Abstract
The yeast YLR209c (PNP1) gene encodes a protein highly similar to purine nucleoside phosphorylases. This protein specifically metabolized inosine and guanosine. Disruption of PNP1 led to inosine and guanosine excretion in the medium, thus showing that PNP1 plays an important role in the metabolism of these purine nucleosides in vivo.
Purine salvage is a complex pathway allowing interconversion of bases, nucleosides, and nucleotides. In yeast, major attention has been paid to the conversion of bases into nucleotides by phosphoribosyltransferases (PRTs): adenine-PRT, hypoxanthine-guanine-PRT, and xanthine-PRT activities have been reported (19, 20), and the cognate genes have been identified (1, 5, 6). Yeast purine nucleoside metabolism has received far less attention, and only very recently was the first yeast gene encoding a purine nucleoside metabolizing enzyme identified (11). This gene, named ADO1, encodes adenosine kinase allowing synthesis of AMP from adenosine. Although several other enzymatic activities involved in yeast purine nucleoside metabolism have been described in the past, the corresponding genes have not yet been identified. Enzymatic activities responsible for the synthesis of inosine either from adenosine by adenosine deaminase (14) or from IMP by an IMP-specific 5′ nucleotidase have been reported (8). Also, two distinct enzymatic activities (purine nucleoside hydrolase and purine nucleoside phosphorylase [PNP]) responsible for the degradation of inosine into hypoxanthine have been reported (7). The latter two enzymes catalyze the conversion of nucleosides to bases, although through distinct enzymatic mechanisms: (i) for nucleoside hydrolase, nucleoside + H2O→base + ribose and (ii) for nucleoside phosphorylase, nucleoside + Pi→base + ribose-1P.
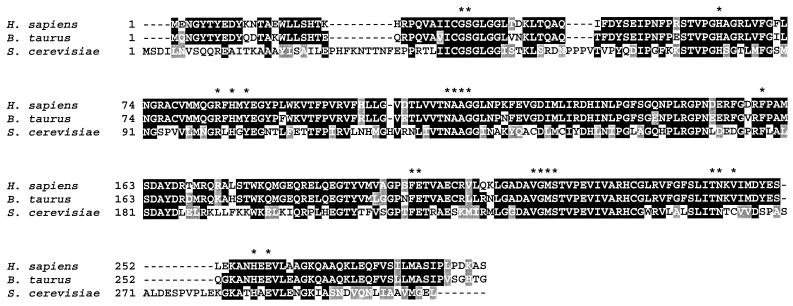
As a further step toward understanding yeast purine nucleoside metabolism, we searched for open reading frames (ORFs) in the complete yeast genome sequence that would encode candidate PNP. We found an uncharacterized ORF (YLR209c) that encodes a putative polypeptide highly similar to human and bovine PNP (Fig. 1). This enzyme has been thoroughly studied, and the three-dimensional structures of the trimeric human and bovine PNPs have been solved (3, 10). Important residues for substrate binding and catalysis have been identified (4, 12), all of which (except Val263) are conserved in the yeast enzyme (shown by asterisks in Fig. 1).
FIG. 1.
Sequence comparison of PNPs from various species. The amino acid sequence deduced from the nucleotide sequence of the YLR209c ORF (S. cerevisiae) was compared with the PNP sequences from Homo sapiens (18) and Bos taurus (2). Identities and similarities are highlighted in black and gray boxes, respectively. Dashes indicate the gaps created for alignment. Asterisks indicate residues important for substrate binding and catalysis.
To gain insight into the precise function of the yeast ORF YLR209c, the protein encoded by this ORF was tagged with 10 histidine residues at its N terminus and expressed in Escherichia coli. The PNP expression plasmid was constructed as follows. The YLR209c ORF was amplified by PCR from S288c genomic DNA with the following synthetic oligonucleotides: 359 (5′-CGATGCTCGAGATGAGTGATATCTTGAACGT-3′) and 360 (5′-GGACCCGGGTTATAATTCCCCCATTACGG-3′). The amplification product was then digested with XhoI and SmaI and inserted in the pJC20-HisN vector (15) digested with XhoI and SmaI. The BL21(DE3) E. coli strain carrying the resulting plasmid was induced at 25°C for 6.5 h in the presence of 0.1 mM IPTG (isopropyl-β-d- thiogalactopyranoside). The His-tagged PNP was then purified by following the native protein purification protocol from the QIAexpressionist kit (Qiagen), with the following two modifications: phosphate buffers were replaced by Tris-HCl, and elution was done using 300 mM imidazole. The purified protein was then assayed for its ability to metabolize the various purine and pyrimidine nucleosides. In this assay, the reaction was performed at 30°C and stopped by freezing the samples after 30 min. The disappearance of the substrates (present at a concentration of 20 μM) and the appearance of the products were monitored by high-pressure liquid chromatography (HPLC) as described previously (11).
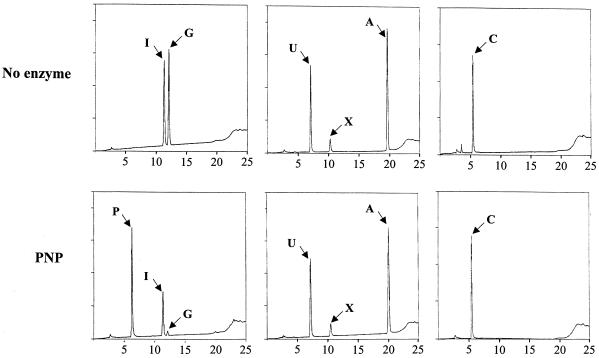
Ylr209cp was unable to metabolize the purine nucleosides adenosine and xanthosine or the pyrimidine nucleosides uridine and cytidine (Fig. 2, middle and right panels), while inosine and guanosine were substrates for Ylr209p (Fig. 2, left panel). Therefore, the YLR209c ORF clearly encodes a PNP and was renamed PNP1. The yeast PNP is thus closely related to the human enzyme both structurally (50% sequence identity) and functionally, since both enzymes only accept inosine and guanosine as substrates. In contrast, yeast PNP appears more distant to the E. coli enzyme (<10% identity), which is hexameric (13) and also accepts adenosine as a substrate (9).
FIG. 2.
Substrate specificity of the yeast PNP. The yeast PNP purified from E. coli was assayed for catalysis of various purine and pyrimidine nucleosides (inosine + guanosine, left panels; uridine + xanthosine + adenosine, middle panels; cytidine, right panels). Abbreviations: A, adenosine; C, cytidine; G, guanosine; I, inosine; P, hypoxanthine + guanine; U, uridine. The nucleosides treated with the enzyme (bottom panels) or untreated (top panels) were separated by HPLC.
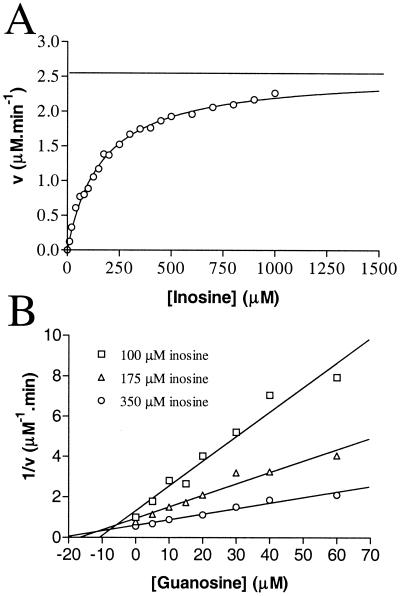
The PNP activity was then measured using a coupled reaction with xanthine oxidase. The principle of the assay is that hypoxanthine produced from degradation of inosine by PNP is followed by its two-step transformation into uric acid by commercial (Roche 110442) xanthine oxidase (0.5 U/reaction). Appearance of uric acid in the reaction is then monitored by its specific absorbance at 293 nm (ɛ = 12 mM−1 cm−1). The reaction was done at 25°C in potassium phosphate buffer (pH 7.5; 100mM) with 33 ng of purified PNP enzyme in a final volume of 0.8 ml. Using this assay, the Km for inosine was found to be 166 ± 8 μM (Fig. 3A). Addition of adenosine (up to 800 μM), xanthosine (up to 600 μM), uridine (up to 800 μM), and cytidine (up to 400 μM) did not result in any detectable inhibition of Pnp1p activity measured using inosine as the substrate at a final concentration of 175 μM. In contrast, guanosine had a very strong competitive inhibitory effect, and its Ki was estimated at 8.4 ± 2.7 μM (Fig. 3B). This value also refers to the Km for guanosine since this substance is a strict competitive inhibitor of inosine (Fig. 3B). Guanosine can therefore be bound more efficiently than inosine by yeast PNP, a situation similar to that reported for human PNP for which the Kms for guanosine and inosine are 12 and 45 μM, respectively (17). Finally, the Vmax and kcat of PNP, with inosine as substrate, were 2.6 μM min−1 and 38 s−1, respectively. This latter parameter of the yeast enzyme is therefore similar to the one reported for human PNP and inosine: 57 s−1 for kcat (17).
FIG. 3.
Kinetic parameters of the yeast purine nucleoside phosphorylase. (A) Determination of the Km for inosine using a xanthine oxidase-coupled assay. Vmax (horizontal line) and Km were calculated by nonlinear regression analysis of the saturation curve. (B) The Ki for guanosine was determined by linear regression of the data presented in the Dixon plot.
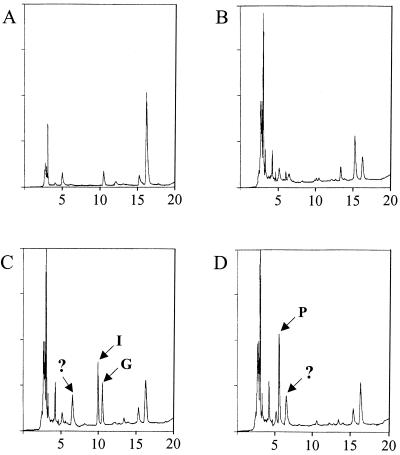
A pnp1 null mutant (purchased from Euroscarf) did not show any obvious growth defect. We reasoned that if PNP activity is important for purine metabolism, the pnp1 mutant should accumulate the natural substrates of the enzyme and that some of them could be excreted in the growth medium. Wild-type (BY4741: MATa ura3Δ0 leu2Δ0 met15Δ0 his3Δ1) and pnp1 (Y04158:MATa ura3Δ0 leu2Δ0 met15Δ0 his3Δ1 ylr209c::KanMX4) strains were grown in adenine- and uracil-free SC medium (16) to mid-exponential phase. Cells were then harvested, and the medium was filtered through a 0.2-μm (pore-size) filter. Separation of compounds in the growth medium was done by chromatography as described previously (11). The inosine and guanosine peaks in the pnp1 strain growth medium were identified by their respective retention times and confirmed by treating the growth medium with commercial E. coli PNP as recommended by the supplier (Sigma). Analysis of the medium after growth of the wild-type and mutant pnp1 strains did indeed reveal that the mutant strain excretes both inosine and guanosine in the medium (Fig. 4). This result strongly supports the idea that inosine and guanosine are the natural substrates of yeast PNP in vivo.
FIG. 4.
Excretion of inosine and guanosine by the pnp1 mutant. Purine derivatives in the growth medium were separated by HPLC. Compound separation was monitored by following absorbance at 260 nm. HPLC profiles are presented as a function of retention time on the column. (A) SC medium. (B) SC medium after growth of the wild-type strain (BY4741). (C) SC medium after growth of the pnp1 mutant strain (Y04158). (D) Same medium as in panel C but treated with commercial PNP. Arrows indicate the peaks specifically found in the pnp1 growth medium. Abbreviations: G, guanosine; I, inosine; P, hypoxanthine + guanine. Unidentified peaks are indicated by a question mark.
Inosine and guanosine excretion by the pnp1 mutant also suggests that the alternative purine nucleoside hydrolase activity described by Heppel and Hilmoe (7) cannot compensate for the nucleoside accumulation due to the lack of PNP. Our attempt to identify a candidate ORF for such a purine nucleoside hydrolase activity was unsuccessful. The best candidate for inosine hydrolase was ORF YDR400w, which was found to encode a cytidine-uridine hydrolase but could not metabolize any of the purine nucleosides (K.L. and B.D.-F., unpublished results). The identification of PNP1 further improves our understanding of purine metabolism in yeast, although several genes encoding purine nucleoside-metabolizing enzymes remain to be identified.
Acknowledgments
We thank B. Pinson for helpful discussions and critical reading of the manuscript. We also thank F. Borne for technical help and O. Spangenberg for helpful advice.
This work was supported by grants from Conseil Régional d'Aquitaine, Procope 99016 European Collaboration Programme, and CNRS (UMR5095). K.L. was supported by a Ministère de la Recherche training fellowship.
REFERENCES
- 1.Alfonzo J, Sahota A, Deeley M C, Ranjekar P, Taylor M W. Cloning and characterization of the adenine phosphoribosyltransferase-encoding gene (APT1) from Saccharomyces cerevisiae. Gene. 1995;161:81–85. doi: 10.1016/0378-1119(95)00239-3. [DOI] [PubMed] [Google Scholar]
- 2.Bzowska A, Luic M, Schröder W, Shugar D, Saenger W, Koellner G. Calf spleen nucleoside phosphorylase: purification, sequence and crystal structure of its complex with an N(7)-acycloguanosine inhibitor. FEBS Lett. 1995;367:214–218. doi: 10.1016/0014-5793(95)00540-p. [DOI] [PubMed] [Google Scholar]
- 3.Ealick S E, Rule S A, Carter D C, Greenhough T J, Babu Y S, Cook W J, Habash J, Helliwell J R, Stoeckler J D, Parks R E, Jr, Chen S-F, Bugg C E. Three-dimensional structure of human erythrocytic purine nucleoside phosphorylase at 3.2 Å resolution. J Biol Chem. 1990;265:1812–1820. doi: 10.2210/pdb2pnp/pdb. [DOI] [PubMed] [Google Scholar]
- 4.Erion M D, Takabayashi K, Smith H B, Kessi J, Wagner S, Honger S, Shames S L, Ealick S E. Purine nucleoside phosphorylase. 1. Structure-function studies. Biochemistry. 1997;36:11725–11734. doi: 10.1021/bi961969w. [DOI] [PubMed] [Google Scholar]
- 5.Guetsova M L, Lecoq K, Daignan-Fornier B. The isolation and characterization of Saccharomyces cerevisiae mutants that constitutively express purine biosynthetic genes. Genetics. 1997;147:383–397. doi: 10.1093/genetics/147.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guetsova M L, Crother T R, Taylor M W, Daignan-Fornier B. Isolation and characterization of the Saccharomyces cerevisiae XPT1 gene encoding xanthine phosphoribosyl transferase. J Bacteriol. 1999;181:2984–2986. doi: 10.1128/jb.181.9.2984-2986.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heppel L A, Hilmoe R J. Phosphorolysis and hydrolysis of purine ribosides by enzymes from yeast. J Biol Chem. 1952;198:683–694. [PubMed] [Google Scholar]
- 8.Itoh R. Purification and some properties of an IMP-specific 5′-nucleotidase from yeast. Biochem J. 1994;298:593–598. doi: 10.1042/bj2980593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen K F, Nygaard P. Purine nucleoside phosphorylase from Escherichia coli and Salmonella typhimurium. Purification and some properties. Eur J Biochem. 1975;51:253–265. doi: 10.1111/j.1432-1033.1975.tb03925.x. [DOI] [PubMed] [Google Scholar]
- 10.Koellner G, Lui M, Shugar D, Saenger W, Bzowska A. Crystal structure of calf spleen purine nucleoside phosphorylase in a complex with hypoxanthine at 2.15 Ä resolution. J Mol Biol. 1997;265:202–216. doi: 10.1006/jmbi.1996.0730. [DOI] [PubMed] [Google Scholar]
- 11.Lecoq K, Belloc I, Desgranges C, Daignan-Fornier B. Role of adenosine kinase in Saccharomyces cerevisiae: identification of the ADO1 gene and study of the mutant phenotypes. Yeast. 2001;18:335–342. doi: 10.1002/1097-0061(20010315)18:4<335::AID-YEA674>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 12.Mao C, Cook W J, Zhou M, Federov A A, Almo S C, Ealick S E. Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues. Biochemistry. 1998;37:7135–7146. doi: 10.1021/bi9723919. [DOI] [PubMed] [Google Scholar]
- 13.Mao C, Cook W J, Zhou M, Koszalka G W, Krenitsky T A, Ealick S E. The crystal structure of Escherichia coli purine nucleoside phosphorylase: a comparison with the human enzyme reveals a conserved topology. Structure. 1997;5:1373–1383. doi: 10.1016/s0969-2126(97)00287-6. [DOI] [PubMed] [Google Scholar]
- 14.Marmocchi F, Lupidi G, Vernardi G, Riva F. Adenosine deaminase from Saccharomyces cerevisiae: purification and characterization. Biochem Int. 1987;14:569–580. [PubMed] [Google Scholar]
- 15.Schaertl S, Geeves M A, Konrad M. Human nucleoside diphosphate kinase B (Nm23–H2) from melanoma cells shows altered phosphoryl transfer activity due to the S122P mutation. J Biol Chem. 1999;274:20159–20164. doi: 10.1074/jbc.274.29.20159. [DOI] [PubMed] [Google Scholar]
- 16.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 17.Stoeckler J D, Poirot A F, Smith R M, Parks Jr R E, Ealick S E, Takabayashi K, Erion M D. Purine nucleoside phosphorylase. 3. Reversal of purine base specificity by site-directed mutagenesis. Biochemistry. 1997;36:11749–11756. doi: 10.1021/bi961971n. [DOI] [PubMed] [Google Scholar]
- 18.Williams S R, Goddard J M, Martin D W., Jr Human purine nucleoside phophorylase cDNA sequence and genomic clone characterization. Nucleic Acids Res. 1984;12:5779–5787. doi: 10.1093/nar/12.14.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woods R A, Roberts D G, Friedman T, Jolly D, Filpula D. Hypoxanthine:guanine phosphoribosyltransferase mutants in Saccharomyces cerevisiae. Mol Gen Genet. 1983;191:407–412. doi: 10.1007/BF00425755. [DOI] [PubMed] [Google Scholar]
- 20.Woods R A, Roberts D G, Sein D S, Filpula D. Adenine phosphoribosyltransferase mutants in Saccharomyces cerevisiae. J Gen Microbiol. 1984;130:2629–2637. doi: 10.1099/00221287-130-10-2629. [DOI] [PubMed] [Google Scholar]