Abstract
Omp2a and Omp2b are highly homologous porins present in the outer membrane of the bacteria from the genus Brucella, a facultative intracellular pathogen. The genes coding for these proteins are closely linked in the Brucella genome and oriented in opposite directions. In this work, we present the cloning, purification, and characterization of four Omp2b size variants found in various Brucella species, and we compare their antigenic and functional properties to the Omp2a and Omp2b porins of Brucella melitensis reference strain 16M. The variation of the Omp2a and Omp2b porin sequences among the various strains of the genus Brucella seems to result mostly from multiple gene conversions between the two highly homologous genes. As shown in this study, this phenomenon has led to the creation of natural Omp2a and Omp2b chimeric proteins in Omp2b porin size variants. The comparison by liposome swelling assay of the porins sugar permeability suggested a possible functional differences between Omp2a and Omp2b, with Omp2a showing a more efficient pore in sugar diffusion. The sequence variability in the Omp2b size variants was located in the predicted external loops of the porin. Several epitopes recognized by anti-Omp2b monoclonal antibodies were mapped by comparison of the Omp2b size variants antigenicity, and two of them were located in the most exposed surface loops. However, since variations are mostly driven by simple exchanges of conserved motifs between the two genes (except for an Omp2b version from an atypical strain of Brucella suis biovar 3), the porin variability does not result in major antigenic variability of the Brucella surface that could help the bacteria during the reinfection of a host. Porin variation in Brucella seems to result mainly in porin conductivity modifications.
Porins are abundant proteins in the outer membrane of gram-negative bacteria. They create a controlled permeability of the outer membrane toward small hydrophilic solutes such as sugars that are otherwise not allowed to diffuse through the outer membrane (for a review, see references 21, 22, and 24). In a sense, porins are the Achilles' heel of this protective barrier, since they can act as receptors for bacteriophage and colicins (13), surface-exposed antigens (37), complement-binding sites (29), and the gate of entry for some antibiotics (27, 28). In some pathogenic bacteria, such as Neisseria gonorrhoeae, porin sequence variations between serotypes are well described and probably allow successive infection by different serotypes presenting different surface-exposed epitopes (20). Neisseria porin genes are subject to frequent horizontal transfer movements of genetic material, allowing for numerous changes in sequence that can hamper vaccine development (18).
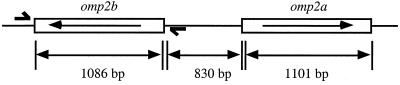
Brucellae are gram-negative pathogens infecting numerous mammalian species and causing economic losses in domestic cattle, sheep, and goats, as well as human health problems in zones where they are endemic. The mechanisms of pathogenicity of Brucella spp. are still poorly understood. Brucella outer membrane proteins (OMPs) have been extensively studied because of their potential role as virulence factors, antigenic factors, and molecular typing tools (7). Among the Brucella OMPs, the Omp2a and Omp2b porin proteins behave biochemically much as the classical nonspecific trimeric Escherichia coli porins, but they are only remotely related to other known porins by their sequence (25, 26). The pore-forming activities of Omp2a and Omp2b differs slightly, B. melitensis 16M Omp2a showing characteristics of a larger pore than B. melitensis 16M Omp2b (J.-Y. Paquet et al., unpublished data). The predicted topology for both Brucella porins is a 16-stranded β-barrel with large surface-exposed loops (30). The two Brucella porins share 85% sequence identity and are encoded in the same genetic locus. The omp2a and omp2b genes are only 850 bp apart and are oriented in opposite directions (Fig. 1). In Brucella abortus, only omp2b has been shown to be expressed, and the presence of Omp2a has never been detected (15). The pattern of porin gene expression in the other Brucella strains is still unknown. The polymorphism of both porin genes has been extensively studied, and restriction fragment length polymorphism (RFLP) studies have identified 11 omp2b variants and 8 omp2a variants (6, 16). The omp2 locus sequences in the different Brucella species show complex variability, involving gene conversion phenomenon between the closely related inverted gene copies (17). RFLP also revealed that omp2b variants in several Brucella strains are not only variable in sequence but also variable in size, indicating that insertions and/or deletions had occurred in these genes (6). These insertion/deletion variants are of particular interest because their antigenic and functional characteristics could have been drastically modified compared to the previously characterized Omp2b porins from B. melitensis 16M or B. abortus S19 (25, 26; Paquet et al., unpublished). The study of their antigenic and pore-forming properties in relation to their sequence could allow epitope mapping by use of anti-Omp2b monoclonal antibodies (MAbs) (5) and give new clues about Omp2b topology.
FIG. 1.
Genetic organization of the omp2 locus in Brucella spp. The two opposite coding sequences are represented by the white boxes. The primers used in this work to amplify omp2b variant genes are represented by the short bold arrows.
Throughout the following work, Omp2a and Omp2b porins from B. melitensis 16M and B. abortus S19 (15) will be used as references since these strains are widely used in different laboratories and they show the highest degree of divergence among all Omp2a-Omp2b pairs sequenced.
MATERIALS AND METHODS
Plasmids and strains.
The following Brucella strains were used in this study: B. melitensis 16M (reference strain), B. abortus S19 (vaccine strain), B. abortus 45/20, B. suis 83-210 (biovar 3 atypical strain), B. ovis 63/290, and B. ovis 76-250. The porin variants of strains B. abortus 45/20, B. ovis 63/290, and B. ovis 76-250 have been previously characterized by PCR-RFLP (6). B. abortus 45/20 has been used in the past as vaccine strain. B. ovis 63/290 is the reference strain for B. ovis. B. ovis 76-250 is a field isolate from a ram in France. B. suis 83-210 has been isolated from a wild rodent in Australia. Culture of Brucella strains and DNA extraction were performed as described previously (6). Recombinant porin production was obtained by cloning the genes into the pET3a plasmid vector (Novagen, Madison, Wis.) and transforming the E. coli strain BL21(DE3) harboring the pLyS plasmid (Novagen).
Cloning and sequencing of omp2b size variants and their corresponding omp2a gene copies.
Porin genes were PCR-amplified from genomic DNA of each strain and checked by RFLP as described previously (6). Nucleotide sequencing of the PCR products was performed as described previously (4).
To construct the expression vectors, the omp2b genes were PCR amplified from the genomic DNA of the strain using the proofreading Pwo polymerase (Boehringer Mannheim) and the primers 2AF (5′-GGCGGATCCCATATGGACGCAATCGTCGCGCCA-3′) and 2BR (5′-GGATCCGGTCAGCATAAAAAGCAAGC-3′). Since the signal peptide hampers the high-level production of outer membrane proteins (32, 36), the fragments amplified with these primers contain the omp2b open reading frame without the signal peptide coding sequence. The first codon of the mature polypeptide (coding for an alanine) is replaced by the ATG start codon. The resulting recombinant protein is thus equivalent to the mature wild-type protein with the mutation A1M. Conservation of the correct sequence of the insert in the expression vector was checked by nucleotide sequencing.
Purification of recombinant Omp2b size variants.
The following protocol was modified from (32). Single fresh colonies of BL21(DE3)/pLyS containing an expression vector were inoculated into a 10 ml of Luria-Bertani (LB) culture (containing 50 μg of ampicillin per ml) and grown overnight at 37°C with shaking. A 100-ml LB growth culture (containing 50 μg of ampicillin per ml) was inoculated with 1 ml of the overnight preculture. The culture was shaken at 37°C until an optical density at 600 nm of 0.7 was reached. IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 0.4 mM was added, and incubation resumed for an additional 3 h. The cells were harvested by centrifugation at 1,500 × g for 20 min at 4°C. About 1 g of bacterial pellet was obtained. The cells were resuspended in 3 ml of TEN buffer (50 mM Tris, 1 mM EDTA, 1 g of NaCl per liter; pH 8.0), with phenylmethylsulfonyl fluoride (PMSF) at a final concentration of 125 μM and lysozyme at a final concentration of 250 μg/ml. The mixture was stirred for 20 min at room temperature, and then 4 mg of deoxycholate sodium salt was added. The mixture was then shaken vigorously at 37°C for 1 h. DNase I was added (final concentration, 7 μg/ml), and the mixture was incubated at room temperature for 1 h or until it was no longer viscous. The cell lysate was then centrifuged at 14,000 × g for 20 min at 4°C. The pellet was washed once in TEN buffer containing 125 μM PMSF and resuspended in freshly made TEN buffer containing 125 μM PMSF and 8 M urea. Resuspension was helped by sonication (four times for 30 s each time at 4°C). The protein concentration was determined with a Bio-Rad protein assay mixture (Bio-Rad, Munich, Germany). Samples were diluted 1:1 in 10% (wt/vol) Zwittergent 3-14 (Calbiochem, La Jolla, Calif.), placed in a sonicator bath (Bransonic Ultrasonic Cleaner B1200; Branson, Danbury, Conn.) for 1 h, and then concentrated up to five times using an Ultrafree centrifugal filter tube (Millipore, Bedford, Mass.) with an exclusion limit of 10,000 Da. Samples were loaded onto a Superose-12 column (Pharmacia, Uppsala, Sweden) previously equilibrated with TEN containing 0.01% (wt/vol) Zwittergent 3-14, with a column flow rate of 0.5 ml/min. The protein-containing peaks were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue.
Circular dichroism spectroscopy.
Far-UV circular dichroism spectra were measured at between 190 and 250 nm using a 0.1-mm cell on a Jobin-Yvon CD6 spectrometer at 20°C at a protein concentration of 200 to 400 μg/ml (20 single spectra were accumulated with a scan rate of 0.5 nm/s).
Sequence analysis and topology prediction.
Multiple sequence alignments were performed using the CLUSTALW alignment server (38) and the MATCH-BOX program (9). The Omp2b topology model has been described previously (31). Briefly, PHD (33), Dsc (23), and Sopma (19), three secondary structure prediction methods previously tested for porin topology prediction, were applied on the Omp2b amino acid mature sequence. A porin-specific transmembrane β-strand prediction was also performed by using the method described by Schirmer and Cowan (35). All four predictions were used to establish a consensus prediction, where a prediction for a β-strand was only taken as valid if it was given simultaneously by several of the independent methods. The topology was deduced from the alternance pattern of transmembrane β-strands: predicted strands are connected either by short turns or long loops. Turns are periplasmic, and loops are predicted to be exposed at the surface, as observed in all outer membrane proteins to date (2). This prediction method has previously been validated with a set of porins of known structure (31).
Antigenic analysis.
The anti-Omp2b specific MAbs A68/25G05/A05, A68/15B06/C08, A63/05A07/A08, A63/04D11/G01, A63/03H02/B01, A63/13G02/C08, and A63/08D08/C07 were previously described (5).
For the Western blot experiments, the purified recombinant proteins or Brucella sonicated whole-cell extracts were run on SDS-PAGE and subsequently transferred to nitrocellulose (Millipore) using a Mini-Protean II cell apparatus (Bio-Rad) at 100 mV for 1 h. Blocking was done by incubating the membrane in Tris-buffered saline (TBS; 0.15 M NaCl, 10 mM Tris; pH 7.5) containing 3% bovine serum albumin (BSA) at room temperature for 2 h. The membranes were incubated overnight at room temperature with ascitic fluids of the MAbs diluted 500 times in TBS containing 1% BSA and 0.05% Tween 20 and then washed twice in TBS containing 0.05% Tween 20. Horseradish peroxidase conjugated with goat anti-mouse antibodies (GAM-HRP; Daxo A/S) was used as the secondary antibody. Membranes were incubated with GAM-HRP diluted 1,000 times in TBS-BSA-Tween for 1 h at room temperature and then washed twice with TBS. Revelation was achieved by incubating the washed membrane in a freshly made TBS solution containing 0.06% 4-chloro-1-naphthol (Bio-Rad), first diluted in methanol, and 5 mM H2O2. The reaction was stopped with distilled water.
For the enzyme-linked immunosorbent assay (ELISA), 96-well microplates (Nunc) were coated with the purified proteins at 2 μg/ml or the sonicated Brucella cells (at an absorbance value [600 nm] of 1.0) in phosphate-buffered saline (PBS) overnight at 4°C. In the case of the purified protein only, the plates were saturated with a phosphate buffer with 4% casein hydrolysate. MAbs (500-fold-diluted ascite fluid or 2-fold-diluted hybridoma spernate in PBS with 0.01% Tween 20) were incubated in the plates for 1 h at 37°C. Binding of the antibody was detected by further incubation for 1 h at 37°C with GAM-HRP (Daxo A/S) diluted 1,000-fold in PBS with 0.01% Tween 20, and subsequent revelation with K-blue substrate (Neogen, Lexington, Ky.). The reaction was stopped with 2N H2SO4, and the plates were read at 450 nm with an automated plate reader (model EL340; Bio-Tek).
Liposome swelling assays.
The following procedure is modified from that of Douglas et al. (12). Soy bean phosphatidylcholine (20 mg) was dried as a film at the bottom of a glass round-bottom flask, and 500 μl of the porin sample (50 μg/ml in TEN containing 0.01% Zwittergent 3-14) was added. The suspension was homogenized by sonication and dried again at 50°C under vacuum (in a rotating evaporator). The dried mixture was resuspended by briefly vortexing in 2 ml of a solution of 5 mM Tris–10% (wt/vol) Dextran T40 (Pharmacia) (pH 7.5). Liposomes were diluted in isotonic solutions of arabinose, glucose, or N-acetylglucosamine, and the initial rate of swelling was measured turbidimetrically at 450 nm with a Uvikon 820 spectrophotometer. All rates were compared to the arabinose swelling rate which was taken as 100%. The isotonicity of the sugar solution was checked using a porin-free liposome preparation.
Nucleotide sequence accession numbers.
The nucleotide sequences have been deposited in the GenBank database under accession numbers AF268033, AF268034, AF268035, AY008719, AY008720, and AY008721.
RESULTS
Selection and sequence comparison of Omp2b size variants.
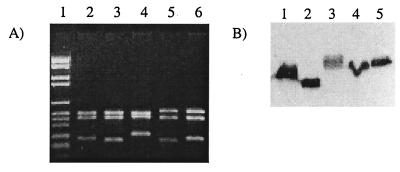
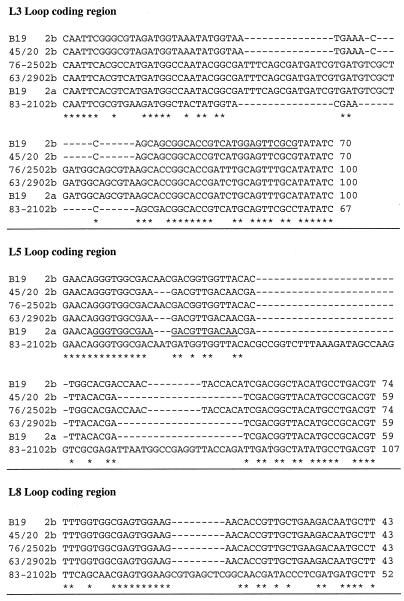
Figure 2A shows the HinfI restriction patterns obtained for the omp2b genes PCR amplified from B. abortus S19 (used as a reference), B. abortus 45/20, B. suis 83-210 (atypical biovar 3 isolate), B. ovis 63/290, and B. ovis 76-250. All restriction patterns show three fragments, but at least one fragment for each pattern has a modified size compared to that of omp2b of B. abortus S19. These size modifications are also obvious when Omp2b is detected by a specific MAb in a Western blot experiment using Brucella whole-cell extracts (Fig. 2b). Omp2b of B. abortus 45/20 is shorter than that of B. abortus S19, while those of B. suis 83-210, B. ovis 76-250, and B. ovis 63/290 are clearly longer in size. All PCR-amplified omp2b genes were sequenced. The aligned nucleotide sequences, for the positions where insertion or deletion explain gene size variation are shown in Fig. 3. The sequences inserted in the longer omp2b genes or deleted in the shorter version (B. ovis 76-250, B. ovis 63-290, and B. abortus 45/20, respectively) correspond to B. abortus S19 omp2a motifs. Interestingly, omp2b of B. suis 83-210 shows unique sequence motifs, different from both reference omp2b and omp2a genes, but appearing at positions identical to other omp2b and omp2a divergences.
FIG. 2.
(A) Size variation of omp2b shown by RFLP. Agarose gel electrophoresis of the PCR-amplified omp2b genes digested with HinfI of B. abortus S19 (lane 2), B. abortus 45/20 (lane 3), B. suis 83-210 (lane 4), B. ovis 63/290 (lane 5), and B. ovis 76-250 (lane 6) was done. Lane 1, DNA marker kit VI (Boehringer Mannheim). (B) Size variation of the corresponding Omp2b protein shown by Western blot. A Western blot with anti-Omp2b MAb A63/05A07/A08 after SDS-PAGE of B. abortus S19 (lane 1), B. abortus 45/20 (lane 2), B. suis 83-210 (lane 3), B. ovis 63/290 (lane 4), and B. ovis 76-250 (lane 5) is shown.
FIG. 3.
Multiple alignment of the nucleotide sequences of the omp2b size variants. Only the part of the sequence explaining the size differences between the variants are shown. These variable regions are denominated from the topological features that they encode (L3, L5, or L8) according to the topology model proposed by Paquet et al. (31) (see also Fig. 5). The omp2b sequences are named from their respective Brucella strain. The sequences of the oligonucleotide probes used to differentiate omp2a from omp2b (16) are underlined.
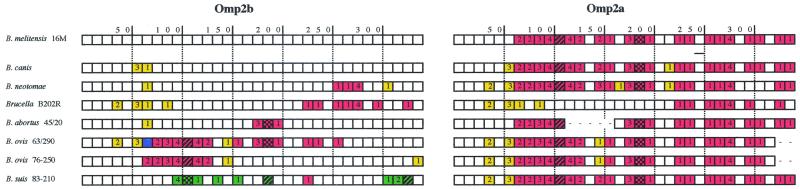
Presence of omp2a motifs in omp2b variants extends further the region implicated in size variation. Moreover, omp2b genes from other Brucella strains, present in the databases, also bear omp2a motifs in their sequence. This is summarized in Fig. 4 with a schematic representation of the deduced amino acid sequences, including the corresponding Omp2a amino acid sequences, which actually show very little sequence divergence with reference Omp2a. This schematic alignment shows that Omp2b of B. ovis 63/290 is almost similar to a typical Omp2a protein as previously described (17), whereas Omp2b of B. abortus 45/20 and B. ovis 76-250, although mostly Omp2b-like, show Omp2a-specific motifs on their sequences. Thus, the B. abortus 45/20, B. ovis 63/290, and B. ovis 76-250 Omp2b size variants appear to be natural chimeric proteins of the Omp2b and Omp2a pair previously characterized in B. melitensis 16M or in B. abortus S19 (25, 26).
FIG. 4.
Schematic representation of multiple amino acid sequence alignment of the Omp2b size variants along with their corresponding Omp2a. This alignment includes sequences not obtained in this study but found in sequence databases. Sequences are subdivided in stretches of 10 residues, numbered following the B. melitensis 16M Omp2b sequence. A change in color indicates the presence of substitutions (the number inside the box indicates the number of substitutions compared to Omp2b of B. melitensis 16M among the 10 residues). The red color always indicates identity with motifs specific for Omp2a. The yellow color indicates sequence motifs not present in reference Omp2b nor in reference Omp2a but conserved between variants that share this color. The blue and green colors indicate motifs specific, respectively, of B. ovis 63/290 Omp2b and B. suis 83-210 Omp2b. Hatching in the box symbolizes a deletion; double hatching symbolizes an insertion, always compared to the Omp2b sequence of B. melitensis 16M. This figure clearly shows that motif exchanges between Omp2a and Omp2b are common among all Brucella strains, suggesting that gene conversion as a phenomenon occurred several times during Brucella evolution, thus explaining most of the Omp2b variation between strains, except the variation found in the B. suis 83-210 Omp2b porin. Interestingly, as shown in this figure, Omp2a of both B. ovis strains is truncated in its C-terminal end due to the presence of a stop codon, leading probably to a nonfunctional porin. B. abortus 45/20 Omp2a shows the same deletion as in B. abortus S19 Omp2a characterized by others (15, 16, 17, 25, 26).
Topological analysis of Omp2b size variants.
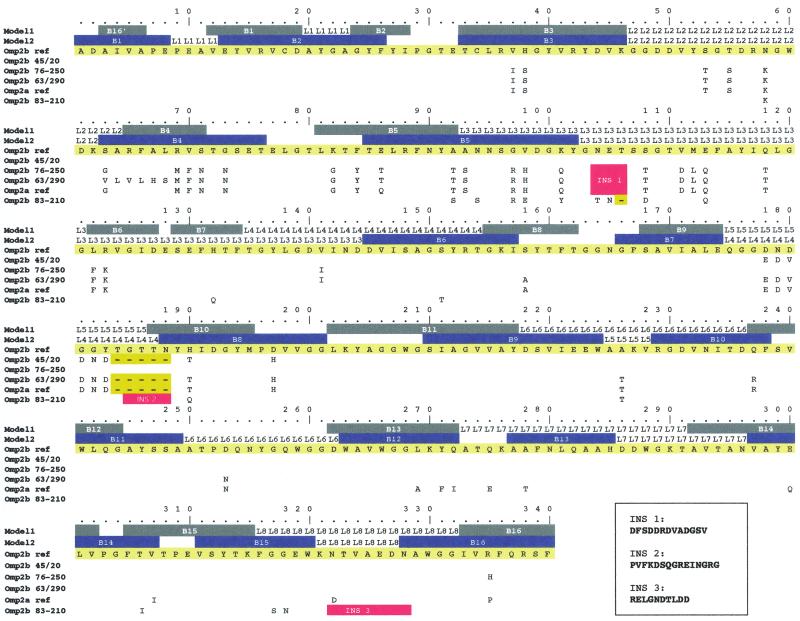
In Fig. 5, the locations of the insertions, deletions, and substitutions between Omp2b variants and Omp2a have been shown on the Omp2b topology model (31). Substitutions were also positioned on another topology model for Omp2b, which was based on alternative prediction methods and published by Mobasheri et al. (26). The main substitution positions on both models are the following.
FIG. 5.
Topology of the Omp2b size variants. The Omp2b sequence of B. abortus S19 is presented in yellow as a reference sequence and is numbered according to the mature protein. The first two lines present two different Omp2b topology models (model 1 is from Paquet et al. [31] and model 2 is from Mobasheri et al. [26]). Colored boxes represent predicted transmembrane β-strands. L, segments that symbolize surface-exposed loops numbered from 1 to 8, the L3 segment being the porin predicted constriction loop. Each substitution for all the sequences examined is figured (the conserved residues are not shown). Deletions are symbolized by yellow “-” boxes. Insertions are detailed beneath the figure and are indicated on the topology as red boxes.
In the Omp2b size variants, numerous amino acid substitutions (not deletions or insertions) occur in the region between residues 60 and 100. On both models, this region includes the B4 and B5 β-strands, as well as the predicted T3/T2 periplasmic turns. A 13-residue insertion, substituting an N-E-T motif at position 103 in the reference Omp2b, is present in Omp2a and in Omp2b from B. ovis 63/290 and B. ovis 76-250. Both topology models locate this insertion at the beginning of the L3 loop. In all porins of known structure, the L3 loop is folded into the β barrel, constricting the pore, and is thus a crucial determinant of the porin pore-forming characteristics. Another variable region in Brucella Omp2 porins is located between residues 175 and 190. Both topology models locate this variable region in a surface-exposed loop, although the numbering of this loop varies according to the model (L4 loop in Mobashery's model, L5 loop in our study). On this loop, reference Omp2a and Omp2b of B. abortus 45/20 and Omp2b of B. ovis 63/290 all show a deletion along with six residue substitutions, compared to reference Omp2b. In the case of B. abortus 45/20, this is the only “Omp2a-specific” signature, whereas Omp2b of B. ovis 63/290 presents other Omp2a-specific motifs in its N-terminal part. The B. suis 83-210 Omp2b porin shows a 15-residue insertion at position 184, modifying drastically the length and physicochemical properties of the L4/L5 loop in this porin. Omp2b of B. suis 83-210 shows also a large replacement of a part of the L8 loop, leading to an addition of negatively charged residues. This replacement does not affect the C-terminal transmembrane strand.
In summary, the major sequence differences (insertion and/or deletion) between the Omp2b size variants, reference Omp2a, and reference Omp2b all occur in external loops, but minor substitutions are distributed between residues 35 and the C terminus.
Antigenic analysis and partial epitope mapping of Omp2b size variants.
Reactivity of anti-Omp2b MAbs against total cellular protein extract of B. abortus S19, B. abortus 45/20, B. suis 83-210, B. ovis 63/290, and B. ovis 76-250 was assessed by Western blot and ELISA (Table 1). Only Omp2b is supposed to be produced in Brucella, the omp2a gene being only expressed when the intergenic region is artificially reversed (15). Both MAbs A68/25G05/A05 and A68/15B06/C08 recognize only folded Omp2b and react thus only in the ELISA, and their reactivity is lost in the Western blot. Conversely, some MAbs can only recognize Omp2b in denatured extracts (Western blot).
TABLE 1.
Reactivity of the MAbs to the Brucella Omp2b porin size variants
| MAb | Western blot results after SDS-PAGE with:
|
ELISA resultsa (mean absorbance ± SD) with:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
B. abortus
|
B. suis 83-210 |
B. ovis
|
B. abortus
|
B. suis 83-210 |
B. ovis
|
|||||
| S19 | 45/20 | 63/290 | 76-250 | S19 | 45/20 | 63/290 | 76-250 | |||
| A63/04D11/G01 | + | + | − | + | + | 0.97 ± 0.04 | 1.08 ± 0.05 | 0.26 ± 0.1 | 0.27 ± 0.07 | 0.18 ± 0.01 |
| A63/05A07/A08 | + | + | + | + | + | 0.16 ± 0.01 | 0.20 ± 0.01 | 0.11 ± 0.01 | 0.02 ± 0.01 | 0.05 ± 0.01 |
| A63/08D08/C07 | + | + | + | + | + | 0.49 ± 0.01 | 0.54 ± 0.01 | 0.38 ± 0.01 | 0.09 ± 0.01 | 0.10 ± 0.01 |
| A63/11E05/D11 | + | + | + | − | − | 0.29 ± 0.02 | 0.45 ± 0.01 | 0.27 ± 0.02 | 0.04 ± 0.01 | 0.08 ± 0.11 |
| A63/13G02/C04 | + | + | − | + | + | 0.75 ± 0.02 | 1.13 ± 0.19 | 0.16 ± 0.10 | 0.29 ± 0.07 | 0.19 ± 0.01 |
| A68/15B06/C08 | − | − | − | − | − | 2.01 ± 0.02 | 2.43 ± 0.01 | 0.11 ± 0.01 | 1.91 ± 0.25 | 2.39 ± 0.01 |
| A68/25G05/A05 | − | − | − | − | − | 2.37 ± 0.01 | 2.48 ± 0.01 | 0.27 ± 0.01 | 0.34 ± 0.02 | 2.40 ± 0.01 |
ELISA was done on sonicated bacteria. Absorbance values (mean ± the standard deviations) MAbs at one-half dilution are shown.
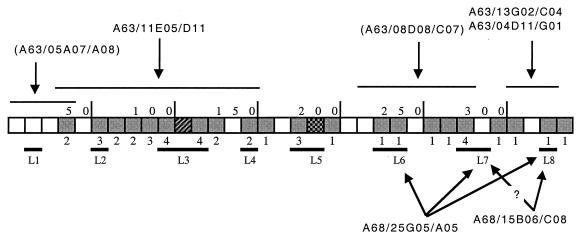
Loss of MAb reactivity correlated with sequence variability. The most probable locations on the Omp2b sequence of the epitopes recognized by MAbs A63/11E05/D11, A63/13G02/C04, A63/04D11/G01, A68/25G05/A05, and A68/15B06/C08 are shown in Fig. 6. The epitope recognized by MAb A63/11E05/D11 has been located between residues 30 and 150 because this is where the two B. ovis strains diverge from all the other strains, and they are not recognized by the antibody. The epitopes recognized by MAbs A63/04D11/G01 and A63/13G02/C04 are probably located in the only significant region of variation between B. suis 83-210 Omp2b and all other Brucella porins, i.e., the L8 loop.
FIG. 6.
Localization of Omp2b epitopes recognized by the MAbs which present differences of reactivity between the size variants. The boxes shaded in gray show places where substitutions, deletions, or insertions between Omp2b and Omp2a occur (the number of the substitution is indicated beneath the box). The external loops of the predicted topology are also indicated. The linear epitope recognized by the two MAbs between brackets have been identified with truncated Omp2b in another study (11).
MAbs A68/25G05/A05 and A68/15B06/C08 recognize nonlinear surface-exposed epitopes (5). Size variant porins are chimeric porins between Omp2b (recognized by both MAbs [30]) and Omp2a (not recognized by any of these two MAbs [30]). B. ovis 76-250 and B. abortus 45/20 are recognized by both MAbs, which indicates that the epitopes are located in the C-terminal loops of the protein. B. ovis 63/290 is not recognized by MAb A68/25G05/A05, which indicates that region 220 to 260, with only 3 substitions between Omp2b and Omp2a-type porin is important for the recognition by this antibody. This region encompasses the predicted L6 and L7 external loops. It is also probable that the L8 loop is implicated in the A68/25G05/A05-recognized epitope, because B. suis 83-210 is not recognized by this antibody. This indicated that several surface loops are probably involved in this nonlinear epitope.
The A68/15B06/C08 binding site probably does not include the L6 loop since this MAb does recognize B. ovis 63/290. From the fact that B. suis 83-210 is not recognized by MAb A68/15B06/C08, we can deduce that the epitope recognized by this antibody includes the L8 external loop. We cannot exclude the involvement of loop L7 since reference Omp2a is not recognized and shows only a few substitutions in the L8 loop. Thus, for these latter two MAbs, it should be noted that the true epitope may also include other parts of the protein because if several loops form the conformational epitope, the modification of just one of these loops could lead to the absence of this epitope.
Purification and refolding of Omp2b size variants.
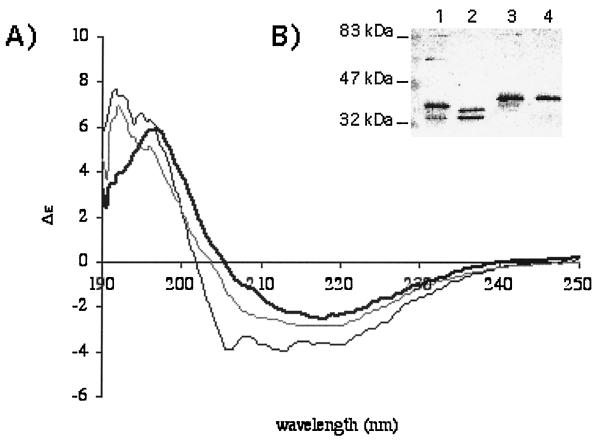
In order to prepare large amounts of purified porins, Omp2b size variants from the following strains were produced in the T7 expression system in E. coli: B. abortus 45/20, B. ovis 76-250, and B. suis 83-210, along with Omp2b and Omp2a from B. melitensis 16M. All porin size variants were successfully purified, with a final yield of 1 to 4 mg of pure protein by 100 ml of culture. The purity of the protein sample was greater than 95% as determined by SDS-PAGE. Contamination with native E. coli porin was excluded by a Western blot experiment realized with the purified porin and chicken egg polyclonal antibodies directed against total extract of the strain BL21(DE3) (data not shown). This is understandable because recombinant Omp2b is produced without signal peptide and is accumulated in inclusion bodies instead of being exported to the outer membrane along with the endogenous E. coli porins. The B. melitensis 16M reference porins were refolded into a native trimeric form as shown by circular dichroism spectroscopy (Fig. 7A), analysis of antigenic properties (30), and quaternary structure (Fig. 7B). Omp2b size variants were also refolded at least partly as native trimers (Fig. 7B). The only exception was the B. suis 83-210 Omp2b porin, which was never observed in an oligomeric form in our experiment; data on functionnal properties must be taken with caution for this particular variant. The Omp2b refolded trimers are recognized by both A68/15B06/C08 and A68/25G05/A05 MAbs, suggesting that conformational epitopes were recovered by the refolded porins (30).
FIG. 7.
(A) Circular dichroim spectroscopy of OmpF, the archetype of porins purified from E. coli (bold line), refolded B. melitensis 16M Omp2b (gray line), or Omp2a (black line). All three proteins show a very high content of β-strands, although Omp2a is comparatively richer in the α-helix component. These spectra suggest a correct recovery of secondary structure for the refolded Brucella porins. (B) Western blot with purified Brucella porins (lane 1, B melitensis 16M Omp2b; lane 2, B. abortus 45/20 Omp2b; lane 3, B. ovis 76-250 Omp2b; lane 4, B. suis 83-210 Omp2b) and detection with a mix of MAbs directed against Omp2b. Purified trimers are unstable in the presence of SDS, and several quaternary states are visible: trimers at about 90 kDa, dimers, and several forms of monomers. The refolded Omp2b size variants, with the exception of that of B. suis 83–210, show a pattern with trimeric state, suggesting that correct refolding also occurred for the variant porins, although quantification was not possible with this experiment.
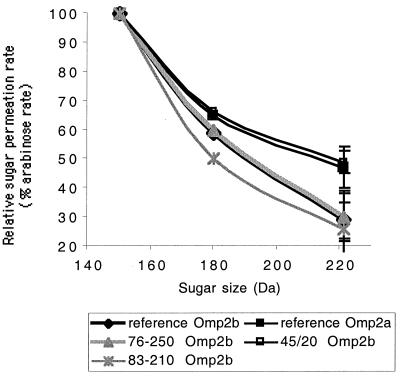
Differential sugar permeation of Omp2b size variants.
Purified recombinant Omp2b size variants and reference porins were tested for their permeability toward small sugars (Fig. 8). An ANOVA2 model was used for statistical analysis. Permeation rates for glucose and N-acetylglucosamine relative to arabinose were significantly lower for B. suis 83-210 Omp2b than for reference B. melitensis Omp2b (Fobs = 6.2908, P < 0.0005). Omp2b of B. ovis 76-250 showed no strong modification of its permeation rate compared to the reference Omp2b porin, whereas Omp2b of B. abortus 45/20 had a significantly higher permeation rate for N-acetylglucosamine (Fobs = 17.0087, P < 0.0005). This points out the crucial role of the 170 to 200 region, which is the only region of divergence between the Omp2b of B. abortus 45/20 and the other Omp2b porins, in the determination of pore characteristics. This 170 to 200 region is subject to a small deletion in the B. abortus 45/20 Omp2b porin as in the reference Omp2a porin, this deletion being located in the L5 loop in our topology model. The L5 loop of B. abortus 45/20 Omp2b and reference Omp2a is shorter by five residues but still richer in negatively charged residues (three Asps in Omp2a against two Asps in Omp2b) than the L5 loop of reference Omp2b.
FIG. 8.
Comparison of sugar permeability of the Omp2b size variants with the reference Omp2b and Omp2a porins of B. melitensis 16M. The graph shows the relative permeation rate of glucose (180 Da) and N-acetylglucosamine (221 Da) compared to arabinose (150 Da) rate, taken as 100%, for all purified variants. The experiments were conducted in quintuplate for every point, and standard deviations are shown for the 221-Da point.
DISCUSSION
Omp2b porin size variants, identified by PCR-RFLP and Western blot analysis, have been cloned and sequenced in order to understand the mechanisms by which such a polymorphism occurs. Furthermore, the effect of the variation on porin structure, function, and antigenicity has been assessed.
The nature of the polymorphism in this group of Omp2b size variants is of particular interest. It is neither due to free sequence variation, as observed for other porins such as the PorA and PorB porins from Neisseria spp. (10, 18), nor to horizontal transfers of sequence motifs from one strain to another as observed in Neisseria meningitidis porins (14). Variability in Brucella porins seems to be mainly the result of a variable assembly of defined sequence motifs found in the two omp2 porin gene copies. The peculiar genomic organization of the omp2 locus has probably allowed for genetic conversions between omp2b and omp2a, as has already been proposed for B. ovis 63/290 and B. neotomae 5K33 (17). The proposed mechanisms would consist of a correction of part of the omp2b coding sequence using omp2a as template. The present work shows that such conversions may have occurred at several distinct moments in the evolution of the genus Brucella, resulting in the observed size variations of the genes.
In terms of evolution, the omp2 locus recently reported from a Brucella strain (B202R) isolated from a Minke whale is of particular interest (4). The Omp2b sequence from this strain shows first an “Omp2a-type” N-terminal part, then a long “Omp2b-type” sequence, and then again an “Omp2a-type” C terminus (Fig. 4). Also, its Omp2a sequence is almost identical to this composite Omp2b sequence, which makes this locus a good candidate to represent the “primitive” omp2 locus.
Only in Omp2b of B. suis 83-210 is the size variation truly the result of mere sequence variability. The 15- and 10-residue insertions in its Omp2b loops L5 and L8 are not found in its Omp2a sequence, suggesting that no gene conversion occurred in this strain.
The most important sequence divergences observed between Brucella porins are two insertions and/or deletions located in the beginning of the loops L3 and L5 by the topology models. In the Neisseria PorB porin, sequence variability is also found in external loops but not in loop L3, the pore-constricting loop, as suggested by homology modeling (10). Neisseria porin variants are thus supposed to differ highly in antigenicity but in pore-forming activity (10). In contrast, as shown in this study Brucella porins appear to be not truly variable in antigenicity, since their variation is mostly due to exchanges of only two different types of motifs.
Comparison of MAb reactivity patterns of the Omp2b porin size variants and their sequences allowed us to partially map the epitopes for five of the MAbs. A partial epitope mapping has been previously realized using Omp2b truncations (11), and our results are in agreement with this approach. The results obtained with the truncated proteins showed, in addition to the data presented here, that MAbs A63/13G02/C04 and A63/04D11/G01 recognize two different epitopes in the C-terminal part of Omp2b. Due to the misfolded nature of the truncated protein, this approach failed to identify nonlinear epitopes recognized by MAbs A68/25G05/A05 and A68/15B06/C08. In contrast, the use of natural chimeric porins proved to be useful in determining which parts of the protein are implicated in these nonlinear surface-exposed epitopes, namely, the predicted loops L6, L7, and L8. The strong binding of MAbs A68/25G05/A05 and A68/15B06/C08 to the Brucella cell surface (1, 5) suggests a strong surface exposure of these loops, which is in agreement with the proposed role in bacterial adhesion of the RGD-containing L6 loop (3).
Comparison of pore-forming activity of the Omp2b size variants led to interesting results concerning the structural basis of Omp2a and Omp2b difference in sugar permeability, even if caution must be taken while interpreting results obtained with refolded porins, which probably need confirmation with native porins. Because of the crucial role in pore-forming activity played by the constriction L3 loop in most porins (21), it has been hypothetically proposed that the strongly modified and notably more negatively charged L3 loop of Omp2a would be responsible for a change in the permeability of the porin (31). Unexpectedly, Omp2b of B. ovis 76-250, a chimeric Omp2b with an Omp2a-type L3 loop, did not show any increase in sugar permeability. A sugar permeability comparable to Omp2a permeability was observed with Omp2b of B. abortus 45/20, which is identical to reference Omp2b but with an Omp2a-type L5 loop, which actually makes this Omp2b size variant the shortest known Omp2b porin. The L5 loop appears thus to be a critical determinant in Brucella porin sugar permeability. This is confirmed by the lower permeability of Omp2b of B. suis 83-210, which has a large insertion in the L5 loop, although in this case a large insertion also occurred in the L8 loop. The L5 loop could possibly participate in the formation of the pore “external mouth,” which serves to prescreen the solute in porins of known structure (8, 21). The small deletion present in Omp2a and Omp2b of B. abortus 45/20 could possibly enlarge this external mouth of the pore.
It must be stressed that the results obtained here on sugar permeability do not preclude the possibility that the L3 loop could have a strong influence on ion movements through the Brucella porins. The influence of charged residues of the L3 and L5 loops on ion conductance and ion selectivity remains to be elucidated. On the other hand, the constriction of the pore may rather be exerted by the L5, instead of the L3 loop, in Brucella porins.
The question remains as to why the omp2b gene has sometimes been corrected with the omp2a template during Brucella evolution. Has sugar permeability modification provided a selective advantage to some Brucella strains? It must be stressed that Brucella membrane permeability is probably not solely dependent on Omp2b and Omp2a permeability, but that other potentially strain-specific factors, such as other outer membrane components (34, 39), could be implicated as well. Since the expression of omp2a has never been observed, the conservation of this porin gene copy in all Brucella strains remains also to be explained. Comparisons between Brucella strains of omp2a and omp2b gene expression patterns during the infectious cycle and the related variation in outer membrane permeability constitute exciting new questions to be resolved.
ACKNOWLEDGMENTS
J.-Y. Paquet and S. Genevrois were recipients of a fellowship from the Fond pour la Formation à la Recherche dans l'Industrie et l'Agriculture (FRIA, Brussels, Belgium).
REFERENCES
- 1.Bowden R A, Cloeckaert A, Zygmunt M S, Bernard S, Dubray G. Surface exposure of outer membrane protein and lipopolysaccharide epitopes in Brucella species studied by enzyme-linked immunosorbent assay and flow cytometry. Infect Immun. 1995;63:3945–3952. doi: 10.1128/iai.63.10.3945-3952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchanan S K. β-Barrel proteins from bacterial outer membranes: structure, function and refolding. Curr Opin Struct Biol. 1999;9:455–461. doi: 10.1016/S0959-440X(99)80064-5. [DOI] [PubMed] [Google Scholar]
- 3.Campbell G A, Adams L G, Sowa B A. Mechanisms of binding of Brucella abortus to mononuclear phagocytes from cows naturally resistant or susceptible to brucellosis. Vet Immunol Immunopathol. 1994;41:295–306. doi: 10.1016/0165-2427(94)90103-1. [DOI] [PubMed] [Google Scholar]
- 4.Clavareau C, Wellemans V, Walravens K, Tryland M, Verger J M, Grayon M, Cloeckaert A, Letesson J J, Godfroid J. Phenotypic and molecular characterization of a Brucella strain isolated from a minke whale (Balaenoptera acutorostrata) Microbiology. 1998;144:3267–3273. doi: 10.1099/00221287-144-12-3267. [DOI] [PubMed] [Google Scholar]
- 5.Cloeckaert A, de Wergifosse P, Dubray G, Limet J N. Identification of seven surface-exposed Brucella outer membrane proteins by use of monoclonal antibodies: immunogold labeling for electron microscopy and enzyme-linked immunosorbent assay. Infect Immun. 1990;58:3980–3987. doi: 10.1128/iai.58.12.3980-3987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloeckaert A, Verger J M, Grayon M, Grépinet O. Restriction site polymorphism of the genes encoding the major 25 kDa and 36 kDa outer-membrane proteins of Brucella. Microbiology. 1995;141:2111–2121. doi: 10.1099/13500872-141-9-2111. [DOI] [PubMed] [Google Scholar]
- 7.Cloeckaert A, Verger J M, Grayon M, Vizcaino N. Molecular and immunological characterization of the major outer membrane proteins of Brucella. FEMS Microbiol Lett. 1996;145:1–8. doi: 10.1111/j.1574-6968.1996.tb08547.x. [DOI] [PubMed] [Google Scholar]
- 8.Cowan S W, Schirmer T, Rummel G, Steirt M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Crystal structure explains functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 9.Depiereux E, Feytmans E. MATCH-BOX: a fundamentally new algorithm for the simultaneous alignment of several protein sequences. Comput Appl Biosci. 1992;8:501–509. doi: 10.1093/bioinformatics/8.5.501. [DOI] [PubMed] [Google Scholar]
- 10.Derrick J P, Urwin R, Suker J, Feavers I M, Maiden M C. Structural and evolutionary inference from molecular variation in Neisseria porins. Infect Immun. 1999;67:2406–2413. doi: 10.1128/iai.67.5.2406-2413.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Wergifosse P. Analyse génétique et immunologique de deux protéines de la membrane externe de Brucella abortus: l'Omp25 et l'Omp36. Ph.D. thesis. Louvain, Belgium: Catholic University of Louvain; 1992. [Google Scholar]
- 12.Douglas J T, Rosenberg E Y, Nikaido H, Verstreate D R, Winter A J. Porins of Brucella species. Infect Immun. 1984;44:16–21. doi: 10.1128/iai.44.1.16-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans L J, Cooper A, Lakey J H. Direct measurement of the association of a protein with a family of membrane receptors. J Mol Biol. 1996;255:559–563. doi: 10.1006/jmbi.1996.0047. [DOI] [PubMed] [Google Scholar]
- 14.Feavers I M, Heath A B, Bygraves J A, Maiden M C. Role of horizontal genetic exchange in the antigenic variation of the class 1 outer membrane protein of Neisseria meningitidis. Mol Microbiol. 1992;6:489–495. doi: 10.1111/j.1365-2958.1992.tb01493.x. [DOI] [PubMed] [Google Scholar]
- 15.Ficht T A, Bearden S W, Sowa B A, Adams L G. DNA sequence and expression of the 36-kilodalton outer membrane protein gene of Brucella abortus. Infect Immun. 1989;57:3281–3291. doi: 10.1128/iai.57.11.3281-3291.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ficht T A, Bearden S W, Sowa B A, Marquis H. Genetic variation at the omp2 porin locus of the brucellae: species-specific markers. Mol Microbiol. 1990;4:1135–1142. doi: 10.1111/j.1365-2958.1990.tb00688.x. [DOI] [PubMed] [Google Scholar]
- 17.Ficht T A, Husseinen H S, Derr J, Bearden S W. Species-specific sequences at the omp2 locus of Brucella type strains. Int J Syst Bacteriol. 1996;46:329–331. doi: 10.1099/00207713-46-1-329. [DOI] [PubMed] [Google Scholar]
- 18.Fudyk T C, Maclean I W, Simonsen J N, Njagi E N, Kimani J, Brunham R C, Plummer F A. Genetic diversity and mosaicism at the por locus of Neisseria gonorrhoeae. J Bacteriol. 1999;181:5591–5599. doi: 10.1128/jb.181.18.5591-5599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georgeon C, Deleage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. CABIOS. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 20.Hobbs M M, Alcorn T M, Davis R H, Fischer W, Thomas J C, Martin I, Ison C, Sparling P F, Cohen M S. Molecular typing of Neisseria gonorrhoeae causing repeated infections: evolution of porin during passage within a community. J Infect Dis. 1999;179:371–381. doi: 10.1086/314608. [DOI] [PubMed] [Google Scholar]
- 21.Jap B K, Walian P J. Structure and functional mechanism of porins. Physiol Rev. 1996;76:1073–1088. doi: 10.1152/physrev.1996.76.4.1073. [DOI] [PubMed] [Google Scholar]
- 22.Jeanteur D, Schirmer T, Fourel D, Simonet V, Rummel G, Widmer C, Rosenbusch J P, Pattus F, Pagès J M. Structural and fonctional alterations of a colicin-resistant mutant of OmpF porin from Escherichia coli. Proc Natl Acad Sci USA. 1994;91:10675–10679. doi: 10.1073/pnas.91.22.10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King R D, Sternberg J E. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci. 1996;5:2298–2310. doi: 10.1002/pro.5560051116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klebba P E, Newton S M. Mechanisms of solute transport through outer membrane porins: burning down the house. Curr Opin Microbiol. 1998;1:238–247. doi: 10.1016/s1369-5274(98)80017-9. [DOI] [PubMed] [Google Scholar]
- 25.Marquis H, Ficht T A. The omp2 gene locus of Brucella abortus encodes two homologous outer membrane proteins with properties characteristic of bacterial porins. Infect Immun. 1993;61:3785–3790. doi: 10.1128/iai.61.9.3785-3790.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mobasheri H, Ficht T A, Marquis H, Lea E J, Lakey J H. Brucella Omp2a and Omp2b porins: single channel measurements and topology prediction. FEMS Microbiol Lett. 1997;155:23–30. doi: 10.1111/j.1574-6968.1997.tb12681.x. [DOI] [PubMed] [Google Scholar]
- 27.Mortimer P G, Piddock L J. The accumulation of five antibacterial agents in porin-deficient mutants of Escherichia coli. J Antimicrob Chemother. 1993;32:195–213. doi: 10.1093/jac/32.2.195. [DOI] [PubMed] [Google Scholar]
- 28.Nikaido H, Liu W, Rosenberg E Y. Outer membrane permeability and beta-lactamase stability of dipolar ionic cephalosporins containing methoxyimino substituents. Antimicrob Agents Chemother. 1990;34:337–342. doi: 10.1128/aac.34.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nogueras M M, Merino S, Aguilar A, Benedi V J, Tomas J M. Cloning, sequencing, and role in serum susceptibility of porin II from mesophilic Aeromonas hydrophila. Infect Immun. 2000;68:1849–1854. doi: 10.1128/iai.68.4.1849-1854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paquet J Y, De Bolle X, Mertens P, Genevrois S, Tibor A, Depireux E, Lakey J H, Letesson J J. Antigenic properties of Brucella Omp2a and Omp2b porins purified from recombinant Escherichia coli and renatured in vitro. Arch Physiol Biochem. 1999;107:B20. [Google Scholar]
- 31.Paquet J Y, Vinals C, Wouters J, Letesson J J, Depiereux E. Topology prediction of Brucella abortus Omp2b and Omp2a porins after critical assessment of transmembrane beta strands prediction by several secondary structure prediction methods. J Biomol Struct Dyn. 2000;17:747–757. doi: 10.1080/07391102.2000.10506564. [DOI] [PubMed] [Google Scholar]
- 32.Qi H L, Tai J Y, Blake M S. Expression of large amounts of neisserial porin proteins in Escherichia coli and refolding of the proteins into native trimers. Infect Immun. 1994;62:2432–2439. doi: 10.1128/iai.62.6.2432-2439.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 34.Samartzidou H, Delcour A H. Excretion of endogenous cadaverine leads to a decrease in porin-mediated outer membrane permeability. J Bacteriol. 1999;181:791–798. doi: 10.1128/jb.181.3.791-798.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schirmer T, Cowan S W. Prediction of membrane-spanning β-strands and its application to maltoporin. Protein Sci. 1993;2:1361–1363. doi: 10.1002/pro.5560020820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmid B, Krömer M, Schulz G E. Expression of porin from Rhodopseudomonas blastica in Escherichia coli inclusion bodies and folding into exact native structure. FEBS Lett. 1996;381:111–114. doi: 10.1016/0014-5793(96)00080-4. [DOI] [PubMed] [Google Scholar]
- 37.Singh S P, Singh S R, Williams Y U, Jones L, Abdullah T. Antigenic determinants of the OmpC porin from Salmonella typhimurium. Infect Immun. 1995;63:4600–4605. doi: 10.1128/iai.63.12.4600-4605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vizcaino N, Cloeckaert A, Zygmunt M S, Dubray G. Cloning, nucleotide sequence, and expression of the Brucella melitensis omp31 gene coding for an immunogenic major outer membrane protein. Infect Immun. 1996;64:3744–3751. doi: 10.1128/iai.64.9.3744-3751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]