Abstract
The presence of excess glucose in growth media prevents normal sporulation of Bacillus subtilis. The crsA47 mutation, located in the gene for the vegetative phase sigma factor (ςA) results in a glucose-resistant sporulation phenotype. As part of a study of the mechanisms whereby the mutation in ςA overcomes glucose repression of sporulation, we examined the expression of genes involved in sporulation initiation in the crsA47 background. The crsA47 mutation had a significant impact on a variety of genes. Changes to stage II gene expression could be linked to alterations in the expression of the sinI and sinR genes. In addition, there was a dramatic increase in the expression of genes dependent on the minor sigma factor ςH. This latter change was paralleled by the pattern of spo0H gene transcription in cells with the crsA47 mutation. In vitro analysis of RNA polymerase containing ςA47 indicated that it did not have unusually high affinity for the spo0H gene promoter. The in vivo pattern of spo0H expression is not predicted by the known regulatory constraints on spo0H and suggests novel regulation mechanisms that are revealed in the crsA47 background.
The activation of genes that are induced late in the growth cycle of Bacillus subtilis involves a network of interacting regulatory pathways. These pathways control the cellular response to conditions that include nutritional stress and high cell density (reviewed in references 13, 19, 24, 36, and 47). Under the appropriate environmental stimuli, B. subtilis will differentiate to form dormant endospores. The key factor in initiating sporulation is the accumulation and phosphorylation of the response regulator and transcription factor Spo0A (reviewed in references 13, 19, and 24). Phosphorylation of Spo0A takes place through a multicomponent pathway (the phosphorelay) (3) that appears to integrate multiple signals that act positively or negatively to regulate sporulation (24). Once a sufficient level of phosphorylated Spo0A (Spo0A∼P) is reached, a complex series of feedback loops will drive differentiation forward (19, 24, 36).
One critical component of sporulation is the minor sigma factor, ςH, encoded by a gene originally found as a stage zero sporulation mutant, spo0H (12, 20, 46). ςH is required for the transcription of a variety of sporulation genes, including spo0A, spo0F, and kinA (20, 41). The activity of ςH, which increases as cells enter stationary phase, is under complex, still-undefined regulatory controls (19, 45, 46). Transcription of the spo0H gene is repressed by the transition state regulator AbrB (11, 48, 50), and so the induction of spo0H seen at the transition between log growth and sporulation is influenced by Spo0A∼P repression of the abrB gene (4, 11). Transcriptional and translational regulation of spo0H by the presence of nutrients has also been reported (8, 14).
A variety of experiments have shown that the increase in ςH activity as measured by transcription of ςH-dependent genes does not match the accumulation of the ςH protein, implying the existence of posttranslational controls (8, 16, 17, 21, 23, 45, 50). Furthermore, evidence has been presented that external pH (8) and the activity of the tricarboxylic acid cycle (26) affect ςH activity and that ςH protein levels are affected by a Lon-type protease during the onset of sporulation and during stress responses (30). Recently, ClpX has been implicated in ςH-dependent transcription activity. In vivo and in vitro experiments have suggested that ClpX may interact directly with RNA polymerase containing ςH and stimulate transcription (31, 37). The level of ςH-dependent transcription increases for 1 to 2 h after sporulation initiation and then declines (4, 11, 46). The decline of activity has been linked to another Clp protein, ClpP, and the decline is associated with the loss of ςH protein (38).
Excess glucose in the growth medium represses sporulation, and some of the effects on sporulation have been linked to specific repression of ςH activity (1, 7, 8, 16, 17, 50, 52). The crsA47 mutation renders sporulation resistant to repression by glucose (49). The molecular basis for this phenotype is unknown, but the crsA47 mutation is located within the gene for the ςA subunit of RNA polymerase (sigA or rpoD) (27).
In this report, we describe experiments that suggest that the crsA47 mutation in ςA affects the expression of ςH-dependent genes through its effect on the expression of spo0H. The effects include overexpression of the spo0H gene during the onset of sporulation and unusual extended expression past the time of normal shutoff during differentiation. As the pattern cannot be explained solely by increased promoter affinity, these effects suggest novel regulation of the spo0H gene.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains used in this study are shown in Table 1. The promoter-lacZ fusions used were created using a B. subtilis amyE integrative vector, with the exception of the kinA-lacZ fusions that were inserted in the kinA gene. Promoter-lacZ fusions provided in the JH642 background were transferred into GBS10 (containing the crsA47 mutation in the rpoD gene but otherwise isogenic to JH642) by transformation with chromosomal DNA, and selection for both the antibiotic resistance and the amyE mutant phenotype was conferred with the acquisition of the construct. The plasmid pGS0H was created by the ligation of the EcoRI/BamHI-digested Vent polymerase (New England Biolabs, Inc.) PCR product generated from chromosomal DNA and the primer pair 5′-AAGGATCCTGTTTCTGGCGAGTAG-3′ and 5′-ACGAATTCGGCACGGACGTTAGAA-3′ that targets the spo0H gene into the EcoRI/BamHI sites of the B. subtilis integrative vector pDH32 (44). The pDH32-based clone was linearized with PstI prior to transformation into B. subtilis. Chloramphenicol-resistant transformants generated were confirmed to be amylase negative by using 1% starch agar plates prior to β-galactosidase analysis.
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype or descriptiona | Source or construction |
|---|---|---|
| JH642 | trpC2 phe-1 | J. Hoch |
| GLU-47 | crsA47 strA | Bacillus Genetic Stock Center |
| JH12862 | trpC2 phe-l amyE::(spo0F-lacZ Cmr) | M. Perego |
| JH12866 | trpC2 phe-l amyE::(rapB-lacZ Cmr) | M. Perego |
| JH12961 | trpC2 phe-l amyE::(rapA-lacZ Cmr) | M. Perego |
| JH16124 | trpC2 phe-l amyE::(spoIIA-lacZ Cmr) | M. Perego |
| JH12664 | trpC2 phe-l kinA::(1.7-kb kinA-lacZ Cmr) | M. Perego |
| JH12638 | trpC2 phe-l kinA W168::pJM8115 Cmr | M. Perego |
| JH16304 | trpC2 phe-l amyE::(spoIIG-lacZ Kmr) | M. Perego |
| IS688 | leuA8 metB5 hisA1 spoVG::(spoVG-lacZ Cmr) | I. Smith |
| IS875 | leuA8 metB5 hisA1 ΔsinR:: Cmr | I. Smith |
| IS423 | leuA8 metB5 hisA1 sinI::(pIS135 Cmr) | I. Smith |
| IS424 | leuA8 metB5 hisA1 sinR::(pIS142 Cmr) | I. Smith |
| GBS10 | crsA47 trpC2 phe-l | GLU-47DNA→JH642 |
| GBS101 | crsA47 trpC2 phe-1 amyE::(spoIIG-lacZ Kmr) | JH16304DNA→GBS10 |
| GBS103 | crsA47 trpC2 phe-1 amyE::(rapB-lacZ Kmr) | JH12866DNA→GBS10 |
| GBS104 | crsA47 trpC2 phe-1 amyE::(rapA-lacZ Kmr) | JH12961DNA→GBS10 |
| GBS105 | crsA47 trpC2 phe-1 amyE::(spo0F-lacZ Cmr) | JH12862DNA→GBS10 |
| GBS106 | crsA47 trpC2 phe-1 amyE::(spoIIA-lacZ Cmr) | JH16124DNA→GBS10 |
| GBS107 | crsA47 trpC2 phe-1 kinA::(1.7-kb kinA-lacZ Cmr) | JH12664DNA→GBS10 |
| GBS108 | crsA47 trpC2 phe-1 kinA::(pJM8115 Cmr) | JH12638DNA→GBS10 |
| GBS109 | crsA47 trpC2 phe-1 spoVG::(spoVG-lacZ Cmr) | IS688 DNA→GBS10 |
| GBS110 | trpC2 phe-1 spoVG::(spoVG-lacZ Cmr) | IS688 DNA→JH642 |
| GBS111 | crsA47 trpC2 phe-1 ΔsinR::Cmr | IS875 DNA→GBS10 |
| GBS112 | trpC2 phe-1 ΔsinR::Cmr | IS875 DNA→JH642 |
| GBS113 | crsA47 trpC2 phe-1 sinI::(pIS135 Cmr) | IS423 DNA→GBS10 |
| GBS114 | trpC2 phe-1 sinI::(pIS135 Cmr) | IS423 DNA→JH642 |
| GBS115 | crsA47 trpC2 phe-1 sinR::(pIS142 Cmr) | IS424 DNA→GBS10 |
| GBS116 | trpC2 phe-1 sinR::(pIS142 Cmr) | IS424 DNA→JH642 |
| GBS150 | crsA47 trpC2 phe-1 amyE::(spo0H-lacZ Cmr) | pGS0H→GBS10 |
| GBS151 | trpC2 phe-1 amyE::(spo0H-lacZ Cmr) | pGS0H→JH642 |
Km, kanamycin; Cm, chloramphenicol.
Bacterial transformation and growth conditions.
B. subtilis transformations were performed by the method of Hoch (24) with 1 to 2 μg of plasmid DNA or 20 to 100 ng of chromosomal DNA. Transformants were selected on Schaeffer sporulation agar plates supplemented with 5 μg of chloramphenicol or kanamycin/ml. Bacillus cultures used to determine sporulation frequency were grown in Schaeffer spore media (SSM), pH 7.5, supplemented with tryptophan and phenylalanine (each 10 μg/ml), chloramphenicol or kanamycin (5 μg/ml), and, when appropriate, SSM with 1% glucose (SSMG). Cells were grown for 22 to 24 h, serially diluted in SSM, and plated before and after treatment with 0.1 volume of chloroform to obtain a viable cell count and a spore count.
β-Galactosidase assay.
B. subtilis strains used for analysis of promoter-lacZ activity were inoculated into SSM, pH 7.5, containing 5 μg of the appropriate antibiotic/ml and supplemented with tryptophan and phenylalanine (each, 10 μg/ml) and, when appropriate, 0.2% glucose. Aliquots (0.5 ml) were removed hourly, the cells were collected by centrifugation, and cell pellets were frozen at −70°C until analyzed. β-Galactosidase assays and designation of time zero (T0) in sporulation were done as previously described (15). Enzyme specific activity was expressed in Miller units (35). Assays of a minimum of three independent cultures were performed, and one representative pattern for each strain is shown. Each point in the assays is an average of duplicate samples that differed by no more than 5% from the mean.
In vitro transcription assay.
The RNA polymerase preparations were isolated as described by Dobinson and Spiegelman (10) from logarithmic-stage cultures of JH642 and GBS10, except that the heparin-Sepharose column set was eliminated. Transcription assays used fractions from the glycerol gradient. Two DNA templates were used. The plasmid pUCA2trpA (6) that contains the bacteriophage φ29A2 promoter was treated with PvuII, and the 600-bp fragment containing the promoter was isolated by electrophoresis through agarose and extracted from the agarose with a GeneClean kit from Qiagen. The spo0H promoter was isolated by amplifying a DNA fragment using 20 pmol (each) of two specific primers described above, 800 ng of chromosomal DNA from JH642, and an amplification protocol as follows: preincubation of the template and primers at 95°C for 5 min, and after the addition of 2.5 U of Taq polymerase (Promega), 30 cycles of 95°C for 1 min, 61°C for 1 min (with descending annealing temperatures of 0.3°C per cycle), and 72°C for 1 min. The product was purified by electrophoresis through 1% agarose and extracted from the gel as described above for the A2 promoter. The extracted product was digested with HindIII to provide a fixed endpoint for the transcription assays and then precipitated. The concentrations of promoter fragments were determined by measuring the absorbance at 260 nM.
The in vitro transcription assay followed published protocols (6). In brief, reaction tubes containing 16 μl of reaction mixture with template, transcription buffer (6), ATP, and [α-32P]GTP (3 μCi/reaction mixture) were warmed to 37°C. The polymerase (2 μl of an appropriate dilution) was added. Two minutes later, 2 μl of a mixture of heparin (100 μg/ml, final concentration), UTP, and CTP was added to inactivate noninitiated RNA polymerase molecules and allow those that had initiated to elongate either to the terminator (in the case of the φ29A2 promoter) or to the end of the DNA fragment (in the case of the spo0H promoter). After 5 min, a stop buffer containing 7 M urea was added, and the reaction products were separated from free nucleotides on 5% polyacrylamide gels containing 7 M urea and 0.5× Tris-borate-EDTA (TBE). The gels were exposed to a Molecular Dynamics PhosphorImager screen, and the data were collected and analyzed with the ImageQuant 1.0 software on the instrument.
RESULTS
The crsA47 mutation results in overexpression of ςH-dependent spo0 genes in glucose-containing media.
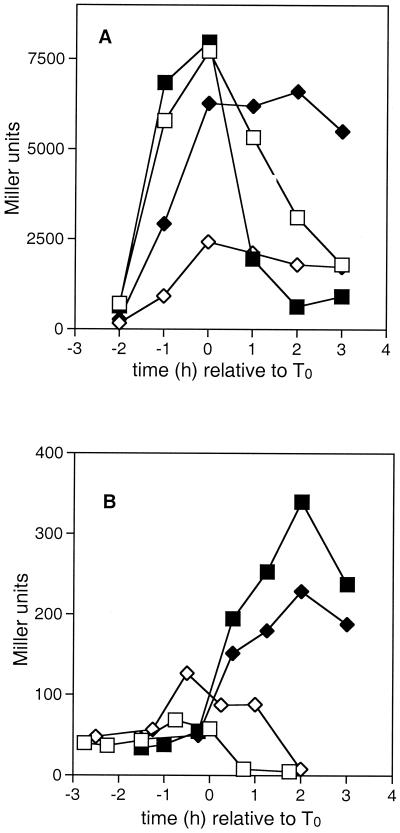
To investigate the mechanism by which the crsA47 mutation resulted in catabolite-resistant sporulation, we examined the expression of a variety of genes important in the sporulation initiation pathway in the presence and absence of glucose. Among these genes, the ones with ςH-dependent promoters showed very unusual profiles of activity. Figure 1 shows the expression of the kinA and spoVG promoter fusions in wild-type (JH642) and crsA47 (GBS10) backgrounds. The kinA promoter has not been previously shown to be subject to regulation other than that imposed by ςH activity (1, 41). The spoVG gene is preceded by a ςH promoter that is repressed during logarithmic growth by the transition state regulator AbrB (43, 48).
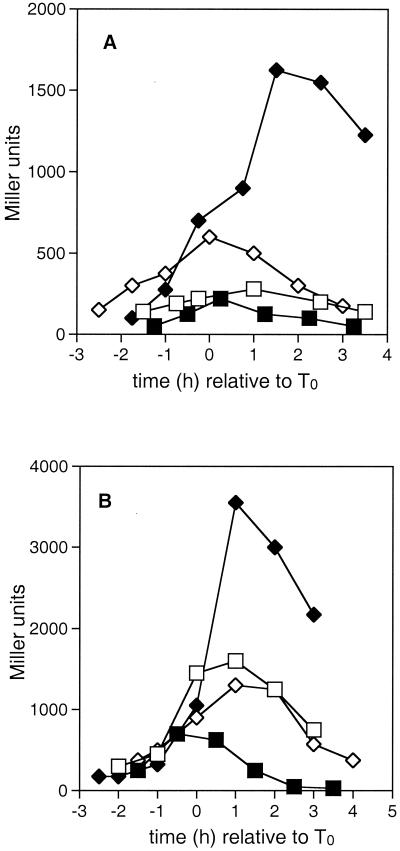
FIG. 1.
The effect of the crsA47 mutation on expression of kinA and spoVG. β-Galactosidase activities in strains carrying kinA-lacZ (A) or spo0VG-lacZ (B) fusions were measured as described in Materials and Methods. T0 represents the onset of sporulation. Strains contained the wild-type ςA gene (squares) or the crsA47 mutation (diamonds) and were grown in either SSM (open symbols) or SSMG (filled symbols). (A) JH12664 (kinA-lacZ) and GBS107 (crsA47 kinA-lacZ); (B) GBS110 (spoVG-lacZ) and GBS109 (crsA47 spoVG-lacZ).
Three observations were common to the activity of the kinA and spoVG promoters. First, in a wild-type ςA background promoter activity was depressed by the presence of glucose. Second, in a crsA47 background the expression was elevated by the presence of glucose compared to that seen in the wild type in the absence of glucose, with transcription levels persisting long after the point of maximum activity in the wild type. Third, promoter activity in a crsA47 background in the absence of glucose was only marginally affected compared to the effect in the wild type. The onset of promoter activity was not affected in the same way for the two promoters. kinA promoter activity (Fig. 1A) in GBS107 grown in the absence of glucose began earlier than in JH12664, whereas the timing of spoVG transcription activity (Fig. 1B) appeared similar in both GBS110 and GBS109. The logarithmic-phase repression of spoVG by AbrB did not differ between wild-type and crsA47 mutants. The suggestion that AbrB regulation was not altered in the GBS109 was supported by the expression of an abrB-lacZ fusion which showed similar patterns in GBS10 and JH642 (data not shown).
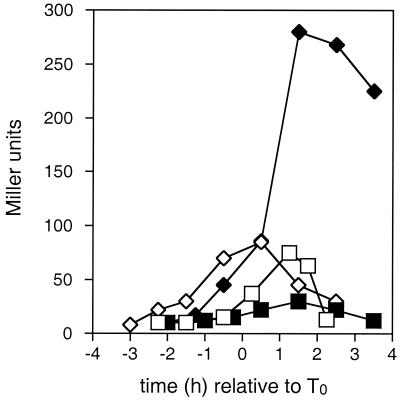
The expression patterns of spo0F promoter-lacZ fusions are shown in Fig. 2. The spo0F gene is preceded by dual ςA ςH promoters, with the ςH-dependent promoter requiring Spo0A∼P as a transcription activator (28, 41, 51). In JH12862, grown in SSM, expression from the spo0F promoter-lacZ fusion increased during late-exponential-phase growth, peaked at roughly T1, and decreased thereafter. The addition of glucose to the media resulted in a decrease in overall expression. In GBS105, the expression from spo0F began earlier and peaked at higher levels than were seen in JH12862, both with and without added glucose. As was seen with both the kinA and spoVG promoters (Fig. 1), transcription of spo0F in GBS105 in cells grown with added glucose appeared to be stimulated after T0 compared to levels seen in cells grown without glucose.
FIG. 2.
The effect of the crsA47 mutation on expression of spo0F. β-Galactosidase activities in strains carrying spo0F-lacZ fusions were measured as described in Materials and Methods. T0 represents the onset of sporulation. Strains contained the wild-type ςA gene (JH12862; squares) or the crsA47 mutation (GBS105; diamonds) and were grown in either SSM (open symbols) or SSMG (filled symbols).
The crsA47 mutation results in the expression of spoII genes despite the presence of glucose. The late- and post-exponential phase expression of ςH-dependent spo0 genes appeared to be elevated in the cells containing the crsA47 mutation (Fig. 1 and 2), suggesting that ςH was active in these cells despite the presence of glucose. The activation of ςH during the transition state has been shown to be critical for the initiation of sporulation and is required for the maximal expression of genes encoding phosphorelay proteins and accumulation of a sufficient level of Spo0A∼P to activate the transcription of stage II spo genes. As a test for whether or not Spo0A∼P was fully activated, we examined the expression of the stage II genes spoIIA and spoIIG (Fig. 3).
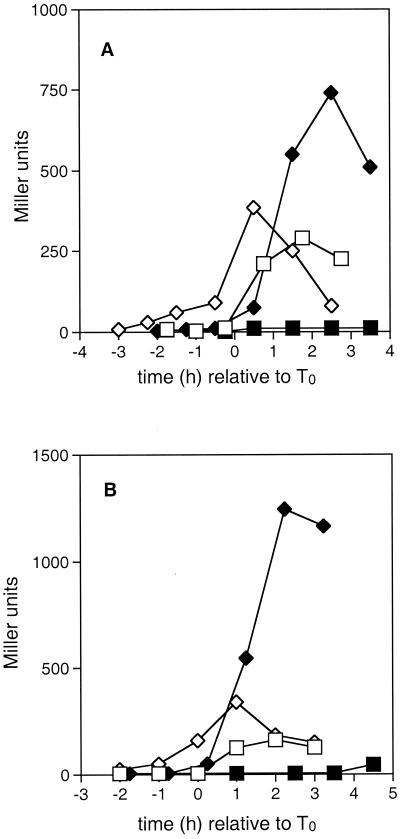
FIG. 3.
The effect of the crsA47 mutation on expression of spoIIA and spoIIG. β-Galactosidase activities in strains carrying spoIIA-lacZ (A) or spoIIG-lacZ (B) fusions were measured as described in Materials and Methods. T0 represents the onset of sporulation. Strains contained the wild-type ςA gene (squares) or the crsA47 mutation (diamonds), and were grown in either SSM (open symbols) or SSMG (filled symbols). (A) JH16124 (spoIIA-lacZ) and GBS106 (crsA47 spoIIA-lacZ); (B) JH16304 (spoIIG-lacZ) and GBS101 (crsA47 spoIIG-lacZ).
In strain JH16124 cells grown in SSM (Fig. 3A), the expression from the ςH-dependent spoIIA promoter-lacZ fusion began roughly at T0 and peaked at T2, dropping thereafter. In the presence of glucose, spoIIA-lacZ expression was depressed in stationary phase. In GBS106 grown in SSM (Fig. 3A), transcription activity from the spoIIA promoter began to increase at the same time as that observed in JH16124 and peaked at levels not substantially different from that in JH16124. In the presence of glucose, the expression of spoIIA-lacZ in GBS106 began immediately after the onset of stationary phase and rose to a level more than twice that seen in JH16124 grown in the absence of glucose.
Figure 3B depicts the expression from the ςA-dependent spoIIG promoter-lacZ fusion in the wild type (JH16304) and cells containing the crsA47 mutation (GBS101). As seen in Fig. 3A for the spoIIA-lacZ fusion, promoter activity from the spoIIG promoter was repressed by glucose in the wild type but not repressed in GBS101. Since effective expression of these two stage II operons directly requires high Spo0A∼P levels, these cells must contain high levels of Spo0A∼P. The spo0A gene is expressed from two promoters, one ςA dependent and one ςH dependent. It has been shown elsewhere that expression for the ςH-dependent promoter is enhanced in crsA47 mutants (9).
The presence of the crsA47 mutation results in an alteration in the pattern of transcription from sinI and sinR promoters in the presence of glucose.
SinR inhibits the expression of several spo genes, including spo0A (34), spoIIG, and spoIIA (5, 32, 33). The sinR gene is constitutively expressed from a ςA-dependent promoter throughout exponential and post-exponential growth of B. subtilis (18). SinR inhibition of transcription is negatively regulated by Spo0A∼P levels, which stimulate increased transcription of the ςH-dependent sinI gene (the gene directly upstream of sinR) (18). SinI sequesters SinR via protein-protein interaction preventing SinR-mediated repression of promoter activity (2, 29). Gaur et al. showed that the presence of excess glucose in the media inhibits the transcription of the sinI gene (18). Presumably, inadequate transcription of sinI results in a SinI/SinR protein ratio insufficient to fully sequester SinR and relieve repression of sporulation genes (2, 29).
The data in Fig. 3 showed that ςH- and ςA-dependent stage II promoters were deregulated in cells containing the crsA47 mutation grown in glucose. Because SinR is central to regulation of both the spoIIG and spoIIA operons, we examined the regulation of the sinR and sinI promoters. The data shown in Fig. 4 indicate that in cells containing wild-type ςA (GBS114) grown in the absence of glucose, sinI transcription levels increased throughout late logarithmic growth to peak at T0 (Fig. 4A). Transcription of sinR increased throughout late logarithmic growth and into stationary phase (GBS116) (Fig. 4B), presumably in part due to readthrough from the sinI promoter (18) as well as from the sinR promoter. The expression patterns in Fig. 4 are similar to those observed by others (18, 33). When glucose was added, sinI transcription remained relatively low during stationary phase in GBS114 (Fig. 4A), whereas sinR transcription in GBS116 (Fig. 4B) remained roughly the same as in the absence of glucose.
FIG. 4.
The effect of the crsA47 mutation on expression of sinI and sinR. β-Galactosidase activities in strains carrying sinI-lacZ (A) or sinR-lacZ (B) fusions were measured as described in Materials and Methods. T0 represents the onset of sporulation. Strains contained the wild-type ςA gene (squares) or the crsA47 mutation (diamonds) and were grown in either SSM (open symbols) or SSMG (filled symbols). (A) GBS114 (sinI-lacZ) and GBS113 (crsA47 sinI-lacZ); (B) GBS116 (sinR-lacZ) and GBS115 (crsA47 sinR-lacZ).
If transcription of the sinI and sinR genes reflects protein levels, then cells with wild-type ςA grown in the absence of glucose would contain a roughly 20-fold excess of SinI over SinR. Since these cells sporulate efficiently, this ratio should indicate the level of SinI needed to complex SinR between T0 and T1.5. In the presence of glucose, expression of the sinI promoter in cells with wild-type ςA was reduced, with the implication that the ratio of SinI to SinR would not block SinR repression of sporulation.
sinI promoter activity in cells containing the crsA47 mutation (GBS113) is also shown in Fig. 4A. In these cells, the activity from the sinI promoter in the absence of glucose rose slowly during logarithmic growth to peak at T0.5 at levels 25 to 30% of that seen in GBS114. The sinI-lacZ activity was also altered in GBS113 grown in the presence of glucose (Fig. 4A), with the observed pattern of transcription similar to that seen in other ςH-dependent promoters examined in the crsA47 background. Transcription from the sinI promoter rose from T−1.5 to peak after T2 at levels five times higher than was seen in cells with wild-type ςA.
Figure 4B shows sinR-lacZ activity in cells containing the crsA47 mutation (GBS115). Without excess glucose, transcription of the sinR gene was reduced from that seen in cells containing wild-type ςA (GBS116), peaking at roughly T0 and decreasing thereafter. In the presence of glucose, transcription rose sharply from T−2 to peak at T0 at levels similar to those achieved in GBS116 in the presence or absence of glucose.
Continuing the assumption that the activity of the sinI and sinR promoter fusions reflects protein levels, then even though SinR and SinI levels were reduced in cells containing the crsA47 mutation (in media without excess glucose), the ratio would be similar to that seen in wild-type cells. Thus, SinR activity would be blocked by SinI during early stationary phase. When glucose was added, the large induction of the sinI promoter would further reduce SinR activity. Thus, unlike cells with wild-type ςA where the addition of glucose decreased the ratio of expression of sinI/sinR, in the cells with the crsA47 mutation the ratio of sinI/sinR would increase with added glucose. This alteration in sin operon transcription in the crsA47 mutant could contribute to the ability of these cells to express the spoIIA and spoIIG operons in the presence of glucose.
The importance of SinR negative regulation in glucose repression of sporulation was examined by determining the sporulation efficiency of a ΔsinR mutant, as shown in Table 2. In the crsA47 mutant (GBS10), the sporulation efficiencies of cells grown in the presence and absence of glucose were comparable, clearly indicating a glucose-resistant sporulation phenotype. In JH642, the addition of excess glucose to the medium resulted in a 104-fold decrease in sporulation efficiency. However, in GBS112 (sigA+ ΔsinR) the sporulation efficiency in medium with excess glucose was only three-fold less than that observed in the absence of glucose. These results, combined with those shown in Fig. 2 through 4, suggest that a decrease in SinR repression whether by sinR gene deletion or by altering the ratios of sinI and sinR transcription increased expression of spoII genes and contributed to the glucose-resistant sporulation phenotype.
TABLE 2.
Sporulation efficiencies of sinl-lacZ-, sinR-lacZ-, and ΔsinR-containing strains
| Strain | Sporulation efficiencya
|
|
|---|---|---|
| SSMb | SSMGc | |
| JH642 | 6.5 × 10−1 | 1.0 × 10−5 |
| GBS114 | 7.1 × 10−1 | 7.9 × 10−6 |
| GBS116 | 6.2 × 10−1 | 9.3 × 10−6 |
| GBS112 | 7.4 × 10−1 | 2.5 × 10−1 |
| GBS10 | 5.0 × 10−1 | 1.0 |
| GBS113 | 8.9 × 10−1 | 9.8 × 10−1 |
| GBS115 | 9.1 × 10−1 | 9.3 × 10−1 |
Sporulation efficiency was calculated as the number of chloroform-resistant cells per total cells.
SSM, strain grown in Schaeffer's spore media, pH 7.5.
SSMG, strain grown in Schaeffer's spore media (pH 7.5) plus 1.0% glucose.
The crsA47 mutation results in increased transcription from the spo0H gene in late- and post-exponential-phase growth.
The results shown in Fig. 3 and 4 demonstrate increased expression of both ςA- and ςH-dependent stage II genes in the crsA47 background when the cells were grown in the presence of excess glucose. The implication from the analysis of the sinI-sinR operon was that the increase in stage II gene expression reflected the increase in ςH activity, which allowed SinI to inactivate SinR. Thus, overexpression of the spoIIG operon (ςA dependent) has the same fundamental mechanism as overexpression of the spoIIA operon (ςH dependent); that is, increased activity of ςH. However, the reason for the high levels of stationary-phase transcription from all of the ςH-dependent promoters examined in the crsA47 mutant in the presence of glucose was not clear. Previous studies have shown higher-than-normal ςH-dependent promoter activity during the stationary phase either by using an increased copy number of spo0H or by placing spo0H under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible PSPAC promoter (21). Thus, the increased ςH activity could be due to overexpression of the spo0H gene in the crsA47 mutant, and this was examined using a spo0H-lacZ fusion in the presence and absence of added glucose.
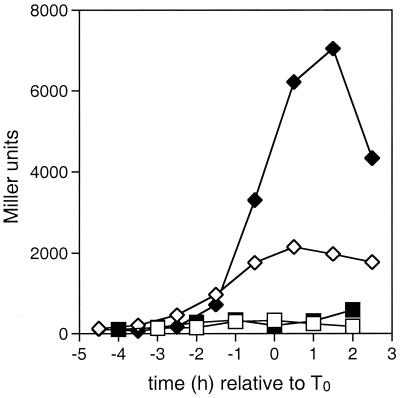
Figure 5 shows the expression patterns of the ςA-dependent spo0H-lacZ fusion in strains containing wild-type sigA (GBS151) or the crsA mutation (GBS150). In GBS151 the activity of the spo0H promoter began to increase at T−2 to peak at roughly T1, with the maximum activity not substantially different in the presence or absence of glucose. This pattern of transcription is similar to that seen elsewhere (1, 50). In GBS150 the timing of transcription from the spo0H promoter was the same as in GBS151, but peak activity was roughly eight times higher in the absence of glucose and 19 times higher in the presence of glucose than was seen in the GBS151.
FIG. 5.
Effect of the crsA47 mutation on the expression of spo0H. β-Galactosidase activities in a strain carrying spo0H-lacZ fusions were measured as described in Materials and Methods. T0 represents the onset of sporulation. Strains contained the wild-type ςA gene (GBS151; squares) or the crsA47 mutation (GBS150; diamonds) and were grown in either SSM (open symbols) or SSMG (filled symbols).
The high level of transcription of the spo0H gene seen in GBS150 grown in the presence of glucose could lead to excess ςH protein and thus explain the unusual patterns of ςH-dependent transcription seen in cells containing the crsA47 mutation. These data raise the question whether the extended transcription from the ςA-dependent promoter of the spo0H gene reflected a general phenomenon or whether it was specific to the spo0H promoter. The only known regulator of the spo0H promoter is AbrB. We would not expect AbrB levels to play a role in this regulation of spo0H beyond T1 as the gene is repressed earlier, and this repression was not changed in cells containing the crsA47 mutation (9). However, it was possible that the crsA47 mutation altered the reduction of ςA activity that is normally seen in sporulation (46).
As part of the characterization of the crsA47 mutation, we examined several other ςA-dependent promoters. The rapA-encoded phosphatase removes phosphate from the phosphorelay by dephosphorylating phosphorylated Spo0F (reviewed in reference 40). Transcription from the rapA gene promoter also showed a crsA47- and excess-glucose-specific increase in transcription after T0 (Fig. 6A). In other work, the same effect for the ςA-dependent spo0APv promoter has been seen (9). In contrast, expression of a second ςA-dependent gene encoding a phosphatase, rapB, did not show increased transcription in strains containing the crsA47 mutation and grown in excess glucose (Fig. 6B). Thus, the effect of the crsA47 mutation was promoter specific as well as being specific to growth conditions.
FIG. 6.
Effect of the crsA47 mutation on the expression of rapA and rapB. β-Galactosidase activities in strains carrying rapA-lacZ (A) or rapB-lacZ (B) fusions were measured as described in Materials and Methods. T0 represents the onset of sporulation. Strains contained the wild-type ςA gene (squares) or the crsA47 mutation (diamonds) and were grown in either SSM (open symbols) or SSMG (filled symbols). (A) JH12961 (rapA-lacZ) and GBS104 (crsA47 rapB-lacZ); (B) JH12866 (rapB-lacZ) and GBS103 (crsA47 rapB-lacZ).
In vitro analysis of transcription from the spo0H promoter by RNA polymerase containing ςA47.
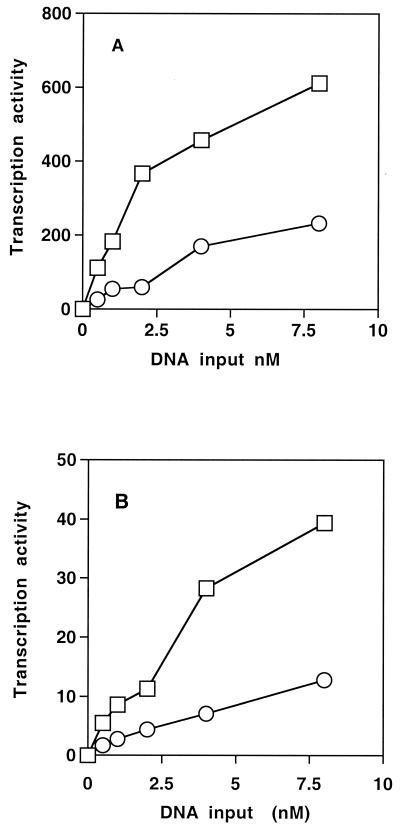
One possible explanation for the increase in ςH activity seen in strains containing the crsA mutation is that the mutation in ςA increases the affinity of the polymerase for the spo0H gene promoter. To directly measure this affinity, we purified RNA polymerase from wild-type and GBS10 strains and tested them in vitro in single-round transcription assays (as described in Materials and Methods) (Fig. 7). As a means of measuring the specific activity of the RNA polymerase preparations, we compared the activity of the two preparations on a promoter from bacteriophage φ29 that has been cloned into the plasmid pUCA2trpA (Fig. 7A). Aliquots of the polymerase preparations were mixed with DNA and initiating nucleotides (ATP plus GTP) and challenged with a mixture of heparin and CTP plus UTP. The heparin inactivates noninitiated RNA polymerase molecules while the CTP plus UTP allow enzymes that have initiated to elongate. Over a range of DNA concentrations of 0.5 to 8 nM, the wild-type RNA polymerase was three- to five-fold more active than the preparation from GBS10. We then repeated this experiment using a DNA fragment containing the spo0H promoter. This DNA fragment was produced by amplification of the promoter region. Transcription from this promoter produced a single transcript (data not shown). The activity of both polymerase preparations was significantly lower on the spo0H promoter than on the phage promoter. However, as with the φ29A2 promoter, the polymerase containing ςA47 was three- to five-fold less active on this template than was the wild-type polymerase. By this assay, the change in ςA caused by the crsA mutation did not increase the affinity of the polymerase for the spo0H promoter.
FIG. 7.
In vitro transcription activity of RNA polymerase isolated from JH642 and GBS10. In vitro transcription reactions were carried out using a control promoter (pUCA2trpA) (A) or a DNA fragment containing the spo0H gene promoter (B). Reaction mixtures contained a constant amount of RNA polymerase from either JH642 (wild type; squares) or GBS10 (crsA; circles) and increasing amounts of template. Transcription products were separated by electrophoresis through polyacrylamide gels containing 7 M urea. The level of product produced was determined using a Molecular Dynamics PhosphorImager and ImageQuant 1.0 software and is reported in arbitrary units.
DISCUSSION
We began the study of the crsA47 mutation to uncover the mechanism by which it makes sporulation resistant to the presence of excess glucose in the growth medium. During the course of these studies, we observed the consistent overexpression of ςH-dependent genes in strains carrying crsA47 that we have described in this paper. The overexpression occurred after T0 and only in strains grown in the presence of excess glucose. The overexpression peaked 2 to 3 h later than the normal peak of ςH-dependent expression and then declined. This combination of features suggests a novel feature of ςH transcription regulation that is revealed in the crsA47 genetic background.
The regulation of ςH activity appears to be complex. There is a low level of transcription of the spo0H gene during vegetative growth, and at least a few ςH-dependent genes are transcribed during this time (19, 42). In wild-type cells, transcription dependent on ςH is induced during the transition stage, and it normally peaks within 1 to 2 h after the onset of sporulation and decays after that time (46). Comparison of the levels of ςH protein with ςH-dependent transcription in vegetative growth and in early sporulation indicates the presence of posttranscriptional regulation of ςH (1, 8, 16, 17, 21, 23, 42, 50). Later in sporulation, the decrease in ςH activity is associated with loss of ςH protein due to the activity of ClpC protease (38). Another Clp family protein, ClpX, influences the induction of activity of ςH during early sporulation, although not through changes in the level of ςH protein. It has been reported that ClpX stimulates ςH-dependent transcription by direct interaction with the polymerase containing ςH (31, 37). It is known that ςH activity is regulated at transcription, since spo0H gene transcription is repressed by AbrB (11, 43). It has been recently shown that ςH-dependent expression of a limited set of genes is enhanced by amino acid starvation and that this enhancement requires the relA gene product (14). While this finding is yet another illustration of the complexity of controls over ςH activity, it is probably not related to our findings, which appeared to be specific to the presence of glucose.
It seems likely to us that the overexpression of ςH-dependent genes in GBS10 and its derivatives grown in the presence of glucose cannot be explained by loss of repression by AbrB, since AbrB repression was normal in GBS10 (9), and ςH-dependent transcription continues to increase after the time when AbrB is repressed (9). Given the similarity of the expression patterns of the spo0H gene (Fig. 5) and the ςH-dependent genes (Fig. 1 and 3), we suggest that overexpression of spo0H in cells containing the crsA47 mutation is a sufficient explanation for the unusually high transcription of ςH-dependent genes.
In a direct in vitro test of the activity of RNA polymerase isolated from GBS10 and wild-type cells (Fig. 7), there was no evidence that the crsA47 mutation increased the activity at the spo0H promoter. Furthermore, several lines of in vivo evidence support the finding that the crsA47 mutation does not simply increase the affinity of the polymerase for the spoH promoter. First, GBS10 does not grow unusually slowly, as might be expected if the crsA47 mutation altered the polymerase specificity. Second, the overexpression of the spo0H promoter in GBS10 happened only in stationary phase and only in the presence of excess glucose in the growth medium, suggesting a specific regulatory mechanism. Third, two other ςA-dependent promoters studied (for the abrB and rapB genes) did not show unusual expression patterns in GBS10 (9). We note in passing that the regulation of AbrB, which is known to control expression of a number of genes (48), must be particularly important to the overall physiology of cells with a crsA47 mutation, because it was found that a crsA47 abrB double mutant grew so poorly even in rich media that regulation in the strain could not be studied (9).
Our results indicate that there must be a regulator of the spo0H promoter (and possibly other similar promoters) whose activity is changed in GBS10 grown in excess glucose. We cannot provide any indication whether the regulator is an activator of the spo0H promoter that is hyperactive or a repressor whose activity is reduced. The latter seems more likely, since the extended high expression can be viewed as a lack of shutoff of spo0H transcription. The existence of such a regulator implies that other controls of ςH are yet to be discovered.
A consequence of ςH overexpression in the presence of glucose was the change in the SinI-to-SinR ratio, with the predicted result that SinR repression of stage 0 and stage II sporulation genes would be reduced. There is little question that reduction of SinR in the cell would increase the glucose resistance of sporulation (32). This is likely to be a major contributor to the glucose-resistant sporulation in GBS10 and illustrates the redundant pathways that regulate entry into sporulation.
ACKNOWLEDGEMENTS
We thank M. Perego, J. A. Hoch, and I. Smith for their continuous generosity in providing strains and insights during the course of this study.
The research was supported by grants from the Natural Sciences and Engineering Research Council of Canada and the Canadian Institutes of Health Research to G.B.S.
REFERENCES
- 1.Asai K, Kawamura F, Yoshikawa H, Takahashi H. Expression of kinA and accumulation of ςH at the onset of sporulation in Bacillus subtilis. J Bacteriol. 1995;177:6679–6683. doi: 10.1128/jb.177.22.6679-6683.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai U, Mandic-Mulec I, Smith I. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev. 1993;7:139–148. doi: 10.1101/gad.7.1.139. [DOI] [PubMed] [Google Scholar]
- 3.Burbulys D, Trach K A, Hoch J A. Initiation of sporulation in Bacillus subtilis is controlled by a multi-component phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 4.Carter H L, III, Moran C P., Jr New RNA polymerase sigma factor under spo0 control in Bacillus subtilis. Proc Natl Acad Sci USA. 1986;83:9438–9442. doi: 10.1073/pnas.83.24.9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cervin M A, Lewis R J, Brannigan J A, Spiegelman G B. The Bacillus subtilis regulator SinR inhibits spoIIG promoter transcription in vitro without displacing RNA polymerase. Nucleic Acids Res. 1998;26:3806–3812. doi: 10.1093/nar/26.16.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cervin M A, Spiegelman G B, Raether B, Ohlsen K, Perego M, Hoch J A. A negative regulator linking chromosome segregation to developmental transcription in Bacillus subtilis. Mol Microbiol. 1998;29:85–95. doi: 10.1046/j.1365-2958.1998.00905.x. [DOI] [PubMed] [Google Scholar]
- 7.Chibazakura T, Kawamura F, Takahashi H. Differential regulation of spo0A transcription in Bacillus subtilis: glucose represses promoter switching at the initiation of sporulation. J Bacteriol. 1991;173:2625–2632. doi: 10.1128/jb.173.8.2625-2632.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosby W M, Zuber P. Regulation of Bacillus subtilis ςH (Spo0H) and AbrB in response to changes in external pH. J Bacteriol. 1997;179:6778–6787. doi: 10.1128/jb.179.21.6778-6787.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon L G. Ph.D. thesis. Vancouver, Canada: University of British Columbia; 2000. [Google Scholar]
- 10.Dobinson K D, Spiegelman G B. Effect of the delta subunit of Bacillus subtilis RNA polymerase on initiation of RNA synthesis at two bacteriophage phi 29 promoters. Biochemistry. 1987;26:8206–8217. doi: 10.1021/bi00399a028. [DOI] [PubMed] [Google Scholar]
- 11.Dubnau E J, Cabane K, Smith I. Regulation of spo0H, an early sporulation gene in bacilli. J Bacteriol. 1987;169:1187–1191. doi: 10.1128/jb.169.3.1182-1191.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubnau E J, Weir J, Nair G, Carter III H L, Moran C P, Jr, Smith I. Bacillus sporulation gene spo0H codes for ς30 (ςH) J Bacteriol. 1988;170:1054–1062. doi: 10.1128/jb.170.3.1054-1062.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eymann C, Mittenhuber G, Hecker M. The stringent response, ςH-dependent gene expression and sporulation in Bacillus subtilis. Mol Gen Genet. 2001;264:913–923. doi: 10.1007/s004380000381. [DOI] [PubMed] [Google Scholar]
- 15.Ferrari E, Henner D J, Perego M, Hoch J A. Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulation mutants. J Bacteriol. 1988;170:289–295. doi: 10.1128/jb.170.1.289-295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frisby D, Zuber P. Analysis of the upstream activating sequence and site of carbon and nitrogen repression in the promoter of an early-induced sporulation gene of Bacillus subtilis. J Bacteriol. 1991;173:7557–7564. doi: 10.1128/jb.173.23.7557-7564.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frisby D, Zuber P. Mutations in pts cause catabolite-resistant sporulation and altered regulation of spo0H in Bacillus subtilis. J Bacteriol. 1994;176:2587–2595. doi: 10.1128/jb.176.9.2587-2595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaur N K, Cabane K, Smith I. Structure and expression of the Bacillus subtilis sin operon. J Bacteriol. 1988;170:1046–1053. doi: 10.1128/jb.170.3.1046-1053.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grossman A D. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 20.Haldenwang W G. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Healy J, Weir J, Smith I, Losick R. Post-transcriptional control of a sporulation regulatory gene encoding transcription factor ςH in Bacillus subtilis. Mol Microbiol. 1991;5:477–487. doi: 10.1111/j.1365-2958.1991.tb02131.x. [DOI] [PubMed] [Google Scholar]
- 22.Helmann J D, Chamberlin M J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 23.Hicks K A, Grossman A D. Characterization of csh203::Tn916lac, a mutation in Bacillus subtilis that makes the sporulation sigma factor sigma-H essential for normal vegetative growth. J Bacteriol. 1995;177:3736–3742. doi: 10.1128/jb.177.13.3736-3742.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoch J A. Genetic analysis in Bacillus subtilis. In: Miller J H, editor. Methods in enzymology. 204. Bacterial genetic systems. San Diego, Calif: Academic Press, Inc.; 1991. pp. 305–320. [DOI] [PubMed] [Google Scholar]
- 25.Hoch J A. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu Rev Microbiol. 1993;47:441–465. doi: 10.1146/annurev.mi.47.100193.002301. [DOI] [PubMed] [Google Scholar]
- 26.Ireton K, Jin S, Grossman A D, Sonenshein A L. Krebs cycle function is required for activation of the Spo0A transcription factor in Bacillus subtilis. Proc Natl Acad Sci USA. 1995;92:2845–2849. doi: 10.1073/pnas.92.7.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawamura F, Wang L-F, Doi R H. Catabolite-resistant sporulation (crsA47) mutations in the Bacillus subtilis RNA polymerase ς43 gene (rpoD) can suppress and be suppressed by mutations in spo0 genes. Proc Natl Acad Sci USA. 1985;82:8124–8128. doi: 10.1073/pnas.82.23.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewandoski M, Dubnau E, Smith I. Transcriptional regulation of the spo0F gene of Bacillus subtilis. J Bacteriol. 1986;168:870–877. doi: 10.1128/jb.168.2.870-877.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis R J, Brannigan J A, Offen W A, Smith I, Wilkinson A J. An evolutionary link between sporulation and prophage induction in the structure of a repression: anti-repressor complex. J Mol Biol. 1998;283:907–913. doi: 10.1006/jmbi.1998.2163. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Cosby W M, Zuber P. Role of Lon and CplX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation in Bacillus subtilis. Mol Microbiol. 1999;33:415–428. doi: 10.1046/j.1365-2958.1999.01489.x. [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Zuber P. The CplX protein of Bacillus subtilis indirectly influences RNA polymerase holoenzyme composition and directly stimulates ςH-dependent transcription. Mol Microbiol. 2000;37:885–897. doi: 10.1046/j.1365-2958.2000.02053.x. [DOI] [PubMed] [Google Scholar]
- 32.Louie P, Lee A, Stansmore K, Grant R, Ginther C, Leighton T. Roles of rpoD, spoIIF, spoIIJ, spoIIN, and sin in regulation of Bacillus subtilis stage II sporulation-specific transcription. J Bacteriol. 1992;174:3570–3576. doi: 10.1128/jb.174.11.3570-3576.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandec-Mulec I, Gaur N, Bai U, Smith I. Sin, a stage-specific repressor of cellular differentiation. J Bacteriol. 1992;174:3561–3569. doi: 10.1128/jb.174.11.3561-3569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandec-Mulec I, Doukhan L, Smith I. The Bacillus subtilis SinR protein is a repressor of the key sporulation gene spo0A. J Bacteriol. 1995;177:4619–4627. doi: 10.1128/jb.177.16.4619-4627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 36.Msadek T. When the going gets tough: survival strategies and environmental signaling networks in Bacillus subtilis. Trends Microbiol. 1999;7:201–207. doi: 10.1016/s0966-842x(99)01479-1. [DOI] [PubMed] [Google Scholar]
- 37.Nakano M M, Zhu Y, Liu J, Reyes D Y, Yoshikawa H, Zuber P. Mutations conferring amino acid residue substitutions in the carboxy-terminal domain of RNA polymerase α can suppress clpX and clpP with respect to developmentally regulated transcription in Bacillus subtilis. Mol Microbiol. 2000;37:869–884. doi: 10.1046/j.1365-2958.2000.02052.x. [DOI] [PubMed] [Google Scholar]
- 38.Nanamiya H, Asai K, Fujita M, Moriya S, Ohashi Y, Sadaie Y, Ogasawara N, Kawamura F. ClpC regulates the fate of a sporulation initiation sigma factor, sigmaH protein, in Bacillus subtilis. Mol Microbiol. 1998;29:505–513. doi: 10.1046/j.1365-2958.1998.00943.x. [DOI] [PubMed] [Google Scholar]
- 39.Perego M. Kinase-phosphatase competition regulates Bacillus subtilis development. Trends Microbiol. 1998;6:366–370. doi: 10.1016/s0966-842x(98)01350-x. [DOI] [PubMed] [Google Scholar]
- 40.Perego M, Spiegelman G B, Hoch J A. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol. 1998;2:689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 41.Predich M, Nair G, Smith I. Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing ςH. J Bacteriol. 1992;174:2771–2778. doi: 10.1128/jb.174.9.2771-2778.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price V A, Feavers I M, Moir A. Role of sigmaH in expression of the fumarase gene (citG) in vegetative cells of Bacillus subtilis 168. J Bacteriol. 1989;171:5933–5939. doi: 10.1128/jb.171.11.5933-5939.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robertson J B, Gocht M, Marahiel M A, Zuber P. AbrB, a regulator of gene expression in Bacillus, interacts with the transcription initiation regions of a sporulation gene and an antibiotic biosynthesis gene. Proc Natl Acad Sci USA. 1989;86:8457–8461. doi: 10.1073/pnas.86.21.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimotsu H, Henner D. Construction of a single copy integration vector and its use in analysis of regulation of the trp operon in Bacillus subtilis. Gene. 1986;4:85–94. doi: 10.1016/0378-1119(86)90011-9. [DOI] [PubMed] [Google Scholar]
- 45.Smith I. Regulatory proteins that control late-growth development. In: Sonenshein A L, Losick R, Hoch J A, editors. Bacillus subtilis and other gram-positive bacteria: physiology, biochemistry, and molecular biology. Washington, D.C.: American Society for Microbiology; 1993. pp. 785–800. [Google Scholar]
- 46.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 47.Strauch M A, Spiegelman G B, Perego M, Johnson W C, Burbulys D, Hoch J A. The transition state regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 1989;8:1615–1621. doi: 10.1002/j.1460-2075.1989.tb03546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strauch M A, Hoch J A. Transition-state regulators: sentinels of Bacillus subtilis post-exponential gene expression. Mol Microbiol. 1993;7:337–342. doi: 10.1111/j.1365-2958.1993.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi I. Catabolite repression-resistant mutants of Bacillus subtilis. Can J Microbiol. 1979;35:1283–1287. doi: 10.1139/m79-202. [DOI] [PubMed] [Google Scholar]
- 50.Weir J, Predich M, Dubnau E, Nair G, Smith I. Regulation of spo0H, a gene coding for the Bacillus subtilis ςH factor. J Bacteriol. 1991;173:521–529. doi: 10.1128/jb.173.2.521-529.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamashita S, Yoshikawa H, Kawamura F, Takahashi H, Yamamoto T, Kobayashi Y, Saito H. The effect of spo0 mutations on the expression of spo0A- and spo0F-lacZ fusions. Mol Gen Genet. 1986;205:28–33. doi: 10.1007/BF02428029. [DOI] [PubMed] [Google Scholar]
- 52.Yamashita S, Kawamura F, Yoshikawa H, Takahashi H, Kobayashi Y, Saito H. Dissection of the expression signals of the spo0A gene of Bacillus subtilis: glucose represses sporulation-specific expression. J Gen Microbiol. 1989;135:1335–1345. doi: 10.1099/00221287-135-5-1335. [DOI] [PubMed] [Google Scholar]