Abstract
The specificity of XapB permease was compared with that of the known nucleoside transporters NupG and NupC. XapB-mediated xanthosine uptake is abolished by 2,4-dinitrophenol and exhibits saturation kinetics with an apparent Km of 136 μM. A 12-transmembrane-segment model was confirmed by translational fusions to alkaline phosphatase and the α fragment of β-galactosidase.
Escherichia coli can utilize nucleosides as the sole carbon source. Two nucleoside permeases, NupC and NupG, are known and have been characterized in some detail (15). The NupG protein can facilitate uptake of all tested purine and pyrimidine nucleosides, while NupC has specificity towards the pyrimidine nucleosides, adenosine, and their deoxy derivatives. They are both powered by the proton motive force. Specificity towards xanthosine has not been examined in detail, but xanthosine has been found to inhibit the uptake of adenosine in the wild type and in a nupC mutant (11). Moreover, a nupC nupG double mutant was unable to grow on xanthosine as the sole carbon source, whereas a nupC or nupG mutant grew well (8). This indicates that both permeases can transport xanthosine.
The presence of several genes (xapAB and xapR) dedicated to the metabolism of the purine nucleoside xanthosine was indicated by Buxton et al. (4, 8). They suggested that together with the catabolizing enzyme xanthosine phosphorylase (xapA), a new uptake system could exist. When the operon was sequenced (20), it became clear that this function at least in part could be assigned to the presumed product of the xapB gene. Supporting its role as a nucleoside transporter, XapB showed 58% identity to NupG and was found to be enriched in the membrane fraction in minicell experiments (20). The hydropathy profile of the protein indicates that it is a membrane protein with 12 transmembrane segments. Finally, xanthosine phosphorylase is induced to less than 10% of the wild-type level by the inducer xanthosine in a xapB mutant, emphasizing the importance of XapB (20).
Our goal in the present work was to characterize XapB and compare it with NupG and NupC. We cloned the corresponding genes into the low-copy-number plasmid pSU18 (10 copies/cell) (2). The resulting plasmids, pGD227, pGD253, and pGD257, express xapB, nupG, and nupC, respectively. Since expression of all three permeases is under complex control and is induced by nucleosides, we cloned the genes without their normal promoters and expressed them from the plasmid-borne lac promoter under derepressed conditions (lacI).
Purine nucleoside phosphorylase and xanthosine phosphorylase catalyze the phosphorylytic breakdown of nucleosides, releasing ribose-1-phosphate (ribose-1-P) and the nucleobase moiety. Ribose-1-P is converted to ribose-5-P by phosphoribomutase and can be used as an energy source. We exploited this by examining the requirement for nucleoside permeases to grow on nucleosides as the sole carbon source (Table 1). The XapB, NupG, and NupC permeases cloned in pSU18 were transformed into the transport-negative E. coli strain SØ6687 (araD139 ΔlacU169 strA thi codAB ΔnupC ΔnupG ΔxapB::Kanr). Since growth on xanthosine requires induction of xanthosine phosphorylase, which is a slow process, we also expressed xapAB for comparison. Fast-growing mutants which outgrew their parents in liquid cultures appeared frequently; therefore, growth was monitored on plates, where mutants are easily distinguished. As shown in Table 1, expression of xapB results in growth on all nucleosides tested except guanosine. When both xapA and xapB are expressed, the same growth pattern is seen except that cells seem to grow better on purine nucleosides in the presence of XapA. XapA expressed alone did not result in growth on any of the nucleosides (data not shown). Expression of nupG results in growth on all nucleosides except adenosine and xanthosine, although microcolonies appear on xanthosine after 5 days of incubation. When nupC is expressed, the cells grow well on all tested nucleosides except xanthosine and guanosine.
TABLE 1.
Growth on nucleosides as sole carbon sourcesa
| Nucleoside | Growth
|
||||
|---|---|---|---|---|---|
| Control | XapB | XapAB | NupC | NupG | |
| Xanthosine | − | ++ | +++ | − | (+) |
| Guanosine | − | − | − | − | +++ |
| Inosine | − | +++ | ++++ | ++ | +++ |
| Adenosine | − | ++ | +++ | +++ | − |
| Uridine | − | + | + | + | + |
| Cytidine | − | +++ | +++ | +++ | +++ |
| Thymidine | − | +++ | +++ | ++ | +++ |
SØ6687 harboring the indicated transporters on plasmids was grown with nucleosides as the sole carbon source on AB minimal plates (5). ++++, excellent growth after 3 days at 37°C; +++, good growth; ++, poor growth; +, barely visible growth; −, no growth; (+), microcolonies appeared after 5 days.
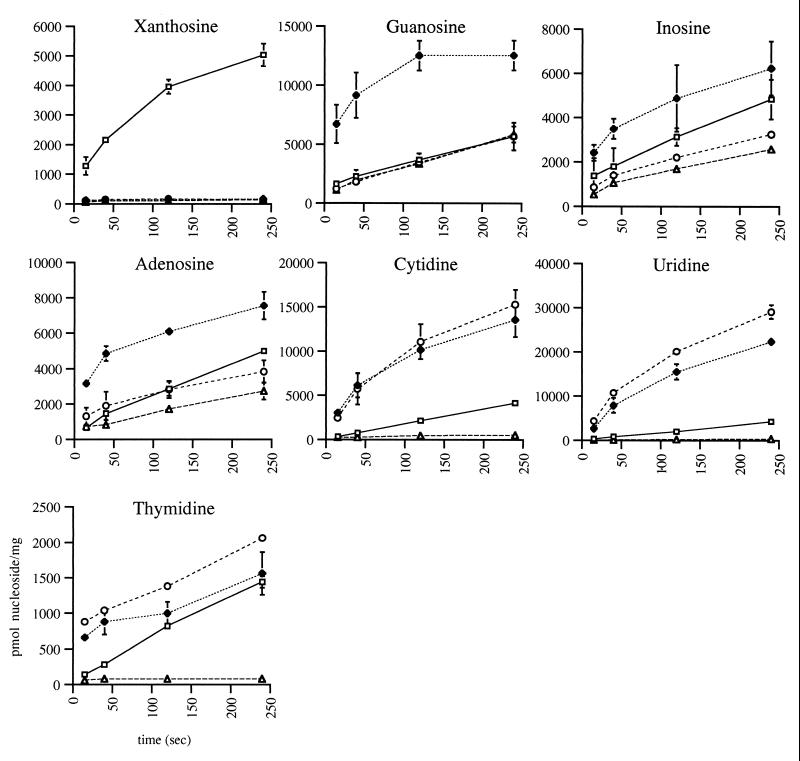
Next, we tested the uptake of 14C-labeled nucleosides in transport-negative strain GD1333 (araD139 ΔlacU169 strA thi cod ΔnupC ΔnupG ΔxapABR::Kanr cytR deoD zjj::Tn10) containing pGD227, pGD253, pGD257, or pSU18. As shown in Fig. 1, XapB can transport all tested nucleosides except guanosine, although the uptake of uridine and cytidine is only slightly above the background level. A similar specificity is seen with NupG, with the main difference being that NupG can transport all nucleosides except xanthosine. NupC is the most efficient permease for the uptake of pyrimidine nucleosides, while inosine and adenosine uptake is only slightly above the control level and xanthosine and guanosine cannot be taken up at all. It should be noted, however, that we do not know the concentration of each permease, and only differences in nucleoside specificity can be tested. Thus, at a low nucleoside concentration (2 μM), xanthosine can only be transported by XapB and guanosine can only be transported by NupG. With all purine nucleosides except xanthosine, a small amount of uptake is seen with the control plasmid. A similar background growth is seen in growth experiments when a cytR mutant is used as the host (data not shown). Since the strain used for the uptake experiments is cytR, a likely explanation for the background is that another purine-specific, low-affinity, cytR-regulated permease exists. Such a permease has recently been identified by Bente Mygind (personal communication). The presence of another cytR-regulated permease can also explain the appearance of fast-growing mutants in the growth experiments, since cytR mutants easily appear under selection pressure (7).
FIG. 1.
Uptake of 14C-radiolabeled nucleosides by GD1333. The time course of the activity of recombinant XapB (open square), NupG (solid diamond), NupC (open circle), and the control (open triangle) towards the ribonucleosides xanthosine, guanosine, inosine, adenosine, uridine, and cytidine and the deoxy ribonucleoside thymidine was assayed. Error bars indicate the standard deviation. Cells were grown exponentially in AB, B1, and glucose minimal medium. Radioactively labeled nucleoside was added (2.5 to 5 mCi/mmol) to a final concentration of 2 μM. Samples were withdrawn at different times and filtered through a 0.45-μm nitrocellulose membrane (Schleicher & Schuell). The filter was washed and dried, and radioactivity was counted in a liquid scintillation counter (17).
It has previously been reported that the initial nucleoside uptake rate, mediated by NupG and NupC, equals the exchange rate (17). This also applies to XapB, since similar time curves were seen for xanthosine uptake in cultures grown with and without unlabeled xanthosine added prior to the addition of labeled xanthosine (data not shown). The initial uptake rate (or the exchange rate) of xanthosine was measured by taking samples 10, 20, 30, and 40 s after addition of radiolabeled xanthosine. The initial uptake rate is constant for up to approximately 40 s. By varying the substrate concentration, we determined the saturation kinetic parameters of XapB. Because of the very low solubility of xanthosine, we took advantage of the finding that the uptake rate equals the exchange rate. Thus, when the initial uptake rate was measured at high xanthosine concentrations, unlabeled xanthosine was added prior to the assay, and only trace amounts of labeled xanthosine were added to start the assay. The data were fitted to the Michaelis-Menten equation using the Ultrafit 3.0 software (BioSoft), and the Km was found to be 136 ± 26 μM. This value is much higher than the values of 0.3 to 1 μM determined for NupC- and NupG-mediated cytidine transport (12, 15, 16). It is, however, in the same range as many eukaroytic nucleoside transporters, e.g., the NupC homologue in rat of 37 μM (10). Moreover, the high Km is in good agreement with the finding that full induction of xapAB requires an extracellular concentration of xanthosine above 1 mM (G. Dandanell, unpublished data). The low solubility of xanthosine is a technical problem only in the uptake assays and not in the induction and growth experiments. It could be argued that the outer membrane limits the uptake of xanthosine in strains in which xapB is overexpressed. Nucleosides enter the outer membrane through OmpC, OmpF, and Tsx. Although Tsx is a nucleoside-specific porin, it is only necessary at a very low nucleoside concentration (<0.1 μM) (15). When we compared xanthosine uptake in wild-type cells with uptake in a cytR mutant, which has sevenfold-higher expression of tsx (3), we found no difference in xanthosine uptake, strongly indicating that diffusion through the outer membrane is not limiting in our experiments (data not shown).
To find out if the xanthosine permease is energized by the proton motive force, we examined the effect of the uncoupler 2,4-dinitrophenol on xanthosine uptake. When we added 2,4-dinitrophenol to a final concentration of 1 mM, it completely abolished uptake, whether added 10 min prior to addition of 14C-labeled xanthosine or 60 s after some uptake had taken place (data not shown).
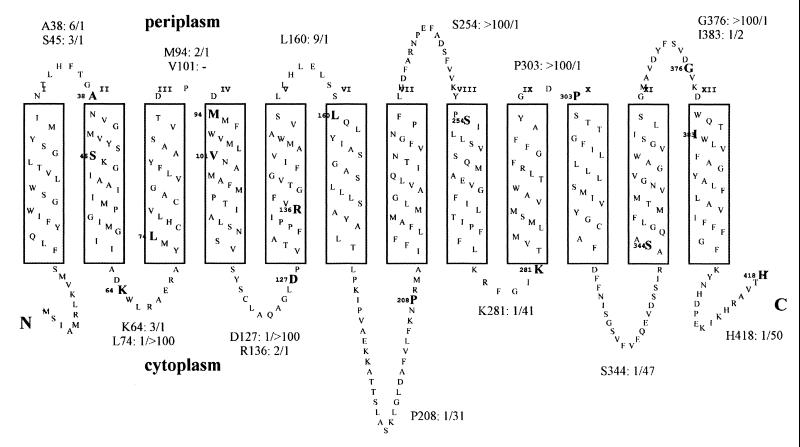
The hydropathy profile of XapB indicates that xanthosine permease has 12 transmembrane segments (TMs) (20). We used the TopPred2 program (21) and the TMHMM program (18) to predict the topology of XapB (the TMHMM model is shown in Fig. 2). Both programs predict that XapB has 12 TMs, and to confirm this model, we made xapB fusions to a dual phoA-lacZ(α) reporter, a method developed by Alexeyev and Winkler (1). The combination of alkaline phosphatase (phoA) and β-galactosidase (lacZ) to study membrane topology has been controversial. However, by fusing all three proteins into a single polypeptide and measuring the ratio of alkaline phosphatase and β-galactosidase activity, differences in expression levels and protein stability of different fusions can largely be ignored. Fusion points just preceding a TM segment were designed so that the phoA-lacZ(α) part would not be buried in the lipid bilayer and putative topogenic signals in the loop would be preserved. In addition, four fusions predicted to be positioned in a TM were constructed. First, dual reporter vector pMN8h was constructed by inserting the phoA gene lacking the N-terminal export signal sequence into pSU19 (a BamHI-BanII phoA fragment was isolated from pUI310 [9]). Next a BanII (Klenow)-BsrBI fragment was deleted, creating an in-frame fusion between phoA (lacking the eight C-terminal amino acids) and the α-fragment of β-galactosidase with a 6-amino-acid linker (SSNSLA) between. All xapB-phoA-lacZ(α) fusions were constructed by amplifying the xapB fragment by PCR and ligating the PCR product into pMN8h. These plasmids were transformed into TG1 (19), which express the ω-subunit of β-galactosidase, and the activity of alkaline phosphatase and β-galactosidase was determined in exponentially growing cells (TG1 is not phoA; however, the background alkaline phosphatase activity level was negligible in all our experiments). As shown in Fig. 2, all fusions predicted to be positioned in periplasmic loops give high alkaline phosphatase/β-galactosidase ratios, whereas all fusions predicted to be in cytoplasmic loops give high β-galactosidase/alkaline phosphatase ratios. The four fusions predicted to be in the membrane all give very low ratios (less than 3). These data correlate with the model shown in Fig. 2. The only exceptions are K64 in the first cytoplasmatic loop and M94 in the second periplasmic loop. The normalized alkaline phosphatase/β-galactosidase ratio is high for K64 because the absolute alkaline phosphatase activity is high. Unexpectedly high activities have previously been observed for phoA fusions positioned N-terminally in cytoplasmic loops (6). The absolute activities of M94 are very low (data not shown), and the fusion protein could be poorly expressed or unstable. Another possibility is that the reporters might be buried in the lipid bilayer, since the second periplasmic loop is predicted to be very short (3 residues).
FIG. 2.
Topological model of XapB. The model was constructed by the TMHMM software (18). The highlighted amino acids indicate the C-terminal amino acids of XapB and fusion points to PhoA and LacZ(α). β-Galactosidase and alkaline phosphatase activities were measured in exponentially growing cells (AB, B1, and glucose minimal medium) on whole sodium dodecyl sulfate-CHCl3-permeabilized cells as described (13, 14) and normalized to the highest value. The alkaline phosphatase/β-galactosidase ratios are indicated at the loops.
Concluding remarks.
Both the growth experiments and the uptake assays clearly show that XapB is a permease that can transport both purine and pyrimidine nucleosides. The interchanging pattern of high and low PhoA-LacZ activity ratios in our xapB-phoA-lacZ(α) fusions points to a 12-TM topology, with N and C termini on the cytoplasmic side, as predicted by both TMHMM and TopPredII. Cells expressing only xapB grow well on (and can concentrate) all the nucleosides tested except guanosine (Table 1 and Fig. 1). Our growth experiments with recombinant NupC and NupG support the previous finding, with the chromosomal genes, that NupC can transport all nucleosides except guanosine and inosine, whereas NupG can transport all nucleosides (15). However, two exceptions are noted. First, NupG does not seem to support growth on adenosine despite the fact that adenosine is effectively taken up by NupG in the transport assay (Table 1 and Fig. 1). The reason is most likely that NupG transport adenosine very effectively and that the high concentration of adenosine inhibits growth. Such inhibition has also been seen in liquid cultures after addition of adenosine to exponentially growing cultures (data not shown). Second, we find that when NupC is expressed from plasmids, cells grow well on inosine (Table 1). A similar result has been observed with different plasmid constructs carrying nupC (B. Mygind, personal communication). This indicates that NupC does transport inosine; however, the affinity for inosine is too low to support growth in wild-type cells, in which only a single copy of nupC is expressed. Growth on uridine was very poor for all the constructs. We have no explanation for this finding, since they all grow well on cytidine. In E. coli, cytidine is deaminated to uridine by cytidine deaminase, and the cell must therefore contain all the enzymes required for the catabolism of uridine.
In our growth experiment, a strain expressing only nupC does not grow on xanthosine, as previously reported (8). Since the strain used here is xapB, it indicates that although NupC is necessary for induction of xapAB, it is XapB that takes up the xanthosine. These observations and our growth experiment indicate that both NupC and NupG can transport xanthosine but only with a very low affinity. We have not been able to show uptake of 14C-labeled xanthosine by NupC or NupG even though they are expressed from approximately 10 gene copies. Based on our results, we suggest that NupC and NupG are necessary for the initial induction of xapAB by transporting small amounts of xanthosine into the cell. This transport, however, is not sufficient to obtain full induction of xapAB but is sufficient to induce xapAB, after which XapB can efficiently transport xanthosine into the cell. This model explains why induction of xapAB takes several generations.
Acknowledgments
This work was supported by a scholarship to M.H.H.N. from the Peter and Emma Thomsen foundation.
We thank Jan Neuhard and Bente Mygind for helpful comments on the manuscript.
REFERENCES
- 1.Alexeyev M F, Winkler H H. Membrane topology of the Rickettsia prowazekii ATP/ADP translocase revealed by novel dual pho-lac reporters. J Mol Biol. 1999;285:1503–1513. doi: 10.1006/jmbi.1998.2412. [DOI] [PubMed] [Google Scholar]
- 2.Bartolomé B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 3.Bremer E, Gerlach P, Middendorf A. Double negative and positive control of tsx expression in Escherichia coli. J Bacteriol. 1988;170:108–116. doi: 10.1128/jb.170.1.108-116.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buxton R S, Hammer-Jespersen K H, Valentin-Hansen P. A second purine nucleoside phosphorylase in Escherichia coli K-12. I. Xanthosine phosphorylase regulatory mutants isolated as secondary-site revertants of a deoD mutant Mol. Gen Genet. 1980;179:331–340. doi: 10.1007/BF00425461. [DOI] [PubMed] [Google Scholar]
- 5.Clark D J, Maaløe O. DNA replication and the division cycle of Escherichia coli. J Mol Biol. 1967;23:99–112. [Google Scholar]
- 6.Ehrmann M, Boyd D, Beckwith J. Genetic analysis of membrane protein topology by a sandwich gene fusion approach. Proc Natl Acad Sci USA. 1990;87:7574–7578. doi: 10.1073/pnas.87.19.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammer-Jespersen K. Nucleoside catabolism. In: Munch-Petersen A, editor. Metabolism of nucleotides, nucleosides and nucleobases in microorganisms. London, England: Academic Press; 1983. pp. 203–258. [Google Scholar]
- 8.Hammer-Jespersen K, Buxton R S, Hansen T D H. A second Purine nucleoside phosphorylase in Escherichia coli K-12. II. Properties of xanthosine phosphorylase and its induction by xanthosine. Mol Gen Genet. 1980;179:341–348. doi: 10.1007/BF00425462. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman C S, Wright A. Fusions of secreted proteins to alkaline phosphatase: an approach for studying protein secretion. Proc Natl Acad Sci USA. 1985;82:5107–5111. doi: 10.1073/pnas.82.15.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Q Q, Yao S Y, Ritzel M W, Paterson A R, Cass C E, Young J D. Cloning and functional expression of a complementary DNA encoding a mammalian nucleoside transport protein. J Biol Chem. 1994;269:17757–17760. [PubMed] [Google Scholar]
- 11.Komatsu Y. Adenosine uptake by isolated membrane vesicles from Escherichia coli K-12. Biochim Biophys Acta. 1973;330:206–221. doi: 10.1016/0005-2736(73)90226-5. [DOI] [PubMed] [Google Scholar]
- 12.Komatsu Y, Tanaka K. Deoxycytidine uptake by isolated membrane vesicles from Escherichia coli K 12. Biochim Biophys Acta. 1973;311:496–506. doi: 10.1016/0005-2736(73)90125-9. [DOI] [PubMed] [Google Scholar]
- 13.Manoil C. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 1991;34:61–75. doi: 10.1016/s0091-679x(08)61676-3. [DOI] [PubMed] [Google Scholar]
- 14.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 15.Munch-Petersen A, Mygind B. Transport of nucleic acid precursors. In: Munch-Petersen A, editor. Metabolism of nucleotides, nucleosides and nucleobases in microorganisms. London, England: Academic Press; 1983. pp. 259–305. [Google Scholar]
- 16.Munch-Petersen A, Mygind B, Nicolaisen A, Pihl N J. Nucleoside transport in cells and membrane vesicles from Escherichia coli K12. J Biol Chem. 1979;254:3730–3737. [PubMed] [Google Scholar]
- 17.Mygind B, Munch-Petersen A. Transport of pyrimidine nucleosides in cells of Escherichia coli K 12. Eur J Biochem. 1975;59:365–372. doi: 10.1111/j.1432-1033.1975.tb02463.x. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst. 1997;8:581–599. doi: 10.1142/s0129065797000537. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Seeger C, Poulsen C, Dandanell G. Identification and characterization of genes (xapA, xapB, and xapR) involved in xanthosine catabolism in Escherichia coli. J Bacteriol. 1995;177:5506–5516. doi: 10.1128/jb.177.19.5506-5516.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Heijne G. Membrane protein structure prediction: hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]