Abstract
The virulence mechanisms of the facultative intracellular parasite Rhodococcus equi remain largely unknown. Among the candidate virulence factors of this pathogenic actinomycete is a secreted cholesterol oxidase, a putative membrane-damaging toxin. We identified and characterized the gene encoding this enzyme, the choE monocistron. Its protein product, ChoE, is homologous to other secreted cholesterol oxidases identified in Brevibacterium sterolicum and Streptomyces spp. ChoE also exhibits significant similarities to putative cholesterol oxidases encoded by Mycobacterium tuberculosis and Mycobacterium leprae. Genetic tools for use with R. equi are poorly developed. Here we describe the first targeted mutagenesis system available for this bacterium. It is based on a suicide plasmid, a selectable marker (the aacC4 apramycin resistance gene from Salmonella), and homologous recombination. The choE allele was disrupted by insertion of the aacC4 gene, cloned in pUC19 and introduced by electroporation in R. equi. choE recombinants were isolated at frequencies between 10−2 and 10−3. Twelve percent of the recombinants were double-crossover choE mutants. The choE mutation was associated with loss of cooperative (CAMP-like) hemolysis with sphingomyelinase-producing bacteria (Listeria ivanovii). Functional complementation was achieved by expression of choE from pVK173-T, a pAL5000 derivative conferring hygromycin resistance. Our data demonstrate that ChoE is an important cytolytic factor for R. equi. The highly efficient targeted mutagenesis procedure that we used to generate choE isogenic mutants will be a valuable tool for the molecular analysis of R. equi virulence.
The nocardioform actinomycete Rhodococcus equi is a primary pathogen of horses. Rhodococcal infection occurs in foals aged under 6 months old and results in severe pyogranulomatous bronchopneumonia and a high mortality rate. Respiratory disease is sometimes accompanied by mesenteric lymphadenitis and ulcerative enterocolitis. R. equi is widespread in its natural habitat, the soil, and rhodococcal infection is endemic in some horse farms. R. equi has recently emerged as an opportunistic pathogen in humans, especially in association with human immunodeficiency virus infection. Like in foals, human R. equi infection mainly affects the lungs, with clinical and pathological characteristics similar to pulmonary tuberculosis in immunocompromised patients. Although rare, granulomatous pneumonia, lymphadenitis, and abscesses caused by R. equi have been reported in a variety of mammals other than horses and humans (11, 17, 32, 40).
Despite its importance in veterinary medicine and as an emerging AIDS-associated pathogen, nothing is known about the virulence mechanisms that R. equi uses to colonize host tissues. The capacity of these bacteria to survive and to multiply inside the vacuolar compartment of macrophages is central to rhodococcal pathogenesis (16). Virulence in the natural host and in the mouse experimental model and the ability to replicate in macrophages has been related to the presence of an 80- to 90-kb plasmid (47). This plasmid is present in virtually all clinical isolates from foals, but it is absent from most environmental strains (10, 14, 45). The plasmid carries a cluster of seven vap genes encoding surface-associated proteins which react in the form of 15- to 18-kDa antigens with sera from pneumonic foals or from foals exposed to plasmid-containing, virulent R. equi isolates (46, 47). Because Vap antigens are upregulated at elevated temperatures (34 to 41°C) (46, 47) and have a role in the protective immune response against R. equi in foals (39), they are believed to play an important role in pathogenesis. To date, only one attempt has been made to assess the role of Vap proteins in virulence by using a genetic approach. The vapA gene was expressed in a plasmid-cured isogenic strain of R. equi, but this was not sufficient to restore the capacity to proliferate in macrophages and to colonize the lungs of experimentally infected foals (10), questioning a role for VapA in virulence. The virulence plasmid itself does not appear to be essential for pathogenesis in non-horse hosts, as it is not always detected in clinical isolates from humans and other animal species (4, 7, 33, 48, 49, 52). This suggests that chromosomally determined factors are involved in R. equi pathogenicity. Candidates for such chromosomal virulence factors include the following: the capsular polysaccharide, which might interfere with phagocytosis; mycolic acid-containing glycolipids, which are thought to be involved in granuloma formation; and, especially, cholesterol oxidase, a secreted enzyme that may act on eukaryotic membranes and be responsible for the observed cytotoxicity and macrophage destruction that accompany rhodococcal infection (17, 40). However, as for VapA, there is no direct proof that any of these putative virulence factors are involved in pathogenesis.
A major reason why the molecular mechanisms of R. equi pathogenesis remain unknown is the absence of genetic tools for creating isogenic mutants affected in individual loci in these bacteria. We have developed the first site-directed mutagenesis system that is functional in R. equi. This system is based on homologous recombination and on a suicide vector carrying a cassette that confers apramycin resistance. This system allowed us to generate cholesterol oxidase null mutants by insertional disruption of the structural gene of the enzyme, choE, which was also identified and characterized in this study.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. R. equi and Escherichia coli were routinely grown at 37°C in Luria-Bertani medium, with rotary agitation in the case of fluid cultures. When required, antibiotics were added to culture media at the following concentrations: apramycin, 30 μg/ml; hygromycin, 150 μg/ml; ampicillin, 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Description | Source or reference |
|---|---|---|
| R. equi | ||
| ATCC 6939 | Type strain | 30 |
| MAD | Cholesterol oxidase-hyperproducing strain | 42 |
| 103+ | Clinical isolate with virulence plasmid | 8 |
| 103− | 103 derivative lacking virulence plasmid | 8 |
| RHE3-15+ | choE knockout mutant of 103+ | This work |
| RHE3-19 | choE knockout mutant of 103− | This work |
| L. ivanovii | ||
| ATCC 19119 | Type strain | Collection |
| E. coli | ||
| DH5α | Cloning host strain | Our laboratory |
| HB101 | Cloning host strain | Our laboratory |
| TG1 | Cloning host strain | Our laboratory |
| Plasmids | ||
| pUC19 | Cloning vector | 54 |
| pGEM-Te | T-vector for cloning of PCR products | Promega |
| pPE207 | E. coli-Mycobacterium shuttle vector containing apramycin resistance marker | 37 |
| pVK173-T | E. coli-Mycobacterium shuttle vector containing hygromycin resistance marker | 37 |
| pRE7 | E. coli-R. equi shuttle vector | 55 |
| pRHE1 | pUC19 inserted with choE | This work |
| pRHE2 | pUC19 inserted with accC4 | This work |
| pRHE3 | pUC19 with choE::accC4 mutant allele (suicide vector for choE mutagenesis by gene replacement) | This work |
| pRHE4 | pGEM-Te inserted with choE and its natural promoter | This work |
| pRHE5 | pVK173-T inserted with choE and its natural promoter | This work |
| pRHE6A | pRE7 inserted with accC4 | This work |
DNA techniques.
R. equi genomic DNA was prepared using a modification of a previously described protocol (2). Bacteria from 5-ml aliquots of a stationary-phase broth culture were collected by centrifugation at 10,000 × g for 10 min, washed in distilled water, resuspended in 0.25 ml of Tris-EDTA buffer containing 20 mg of lysozyme/ml and 50 mg of proteinase K/ml and incubated at 37°C for 2 h. Bacterial cells were then lysed by the addition of 0.25 ml of 0.1 M Tris containing 1% sodium dodecyl sulfate (SDS) and 400 μg of proteinase K/ml and incubated at 55°C for 1 h. The lysate was mixed with 0.1 ml of 5 M NaCl and 100 μl of cetyltrimethylammonium bromide-NaCl and incubated at 65°C for 10 min. DNA was then extracted with choloroform-isoamyl alcohol and phenol-choloroform, precipitated with isopropanol, and resuspended gently in distilled water. Plasmid DNA was extracted from E. coli using Qiagen plasmid purification kit. Single-stranded DNA (ssDNA) was prepared by mixing 1 μg of plasmid DNA with 20 μl of 0.2 M NaOH and 0.2 mM EDTA in distilled water. The mixture was incubated at 37°C for 30 min and DNA was precipitated with ethanol according to standard methodology. PCR products were purified from agarose gels with the Qiaquick purification system (Qiagen). Restriction enzymes and ligase were purchased from New England Biolabs and used according to the manufacturer's instructions. DNA was amplified using the Expand High-Fidelity PCR System (Roche). For Southern blotting, restriction endonuclease-treated total DNA was separated by agarose gel electrophoresis and immobilized on nylon membranes (Roche) by capillary blotting. Radiolabeling was performed with [α-32P]dCTP (Amersham) by random priming using the Ready-to-Go kit from Pharmacia. Hybridization was performed at 65°C in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt's solution, 0.5% SDS, and 20 mg of salmon sperm DNA/ml. Blots were washed with 2× SSC, 0.5% SDS at room temperature for 5 min and then with 2× SSC, 0.1% SDS at 37°C for 30 min. DNA sequencing was performed on PCR products at the “Unidad de Secuenciación Automatizada de DNA” of the Universidad Complutense de Madrid using an Applied Biosystems 377 apparatus. Homology searches were performed with BLAST at the National Center for Biotechnology Information (Bethesda, Md.) website and with the Pfam (protein family database) search tool on the Sanger Centre (Cambridge, United Kingdom) internet server.
Electroporation of R. equi.
Bacteria from 50 ml of a culture grown to an optical density at 600 nm (OD600) of 0.6 were harvested by centrifugation, washed two times in 25 ml of washing buffer (cold 10% glycerol in distilled water), and resuspended in 0.2 ml of the same solution. Aliquots (40 μl) of the R. equi cell suspension were mixed with 200 ng of DNA in a prechilled 0.2-cm chamber and electroporated using the Gene Pulser apparatus (Bio-Rad) set at 2.5 kV/cm, 25 μF, and 1,000 Ω. After electroporation, the bacterial suspension was diluted with 1 ml of Luria-Bertani medium, incubated at 37°C for 2 h, and plated onto solid medium containing the appropriate antibiotics.
RNA techniques.
Total RNA was extracted from 50-ml broth cultures grown to an OD600 of 0.6. Bacteria were harvested by centrifugation, resuspended in 2.5 ml of lysis buffer (0.02 M sodium acetate [pH 5.0], 1 mM EDTA, 20 mg of lysozyme/ml), and transferred to a 100-ml Erlenmeyer flask. The bacterial suspension was immediately frozen by placing the flask on dry ice and then thawed at 37°C. This treatment was repeated five times. After the last thawing, 0.25 ml of 10% SDS and 2.5 ml of 0.02 M sodium acetate-saturated phenol were added. The mixture was incubated at 70°C for 5 min with shaking and spun for 5 min in a microcentrifuge, and the aqueous phase was transferred to a fresh 100-ml flask. Phenol extractions were repeated two more times. Aliquots of 0.4 ml of the final aqueous phase were transferred to 2-ml microcentrifuge tubes, mixed with 40 μl of sodium acetate (pH 7), and precipitated with 3 volumes of ethanol. The RNA precipitate was collected by centrifugation at 12,000 × g for 15 min at 4°C, washed with 70% ethanol, and dried. The RNA pellet was resuspended in 100 μl of diethyl pyrocarbonate-treated distilled water and stored at −70°C. Northern blotting was carried out using 10 μg of RNA as previously described (12).
Cholesterol oxidase determinations.
An assay based on the method described by Sojo et al. (43) was used. Cultures were grown to an OD600 of 0.6. After centrifugation, 0.1 ml of culture supernatant was mixed with 0.9 ml of reaction buffer (50 mM potasium phosphate [pH 7.0], 5 mM sodium cholate, 7 mM phenol, 0.4 mM 4-amino-antiprine, 1 mM cholesterol, 0.33% Triton X-100, and 6.7 U of horseradish peroxidase) and incubated at 37°C until a red color developed. Cholesterol oxidase activity units were calculated from the ratio between A500 × 104 and A600 × d × t, where d is the dilution factor and t is the reaction time expressed in minutes. choE-derived cholesterol oxidase activity was calculated by subtracting the activity of the control (ChoE−) strain from that of the isogenic test strain with functional choE allele.
Cooperative (CAMP-like) hemolysis assays.
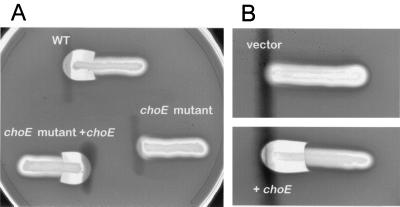
CAMP-like hemolysis tests were performed on sheep blood agar plates with Columbia base medium (Difco) as described previously (42). The test and indicator bacteria were streaked perpendicularly to each other, leaving a distance between streaks of approximately 1 mm. Plates were incubated at 37°C overnight and the appearance of a shovel-shaped patch of hemolysis at the intersection of the streaks (see Fig. 5), due to the hemolytic cooperativity of the sphingomyelinase C produced by the indicator strain (Listeria ivanovii) and the R. equi cholesterol oxidase (9, 42), was recorded as a positive reaction.
FIG. 5.
Cooperative hemolysis assays with the sphingomyelinase-producing indicator species L. ivanovii (horizontal streaks). (A) Wild-type R. equi 103− (WT) and its isogenic derivatives RHE3-19 (choE/pRHE5 mutant) and RHE3-19 (choE mutant choE). The choE mutant has lost the capacity to produce a shovel-shaped CAMP-like reaction and this property is recovered upon complementation with choE. (B) E. coli K-12 with the control vector pRHE2 (vector) and with the choE-containing plasmid pRHE1 (+choE); complementation with choE confers CAMP-like activity similar to that of wild-type R. equi.
Nucleotide sequence accession number.
The sequence for the R. equi choE gene has been deposited in the EMBL database under accession number AJ242746.
RESULTS
Identification of the R. equi cholesterol oxidase gene choE.
A number of gram-positive bacteria with a high G+C content, such as Brevibacterium sterolicum, Streptomyces spp., Mycobacterium spp., and Rhodococcus spp. (R. equi, R. erythropolis, and R. rhodochrous), have a cholesterol oxidase activity (29). This suggests that a common cholesterol oxidase genetic determinant is widespread among actinomycetes and related bacteria. Supporting this notion, the sequences of the cholesterol oxidase genes cloned to date, choA and choM from two Streptomyces spp. isolates (5, 19) and choB from B. sterolicum (35), show a high degree of similarity at both the nucleotide and amino acid levels. To determine whether R. equi contains cho-related sequences, Southern blots of genomic DNA digestions from three strains of this species (103−, ATCC 6939, and MAD) (Table 1) and 20 additional clinical isolates of human and animal origin were probed with a choB DNA fragment. Positive signals were obtained in a 2.3-kb PstI fragment for all of the R. equi strains tested (data not shown). To isolate the hybridizing R. equi DNA, a pair of degenerate oligonucleotide primers {CoXN [5′-AT(CT)TT(CT)TG(CT)GG(GC)ATGCT(AGCT)AA(CT)CC-3′] and CoXC [5′-C(GT)(GC)GC(AG)AA(CT)TT(AG)TACCA(CT)TC(AGCT)GT-3′]} were designed from the aligned amino acid sequences predicted from the known cho genes and used in PCRs with genomic DNA from strain 103−. A 0.3-kb DNA fragment was amplified, cloned in pGEM-Te, and sequenced. The nucleotide sequence obtained was 99% similar to that of the corresponding segment of the choB gene. Several sets of primers were designed from choB, which allowed us to assemble a 3.9-kb region of the R. equi chromosome by direct and inverse PCR and subsequent primer-walking sequencing (Fig. 1). The choB-related sequence in this region belonged to a 1,656-bp open reading frame (ORF). The protein encoded by this ORF showed extensive similarity with the polypeptides encoded by the cholesterol oxidase genes choB from B. sterolicum and choA and choM from Streptomyces spp. (Fig. 2). It also exhibited significant similarity, albeit weaker, to polypeptides encoded by Mycobacterium leprae, Mycobacterium tuberculosis, and Streptomyces coelicolor described as probable cholesterol oxidases and named as ChoD (Fig. 2). The R. equi ORF was designated choE (for cholesterol oxidase of R. equi). choE was preceded at the correct distance by a putative ribosome-binding sequence, GAGG, and its stop codon was followed by a palindromic sequence of 44 nucleotides that might act as a transcription terminator. Putative −35 and −10 sites (GCGACG and CAGACC, respectively) were identified in the intergenic region upstream from choE on the basis of known Rhodococcus promoter sequences (1, 13, 23). The choE gene product, ChoE, is 552 amino acids long and has a predicted signal sequence of 45 residues (Fig. 2). A DNA fragment comprising the entire choE gene plus its putative ribosome-binding sequence and the transcription terminator was amplified by PCR with oligonucleotide primers CoEN (5′-TACCAAGCTTACCAAACCGCCGACAGAGGA-3′) and CoEC (5′-CAGTGAATTCCGCGTGAAGAAAACGTGGTC-3′) and inserted into pUC19, resulting in pRHE1 (Table 1). E. coli TG1 cells transformed with pRHE1 produced cholesterol oxidase activity (Table 2) and displayed cooperative, CAMP-like hemolysis with sphingomyelinase C-producing bacteria (a marker of cholesterol oxidase production; see below and Fig. 5). However, when TG1 was transformed with pRHE3, in which choE is disrupted by an antibiotic resistance cassette, neither cholesterol oxidase activity nor the CAMP-like hemolysis reaction was detected. These results confirmed that choE encodes R. equi cholesterol oxidase.
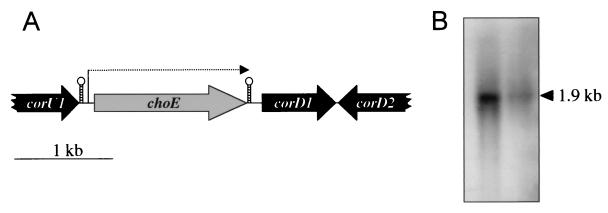
FIG. 1.
Genetic organization of the choE region and transcriptional analysis of choE. (A) Physical map of the 3.9-kb chromosomal region encompassing the choE locus of R. equi. The location of the putative choE promoter and stem-loop transcription terminator is indicated. (B) Northern blot analysis of choE in strains R. equi MAD (left) and 103− (right). In both strains, a single 1.9-kb transcript was detected.
FIG. 2.
Comparison of amino acid sequences of the cholesterol oxidases ChoE from R. equi (R. eq.; accession no. AJ242746) and ChoA from Streptomyces spp. strain SA-COO (S. sp.; accession no. A32260), and related ChoD polypeptides from M. leprae (M. le.; accession no. S72824), M. tuberculosis (M. tu.; accession no. F70736), and S. coelicolor (S. co.; accession no. AL161755). Identical amino acids are shaded in black and similar amino acids are in gray. The putative cleavage site of the signal peptide of ChoE is indicated by an arrowhead.
TABLE 2.
ChoE activity determinations
| Strain | Enzyme activitya |
|---|---|
| E. coli | |
| TG1(pUC19) | 0 |
| TG1(pRHE1) | 100.7 |
| R. equi | |
| 103− | 29.5 |
| RHE3-19 | 0 |
| RHE3-19(pRHE5) | 37.3 |
| 103+ | 32.3 |
| RHE3-15+ | 0 |
| RHE3-15+(pRHE5) | 28.9 |
choE-derived activity (see Materials and Methods).
Genetic structure of the choE region and Northern blot analysis of choE.
Three additional ORFs coding for polypeptides similar to known protein sequences were identified in the 3.9-kb DNA fragment that contains choE (Fig. 1). Upstream from choE, we found the 3′ region of an ORF, cor (for cholesterol oxidase region) U1, which encodes the last 159 residues of a protein with a high degree of similarity to a number of bacterial 3-oxoacyl-(acyl carrier protein) reductases. These enzymes belong to the short-chain dehydrogenase-reductase superfamily that is widespread in bacteria, archaea, and eukaryotes. Immediately downstream from choE and transcribed in the same orientation is corD1, which encodes a putative homolog of the cfp30B gene product (67% identity), a 27.3-kDa antigen from M. tuberculosis. The corD1 product (CorD1) also shows a significant degree of similarity (32 to 39% identity) to polypeptides from Streptomyces and other actinomycetes, which are annotated in the databases as “probable hydrolases” or “hypothetical proteins.” No obvious similarities were detected between CorD1 and any known protein sequence of archaeal or eukaryotic origin. Next to corD1, we found the 3′ portion of a divergently transcribed ORF, corD2. Its product shows significant similarity (26 to 35% identity) along most of its sequence with hypothetical proteins from M. tuberculosis, Streptomyces spp., and archaea, such as Aeropyrum, Pyrococcus, and Sulfolobus.
Thus, the choE gene is encompassed by two ORFs, which are transcribed in the same orientation (Fig. 1). In Streptomyces spp. strain SA-COO, choA and the upstream gene, choP, which encodes a cytochrome P-450-like protein, are cotranscribed (18). To determine whether choE is expressed as part of an operon with corU1 and/or corD1, total RNA from R. equi strains 103− and MAD was subjected to Northern blot analysis using the entire choE gene as a probe (the CoEN-CoEC PCR product). In both strains, a single ≈1.9-kb transcript was detected (Fig. 1). This is consistent with the expected size of the choE message, indicating that choE is transcribed monocistronically.
Construction of choE mutants.
An important factor in the development of a mutagenesis system is the availability of a convenient marker for positive selection of recombinational events. The following antibiotic resistance markers that had been used for selection in rhodococci and related bacteria were tested with R. equi: (i) the spectinomycin resistance gene, aadA2, from the Corynebacterium glutamicum plasmid pCG4 (22); (ii) the kanamycin resistance gene from Tn903 (36) present in the E. coli-R. equi shuttle vector pRE7 (55); (iii) the apramycin resistance gene, aacC4, from the Salmonella enterica serovar Typhimurium plasmid Inc L/M (50), present in the E. coli-Mycobacterium shuttle vector pPE207 (37); and (iv) the chloramphenicol resistance gene from R. erythropolis plasmid pDA71 (41). Spectinomycin produced a high percentage of spontaneous resistant mutants in 103 and other R. equi strains and was discarded. In our hands, kanamycin, which was previously reported to be useful for plasmid selection in R. equi (55), also produced spontaneous mutants, although at a lower frequency than spectinomycin. Apramycin and chloramphenicol at concentrations of 30 and 100 μg/ml, respectively, gave no spontaneous resistant mutants, and the corresponding selection markers were further investigated. pRHE6A was constructed by cloning a NotI fragment containing the aacC4 gene from pPE207 into the unique NotI site of pRE-7 (Table 1). When electroporated into R. equi 103− and 103+, apramycin-resistant transformants (Aprr) appeared at a frequency of 1 × 106 and 1 × 105 per μg of DNA, respectively, which was suitable for the development of a mutagenesis tool based on a suicide vector. The chloramphenicol resistance gene from plasmid pDA71 was similarly tested but could not be expressed in R. equi.
The identified choE locus was used as a target for mutagenesis by allelic exchange in R. equi. A pair of suicide plasmids, pRHE2 and pRHE3, without and with choE target sequences, respectively, was constructed for this purpose (Table 1 and Fig. 3). For pRHE2, a 1.6-kb PstI fragment from pPE207 containing the aacC4 apramycin resistance gene was inserted into the corresponding restriction site of pUC19. For pRHE3, the aacC4 gene was excised from pRHE2 and inserted into the unique BamHI site of pRHE1, thus generating a recombinogenic cassette comprising the aacC4 gene flanked by the 5′ and 3′ regions of choE (Fig. 3). The only origin of replication present in pRHE2/3 was that from pUC19, which is nonfunctional in R. equi. Thus, any Aprr colonies arising after electroporation of these plasmids into R. equi should result from recombination of the plasmid with the genome of the host bacterium. Strains 103− and 103+ were electroporated with 200 ng of each of the plasmids and plated on solid medium containing apramycin. No recombinants were detected with pRHE2, but a large number of recombinants was obtained with pRHE3. These data were compatible with the Aprr colonies resulting from site-specific chromosomal integration of the suicide plasmid via homologous recombination between choE sequences.
FIG. 3.
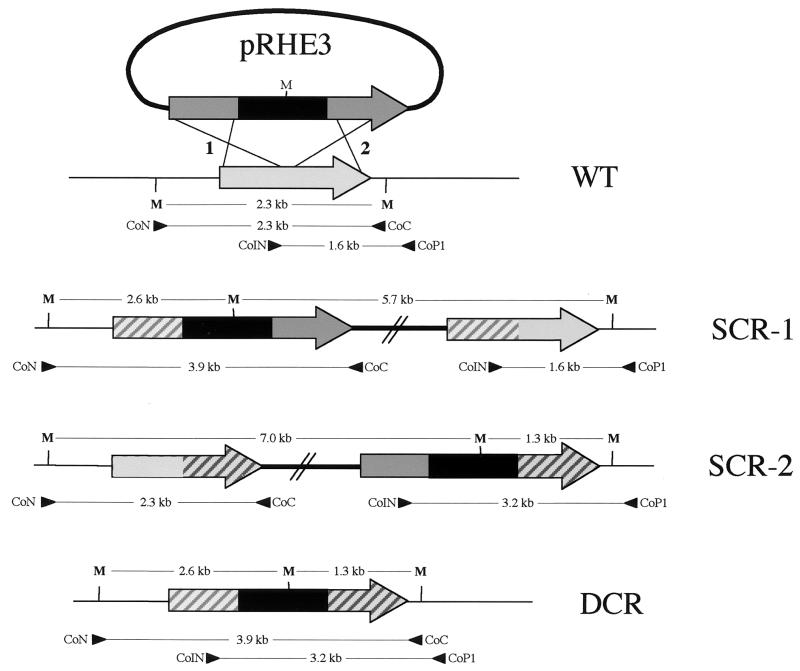
Schematic diagram of the procedure for targeted mutagenesis of choE by homologous recombination and a physical map of the choE locus in the parent strain (WT) and the recombinants (SCR-1 and SCR-2, single crossover recombinants in sites 1 and 2, respectively, and DCR, double crossover recombinant). The recombinogenic cassette in the suicide plasmid, pRHE3, includes the aacC4 apramycin resistance gene, shown in black, surrounded by the flanking choE target sequences, shown in dark gray. The wild-type choE allele is light gray. The crossover target sequences in each type of recombinant are dashed in light and dark gray. CoN-CoC and CoIN-CoP1 primer pairs were used for PCR mapping, and M indicates the position of the MluI restriction sites used in Southern blot analysis of the recombinants (Fig. 4).
The use of ssDNA increased the number of Aprr recombinants six-fold in both host strains. The number of recombinants per microgram of DNA was 10-fold higher in strain 103− than in 103+, consistent with the different transformation frequencies observed with the bifunctional plasmid pRHE6A (see above). Assuming that the suicide plasmids are taken up by R. equi at a frequency similar to that of pRHE6A, the recombination efficiencies would be between 1 × 10−2 and 1 × 10−3 for pRHE3 and between <1 × 10−5 and <1 × 10−6 for pRHE2 (Table 3).
TABLE 3.
Recombination frequencies of the suicide plasmid pRHE3 (containing choE target sequences) or pRHE2 (no choE target sequences) in R. equi
| Suicide plasmid | Recombination frequencya in strain:
|
|
|---|---|---|
| 103+ | 103− | |
| pRHE3 | 1.9 × 10−3 | 7.5 × 10−3 |
| pRHE3ss | 11.5 × 10−3 | 47.5 × 10−3 |
| pRHE2 | <10−5 | <10−6 |
| pRHE2ss | <10−5 | <10−6 |
Recombination frequencies were calculated as the ratio between transformation frequencies (expressed as Aprr colonies per microgram of DNA) obtained with the suicide plasmids and with the pRHE6A vector, which replicates in R. equi.
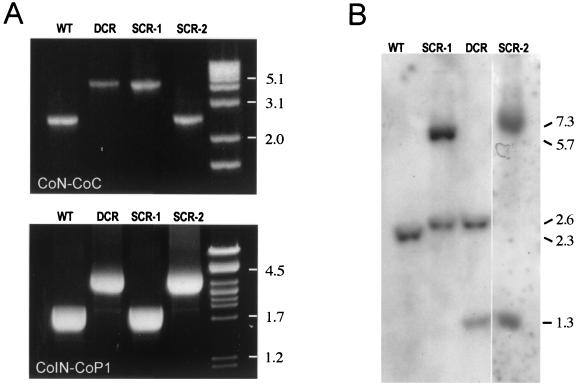
To confirm that homologous recombination events were at the origin of the Aprr phenotype in the pRHE3-transformed R. equi, the structure of the choE region was analyzed by PCR mapping and Southern blotting in 20 recombinants from each of the R. equi strains, 103− and 103+. For PCR mapping, primers CoN (5′-ACGCGTCCCGTCAGGCTCCGCT-3′), CoC (5′-ACGCGTGAAGAAAACGTGGTCG-3′), CoIN (5′-GTCAACAACATCGACCAGGCG-3′), and CoP1 (5′-GACGCTGAATACCGCGTCCT-3′) were used. CoN and CoP1 target sequences were absent from the recombinogenic plasmid (Fig. 3). PCR amplifications with primers CoN and CoC, which normally produce a 2.3-kb product in wild-type R. equi, generated a 3.9-kb DNA fragment in three of the Apr recombinants analyzed from strain 103− and two from the 103+ recombinants (Fig. 3 and 4). Similarly, a 1.6-kb increase in size with respect to the parent strain was also observed when PCR was performed in these recombinants with the primers CoIN and CoP1. This is the expected size for a choE mutant allele resulting from the integration of the aacC4 cassette by double-crossover recombination (DCR) in the choE chromosomal locus (Fig. 3 and 4). The occurrence of a double recombination event at the choE locus was verified by sequencing the junctions between choE and the aacC4 cassette from two DCR mutants, RH3-19 (from 103−) and RH3-15+ (from 103+). The other 35 Aprr transformants were single-crossover recombinants (SCRs), resulting from the integration of pRHE3 either via the upstream (SCR-1) or the downstream (SCR-2) choE target sequences. Of these SCRs, 33 were of the SCR-1 type, characterized by a larger CoN-CoC product of 3.9 kb (1.6-kb increase in size) and a wild-type CoIN-CoP1 product of 1.6 kb, with no PCR product being generated with primers CoN and CoP1 (Fig. 3 and 4). Only two recombinants (both from strain 103+) were of the SCR-2 type, characterized by a CoN-CoC product of wild-type size (2.3 kb) and a larger CoIN-CoP1 product of 3.2 kb (1.6-kb increase), with no product with CoN and CoP1 (Fig. 3 and 4). Southern blot analyses of the insertion mutants were entirely consistent with the results of PCR mapping (Fig. 3 and 4).
FIG. 4.
PCR mapping and Southern blot analysis of choE recombinants. (A) PCR mapping with CoN-CoC (upper panel) and CoIN-CoP1 (lower panel) primer pairs. See text and Fig. 3 legend for details. (B) Southern blot analysis. Genomic DNA from representative mutants of each recombinational type were cut with MluI and hybridized against the CoEN-CoEC probe covering the entire choE gene. Hybridization patterns of R. equi (WT), DCRs, and the two types of SCRs are shown. In R. equi, choE is flanked by two MluI sites separated by 2.3 kb; thus, the WT displays a single 2.3-kb hybridization band. pRHE3 has a single MluI site located at the middle of the aacC4 gene (Fig. 3); therefore, DCR shows two bands of 2.6 and 1.3 kb. In SCR-1, there were two bands of 2.6 and 5.7 kb. The 2.6-kb band is equivalent to the band of the same size observed in DCR and corresponds to the left side of the aacC4-disrupted choE allele; the 5.7-kb band includes the right side of the aacC4-disrupted choE allele, pUC19, and the intact copy of choE (Fig. 3). In SCR-2, two hybridization bands of 7.0 and 1.3 kb were observed (Fig. 3). When membranes were rehybridized with a pUC19 probe, hybridization signals were detected in SCR-1 and SCR-2, confirming the presence of the complete pRHE3 plasmid integrated at the R. equi choE locus as the result of an SCR event (data not shown).
Stability and phenotypic characterization of choE mutants.
To assess the stability of the choE insertion mutants, RHE3-19 and RHE3-15+ were grown for approximately 100 generations in the absence of selective pressure and plated out on antibiotic-free medium. One hundred colonies from each strain were picked onto agar plates containing apramycin. All replica colonies grew in the presence of the antibiotic, indicating that the aacC4 cassette was stably inserted into the R. equi chromosome.
Although R. equi is nonhemolytic on sheep blood agar, it develops a strong patch of synergistic hemolysis if streaked in the vicinity of sphingomyelinase C-producing bacteria, such as L. ivanovii or Staphylococcus aureus (42). Cholesterol oxidase is thought to be the R. equi factor responsible for this cooperative, CAMP-like lytic reaction (9, 26, 42). The five DCRs, in which only a disrupted choE allele is present on the chromosome, were negative in a CAMP-like hemolysis test with L. ivanovii (Fig. 5). This correlated with a loss of cholesterol oxidase activity in the culture supernatant of these mutants (Table 2). All of the SCR mutants, which bear an intact copy of choE in addition to the choE::aacC4 allele (Fig. 3), tested positive in the CAMP-like reaction and produced cholesterol oxidase activity at wild-type levels (data not shown). These results were consistent with the involvement of cholesterol oxidase in the cooperative hemolysis shown by R. equi with L. ivanovii.
The choE mutation had no effects on colony morphology, bacterial growth rate, or biochemical profiles.
Complementation of choE mutants.
To confirm directly that the disruption of choE was responsible for the observed loss of CAMP-like reactivity, we complemented the DCR mutants with choE. A DNA fragment containing choE plus its putative promoter region was inserted into pVK173-T, a pAL5000 derivative, giving rise to pRHE5 (Table 1). We previously verified that pVK173-T replicates and can be selected via its hygromicin resistance marker in R. equi. Introduction of pRHE5 restored cholesterol oxidase activity (Table 2) and CAMP-like reactivity with L. ivanovii (Fig. 5) in both RH3-19 and RH3-15+. These results were entirely consistent with those obtained by complementation of E. coli with choE (see above) and confirmed that cholesterol oxidase is the R. equi membrane-damaging factor responsible for cooperative hemolysis with sphingomyelinase C-producing bacteria.
Finally, we assessed the stability of pRHE5 in R. equi by testing the number of bacteria that retained the hygromycin resistance (Hygr) phenotype after growth for 100 generations without selective pressure, as described above. The percentage of Hygr colonies decreased from 90% after 20 generations to 6% after 60 generations. These data show that the pAL5000-based mycobacterial replicon is largely unstable in the absence of selective pressure in R. equi.
DISCUSSION
We approached the molecular analysis of R. equi virulence by investigating cholesterol oxidase, a putative membrane-active virulence factor that is released into the culture supernatant by all isolates of this pathogenic actinomycete. We identified the gene encoding this enzyme by PCR using degenerate oligonucleotides designed from the known sequences of cholesterol oxidase genes previously identified in several high-G+C-content gram-positive bacteria. The R. equi cholesterol oxidase gene, choE, was almost identical (99.2% identity) to choB from B. sterolicum, and their corresponding protein products differed only in one residue. This high degree of sequence conservation is unusual for two different bacteria, especially if we take into consideration that the primary structures of the cholesterol oxidases from two isolates belonging to the same taxon (strains SA-COO and A19249 of Streptomyces spp.) are 20% divergent. The sequence identity between choE and choB may be explained by a recent horizontal gene transfer event between R. equi and B. sterolicum. Another possibility is that ATCC 21387, the only known isolate of B. sterolicum (a species which, in addition, is not officially recognized as belonging to the genus Brevibacterium [21]), was missclassified and actually belongs to R. equi. Morphological, physiological, and genetic analyses on B. sterolicum ATCC 21387 suggest that this explanation may be correct (our unpublished observations).
The 552-amino-acid-long ChoE protein has a putative signal peptide of 45 residues and is similar (55 to 57% identity) to the above-mentioned cholesterol oxidases from the Streptomyces spp. strains SA-COO (19) and A19249 (5). These Streptomyces cholesterol oxidases were originally designated ChoA and ChoM (respectively) but now both are found in the protein databases with the name ChoD. ChoE is also related, although more distantly (24 to 27% identity), to a group of putative polypeptides found in mycobacteria and in S. coelicolor (Fig. 2). These polypeptides are also designated ChoD in the protein databases and are assumed to be cholesterol oxidases due to their structural similarities with known cholesterol oxidase enzymes. However, unlike the cholesterol oxidases from R. equi and Streptomyces spp. strains SA-COO and A19249, the mycobacterial and S. coelicolor ChoD proteins lack a signal sequence (Fig. 2), suggesting a cytoplasmic localization. This is also the case for the hypothetical proteins Y4NJ from Rhizobium sp. strain NGR234 (EMBL accession no. P55582) and Rv0492c from M. tuberculosis strain H37RV, classified as putative glucose-methanol-choline (GMC)-type oxidoreductases and which also exhibit significant similarity with ChoE (21 and 25% identity in a substantial sequence overlap, respectively). Production of cholesterol oxidase has been reported in Mycobacterium spp. (44), and membrane-bound or cytoplasmic forms of this enzymatic activity have been described in actinomycetes (20, 29). It remains to be determined whether the more distant ChoD polypeptides from Mycobacterium and S. coelicolor are indeed cytoplasmic cholesterol oxidases or enzymes without this activity but which belong to a group of oxidoreductases that share general structural features with cholesterol oxidases.
Targeted gene disruption via homologous recombination provided experimental evidence that choE encodes a cholesterol oxidase. Although there have been some previous attempts to use this genetic strategy in rhodococci, as for example in the plant pathogen Rhodococcus fascians (6) and in various unclassified strains of biodegradative Rhodococcus spp. (e.g., RHA1 and M5) (31, 53), this kind of mutagenesis procedure was not developed for R. equi. The previously described mutagenesis procedures for non-R. equi rhodococci used as selection markers the chloramphenicol resistance gene from R. erythropolis or the kanamycin resistance gene from Tn903 (24). However, in our hands these markers were unsuitable for R. equi due either to the absence of expression (chloramphenicol) or the appearance of an excessively high rate of spontaneous resistance mutations (kanamycin).
We identified apramycin and the apramycin resistance gene, aacC4, from the Salmonella plasmid IncL/M (50) as a clean selection system for use in R. equi. This marker was previously found to be useful in mycobacteria (37). With this selection system, we constructed a suicide recombinogenic plasmid, pRHE3, which allowed us to produce chromosomal choE insertion mutants by allelic exchange following electroporation in R. equi. Specific recombination at the choE locus was confirmed by PCR mapping, Southern blotting, and DNA sequencing. These recombination events were 10-fold more frequently detected in R. equi 103− than in 103+, possibly due to the higher transformation efficiency of the former strain. The reason for this higher transformability of 103− is presently unknown. As in other bacteria, such as Streptomyces (34) and Mycobacterium (15), the frequency of homologous recombination increased if ssDNA was used for transformation. This suggests that homologous recombination in R. equi proceeds through the general recombinational pathway described for E. coli, which involves a ssDNA intermediate (27). The use of ssDNA may be useful to enhance the mutational efficiency in R. equi strains more refractory to genetic manipulation than strain 103.
The choE gene was targeted in all of the Aprr colonies analyzed, and no Aprr transformants were obtained if the suicide plasmid did not contain choE target sequences. This indicates that homologous recombination works perfectly, and it suggests that the frequency of illegitimate rearrangement is negligible in R. equi. This is in contrast with the situation described for other rhodococci, such as the biodegradative Rhodococcus globerulus, in which most of the recombinants obtained resulted from nonhomologous recombination (24). The frequency of DCR we obtained (12% of the Aprr population) was in the range of that reported in mycobacteria using a similar mutational strategy (38). SCR-1-type recombinants may have been obtained more frequently than SCR-2-type recombinants because the choE target sequence upstream from the aacC4 cassette in pRHE3 (1 kb) was significantly longer than that situated downstream (0.6 kb).
Direct evidence that choE encodes a cholesterol oxidase was obtained by trans-complementation in R. equi choE mutants as well as in E. coli. A shuttle vector, pRE-7, containing the origin of replication of the 80-kb virulence plasmid of R. equi and the E. coli CoE1 ori from pBluescript, was available (55). Selection with this plasmid was based on the aph kanamycin resistance gene of pACYC177, originally from Tn903, which was shown by us to be not suitable as it gives rise to spontaneous resistant mutants at a relatively high frequency. Therefore, we tried another shuttle vector, pVK173-T, containing an origin of replication derived from the mycobacterial plasmid pAL5000 and a hygromycin resistance gene (10, 37). The choE-inserted pVK173-T derivative (pRHE5) was able to replicate in R. equi and complemented the choE mutation. However, in contrast to pRE-7, which has been reported to be perfectly stable in R. equi (55), the pAL5000-derived mycobacterial replicon was unstable in R. equi in the absence of selective pressure.
In this study we have, finally, demonstrated that cholesterol oxidase is a major membrane-damaging factor of R. equi. Inactivation of choE abrogated the rhodococcal cohemolytic activity, which was restored upon complementation with choE. In addition, the expression of choE in E. coli K-12 conferred to this nonhemolytic bacterium the same membrane-damaging activity as that of wild-type R. equi. This provides support to the current belief that cholesterol oxidase is a major cytotoxic factor possibly involved in macrophage and leukocyte destruction and in the generation of the pyonecrotic lesions that characterize R. equi infection in humans and animals (26, 28, 40). The membrane-damaging activity of R. equi is observed in vitro on sheep blood agar as a cooperative, CAMP-like hemolytic reaction in the presence of bacterial sphingomyelinase C (12, 42). This requirement for a concomitant exposure of erythrocytes to a sphingomyelinase indicates that the cholesterol oxidase substrate is not directly accessible to the enzyme in intact membranes. Sphingomyelin is exposed on the outer lipid leaflet and its degradation by a sphingomyelinase leads to sublytic damage of the membrane, allowing cholesterol oxidase to reach its target substrate (9). The 3-hydroxyl of cholesterol, thought to mediate sterol-phospholipid interaction, is thus oxidized, leading to the formation of cholest-4-en-3-one and the total disorganization of the membrane (25). The relevance in pathogenesis of this dependency of cholesterol oxidase on another membrane-damaging enzyme for the exertion of a cytolytic effect remains to be determined. In pathogenic bacteria there are precedents for virulence factors that act cooperatively to cause membrane damage, as described for Listeria hemolysin and phospholipases (51). There is evidence that R. equi produces also a phospholipase activity (3), and it is possible that during infection this phospholipase forms part of a bipartite cytolytic system together with ChoE to efficiently alter the host cell membranes and cause cytotoxicity and tissue destruction. The choE isogenic mutants and the choE-complemented bacteria described here will be instrumental in determining the role of ChoE in R. equi virulence.
ACKNOWLEDGMENTS
We thank J. E. Davies for the gift of the pVK173-T and pPE207, J. F. Prescott for supplying the R. equi strains 103− and 103+ and the pRE7, R. Merchante for the Northern blot analysis, and S. Zunzunegui for excellent technical assistance.
The Departamento de Biología Molecular, Facultad de Medicina of the Universidad de Cantabria is a research unit associated with the Centro de Investigaciones Biológicas (CSIC). This study was supported by the DGICYT, Spanish Ministry for Education and Science (grants PB94-0330-C03 and PB97-0327-C03), the Fundación Marqués de Valdecilla, Santander (grants 19/99 and A22/00), and the Dirección General de Investigación of the Madrid Regional Government (grants 08.2/0029/1997 and 08.2/0011/1999.2).
REFERENCES
- 1.Andersen S J, Quan S, Gowan B, Dabbs E R. Monooxygenase-like sequence of a Rhodococcus equi gene conferring increased resistance to rifampin by inactivating this antibiotic. Antimicrob Agents Chemother. 1997;41:218–221. doi: 10.1128/aac.41.1.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moor D D, Seidman J G, Smith J A. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 3.Bernheimer A W, Linder R, Avigad L S. Stepwise degradation of membrane sphingomyelin by corynebacterial phospholipases. Infect Immun. 1980;29:123–131. doi: 10.1128/iai.29.1.123-131.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantor G H, Byrne B A, Hines S A, Richards H M., III VapA-negative Rhodococcus equi in a dog with necrotizing pyogranulomatous hepatitis, osteomyelitis, and myositis. J Vet Diagn Investig. 1998;10:297–300. doi: 10.1177/104063879801000316. [DOI] [PubMed] [Google Scholar]
- 5.Corbin D R, Greenplate J T, Wong E Y, Purcell J P. Cloning of an insecticidal cholesterol oxidase gene and its expression in bacteria and in plant protoplasts. Appl Environ Microbiol. 1994;60:4239–4244. doi: 10.1128/aem.60.12.4239-4244.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crespi M, Messens E, Caplan A B, Van Montagu M, Desomer J. Fasciation induction by the fitopathogen Rhodococcus fascians depends upon a linear plasmid encoding a cytokinin synthase gene. EMBO J. 1992;11:795–804. doi: 10.1002/j.1460-2075.1992.tb05116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis W P, Steficek B A, Watson G L, Yamini B, Madarame H, Takai S, Render J A. Disseminated Rhodococcus equi infection in two goats. Vet Pathol. 1999;36:336–339. doi: 10.1354/vp.36-4-336. [DOI] [PubMed] [Google Scholar]
- 8.De La Peña-Moctezuma A, Prescott J F, Goodfellow M. Attempts to find phenotypic markers of the virulence plasmid of Rhodococcus equi. Can J Vet Res. 1996;60:29–33. [PMC free article] [PubMed] [Google Scholar]
- 9.Fehrenbach F J, Jürgens D. Cooperative membrane-active (lytic) processes. In: Alouf J E, Freer J H, editors. Sourcebook of bacterial protein toxins. London, United Kingdom: Academic Press; 1991. pp. 187–213. [Google Scholar]
- 10.Giguère S, Hondalus M K, Yager J A, Darrah P, Mosser D M, Prescott J F. Role of the 85-kilobase plasmid and plasmid-encoded virulence-associated protein A in intracellular survival and virulence of Rhodococcus equi. Infect Immun. 1999;67:3548–3557. doi: 10.1128/iai.67.7.3548-3557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giguère S, Prescott J F. Clinical manifestations, diagnosis, treatment, and prevention of Rhodococcus equi infections in foals. Vet Microbiol. 1997;56:313–334. doi: 10.1016/s0378-1135(97)00099-0. [DOI] [PubMed] [Google Scholar]
- 12.González-Zorn B, Domínguez-Bernal G, Suárez M, Ripio M T, Vega Y, Novella S, Vázquez-Boland J A. The smcL gene of Listeria ivanovii encodes a sphingomyelinase C that mediates bacterial escape from the phagocytic vacuole. Mol Microbiol. 1999;33:510–523. doi: 10.1046/j.1365-2958.1999.01486.x. [DOI] [PubMed] [Google Scholar]
- 13.Grzeszik C, Lubbers M, Reh M, Schlegel H G. Genes encoding the NAD-reducing hydrogenase of Rhodococcus opacus MR11. Microbiology. 1997;143:1271–1286. doi: 10.1099/00221287-143-4-1271. [DOI] [PubMed] [Google Scholar]
- 14.Haites R E, Muscatello G, Begg A P, Browning G F. Prevalence of the virulence-associated gene of Rhodococcus equi in isolates from infected foals. J Clin Microbiol. 1997;35:1642–1644. doi: 10.1128/jcm.35.6.1642-1644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinds J, Mahenthiralingam E, Kempsell K E, Duncan K, Stokes R W, Parish T, Stoker N G. Enhanced gene replacement in mycobacteria. Microbiology. 1999;145:519–527. doi: 10.1099/13500872-145-3-519. [DOI] [PubMed] [Google Scholar]
- 16.Hondalus M K, Mosser D M. Survival and replication of Rhodococcus equi in macrophages. Infect Immun. 1994;62:4167–4175. doi: 10.1128/iai.62.10.4167-4175.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hondalus M K. Pathogenesis and virulence of Rhodococcus equi. Vet Microbiol. 1997;16:257–268. doi: 10.1016/s0378-1135(97)00094-1. [DOI] [PubMed] [Google Scholar]
- 18.Horii M, Ishizaki T, Paik S Y, Manome T, Murooka Y. An operon containing the genes for cholesterol oxidase and a cytochrome P-450-like protein from a Streptomyces sp. J Bacteriol. 1990;172:3644–3653. doi: 10.1128/jb.172.7.3644-3653.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishizaki T, Hirayama N, Shinkawa H, Nimi O, Murooka Y. Nucleotide sequence of the gene for cholesterol oxidase from a Streptomyces sp. J Bacteriol. 1989;171:596–601. doi: 10.1128/jb.171.1.596-601.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson T, Somkuti G A. Isolation of cholesterol oxidases from Rhodococcus equi ATCC 33706. Biotechnol Appl Biochem. 1991;13:196–204. [PubMed] [Google Scholar]
- 21.Jones D, Keddie R M. Genus Brevibacterium. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 2. Baltimore, Md: Williams & Wilkins; 1986. pp. 1301–1313. [Google Scholar]
- 22.Katsumata R, Ozaki A, Oka T, Furuya A. Protoplast transformation of glutamate-producing bacteria with plasmid DNA. J Bacteriol. 1984;159:306–311. doi: 10.1128/jb.159.1.306-311.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosono S, Maeda M, Fuji F, Arai H, Kudo T. Three of the seven bphC genes of Rhodococcus erythropolis TA421, isolated from a termite ecosystem, are located on an indigenous plasmid associated with biphenyl degradation. Appl Environ Microbiol. 1997;63:3282–3285. doi: 10.1128/aem.63.8.3282-3285.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin M J, De Mot R, Kulakov L A, Nagy I. Applied aspects of Rhodococcus genetics. Antonie Leeuwenhoek. 1998;74:133–153. doi: 10.1023/a:1001776500413. [DOI] [PubMed] [Google Scholar]
- 25.Linder R. Alteration of mammalian membranes by the cooperative and antagonistic actions of bacterial proteins. Biochim Biophys Acta. 1984;779:423–435. doi: 10.1016/0304-4157(84)90019-4. [DOI] [PubMed] [Google Scholar]
- 26.Linder R, Bernheimer A W. Oxidation of macrophage membrane cholesterol by intracellular Rhodococcus equi. Vet Microbiol. 1997;56:269–276. doi: 10.1016/s0378-1135(97)00095-3. [DOI] [PubMed] [Google Scholar]
- 27.Lloyd R G, Low K B. Homologous recombination. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: American Society for Microbiology; 1996. pp. 2236–2255. [Google Scholar]
- 28.Machang'u R S, Prescott J F. Purification and properties of cholesterol oxidase and choline phosphohydrolase from Rhodococcus equi. Can J Vet Res. 1991;55:332–340. [PMC free article] [PubMed] [Google Scholar]
- 29.MacLachlan J, Wotherspoon A T, Ansell R O, Brooks C J. Cholesterol oxidase: sources, physical properties and analytical applications. J Steroid Biochem Mol Biol. 2000;72:169–195. doi: 10.1016/s0960-0760(00)00044-3. [DOI] [PubMed] [Google Scholar]
- 30.Magnusson H. Spezifische Infektiöse Pneumonie beim Fohlen. Ein neuer Eitererreger beim Pferde. Arch Wiss Prakt Tierheilkd. 1923;50:22–38. [Google Scholar]
- 31.Masai E, Sugiyama K, Iwashita N, Shimizu S, Hauschild J E, Hatta T, Kimbara K, Yano K, Fukada M. The bphDEF meta-cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphABC genes in Rhodococcus sp. strain RHA1. Gene. 1997;187:141–149. doi: 10.1016/s0378-1119(96)00748-2. [DOI] [PubMed] [Google Scholar]
- 32.McNeil M M, Brown J M. The medically important aerobic actinomycetes: epidemiology and microbiology. Clin Microbiol Rev. 1994;7:357–417. doi: 10.1128/cmr.7.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicholson V M, Prescott J F. Restriction enzyme analysis of the virulence plasmids of VapA-positive Rhodococcus equi strains isolated from humans and horses. J Clin Microbiol. 1997;35:738–740. doi: 10.1128/jcm.35.3.738-740.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh S H, Chater K F. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J Bacteriol. 1997;179:122–127. doi: 10.1128/jb.179.1.122-127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohta T, Fujishiro K, Yamaguchi K, Tamura Y, Aisaka K, Uwajima T, Hasegawa M. Sequence of gene choB encoding cholesterol oxidase of Brevibacterium sterolicum: comparison with choA of Streptomyces sp. SA-COO. Gene. 1991;103:93–96. doi: 10.1016/0378-1119(91)90397-t. [DOI] [PubMed] [Google Scholar]
- 36.Oka A, Sugisaki H, Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981;147:217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- 37.Paget E, Davies J. Apramycin resistance as a selective marker for gene transfer in mycobacteria. J Bacteriol. 1996;178:6357–6360. doi: 10.1128/jb.178.21.6357-6360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pavelka M S, Jr, Jacobs W R., Jr Comparison of the construction of unmarked deletion mutations in Mycobacterium smegmatis, Mycobacterium bovis bacillus Calmette-Guerin, and Mycobacterium tuberculosis H37Rv by allelic exchange. J Bacteriol. 1999;181:4780–4789. doi: 10.1128/jb.181.16.4780-4789.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prescott J F, Nicholson V M, Patterson M C, Zandona Meleiro M C, Caterino de Araujo A, Yager J A, Holmes M A. Use of Rhodococcus equi virulence-associated protein for immunization of foals against R. equi pneumonia. Am J Vet Res. 1997;58:356–359. [PubMed] [Google Scholar]
- 40.Prescott J F. Rhodococcus equi: an animal and human pathogen. Clin Microbiol Rev. 1991;4:20–34. doi: 10.1128/cmr.4.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quan S, Dabbs E R. Nocardioform arsenic resistance plasmid characterization and improved Rhodococcus cloning vectors. Plasmid. 1993;29:74–79. doi: 10.1006/plas.1993.1010. [DOI] [PubMed] [Google Scholar]
- 42.Ripio M T, Geoffroy C, Domínguez G, Alouf J E, Vázquez-Boland J A. The sulphydryl-activated cytolysin and a sphingomyelinase C are the major membrane-damaging factors involved in cooperative (CAMP-like) haemolysis of Listeria spp. Res Microbiol. 1995;146:303–313. doi: 10.1016/0923-2508(96)81053-9. [DOI] [PubMed] [Google Scholar]
- 43.Sojo M, Bru R, López-Molina D, García-Carmona F, Argüelles J C. Cell-linked and extracellular cholesterol oxidase activities from Rhodococcus erythropolis. Isolation and physiological characterization. Appl Microbiol Biotechnol. 1997;47:583–589. doi: 10.1007/s002530050977. [DOI] [PubMed] [Google Scholar]
- 44.Stadtman T C, Cherkes A, Anfinsen C B. Studies on the microbiological degradation of cholesterol. J Biol Chem. 1954;206:511–523. [PubMed] [Google Scholar]
- 45.Takai S. Epidemiology of Rhodococcus equi infections: a review. Vet Microbiol. 1997;56:167–176. doi: 10.1016/s0378-1135(97)00085-0. [DOI] [PubMed] [Google Scholar]
- 46.Takai S, Hidaka D, Fujii M, Shindoh Y, Murata T, Nakanishi S, Sasaki Y, Tsubaki S, Kamada M. Serum antibody responses of foals to virulence-associated 15- to 17-kilodalton antigens of Rhodococcus equi. Vet Microbiol. 1996;52:63–67. doi: 10.1016/0378-1135(96)00042-9. [DOI] [PubMed] [Google Scholar]
- 47.Takai S, Hines S A, Sekizaki T, Nicholson V M, Alperin D A, Osaki M, Takamatsu D, Nakamura M, Suzuki K, Ogino N, Kakuda T, Dan H, Prescott J F. DNA sequence and comparison of virulence plasmids from Rhodococcus equi ATCC 33701 and 103. Infect Immun. 2000;68:6840–6847. doi: 10.1128/iai.68.12.6840-6847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takai S, Imai Y, Fukunaga N, Uchida Y, Kamisawa K, Sasaki Y, Tsubaki S, Sekizaki T. Identification of virulence-associated antigens and plasmids in Rhodococcus equi from patients with AIDS. J Infect Dis. 1995;172:1306–1311. doi: 10.1093/infdis/172.5.1306. [DOI] [PubMed] [Google Scholar]
- 49.Takai S, Sasaki Y, Ikeda T, Uchida Y, Tsubaki S, Sekizaki T. Virulence of Rhodococcus equi isolates from patients with and without AIDS. J Clin Microbiol. 1994;32:457–460. doi: 10.1128/jcm.32.2.457-460.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tosini F, Visca P, Luzzi I, Dionisi A M, Pezzella C, Petrucca A, Carattoli A. Class 1 integron-borne multiple-antibiotic resistance carried by IncFI and IncL/M plasmids in Salmonella enterica serotype Typhimurium. Antimicrob Agents Chemother. 1998;42:3053–3058. doi: 10.1128/aac.42.12.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vázquez-Boland J A, Kuhn M, Berche P, Chakraborty T, Domínguez-Bernal G, Goebel W, González-Zorn B, Wehland J, Kreft J. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vullo V, Mastroianni C M, Lichtner M, Mengoni F, Chiappini E, D'Agostino C, Delia S. Serologic responses to Rhodococcus equi in individuals with and without human immunodeficiency virus infection. Eur J Clin Microbiol Infect Dis. 1996;15:588–594. doi: 10.1007/BF01709368. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Garnon J, Labbé D, Bergeron H, Lau P C K. Sequence and expression of the bpdC1C2BADE genes involved in the initial steps of the biphenyl/chlorobiphenyl degradation by Rhodococcus sp. M5. Gene. 1995;164:117–122. doi: 10.1016/0378-1119(95)00448-f. [DOI] [PubMed] [Google Scholar]
- 54.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 55.Zheng H, Tkachuk-Saad O, Prescott J F. Development of a Rhodococcus equi-Escherichia coli plasmid shuttle vector. Plasmid. 1997;38:180–187. doi: 10.1006/plas.1997.1311. [DOI] [PubMed] [Google Scholar]