b | n.d. | n.d. | >100 |
| H2B2PF6 | >100 | >100 | >100 | >100 |
| H2B3PF6 | >100 | >100 | >100 | >100 |
| H2C1PF6 | >100 | >100 | >100 | >100 |
| H2C2PF6 | >100 | >100 | >100 | >100 |
| H2C3PF6 | >100 | >100 | >100 | >100 |
| AuB2 | 12.0 ± 0.9 (>8.3) | 52.3 ± 1.5 (>1.9) | >100 | >100 |
| AuB3 | 23.7 ± 1.0 (>4.2) | 53.7 ± 0.1 (>1.9) | >100 | >100 |
| AuC1 | 55.4 ± 2.4 (>1.8) | 52.8 ± 0.7 (>1.9) | >100 | >100 |
| AuC2 | 0.44 ± 0.13 (>227.27) | 28.1 ± 2.0 (>3.5) | 52.9 ± 1.4 (>1.9) | >100 |
| AuC3 | 11.18 ± 1.22 (>8.94) | 13.1 ± 0.2 (>7.6) | >100 | >100 |
Cells were incubated with the respective compounds for 72 h.
Taken from ref (28).
Conclusions
In conclusion, in order to design potent and selective G-quadruplex stabilizers, two cyclic dinuclear Au(I) bis-NHC complexes AuB2–3, in which two xanthine-derived NHC ligands are coupled by an aliphatic bridge, were synthesized and fully characterized, together with three benzimidazole-derived analogues AuC1–3. Most of the compounds have shown high stability in solution and in the presence of cysteine nucleophiles and could be studied for their G4s stabilization properties by FRET DNA melting, showing different selectivity patterns depending on the type of the G4 structure. Interestingly, all the Au(I) complexes can stabilize the selected panel of G4s, with their ΔTm increasing with the length of the linker chain. While the benzimidazole derivatives show higher G4 stabilization activity with respect to the benchmark mononuclear AuTMX2, they are less selective and can also bind to duplex DNA. Instead, the xanthine-based complexes show moderate selectivity for G4s versus duplex DNA, which is relevant for targeted pharmacological applications. The propyl-bridged complex AuB3 results as the best performer, showing a comparable stabilizing capability toward hTelo and cKIT1 with respect to AuTMX2 but significantly higher ΔTm with the promoter cKIT2, BCL2, and hTERT. Furthermore, AuB3 also stabilizes parallel G4s (such as cKIT1 and cKIT2) to a greater extent than the hybrid-mixed hTelo or BCL2 sequences, highlighting a potential preference toward the parallel G4-conformation. Additionally, the appearance of weak ICD bands in the G4s′ CD spectra in the presence of either AuB3 (and also AuC3) suggests the complexes’ top stacking onto the G4 tetrads.
In order to shed light into the mechanisms of binding of the cyclic dinuclear Au(I) bis-NHC complexes with G4s at a molecular/atomic level, metaD simulations were applied to study the adduct of the most potent xanthine derivative AuB3 with cKIT1. The compound interacts with the top tetrad of the G4 monomer establishing noncovalent interactions with some of the nucleobases and oscillating between two energy minima. As expected from the similar experimental Gibbs free energies of G4 stabilization, the cyclic dinuclear AuB3 does not gain any improved stabilizing interaction with cKIT1 with respect to AuTMX2. This result is well justified by the metaD analysis, whereby the compound is shown to escape from a rigid planar structure to achieve a “dynamical docking” on the upper tetrad, enabling only “half” of the molecule to actually interact with some of the guanines in cKIT1. The experimental validation by FRET DNA melting of the binding energy calculated by metaD was also achieved for the AuB3 adducts with cKIT1. Preliminary antiproliferative activity data showed only moderate activity of the xanthine-based Au(I) complexes in cancer cells, while more intriguing cytotoxicity profiles were observed for the benzimidazole derivatives. Nevertheless, all the cyclic dinuclear compounds were nontoxic against nontumorigenic cells.
Overall, our study has evidenced that cyclic dinuclear Au(I) bis-NHC complexes based on xanthine scaffolds may provide some advantages over mononuclear complexes, particularly in the adaptability toward binding to different G4 structures likely due to the flexibility conferred by the alkyl linkers. However, care should be paid to obtain the optimal balance between metallodrug rigidity and conformational adaptability to achieve maximal G4 stabilization. Finally, it is also worth mentioning that metal bis-NHCs complexes derived from purines, including of Pt(II), Pd(II), and Ag(I), have attracted attention for different therapeutic applications in recent years,63−65 and our results point toward a possible common noncovalent binding modes with pharmacological targets of these families of organometallics.
Experimental Section
General
Reactions involving dry solvents were performed under a nitrogen atmosphere and in flame-dried glassware. Syntheses involving silver or gold were carried out in the dark. The remaining manipulations, unless otherwise stated, were conducted under standard conditions. Nitrobenzene was dried by distillation and discarding of the first 5% chloroform by passing it over a plug of basic alumina (75 g for 100 mL of solvent). All other solvents as well as starting materials were commercially obtained and used without further purification. Column chromatography was performed on silica gel 60A (particle size 40–63 μm). Merck Millipore silica gel 60 F-254 plates were employed for thin-layer chromatography (TLC), which were analyzed in UV light (λ = 254 nm) or stained with an alkaline potassium permanganate solution. NMR spectra were recorded on a Bruker AVANCE DPX 400 at ambient temperature (400.13 MHz for 1H NMR, 100.53 MHz for 13C{1H} NMR, and 161.97 MHz for 31P NMR). Chemical shifts δ are reported in parts per million (ppm) with respect to residual 1H and 13C signals of the deuterated solvents as the internal standard, whereas coupling constants J are denoted in Hertz (Hz). 13C{1H} NMR spectra were baseline-corrected and occasionally exponential apodization functions were used to improve resolution. The following abbreviations as well as combinations thereof are employed for NMR signal multiplicities: s = singlet, d = doublet, t = triplet, p = quintet, h = heptet, and b = broad. Elemental analyses (EAs) for C, H, N, and S were obtained from a HEKAtech Euro EA by the microanalytical laboratory at the Technical University of Munich. Measurements that were below the limit of detection are denoted with bdl.
HR-ESI-MS was conducted on a Thermo Fisher Exactive Plus Orbitrap mass spectrometer, which was equipped with an ESI source by the same company. Samples were prepared as 1 mg mL–1 solutions in acetonitrile, syringe filtered to 0.45 μm, and directly injected. With an ionization voltage of 2 kV, ions were detected in the positive mode. Peaks were then fit with Gaussian functions and compared to isotope patterns calculated in enviPat Web 2.4.66 Mass-charge ratios m/z are given in g mol–1 e–1. Chloro(tetrahydrothiophene)gold(I) [Au(THT)Cl],67 the benchmark Au(I) compound AuTMX2,19 ligands, 1,1′-trimethylenebis(benzimidazole)68 and 1,1′-methylenebis(3,3′-dimethylbenzimidazolium) dibromide (H2C1Br),42 were synthesized accordingly to previously published procedures. The syntheses of 7,7′-methylenebis(theophylline) (A1), 7,7′-ethylenebis(theophylline) (A2), and 7,7′-trimethylenebis(theophylline) (A3) were performed according to previously reported procedures.69 The compounds’ purity was assessed to be >95%.
Synthesis of NHC Ligands
General Procedure for the Synthesis of 7,7′-(α,ω-Alkanediyl)-bis(theophylline)
Adapted from a general procedure by Itahara and Imamura.69 Theophylline (2.80 g, 15.5 mmol, 2.00 equiv) was dissolved in 100 mL dry DMF under a N2 atmosphere to give a clear yellow solution and NaH (625 mg of a 60% dispersion in mineral oil, 15.6 mmol, 2.01 equiv) was added, rinsing the powder funnel with another 25 mL dry DMF. After 30 min, 1.00 equiv Br(CH2)nBr (7.77 mmol) was added and the solution was stirred for 22 h at room temperature. A further heating up to 70 °C for another 5 h was applied prior to pouring the suspension into ∼200 mL of distilled water, stirring it for 5 min, and leaving it in the fridge to crystallize. The precipitate was vacuum filtered and washed with 50–80 mL of ice-cold distilled water. It was then dried in high vacuum and subsequently washed with 60–90 mL hexanes or npentane to remove residual mineral oil originating from the NaH dispersion.
Synthesis of 1-Methylbenzimidazole
The title compound was synthesized by modification of literature procedures.70,71 10.0 g benzimidazole (84.7 mmol, 1.00 equiv) were suspended in 40 mL 50% aq NaOH at 0 °C. Upon warming to 30 °C, the solid dissolved almost completely. Over a period of 20 min, 13.7 g MeI (96.4 mmol, 1.14 equiv) were added dropwise to the well-stirred suspension, resulting in a biphasic system. The reaction was completed after stirring at 40 °C for another 70 min as confirmed by TLC. The aqueous phase was extracted with DCM (4 × 30 mL) and the combined organic layers were washed with distilled water (1 × 15 mL). After drying over anhydrous MgSO4, filtration, and evaporation of volatiles in vacuo, 11.15 g of a brown crude oil were obtained. Purification by column chromatography (275 g silica, gradient 3 → 7% MeOH in DCM) gave 9.41 g 1-methylbenzimidazole (71.2 mmol, 84%) as yellow oil that crystallized to a beige solid. NMR was in accordance with the literature.
Synthesis of 7,7′-Methylenebis(9,9′-dimethyltheophyllinium) Bis(methyl sulfate) (H2B1MeSO4)
400 mg A1 (1.07 mmol, 1.00 equiv) were dissolved in 40 mL dry PhNO2 at 90 °C to give a greenish-yellow, clear solution. Me2SO4 (3 mL, ∼4 g, ∼30 equiv) was added dropwise to the stirred solution of A1, resulting in immediate clouding of the reaction mixture. The reaction monitoring by NMR was employed: 0.3 mL were worked up by pouring on 1.7 mL Et2O, centrifuging, decanting, and washing with 1 mL Et2O twice. After a total of 6 days, conversion was satisfactory. Approximately 40% of the initial reaction mixture (the rest was consumed by failed parallel attempts) were worked up by pouring into 50 mL Et2O, filtration, and washing with 3 × 10 mL Et2O. With the described procedure, 244 mg (391 μmol) of the crude title compound were obtained, corresponding to a yield of ∼90% with respect to the partial amount that was worked up. The brown powder, which is highly hygroscopic and extremely sensitive toward alkaline conditions was stored at −30 °C under a N2 atmosphere and used without further purification. In DMSO, H2B1MeSO4 dissociates in less than 2 days. 1H NMR (DMSO-d6): δ 3.28 (s, 6 H, CH3), 3.34 (s, 6 H, CH3), 3.74 (s, 6 H, CH3), 4.26 (s, 6 H, CH3), 7.04 (s, 2 H, CH2), 9.66 (s, 2 H, NCHN).
Synthesis of 7,7′-Ethylenebis(9,9′-dimethyltheophyllinium) Ditosylate (H2B2OTs)
A2 (250 mg, 647 μmol, 1.00 equiv) was united with MeOTs (2.50 g, 13.4 mmol, 20.8 equiv). On heating, the white suspension resolved to a yellow solution around 140 °C and was further warmed to 160 °C, at which temperature the reaction mixture was stirred for 2 h. Once cooled down, the resulting dark amber oil was poured into 40 mL Et2O to form a white to brown, sticky precipitate. The supernatant was decanted off and the solid was recrystallized from iPrOH/EtOH (∼1/1, 20 mL). The white crystals were filtered off and washed with 2 × 4 mL ice-cold iPrOH. Drying in high vacuum afforded the title compound H2B2OTs as 405 mg (534 μmol, 83%) of a white powder. 1H NMR (DMSO-d6): δ 2.28 (s, 6 H, Metosylate), 3.27 (s, 6 H, CH3), 3.70 (s, 6 H, CH3), 4.09 (s, 6 H, CH3), 4.97 (s, 4 H, CH2), 7.10 (d, 3JHH = 7.7, 4 H, CHar, tosylate), 7.42 (d, 3JHH = 7.8, 4 H, CHar, tosylate), 9.26 (s, 2 H, NCHN).
Synthesis of 7,7′-Trimethylenebis(9,9′-dimethyltheophyllinium) Ditosylate (H2B3OTs)
A3 (300 mg, 749 μmol, 1.00 equiv) and MeOTs (3.00 g, 16.1 mmol, 21.5 equiv) were united and heated to 160 °C, at which temperature they formed a clear yellow solution. After 2 h, the resulting dark amber solution was cooled to room temperature and poured into 50 mL Et2O. The supernatant was decanted off, leaving a grayish-orangey precipitate, which was then recrystallized from iPrOH/EtOH (∼1/1, 25 mL, hot filtration). Colorless needles were filtered off, washed with 4 mL cold iPrOH as well as 5 mL Et2O, and dried in vacuo to give the title compound in 80% yield (476 mg, 597 μmol). 1H NMR (DMSO-d6): δ 2.28 (s, 6 H, Metosylate), 2.45 (p, 3JHH = 6.8, 2 H, CH2CH2CH2), 3.25 (s, 6 H, CH3), 3.72 (s, 6 H, CH3), 4.13 (s, 6 H, CH3), 4.53 (t, 3JHH = 6.8, 4 H, NCH2), 7.10 (d, 3JHH = 8.0, CHtosylate), 7.43 (d, 3JHH = 8.1, CHtosylate), 9.43 (s, 2 H, NCHN).
Synthesis of 1,1′-Ethylenebis(3,3′-dimethylbenzimidazolium) Dibromide (H2C2Br)
1-Methylbenzimidazole (1.13 g, 8.53 mmol, 2.46 equiv) was dissolved in 10 mL dry MeCN. 1,2-Dibromoethane (650 mg, 3.46 mmol, 1.00 equiv) was added to give a clear, yellowish solution. The mixture was refluxed for 3 days. The solvent was removed from the resulting white suspension in vacuo and the yellowish solid was washed with 25 mL THF in three portions. Recrystallization from 150 mL EtOH gave the title compound H2C2Br as pure white needles (1.23 g, 2.72 mmol, 79%). NMR was in accordance with the literature.42 1H NMR (DMSO-d6): δ 4.05 (s, 6 H, CH3), 5.14 (s, 4 H, CH2), 7.62 (ddd, JHH = 8.3|7.3|1.2, 2 H, CHar), 7.69 (ddd, JHH = 8.3|7.3|1.1, 2 H, CHar), 7.92 (dt, JHH = 8.2|1.0, 2 H, CHar), 8.03 (dt, JHH = 8.4|1.0, 2 H, CHar), 9.83 (s, 2 H, NCHN). 13C{1H} NMR (DMSO-d6): δ 33.41 (CH3), 45.53 (CH2), 112.99 (CHar), 113.72 (CHar), 126.65 (CHar), 126.72 (CHar), 130.77 (Car), 131.68 (Car), 143.40 (NCHN).
Synthesis of 1,1′-Trimethylenebis(3,3′-dimethylbenzimidazolium) Bis(tetrafluoroborate) (H2C3BF4)
500 mg 1,1′-trimethylene-bis(benzimidazole) (∼90% purity, prior to recryst., 1.63 mmol, 1.00 equiv) were dissolved in 20 mL dry MeCN under slight warming. 600 mg Me3OBF4 (4.06 mmol, 2.49 equiv) were added, leading to immediate clouding of the solution. After 4 h of vigorous stirring, another 25 mL dry MeCN were added to obtain a clear solution and additional 60 mg (0.41 mmol, 0.25 equiv) Me3OBF4 were added. After stirring overnight, the reaction was worked up by the removal of the solvent at reduced pressure. 1.04 g yellowish crude product were washed with 150 mL boiling MeOH. The wash fraction was reduced to 105 mL, out of which further product was obtained by crystallization upon cooling. In total, 563 mg (1.17 mmol, 72%) H2C3BF4 were obtained as a white powder. 1H NMR (DMSO-d6): δ 2.58 (p, 3JHH = 7.2, 2 H, CH2CH2CH2), 4.07 (s, 6 H, CH3), 4.65 (t, 3JHH = 7.3, 4 H, NCH2), 7.67–7.76 (m, 4 H, CHar), 7.99–8.09 (m, 4 H, CHar), 9.66 (s, 2 H, NCHN). 13C{1H} NMR (DMSO-d6): δ 28.04 (CH2CH2CH2), 33.26 (CH3), 43.78 (NCH2), 113.36 (CHar), 113.63 (CHar), 126.54 (CHar), 126.60 (CHar), 130.84 (Car), 131.84 (Car), 142.84 (NCHN). Assigned by HSQC NMR. Calcd for C19H22B2F8N4 (H2C3BF4) [%]: C, 47.54; H, 4.62; N, 11.67. Found [%]: C, 47.36; H, 4.55; N, 11.65. Single crystals suitable for diffraction were obtained by slow diffusion of Et2O into a solution of H2C3BF4 in MeCN.
General Procedure for Ion Exchange to PF6
Benzimidazole ligands: the compound is dissolved in a minimal amount of distilled water (20–75 mM) by careful warming to 60–80 °C under stirring. Five to 6 M equiv. NH4PF6 are dissolved separately in distilled water to give an approximately 1 M solution and heated to the same temperature. The first solution was slowly poured into the second one under vigorous stirring, resulting in the immediate formation of a white suspension. Stirring is continued on cooling to room temperature. The precipitate was collected by filtration and washed thoroughly with cold distilled water. Following drying in high vacuum, if elemental analysis purity was not reached, the product is further resuspended and treated as described above.
Xanthine ligands: the crude product was dissolved in a minimal amount of distilled water at room temperature (∼0.2 M) and cotton-filtered into a vigorously stirred, approximately 2 M aq solution of 10 equiv NH4PF6. The resulting white suspension was stirred for another 15 min. Sometimes, gel-like lumps form initially but resolve to a suspension on continued stirring. The white precipitate was collected by filtration and washed thoroughly with ice-cold water prior to drying in high vacuum.
Synthesis of 7,7′-Methylenebis(9,9′-dimethyltheophyllinium) Bis(hexafluorophosphate) (H2B1PF6)
H2B1MeSO4 (162 mg, 269 μmol, 1.00 equiv) was dissolved in 8 mL MeOH and poured into a solution of NH4PF6 (500 mg, 3.07 mmol, 11.8 equiv) in 4 mL distilled water. Soon after, silver needles started to grow in the solution. Crystallization was aided by cooling in the freezer. The precipitate was collected by filtration, washed (3× 6 mL distilled water), and dried in high vacuum. H2B1PF6 was hereby obtained as an off-white powder in 49% yield (88 mg, 0.13 mmol). The compound was further purified by recrystallization from a mixture of acetone and EtOAc to give a pure white powder. 1H NMR (CD3CN): δ 3.36 (s, 6 H, CH3), 3.73 (s, 6 H, CH3), 4.14 (s, 6 H, CH3), 6.95 (s, 2 H, CH2), 9.05 (s, 2 H, NCHN). 13C{1H} NMR (CD3CN): δ 29.47 (CH3), 32.41 (CH3), 38.97 (CH3), 57.38 (CH2), 108.27, 140.95, 142.13 (NCHN), 151.09, 154.95. Assigned by HSQC NMR. 31P NMR (CD3CN): δ −144.9 (h, 1JPF = 706.9). Calcd for C17H24F12N8O5P2 (H2B1PF6·H2O) [%]: C, 28.74; H, 3.41; N, 15.77. Found [%]: C, 28.57; H, 3.08; N, 15.31.
Synthesis of 7,7′-Ethylenebis(9,9′-dimethyltheophyllinium) Bis(hexafluorophosphate) (H2B2PF6)
Anion exchange of H2B2OTs (500 mg, 659 μmol) according to the general procedure afforded H2B2PF6 as a white powder in 91% yield. 1H NMR (CD3CN): δ 3.33 (s, 6 H, CH3), 3.73 (s, 6 H, CH3), 4.05 (s, 6 H, CH3), 4.98 (s, 4 H, CH2), 8.35 (s, 2 H, NCHN). 13C{1H} NMR (CD3CN): δ 29.31 (CH3), 32.32 (CH3), 38.44 (CH3), 49.30 (CH2), 108.91 (Csp2), 139.99 (NCHN), 141.09 (Csp2), 151.31 (Csp2), 154.63 (Csp2). Assigned by analogy to H2B2OTs. 31P NMR (CD3CN): δ −144.7 (h, 1JPF = 707.3). Calcd for C18H24F12N8O4P2 (H2B2PF6) [%]: C, 30.61; H, 3.42; N, 15.86; S, 0.00. Found [%]: C, 30.45; H, 3.50; N, 15.59, S bdl.
Synthesis of 7,7′-Trimethylenebis(9,9′-dimethyltheophyllinium) Bis(hexafluorophosphate) (H2B3PF6)
Anion exchange of H2B3OTs (383 mg, 496 μmol) according to the general procedure afforded H2B3PF6 as a white powder in 77% yield. 1H NMR (CD3CN): δ 2.48 (p, 3JHH = 7.1, 2 H, CH2CH2CH2), 3.31 (s, 6 H, CH3), 3.74 (s, 6 H, CH3), 4.09 (s, 6 H, CH3), 4.53 (t, 3JHH = 7.1, 4 H, NCH2), 8.52 (s, 2 H, NCHN). 13C{1H} NMR (CD3CN): δ 29.22 (CH3), 30.83 (CH2CH2CH2), 32.30 (CH3), 38.24 (CH3), 46.82 (NCH2), 108.97 (Csp2), 139.56 (NCHN), 141.04 (Csp2), 151.39 (Csp2), 154.55 (Csp2). Assigned by HSQC NMR. 31P NMR (CD3CN): δ −144.68 (h, 1JPF = 705.9). Calcd for C19H28F12N8O5P2 (H2B3PF6·H2O) [%]: C, 30.91; H, 3.82; N, 15.18; S, 0.00. Found [%]: C, 30.63; H, 3.50; N, 14.87, S bdl.
Synthesis of 1,1′-methylenebis(3,3′-dimethylbenzimidazolium) bis(hexafluorophosphate) (H2C1PF6). 1,1′-ethylenebis(3,3′-dimethylbenzimidazolium) bis(hexafluorophosphate) (H2C2PF6) and 1,1′-trimethylenebis(3,3′-dimethylbenzimidazolium) bis(hexafluorophosphate) (H2C3PF6). Anion exchange was carried from the Br- (for H2C1 and H2C2) and BF4 precursors of the ligands according to the general procedure. 1H and 13C{1H} NMR were in accordance with literature42 and purity confirmed by elemental analysis to be >95%.
H2C1PF6: white powder, 82% yield (534 mg). H2C2PF6: white powder, 76% yield (780 mg). H2C3PF6 as white powder in 91% yield (463 mg).
Synthesis of Cyclic Dinuclear Au(I) Bis-NHC Complexes
Synthesis of [Au2(μ-{7,7′-Ethylenebis(9,9′-dimethyltheophyllin-8,8′-ylidene)})2](PF6)2 (AuB2)
66.1 mg H2B2PF6 (93.6 μmol) and 1.01 equiv Au(THT)Cl were dissolved in dry MeCN (2 mL ca), and stirred at 55 °C for an hour in the dark, prior to the addition of 5.0 equiv of oven-dried, freshly crushed K2CO3. After 24 h stirring, the reaction was filtrated over Celite and rinsed with fresh MeCN. The final product was achieved via fractional precipitation, upon addition of a large excess of Et2O, and centrifugation followed by further washes with small aliquots of Et2O. AuB2 was afforded as 52 mg (73%) of a white powder. 1H NMR (CD3CN): δ 3.27 (s, 12 H, CH3), 3.73 (s, 12 H, CH3), 4.15 (s, 12 H, CH3), 5.28 (br s, 8 H, CH2). 13C{1H} NMR (CD3CN): δ 28.98 (CH3), 32.47 (CH3), 40.50 (CH3), 46.69 (NCH2), 108.89 (Csp2), 141.51 (Csp2), 151.72 (Csp2), 154.47 (Csp2), 187.60 (C–Au). Assigned by HSQC NMR. 31P NMR (CD3CN): δ −144.65 (h, 1JPF = 706.2). Calcd for C36H48Au2F12N16O10P2 (AuB2·2H2O) [%]: C 27.92; H 3.12; N, 14.47. Found [%]: C, 27.88; H, 2.94; N 14.07. m/z: 611.1417 (611.1424 calcd for [AuB2-2 PF6]2+), 1367.2482 (1367.2496 calcd for [AuB2-PF6]+). Single crystals suitable for diffraction were obtained by slow diffusion of Et2O into a solution of AuB2 in DMF.
Synthesis of [Au2(μ-{7,7′-Trimethylenebis(9,9′-dimethyltheophyllin-8,8′-ylidene)})2](PF6)2(AuB3)
67.4 mg H2B3PF6 (93.6 μmol) and 1.01 equiv Au(THT)Cl are dissolved in dry MeCN (2 mL ca) and stirred at 55 °C for an hour in the dark, prior to the addition of 5.0 equiv of oven-dried, freshly crushed K2CO3. After 24 h stirring, the reaction was filtered over Celite and rinsed with fresh MeCN. The final product was achieved via fractional precipitation, upon the addition of a large excess of Et2O, and centrifugation followed by further washes with small aliquots of Et2O. AuB3 was afforded as 25 mg (37%) of a white powder. 1H NMR (CD3CN): δ 2.59 (p, 3JHH = 6.9, 4 H, CH2CH2CH2), 3.28 (s, 12 H, CH3), 3.78 (s, 12 H, CH3), 4.19 (s, 12 H, CH3), 4.61 (t, 3JHH = 6.9, 8 H, NCH2). 13C{1H} NMR (CD3CN): δ 29.01 (CH3), 31.23 (CH2CH2CH2), 32.58 (CH3), 39.98 (CH3), 49.48 (NCH2), 109.89 (Csp2), 142.22 (Csp2), 151.89 (Csp2), 154.37 (Csp2), 187.55 (C-Au). 31P NMR (CD3CN): δ −144.66 (h, 1JPF = 707.7). Calcd for C38H50Au2F12N16O9P2 (AuB3·H2O) [%]: C, 29.28; H, 3.23; N, 14.38. Found [%]: C, 29.44; H, 3.23; N, 13.89. m/z: 625.1572 (625.1581 calcd for [AuB3-2 PF6]2+), 1395.2791 (1395.2809 calcd for [AuB3-PF6]+). Single crystals suitable for diffraction were obtained by slow diffusion of Et2O into a solution of AuB3 in DMF.
Synthesis of [Au2(μ-{1,1′-Methylenebis(3,3′-dimethylbenzimidazol-2,2′-ylidene)})2](PF6)2 (AuC1)
100 mg H2C1PF6 (176 μmol) and 1.01 equiv Au(THT)Cl are dissolved in dry MeCN (3 mL ca) and stirred at 60 °C for an hour in the dark, prior to the addition of 5.0 equiv of oven-dried, freshly crushed K2CO3. After a day of stirring, the reaction is filtered over Celite and rinsed with fresh MeCN. The final product was achieved via fractional precipitation, upon the addition of a large excess of dry PhMe and npentane. The precipitate was collected via centrifugation and further washed with small aliquots of npentane to afford AuC1 as 100 mg (92%) of an off-white powder. 1H NMR (CD3CN): δ 4.06 (s, 12 H, CH3), 7.00 (d, 1JHH = 14.8, 1 H, NCHHN), 7.31 (d, 1JHH = 14.7, 1 H, NCHHN), 7.58–7.65 (m, 8 H, CHar), 7.72–7.78 (m, 4 H, CHar), 7.86–7.92 (m, 4 H, CHar). 13C{1H} NMR (CD3CN): δ 36.90 (CH3), 59.62 (CH2), 112.65 (CHar), 113.92 (CHar), 126.74 (CHar), 126.97 (CHar), 133.53 (Car), 135.09 (Car), 191.86 (C-Au). Assigned by HSQC NMR. 31P NMR (CD3CN): δ −144.63 (h, 1JPF = 706.4). Calcd for C34H38Au2F12N8O3P2 (AuC1·3H2O) [%]: C, 31.64; H, 2.97; N, 8.68. Found [%]: C, 31.09; H, 2.39; N, 8.46. m/z: 473.1029 (473.1035 calcd for [AuC1-2 PF6]2+), 1091.1698 (1091.1718 calcd for [AuC1-PF6]+). Single crystals suitable for diffraction were obtained by slow diffusion of Et2O into a solution of AuC1 in MeCN.
Synthesis of [Au2(μ-{1,1′-Ethylenebis(3,3′-dimethylbenzimidazol-2,2′-ylidene)})2](PF6)2 (AuC2)
75.0 mg H2C2PF6 (129 μmol, 1.00 equiv) and 298 mg Ag2O (1.29 mmol, 9.98 equiv) were suspended in 7.0 mL dry acetonitrile. Over the weekend, the reaction mixture was stirred at room temperature in the dark. Excess Ag2O was removed by Celite filtration. Au(THT)Cl (41.8 mg, 130 μmol, 1.01 equiv) was then added, which immediately resulted in the formation of a fine, white precipitate. After stirring overnight, the precipitate of AgCl was removed by centrifugation and decanting. The product was isolated by fractional precipitation with dry PhMe and npentane (impurities precipitate after the product), collection by centrifugation, washing with small aliquots of npentane, and drying in high vacuum. AuC2 was obtained as 28 mg (22 μmol, 34%) of a gray powder. 1H NMR (CD3CN): δ 3.95 (s, 12 H, CH3), 5.29 (s, 8 H, CH2), 7.33 (ddd, JHH = 8.3|7.1|1.1, 4 H, CHar), 7.40–7.47 (m, 8 H, CHar), 7.54 (dt, JHH = 8.3|0.9, 4 H, CHar). 13C{1H} NMR (CD3CN): δ 36.24 (CH3), 47.68 (CH2), 112.18 (CHar), 113.15 (CHar), 125.90 (CHar), 125.99 (CHar), 133.99 (Car), 134.77 (Car), 191.37 (C-Au). 31P NMR (CD3CN): δ −144.60 (h, 1JPF = 707.0). Calcd for C36H38Au2F12N8OP2 (AuC2·H2O) [%]: C, 33.71; H, 2.99; N, 8.74. Found [%]: C, 33.35; H, 2.57; N, 8.94. m/z: 487.1186 (487.1192 calcd for [AuC2-2 PF6]2+), 1119.2017 (1119.2031 calcd for [AuC2-PF6]+). Single crystals suitable for diffraction were obtained by slow evaporation of a solution of AuC2 in MeCN.
Synthesis of [Au2(μ-{1,1′-Trimethylenebis(3,3′-dimethylbenzimidazol-2,2′-ylidene)})2](PF6)2 (AuC3)
59.9 mg H2C3PF6 (100 μmol), and 2.01 equiv Au(THT)Cl were dissolved in dry MeCN (3 mL ca) and stirred at 60 °C for an hour in the dark, prior to the addition of 5.0 equiv of oven-dried, freshly crushed K2CO3. After a day of stirring, the reaction was filtered over Celite and rinsed with fresh MeCN. The final product was achieved via fractional precipitation via the addition of a large excess of dry PhMe and npentane. The precipitate was collected by centrifugation and further washed with small aliquots of npentane to afford AuC3 as 45 mg (69%) of a white powder. 1H and 13C{1H} NMR were in accordance with literature36 and the purity confirmed by elemental analysis to be >95%.
X-ray Crystallography
Data were collected on a Bruker D8 Venture single-crystal X-ray diffractometer equipped with a CMOS detector (Photon-100), a TXS rotating anode with Mo Kα radiation (λ = 0.71073 Å), and a Helios optic (compounds H2C3, AuB2) or a CPAD detector (Photon-II), an IMS microfocus source with Mo Kα radiation and a Helios optic (compounds AuB3, AuC1, AuC2) using the APEX3 software package.72 The measurements were performed on single crystals coated with perfluorinated ether. The crystals were fixed on top of a Kapton micro sampler and frozen under a stream of cold nitrogen. A matrix scan was used to determine the initial lattice parameters. Reflections were corrected for Lorentz and polarization effects, scan speed, and background using SAINT.73 Absorption correction, including odd and even ordered spherical harmonics, was performed using SADABS.73 The space group assignment was based upon systematic absences, E statistics, and successful refinement of the structures. The structures were solved using SHELXT with the aid of successive difference Fourier maps and were refined against all data using SHELXL in conjunction with SHELXLE.74−76 Hydrogen atoms (except on heteroatoms) were calculated in ideal positions as follows: methyl hydrogen atoms were refined as part of rigid rotating groups, with a C–H distance of 0.98 Å and Uiso(H) = 1.5Ueq(C). Non-methyl H atoms were placed in calculated positions and refined using a riding model with methylene, aromatic, and other C–H distances of 0.99, 0.95, and 1.00 Å, respectively, and Uiso(H) = 1.2Ueq(C). Non-hydrogen atoms were refined with anisotropic displacement parameters. Full-matrix least-squares refinements were carried out by minimizing Σw(Fo2 – Fc2)2 with the SHELXL weighting scheme.74 Neutral atom scattering factors for all atoms and anomalous dispersion corrections for the non-hydrogen atoms were taken from International Tables for Crystallography.77 Images of the crystal structure were generated with mercury.78 Deposition numbers 2202916–2202920 contain the supplementary crystallographic data for this paper. These data are provided free of charge by the joint Cambridge Crystallographic Data Centre and Fachinformationszentrum Karlsruhe Access Structures service www.ccdc.cam.ac.uk/structures.
NMR Stability Studies
Dinuclear gold complexes AuB2–3 and AuC1–3 were tested for their stability in a 1/4 mixture of D2O and DMSO-d6 over time and in the presence of the model thiol NAC, in the same solvent mixture. Equal amounts of every gold complex (∼3 mg) were dissolved in 0.5 mL solvent mixture each and 1H NMR spectra were recorded over time, after mixing, and at 1, 3, 8, and 24 h after dissolution. Similarly, 1H NMR spectra of the Au(I) compounds in the presence of an equimolar NAC amount were recorded at the same time intervals.
FRET DNA Melting Assay
FRET experiments were run on an Applied Biosystems QuantumStudio 5 Real-Time PCR thermocycler (Thermo Fisher Scientific, Waltham, USA) equipped with a FAM filter (λex = 492 nm; λem = 516 nm). The thermocycler was set to perform a stepwise increase of 0.3 °C every 30 s, from 25 to 95 °C, and measurements were acquired after each step. All the oligonucleotides were purchased from Sigma-Aldrich (Germany) in HPLC purity grade. The FRET probes used were FAM (6-carboxyfluorescein) and TAMRA (6-carboxy-tetramethylrhodamine), in 5′- and 3′-end, respectively. The lyophilized fluorolabeled hTelo (21-mer), d[GGG(TTAGGG)3], cKIT1, d[GGGAGGGCGCTGGGAGGAGGG], cKIT2, d[CCCGGGCGGGCGCGAGGGAGGGGAGG], BCL2 d[AGGGGCGGGCGCGGGAGGAAGGGGGCGGGAGCGGGGCTG], and hTERT d[GGGGGCTGGGCCGGGGACCCGGGAGGGGTCGGGACGGGGCGGGG] oligonucleotides were first diluted in deionized water to obtain 100 μM stock solutions. Stock solutions were diluted to a concentration of 400 nM in potassium cacodylate buffer (60 mM, pH 7.4) and then annealed to form G-quadruplex (G4) structures by heating to 95 °C for 5 min, followed by cooling to room temperature overnight.
Experiments were carried out in a 96-well plate with a total volume of 30 μL. The final concentration of the G4-oligonucleotide was set to 200 nM in potassium cacodylate buffer (60 mM, pH 7.4). Stock solutions of the gold compounds in DMSO (1 mM) were freshly prepared prior to the experiments. The stock solutions were further diluted to a final concentration of 2 μM (with a total percentage of DMSO of approx. 0.1%) in potassium cacodylate buffer (60 mM, pH 7.4) to achieve the desired G4/gold compound stoichiometry (from 1:1 to 1:5). Competition assays with duplex DNA were performed using calf-thymus DNA (Sigma-Aldrich, Germany) according to a previously reported procedure.58 In all the experiments, to compare different sets of data, FAM emission was normalized (0–1). Melting temperature is defined as the temperature at which the normalized emission is 0.5, and ΔTm is defined as the difference of Tm between treated samples and untreated controls. Independent experiments were run at least in triplicates.
Gibbs Free Energy Analysis from FRET DNA Melting Data and Fitting
In order to determine the energy of binding of the gold compound to the G4, the experimental data were normalized to folded fraction (θ) of G4-DNA and fitted according to eqs 1 and 2,79 where the enthalpy (ΔH) for the process was derived from the resulting fit. In eq 2, T corresponds to the temperature (variable during the melting assay) and R is the gas constant. Subsequently, ΔG300 was derived from eq 3,80 where TMD corresponds to the temperature used in the metaD simulations described above (300 K) and Tm is the melting temperature for each condition, determined from the FRET melting profiles.
| 1 |
where
| 2 |
| 3 |
CD Spectroscopy
CD spectra of G4 structures alone or in the presence of the Au(I) compounds were recorded on a JASCO J-1500 CD spectrometer equipped with a PTC-517 Peltier thermostated cell holder, using the following parameters: range 210–400 nm, bandwidth 1 nm, step 1 nm, accumulation 3, temperature 25 °C. Stock solutions of unlabeled 21-mer hTelo, d[GGG(TTAGGG)3], and cKIT1, d[GGGAGGGCGCTGGGAGGAGGG] (each 100 μM), were prepared by dissolving the lyophilized sample in deionized water and further diluted in a Tris-HCl/KCl buffer (10/50 mM, pH 7.4). Stock solutions of AuB3 and AuC3 (1 mM) in DMSO were freshly prepared prior to the experiments and further diluted in buffer to achieve a G4/gold compound stoichiometry up to 1:10.
For CD G4 melting experiments, a stock solution containing 0.2 nM cKIT1 was diluted to a concentration of 8.0 μM in 60 mM potassium cacodylate buffer (pH 7.4) and then annealed to form G4 structures as described in the FRET paragraph. Stock solutions of AuB2, AuB3, or AuC3 (10 mM in DMSO) were diluted in 60 mM potassium cacodylate buffer to a concentration of 40 μM. The final concentration of G4 in each experiment was set to 4.0 μM with a ratio G4/metal complex of 1:5 (%DMSO = 0.2%). A determination of the melting curve of the pure G4 structure was performed using a solution with ccKIT1 = 4.0 μM, and DMSO (0.2%). The CD spectra of each sample were measured in the range of 25–95 °C with 2 °C temperature steps, the measurements were blank-corrected using in 60 mM potassium cacodylate buffer (pH 7.4) containing 0.2% DMSO. Each spectrum was acquired in the range of 240–400 nm with a 1 nm spectral resolution and integration time of 1 s.
MetaD Simulations
The crystal structures of AuB3 (CCDC number: 2202920) and of the cKIT1 G4 model (PDB ID: 4WO2) were used for the simulations. The system AuB3–G4 adduct was built using the Maestro GUI from Schrödinger (build 2021-1)81 and all metaD calculations were undertaken using the Desmond simulation package from Schrödinger LLC82,83 (ref). The OPLS 2005 force field parameters were used in all simulations.84 The system consisted of cKIT1 with AuB3 placed at a distance of ca. 10 Å from the upper tetrad, the box size being 10 × 10 × 10 nm and solvated using the TIP3P water model (ca. 32,000).85 16 Na+ ions were added to neutralize the charge of the system, with a further addition of 0.6 mol of K+ and Cl– ions added to match FRET DNA melting experimental conditions of pH 7.4. Constraints were added between the potassium ions embedded in the G4 structure and the O6 oxygen of each guanine in the three stacked tetrads to mimic the structurally relevant affinities of the G4. The system was first relaxed using the Desmond minimization application for 100 ps and further equilibrated using the standard relax model option for MD/MetaD before each simulation run. Simulations were run using the NPT ensemble with a Nose–Hoover86 thermostat (300 K) and MTK87 barostat. The time step for all production runs was 1fs with a simulation time of 50 ns (20 production runs in total) for a combined 1 ms of simulation time. A random seed was used for each production run. The chosen CVs were the distance between each of the two gold centers of AuB3 and the upper potassium ion located at the center of cKIT1, with a Gaussian width of 0.05 Å and height of 0.03 kcal/mol and a deposition rate of 0.09 ps. FES were obtained in the form of a heat map for each run with the ΔG taken from the lowest energy minima of each FES. The average ΔG was then calculated, from these values (Table S6). ΔG values from 19 of the 20 simulations were used, while an outlier of both energy and position was omitted.
Cell Culture Maintenance
MDA-MB-231 human breast cancer cells, MCF-7 human breast carcinoma cells, and VERO E6 Cercopithecus aethiops monkey kidney cells were maintained in Dulbecco’s modified Eagle’s medium (high glucose, pyruvate, and no glutamine), which was supplemented with heat-inactivated fetal bovine serum (qualified, South American origin, 10% v/v), gentamicin sulfate solution 50 mg/mL (1% v/v, 50 mg/L end concentration), and l-glutamine solution 200 nM (1% v/v, 2 nM end concentration) and were passaged twice a week. A2780 human ovarian cancer cells were maintained in the RPMI 1640 medium (no glutamine), supplemented with a heat-inactivated fetal bovine serum (qualified, South American origin, 10% v/v), penicillin/streptomycin solution 10,000:10,000 U/mL (1% v/v, 5 mL), and l-glutamine solution 200 nM (1% v/v, 2 nM end concentration) and were also passaged twice a week. All reagents were purchased from Gibco at Thermo Scientific and solvents from Sigma-Aldrich if not stated differently. Ultrapure water (18.2 MΩ/0.56 μS at 25 °C) was provided by a BerryPuremini from Berrytec. MCF-7 and VERO E6 cells were obtained from Cell Line Service (CLS, Eppelheim, Germany), MDA-MB-231 cells were from the Helmholtz Institute for Infection Research (HZI, Braunschweig, Germany), and A2780 cells from the European Collection of Authenticated Cell Cultures (ECACC, United Kingdom/distributed by Sigma Aldrich, Germany).
Antiproliferative Activity Studies
A2780 cells (5000 cells/well), MDA-MB-231 cells (8000 cells/well), MCF-7 cells (10,000 cells/well), or Vero E6 cells (10,000 cells/well) were transferred to a flat-bottom 96-well microtiter plate and incubated at 37 °C/5% CO2 in a humidified atmosphere for 24 h. Stock solutions of the compounds in DMF or DPBS (cisplatin) were freshly prepared and diluted with the respective cell culture medium to graded concentrations (a final concentration of DMF: 0.1% v/v). After 72 h of exposure, the cell biomass of living cells was quantified by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (Sigma-Aldrich) staining and the absorptions were calculated as the delta of two measurements of the same well at wavelengths 570 and 690 nm by an Infinite 200pro plate reader (Tecan). The EC50 values (half-maximal effective concentrations) were determined as the concentration that caused 50% inhibition of cell proliferation compared to the untreated control (0.1% DMF in DMEM/RPMI). The results were calculated as the mean of at least three independent experiments.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.inorgchem.2c03041.
Author Contributions
# C.K. and D.W. shared first authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Zou T.; Lok C.-N.; Wan P.-K.; Zhang Z.-F.; Fung S.-K.; Che C.-M. Anticancer Metal-N-Heterocyclic Carbene Complexes of Gold, Platinum and Palladium. Curr. Opin. Chem. Biol. 2018, 43, 30–36. 10.1016/j.cbpa.2017.10.014. [DOI] [PubMed] [Google Scholar]
- Bertrand B.; Casini A. A Golden Future in Medicinal Inorganic Chemistry: The Promise of Anticancer Gold Organometallic Compounds. Dalton Trans. 2014, 43, 4209–4219. 10.1039/c3dt52524d. [DOI] [PubMed] [Google Scholar]
- Massai L.; Messori L.; Micale N.; Schirmeister T.; Maes L.; Fregona D.; Cinellu M. A.; Gabbiani C. Gold Compounds as Cysteine Protease Inhibitors: Perspectives for Pharmaceutical Application as Antiparasitic Agents. BioMetals 2017, 30, 313–320. 10.1007/s10534-017-0007-0. [DOI] [PubMed] [Google Scholar]
- Harbut M. B.; Vilchèze C.; Luo X.; Hensler M. E.; Guo H.; Yang B.; Chatterjee A. K.; Nizet V.; Jacobs W. R.; Schultz P. G.; Wang F. Auranofin Exerts Broad-Spectrum Bactericidal Activities by Targeting Thiol-Redox Homeostasis. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 4453–4458. 10.1073/pnas.1504022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch R. Ueber Bacteriologische Forschung. Dtsch. Med. Wochenschr. 1890, 16, 756–757. [Google Scholar]
- Nobili S.; Mini E.; Landini I.; Gabbiani C.; Casini A.; Messori L. Gold Compounds as Anticancer Agents: Chemistry, Cellular Pharmacology, and Preclinical Studies. Med. Res. Rev. 2010, 30, 550–580. 10.1002/med.20168. [DOI] [PubMed] [Google Scholar]
- Roder C.; Thomson M. J. Auranofin: Repurposing an Old Drug for a Golden New Age. Drugs R&D 2015, 15, 13–20. 10.1007/s40268-015-0083-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jatoi A.; Radecki Breitkopf C.; Foster N. R.; Block M. S.; Grudem M.; Wahner Hendrickson A.; Carlson R. E.; Barrette B.; Karlin N.; Fields A. P. A Mixed-Methods Feasibility Trial of Protein Kinase C Iota Inhibition with Auranofin in Asymptomatic Ovarian Cancer Patients. Oncology 2015, 88, 208–213. 10.1159/000369257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casini A.; Bonsignore R.; Oberkofler J.. Organometallic Gold-Based Anticancer Therapeutics. Reference Module in Chemistry, Molecular Sciences and Chemical Engineering; Springer, 2018. [Google Scholar]
- Mora M.; Gimeno M. C.; Visbal R. Recent Advances in Gold-NHC Complexes with Biological Properties. Chem. Soc. Rev. 2019, 48, 447–462. 10.1039/c8cs00570b. [DOI] [PubMed] [Google Scholar]
- Porchia M.; Pellei M.; Marinelli M.; Tisato F.; Del Bello F.; Santini C. New Insights in Au-NHCs Complexes as Anticancer Agents. Eur. J. Med. Chem. 2018, 146, 709–746. 10.1016/j.ejmech.2018.01.065. [DOI] [PubMed] [Google Scholar]
- Zhang C.; Maddelein M. L.; Wai-Yin Sun R.; Gornitzka H.; Cuvillier O.; Hemmert C. Pharmacomodulation on Gold-NHC Complexes for Anticancer Applications - Is Lipophilicity the Key Point?. Eur. J. Med. Chem. 2018, 157, 320–332. 10.1016/j.ejmech.2018.07.070. [DOI] [PubMed] [Google Scholar]
- Schmidt C.; Karge B.; Misgeld R.; Prokop A.; Franke R.; Brönstrup M.; Ott I. Gold(I) NHC Complexes: Antiproliferative Activity, Cellular Uptake, Inhibition of Mammalian and Bacterial Thioredoxin Reductases, and Gram-Positive Directed Antibacterial Effects. Chem.—Eur. J. 2017, 23, 1869–1880. 10.1002/chem.201604512. [DOI] [PubMed] [Google Scholar]
- Bertrand B.; Citta A.; Franken I. L.; Picquet M.; Folda A.; Scalcon V.; Rigobello M. P.; Le Gendre P.; Casini A.; Bodio E. Gold(I) NHC-Based Homo- and Heterobimetallic Complexes: Synthesis, Characterization and Evaluation as Potential Anticancer Agents. J. Biol. Inorg Chem. 2015, 20, 1005–1020. 10.1007/s00775-015-1283-1. [DOI] [PubMed] [Google Scholar]
- Oberkofler J.; Aikman B.; Bonsignore R.; Pöthig A.; Platts J.; Casini A.; Kühn F. E. Exploring the Reactivity and Biological Effects of Heteroleptic N-Heterocyclic Carbene Gold(I)-Alkynyl Complexes. Eur. J. Inorg. Chem. 2020, 1040–1051. 10.1002/ejic.201901043. [DOI] [Google Scholar]
- Tolbatov I.; Coletti C.; Marrone A.; Re N. Insight into the Substitution Mechanism of Antitumor Au(I) N-Heterocyclic Carbene Complexes by Cysteine and Selenocysteine. Inorg. Chem. 2020, 59, 3312–3320. 10.1021/acs.inorgchem.0c00106. [DOI] [PubMed] [Google Scholar]
- Bindoli A.; Rigobello M. P.; Scutari G.; Gabbiani C.; Casini A.; Messori L. Thioredoxin Reductase: A Target for Gold Compounds Acting as Potential Anticancer Drugs. Coord. Chem. Rev. 2009, 253, 1692–1707. 10.1016/j.ccr.2009.02.026. [DOI] [Google Scholar]
- Hickey J. L.; Ruhayel R. A.; Barnard P. J.; Baker M. V.; Berners-Price S. J.; Filipovska A. Mitochondria-Targeted Chemotherapeutics: The Rational Design of Gold(I) N-Heterocyclic Carbene Complexes That Are Selectively Toxic to Cancer Cells and Target Protein Selenols in Preference to Thiols. J. Am. Chem. Soc. 2008, 130, 12570–12571. 10.1021/ja804027j. [DOI] [PubMed] [Google Scholar]
- Bertrand B.; Stefan L.; Pirrotta M.; Monchaud D.; Bodio E.; Richard P.; Le Gendre P.; Warmerdam E.; de Jager M. H.; Groothuis G. M. M.; Picquet M.; Casini A. Caffeine-Based Gold(I) N-Heterocyclic Carbenes as Possible Anticancer Agents: Synthesis and Biological Properties. Inorg. Chem. 2014, 53, 2296–2303. 10.1021/ic403011h. [DOI] [PubMed] [Google Scholar]
- Meier-Menches S. M.; Neuditschko B.; Zappe K.; Schaier M.; Gerner M. C.; Schmetterer K. G.; Del Favero G.; Bonsignore R.; Cichna-Markl M.; Koellensperger G.; Casini A.; Gerner C. An Organometallic Gold(I) Bis-N-Heterocyclic Carbene Complex with Multimodal Activity in Ovarian Cancer Cells. Chem.—Eur. J. 2020, 26, 15528–15537. 10.1002/chem.202003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney D.; Spiegel J.; Zyner K.; Tannahill D.; Balasubramanian S. The Regulation and Functions of DNA and RNA G-Quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. 10.1038/s41580-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle S. Quadruplex Nucleic Acids as Targets for Anticancer Therapeutics. Nat. Rev. Chem. 2017, 1, 41. 10.1038/s41570-017-0041. [DOI] [Google Scholar]
- Palma E.; Carvalho J.; Cruz C.; Paulo A. Metal-Based g-Quadruplex Binders for Cancer Theranostics. Pharmaceuticals 2021, 14, 605. 10.3390/ph14070605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q.; Li Y.; Freisinger E.; Qin P. Z.; Sigel R. K. O.; Mao Z. W. G-Quadruplex DNA Targeted Metal Complexes Acting as Potential Anticancer Drugs. Inorg. Chem. Front. 2017, 4, 10–32. 10.1039/c6qi00300a. [DOI] [Google Scholar]
- Caracelli I.; Zukerman-Schpector J.; Tiekink E. R. T. Supra-molecular synthons based on gold...π(arene) interactions. Gold Bull. 2013, 46, 81–89. 10.1007/s13404-013-0088-7. [DOI] [Google Scholar]
- Meier-Menches S. M.; Aikman B.; Döllerer D.; Klooster W. T.; Coles S. J.; Santi N.; Luk L.; Casini A.; Bonsignore R. Comparative Biological Evaluation and G-Quadruplex Interaction Studies of Two New Families of Organometallic Gold(I) Complexes Featuring N-Heterocyclic Carbene and Alkynyl Ligands. J. Inorg. Biochem. 2020, 202, 110844. 10.1016/j.jinorgbio.2019.110844. [DOI] [PubMed] [Google Scholar]
- Bazzicalupi C.; Ferraroni M.; Papi F.; Massai L.; Bertrand B.; Messori L.; Gratteri P.; Casini A. Determinants for Tight and Selective Binding of a Medicinal Dicarbene Gold(I) Complex to a Telomeric DNA G-Quadruplex: A Joint ESI MS and XRD Investigation. Angew. Chem. 2016, 128, 4328–4331. 10.1002/ange.201511999. [DOI] [PubMed] [Google Scholar]
- Wragg D.; de Almeida A.; Bonsignore R.; Kühn F. E. F. E.; Leoni S.; Casini A. On the Mechanism of Gold/NHC Compounds Binding to DNA G-Quadruplexes: Combined Metadynamics and Biophysical Methods. Angew. Chem., Int. Ed. 20718, 57, 14524–14528. 10.1002/anie.201805727. [DOI] [PubMed] [Google Scholar]
- Smargiasso N.; Gabelica V.; Damblon C.; Rosu F.; De Pauw E.; Teulade-Fichou M. P.; Rowe J. A.; Claessens A. Putative DNA G-Quadruplex Formation within the Promoters of Plasmodium Falciparum Var Genes. BMC Genom. 2009, 10, 362. 10.1186/1471-2164-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven H. M.; Bonsignore R.; Lenis V.; Santi N.; Berrar D.; Swain M.; Whiteland H.; Casini A.; Hoffmann K. F. Identifying and Validating the Presence of Guanine-Quadruplexes (G4) within the Blood Fluke Parasite Schistosoma Mansoni. PLoS Neglected Trop. Dis. 2021, 15, e0008770 10.1371/journal.pntd.0008770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saranathan N.; Vivekanandan P. G.-Q. G-Quadruplexes: More Than Just a Kink in Microbial Genomes. Trends Microbiol. 2019, 27, 148–163. 10.1016/j.tim.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minori K.; Rosa L. B.; Bonsignore R.; Casini A.; Miguel D. C. Comparing the Antileishmanial Activity of Gold(I) and Gold(III) Compounds in L. Amazonensis and L. Braziliensis in Vitro. ChemMedChem 2020, 15, 2146–2150. 10.1002/cmdc.202000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobialka S.; Müller-Tautges C.; Schmidt M. T. S.; Schnakenburg G.; Hollóczki O.; Kirchner B.; Engeser M. Stretch Out or Fold Back? Conformations of Dinuclear Gold(I) N-Heterocyclic Carbene Macrocycles. Inorg. Chem. 2015, 54, 6100–6111. 10.1021/ic502751s. [DOI] [PubMed] [Google Scholar]
- Guevara-Vela J. M.; Hess K.; Rocha-Rinza T.; Martín Pendás Á.; Flores-Álamo M.; Moreno-Alcántar G. Stronger-Together: The Cooperativity of Aurophilic Interactions. Chem. Commun. 2022, 58, 1398–1401. 10.1039/d1cc05241a. [DOI] [PubMed] [Google Scholar]
- Laio A.; Parrinello M. Escaping Free-Energy Minima. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 12562–12566. 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubaro C.; Baron M.; Costante M.; Basato M.; Biffis A.; Gennaro A.; Isse A. A.; Graiff C.; Accorsi G. Dinuclear Gold(i) Complexes with Propylene Bridged N-Heterocyclic Dicarbene Ligands: Synthesis, Structures, and Trends in Reactivities and Properties. Dalton Trans. 2013, 42, 10952–10963. 10.1039/c3dt51260f. [DOI] [PubMed] [Google Scholar]
- Stoppa V.; Scattolin T.; Bevilacqua M.; Baron M.; Graiff C.; Orian L.; Biffis A.; Menegazzo I.; Roverso M.; Bogialli S.; Visentin F.; Tubaro C. Mononuclear and Dinuclear Gold(i) Complexes with a Caffeine-Based Di(N-Heterocyclic Carbene) Ligand: Synthesis, Reactivity and Structural DFT Analysis. New J. Chem. 2021, 45, 961–971. 10.1039/d0nj05906d. [DOI] [Google Scholar]
- Jakob C. H. G.; Dominelli B.; Rieb J.; Jandl C.; Pöthig A.; Reich R. M.; Correia J. D. G.; Kühn F. E. Dinuclear Gold(I) Complexes Bearing N,N′-Allyl-Bridged Bisimidazolylidene Ligands. Chem.—Asian J. 2020, 15, 1848–1851. 10.1002/asia.202000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori M.; Kataoka T.; Shimizu H.; Imai E.; Matsumoto Y.; Kawachi M.; Kuratani K.; Yokomoto M. Generation and properties of N7-xanthinium ylides: Reactions of N7-xanthinium ylides with diphenylcyclopropenone and acetylenic compounds. Chem. Pharm. Bull. 1986, 34, 1328–1332. 10.1248/cpb.34.1328. [DOI] [Google Scholar]
- Hori M.; Kataoka T.; Shimizu H.; Imai E.; Matsumoto Y.; Kawachi M.; Kuratani K.; Ogura H.; Takayanagi H. Reactions of Xanthinium N(7)-Ylides with Olefinic Dipolarophiles. J. Chem. Soc., Perkin Trans. 1 1987, 1211–1219. 10.1039/p19870001211. [DOI] [Google Scholar]
- Hori M.; Kataoka T.; Shimizu H.; Imai E.; Matsumoto Y. Reactions of 8,9-dihydroxanthines with acetylenic compounds. Formation of heteropropellanes. Chem. Pharm. Bull. 1985, 33, 3681–3688. 10.1248/cpb.33.3681. [DOI] [Google Scholar]
- Riederer S. K. U.; Gigler P.; Högerl M. P.; Herdtweck E.; Bechlars B.; Herrmann W. A.; Kühn F. E. Impact of Ligand Modification on Structures and Catalytic Activities of Chelating Bis-Carbene Rhodium(I) Complexes. Organometallics 2010, 29, 5681–5692. 10.1021/om1005897. [DOI] [Google Scholar]
- Nahra F.; Tzouras N. V.; Collado A.; Nolan S. P. Synthesis of N-Heterocyclic Carbene Gold(I) Complexes. Nat. Protoc. 2021, 16, 1476–1493. 10.1038/s41596-020-00461-6. [DOI] [PubMed] [Google Scholar]
- Bruno I. J.; Cole J. C.; Edgington P. R.; Kessler M.; Macrae C. F.; McCabe P.; Pearson J.; Taylor R. New Software for Searching the Cambridge Structural Database and Visualizing Crystal Structures. Acta Crystallogr., Sect. B: Struct. Sci. 2002, 58, 389–397. 10.1107/s0108768102003324. [DOI] [PubMed] [Google Scholar]
- Macrae C. F.; Bruno I. J.; Chisholm J. A.; Edgington P. R.; McCabe P.; Pidcock E.; Rodriguez-Monge L.; Taylor R.; van de Streek J.; Wood P. A. Mercury CSD 2.0- new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. 10.1107/s0021889807067908. [DOI] [Google Scholar]
- Pyykkö P. Theoretical Chemistry of Gold. III. Chem. Soc. Rev. 2008, 37, 1967–1997. 10.1039/B708613J. [DOI] [PubMed] [Google Scholar]
- Fei F.; Lu T.; Chen X. T.; Xue Z. L. Synthesis and Structural Characterization of Metal Complexes with Macrocyclic Tetracarbene Ligands. New J. Chem. 2017, 41, 13442–13453. 10.1039/c7nj02485a. [DOI] [Google Scholar]
- Stefan L.; Bertrand B.; Richard P.; Le Gendre P.; Denat F.; Picquet M.; Monchaud D. Assessing the Differential Affinity of Small Molecules for Noncanonical DNA Structures. ChemBioChem 2012, 13, 1905–1912. 10.1002/cbic.201200396. [DOI] [PubMed] [Google Scholar]
- Wang H. M. J.; Vasam C. S.; Tsai T. Y. R.; Chen S. H.; Chang A. H. H.; Lin I. J. B. Gold(I) N-Heterocyclic Carbene and Carbazolate Complexes. Organometallics 2005, 24, 486–493. 10.1021/om049221u. [DOI] [Google Scholar]
- Cheng Y.; Zhang Y.; You H. Characterization of G-Quadruplexes Folding/Unfolding Dynamics and Interactions with Proteins from Single-Molecule Force Spectroscopy. Biomolecules 2021, 11, 1579. 10.3390/biom11111579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsen R. C.; DeLeeuw L.; Dean W. L.; Gray R. D.; Sabo T. M.; Chakravarthy S.; Chaires J. B.; Trent J. O. The HTERT Core Promoter Forms Three Parallel G-Quadruplexes. Nucleic Acids Res. 2020, 48, 5720–5734. 10.1093/nar/gkaa107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova A. V.; Savitskaya V. Y.; Dolinnaya N. G.; Monakhova M. V.; Litvinova A. V.; Kubareva E. A.; Zvereva M. I. G-Quadruplex Formed by the Promoter Region of the HTERT Gene: Structure-Driven Effects on DNA Mismatch Repair Functions. Biomedicines 2022, 10, 1871. 10.3390/biomedicines10081871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip K. W.; Reed J. C. Bcl-2 Family Proteins and Cancer. Oncogene 2008, 27, 6398–6406. 10.1038/onc.2008.307. [DOI] [PubMed] [Google Scholar]
- del Villar-Guerra R.; Trent J. O.; Chaires J. B. G-Quadruplex Secondary Structure Obtained from Circular Dichroism Spectroscopy. Angew. Chem., Int. Ed. 2018, 57, 7171–7175. 10.1002/anie.201709184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodger A.DNA-Ligand Circular Dichroism. Encyclopedia of Biophysics; Springer, 2013; pp 484–486 [Google Scholar]
- Terenzi A.; Bonsignore R.; Spinello A.; Gentile C.; Martorana A.; Ducani C.; Högberg B.; Almerico A. M.; Lauria A.; Barone G. Selective G-Quadruplex Stabilizers: Schiff-Base Metal Complexes with Anticancer Activity. RSC Adv. 2014, 4, 33245–33256. 10.1039/c4ra05355a. [DOI] [Google Scholar]
- Ang D. L.; Kelso C.; Beck J. L.; Ralph S. F.; Harman D. G.; Aldrich-Wright J. R. A study of Pt(II)-phenanthroline complex interactions with double-stranded and G-quadruplex DNA by ESI-MS, circular dichroism, and computational docking. J. Biol. Inorg Chem. 2020, 25, 429–440. 10.1007/s00775-020-01773-4. [DOI] [PubMed] [Google Scholar]
- Ruehl C. L.; Lim A. H. M.; Kench T.; Mann D. J.; Vilar R. An Octahedral Cobalt(III) Complex with Axial NH 3 Ligands that Templates and Selectively Stabilises G-quadruplex DNA. Chem.—Eur. J. 2019, 25, 9691–9700. 10.1002/chem.201901569. [DOI] [PubMed] [Google Scholar]
- Bussi G.; Laio A. Using Metadynamics to Explore Complex Free-Energy Landscapes. Nat. Rev. Phys. 2020, 2, 200–212. 10.1038/s42254-020-0153-0. [DOI] [Google Scholar]
- Wragg D.; de Almeida A.; Casini A.; Leoni S. Unveiling the Mechanisms of Aquaglyceroporin-3 Water and Glycerol Permeation by Metadynamics. Chem.—Eur. J. 2019, 25, 8713–8718. 10.1002/chem.201902121. [DOI] [PubMed] [Google Scholar]
- Wragg D.; Leoni S.; Casini A. Aquaporin-Driven Hydrogen Peroxide Transport: A Case of Molecular Mimicry?. RSC Chem. Biol. 2020, 1, 390–394. 10.1039/d0cb00160k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farine G.; Migliore C.; Terenzi A.; Lo Celso F.; Santoro A.; Bruno G.; Bonsignore R.; Barone G. On the G-Quadruplex Binding of a New Class of Nickel(II), Copper(II), and Zinc(II) Salphen-Like Complexes. Eur. J. Inorg. Chem. 2021, 1332–1336. 10.1002/ejic.202100067. [DOI] [Google Scholar]
- Kascatan-Nebioglu A.; Melaiye A.; Hindi K.; Durmus S.; Panzner M. J.; Hogue L. A.; Mallett R. J.; Hovis C. E.; Coughenour M.; Crosby S. D.; Milsted A.; Ely D. L.; Tessier C. A.; Cannon C. L.; Youngs W. J. Synthesis from Caffeine of a Mixed N-Heterocyclic Carbene–Silver Acetate Complex Active against Resistant Respiratory Pathogens. J. Med. Chem. 2006, 49, 6811–6818. 10.1021/jm060711t. [DOI] [PubMed] [Google Scholar]
- Valdés H.; Canseco-González D.; Germán-Acacio J. M.; Morales-Morales D. Xanthine Based N-Heterocyclic Carbene (NHC) Complexes. J. Organomet. Chem. 2018, 867, 51–54. 10.1016/j.jorganchem.2018.01.008. [DOI] [Google Scholar]
- Scattolin T.; Caligiuri I.; Canovese L.; Demitri N.; Gambari R.; Lampronti I.; Rizzolio F.; Santo C.; Visentin F. Synthesis of New Allyl Palladium Complexes Bearing Purine-Based NHC Ligands with Antiproliferative and Proapoptotic Activities on Human Ovarian Cancer Cell Lines. Dalton Trans. 2018, 47, 13616–13630. 10.1039/c8dt01831f. [DOI] [PubMed] [Google Scholar]
- Loos M.; Gerber C.; Corona F.; Hollender J.; Singer H. Accelerated Isotope Fine Structure Calculation Using Pruned Transition Trees. Anal. Chem. 2015, 87, 5738–5744. 10.1021/acs.analchem.5b00941. [DOI] [PubMed] [Google Scholar]
- Price G. A.; Brisdon A. K.; Flower K. R.; Pritchard R. G.; Quayle P. Solvent Effects in Gold-Catalysed A3-Coupling Reactions. Tetrahedron Lett. 2014, 55, 151–154. 10.1016/j.tetlet.2013.10.141. [DOI] [Google Scholar]
- Wong W. W. H.; Vickers M. S.; Cowley A. R.; Paul R. L.; Beer P. D. Tetrakis(Imidazolium) Macrocyclic Receptors for Anion Binding. Org. Biomol. Chem. 2005, 3, 4201–4208. 10.1039/b510068b. [DOI] [PubMed] [Google Scholar]
- Itahara T.; Imamura K. Preparation and NMR Study of 7,7′-(α,ω-Alkanediyl)bis[theophylline], 1,1′-(α,ω-Alkanediyl)bis[theobromine], and 1,1′-(α,ω-Alkanediyl)bis[3-methyluracil]. Bull. Chem. Soc. Jpn. 1994, 67, 203–209. 10.1246/bcsj.67.203. [DOI] [Google Scholar]
- Pilarski B. A. A new method forN-alkylation of imidazoles and benzimidazoles. Liebigs Ann. Chem. 1983, 1983, 1078–1080. 10.1002/jlac.198319830619. [DOI] [Google Scholar]
- Peppel T.; Hinz A.; Köckerling M. Salts with the [NiBr3(L)]– complex anion (L=1-methylimidazole, 1-methylbenzimidazole, quinoline, and triphenylphosphane) and low melting points: A comparative study. Polyhedron 2013, 52, 482–490. 10.1016/j.poly.2012.08.047. [DOI] [Google Scholar]
- Bruker AXS Inc APEX Suite of Crystallographic Software; APEX3: Madison, Wisconsin, USA, 2015.
- SAINT; Version 8.40A and SADABS, Version 2016/2; Bruker AXS Inc.: Madison, Wisconsin, USA, 2016.
- Sheldrick G. M. SHELXT- Integrated space-group and crystal-structure determination. Acta Crystallogr., Sect. A: Found. Crystallogr. 2015, 71, 3–8. 10.1107/s2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G. M. Crystal structure refinement withSHELXL. Acta Crystallogr., Sect. C: Struct. Chem. 2015, 71, 3–8. 10.1107/s2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübschle C. B.; Sheldrick G. M.; Dittrich B. ShelXle: a Qt graphical user interface for SHELXL. J. Appl. Crystallogr. 2011, 44, 1281–1284. 10.1107/S0021889811043202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. J. C.; Geist V. International Tables for Crystallography. Volume C: Mathematical, Physical and Chemical Tables. Kluwer Academic Publishers, Dordrecht/Boston/London 1992 (Published for the International Union of Crystallography), 883 Seiten, ISBN 0-792-3-16-38X. Cryst. Res. Technol. 1993, 28, 110. 10.1002/crat.2170280117. [DOI] [Google Scholar]
- Macrae C. F.; Bruno I. J.; Chisholm J. A.; Edgington P. R.; McCabe P.; Pidcock E.; Rodriguez-Monge L.; Taylor R.; van de Streek J.; Wood P. A. Mercury CSD 2.0- new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. 10.1107/s0021889807067908. [DOI] [Google Scholar]
- Böttcher A.; Kowerko D.; Sigel R. K. O. Explicit Analytic Equations for Multimolecular Thermal Melting Curves. Biophys. Chem. 2015, 202, 32–39. 10.1016/j.bpc.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Lane A. N.; Chaires J. B.; Gray R. D.; Trent J. O. Stability and Kinetics of G-Quadruplex Structures. Nucleic Acids Res. 2008, 36, 5482–5515. 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger Maestro; Schrödinger: New York, 2021.
- Shaw D. E.Schrödinger: Desmond Molecular Dynamics System; Schrödinger Release: New York, NY, USA, 2021; p 4. [Google Scholar]
- Govindaraju N. K.; Larsen S.; Gray J.; Manocha D.. A Memory Model for Scientific Algorithms on Graphics Processors. Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, SC’06; ACM: New York, NY, USA, 2006; p 746.
- Banks J. L.; Beard H. S.; Cao Y.; Cho A. E.; Damm W.; Farid R.; Felts A. K.; Halgren T. A.; Mainz D. T.; Maple J. R.; Murphy R.; Philipp D. M.; Repasky M. P.; Zhang L. Y.; Berne B. J.; Friesner R. A.; Gallicchio E.; Levy R. M. Integrated Modeling Program, Applied Chemical Theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark P.; Nilsson L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. 10.1021/jp003020w. [DOI] [Google Scholar]
- Nosé S. A Molecular Dynamics Method for Simulations in the Canonical Ensemble. Mol. Phys. 1984, 52, 255–268. 10.1080/00268978400101201. [DOI] [Google Scholar]
- Martyna G. J.; Tobias D. J.; Klein M. L. Constant Pressure Molecular Dynamics Algorithms. J. Chem. Phys. 1994, 101, 4177–4189. 10.1063/1.467468. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.