Abstract
Simple Summary
In our study, we analyzed the role of CD44 expression as a prognostic marker in patients with metastatic colorectal cancer. CD44 is a multi-structural and multifunctional non-kinase transmembrane glycoprotein. Its expression is recognized as a molecular marker of cancer stem cells. Due to its role in the maintenance of stemness and the function of CSCs in tumor progression, CD44 could be an important prognostic marker. In our study, elevated CD44 expression correlated with more aggressive tumor behavior and poor prognostic features, suggesting its role as a novel prognostic marker and potential therapeutic target for mCRC patients.
Abstract
Background: The transmembrane glycoprotein CD44, the major hyaluronan (HA) receptor, has been proven to regulate cell growth, survival, differentiation, and migration. It is therefore widely considered to be involved in carcinogenesis. Its role as a new therapeutic target in solid tumors is under evaluation in clinical trials. The prognostic value remains controversial. Here, we aimed to investigate the correlation between CD44 expression and the clinicopathological features and survival in metastatic colorectal cancer (mCRC) patients. Methods: Data from 65 mCRC patients of the Medical Oncology Unit, University Hospital and University of Cagliari were retrospectively collected from 2008 to 2021. Immunohistochemical analysis was performed at the Pathology Division, University Hospital of Cagliari on 3 μm thick sections obtained from paraffin blocks. The intensity of immunohistochemical staining was subclassified into four groups: score 0 if negative or weak membrane staining in less than 10% of tumor cells; score 1+ if weak membrane staining in at least 10% of tumor cells or moderate membrane staining in less than 10% of tumor cells; score 2+ if moderate membrane staining in at least 10% of tumor cells or intensive membrane staining in less than 10% of tumor cells; score 3+ if intense membrane staining in at least 10% of tumor cells. Based on this score, we distinguished patients into low CD44 expression (score 0, 1+, 2+) and high CD44 expression (score 3+). Statistical analysis was performed with MedCalc (survival distribution: Kaplan–Meier; survival comparison: log-rank test; association between categorical variables: Fisher’s exact test). Results: Patients’ median age was 66 years (range 49–85). Regarding CD44 expression, score was 0 in 18 patients, 1+ in 15 patients, 2+ in 18 patients, and 3+ in 14 patients. Median overall survival (mOS) was 28.1 months (95%CI: 21.3–101). CD44 overexpression (3+) was correlated with poor prognosis (p = 0.0011; HR = 0.2), with a mOS of 14.5 months (95%CI 11.7 to 35.9) versus 30.7 months (95%CI 27.8 to 101) in lower CD44 expression. Higher CD44 expression was associated with clinically poor prognostic features: age ≥ 70 years (p = 0.0166); inoperable disease (p = 0.0008); stage IV at diagnosis (p = 0.0241); BRAF mutated (p = 0.0111), high-grade tumor (p = 0.0084). Conclusions: CD44 markedly correlated with aggressive tumor behavior and contributed to the earlier progression of disease, thus suggesting its role as a novel prognostic marker and potential therapeutic target for mCRC patients.
Keywords: metastatic colorectal cancer, biomarkers, CD44 expression, cancer stem cells
1. Introduction
GLOBOCAN epidemiological data show nearly 2 million new colorectal cancer (CRC) cases worldwide in 2020, accounting for about 10% of all cancer cases. It represents the third most common neoplasm in men and the second most common in women [1]. Despite advances in surgical techniques, chemotherapy, and radiotherapy, CRC remains the second leading cause of cancer death, accounting for nearly 1 million deaths annually [1]. Among new CRC diagnoses, 20% of patients have metastatic disease at presentation, and up to 25% who have been diagnosed as having localized disease will later develop metastases. The 5-year survival for stage IV disease is only 12% [2].
Over the last decade, the management of CRC patients changed considerably, particularly in metastatic disease, mainly due to the introduction of combination chemotherapy with targeted agents, leading to more curative resections and prolonging survival in patients with unresectable disease. A deeper understanding of the tumorigenesis mechanisms facilitated tumor characterization, prognosis, and patient stratification, with a view to personalized medicine. Indeed, like other solid tumors, CRC is a heterogeneous disease in which different subtypes can be distinguished by their specific clinical and/or molecular features [3,4,5,6]. Therefore, understanding the molecular pathways underlying the initiation and development of CRC is essential to identify new biomarkers for diagnosis and prognosis, thereby improving outcomes.
Ongoing efforts to investigate new treatment strategies for CRC include understanding the involvement of cancer stem cells (CSCs) [7]. Greater knowledge of the CSCs’ metabolic characteristics is undoubtedly related to discovering a subpopulation of tumor-initiating cells in CRC. Indeed, in 2007 the existence of tumorigenic and non-tumorigenic cells within CRC was demonstrated, implying that not all cells within a tumor are capable of initiating and sustaining the neoplastic growth [8]. This concept has important therapeutic implications and suggests that targeting CSCs is necessary to increase the efficacy of therapeutic strategies. CSC features include the ability to self-renew, resistance to chemotherapy and/or radiotherapy, and increased metastatic potential that the tumor microenvironment can shape [9].
Specific surface markers of CSCs appear to have a prognostic role [10,11,12]. However, their clinical relevance in CRC is currently controversial and has been reported only in small institutional studies [12,13,14,15,16,17,18,19,20,21,22]. Among stem cell surface markers, CD44 is of great interest. Indeed, in vitro studies have shown that a single CRC cell expressing CD44 can produce highly heterogeneous CRCs [23,24].
CD44 is a multistructural and multifunctional non-kinase transmembrane glycoprotein expressed in embryonic stem cells and at various levels in other cell types, including connective tissues and bone marrow [25,26]. The expression of CD44 is recognized as a molecular marker of CSCs [27]. It is encoded by a single gene containing 19 exons. The first five and last five exons are constant and encode the shortest isoform of CD44 (85–95 kDa) called standard CD44 (CD44s). Variant isoforms (CD44v) are generated by alternative splicing and possess the ten constant exons and any combination of the remaining nine variant exons [28,29]. The main ligand of CD44 is hyaluronic acid (HA), an abundant component of the extracellular matrix (ECM) expressed by stromal and tumor cells [30]. The HA-CD44 complex induces conformational changes by binding adaptor proteins or cytoskeletal elements to the intracellular domain. This binding activates several signaling pathways that lead to cell proliferation, adhesion, migration, and invasion [31,32].
Therefore, the role of CD44 in maintaining stemness and the CSC function in tumor progression suggests that CD44 might be an important prognostic marker. Here, we aimed to investigate the correlation between CD44 expression and the clinicopathological features and survival of metastatic colorectal cancer (mCRC) patients.
2. Materials and Methods
We retrospectively collected data from 65 cases of mCRC patients diagnosed between 2008 and 2021, referred to the Medical Oncology Unit of the University Hospital of Cagliari. Baseline demographic and clinical characteristics, treatment, and survival information were collected from clinical charts. Pathological and molecular features were retrieved from histological reports. The following data were collected: gender, age, Eastern Cooperative Oncology Group (ECOG) performance status (PS) at diagnosis of metastatic disease, the onset of metastatic disease, primary tumor location, sites of metastases, mucinous histology, grade of differentiation, CD44 and CDX2 tumor expression, BRAF/RAS mutational status, MSI/MMR status, and treatment outcome. For study purposes, right-sided and left-sided CRC primary tumors were defined as proximal or distal to the splenic flexure. Ethics Committee approval was obtained for the study (Protocol number 2020/10912—code: EMIBIOCCOR) and written informed consent was obtained from all participants for their tissues to be utilized for this work.
Tumor samples were retrospectively tested for CD44 immunohistochemical (IHC) expression with the aim of evaluating the correlation with clinical outcome in terms of overall survival (OS), progression-free survival (PFS), response rate (RR), disease control rate (DCR). The primary endpoint was the OS rate at 24 months. The secondary endpoints were overall survival (OS), progression-free survival (PFS), objective response rate (ORR), and disease control rate (DCR) in the first and second line.
2.1. Histological, Immunohistochemical, and Molecular Analysis
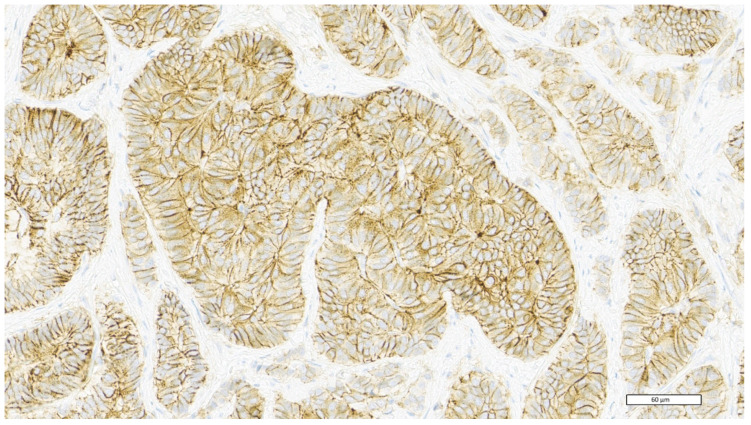
Tumor samples were routinely processed for histological observation and stained with haematoxylin–eosin (H&E). For immunohistochemical analysis, 3 μm thick sections were obtained from the paraffin block. All reagents were purchased from Ventana Medical Systems Inc. 1910 E. Innovation Park Drive Tucson, Arizona 85755 USA. The sections were automatically dewaxed and rehydrated with EZ Prep 1X (Ref. 950–102) and pre-treated with heat-induced epitope retrieval in Ultra CC1 (Ref. 950–224), following the manufacturer’s instructions. Slides were then incubated at room temperature with anti-human CD44 rabbit monoclonal antibody—clone SP37 (Ref. 790–4537) and with anti-human CDX2 rabbit monoclonal antibody—clone EPR2764Y (Ref. 760–4380). All immunostaining procedures were performed using the UltraView Universal DAB Detection Kit (Ref. 760–5000) on the BenchMark Ultra (Ventana Medical Systems Inc. 1910 E. Innovation Park Drive Tucson, Arizona 85755 USA) instrument, according to the manufacturer’s instructions. For CD44 interpretation, we used the following grading score system, based on the HER2/neu scheme (Table 1) (22). Based on this score, we distinguished patients as low CD44 expression (score 0, 1+, 2+) and high CD44 expression (score 3+). The presence of intense membrane staining in at least 10% of tumor cells is classified as score 3+ (Figure 1). RAS and BRAF gene mutational status was assessed by pyrosequencing of formalin-fixed, paraffin-embedded (FFPE) archival tumor tissue samples from primary tumors or metastases. Expression of MMR proteins (MLH1, MSH2, MSH6, and PMS2) was measured by immunohistochemistry. Histological, immunohistochemical, and molecular analysis were conducted at the Division of Pathology of the University Hospital of Cagliari.
Table 1.
CD44 scoring system.
| CD44 Expression | Score |
|---|---|
| Negative or weak membrane staining in less than 10% of tumour cells | 0 |
| Weak membrane staining in at least 10% of tumour cells or moderate membrane staining in less than 10% of tumour cells | 1+ |
| Moderate membrane staining in at least 10% of tumour cells or intense membrane staining in less than 10% of tumour cells | 2+ |
| Intense membrane staining in at least 10% of tumour cells | 3+ |
Figure 1.
CD44 high expression—score 3+: intense membrane staining in at least 10% of tumor cells.
2.2. Statistical Analysis
Statistical analysis was performed with the MedCalc® Statistical Software version 20.008 (MedCalc Software Ltd, Ostend, Belgium; https://www.medcalc.org; 2021; accessed on 1 June 2022). The association between categorical variables was estimated by the Fisher exact test for categorical binomial variables or by the chi-square test in all other instances. Survival probability over time was estimated by the Kaplan–Meier method. Significant differences in the probability of survival between the strata were evaluated by the log-rank test. The independent role of variables that were statistically significant in a univariate analysis was assessed with a logistic regression analysis. OS was defined as the time interval between the date of the beginning of the first-line treatment to death or the last follow-up visit for patients who were lost to follow-up. PFS-I was defined as the interval between the date of the beginning of the first line of treatment to death, first sign of clinical progression, or the last follow-up visit for patients who were lost to follow-up. PFS-II was defined as the interval between the date of the beginning of the second line of treatment to death, first sign of clinical progression, or the last follow-up visit for patients who were lost to follow-up. ORR was defined as the percentage of patients who achieved a partial or complete response to treatment according to RECIST version 1.1. DCR was defined as the percentage of patients with stable disease or partial/complete response to treatment.
In order to detect a difference in the effect size with statistical significance in the proportion of patients alive at 24 months according to CD44 status and assuming a 24-month OS of 65% in the low CD44 expression group and 25% in patients with high CD44 expression, at least 52 patients were necessary with α = 0.2 and β = 0.2, using a “comparison of proportion test.” A p-value <0.05 was considered statistically significant.
3. Results
Data from 65 mCRC patients diagnosed between 2008 and 2021, referred to the Medical Oncology Unit of the University Hospital of Cagliari, were collected. Patients’ characteristics were consistent with a stage IV CRC population (Table 2). The median age was 66 years (range, 49–85), 37 were male (57%) and 28 were female (43%). Negative or weak CD44 membrane staining in less than 10% of tumor cells (score 0) was observed in 18 (27.7%) patients, 15 (23.1%) showed weak membrane staining in at least 10% of tumor cells or moderate staining in less than 10% of tumor cells (score 1+), 18 (27.7%) showed moderate membrane staining in at least 10% of tumor cells or intense staining in less than 10% of tumor cells (score 2+), and 14 (21.5%) showed intense membrane staining in at least 10% of tumor cells (score 3+).
Table 2.
Patient’s characteristic. Abbreviations: ECOG PS = Eastern Cooperative Oncology Group performance status; K-RAS = Kirsten rat sarcoma viral oncogene homologue; N-RAS = Neuroblastoma RAS viral oncogene homologue B1; Anti-VEGF: anti-vascular endothelial growth factor antibodies; Anti-EGFR: anti-epidermal growth factor antibodies.
| Low CD44 Expression | High CD44 Expression | |
|---|---|---|
Gender
|
28 (54.9%) 23 (45.1%) |
11 (78.6%) 3 (21.4%) |
Age
|
39 (76.5%) 12 (23.5%) |
8 (57.2%) 6 (42.8%) |
Stage at diagnosis
|
20 (39.2%) 31 (60.8%) |
1 (7.1%) 13 (92.9%) |
CDX-2
|
48 (94.1%) 3 (5.9%) |
11 (78.6%) 3 (21.4%) |
Site of primary tumour
|
38 (74.5%) 13 (25.5%) |
10 (71.4%) 4 (28.6%) |
Surgery of the primary tumour
|
46 (90.2%) 5 (9.8%) |
7 (50%) 7 (50%) |
Tumour Grade
|
41 (80.4%) 10 (19.6%) |
6 (42.8%) 8 (57.2%) |
Metastases sites
|
19 (37.3%) 32 (62.7%) |
3 (21.4%) 11 (78.6%) |
Liver Metastases
|
34 (66.7%) 17 (33.3%) |
12 (85.7%) 2 (14.3%) |
Peritoneal Metastases
|
16 (31.4%) 35 (68.6%) |
6 (42.9%) 8 (57.1%) |
K-RAS/N-RAS mutational status
|
27 (52.9%) 24 (47.1%) |
7 (50%) 7 (50%) |
B-RAF mutational status
|
46 (90.2%) 5 (9.8%) |
9 (64.3%) 5 (35.7%) |
First-line chemotherapy
|
6 (11.8%) 42 (82.3%) 3 (5.9%) |
2 (14.3%) 8 (57.1%) 4 (28.6%) |
First-line biological drug
|
32 (62.8%) 7 (13.7%) 12 (23.5%) |
9 (64.3%) 2 (14.3%) 3 (21.4%) |
Second-line chemotherapy
|
3 (5.9%) 32 (62.7%) 16 (31.4%) |
1 (7.1%) 7 (50%) 6 (42.9%) |
Second-line biological drug
|
22 (43.1%) - 29 (56.9%) |
5 (35.7%) 1 (7.1%) 8 (57.2%) |
As of the data cut off, 31 December 2021, 13 (20%) patients were alive, while the remaining 52 (80%) had died.
Clinical Outcomes
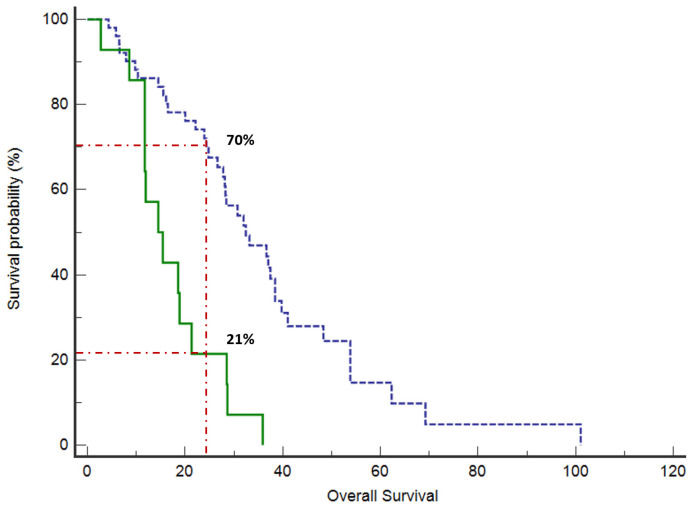
At a median follow-up of 26.6 months (95% confidence interval (CI) 21.4 to 28.8), the median OS was 28.3 months (95%CI 21.3 to 101). The proportion of patients alive at 24 months was 70% in the low CD44 expression group versus 21% of high CD44 expression patients (p = 0.0001) (Figure 1). Median OS was 30.7 months (95%CI 27.8 to 101) in patients with low CD44 expression versus 14.5 months (95%CI 11.7 to 35.9) in the high CD44 expression group (p = 0.0001) (Figure 2).
Figure 2.
Median OS was 30.7 months (95%CI 27.8 to 101) in patients with low CD44 expression (blue line, dashed) versus 14.5 months (95%CI 11.7 to 35.9) in high CD44 expression group (green line, continuous) (p = 0.0001). The proportion of patients alive at 24 months was 70% in the low CD44 expression group versus 21% of high CD44 expression patients (p = 0.0001).
Patients involved in our analysis received first- and second-line chemotherapy treatment based on the patient’s clinical features, tumor-related biological characteristics, and clinician preference (Table 2). There is substantial uniformity between the populations with low and high CD44 expression. However, it is useful to note a trend towards greater first-line triplet use in highly expressed CD44. In low CD44 expression, the chemotherapy regimen used (monotherapy, doublet, or triplet) did not impact first-line (p = 0.3) or second-line treatment (p = 0.4). The same applies to highly expressed CD44 (p = 0.3 and p = 0.1, respectively). The biological drug used (anti-VEGF, anti-EGFR, or nothing) in low CD44 expression also did not impact the first line (p = 0.2) nor the second line (p = 0.3). The same applies to highly expressed CD44 (p = 0.6 and p = 0.3, respectively).
Furthermore, regarding first- and second-line responses in terms of PFS and RR, our study showed no statistically significant difference in patients with high and low CD44 expression. However, we can see a trend towards higher treatment efficacy in patients with low CD44 expression compared to those with high expression (Table 3).
Table 3.
Response to first and second line. Abbreviations: ORR = overall response rate; DCR = disease control rate; CR = complete response; PR = partial response; SD = stable disease; PD = progression disease; PFS-1 = progression-free survival in first line; PFS-2 = progression-free survival in second line; mo = months.
| Response | Low CD44 Expression | High CD44 Expression | p-Value |
|---|---|---|---|
| ORR first line, n (%) ORR second line, n (%) |
21 (41.2%) 4 (12.1%) |
6 (42.9%) - |
p = 0.9 p = 0.3 |
| DCR first line, n (%) DCR second line, n (%) |
43 (87.8%) 26 (78.8) |
10 (71.4%) 4 (50%) |
p = 0.14 p = 0.1 |
| Median PFS-1, mo (range) | 11.6 mo (7 to 93.5) | 9.5 mo (3.9 to 19.4) | p = 0.17 |
| Median PFS-2, mo (range) | 5.6 mo (3.5 to 26) | 2.9 (2.1 to 9.6) | p = 0.12 |
We analyzed the impact of different clinicopathological features on OS and TTP. In the univariate analysis, it was interesting to note that clinicopathological features associated with poor prognosis were more frequent in patients with highly expressed CD44 (Table 4). We recorded an age ≥70 years in 42.8% of patients with highly expressed CD44 versus 23.5% of those with low CD44 expression (p = 0.0166). Of patients with highly expressed CD44, 58.3% had an inoperable disease at diagnosis versus 9.8% of those with low CD44 expression (p = 0.0008). Furthermore, at diagnosis, 92.9% of patients with high expression of CD44 presented with stage IV versus 60.8% of patients with low CD44 expression (p = 0.0241). Of patients with highly expressed CD44, 57.2% had a grade 3 tumor compared with 19.6% of patients with low CD44 expression (p = 0.0084). Finally, BRAF was mutated in 9.8% of patients with non-overexpressed CD44 compared to 35.7% of patients with CD44 3+ (p = 0.0111).
Table 4.
Correlation between high CD44 expression and clinically poor prognostic features.
| Variable | OS—p-Value |
|---|---|
| Age (≥70 y) | 0.0166 |
| CDX2 negative | 0.09 |
| Stage IV at diagnosis | 0.0241 |
| Right-sided colon cancer | 0.8 |
| Inoperable colon cancer | 0.0008 |
| High-grade tumour | 0.0084 |
| Metastasis sites (single vs. multiple sites) |
0.27 |
| Liver Metastases | 0.16 |
| Peritoneal Metastases | 0.4 |
| B-RAF mutational status | 0.0111 |
4. Discussion
Our study analyzed the correlation between CD44 expression and clinically relevant CRC outcomes using a large cohort of patients. CD44 is a cell surface glycoprotein involved in cell–cell interaction, adhesion, and migration [33]. It participates in multiple physiological processes, and its aberrant expression and dysregulation contribute to tumor initiation and progression [34]. The different functional roles of CD44s and specific CD44v still need to be fully elucidated. CD44 and the various CD44v isoforms have partly overlapping and distinct functional roles. CD44v isoforms have additional binding sites that promote the interaction of CD44 with molecules in the microenvironment. CD44v can act as co-receptors by binding/sequestering growth factors on the cell surface and presenting them to their specific receptors [35,36]. Moreover, CD44v has been reported to be expressed in metastatic tumors [37,38], whereas switching between CD44v and CD44s may regulate epithelial to mesenchymal transition (EMT). CD44 can undergo isoform switching in cancer cells [39]. Recently, it has been seen that in pancreatic cancer cells, EMT phenotype induction seems to require the upregulation of CD44 expression with isoform switching of CD44v to CD44s expression [40].
Cells with CD44 overexpression appear to have less spontaneous apoptosis and are more resistant to drug-induced cell death, inducing an anti-apoptotic effect. High expression of CD44 confers a selective advantage to resist apoptosis, thereby promoting cell transformation into a malignant phenotype in conjunction with other anti-apoptotic factors [41,42,43,44].
Recent studies have shown that CD44 is the most common CSC surface marker and plays an essential role in communication between CSCs and the tumor microenvironment [45]. CSCs represent a cohort of long-lived cells that can generate lifelong cell progeny and fuel tumor growth [46,47]. Their existence may explain the ineffective response of many therapies, leading to poor prognosis and recurrence in patients with different types of cancer. CSCs reside in a niche where immune cells, microvesicles, and cytokines can stimulate self-renewal and promote metastasis. These microvesicles and cytokines may also exert immunosuppressive functions. Some studies have found that CSCs can weaken T-cell differentiation, proliferation, and anti-tumor effects [48]. The CD44 molecular structure performs as a receptor for hyaluronic acid and other components of the ECM, enabling CSCs to identify changes in the environment and adapt their effects according to those changes and the new environment [45]. Indeed, CD44 is a critical cell surface molecule capable of detecting, integrating, and transducing signals from the cell microenvironment to cytoskeletal proteins or the nucleus to regulate a range of gene expressions that govern cell behavior. Aberrant expression and dysregulation of CD44 contribute to tumor initiation, progression, and metastasis development. Therefore, CD44 overexpression represents an unfavorable prognostic factor for CRC patients and could be used to predict tumor-associated poor prognostic features.
Several studies have revealed the central role of high CD44 expression in carcinogenesis, tumor growth, differentiation, and tumor metastasis in CRC [49,50,51,52]. In a study by Weber et al., knockout of the CD44 gene prevented tumor metastasis, even though the tumor continued to exist in mice [48]. Du et al. found that a single CD44-positive cell from a CRC tissue can grow to form a mass of tissue similar to a primary tumor [23,53]. The results of our study are substantially in line with literature data. Our analysis showed that patients with overexpressed CD44 have a worse prognosis in terms of overall survival.
Furthermore, our work demonstrated the association of high CD44 expression with clinically poor prognostic features. Some previous research reported that the expression level of CD44 was higher in high-grade CRCs than in low-grade tumors, and this overexpression was associated with reduced patient survival [53,54]. In line with that study, we also found that patients overexpressing CD44 more frequently had a high-grade tumor than patients with low CD44 expression. In this regard, there are conflicting data in the literature; some studies showed that loss of CD44 expression was associated with increased tumour aggressiveness [55,56]. This inconsistency in results may be due to alternative splicing of CD44 pre-RNA. Higher expression of CD44 in advanced tumors has been recorded in several types of malignancies, such as in lymphomas, and gastric and cervical carcinomas, possibly associated with poor prognosis [57,58]. Consistent with these data, increased expression of CD44 also was found to correlate with advanced tumor stage in the current study.
Moreover, in previous studies, a statistically significant association was found between positive CD44 expression and left-sided tumors in an Egyptian population [59], which tended to be associated with a better prognosis than right-sided CRC localization [60,61]. In the current study, there was no significant association between CD44 and tumor site, which could be due to the difference in the number and type of patients.
Similar to the literature data, our analysis confirms the association between CD44 expression and poor prognosis. However, our study documented that differential CD44 expression impacts patient prognosis differently. This finding is certainly new. We thus obtained that mild or moderate expression of CD44 is not associated with a worse prognosis in the same way as no expression. On the contrary, high expression is associated with a significantly worse prognosis. Furthermore, high CD44 expression is also associated with poor clinical prognostic features.
Interestingly, we used a simple and inexpensive method, immunohistochemistry. Specifically, we used an already known and established expression scoring scale for HER2 in breast cancer. We assessed the number of cells expressing CD44 and the intensity of CD44 expression in the different samples during immunohistochemical evaluations. We realized that the variability within our case series was much more related to the intensity of staining than the percentage of positive cells. This observation led us to choose a scoring system based on the HER2/neu scheme. The choice of this score was positively correlated with the patient’s prognosis and clinical–pathological features. For this reason, it might play a role in identifying poor prognosis mCRC patients.
Finally, many studies have evaluated the different isoforms in the literature or used very complex methods that are difficult to implement in standard clinical practice. For these reasons, the results of our work and the methodology used appear innovative.
Concerning first- and second-line responses in terms of PFS and RR, our study showed no statistically significant difference in patients with high and low CD44 expression. However, we can see a trend towards higher treatment efficacy in patients with low CD44 expression compared to those with high expression. The lack of statistical significance could be related to the study’s retrospective nature and a poorly selected and non-homogeneous population. The patients involved in our analysis received first- and second-line chemotherapy treatment according to each patient’s clinical characteristics, the tumor’s biological features, and the physician’s preferences. There is substantial uniformity between the treatment used in low and high CD44 expression mCRC patients. However, it is useful to note a trend towards greater first-line triplet use in highly expressed CD44. This is likely related to the poor clinical features in this subgroup. Furthermore, the regimens used did not impact the differences in outcomes between patients with high and low CD44 expression.
Therefore, it is difficult to draw definitive conclusions on resistance to anti-cancer treatments. However, the trend towards a better response in patients with low CD44 expression, together with data on overall survival and solid biological basis, suggests that CD44 could be used not only as a marker of poor prognosis but also of chemoresistance. Prospective studies will help confirm the role of CD44, as a marker of both poor prognosis and poorer response to anti-cancer treatments. Moreover, due to the ease and low cost of immunohistochemical analysis, CD44 could be an excellent marker of poor prognosis and chemoresistance in our clinical practice.
Due to its retrospective nature, this study carries some critical issues as it included the analysis of the outcomes of a relatively small number of patients, and its results should be handled with caution. To obtain more robust results, it will be useful to prospectively evaluate the expression of CD44 in a larger sample size of CRC patients. In this context, the analysis of a more uniform population could be helpful, making more information available on the importance of the molecular profile and treatments performed on the patient’s prognosis.
Finally, in light of the above, we think that CD44 research may have further fields of use. In fact, several pre-clinical and clinical studies have been conducted to evaluate the pharmacokinetics, efficacy, and toxicity of anti-CD44 therapies in CD44-expressing tumors. The main categories of CD44-targeted therapies currently under investigation include neutralizing antibodies, mimetic peptides, aptamers, natural compounds that suppress CD44 expression, direct targeting of HA by bioconjugates and nanoparticles, HA oligomers, and CD44 decoys [62]. The data from these studies seem encouraging from both an efficacy and safety perspective. Indeed, these studies support the continued development of therapeutic strategies to target CD44 expression in patients. However, it remains paramount to investigate the possible short- and long-term consequences of CD44-directed drugs in future studies, given the numerous implications of CD44. These include its role in T-cell activation [63].
5. Conclusions
In conclusion, our work confirmed the prognostic value of CD44 expression in CRC. Specifically, high CD44 expression was significantly associated with higher proliferative activity of CRC and poor prognosis. Furthermore, CD44 overexpression was also associated with clinically poor prognostic features: older age, inoperable disease, stage IV at diagnosis, mutated BRAF, and high-grade tumor. Therefore, our results suggest that CD44 induced by malignant cells might downregulate tumor cells’ apoptosis, leading to a decline in growth inhibition and increased ability to resist chemotherapy. However, in our analysis, it is difficult to draw definitive conclusions on resistance to cancer treatments. Given the tendency for a better response in patients with low CD44 expression, the data on overall survival, and the solid biological basis, our work suggests that CD44 could also be a predictive marker of chemoresistance. Future prospective studies on larger sample sizes are needed to confirm our results and investigate the possible predictive role of resistance to anti-cancer therapy. Finally, in addition to its role as a predictive biomarker of poor prognosis and an independent predictor of tumor development, CD44 may be an effective therapeutic target. Potential therapeutic strategies targeting CD44-positive tumors, effectively blocking CD44, might provide ample opportunities to overcome chemoresistance in various cancer types.
Acknowledgments
We thank all the investigators and patients for participating in this study.
Author Contributions
Conceptualization: P.Z., V.A., A.P., G.F., and M.S.; data curation: P.Z., V.A., A.P., M.D., N.L., M.P. (Mara Persano), M.M., and M.S.; formal analysis: P.Z., V.A., A.P., R.M., F.C., G.F., and M.S.; investigation: P.Z., V.A., A.P., M.D., G.F., and M.S.; software: P.Z., V.A., A.P., R.M., F.C., G.F., and M.S.; validation: P.Z., V.A., A.P., G.F., and M.S.; writing—original draft: P.Z., V.A., A.P., M.D., R.M., N.L., F.C., M.P. (Mara Persano), G.D., E.P., F.L., M.M., V.P., M.P. (Marco Puzzoni), E.L., S.C., G.F., and M.S.; writing—review and editing: P.Z., V.A., A.P., M.D., R.M., N.L., F.C., M.P. (Mara Persano), G.D., E.P., F.L., M.M., V.P., M.P. (Marco Puzzoni), E.L., S.C., G.F., and M.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of University Hospital of Cagliari (Protocol number 2020/10912—code: EMIBIOCCOR, approved on 28 April 2021).
Informed Consent Statement
All patients signed a written informed consent form before study entry. Adequate information was given to eligible patients by the principal investigator or co-investigators in accordance with local regulations. The declaration of informed consent was personally signed and dated by the subject, and by the investigator/person designated by the investigator to conduct the informed consent discussion. Patients signed an informed consent form regarding the publication of their data.
Data Availability Statement
Datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. The data are not publicly available due to privacy restrictions.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
No external funding was used for the conduct of this study or the preparation of this article.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Morgan E., Arnold M., Gini A., Lorenzoni V., Cabasag C.J., Laversanne M., Vignat J., Ferlay J., Murphy N., Bray F. The global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut. 2023;72:338–344. doi: 10.1136/gutjnl-2022-327736. [DOI] [PubMed] [Google Scholar]
- 2.Miller K.D., Nogueira L., Mariotto A.B., Rowland J.H., Yabroff K.R., Alfano C.M., Jemal A., Kramer J.L., Siegel R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019;69:363–385. doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- 3.Ziranu P., Lai E., Schirripa M., Puzzoni M., Persano M., Pretta A., Munari G., Liscia N., Pusceddu V., Loupakis F., et al. The Role of p53 Expression in Patients with RAS/BRAF Wild-Type Metastatic Colorectal Cancer Receiving Irinotecan and Cetuximab as Later Line Treatment. Target. Oncol. 2021;16:517–527. doi: 10.1007/s11523-021-00816-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai E., Liscia N., Donisi C., Mariani S., Tolu S., Pretta A., Persano M., Pinna G., Balconi F., Pireddu A., et al. Molecular-Biology-Driven Treatment for Metastatic Colorectal Cancer. Cancers. 2020;12:1214. doi: 10.3390/cancers12051214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puzzoni M., Ziranu P., Demurtas L., Lai E., Mariani S., Liscia N., Soro P., Pretta A., Impera V., Camera S., et al. Why precision medicine should be applied across the continuum of care for metastatic colorectal cancer patients. Future Oncol. 2020;16:4337–4339. doi: 10.2217/fon-2019-0624. Erratum in Future Oncol. 2020, 16, 219. [DOI] [PubMed] [Google Scholar]
- 6.Giampieri R., Lupi A., Ziranu P., Bittoni A., Pretta A., Pecci F., Persano M., Giglio E., Copparoni C., Crocetti S., et al. Retrospective Comparative Analysis of KRAS G12C vs. Other KRAS Mutations in mCRC Patients Treated With First-Line Chemotherapy Doublet + Bevacizumab. Front. Oncol. 2021;11:736104. doi: 10.3389/fonc.2021.736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamas K., Walenkamp A.M., de Vries E.G., van Vugt M.A., Beets-Tan R.G., van Etten B., de Groot D.J., Hospers G.A. Rectal and colon cancer: Not just a different anatomic site. Cancer Treat. Rev. 2015;41:671–679. doi: 10.1016/j.ctrv.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien C.A., Pollett A., Gallinger S., Dick J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 9.Ricci-Vitiani L., Lombardi D.G., Pilozzi E., Biffoni M., Todaro M., Peschle C., De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 10.Zhu L., Gibson P., Currle D.S., Tong Y., Richardson R.J., Bayazitov I.T., Poppleton H., Zakharenko S., Ellison D.W., Gilbertson R.J. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–607. doi: 10.1038/nature07589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Todaro M., Gaggianesi M., Catalano V., Benfante A., Iovino F., Biffoni M., Apuzzo T., Sperduti I., Volpe S., Cocorullo G., et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014;14:342–356. doi: 10.1016/j.stem.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 12.de Sousa e Melo F., Kurtova A.V., Harnoss J.M., Kljavin N., Hoeck J.D., Hung J., Anderson J.E., Storm E.E., Modrusan Z., Koeppen H., et al. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature. 2017;543:676–680. doi: 10.1038/nature21713. [DOI] [PubMed] [Google Scholar]
- 13.Choi D., Lee H.W., Hur K.Y., Kim J.J., Park G.S., Jang S.H., Song Y.S., Jang K.S., Paik S.S. Cancer stem cell markers CD133 and CD24 correlate with invasiveness and differentiation in colorectal adenocarcinoma. World J. Gastroenterol. 2009;15:2258–2264. doi: 10.3748/wjg.15.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S., Song X., Chen Z., Li X., Li M., Liu H., Li J. CD133 expression and the prognosis of colorectal cancer: A systematic review and meta-analysis. PLoS ONE. 2013;8:e56380. doi: 10.1371/journal.pone.0056380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jao S.W., Chen S.F., Lin Y.S., Chang Y.C., Lee T.Y., Wu C.C., Jin J.S., Nieh S. Cytoplasmic CD133 expression is a reliable prognostic indicator of tumor regression after neoadjuvant concurrent chemoradiotherapy in patients with rectal cancer. Ann. Surg. Oncol. 2012;19:3432–3440. doi: 10.1245/s10434-012-2394-3. [DOI] [PubMed] [Google Scholar]
- 16.Jing F., Kim H.J., Kim C.H., Kim Y.J., Lee J.H., Kim H.R. Colon cancer stem cell markers CD44 and CD133 in patients with colorectal cancer and synchronous hepatic metastases. Int. J. Oncol. 2015;46:1582–1588. doi: 10.3892/ijo.2015.2844. [DOI] [PubMed] [Google Scholar]
- 17.Lin E.H., Hassan M., Li Y., Zhao H., Nooka A., Sorenson E., Xie K., Champlin R., Wu X., Li D. Elevated circulating endothelial progenitor marker CD133 messenger RNA levels predict colon cancer recurrence. Cancer. 2007;110:534–542. doi: 10.1002/cncr.22774. [DOI] [PubMed] [Google Scholar]
- 18.Iinuma H., Watanabe T., Mimori K., Adachi M., Hayashi N., Tamura J., Matsuda K., Fukushima R., Okinaga K., Sasako M., et al. Clinical significance of circulating tumor cells, including cancer stem-like cells, in peripheral blood for recurrence and prognosis in patients with Dukes’ stage B and C colorectal cancer. J. Clin. Oncol. 2011;29:1547–1555. doi: 10.1200/JCO.2010.30.5151. [DOI] [PubMed] [Google Scholar]
- 19.Sprenger T., Conradi L.C., Beissbarth T., Ermert H., Homayounfar K., Middel P., Ruschoff J., Wolff H.A., Schuler P., Ghadimi B.M., et al. Enrichment of CD133-expressing cells in rectal cancers treated with preoperative radiochemotherapy is an independent marker for metastasis and survival. Cancer. 2013;119:26–35. doi: 10.1002/cncr.27703. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q., Chen Z.G., Du C.Z., Wang H.W., Yan L., Gu J. Cancer stem cell marker CD133+ tumour cells and clinical outcome in rectal cancer. Histopathology. 2009;55:284–293. doi: 10.1111/j.1365-2559.2009.03378.x. [DOI] [PubMed] [Google Scholar]
- 21.Weichert W., Denkert C., Burkhardt M., Gansukh T., Bellach J., Altevogt P., Dietel M., Kristiansen G. Cytoplasmic CD24 expression in colo-rectal cancer independently correlates with shortened patient survival. Clin. Cancer Res. 2005;11:6574–6581. doi: 10.1158/1078-0432.CCR-05-0606. [DOI] [PubMed] [Google Scholar]
- 22.Aimola V., Fanni D., Gerosa C., Cerrone G., Ziranu P., Pretta A., Murru R., Piras M., Cau F., Zorcolo L., et al. Balance between the stem cell marker CD44 and CDX2 expression in colorectal cancer. Ann. Res. Oncol. 2022;2:160–166. doi: 10.48286/aro.2022.43. [DOI] [Google Scholar]
- 23.Du L., Wang H., He L., Zhang J., Ni B., Wang X., Jin H., Cahuzac N., Mehrpour M., Lu Y., et al. CD44 is of functional importance for colorectal cancer stem cells. Clin. Cancer Res. 2008;14:6751–6760. doi: 10.1158/1078-0432.CCR-08-1034. [DOI] [PubMed] [Google Scholar]
- 24.Vermeulen L., Todaro M., de Sousa Mello F., Sprick M.R., Kemper K., Perez Alea M., Richel D.J., Stassi G., Medema J.P. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc. Natl. Acad. Sci. USA. 2008;105:13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gronthos S., Franklin D.M., Leddy H.A., Robey P.G., Storms R.W., Gimble J.M. Surface protein characterization of human adipose tissue-derived stromal cells. J. Cell. Physiol. 2001;189:54–63. doi: 10.1002/jcp.1138. [DOI] [PubMed] [Google Scholar]
- 26.Domev H., Amit M., Laevsky I., Dar A., Itskovitz-Eldor J. Efficient engineering of vascularized ectopic bone from human embryonic stem cell-derived mesenchymal stem cells. Tissue Eng. Part A. 2012;18:2290–2302. doi: 10.1089/ten.tea.2011.0371. [DOI] [PubMed] [Google Scholar]
- 27.Yin T., Wang G., He S., Liu Q., Sun J., Wang Y. Human cancer cells with stem cell-like phenotype exhibit enhanced sensitivity to the cytotoxicity of IL-2 and IL-15 activated natural killer cells. Cell. Immunol. 2016;300:41–45. doi: 10.1016/j.cellimm.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Screaton G.R., Bell M.V., Bell J.I., Jackson D.G. The identification of a new alternative exon with highly restricted tissue expression in transcripts encoding the mouse Pgp-1 (CD44) homing receptor. Comparison of all 10 variable exons between mouse, human, and rat. J. Biol. Chem. 1993;268:12235–12238. doi: 10.1016/S0021-9258(18)31376-0. [DOI] [PubMed] [Google Scholar]
- 29.Prochazka L., Tesarik R., Turanek J. Regulation of alternative splicing of CD44 in cancer. Cell. Signal. 2014;26:2234–2239. doi: 10.1016/j.cellsig.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Banerjee S., Modi S., McGinn O., Zhao X., Dudeja V., Ramakrishnan S., Saluja A.K. Impaired synthesis of stromal components in response to Minnelide improves vascular function, drug delivery, and survival in pancreatic cancer. Clin. Cancer Res. 2016;22:415–425. doi: 10.1158/1078-0432.CCR-15-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponta H., Sherman L., Herrlich P.A. CD44: From adhesion molecules to siignaling regulators. Nat. Rev. Mol. Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 32.Zoller M. CD44: Can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer. 2011;11:254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 33.Park J.W., Jung K.-H., Lee J.H., Moon S.H., Cho Y.S., Lee K.-H. 89Zr anti-CD44 immuno-PET monitors CD44 expression on splenic myeloid cells and HT29 colon cancer cells. Sci. Rep. 2021;11:3876–3886. doi: 10.1038/s41598-021-83496-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu H., Niu M., Yuan X., Wu K., Liu A. CD44 a tumor biomarker and therapeutic target. Exp. Hematol. Oncol. 2020;9:36–49. doi: 10.1186/s40164-020-00192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamano S., Gi M., Tago Y., Doi K., Okada S., Hirayama Y., Tachibana H., Ishii N., Fujioka M., Tatsumi K., et al. Role of deltaNp63(pos)CD44v(pos) cells in the development of N-nitroso-tris-chloroethylurea-induced peripheral-type mouse lung squamous cell carcinomas. Cancer Sci. 2016;107:123–132. doi: 10.1111/cas.12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orian-Rousseau V., Chen L., Sleeman J.P., Herrlich P., Ponta H. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev. 2002;16:3074–3086. doi: 10.1101/gad.242602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozawa M., Ichikawa Y., Zheng Y.W., Oshima T., Miyata H., Nakazawa K., Guan H.B., Shiozawa M., Akaike M., Watanabe K., et al. Prognostic significance of CD44 variant 2 upregulation in colorectal cancer. Br. J. Cancer. 2014;111:365–374. doi: 10.1038/bjc.2014.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z., Chen K., Jiang P., Zhang X., Li X., Li Z. CD44v/CD44s expression patterns are associated with the survival of pancreatic carcinoma patients. Diagn. Pathol. 2014;9:79. doi: 10.1186/1746-1596-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown R.L., Reinke L.M., Damerow M.S., Perez D., Chodosh L.A., Yang J., Cheng C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J. Clin. Investig. 2011;121:1064–1074. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao S., Chen C., Chang K., Karnad A., Jagirdar J., Kumar A.P., Freeman J.W. CD44 Expression Level and Isoform Contributes to Pancreatic Cancer Cell Plasticity, Invasiveness, and Response to Therapy. Clin. Cancer Res. 2016;22:5592–5604. doi: 10.1158/1078-0432.CCR-15-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang C., Xie J., Guo J., Manning H.C., Gore J.C., Guo N. Evaluation of CD44 and CD133 as cancer stem cell markers for colorectal cancer. Oncol. Rep. 2012;28:1301–1308. doi: 10.3892/or.2012.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madjd Z., Mehrjerdi A.Z., Sharifi A.M., Molanaei S., Shahzadi S.Z., Asadi-Lari M. CD44+ cancer cells express higher levels of the anti-apoptotic protein Bcl-2 in breast tumors. Cancer Immun. 2009;9:4–10. [PMC free article] [PubMed] [Google Scholar]
- 43.El-Emshaty H., Hassan D., El-Hemaly M., Ismail H. Clinical Association of CD44 Expression with Proliferative Activity and Apoptotic State in Egyptian Patients Suffering from Ulcerative Colitis and Colorectal Carcinoma. Asian Pac. J. Cancer Prev. 2021;22:3577–3583. doi: 10.31557/APJCP.2021.22.11.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lakshman M., Subramaniam V., Rubenthiran U., Jothy S. CD44 promotes resistance to apoptosis in human colon cancer cells. Exp. Mol. Pathol. 2004;77:18–25. doi: 10.1016/j.yexmp.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 45.Naor D., Wallach-Dayan S.B., Zahalka M.A., Sionov R.V. Involvement of CD44, a molecule with a thousand faces, in cancer dissemination. Semin. Cancer Biol. 2008;18:260–267. doi: 10.1016/j.semcancer.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 46.Prasetyanti P.R., Medema J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer. 2017;16:41. doi: 10.1186/s12943-017-0600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vahidian F., Duijf P.H.G., Safarzadeh E., Derakhshani A., Baghbanzadeh A., Baradaran B. Interactions between cancer stem cells, immune system and some environmental components: Friends or foes? Immunol. Lett. 2019;208:19–29. doi: 10.1016/j.imlet.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 48.Fernández J.C., Vizoso F.J., Corte M.D., Gava R.R., Corte M.G., Suárez J.P., García-Muñíz J.L., García-Morán M. CD44s expression in resectable colorectal cancers and surrounding mucosa. Cancer Investig. 2004;22:878–885. doi: 10.1081/CNV-200039658. [DOI] [PubMed] [Google Scholar]
- 49.Wielenga V.J., van der Neut R., Offerhaus G.J. Pals ST: CD44 glycoproteins in colorectal cancer expression function and prognostic value. Adv. Cancer Res. 2000;77:169–187. doi: 10.1016/s0065-230x(08)60787-3. [DOI] [PubMed] [Google Scholar]
- 50.Bendardaf R., Elzagheid A., Lamlum H., Ristamäki R., Collan Y., Pyrhönen S. E-cadherin CD44s and CD44v6 correlate with tumour differentiation in colorectal cancer. Oncol. Rep. 2005;13:831–835. doi: 10.3892/or.13.5.831. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y.J., Yan P.S., Li J., Jia J.F. Expression and significance of CD44s CD44v6 and nm23 mRNA in human cancer. World J. Gastroenterol. 2005;11:6601–6606. doi: 10.3748/wjg.v11.i42.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weber G.F., Bronson R.T., Ilagan J., Cantor H., Schmits R., Mak T.W. Absence of the CD44 gene prevents sarcoma metastasis. Cancer Res. 2002;62:2281–2286. [PubMed] [Google Scholar]
- 53.Clara J.A., Monge C., Yang Y., Takebe N. Targeting signalling pathways and the immune microenvironment of cancer stem cells—A clinical update. Nat. Rev. Clin. Oncol. 2020;17:204–232. doi: 10.1038/s41571-019-0293-2. [DOI] [PubMed] [Google Scholar]
- 54.Ropponen K.M., Eskelinen M.J., Lipponen P.K., Alhava E., Kosma V.M. Expression of CD44 and variant proteins in human colorectal cancer and its relevance for prognosis. Scand J. Gastroenterol. 1998;33:301–309. doi: 10.1080/00365529850170900. [DOI] [PubMed] [Google Scholar]
- 55.Ylagan L.R., Scholes J., Demopoulos R. CD44 a marker of squamous differentiation in adenosquamous neoplasms. Arch. Pathol. Lab. Med. 2000;124:212–215. doi: 10.5858/2000-124-0212-C. [DOI] [PubMed] [Google Scholar]
- 56.Dallas M.R., Liu G., Chen W., Thomas S.N., Wirtz D., Huso D.L., Konstantopoulos K. Divergent roles of CD44 and carcinoembryonic antigen in colon cancer metastasis. FASEB J. 2012;26:2648–2656. doi: 10.1096/fj.12-203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carr N.J., Emory T.S., Sobin L.H. Epithelial neoplasms of the appendix and colorectum: An analysis of cell proliferation, apoptosis and expression of p53, CD44 and bcl-2. Arch. Pathol. Lab. Med. 2002;126:837–841. doi: 10.5858/2002-126-0837-ENOTAA. [DOI] [PubMed] [Google Scholar]
- 58.Zhao L.H., Lin Q.L., Wei J., Huai Y.L., Wang K.J., Yan H.Y. CD44v6 expression in patients with stage II or stage III sporadic colorectal cancer is superior to CD44 expression for predicting progression. Int. J. Clin. Exp. Pathol. 2015;8:692–701. [PMC free article] [PubMed] [Google Scholar]
- 59.Holah N.S., Aiad H.A., Asaad N.Y., Elkhouly E.A., Lasheen A.G. Evaluation of the role of CD44 as a cancer stem cell marker in colorectal carcinoma: Immunohistochemical study. Menoufia Med. J. 2017;30:174–183. doi: 10.7860/JCDR/2017/22671.9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meguid R.A., Slidell M.B., Wolfgang C.L., Chang D.C., Ahuja N. Is there a difference inn survival between right-versus left-sided colon cancers? Ann. Surg. Oncol. 2008;15:2388–2394. doi: 10.1245/s10434-008-0015-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Demurtas L., Puzzoni M., Giampieri R., Ziranu P., Pusceddu V., Mandolesi A., Cremolini C., Masi G., Gelsomino F., Antoniotti C., et al. The role of primary tumour sidedness, EGFR gene copy number and EGFR promoter methylation in RAS/BRAF wild-type colorectal cancer patients receiving irinotecan/cetuximab. Br. J. Cancer. 2017;117:315–321. doi: 10.1038/bjc.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen C., Zhao S., Karnad A., Freeman J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018;11:64. doi: 10.1186/s13045-018-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schumann J., Stanko K., Schliesser U., Appelt C., Sawitzki B. Differences in CD44 Surface Expression Levels and Function Discriminates IL-17 and IFN-γ Producing Helper T Cells. PLoS ONE. 2015;10:e0132479. doi: 10.1371/journal.pone.0132479. Erratum in PLoS ONE 2015, 10, e0143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. The data are not publicly available due to privacy restrictions.