Abstract
Purpose:
To clinically and molecularly study a newly found family with North Carolina macular dystrophy (NCMD/MCDR1) from Mexico.
Methods:
This retrospective study comprised 6 members of a 3-generation Mexican family with NCMD. Clinical ophthalmic examinations, including fundus imaging, spectral-domain optical coherence tomography, electroretinography, and electrooculography, were performed. Genotyping with polymorphic markers in the MCDR1 region was performed to determine haplotypes. Whole-genome sequencing (WGS) was performed followed by variant filtering and copy number variant analysis.
Results:
Four subjects from 3 generations were found to have macular abnormalities. The proband presented with lifelong bilateral vision impairment with bilaterally symmetric vitelliform Best disease-like appearing macular lesions. Her 2 children had bilateral large macular coloboma-like malformations, consistent with autosomal dominant NCMD. The 80-year-old mother of the proband had drusen-like lesions consistent with grade 1 NCMD. WGS and subsequent Sanger sequencing found a point mutation at chr6:99593030G>C (hg38) in the noncoding region of the DNase I site thought to be a regulatory element of the retinal transcription factor gene PRDM13. This mutation is the identical site/nucleotide as in the original NCMD family (#765) but is a guanine to cytosine change rather than a guanine to thymine mutation, as found in the original NCMD family.
Conclusions:
We report a new noncoding mutation at the same locus (chr6:99593030G>C) involving the same DNase I site regulating the retinal transcription factor gene PRDM13. This suggests that this site, chr6:99593030, is a mutational hotspot.
Keywords: North Carolina macular dystrophy, NCMD, MCDR1, PRDM13, mutational hotspot, whole-genome sequencing, copy number variant, DNase I site, chromosome 6, single nucleotide variants, SV, inherited retinal diseases
Introduction
North Carolina macular dystrophy (NCMD/MCDR1 [OMIM #136,550]) is an autosomal dominant, completely penetrant, congenital, nonprogressive macular malformation first reported in a large family in North Carolina as described by Lefler et al 50 years ago.1,2 The disease was initially and inappropriately named after the founder effect; that is, in the western part of North Carolina. 3 Although rare, NCMD has been found worldwide in more than 50 families in the United States, Europe, Central America, Australia, New Zealand, Korea, and China.4–19 Therefore, North Carolina macular dystrophy is a misleading misnomer.
One of the most striking clinical features of NCMD is the intrafamilial phenotypic variability and the relatively good vision despite severe-appearing macular malformations in some patients. Approximately one-third of individuals with NCMD have a few drusen centrally (grade 1); one-third have confluent drusen, some with subretinal fibrosis (grade 2); and another one-third have a choroidal coloboma-like excavation with an absence of retinal pigment epithelium and the presence of underlying choroidal lacunae and surrounding subretinal fibrosis (grade 3). Affected individuals have a particular grade of NCMD at birth and do not progress from one grade to the next, despite this disease being named dominant progressive foveal dystrophy 50 years ago. 20
There can be some vision decline secondary to the development of choroidal neovascular membranes (CNVMs). 21 Because fixation is along the nasal edge of the lesion and submacular fibrosis, presumably caused by CNVMs, typically occurs along the temporal edge, visual acuity (VA) is not affected. The nasal fixation also can cause a slight appearance of a positive angle kappa with exotropia. When CNVMs occur in grade 2 coloboma or along the nasal edge of grade 3 coloboma, vision loss can occur in cases of NCMD. Antivascular endothelial growth factor injections have been beneficial in these situations. 21
Phenocopies of NCMD include drusen of age-related macular degeneration (AMD), Stargardt macular dystrophy, Best vitelliform macular dystrophy (BVMD [OMIM #153700]), torpedo maculopathy, and toxoplasmosis.22,23 To diagnose NCMD clinically, it is imperative to examine additional family members to appreciate the full spectrum of the disease. Since 1990, Small et al 14 hypothesized that the causative gene(s) for NCMD are involved in embryonic macular development based on the congenital and nonprogressive clinical features. This would help explain why vision is relatively good considering the severe-appearing lesions present in many cases.11–16 Electroretinogram (ERG) and electrooculogram (EOG) findings have been reported in a few subjects and are typically normal, as is the color vision test. However, abnormal oscillatory potentials (OPs) have been reported in one case despite the lack of standardized normative values for OPs. 24 As expected, multifocal ERG recordings show significant amplitude reductions in the central maculae, but only in severely affected individuals. 25
In-depth genealogy studies by Small et al26,27 found that families were reported as having distinct clinical entities such as the “Lefler Wadsworth Sidbury syndrome”, “dominant progressive foveal dystrophy”, “central areolar pigment epithelial dystrophy”, and “central pigment and choroidal degeneration”; all were branches of the same family from North Carolina. Genetic linkage analysis by Small et al in this family showed a locus on chromosome 6q16 (NCMD/MCDR1 [OMIM #136550]).28–31 Most of the families from the US were found to have a shared haplotype across this region, suggesting a single mutation from a single founder for these ethnically diverse families. Subsequent linkage analysis of additional families yielded a logarithm of the odds ratio greater than 40, one of the highest recorded in human genetics.28–31
After exome sequencing was unsuccessful, targeted genomic sequencing of the 880 kb candidate region by Small et al eventually identified 5 mutations. 13 Three were single nucleotide variants (SNVs) in a noncoding region (MCDR1 locus). Two were large duplications, 1 on chromosome 6 (MCDR1 locus) and 1 on chromosome 5 (MCDR3 locus). The 3 noncoding SNVs in a DNase I hypersensitivity site were 12 kb from the nearest gene. These SNVs potentially alter regulation of expression of the neighboring gene encoding the PR/SET domain-containing zinc finger protein 13 (PRDM13 [OMIM #616741]). 13 Ellingford et al 32 later confirmed 1 of the SNVs (V2) in a small independent family with NCMD in the UK.
More recently, another unique noncoding SNV within the same DNase 1 site as in the original NCMD family, upstream of PRDM13, was reported. 33 This was found in a single, small, genetically isolated Georgian Jewish family with probable NCMD. However, diagnostic inconsistencies and molecular confounding factors with a complement factor H duplication were found in some possibly affected subjects. 33 This must be corroborated before definitive conclusions can be made.
In addition, Small et al 13 found a 123 kb duplication in the MCDR1 locus in a Mayan Belizean family involving the same upstream DNase I hypersensitivity site and the PRDM13 gene. Bowne et al 34 subsequently and independently reported a distinct large duplication involving the same DNase I site and PRDM13. Another unique duplication in the same genomic region was found by Manes et al 24 in a family from northern Italy. PRDM13 is expressed in the fetal retina and dorsal spinal columns and is not expressed in adult tissues. 35 Both the duplication in families with NCMD and overexpression experiments in Drosophila suggest that the malformation of the macula, including drusen, is the result of overexpression of PRDM13.13,35
Rosenberg et al 11 previously mapped a Danish family with the NCMD phenotype to a second locus on chromosome 5 (5p21, MCDR3), showing genetic heterogeneity. Using this positional information, a large duplication involving another DNase I site and the IRX1 gene was found by Small et al. 13 Subsequently, Cipriani et al 36 reported several European families with NCMD in which 2 different overlapping, smaller tandem duplications were located in a noncoding region 791 kb downstream of the IRX1 gene, involving the same DNase I site on chromosome 5p21.
A single family with nystagmus and severe congenital developmental anomaly/coloboma of the maculae, representing progressive bifocal chorioretinal atrophy (PBCRA [OMIM #600790]), was mapped to a large genomic region on chromosome 6 overlapping MCDR1 in 1996.37,38 Recently, Silva et al found in this family and in 2 additional families 2 distinct noncoding SNVs located in a different DNase I site 7.8 kb upstream of PRDM13.36,39 One of these 2 families had a phenotype more consistent with NCMD than with PBCRA because there was no progression and it was not “bifocal” with no chorioretinal atrophy nasal to the optic nerve.
Here, we report a new SNV at the identical noncoding site/nucleotide as in the original NCMD family (#765). As in the original family, this newly found mutation is at the MCDR1 chromosome 6 locus involving the same DNase I site. This family originated from Mexico and denies a genealogical connection with North Carolina or the US.
Methods
Clinical Assessment
Comprehensive ophthalmic examinations were performed on 7 family members of a 3-generation family originating from Mexico. A detailed family history was obtained and a pedigree developed as shown in Figure 1. The family denied any ancestral connections with North Carolina or the US in general. The family members examined lived in New York and Chicago and were examined in an office setting.
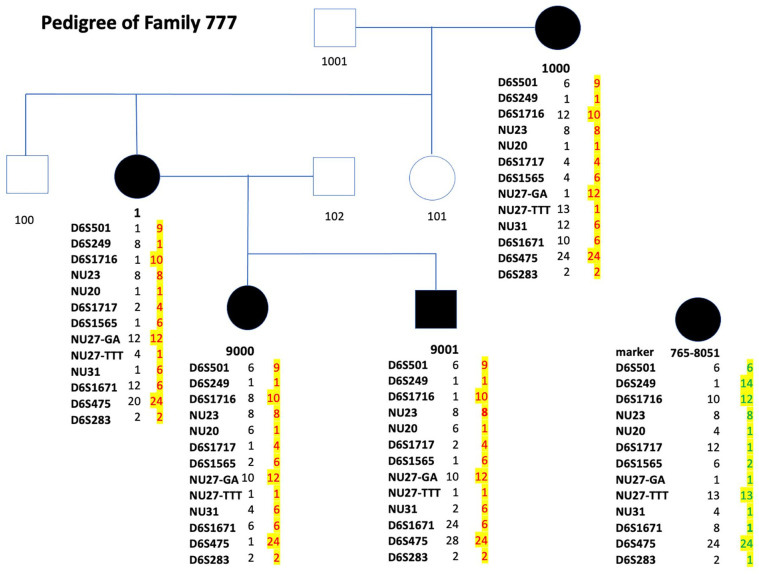
Figure 1.
Pedigree of family #777.Three-generation pedigree with affected individuals represented by black-filled symbols. The subject number is beneath the symbol. Subject #1 is the proband. Beneath the subject number are the numbered alleles for the polymorphic markers. The number of each allele was assigned to the base pair size of the allele. The base pair sizes of the alleles are available by request. The allele of the affected haplotypes that segregate with the disease in family #777 are in red type and highlighted in yellow. At the lower right side of the image is individual #765-8051. This affected individual is from the original NCMD family #765l; beneath #765-8051 is the haplotype of this original NCMD family. Note the affected haplotype in family #777 is different than the affected haplotype in the original NCMD family (#765), suggesting that the 2 families do not share a common founder. The order of the markers is centromere to D6S501, D6S249, D6S1716, NU23, NU20, D6S1717, D6S1565, NU27-GA, NU27-TTT, NU31, D6S1671, D6S475, and D6S283 to telomere. The D6S. . . numbers are polymorphic markers in the MCDR1 locus that are publicly available. The markers noted with “NU. . .” are proprietary markers developed in the authors’ laboratory and are available by request.
Abbreviation: NCMD, North Carolina macular dystrophy.
Fundus photographs were obtained using a Zeiss Visucam NM/FA and Optos California camera. Spectral-domain optical coherence tomography (SD-OCT) was performed using a Heidelberg Spectralis OCT device. Blood, saliva, or both were collected from consenting family members. Institutional review board approval (#94-07-241-21) was obtained from all participating individuals.
Genetic Assessment
DNA was extracted using standard methods and banked with pseudonymous identifiers. Whole-genome sequencing (WGS) was performed on DNA from 2 affected individuals (#1 and #9001) (pedigree shown in Figure 1) using standard protocols (NovoGene 6000, Illumina, Inc). 40 In short, after sample quality control (QC), sequencing libraries were prepared using the NEBNext DNA Library Prep Kit (New England Biolabs) according to manufacturer’s recommendations. Libraries were then sequenced using paired-end 2 × 125 bp format at >30× (Illumina, Inc). After adaptor and quality trimming, QC’d FASTQ files were used in alignment with Burrows-Wheeler Aligner (BWA) software to the reference human genome (hg38).40–44 At this stage, final binary alignment/map (BAM) files were obtained at the John P. Hussman Institute for Human Genomics and Pindel analyses were performed on the final BAM files using default settings to detect large structural variants (SVs).40–46
To exclude the presence of pathogenic variants in the genes known to be associated with inherited retinal diseases (IRDs), an analysis of the RetNet panel (https://sph.uth.edu/retnet) was performed on the WGS data of the 2 affected family members (#1 and #9000). Therefore, BWA was used for alignment to the reference human genome (hg38) and Alamut Batch was used for annotation of variants located in RetNet genes. Filters were applied based on population frequency (<0.02) and coding effect, and the remaining variants were eventually filtered based on associated phenotype so that only variants associated with a macular phenotype remained. For targeted testing of the known pathogenic variants (SNVs) implicated in NCMD (Table 1),13,24,32,33,36,39,47 polymerase chain reaction and Sanger sequencing was performed.
Table 1.
Known Genetic Defects in the (MCDR1) PRDM13 and (MCDR3) IRX1 Regions Found in NCMD and Possibly Related Diseases.
| Region/Variant Number | Type of Variant | Chromosomal Position (hg19) | Chromosomal Position (hg38) | Nucleotide Change | Phenotype | Reference a |
|---|---|---|---|---|---|---|
| MCDR1 locus(PRDM13),chr6q16V1 | SNV | chr6:100040906 | chr6:99593030 | G>T | NCMD | Small et al 13 |
| V2 | SNV | chr6:100040987 | chr6:99593111 | G>C | NCMD | Small et al 13 |
| V3 | SNV | chr6:100041040 | chr6:99593164 | C>T | NCMD | Small et al 13 |
| V4 | Tandem DUP | chr6:100020205-100143306 | chr6:99572329-99695430 | 123,101 bp DUP | NCMD | Small et al 13 |
| V6 | Tandem DUP | chr6:99996226-100065137 | chr6:99548350-99617261 | 69,912 bp DUP | NCMD | Bowne et al 33 |
| V7 | Tandem DUP | chr6:99984309-100082698 | chr6:99536433-99634822 | 98,389 bp DUP | NCMD | Manes et al 24 |
| V10 | SNV | chr6:100046804 | chr6:99598907 | T>C | PBCRA | Silva et al 39 |
| V11 | SNV | chr6:100046783 | chr6:99598928 | A>C | NCMD PBCRA |
Silva et al 39 |
| V12 | SNV b | chr6:100040974 | chr6:99593098 | A>C | Possible NCMD | Namburi et al 32 |
| V13 | Tandem DUP | chr6:100008141-100064368 | chr6:99560265-99616492 | 56,228 bp DUP | NCMD | Small 47 |
| V14 | SNV | chr6:100040906 | chr6:99593030 | G>C | NCMD | Current |
| MCDR3 locus(IRX1),chr5p21 | ||||||
| V5 | Tandem DUP | chr5:3587901-4486027 | chr5:3587787-4485914 | 898,126 bp DUP | NCMD | Small et al 13 |
| V8 | Tandem DUP | chr5:4391377–4436535 | chr5:4391264-4436422 | 45,158 bp DUP | NCMD | Cipriani et al 36 |
| V9 | Tandem DUP | chr5:4396927–4440442 | chr5:4396814-4440329 | 43,515 bp DUP | NCMD | Cipriani et al 36 |
Abbreviations: A, adenine; bp, base pair; C, cytosine; chr, chromosome; DUP, duplication; G, guanine; NCMD, North Carolina macular dystrophy; PBCRA, progressive bifocal chorioretinal atrophy; SNV, single nucleotide variation; T, thymine
First author.
This reported SNV is in a small family in an isolated population with diagnostic inconsistencies and molecular confounding factors and must be corroborated.
To determine the possible ancestral relationship with any of the other NCMD families ascertained by Small et al, haplotype analysis was performed by genotyping with polymorphic genetic markers in the MCDR1 region in this family and these were compared with those of the original NCMD family. 31 In this family as well as in the original family #765, genotyping was performed with 13 polymorphic markers spanning the MCDR1 locus. The polymorphic markers used were between D6S501 to D6S283. 31 (The primers are available on request.)
Results
Clinical Study
Table 2 shows a summary of the clinical status, sex, age at examination, and VA of the 7 members of the family living in New York and Chicago but originating from Mexico (Figure 1). The proband (#1) was a 41-year-old woman presenting with lifelong bilateral vision impairment. The Snellen VA was 20/40 OD and 20/40 OS. The anterior segments and the intraocular pressures were normal. Fundus assessment and SD-OCT showed symmetric, elevated, subfoveal yellow vitelliform-like lesions consistent with BVMD (Figure 2). In addition, an EOG showed an Arden ratio equal to 1.0 with a normal full-field ISEV ERG (following International Society for Clinical Electrophysiology of Vision standard protocol), consistent with BVMD. The initial diagnosis was BVMD.
Table 2.
Clinical Characteristics of the Individuals in Family #777.
| Individual # | Sex | Macular Dystrophy (Bilateral) | Age at Exam (Y) | Visual Acuity (Snellen) | |
|---|---|---|---|---|---|
| OD | OS | ||||
| 1000 | F | Affected grade 1 | 80 | 20/50 | 20/63 |
| 1 | F | Affected grade 2 | 41 | 20/40 | 20/40 |
| 101 | F | Unaffected | 54 | 20/20 | 20/20 |
| 102 | M | Unaffected | 54 | 20/40 | 20/63 |
| 9000 | F | Affected grade 3 | 15 | 20/50 | 20/63 |
| 9001 | M | Affected grade 3 | 12 | 20/70 | 20/80 |
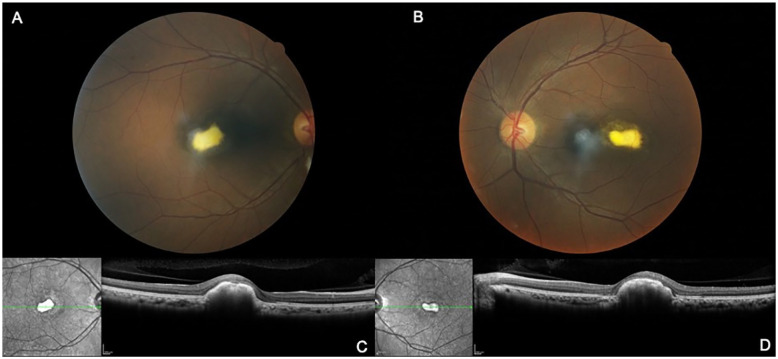
Figure 2.
Fundus and SD-OCT images of the proband (#1). Fundus photographs of the right eye (A) and left eye (B) and SD-OCT images of the right eye (C) and left eye (D) show focal and elevated white–yellow lesions with central dense material.
Abbreviation: SD-OCT, spectral-domain optical coherence tomography.
The woman’s 2 children (#9000 and #9001) were found to have bilaterally symmetric coloboma-like macular malformations consistent with NCMD grade 3 and a variable degree of decreased VA ranging from 20/50 to 80/80 (Figures 3 and 4). Their father was examined and found to be clinically unaffected. The proband’s mother (#1000) was found to have only macular drusen lesions consistent with NCMD grade 1 (Figure 5).
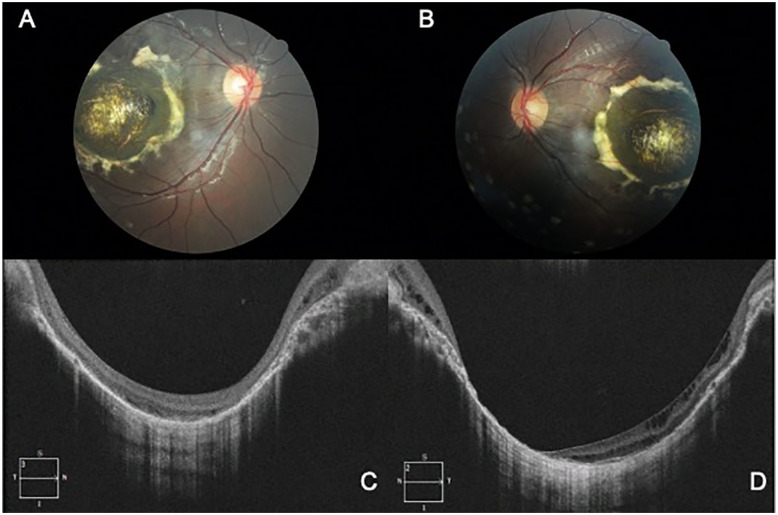
Figure 3.
Fundus and SD-OCT images of subject #9001, a 12-year-old son of the proband with grade 3 NCMD. Fundus photographs of the right eye (A) and left eye (B) show symmetric focal and elevated white–yellow lesions with central dense material. Spectral-domain optical coherence tomography of the right eye (C) and left eye (D) show a macular coloboma-like lesion.
Abbreviation: NCMD, North Carolina macular dystrophy.
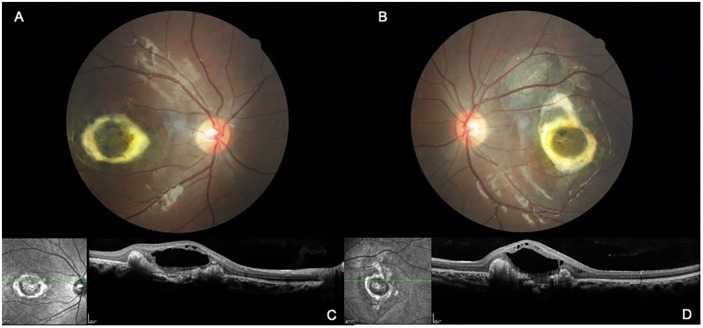
Figure 4.
Fundus and SD-OCT images of subject #9000, a 15-year-old daughter of the proband with grade 3 NCMD. Fundus photographs of the right eye (A) and left eye (B) show elevated dense material circumscribing the maculae and with intraretinal fluid. SD-OCT images of the right eye (C) and left eye (D) show serous elevation of the central macula.
Abbreviations: NCMD, North Carolina macular dystrophy; SD-OCT, spectral-domain optical coherence tomography.
Figure 5.
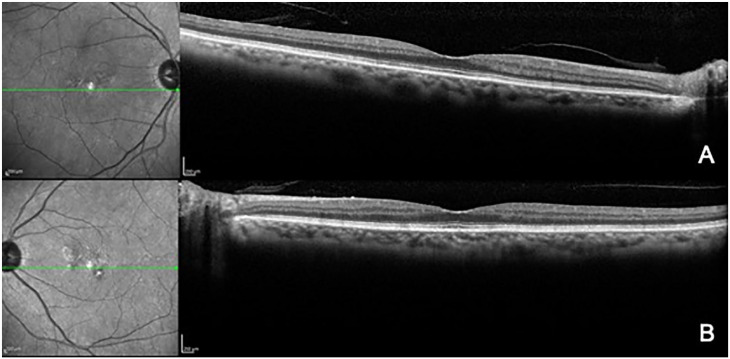
SD-OCT images of subject #1000, the 80-year-old mother of the proband with grade 1 NCMD. SD-OCT of the right eye (A) and left eye (B) show pigment irregularities.
Abbreviations: NCMD, North Carolina macular dystrophy; SD-OCT, spectral-domain optical coherence tomography.
Genetic Study
Initially, an outside laboratory tested the proband (#1) for known pathogenic variants previously found in NCMD; the reported the identical mutation (chr6:99593030G>T, V1) as the in original NCMD family (#765). This created suspicion that this Mexican family (#777) was genealogically related to the original NCMD family (#765).
Subsequently, while investigating the possible genealogical relationships of the 50 NCMD families being studied by Small et al (unpublished data), haplotype studies were initiated. Genotyping of the polymorphic markers in the MCDR1 locus of all of the V1 mutation families showed that this family (#777) did not share the same haplotypes as all the other families with the V1 mutation. This suggested that family #777 had a new but identical mutation as in the original NCMD family (#765) or that an error was present in the initial sequencing data analysis. Thus, WGS of subject #1 and subject #9001 was performed using 2 independent laboratories/collaborators. These 2 laboratories independently found the same mutation locus but with a new chr6:99593030 G>C (hg38) mutation rather than the V1 G>T mutation of the original NCMD family. Sanger sequencing in the remaining family members showed segregation of the mutation in all affected subjects. Finally, RetNet analysis on the available WGS data from the 2 affected family members (#1 and #9001) did not show other (likely) pathogenic variants that could explain the retinal phenotype in this family.
Conclusions
We clinically and genetically studied a 3-generation NCMD family that originated from Mexico, a previously unreported geographic region for NCMD. Because many NCMD families are found outside of North Carolina, the name of the disease is very misleading. Although the Mexican family in our study (#777) claimed no genealogical connection to North Carolina, our impression is that such negative family history information is rarely useful. Many families contacting us over the decades for consideration as being diagnosed with NCMD because they have a North Carolina family connection rarely have NCMD. A family history of a North Carolina association is generally confounding; thus, the name NCMD is a misnomer and should be changed.
Most families with NCMD studied to date have a considerable intrafamilial variable expressivity. Clinically, the Mexican family (#777) reported here has this variable expressivity despite only 4 affected subjects. Within this small family, the phenotypes and phenocopies range from drusen/AMD phenocopy, BVMD phenocopy, and toxoplasmosis phenocopy.23,47 Indeed, even an EOG of subject #1 was abnormal and was consistent with BVMD. This strongly suggests that EOG is not as helpful as we were taught in diagnosing BVMD, which is a phenocopy of NCMD. 48
Even more noteworthy is that the severity grade of the NCMD phenotype worsened with each successive generation within this family. If this were the only family with NCMD, a researcher might be tempted to claim genetic anticipation as a characteristic of NCMD. However, Small et al 49 evaluated the NCMD families for genetic anticipation 25 years ago and found no statistical evidence of it.
A new finding in this family regarding NCMD was the presence of a blister of submacular fluid that appeared exudative in nature. Subject #9000, a 10-year-old girl with 20/50 acuity, had macular lesions that on fundus photography appeared to be coloboma-like. However, the SD-OCT image showed little excavation into the choroid and more fibrotic elevation surrounding the maculae. In addition, this finding was bilaterally symmetric. More dramatically and never before described in NCMD is the blister of fluid that the fibrosis circumscribed. This submacular fluid was presumably exudative in nature and could have originated from the surrounding fibrosis, or less likely, from the choroid. Observing these lesions over time should add insight into whether this was a lesion that evolved and collapsed into the choroidal coloboma-like defect. It seems unlikely that a large choroidal excavation could develop after 10 years of age; however, it is conceivable that the submacular fluid could resolve, collapsing the elevation.
The initial molecular misdiagnosis was a result of misinterpreting the original automated sequencing spherograms. In the process of reviewing the haplotype data of all our families with NCMD, it became apparent that this Mexican family (#777) had a unique haplotype and a distinct founder from the original NCMD family #765. This suggests that family #777 had a new mutation that was identical in location to the originally reported SNV V1 chr6:99593030 G>T or that there was an error in the sequencing data. This caused us to evaluate the sequencing data more critically and to repeat sequencing with WGS and Sanger sequencing using 2 independent laboratories/collaborators. Both laboratories found the SNV chr6:99593030 G>C rather than the G>T. Therefore, this noncoding DNase I hypersensitivity site, chr6:99593030 (MCDR1), can have 2 different mutations (first one G>T and herein G>C), suggesting this is a mutational hotspot. 50
Reviewing the known mutations along with this newly found mutation might provide insight into common underlying mechanisms causing NCMD and possibly related diseases such as PBRCA.13,35–39 This brings the total number of distinct pathogenic variants to 11 for PRDM13 (MCDR1) (Table 1). Seven are unique noncoding SNVs located in 2 distinct DNase I sites, and 4 are unique tandem duplications overlapping the originally reported DNase I site and the PRDM13 gene.13,24,34,39 Thus far, a common feature of these variants, SNVs, and SVs, is that they involve DNase I sites. Interestingly, this is also the case for the 3 overlapping duplications on chromosome 5 of the IRX1 region (MCDR3 locus).13,33 One hypothesis is that these SNVs in these DNase I sites might represent a new type of SV. Although in general an SV is defined as a region of DNA approximately 1 kb and larger, one should consider that a single nucleotide polymorphism caused a conformational change in the chromatin, which could lead to altered expression of genes. These DNase I sites might become disproportionately uncoiled, exposing it to more frequent mutation pressures (ie, a mutational hotspot).
The variable expressivity of the severity of NCMD has yet to be fully understood. Disease severity might depend on the spatiotemporal expression of PRDM13 during development rather than on the PRDM13 dosage itself. This hypothesis is supported by the fact that PRDM13 is not expressed in the adult retina. 35 However, there continues to be no evidence to support a genotype–phenotype correlation.
Moreover, it is known that SVs such as duplications do not only affect gene dosage but can also affect gene regulation. They can change the copy number of regulatory elements or alter the 3-dimensional (3D) genome by disrupting higher-order chromatin organization, such as topologically associating domains (TADs).51,52 The impact of SVs on the 3D genome and on gene expression regulation has to be considered when interpreting the consequences of these variant types.51,52 Potential pathomechanisms resulting from an SV, such as the formation of neo-TADs, can become apparent by high-throughput chromosome conformation capture (Hi-C) generated from cultured patient cells in comparison with a wild-type reference Hi-C map.51,52
Access to relevant patient material is not possible for developmental retinal diseases. However, with the development of the induced pluripotent stem cell and retinal organoid reprogramming techniques, alternatives are becoming available. These could give insight into retinal development and how it is affected by patient-specific variants. At this point, however, cultured retinal organoids do not have a macula, which might prevent us from obtaining the full picture.
In general, an increasing number of coding and noncoding SNVs have been shown to underlie IRDs.53–58 The majority of the reported noncoding SNVs have an effect on splicing. However, only a handful of noncoding cis-regulatory variants have been identified in IRDs.54,55 NCMD can be considered a model for noncoding regulatory SNVs and SVs in IRDs. Future studies of patient-derived cells might help elucidate the underlying mechanisms of this cis-regulatory disease.
Footnotes
Ethical Approval: This study was performed in accordance with the Declaration of Helsinki. The collection and evaluation of all protected patient health information was performed in a Health Insurance Portability and Accountability Act (HIPAA)–compliant manner.
Statement of Informed Consent: Informed consent was obtained prior to performing the procedure, including permission for publication of all photographs and images included herein.
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article. Pending North Carolina Macular Dystrophy (NCMD/MCDR1) patent: Kent Small, MD.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Foundation Fighting Blindness (grant no. BR-GE-1216-0715-CSH) to K.S. E.D.B. is senior clinical investigator of the Research Foundation-Flanders (FWO) (1802220N) and S.V.S. is PhD fellow of the FWO (1145719N). E.D.B. is members of ERN-EYE which is co-funded by the Health Program of the European Union under the Framework Partnership Agreement No 739534 ‘ERN-EYE’.
ORCID iD: Kent W. Small  https://orcid.org/0000-0002-5204-4959
https://orcid.org/0000-0002-5204-4959
References
- 1. Lefler WH, Wadsworth JA, Sidbury JB. Hereditary macular degeneration and amino-aciduria. Am J Ophthalmol. 1971;71:224-230. [DOI] [PubMed] [Google Scholar]
- 2. Frank HR, Landers MB, Williams RJ, Sidbury JB. A new dominant progressive foveal dystrophy. Am J Ophthalmol. 1974;78(6):903-916. [DOI] [PubMed] [Google Scholar]
- 3. Agarwal A. Gass’ Atlas of Macular Diseases. 5th ed. Elsevier Saunders; 2012. [Google Scholar]
- 4. Pauleikhoff D, Sauer CG, Müller CR, et al. Clinical and genetic evidence for autosomal dominant North Carolina macular dystrophy in a German family. Am J Ophthalmol. 1997;124(3):412-415. [DOI] [PubMed] [Google Scholar]
- 5. Small KW, Puech B, Mullen L, Yelchits S. North Carolina macular dystrophy phenotype in France maps to the MCDR1 locus. Mol Vis. 1997;3:1. [PubMed] [Google Scholar]
- 6. Small KW, Garcia CA, Gallardo G, Udar N, Yelchits S. North Carolina macular dystrophy (MCDR1) in Texas. Retina. 1998;18(5):448-452. [PubMed] [Google Scholar]
- 7. Rohrschneider K, Blankenagel A, Kruse FE, Fendrich T, Völcker HE. Macular function testing in a German pedigree with North Carolina macular dystrophy. Retina. 1998;18(5):453-459. [DOI] [PubMed] [Google Scholar]
- 8. Rabb MF, Mullen L, Yelchits S, Udar N, Small KW. A North Carolina macular dystrophy phenotype in a Belizean family maps to the MCDR1 locus. Am J Ophthalmol. 1998;125(4):502-508. [DOI] [PubMed] [Google Scholar]
- 9. Reichel MB, Kelsell RE, Fan J, et al. Phenotype of a British North Carolina macular dystrophy family linked to chromosome 6q. Br J Ophthalmol. 1998;82(10):1162-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Michaelides M, Johnson S, Tekriwal AK, et al. An early-onset autosomal dominant macular dystrophy (MCDR3) resembling North Carolina macular dystrophy maps to chromosome 5. Invest Ophthalmol Vis Sci. 2003;44(5):2178-2183. [DOI] [PubMed] [Google Scholar]
- 11. Rosenberg T, Roos B, Johnsen T, et al. Clinical and genetic characterization of a Danish family with North Carolina macular dystrophy. Mol Vis. 2010;16:2659-2668. [PMC free article] [PubMed] [Google Scholar]
- 12. Kim SJ, Woo SJ, Yu HG. A Korean family with an early-onset autosomal dominant macular dystrophy resembling North Carolina macular dystrophy. Korean J Ophthalmol. 2006;20(4): 220-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Small KW, DeLuca AP, Whitmore SS, et al. North Carolina macular dystrophy is caused by dysregulation of the retinal transcription factor PRDM13. Ophthalmology. 2016;123(1):9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Small KW. North Carolina macular dystrophy: revisited. Ophthalmology. 1989;96(12):1747-1754. [DOI] [PubMed] [Google Scholar]
- 15. Rabb M, Small KW, Mullen L, Yelchits L, Udar N. North Carolina macular dystrophy maps to the MCDR1 locus (MCDR1) phenotype in Central America. Am J Ophthalmol. 1998;125(4):502-508. [DOI] [PubMed] [Google Scholar]
- 16. Small KW. North Carolina macular dystrophy. Trans Am Ophthalmol Soc. 1998;96:926-961. [PMC free article] [PubMed] [Google Scholar]
- 17. Voo I, Glasgow B, Flannery J, Udar U, Small KW. North Carolina macular dystrophy: clinicopathologic correlation. Am J Ophthalmol. 2001;132(6):933-935. [PubMed] [Google Scholar]
- 18. Small KW, Voo I, Glasgow B, Flannery J, Udar N. Clinicopathologic correlation of North Carolina macular dystrophy. Trans Am Ophthalmol Soc. 2001;99:233-238. [PMC free article] [PubMed] [Google Scholar]
- 19. Kiernan DF, Shah RS, Hariprasad SM, et al. Thirty year follow-up of an African-American family with macular dystrophy of the retina, locus 1 (North Carolina macular dystrophy) (MCDR1). Ophthalmology. 2011;118(7):1435-1443. [DOI] [PubMed] [Google Scholar]
- 20. Freund KB, Sarraf D, Mieler WF, Yannuzzi LA, Shields CL. The Retinal Atlas. Elsevier; 2017. [Google Scholar]
- 21. Bakall B, Bryan JS, Stone E, Small KW. Choroidal neovascularization in North Carolina macular dystrophy responsive to anti–vascular endothelial growth factor therapy. Retin Cases Brief Rep. 2021;15(5):509-513. [DOI] [PubMed] [Google Scholar]
- 22. Small K, Small L, Tran E, Rao R, Shaya F. Multimodal imaging and functional testing in a North Carolina macular disease family: toxoplasmosis, fovea plana, and torpedo maculopathy are phenocopies. Ophthalmol Retina. 2019;3(7):607-614. [DOI] [PubMed] [Google Scholar]
- 23. Small KW, Vincent A, Knapper CL, Shaya F. Congenital toxoplasmosis as one phenocopy of North Carolina Macular Dystrophy (NCMD/MCDR1). Am J Ophthalmol Case Rep. 2019;15:100521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manes G, Joly T, Smirnov S, et al. A novel duplication of PRMD13 causes North Carolina macular dystrophy: overexpression of PRDM13 orthologue in Drosophila eye reproduces the human phenotype. Hum Mol Genet. 2017;26(22):4367-4374. [DOI] [PubMed] [Google Scholar]
- 25. Seiple W, Szlyk JP, Paliga J, Rabb MF. Perifoveal function in patients with North Carolina macular dystrophy: the importance of accounting for fixation locus. Invest Ophthalmol Vis Sci. 2006;47(4):1703-1709. [DOI] [PubMed] [Google Scholar]
- 26. Small KW, Killian J, McLean W. North Carolina’s dominant progressive foveal dystrophy: how progressive is it? Br J Ophthalmol. 1991;75(7):401-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Small KW, Agemy S, Shaya FS. Terminology of MCDR1: what’s in a name? JAMA Ophthalmol. 2016;134(4):355-356. [DOI] [PubMed] [Google Scholar]
- 28. Small KW, Weber JL, Roses AD, et al. North Carolina macular dystrophy is assigned to chromosome 6. Genomics. 1992;13(3):681-685. [DOI] [PubMed] [Google Scholar]
- 29. Small KW, Weber J, Roses A, Pericak-Vance MA. North Carolina macular dystrophy (MCDR1). A review and refined mapping to 6q14-q16.2. Ophthalmic Paediatr Genet. 1993;14(4):143-150. [DOI] [PubMed] [Google Scholar]
- 30. Small KW, Weber JL, Hung WY, et al. North Carolina macular dystrophy: exclusion map using RFLPs and microsatellites. Genomics. 1991;11(3):763-766. [DOI] [PubMed] [Google Scholar]
- 31. Small KW, Udar N, Yelchits S, et al. North Carolina macular dystrophy (MCDR1) locus: a fine resolution genetic map and haplotype analysis. Mol Vis. 1999;5:38. [PubMed] [Google Scholar]
- 32. Ellingford JM, Sergouniotis PI, Jenkins E, Black GC. Genome sequencing identifies a non-coding variant in the MCDR1 locus as a cause of macular dystrophy. Clin Exp Ophthalmol. 2017;45(3):297-299. [DOI] [PubMed] [Google Scholar]
- 33. Namburi P, Khateb S, Meyer S, et al. A unique PRDM13-associated variant in a Georgian Jewish family with probable North Carolina macular dystrophy and the possible contribution of a unique CFH variant. Mol Vis. 2020;26:299-310. [PMC free article] [PubMed] [Google Scholar]
- 34. Bowne SJ, Sullivan LS, Wheaton DK, et al. North Carolina macular dystrophy (MCDR1) caused by a novel tandem duplication of the PRDM13 gene. Mol Vis. 2016;22:1239-1247. [PMC free article] [PubMed] [Google Scholar]
- 35. Watanabe S, Sanuki R, Sugita Y, et al. Prdm13 regulates subtype specification of retinal amacrine interneurons and modulates visual sensitivity. J Neurosci. 2015;35(20):8004-8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cipriani V, Silva RS, Arno G, et al. Duplication events downstream of IRX1 cause North Carolina macular dystrophy at the MCDR3 locus. Sci Rep. 2017;7(1):7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kelsell RE, Godley BF, Evans K, et al. Localization of the gene for progressive bifocal chorioretinal atrophy (PBCRA) to chromosome 6q. Hum Mol Genet. 1995;4(9):1653-1656. [DOI] [PubMed] [Google Scholar]
- 38. Godley BF, Tiffin PA, Evans K, et al. Clinical features of progressive bifocal chorioretinal atrophy: a retinal dystrophy linked to chromosome 6q. Ophthalmology. 1996;103(6):893–898. [DOI] [PubMed] [Google Scholar]
- 39. Silva RS, Arno G, Cipriani V, et al. Unique noncoding variants upstream of PRDM13 are associated with a spectrum of developmental retinal dystrophies including progressive bifocal chorioretinal atrophy. Hum Mutat. 2019;40(5):578–587. [DOI] [PubMed] [Google Scholar]
- 40. 1000 Genomes Project Consortium, Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen K, Wallis JW, McLellan MD, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6(9):677-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boeva V, Popova T, Bleakley K, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28(3):423-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Small KW, Van de Sompele S, Nuytemans K, et al. A novel duplication involving PRDM13 in a Turkish family supports its role in North Carolina macular dystrophy (NCMD/MCDR1). Mol Vis. 2021;27:518-527. [PMC free article] [PubMed] [Google Scholar]
- 48. Small KW, Jampol LM, Bakall B, et al. Best Vitelliform Macular Dystrophy (BVMD) is a phenocopy of North Carolina Macular Dystrophy (NCMD/MCDR1). Ophthalmic Genet. Published online December 13, 2021. doi: 10.1080/13816810.2021.2010771 [DOI] [PubMed] [Google Scholar]
- 49. Small KW: Genetics segregation distortion in MCDR1 (North Carolina macular dystrophy). Invest Ophthalmol Vis Sci. 1993; 34(suppl):1305. [Google Scholar]
- 50. Van de Sompele S, Small KW, Cicekdal MB, et al. Multi-omics profiling, in vitro and in vivo enhancer assays dissect the cis-regulatory mechanisms underlying North Carolina macular dystrophy, a retinal enhanceropathy. Preprint. Posted online March 9, 2022. bioRxiv 481329. doi: 10.1101/2022.03.08.481329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131(7):1235-1247. [DOI] [PubMed] [Google Scholar]
- 52. Spielmann M, Lupiáñez DG, Mundlos S. Structural variation in the 3D genome. Nat Rev Genet. 2018;19(7):453-467. [DOI] [PubMed] [Google Scholar]
- 53. Melo US, Schöpflin R, Acuna-Hidalgo R, et al. Hi-C identifies complex genomic rearrangements and TAD-shuffling in developmental diseases. Am J Hum Genet. 2020;106(6):872-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Carss KJ, Arno G, Erwood M, et al. Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. Am J Hum Genet. 2017;100(1):75-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Van Cauwenbergh C, Van Schil K, Cannoodt R, et al. arrEYE: a customized platform for high-resolution copy number analysis of coding and noncoding regions of known and candidate retinal dystrophy genes and retinal noncoding RNAs. Genet Med. 2017;19(4):457-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Van Schil K, Naessens S, Van de Sompele S, et al. Mapping the genomic landscape of inherited retinal disease genes prioritizes genes prone to coding and noncoding copy-number variations. Genet Med. 2018;20(2):202-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ellingford JM, Horn B, Campbell C, et al. Assessment of the incorporation of CNV surveillance into gene panel next-generation sequencing testing for inherited retinal diseases. J Med Genet. 2018;55(2):114-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zampaglione E, Kinde B, Place EM, et al. Copy-number variation contributes 9% of pathogenicity in the inherited retinal degenerations. Genet Med. 2020;22(6):1079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]